Abstract
Identifying and separating a subpopulation of cells from a heterogeneous mixture are essential elements of biological research. Current approaches require detailed knowledge of unique cell surface properties of the target cell population. A method is described that exploits size differences of cells to facilitate selective intracellular delivery using a high throughput microfluidic device. Cells traversing a constriction within this device undergo a transient disruption of the cell membrane that allows for cytoplasmic delivery of cargo. Unique constriction widths allow for optimization of delivery to cells of different sizes. For example, a 4 µm wide constriction is effective for delivery of cargo to primary human T-cells that have an average diameter of 6.7 µm. In contrast, a 6 or 7 µm wide constriction is best for large pancreatic cancer cell lines BxPc3 (10.8 µm) and PANC-1 (12.3 µm). These small differences in cell diameter are sufficient to allow for selective delivery of cargo to pancreatic cancer cells within a heterogeneous mixture containing T-cells. The application of this approach is demonstrated by selectively delivering dextran-conjugated fluorophores to circulating tumor cells in patient blood allowing for their subsequent isolation and genomic characterization.
1. Introduction
Human tissue largely consists of heterogeneous cell populations with varying properties. A major focus of biological research is to understand the contribution of specific cell populations in normal homeostasis and response to disease. Subpopulations of cells are commonly identified by immunological labeling or by genetic tagging with reporter proteins such as green fluorescent protein (GFP).[1] Once identified, these cells are often purified by fluorescence activated cell sorting (FACS). However, such methods require prior knowledge of surface markers or genes specific to the targeted subpopulations of cells. In addition, the dependence on these specific factors may introduce an inherent bias when attempting to perform broad analysis of a cell type. For example, some techniques for circulating tumor cell (CTC) isolation depend on the epithelial marker EpCAM, but recent studies have also demonstrated the presence and biological significance of EpCAM-negative CTCs.[2,3] Indeed it is still largely unknown which subset of CTCs is responsible for metastasis or which surface markers best characterize such cells.
Methods have been developed to isolate cells based on their physical properties such as size, deformability, and photoacoustic properties, thus bypassing the use of cell surface markers and genes.[4–7] Techniques based on physical properties have the potential to overcome biases inherent to marker or gene-based methods. While the selective tagging or manipulation of target cells within a mixture of cells is of great value for research, diagnostics, and therapeutics, the physical properties of cells have yet to be used as differentiating factors for selective delivery of payload to cells.
Current methods for selective delivery largely rely on interactions at the surfaces of cells, such as ones dependent on electrostatic properties and hydrogen bonds, or cell surface receptors and molecules, such as the cluster of differentiation molecules and proteoglycans.[8,9] Moreover, these methods rely on endocytosis or pinocytosis for delivery, which can lead to subsequent undesired lysosomal degradation of all or significant portions of the delivered payload. The efficacy of these delivery methods can be enhanced by decreasing lysosomal degradation or avoiding lysosomal uptake, but perhaps the most effective way to reduce lysosomal degradation is to avoid the dependence on cell surface interactions altogether.[10–12] Methods that transiently induce cell membrane disruption allow diffusion of payload into the cells. A commonly employed technique of plasma membrane disruption is by means of electrical forces, such as electroporation. However, these electrical methods are limited by their inability to deliver materials selectively among a heterogeneous mixture of cells and are often restricted to nucleic acids.
A method for cytosolic delivery that is independent of surface-expressed markers on cells has been demonstrated using a microfluidic device that disrupts the cell membrane using mechanical forces as cells traverse through tight constrictions.[13–15] The delivery of a wide range of cargo sizes (3 kDa to 2 MDa) can be achieved with high efficiency and uniform distributions, suggesting that the size of holes resulting from the membrane disruption are homogeneous within that population of cells.[15,16] While this transient disruption of the cell membrane allows for bidirectional movement of material across the membrane, cells remain viable and retain their proliferative capacity and biological activity.[13,15–17] Here we demonstrate the use of a microfluidic system to confer selective cytosolic delivery within a mixture of cells based on cell size. We further validated this approach by demonstrating size-selective delivery to the large circulating tumor cells found in the blood of pancreatic cancer patients, allowing for their identification and genomic analysis.
