Abstract
Background
The peripheral blood neutrophil–to-lymphocyte ratio (NLR) is a cytological marker of both inflammation and poor outcomes in cancer patients. DNA methylation is a key element of the epigenetic program defining different leukocyte subtypes and may provide an alternative to cytology in assessing leukocyte profiles. Our aim was to create a bioinformatic tool to estimate NLR using DNA methylation, and to assess its diagnostic and prognostic performance in human populations.
Methods
We developed a DNA methylation-derived NLR (mdNLR) index based on normal isolated leukocyte methylation libraries and established cell-mixture deconvolution algorithms. The method was applied to cancer case-control studies of the bladder, head and neck, ovary and breast, as well as publicly available data on cancer-free subjects.
Results
Across cancer studies, mdNLR scores were either elevated in cases relative to controls, or associated with increased hazard of death. High mdNLR values (>5) were strong indicators of poor survival. Additionally, mdNLR scores were elevated in males, in non-Hispanic white versus Hispanic ethnicity, and increased with age. We also observed a significant interaction between cigarette smoking history and mdNLR on cancer survival.
Conclusions
These results mean that our current understanding of mature leukocyte methylomes is sufficient to allow researchers and clinicians to apply epigenetically-based analyses of NLR in clinical and epidemiologic studies of cancer risk and survival.
Impact
As cytological measurements of NLR are not always possible (i.e., archival blood), mdNLR, which is computed from DNA methylation signatures alone, has the potential to expand the scope of epigenome-wide association studies (EWAS).
Keywords: DNA methylation, neutrophil lymphocyte ratio, cancer survival, EWAS
1. Introduction
Systemic inflammation in cancer is associated with altered myelopoiesis and the production of myeloid suppressor cells (MDSCs), contributing to an immunosuppressive network that adversely affects cancer survival (1-3). MDSCs are aberrantly activated immature myeloid cells that are functionally distinct from terminally differentiated myeloid cells, although they are morphologically similar to their normal mature counterparts (i.e. mononuclear cells, neutrophils) (4). Epigenetic reprograming is implicated in the altered differentiation pathway leading to MDSCs (5), which has been shown to target the retinoblastoma cell cycle (6), and Notch signaling pathways (7) as well as other transcriptional networks (8).
The best methods to assess altered myeloid populations or systemic inflammation, more generally, are still evolving and as a result, large-scale studies are lacking. However, the peripheral blood neutrophil to lymphocyte ratio (NLR), derived from the common five-part white blood cell differential count has emerged as a robust marker of cancer associated inflammation (9-14). Increases in the blood NLR have been remarkably consistent in their association with poor cancer survival. A recent meta-analysis including 100 independent studies encompassing over 40,000 subjects demonstrated that an elevated NLR was a statistically significant predictor of poor overall survival, cancer specific survival, as well as progression free and disease free survival, even after adjustment for established risk predictors (15). While the NLR is an index of systemic inflammation, the biology underlying its strong connection with cancer outcomes remains obscure. Intriguingly, it has recently been shown that the mutational landscape of some cancers (associated with tobacco carcinogen exposure) engender an immune response detectable in the periphery, strongly supporting the concept that the blood harbors phenotypically active subsets of immune cells (16).
One plausible explanation for the association of elevated NLRs with cancer mortality is its presumed correlation with altered myeloid differentiation and production of MDSCs. While this idea is largely untested, the crucial role that epigenetic modifications (including DNA methylation) play in programming myeloid and lymphoid cell differentiation is recognized (17-20). Remodeling the epigenome during hematopoiesis leads to progressively restricted immune subtypes and DNA methylation provides a chemically stable mark for these cell fate decisions (21). The DNA methylomes of circulating myeloid (monocytes, neutrophil, basophils, eosinophil) and lymphoid (CD4, CD8 T cells, B-cells, NK cells) cells have been extensively studied, revealing that lineage specific peripheral blood immune cells can be distinguished by a signature or “fingerprint” of leukocyte differentially methylated regions (L-DMRs) (22-24). Using a bioinformatic approach (a cell-mixture deconvolution algorithm), specific L-DMR libraries accurately estimate the proportion of each cell type in complex mixtures such as whole blood (25). Although the transition from immature to mature myeloid cells has been shown to involve changes in DNA methylation (26,27), the diagnostic L-DMRs of immature myeloid cells and more specifically, cancer-induced MDSCs, have not been defined. Given that epigenetic mechanisms specify both normal and cancer related myelopoiesis, the possibility exists that without specific DMRs for cancer related leukocytes, previously identified immune methylation profiles may not predict cancer patient outcomes as they do with the cytological NLR. We questioned whether - similar to the cytological NLR - established DMR signatures of blood neutrophils and lymphocytes used to estimate the NLR would be predictive of cancer patient survival and correlate with other risk factors.
Here, we develop and evaluate a methylation derived NLR (mdNLR) index using L-DMR cell libraries and our validated bioinformatic approach (23,28). Often times cytological NLR data are not available in clinical and research studies, including epigenome-wide association (EWAS) studies; an mdNLR index, based solely on archival blood DNA, would expand the scope of studies that seek to evaluate immune parameters in cancer to include large, prospective studies.
2. Materials and Methods
Computing the mdNLR
Estimation of the mdNLR required three main steps: (i) identify differentially methylated CpGs among leukocyte subtypes, (L-DMRs), (ii) perform cell-mixture deconvolution (23) to estimate the proportion of leukocyte subtypes using the L-DMRs identified in step 1, and (iii) compute the ratio of the predicted proportion of neutrophil granulocytes to lymphocytes (Figure 1). Using DNA methylation data from isolated leukocyte subtypes (22,23), we identified CpG-specific patterns of DNA methylation among monocytes, granulocytes, and lymphocytes using a series of t-tests fit independently to each CpG. For each of the three pairs of comparisons, the top 50 CpGs with the smallest and largest t-statistics were combined to create a single list of J non-overlapping L-DMRs (Supplementary Tables 1 and 2). The rationale for selecting only the top 50 CpGs with the smallest and largest t-statistics to create the L-DMR library was based on previous work (29) and empirical analyses that suggested that the inclusion of additional CpGs offered only marginal improvements in the accuracy of our estimates of NLR (data not shown). Using the J L-DMRs and cell-mixture deconvolution we estimated the fractions of monocytes, granulocytes, and lymphocytes for the ith study sample, . Finally, the mdNLR was computed for each sample by taking the ratio of its predicted granulocyte and lymphocyte fractions, . A publicly available implementation of this method is available in the IDOL R package (http://r-project.org)*.
Figure 1. Conceptual diagram illustrating the various steps involved in estimating the mdNLR.