2. Results
2.1. Delivery Approach
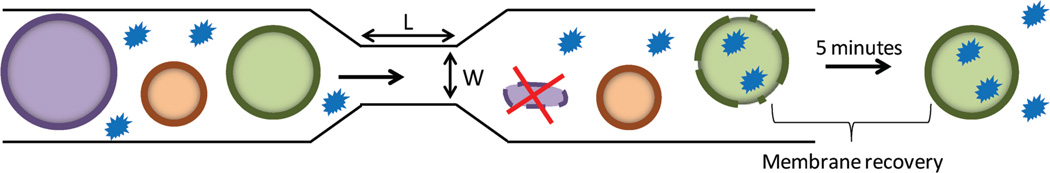
We previously described a microfluidic platform that allows intracellular delivery of macromolecules to cells based on membrane deformation.[15] We hypothesized that by selecting the appropriate deformation parameters, one could selectively disrupt the membrane of target cells while leaving those of nontarget cells intact. To this end, we employed microfluidic chips comprising 75 parallel channels etched onto a silicon chip sealed by a Pyrex layer.[15] Each channel had a central constriction of a specific length and width. We posited that during transit through this constriction, the cell membranes would be disrupted by mechanical deformation and this temporary disruption would facilitate diffusion of small molecules into the cytoplasm of the cells. The degree of deformation is a function of the cell diameter relative to constriction width. Figure 1 illustrates the effect encountered by cells of different sizes as they traverse through a constriction of specific width.
Figure 1.
Microfluidic delivery platform. Each microfluidic chip has 75 parallel channels, and each channel has a central constriction of 10 µm in length (L), and 4 to 9 µm in width (W). This illustration demonstrates the transit of cells through a single channel and its corresponding central constriction. As the cells pass through the constriction, there is temporary disruption of the cell membrane, and this allows for intracellular diffusion of materials (green cell). When a cell traverses a constriction whose width is too narrow it results in cell death (purple cell). Conversely, when the constriction width is too large the mechanical deformation is insufficient to cause sufficient membrane disruption to enable delivery (brown cell).
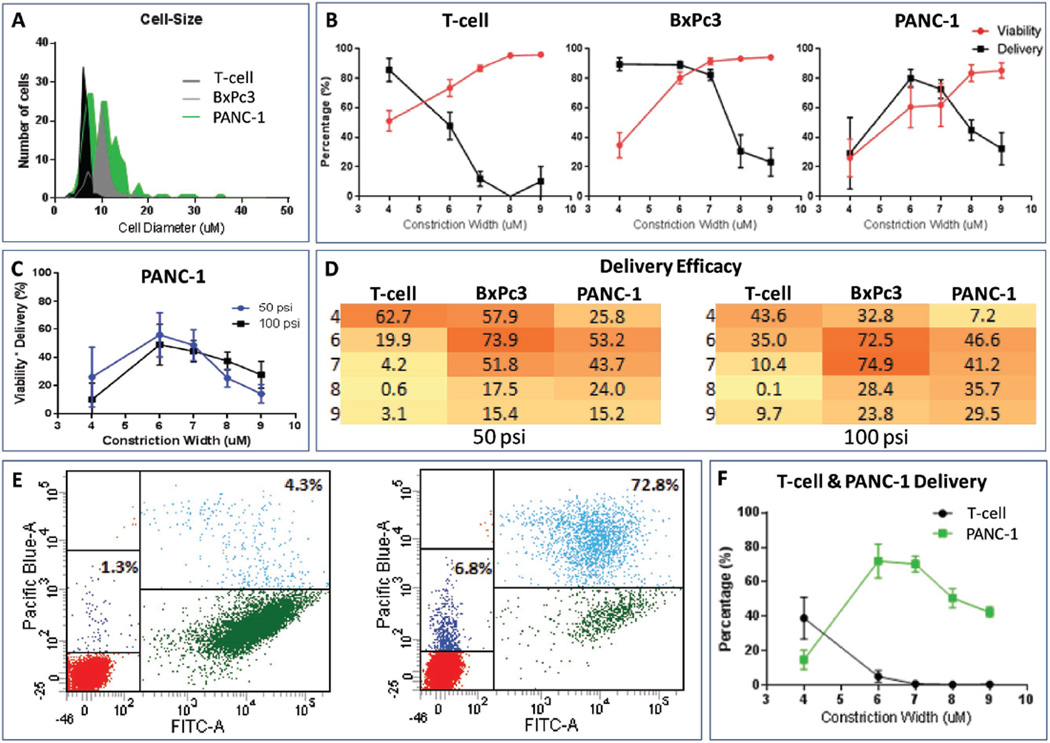
To evaluate the size-selectivity of the delivery platform, we employed cells with different average cell diameters: primary human T-cells (6.7 µm) and two pancreatic cancer cell lines BxPc3 (10.8 µm) and PANC-1 (12.3 µm) (Figure 2A). A suspension of each cell type along with a fluorophore-conjugated macromolecule (cascade blue-conjugated 3 kDa dextran polymer) was delivered through devices with varying constrictions (4 to 9 µm wide). Endocytic uptake and surface binding are not observed when cells are incubated with the fluorophore-conjugated dextran polymer (data not shown).[13,15] However, efficient intracellular delivery of the dextran polymer occurs after passage through the microfluidic device (Figure S1, Supporting Information). The percentage of cells with intracellular dextran was determined using FACS based on the fluorescence of the labeled dextran. Constrictions that were several micrometers less than the average width of the cell allowed the maximum delivery of the dextran polymer (Figure 2B). For example, an average of 85.5% T-cells received intracellular dextran when the constriction was 4 µm. As the constriction width increased, the average delivery progressively decreased to less than 12% when the constriction exceeded the average diameter of the T-cells (≥7 µm constriction widths compared to cell diameters of 6.7 µm, Figure 2B). A similar trend was observed for BxPc3 and PANC-1 cells, with highest intracellular delivery observed with 6 µm constrictions and progressively lower levels of delivery as the width of the constriction approached the average cell diameter.
Figure 2.
Cell size specific delivery. A) Diameters of the three different cell types (T-cells, BxPc3, PANC-1 cells). B) Percentage delivery and viability of T-cell (left), BxPc3 (center), and PANC-1 (right) when passed through chips with different constriction widths at 100 psi. The average and standard deviation of 3 to 5 independent experiments are shown. C) Delivery efficacy of PANC-1, where delivery efficacy is the product of percent viability and percent delivery at each condition (constriction width and pressure). D) Heat map of delivery efficacy when a suspension of one cell type (x-axis) is passed through chips with different constriction widths (y-axis) at 50 and 100 psi. Darker colors indicate higher delivery efficacy and lighter colors indicates lower delivery efficacy. E) Flow cytometry plot of a mixture of T-cells (low FITC region) and PANC-1 (high FITC region) cells incubated with the dextran blue dye (Pacific Blue), but not processed through the microfluidic delivery platform (left panel) demonstrating negative controls to establish the basal levels of surface receptor-mediated binding and nonspecific uptake of dye. Flow cytometry plot of a mixture of T-cell and PANC-1 cells passed through a chip with constrictions that were 6 µm wide (and 10 µm long) at 100 psi (right panel). F) Percentage delivery of a mixture of T-cell and GFP-labeled PANC-1 cells when passed through chips with different constriction widths at 100 psi. Each condition represents the average and standard deviation of three independent experiments.