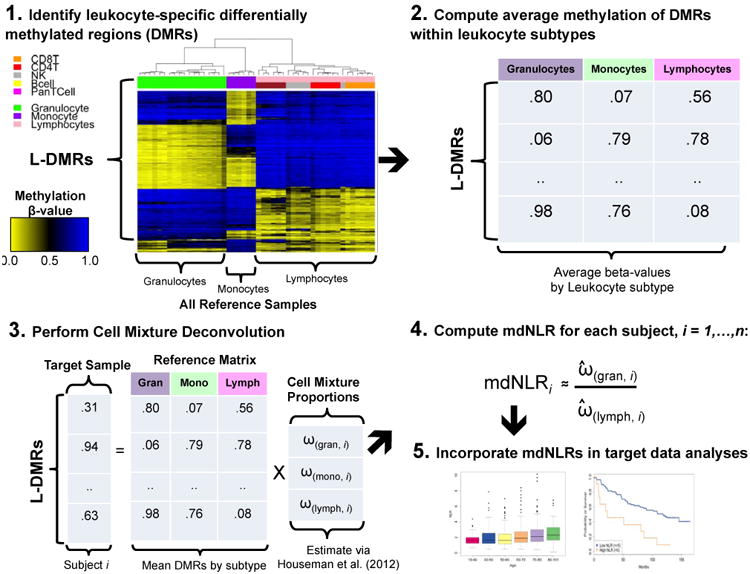
In Step 1, putative leukocyte differentially methylated regions (L-DMRs) are identified between monocytes, granulocytes, and lymphocytes. Step 2 involves computing the within cell type mean methylation beta-values for each of the putative L-DMRs identified in Step 1. In Step 3, the within cell type mean methylation beta-values are used in conjunction with Houseman et al., (23) to predict the proportion monocytes, granulocytes, and lymphocytes for a sample consisting of DNA methylation signatures profiled in whole blood. In Step 4, the mdNLR is calculated as the ratio of the predicted proportions of granulocytes and lymphocytes and finally, in Step 5 the estimated mdNLR is examined with respect to its association with cancer risk, outcomes, or other clinical variables of interest.
Validating the mdNLR
As a validation of the mdNLR, we performed an analysis comparing mdNLR to cytological NLR within an independent set consisting of whole blood (WB) DNA methylation measurements across 18 samples (29). These data are publicly available in Gene Expression Omnibus (GEO Accession: GSE77797). Of the 18 samples, 12 were artificial WB reconstructions for which the mixing proportions of leukocyte cell types (i.e., granulocytes, monocytes, CD4T, CD8T, natural killer cells, and B cells) were known exactly. The remaining 6 WB samples were collected from disease-free adult donors with available immune cell profiling data from flow cytometry (FC). Further details on the 18 WB samples can be found elsewhere (29). The mdNLR and cytological NLR were first determined for each of the 18 samples and subsequently compared by computing the coefficient of determination (R2) and root mean squared prediction error (RMSPE).
Statistical analyses of the mdNLR and clinical outcomes
Associations between mdNLR and clinical covariates were assessed using either logistic regression or linear regression models. Both univariate and multivariable regression models adjusted for potential confounders were employed. Cox-proportional hazards regression models adjusted for potential confounding covariates were used to examine the association between mdNLR and survival time. In our survival analyses mdNLR was modeled both as a continuous predictor and by dichotomizing subjects into high and low mdNLR groups. High and low mdNLR groups were determined by first identifying mdNLR cut-point that maximized the log-rank (LR) test statistic within each data set separately (Δd, d = {Bladder, HNSCC}), followed by group assignment:
To determine the mdNLR cut-point (Δd) for each data set, we computed the LR test statistic that resulted from comparing survival profile between subjects in the high and low mdNLR groups as a function of varying thresholds for defining membership in those groups, i.e., LRdt = fd (Sdi, Cdi |δt), where Sdi is the time to death or censoring for subject i in data set d = {Bladder, HNSCC}, Cdi is the censoring indicator for subject i in data set d, and δt t = 1,…,T represents the cut-point for determining the high and low NLR groups). Finally, the optimal cut-point was obtained by finding the δt that resulted in the maximum LR test statistic, i.e., Δd = arg maxδt(LRdt). The purpose of dichotomizing mdNLR into high and low groups was two-fold: first, as a preventative measure due to the potential non-linear effect of mdNLR on the log-hazard and second, to enable straightforward comparisons across data sets and previously published studies.
Target DNA methylation data sets
We derived and investigated mdNLR scores in five published DNA methylation data sets. The study sample sizes, clinical characteristics, and available demographic/epidemiological information is given in Table 1. The data sets used here included case-control EWAS of bladder cancer (30), head and neck squamous cell carcinoma (HNSCC) (31), ovarian cancer (32), and breast cancer (33). Treatment status was available for the patients in the ovarian cancer data set and separate analyses were performed on pre- and post-treatment cases. For the HNSCC data set, blood was drawn pre-treatment. Conversely blood was drawn post-treatment for cases in the bladder cancer data set. Treatment was fairly consistent among non-muscle invasive bladder cancer cases, which comprised the vast majority (70%) of bladder cancer cases. Across all cases, approximately 75% received surgery only (i.e., transurethral resection of the bladder tumor (TURBT)) and approximately 13% received bacillus Calmette-Guérin (BCG) at the time of blood-draw. Treatment information was not available for the six blood samples collected post-diagnosis in the breast cancer data set.
Table 1. Characteristics of target study populations.
| Study (Array) | Characteristics | Cases | Controls | pdifference |
|---|---|---|---|---|
| HNSCC (27K) | Total samples | 92 | 92 | |
| mdNLR, mean (sd) | 3.0 (1.6) | 1.7 (.7) | 1.2 × 10-10 | |
| Age,median years (range) | 58 (31-84) | 59 (32-86) | 0.54a | |
| Gender | 0.99b | |||
| Male | 64 (70%) | 64 (70%) | ||
| Female | 28 (30%) | 28 (30%) | ||
| Race | 0.99b | |||
| White | 84 (91%) | 85 (92%) | ||
| Non-White | 8 (9%) | 7 (8%) | ||
| Smoking History | 0.04b | |||
| Never | 17 (19%) | 32 (35%) | ||
| Former | 59 (64%) | 47 (51%) | ||
| Current | 16 (17%) | 13 (14%) | ||
| HPV16 (E6, E7, or L1) | .0002b | |||
| Negative | 66 (72%) | 83 (90%) | ||
| Positive | 26 (28%) | 9 (10%) | ||
| Tumor site | ||||
| Laryngeal | 18 (20%) | N/A | ||
| Oral cavity | 47 (50%) | N/A | ||
| Oropharyngeal | 25 (27%) | N/A | ||
| Bladder Ca. (27K) | Total samples | 223 | 237 | |
| mdNLR, mean (sd) | 2.6 (1.9) | 2.8 (2.2) | 0.33 | |
| Age, median years (range) | 66 (25-74) | 65 (28-74) | 0.05a | |
| Gender | 0.06b | |||
| Male | 171 (77%) | 158 (67%) | ||
| Female | 52 (23%) | 79 (33%) | ||
| Race | 0.99b | |||
| White | 223 (100%) | 237 (100%) | ||
| Non-White | ||||
| Smoking History | <0.001b | |||
| Never | 40 (18%) | 72 (30%) | ||
| Former | 111 (50%) | 126 (53%) | ||
| Current | 72 (32%) | 39 (16%) | ||
| Tumor Stage | ||||
| T0a | 156 (70%) | N/A | ||
| Tis | 6 (2.7%) | N/A | ||
| T1 | 37 (17%) | N/A | ||
| T2 | 12 (5.4%) | N/A | ||
| T3 | 6 (2.7%) | N/A | ||
| T4 | 6 (2.7%) | N/A | ||
| Ovarian Ca. (27K) | Total samples | 266 | 274 | |
| Pre-treatment cases | 131 (49%) | N/A | ||
| Post-treatment cases | 135 (51%) | N/A | ||
| mdNLR, mean (sd) | 3.15 (2.24) | 2.08 (1.01) | 2.2×10-16 a | |
| Age group | 3.7×10-6 b | |||
| 50-55 | 34 (13%) | 14 (5%) | ||
| 55-60 | 47 (18%) | 68 (25%) | ||
| 60-65 | 42 (16%) | 67 (24%) | ||
| 65-70 | 43 (16%) | 39 (14%) | ||
| 70-75 | 50 (19%) | 66 (24%) | ||
| > 75 | 50 (19%) | 20 (7%) | ||
| Histology | ||||
| Serous | 150 (56%) | N/A | ||
| Endometriod | 34 (13%) | N/A | ||
| Mucinous | 30 (11%) | N/A | ||
| Clear cell | 25 (9%) | N/A | ||
| Other | 27 (10%) | N/A | ||
| Breast Twin Study (450K) | Total Samples | 15 | 15 | |
| mdNLR, mean (sd) | 2.7 (1.4) | 2.4 (1.2) | .08c | |
| Healthy aging study (450K) | Total | 656 | ||
| mdNLR, mean (sd) | 3.0 (5.6)d | |||
| Age, median years | 65 (19-101) | |||
| Race | ||||
| Caucasian - European | 426 (65%) | |||
| Hispanic - Mexican | 230 (35%) | |||
| Gender | ||||
| Female | 338 (52%) | |||
| Male | 318 (48%) |
Abbreviations:
Wilcoxox rank-sum test for a difference between cases and control;
Fisher's exact or Chi-square test for a difference between cases and controls.