Interestingly, we also observed poor delivery of intracellular dextran in BxPc3 and PANC-1 cells when the constriction width was very narrow. To determine the mechanism of this effect, cell viability was measured using the viability dye propidium iodide (PI). These experiments demonstrated that the narrower constrictions lowered cell viability and, as a result, there was an optimal range of widths for the constriction for which maximum cell viability and dextran delivery were achieved for each cell type (Figure 2B). As these experiments demonstrate, the constriction widths that maximize delivery and cell viability are not necessarily the same, and therefore one must balance these two objectives. To determine the optimal channel sizing we developed a metric called delivery efficacy, a product of delivery (%) and viability (%). The highest delivery efficacy was achieved with a 4 µm wide constriction for T-cells (Figure 2D), with an average delivery of 85.3% and average viability of 73.5% when delivered at 50 psi (Figure S2, Supporting Information), and average delivery of 85.5% and average viability of 51.0% when delivered at 100 psi (Figure 2B). Since the BxPc3 and PANC-1 cells were close in cell diameter, their optimal constrictions were similar, 6 or 7 µm wide, depending on the delivery pressure (Figure 2D). As noted in a previous study, the applied pressure also affected the delivery efficacy (Figure 2C,D).[15] We observed that higher pressure correlated with higher delivery but lower viability for a given constriction width (Figure S2, Supporting Information). When the constriction width was narrower than its optimal value for a given cell size, the higher delivery pressure likely led to extensive membrane disruption that was beyond the ability of the cell to repair and reseal.
We then assessed the cell size selectivity of the microfluidic device in a heterogeneous cell population. In order to verify the size-selective delivery in a mixed population, these experiments required one cell population to express a marker that would readily allow for the differentiation of the two cell types by FACS. The PANC-1 cells employed here express GFP and thus could be differentiated from the T-cells during FACS based on their green fluorescence. While BxPc3 cells were not analyzed in these experiments, BxPc3 and PANC-1 cells share a common effective constriction width and therefore we expected these cells to exhibit a similar performance in these experiments. A suspended mixture of T-cells and PANC-1 cells along with the cascade blue-conjugated 3 kDa dextran fluorophore was passed through the device using chips with different constriction widths. As expected, the constriction width that allows for maximum fluorophore delivery for each cell type remained the same (Figure 2E,F).
Interestingly, the delivery into T-cells was low when delivered together with PANC-1 cells as compared to when delivered alone, even at their optimal constriction width of 4 µm (39.0% versus 85.5% delivery at 100 psi, respectively). The reason is unknown, but since we noticed that the flow rate within the chip was reduced during delivery, we hypothesize that the flow characteristics within the chip may have been altered due to accumulation of debris from the PANC-1 cells that were destroyed while traversing through constriction widths that were too narrow for their cell diameter. Importantly, at their optimal constriction width, larger cells achieved fluorophore delivery at a significantly higher percentage than smaller cells; relative to T-cells, delivery into PANC-1 cells was 14-fold higher with 6 µm wide constrictions and 82-fold higher with 7 µm wide constrictions. Figure 2E shows such an example where when a mixture of T-cells and PANC-1 was passed into a chip with a 6 µm constrictions widths, 72.8% of the viable PANC-1 cells (high FITC region) clustered in the high dextran blue region and only 6.8% of the viable T-cells (low FITC region) clustered in the high dextran blue region.
2.2. Selective Delivery to Circulating Tumor Cells
CTCs are the rare tumor cells found in the bloodstreams of cancer patients and believed to be responsible for metastasis.[18] Numerous methods have been developed to detect or enrich CTCs based on size or surface-expressed markers, but none have demonstrated selective intracellular delivery to CTCs. Such an approach could have a range of implications for research, diagnostics, and therapeutic development. Considering our ability to label specific cell types within a heterogeneous population, we explored whether we could selectively deliver material to CTCs directly.