One-sided paired t-test to assess difference in cases and controls.
Average and standard deviation driven up by several large mdNLR outlier values in the data.
To explore the relationship between mdNLR and age, ethnic origin, and gender in healthy adults, we used a large (n=656), publicly available blood-derived DNA methylation data set (34) (GEO Accession: GSE40279). None of the methylation studies included direct cytological measurements of leukocyte proportions.
Reference leukocyte-specific DNA methylation data sets
We made use of two previously published leukocyte-specific DNA methylation data sets as the basis for mdNLR estimation as the target DNA methylation data sets used in our analysis spanned two different array technologies (HM27 and HM450 arrays) (22,23). Specifically, HM27 methylation data for leukocyte subtypes isolated from the peripheral blood 46 different non-diseased human adults (B-cells (n=6), natural killer cells (n=11), CD4+ T cells (n=8), CD8+ T cells (n=2), Pan-T cells (n=6), monocytes (n=5), granulocytes (n = 8)) (23), and a data set (GEO Accession: GSE35069), that profiled the same leukocyte subtypes in each of 6 healthy male adults using the Illumina HM450 BeadChip (22).
Quality control and preprocessing of the DNA methylation data sets
For each of the DNA methylation data sets, preprocessing and quality control was accomplished using the minfi Bioconductor package (35). To ensure high-quality methylation data, CpG loci having a sizable fraction (>25%) of detection P-values above a predetermined threshold (detection P>10E-5) were excluded (36). For the HM450 data sets, Subset Quantile Within Array (SWAN) normalization was performed for type 1/2 probe adjustment (37). The presence of technical sources of variability induced by plate and/or BeadChip was examined using principal components analysis (PCA) and the top K principal components (38) were examined in terms of their association with plate and BeadChip. If plate and/or BeadChip was found to be significantly associated with any of the top K principal components, we applied ComBat method (39) for normalization using the sva Bioconductor package.
3. Results
mdNLR validation analysis
As a validation of our proposed mdNLR, we first performed an analysis comparing mdNLR to cytological NLR using an independent set consisting of whole blood (WB) DNA methylation measurements across 18 samples with observed cell proportions. The results of this analysis showed a high correlation between mdNLR and cytological NLR (R2 = 0.99) (Supplementary Figure 1). While a small downward bias in our estimates of NLR was observed, the average difference between mdNLR and NLR across the 18 samples was minimal, RMSPE = 0.60 (in NLR units); that is, on average, mdNLR and cytological NLR differed by only 0.60 units.
Head and neck squamous cell carcinoma
A univariate comparison of the mdNLR between cases and controls revealed a statistically significant inflation in the mdNLR among head and neck squamous cell carcinoma (HNSCC) cases (P=1.2×10-10) with the mean mdNLR of cases estimated at 2.99 compared with 1.75 for controls (Figures 2a and 2b). In a multivariable linear regression model adjusted for patient age, gender, race, smoking status, and HPV16 status, HNSCC cases exhibited a significantly larger mdNLR compared to controls (P=2.6×10-10). Within the same model, age was also observed to be positively associated with mdNLR (P=0.009), with each 10-year increment in age being associated with an expected increase of 0.20 in the mdNLR.
Figure 2. Results obtained from examining the association between mdNLR and HNSCC case-control status and survival.
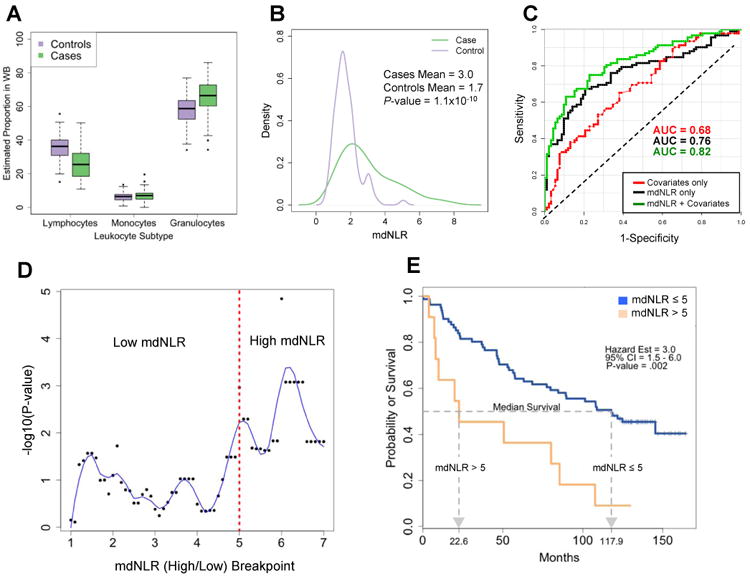
(A) Distribution of the predicted proportion of lymphocytes, monocytes, and granulocytes between HNSCC cases and age-matched cancer-free controls. (B) Distribution of the mdNLR between HNSCC cases and age-matched cancer-free controls. (C) ROC curves demonstrating the ability of mdNLR to correctly classify HNSCC cases from cancer-free controls. Each ROC curve was generated from a different classifier: red ROC curve was based on a classifier that used covariates only (i.e., age, gender, smoking history, and HPV16 status), black ROC curve was from a classifier based on mdNLR only, and green curve was based on a classifier using both mdNLR and the aforementioned covariates. (D) Scatter plot of the –log10 log-rank P-value as a function of the mdNLR breakpoint used to determine mdNLR low and high groups. Blue line represents the estimated lowess smoothed curve. (E) Kaplan-Meier survival curves for the HNSCC cases in the mdNLR high and low groups.