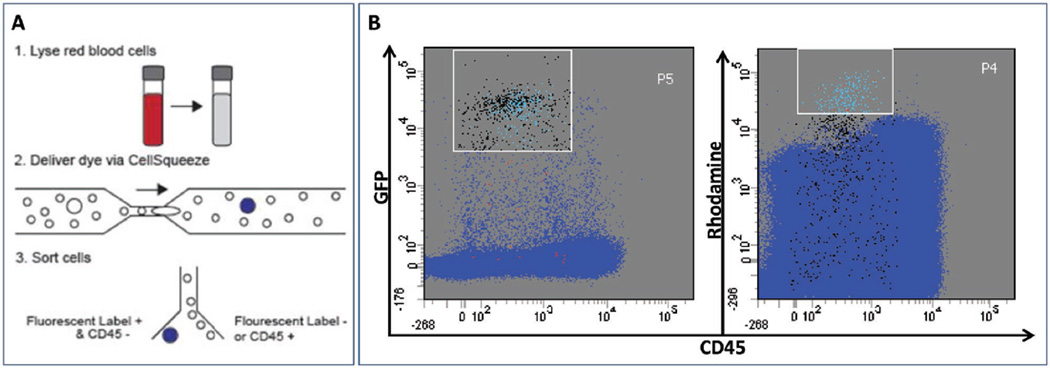
To assess the potential performance of our approach for labeling tumor cells in clinical samples, healthy patient whole blood was spiked with various amounts of GFP-expressing PANC-1 cells. These samples were depleted of red blood cells by lysis, resulting in a cell suspension consisting primarily of leukocytes and PANC-1 cells. This cell suspension was mixed with a tetramethylrhodamine-conjugated 10 kDa dextran fluorescent dye and passed through a chip whose constrictions were 7 µm wide (Figure 3A). Tetramethylrhodamine-conjugated dextran was chosen for these experiments for compatibility with downstream FACS analysis; while larger than the 3 kDa cascade blue-conjugated dextran employed earlier, delivery efficiency is not affected by the molecular weight of dextran.[15,16] GFP expression in PANC-1 cells provided an independent signal to confirm successful labeling of tumor cells. To further enhance the selectivity of the method, cells were stained with an antibody specific to CD45 to exclude contaminating dextran-positive leukocytes during FACS sorting. Analysis of the treated samples by FACS indicated that most of the cells in the high dextran, low CD45 region also displayed high green fluorescence, indicating that we were able to deliver the tetramethylrhodamine dextran selectively into the GFP-expressing PANC-1 cells with high specificity. Experiments performed with blood spiked with varying concentrations of PANC-1 cells demonstrated highly specific delivery into PANC-1 cells, with 75% (Figure S2, Supporting Information) and 92% (Figure 3B) specificity observed with low (200 cells per mL) and high (2000 cells per mL) concentrations of PANC-1 cells, respectively.
Figure 3.
Selective labeling of blood spiked with tumor cells. A) Schematic diagram for CTC isolation from whole blood. 1. GFP-expressing PANC-1 cells were spiked into whole blood and then depleted of red blood cells. 2. Red blood cell-depleted sample was delivered through device in the presence of tetramethylrhodamine dextran-labeled dye. 3. Cells were counterstained with an anti-CD45 antibody (APC) and GFP-positive CD45-negative cells were isolated by FACS. B) FACS plot demonstrating high specificity in tagging PANC-1 cells when PANC-1 cells are spiked into whole blood at high concentration (2000 cells per mL). GFP-expressing PANC-1 cells tagged with the rhodamine fluorophore were independently verified based on GFP fluorescence. The P4 gate [high rhodamine, low CD45 region] was used as a basis for sorting high candidate CTCs, and the P5 gate [high GFP, low CD45 region] was used to sort for GFP-expressing PANC-1 cells. The light blue dots within P5 are accurate hits (i.e., cells that are present within both P4 & P5 gates), such that those are GFP-expressing PANC-1 cells with intracellular rhodamine; 92% of the cells within P4 are accurate hits. False positive hits are red, and false negative hits are black. Within a mixture of about 1.2 million viable cells, we were able to isolate 227 GFP-expressing PANC-1 cells using the size-selective delivery platform.
We further validated that the sorted cells were indeed PANC-1 cells by performing targeted genomic sequencing. PANC-1 cells have a loss of heterozygosity in TP53 and a missense mutation at codon 273 (p.R273H), and a heterozygous mutation in KRAS codon 12 (p.G12D).[19] Replicate populations of 50 recovered PANC-1 cells from FACS sorting were amplified by multiple displacement amplification (MDA) and sequenced for a panel of 20 oncogenes representative of mutations commonly observed across multiple cancer types. Bulk genomic DNA (gDNA) from the blood of the healthy donor and the PANC-1 cell line were also sequenced. Missense TP53 (p.R273H) and KRAS (p.G12D) somatic mutations were evident in the cells recovered by intracellular labeling in the spiking studies and the PANC-1 controls but not in the healthy donor blood (Figure S4, Supporting Information). Preferential amplification of alleles in whole-genome amplification by MDA limits the ability to estimate the fraction of tumor cells relative to normal cells based purely on alternate and reference allele counts.[20] Nonetheless, the identification of the KRAS and p53 variants found in PANC-1 cells within the cell population isolated based on intracellular tetramethylrhodamine dextran demonstrate the capability of the size-selective delivery method to sort CTCs based on high dextran, low CD45 gates.
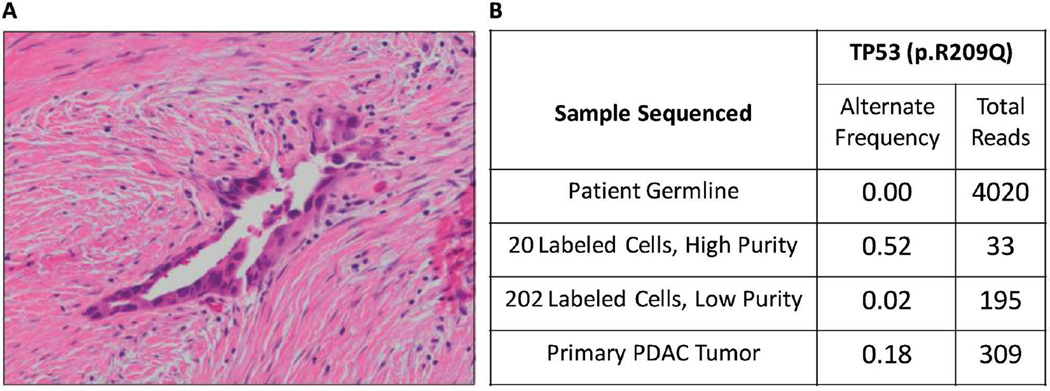
CTCs in patients are extraordinarily rare, with generally less than 10 cells found per mL of blood.[21] To illustrate the utility of this size-selective delivery method for targeting such rare cells, we processed blood from a patient with pancreatic duct adenocarcinomas (PDAC). A blood draw was obtained from a patient (HTB1760) prior to surgical resection of their tumor (Figure 4A). After RBC lysis, the remaining cells were processed through our microfluidic platform along with the tetramethylrhodamine-conjugated 10 kDa dextran polymer. Cells were sorted and recovered with both low and high purity gates, indicative of likely high and low leukocyte contamination by anti-CD45 antibody labeling, respectively. Targeted sequencing of the recovered cells identified a missense mutation in TP53 (R209Q) in both high and low purity samples (Figure 4B) that was absent in the bulk gDNA from the blood. The genetic alterations found in CTCs have been demonstrated to accurately reflect those of primary and metastatic tumors.[22] Therefore, parallel sequencing of DNA isolated from laser capture microdissected cells from the primary tumor was performed to validate the nature of the size-selected cells. This analysis revealed the same TP53 mutation in the primary tumor, confirming that tumor cells were present among the recovered population.