We computed receiver operating characteristic (ROC) curves and corresponding area under the curve (AUC) based on covariate data only (age, gender, race, smoking status, and HPV16 status), mdNLR, and their combination (Figure 2c). The classifier with mdNLR alone was sufficient to distinguish HNSCC cases from controls with an AUC=0.76 (95% CI = [0.69, 0.83]), and including the covariates with mdNLR resulted in an AUC=0.82 (95% CI = [0.76, 0.88]), a statistically significant improvement in the AUC compared to the covariate only classifier (P=0.002).
Among HNSCC cases there was no significant difference in the mdNLR based on the site of the tumor (oral, pharyngeal, and laryngeal: P=0.83), however the mdNLR was elevated in subjects who died during follow-up period compared to those that survived or were censored, mean mdNLR = 3.23 and 2.62, respectively (P=0.07). We next compared survival between HNSCC cases whose mdNLR≤5 (88% of cases) and cases whose mdNLR>5 (12% of cases), and observed that cases in the low mdNLR group had a median survival time that was ∼5.5 times longer than subjects in the high group (Figure 2e; log-rank P=0.002). In a Cox-regression model adjusted for age, gender, smoking status (never versus ever), HPV16 status and tumor stage (I versus II, III, and IV), cases in the high mdNLR group had an approximately 2-fold increase hazard of death compared to those in the low mdNLR group (HR=2.04, 95% CI = [0.97, 4.29]) (Supplementary Table 3). Since it has been shown that tobacco carcinogens alter the blood immune profile associated with cancer survival (16), we tested for and found a statistically significant interaction between mdNLR and survival with smoking status (Table 2; P=0.014). Among those with a high mdNLR, the never-smokers exhibited a 3-fold increased hazard of death compared with ever-smokers (HR = 3.19, 95% CI = [0.71, 14.34]); among those with a low mdNLR, the never-smokers exhibited a 3-fold decreased hazard of death compared to ever-smokers (HR = 0.33, 95% CI = [0.12, 0.91]). Further, a Cox-proportional hazard model treating mdNLR as a continuous predictor also revealed significant effects of both mdNLR and smoking status on overall survival as well as a significant interaction between smoking status and mdNLR (Table 2).
Table 2. Results obtained from examining the association between survival and mdNLR among the Bladder and HNSCC studies.
| Study | Variable | Hazard Ratio (HR) | HR 95% CI | P-value |
|---|---|---|---|---|
| HNSCC | ||||
| mdNLR (cont.) | 2.58 | (1.41, 4.71) | 0.002*** | |
| Age | 1.05 | (1.02, 1.08) | 2.9×10-4*** | |
| Gender | ||||
| Female | (ref) | N/A | N/A | |
| Male | 0.71 | (0.38, 1.31) | 0.270 | |
| Smoking History | ||||
| Never | (ref) | N/A | N/A | |
| Ever | 45.72 | (3.22, 648.56) | 0.005*** | |
| Tumor site | ||||
| Laryngeal | (ref) | N/A | N/A | |
| Oral cavity | 1.13 | (0.53, 2.43) | 0.744 | |
| Oropharyngeal | 2.7 | (1.13, 6.43) | 0.025* | |
| HPV16 | 0.17 | (0.07, 0.42) | 1.2×10-4*** | |
| Tumor stage | ||||
| Stage I | (ref) | N/A | N/A | |
| Stages II, III, IV | 2.53 | (0.97, 6.6) | 0.058+ | |
| Smoking × mdNLR | 0.41 | (0.22, 0.78) | 0.006** | |
| HNSCC | ||||
| mdNLR | ||||
| mdNLR ≤ 5 | ||||
| mdNLR > 5 | 13.87 | (2.76, 69.66) | 0.001*** | |
| Age | 1.05 | (1.02, 1.08) | 4.9×10-4*** | |
| Gender | ||||
| Female | ||||
| Male | 0.71 | (0.38, 1.33) | 0.283 | |
| Smoking History | ||||
| Never | ||||
| Ever | 3.05 | (1.1, 8.45) | 0.032* | |
| Tumor site | ||||
| Laryngeal | ||||
| Oral cavity | 1.19 | (0.56, 2.54) | 0.646 | |
| Oropharyngeal | 2.45 | (1.01, 5.92) | 0.047* | |
| HPV16 | 0.2 | (0.08, 0.48) | 2.9×10-4*** | |
| Tumor stage | ||||
| Stage I | ||||
| Stages II, III, IV | 2.31 | (0.88, 6.1) | 0.090+ | |
| Smoking × mdNLR | 0.1 | (0.02, 0.63) | 0.014* | |
| Bladder | ||||
| mdNLR (cont.) | 1.65 | (1.15, 2.37) | 0.007** | |
| Age | 1.07 | (1.04, 1.10) | 2.3×10-6*** | |
| Gender | ||||
| Female | (ref) | N/A | N/A | |
| Male | 1.62 | (0.96, 2.74) | 0.070+ | |
| Smoking History | ||||
| Never | (ref) | N/A | N/A | |
| Ever | 4.98 | (1.44, 17.17) | 0.011* | |
| Tumor Stage | ||||
| Low (T0a & T1-T3) | (ref) | N/A | N/A | |
| High (Tis & T4) | 2.75 | (1.45, 5.20) | 0.002*** | |
| Smoking × mdNLR | 0.67 | (0.46, 0.97) | 0.033* | |
| Bladder | ||||
| mdNLR | ||||
| mdNLR ≤ 5 | (ref) | N/A | N/A | |
| mdNLR > 5 | 33.67 | (6.77, 167.5) | 1.7×10-5*** | |
| Age | 1.08 | (1.05, 1.11) | 1.7×10-7*** | |
| Gender | ||||
| Female | (ref) | N/A | N/A | |
| Male | 1.61 | (0.95, 2.72) | 0.076+ | |
| Smoking History | ||||
| Never | (ref) | N/A | N/A | |
| Ever | 1.97 | (1.09, 3.55) | 0.024* | |
| Tumor Stage | ||||
| Low (T0a & T1-T3) | (ref) | N/A | N/A | |
| High (Tis & T4) | 3.29 | (1.73, 6.27) | 2.8×10-4*** | |
| Smoking × mdNLR | 0.08 | (0.01, 0.42) | 0.003*** |
P ≤ 0.10,
: P≤ 0.05,
: P≤ 0.01,
: P≤ 0.005
Bladder cancer
A multivariable linear regression analysis adjusted for subject age, gender, smoking status (never, former, current) showed no statistically significant difference in mean mdNLR between bladder cancer cases and controls (P=0.23) (Figures 3a and 3b). However, both age and gender were significantly associated with mdNLR among bladder cancer cases, (P=0.009, and P=0.005, respectively), adjusting for tumor stage, grade, age, gender, and smoking history. In particular, among bladder cancer cases, females had a lower mdNLR on average compared to males (1.88 versus 2.78; P=0.005) and each 10-year increase in age was associated with an expected increase of 0.30 in mdNLR (P=0.009).