Figure 4.
Isolation of patient’s PDAC cells. A) Histopathology of the primary tumor (HTB1760) confirmed pancreatic ductal adenocarcinoma. B) Frequency of TP53 mutation (R209Q) identified in DNA from the tumor, high and low purity FACS purified Rhodamine-positive CTCs, and matched germline DNA.
3. Discussion
The work described here demonstrates the ability of a physical property such as cell size to facilitate selective cytosolic delivery of materials in heterogeneous cell populations—a potentially enabling method for selectively tagging and manipulating cells independently of surface markers. The use of physical properties to differentiate cells in a heterogeneous mixture would allow for the manipulation of a subpopulation of cells independent of surface molecules. We demonstrated the use of one such property, cell size, to facilitate selective cytosolic delivery of compounds in heterogeneous cell populations. The mechanism employed here (mechanical disruption) obviates the need to identify surface molecules a priori and can thus reduce the selection bias intrinsic to current techniques.
The microfluidic device has demonstrated applicability in delivering a variety of materials with a wide range of sizes that are retained by the cells, allowing for the manipulation of their biological properties.[13,15–17,23] Here, our use of in vitro CTC models and clinical PDAC patient samples has demonstrated the additional utility of this platform to selectively deliver payload within a heterogeneous population of cells. Moreover, this method may be of particular benefit for delivering payloads that are sensitive to endocytic degradation as the direct delivery of payloads into the cytosol bypasses the endosomal uptake processes and thus expands one’s ability to manipulate cell function.[14] The underlying intracellular delivery technique thus has the potential to facilitate co-delivery of intracellular probes and/or functional materials to enable in vitro and ex vivo cell manipulation.[13–15]
Despite the aforementioned advantages of this delivery platform, it does have a few limitations. Recent studies have demonstrated that mechanical stresses imparted by passage through microfluidic channels can disrupt the nuclear membrane and induce DNA damage.[24] Although not a focus of our prior analysis, we have not observed loss of nuclear integrity or cell function when cells are run on our platform.[13,15,17] It is likely that the larger channels contained in our device do not have significant effects on the nuclear membrane. However, investigation of nuclear integrity may be necessary for studies in which long-term growth of cells is required. There may also be unique situations where analysis of select cellular pathways may be obscured shortly after cytoplasmic membrane disruption. The rapid and Ca+-dependent restoration of cell membrane integrity after passage through the microfluidic device is consistent with active membrane repair.[16,25] This mechanism involves vesicular transport,[26] and while there does not appear to be any long-term biological consequences of these events, components of these pathways may be diverted from their normal cellular functions shortly after delivery of payload.
In its current manifestation, if the difference in cell diameters between subpopulations is too small, the device cannot provide differentiating cytosolic delivery. Another apparent constraint is that only the larger cell population can receive cytosolic materials within a heterogeneous mixture. This constraint is because a chip sized to preferentially deliver to the smaller population would likely lyse the larger population, or potentially clog (Figure 2B). Nevertheless, one can envision step-wise isolation and cytosolic delivery sequentially from larger to smaller cells within a heterogeneous cell population using consecutively smaller constriction widths with each treatment through the device. Finally, there may be other unexamined properties that could affect delivery efficacy. For example, cell stiffness and shape (e.g., spherical vs biconcave), as well as chemical properties such as hygroscopic effects and electrostatic interactions may have significant impacts on delivery performance and these areas could be the subject of future studies.
4. Conclusion
The microfluidic device described here expands the scope of properties that can be exploited for identifying and manipulating cells. Size-selective cytosolic delivery is not only a unique approach, but also provides several potential advantages over current methods of delivery. There are inherent limitations to the technique, but the advantages offered over existing systems have some important implications. We demonstrated its ability to tag with high specificity in in vitro CTC models and in vivo patient samples. This size-dependent method decreases selection bias inherent to surface marker-dependent methods and introduces a method of separating and manipulating cells when cell surface markers and genotype are not known a priori. By allowing one to combine immunological information with size-selective delivery, this approach could potentially enable novel approaches to diagnostics and drug discovery.