Figure 3. Results obtained from examining the association between mdNLR and bladder cancer case-control status and survival.
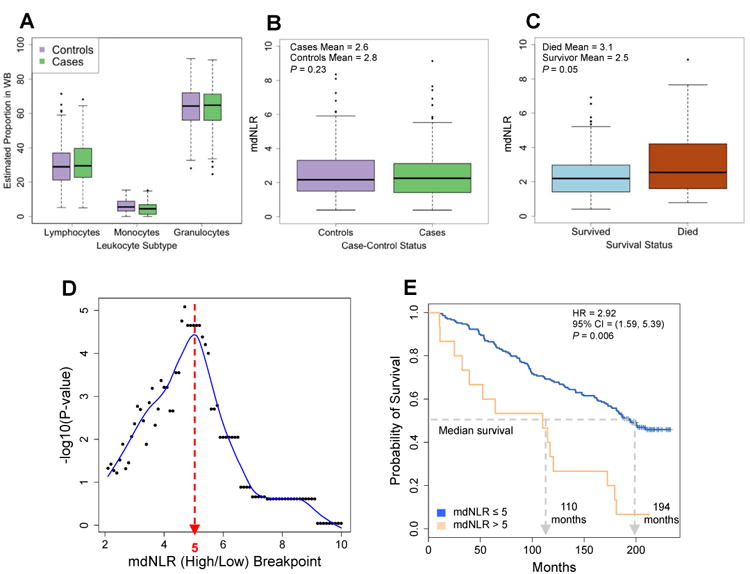
(A) Distribution of the predicted proportion of lymphocytes, monocytes, and granulocytes between bladder cancer cases and cancer-free controls. (B) Distribution of the mdNLR between bladder cancer cases and cancer-free controls. (C) Distribution of the mdNLR among bladder cancer cases that died during the follow-up period compared to those that survived or were censored. (D) Scatter plot of the –log10 log-rank P-value as a function of the mdNLR breakpoint used to determine mdNLR low and high groups. Blue line represents the estimated lowess smoothed curve. (E) Kaplan-Meier survival curves for bladder cancer cases in the mdNLR high and low groups.
Similar to the HNSCC data set, mdNLR was significantly elevated among bladder cancer cases who died during the follow-up period compared to those who were censored or remained alive at the end of the study period (mean=3.14 versus 2.46; P=0.05) (Figure 3c). Also paralleling the HNSCC data set, the optimal cutoff for defining low and high mdNLR groups among bladder cancer cases was found to be 5 (Figure 3d). Based on the optimal cut-point, 7% and 93% of the cases were assigned to high and low mdNLR groups, and a univariate comparison of survival showed that those with an mdNLR≤5 had a median survival nearly twice as long as subjects in the high group (Figure 3e; log-rank P=2.2×10-5). Furthermore, in a model adjusted for age, gender, smoking status (ever versus never) and tumor stage, cases with an mdNLR>5 had an approximately 3-fold increased hazard of death compared to those with an mdNLR≤5 (HR=3.01, 95% CI = [1.69, 5.36]) (Supplementary Table 4). Strikingly, mimicking the results observed in the HNSCC data set, the association between mdNLR and survival also exhibited a statistically significant interaction with smoking status (P=0.003), such that among those with a high mdNLR, never-smokers exhibited a 3.5-fold increased hazard of death compared to ever-smokers (HR = 3.52, P=1.7×10-5) whereas among those with a low mdNLR, never-smokers exhibited a 2-fold decreased hazard of death compared to ever-smokers (HR = 0.51, 95% CI = [0.28, 0.92]). Further, a Cox-proportional hazard model treating mdNLR as a continuous predictor also revealed significant effects of both mdNLR and smoking status on overall survival as well as a significant interaction between smoking status and mdNLR (Table 2). Intriguingly, despite the association between mdNLR and overall survival, there was no significant association between survival and the components used to calculate the mdNLR; granulocytes (HR=2.10; P=0.427) and lymphocytes (HR=0.47; P=0.412).
Ovarian cancer
A comparison of mdNLR between controls and ovarian cancer cases showed that the mdNLR was significantly higher in cases (P=2.2×10-16) (Table 1), and this difference remained significant after adjustment for patient age at blood draw (P=8.0×10-11). Comparing the mean mdNLR of controls versus pre- and post-treatment cases separately revealed a gradient; controls had the smallest average mdNLR (mean=2.08), followed my post-treatment cases (mean=2.47), and pre-treatment cases had the highest mdNLR, (mean=3.84) (Figure 4a). In addition, post-treatment cases had significantly elevated mdNLR compared to cancer-free controls (P=0.002, age-adjusted) and pre-treatment cases had significantly elevated mdNLRs compared to both controls (P=2.2×10-16, age-adjusted) and post-treatment cases (P=1.3×10-6, age-, histology-, and stage-adjusted). To examine the potential of mdNLR to correctly classify controls, pre-treatment cases, and post-treatment cases, we computed ROC curves and corresponding AUCs for each pairwise comparison (Figure 4b). The results from this analysis revealed that mdNLR alone was sufficient to distinguish controls from pre-treatment cases with an AUC=0.79 (95% CI = [0.74, 0.83]). Unexpectedly however, our results showed that the mdNLR was able to better classify pre- and post-treatment cases (AUC=0.69, 95% CI = [0.63, 0.76]) compared to its performance for classifying controls from post-treatment cases (AUC=0.61, 95% CI = [0.55, 0.67]).
Figure 4. Results from the analysis of the mdNLR within the ovarian cancer, breast cancer, and aging data sets.
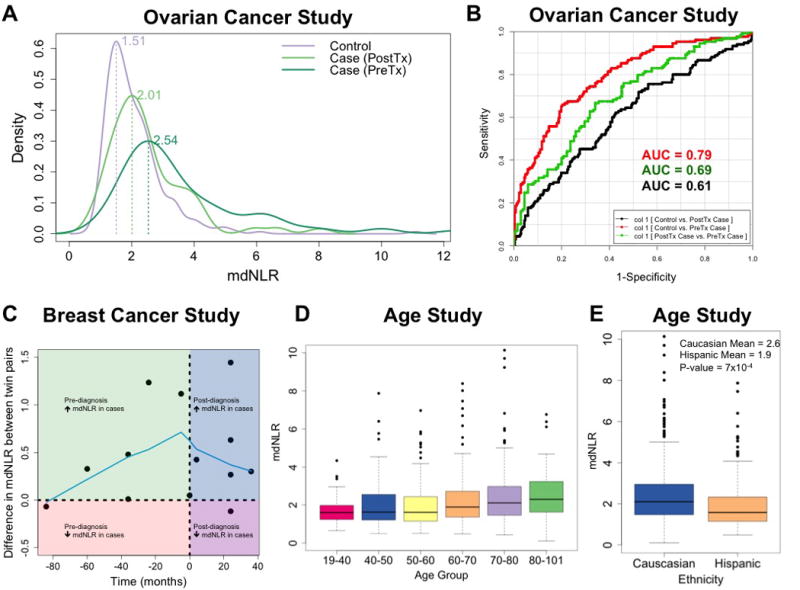
(A) Distribution of the mdNLR between pre-treatment ovarian cancer cases, post-treatment ovarian cancer cases, and age-matched cancer-free controls. (B) ROC curves demonstrating the ability of mdNLR to correctly classify controls versus pre-treatment ovarian cancer cases (red line), controls versus post-treatment ovarian cancer cases (black line), and pre- versus post-treatment ovarian cancer cases (green line). (C) Scatter plot of the within twin-pair difference in the mdNLR between breast cancer cases and controls (y-axis) as a function of whole blood sample collection relative to cancer diagnosis (x-axis). Green quadrant indicates that samples were collected pre-diagnosis and that the mdNLR was higher among the twin-pair member that would eventually be diagnosed with breast cancer. Red quadrant indicates that samples were collected pre-diagnosis and that the mdNLR was lower among the twin-pair member that would eventually be diagnosed with breast cancer. Blue quadrant indicates that samples were collected post-diagnosis and that the mdNLR was higher among the twin-pair member diagnosed with breast cancer. Purple quadrant indicates that samples were collected post-diagnosis and that the mdNLR was lower among the twin-pair member diagnosed with breast cancer. Blue line represents the lowess-smoothed curve estimated using all 13 twin pairs (D) Distribution of mdNLR as a function of age group among the samples in the aging study. (E) Distribution of mdNLR as a function ethnic background among the samples in the aging study.
Breast cancer
Comparing the mdNLR between twins discordant for breast cancer using a Wilcoxon signed rank test showed that subjects with breast cancer had significantly elevated mdNLRs compared to their cancer-free twin (median difference in mdNLR between twin pairs = 0.33) and this difference was statistically significant (P=0.005). We also examined the difference in mdNLR between twin pairs as a function the time pre- versus post-diagnosis at which samples were collected (Figure 4c). A Lowess smoothed curve reflecting the relationship between the twin-pair difference in the mdNLR as a function of sample collection relative to the time of cancer diagnosis was generated based on all 13 twin pairs. Despite limited power due to the small sample size of this study, our results revealed a trend of increasing separation in the mdNLR between twin pairs in the years leading up to cancer diagnosis, which peaked around the time of diagnosis and decreased thereafter (Figure 4c).
Healthy aging
Both the mean and variance of the mdNLR values increased with age (Figure 4d). The mdNLR was, on average, higher among white non-Hispanics (mean=3.49) compared to Mexican Hispanics (mean=2.23) and this difference was statistically significant after adjustment for subject age (P=0.006) (Figure 4e). The mean mdNLRs were also higher for males (mean=3.30) relative to females (mean=2.81), however this difference was not statistically significant (P=0.26).
4. Discussion
Here we demonstrate that a methylation-derived estimate of the NLR displays associations consistent with the simple cytological NLR. Our approach is based on cell mixture deconvolution (23) and only differs from the estimateCellCounts function in the minfi Bioconductor package (35) in the library (i.e., set of L-DMRs) used as the basis for deconvolution. Whereas estimateCellCounts uses a library that is comprised of top 50 hyper- and hypomethylated CpGs between each cell-type (i.e., CD4T, CD8T, natural Killer, B cells, monocytes, and granulocytes) and the remaining five subtypes (J = 600 total L-DMRs comprise the estimateCellCounts library), our library was constructed by identifying the L-DMRs (top 50 hyper- and hypomethylated CpGs) that best discriminated lymphocytes (i.e., CD4T, CD8T, natural Killer, and B cells, collectively), monocytes, and granulocytes. The difference between libraries is subtle, however our decision two select the libraries in this way was based on formulation of NLR (i.e., neutrophil by lymphocyte fraction) and empirical results that suggested more accurate estimates when mdNLR was estimated using the library considered here. Because of the library similarity of these two approaches, comparisons of mdNLR and cytological NLR in our validation analysis were highly similar: R2 = 0.99 when mdNLR was estimated using our approach versus estimateCellCounts.
Our mdNLR estimates were higher in cases across multiple cancer types compared to controls and positively associated with an increased hazard of death in two independent cancer cohorts that included adjustment for established risk predictors. The prognostic importance of the NLR beyond the additive effects of its constituents is underscored by the fact that our analysis of bladder cancer showed a significant association between overall survival and mdNLR, but no associations with the two components of the ratio (i.e. granulocyte and lymphocyte fractions). This is consistent with previous observations using the cytological NLR (40). We further showed that the mdNLR is positively associated with increased aging and varies as expected among ethnic groups (41). Surprisingly, in both bladder and HNSC cancers, cigarette smoking was associated with shorter survival times among patients with non-elevated mdNLR, but not among those with elevated mdNLR scores. Smoking is associated with poor survival in these and other cancers (42,43), however, it has never been shown to interact with any measure of immune status in the manner observed here. This is particularly intriguing given recent observations showing the importance of tobacco and mutagen exposure in shaping treatment response to immunotherapy among tumors with high mutational loads(16). Our data, showing a novel interaction of smoking with the mdNLR may similarly reflect this modification of the immune profile and potentially act as a biomarker of effect for immune therapies. Thus, the mdNLR represents a distinct approach utilizing the leukocyte lineage-determining epigenome that mirrors many established features of systemic inflammation (9-13) and offers promise as an informative survival biomarker.
The mechanisms driving the association of the NLR with cancer survival are not understood. Elevated NLR scores are associated with increases in inflammatory and angiogenic cytokines (44,45) and the levels of MDSCs (46,47). The importance of MDSCs in cancer progression is gaining increased acceptance, as is the fact that MDSCs are major obstacles in the application of new immune checkpoint blockade therapies (48,49). Elevated cytological NLR values are also associated with resistance to checkpoint blockade inhibitors (50,51). It is therefore important to understand the relationships between the NLR and immune dysregulation in cancer. Measuring the mdNLR in the context of genome wide methylation analyses could help identify the epigenetic lineage of phenotypically active cells that are prognostically important; something that has not been possible using the cytological NLR. Discovery of the methylomic features of the lineage of immunomodulatory cells could also provide a path to detecting targets important in cancer inflammation and compromised immune response.
As is the case with all studies, this work is not without some limitations. Similar to examinations into the diagnostic and prognostic potential cytological NLR, the five studies used in this report and their accompanying results, are subject to the same considerations; namely, the external and internal validity of study findings. In this regard, we acknowledge that the mdNLR survival and risk associations reported herein may not be generalizable to the entire population of HNSCC, ovarian, bladder, breast cancer patients. Also, while the data set used to validate mdNLR was relatively small (N=18), 12 of the 18 samples comprising this data set were obtained by mixing leukocyte subtype-specific DNA in known, predetermined proportions. Thus, for these twelve samples, the underlying leukocyte fractions – and, consequently NLR – are known with high confidence and are likely less prone to measurement error associated with cell sorting/counting techniques (i.e., FACS, complete blood cell count (CBC), etc.). Consequently, these twelve samples represent an ideal data set on which to validate the accuracy of our methylation-derived estimates of NLR.
In conclusion, the mdNLR will allow epidemiologists to explore systemic inflammation on an extremely large scale, using archival blood specimens previously not available for this line of inquiry. Our observation of differences in mdNLR among twin pairs discordant for breast cancer suggest that cancer promoting lifestyle and environmental factors that modify host immunity may be revealed through epigenetic analysis of peripheral blood. Such studies aim to integrate environmental and genetic epidemiologic risk factors for cancer, and incorporation of mdNLR opens an opportunity to better understand the etiologic underpinnings of cancer-associated immunomodulation.
Supplementary Material
Acknowledgments
Financial Support: This work was supported by the National Institute of Health (NIH) grants: (1KL2TR000119 to D.C. Koestler), (R01CA52689 and P50CA097257 to J.K. Wiencke), (R01DE022772 and P20GM104416 to B.C. Christensen), (P30CA023108 to C.J. Marsit and M.R. Karagas), Kansas IDeA Network of Biomedical Research Excellence (K-INBRE) Bioinformatics Core supported in part by the National Institute of General Medical Science award P20GM103418 (funded D.C. Koestler), and the Robert Newman Magnin endowment for neurooncology at the University of California, San Francisco (to J.K. Wiencke).
Footnotes
Conflict of Interest: The authors have no conflicts of interest to disclose.
The IDOL R-package has being prepared for submission to the Comprehensive R Archive Network (CRAN). Software is also available by request.
References
- 1.Wu WC, Sun HW, Chen HT, Liang J, Yu XJ, Wu C, et al. Circulating hematopoietic stem and progenitor cells are myeloid-biased in cancer patients. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(11):4221–6. doi: 10.1073/pnas.1320753111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parker KH, Beury DW, Ostrand-Rosenberg S. Myeloid-Derived Suppressor Cells: Critical Cells Driving Immune Suppression in the Tumor Microenvironment. Advances in cancer research. 2015;128:95–139. doi: 10.1016/bs.acr.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindau D, Gielen P, Kroesen M, Wesseling P, Adema GJ. The immunosuppressive tumour network: myeloid-derived suppressor cells, regulatory T cells and natural killer T cells. Immunology. 2013;138(2):105–15. doi: 10.1111/imm.12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marvel D, Gabrilovich DI. Myeloid-derived suppressor cells in the tumor microenvironment: expect the unexpected. The Journal of clinical investigation. 2015;125(9):3356–64. doi: 10.1172/JCI80005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sahakian E, Powers JJ, Chen J, Deng SL, Cheng F, Distler A, et al. Histone deacetylase 11: A novel epigenetic regulator of myeloid derived suppressor cell expansion and function. Molecular immunology. 2015;63(2):579–85. doi: 10.1016/j.molimm.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Youn JI, Kumar V, Collazo M, Nefedova Y, Condamine T, Cheng P, et al. Epigenetic silencing of retinoblastoma gene regulates pathologic differentiation of myeloid cells in cancer. Nature immunology. 2013;14(3):211–20. doi: 10.1038/ni.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng P, Kumar V, Liu H, Youn JI, Fishman M, Sherman S, et al. Effects of notch signaling on regulation of myeloid cell differentiation in cancer. Cancer research. 2014;74(1):141–52. doi: 10.1158/0008-5472.CAN-13-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Condamine T, Gabrilovich DI. Molecular mechanisms regulating myeloid-derived suppressor cell differentiation and function. Trends in immunology. 2011;32(1):19–25. doi: 10.1016/j.it.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yildirim MA, Seckin KD, Togrul C, Baser E, Karsli MF, Gungor T, et al. Roles of neutrophil/lymphocyte and platelet/lymphocyte ratios in the early diagnosis of malignant ovarian masses. Asian Pacific journal of cancer prevention : APJCP. 2014;15(16):6881–5. doi: 10.7314/apjcp.2014.15.16.6881. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Liu P, Xu Y, Zhang W, Tong L, Guo Z, et al. Preoperative neutrophil-to-lymphocyte ratio predicts response to first-line platinum-based chemotherapy and prognosis in serous ovarian cancer. Cancer chemotherapy and pharmacology. 2015;75(2):255–62. doi: 10.1007/s00280-014-2622-6. [DOI] [PubMed] [Google Scholar]
- 11.Salim DK, Mutlu H, Eryilmaz MK, Salim O, Musri FY, Tural D, et al. Neutrophil to lymphocyte ratio is an independent prognostic factor in patients with recurrent or metastatic head and neck squamous cell cancer. Molecular and clinical oncology. 2015;3(4):839–42. doi: 10.3892/mco.2015.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ozcan C, Telli O, Ozturk E, Suer E, Gokce MI, Gulpinar O, et al. The prognostic significance of preoperative leukocytosis and neutrophil-to-lymphocyte ratio in patients who underwent radical cystectomy for bladder cancer. Canadian Urological Association journal = Journal de l'Association des urologues du Canada. 2015;9(11-12):E789–E94. doi: 10.5489/cuaj.3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee SM, Russell A, Hellawell G. Predictive value of pretreatment inflammation-based prognostic scores (neutrophil-to-lymphocyte ratio, platelet-to-lymphocyte ratio, and lymphocyte-to-monocyte ratio) for invasive bladder carcinoma. Korean journal of urology. 2015;56(11):749–55. doi: 10.4111/kju.2015.56.11.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jia W, Wu J, Jia H, Yang Y, Zhang X, Chen K, et al. The Peripheral Blood Neutrophil-To-Lymphocyte Ratio Is Superior to the Lymphocyte-To-Monocyte Ratio for Predicting the Long-Term Survival of Triple-Negative Breast Cancer Patients. PloS one. 2015;10(11):e0143061. doi: 10.1371/journal.pone.0143061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Templeton AJ, McNamara MG, Seruga B, Vera-Badillo FE, Aneja P, Ocana A, et al. Prognostic role of neutrophil-to-lymphocyte ratio in solid tumors: a systematic review and meta-analysis. Journal of the National Cancer Institute. 2014;106(6):1–11. doi: 10.1093/jnci/dju124. [DOI] [PubMed] [Google Scholar]
- 16.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467(7313):338–42. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell stem cell. 2010;6(5):479–91. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calvanese V, Fernandez AF, Urdinguio RG, Suarez-Alvarez B, Mangas C, Perez-Garcia V, et al. A promoter DNA demethylation landscape of human hematopoietic differentiation. Nucleic acids research. 2012;40(1):116–31. doi: 10.1093/nar/gkr685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bocker MT, Hellwig I, Breiling A, Eckstein V, Ho AD, Lyko F. Genome-wide promoter DNA methylation dynamics of human hematopoietic progenitor cells during differentiation and aging. Blood. 2011;117(19):e182–9. doi: 10.1182/blood-2011-01-331926. [DOI] [PubMed] [Google Scholar]
- 21.Bird A. DNA methylation patterns and epigenetic memory. Genes & development. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 22.Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlen SE, Greco D, et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PloS one. 2012;7(7):e41361. doi: 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC bioinformatics. 2012;13(1):86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koestler DC, Marsit CJ, Christensen BC, Accomando W, Langevin SM, Houseman EA, et al. Peripheral blood immune cell methylation profiles are associated with nonhematopoietic cancers. Cancer Epidemiology Biomarkers & Prevention. 2012;21(8):1293–302. doi: 10.1158/1055-9965.EPI-12-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Accomando WP, Wiencke JK, Houseman EA, Nelson HH, Kelsey KT. Quantitative reconstruction of leukocyte subsets using DNA methylation. Genome biology. 2014;15(3):R50. doi: 10.1186/gb-2014-15-3-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Ulm A, Somineni HK, Oh S, Weirauch MT, Zhang HX, et al. DNA methylation dynamics during ex vivo differentiation and maturation of human dendritic cells. Epigenetics & chromatin. 2014;7:21. doi: 10.1186/1756-8935-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alvarez-Errico D, Vento-Tormo R, Sieweke M, Ballestar E. Epigenetic control of myeloid cell differentiation, identity and function. Nature reviews Immunology. 2015;15(1):7–17. doi: 10.1038/nri3777. [DOI] [PubMed] [Google Scholar]
- 28.Koestler DC, Christensen BC, Karagas MR, Marsit CJ, Langevin SM, Kelsey KT, et al. Blood-based profiles of DNA methylation predict the underlying distribution of cell types: a validation analysis. Epigenetics. 2013;8(8):816–26. doi: 10.4161/epi.25430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koestler DC, Jones MJ, Usset J, Christensen BC, Butler RA, Kobor MS, et al. Improving cell mixture deconvolution by identifying optimal DNA methylation libraries (IDOL) BMC Bioinformatics. 2016;17:120. doi: 10.1186/s12859-016-0943-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marsit CJ, Koestler DC, Christensen BC, Karagas MR, Houseman EA, Kelsey KT. DNA Methylation Array Analysis Identifies Profiles of Blood-Derived DNA Methylation Associated With Bladder Cancer. Journal of Clinical Oncology. 2011;29(9):1133–9. doi: 10.1200/jco.2010.31.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langevin SM, Koestler DC, Christensen BC, Butler RA, Wiencke JK, Nelson HH, et al. Peripheral blood DNA methylation profiles are indicative of head and neck squamous cell carcinoma: an epigenome-wide association study. Epigenetics. 2012;7(3):291–9. doi: 10.4161/epi.7.3.19134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Gayther SA, Apostolidou S, et al. An epigenetic signature in peripheral blood predicts active ovarian cancer. PloS one. 2009;4(12):e8274. doi: 10.1371/journal.pone.0008274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heyn H, Carmona FJ, Gomez A, Ferreira HJ, Bell JT, Sayols S, et al. DNA methylation profiling in breast cancer discordant identical twins identifies DOK7 as novel epigenetic biomarker. Carcinogenesis. 2013;34(1):102–8. doi: 10.1093/carcin/bgs321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Molecular cell. 2013;49(2):359–67. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–9. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilhelm-Benartzi CS, Koestler DC, Karagas MR, Flanagan JM, Christensen BC, Kelsey KT, et al. Review of processing and analysis methods for DNA methylation array data. British journal of cancer. 2013;109(6):1394–402. doi: 10.1038/bjc.2013.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maksimovic J, Gordon L, Oshlack A. SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome biology. 2012;13(6):R44. doi: 10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teschendorff AE, Zhuang J, Widschwendter M. Independent surrogate variable analysis to deconvolve confounding factors in large-scale microarray profiling studies. Bioinformatics. 2011;27(11):1496–505. doi: 10.1093/bioinformatics/btr171. [DOI] [PubMed] [Google Scholar]
- 39.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 40.Guthrie GJ, Charles KA, Roxburgh CS, Horgan PG, McMillan DC, Clarke SJ. The systemic inflammation-based neutrophil-lymphocyte ratio: experience in patients with cancer. Critical reviews in oncology/hematology. 2013;88(1):218–30. doi: 10.1016/j.critrevonc.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 41.Azab B, Camacho-Rivera M, Taioli E. Average values and racial differences of neutrophil lymphocyte ratio among a nationally representative sample of United States subjects. PloS one. 2014;9(11):e112361. doi: 10.1371/journal.pone.0112361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Imhoff LC, Kranenburg GG, Macco S, Nijman NL, van Overbeeke EJ, Wegner I, et al. Prognostic value of continued smoking on survival and recurrence rates in patients with head and neck cancer: A systematic review. Head & neck. 2015 doi: 10.1002/hed.24082. [DOI] [PubMed] [Google Scholar]
- 43.Wang LC, Xylinas E, Kent MT, Kluth LA, Rink M, Jamzadeh A, et al. Combining smoking information and molecular markers improves prognostication in patients with urothelial carcinoma of the bladder. Urologic oncology. 2014;32(4):433–40. doi: 10.1016/j.urolonc.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 44.Motomura T, Shirabe K, Mano Y, Muto J, Toshima T, Umemoto Y, et al. Neutrophil-lymphocyte ratio reflects hepatocellular carcinoma recurrence after liver transplantation via inflammatory microenvironment. Journal of hepatology. 2013;58(1):58–64. doi: 10.1016/j.jhep.2012.08.017. [DOI] [PubMed] [Google Scholar]
- 45.Kantola T, Klintrup K, Vayrynen JP, Vornanen J, Bloigu R, Karhu T, et al. Stage-dependent alterations of the serum cytokine pattern in colorectal carcinoma. British journal of cancer. 2012;107(10):1729–36. doi: 10.1038/bjc.2012.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohki S, Shibata M, Gonda K, Machida T, Shimura T, Nakamura I, et al. Circulating myeloid-derived suppressor cells are increased and correlate to immune suppression, inflammation and hypoproteinemia in patients with cancer. Oncology reports. 2012;28(2):453–8. doi: 10.3892/or.2012.1812. [DOI] [PubMed] [Google Scholar]
- 47.Yazawa T, Shibata M, Gonda K, Machida T, Suzuki S, Kenjo A, et al. Increased IL-17 production correlates with immunosuppression involving myeloid-derived suppressor cells and nutritional impairment in patients with various gastrointestinal cancers. Molecular and clinical oncology. 2013;1(4):675–9. doi: 10.3892/mco.2013.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santegoets SJ, Stam AG, Lougheed SM, Gall H, Jooss K, Sacks N, et al. Myeloid derived suppressor and dendritic cell subsets are related to clinical outcome in prostate cancer patients treated with prostate GVAX and ipilimumab. Journal for immunotherapy of cancer. 2014;2:31. doi: 10.1186/s40425-014-0031-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kitano S, Postow MA, Ziegler CG, Kuk D, Panageas KS, Cortez C, et al. Computational algorithm-driven evaluation of monocytic myeloid-derived suppressor cell frequency for prediction of clinical outcomes. Cancer immunology research. 2014;2(8):812–21. doi: 10.1158/2326-6066.CIR-14-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zaragoza J, Caille A, Beneton N, Bens G, Christiann F, Maillard H, et al. Neutrophil to lymphocyte ratio measured before starting ipilimumab treatment is associated with reduced overall survival in patients with melanoma. The British journal of dermatology. 2015 doi: 10.1111/bjd.14155. [DOI] [PubMed] [Google Scholar]
- 51.Ferrucci PF, Gandini S, Battaglia A, Alfieri S, Di Giacomo AM, Giannarelli D, et al. Baseline neutrophil-to-lymphocyte ratio is associated with outcome of ipilimumab-treated metastatic melanoma patients. British journal of cancer. 2015;112(12):1904–10. doi: 10.1038/bjc.2015.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