5. Experimental Section
Cell Culture
The BxPc3 and GFP-expressing PANC-1 cell lines were cultured in DMEM (11965118, ThermoFisher Scientific) with 10% FBS (ThermoFisher Scientific) and 1% Penicillin–Streptomycin (15140122, ThermoFisher Scientific). To maintain the GFP-expressing population of PANC-1 cells, Geneticin (10131027, ThermoFisher Scientific) was also added to PANC-1 cell cultures to a final concentration of 200 µg mL−1. For routine passage and preparation for delivery into the microfluidic device as described below in “Delivery,” sub-confluent cultures of PANC-1 and BxPc3 cells were dissociated from their culture flasks with trypsin-EDTA (25200056, ThermoFisher Scientific).
T-Cell Isolation
Primary T-cell populations were obtained from fresh healthy human whole blood (Research Blood Components) that was drawn the same day as delivery into the microfluidic device. The cells were depleted of RBC with the addition of Ammonium-Chloride-Potassium (ACK) lysing buffer (0.15 m NH4Cl, 10 × 10−3 m KHCO3, 0.1 × 10−3 m Na2EDTA, pH 7.2–7.4) at a 1:10 volume ratio of cell solution to ACK buffer. Chemicals to make ACK were obtained from Sigma-Aldrich. After a 10 min incubation, the cells were centrifuged at 350 RCF for 4 min and washed twice in Human media (Advanced RPMI (12633012, ThermoFisher Scientific) with 10% Human Serum AB (100-318, Gemini Bio Products Inc.) and 1% Penicillin–Streptomycin). T-cells were then isolated by immunomagnetic negative selection with Protocol C in the EasyStep Human T-cell Enrichment Kit (19051, Stemcell Technologies).
Delivery with Microfluidic Device
Microfluidic chips and delivery reservoirs were obtained from SQZ Biotech, and assembled as per their instructions. Chips contained 75 parallel channels with each exhibiting a uniform height of 20 µm across their length. Each channel contained a single central constriction that was 10 µm long, and 4 to 9 µm wide. The cannel on either side of the constriction was 30 µm wide. The microfluidic chip and reservoir were stored in ethanol, so the assembled system was washed with PBS before use with cells. The two cell lines (BxPc3 and GFP-expressing PANC-1) were delivered at room temperature. T-cells were delivered with the reservoir and microfluidic chip sitting on ice, which allowed for improved intracellular delivery for these primary cells. The cells were first suspended in PBS at a concentration of 2 × 106 cells per mL for the two cell lines and 2 × 107 cells per mL for the T-cells. Cascade blue-conjugated 3 kDa dextran polymer (D7132, ThermoFisher Scientific) was added into the cell solution at a final concentration of 0.15 mg mL−1. 100 µL of the cell-dye solution was loaded into the reservoir inlet, and pushed through the microfluidic chip at 50 or 100 psi pressure. After traversing through the chip, the cells were collected from the reservoir outlet, and cell lines were placed in room temperature and T-cells on ice. After a 5 min incubation, 100 µL of the cell-appropriate media was added to the cells (i.e., DMEM-based media for the cell lines and Human media for T-cells) to promote resealing of the pores that formed during transit through the constrictions. The cells were then centrifuged at 400 RCF for 4 min and washed with PBS. Cells were centrifuged again at 400 RCF for 4 min, and prepared for FACS as described below. Note, cells incubated with the dextran dye, but not processed through the microfluidic delivery platform, were used as negative controls to establish the basal levels of surface receptor-mediated binding and nonspecific uptake of dye.
CTC Cell Spike
5 mL of healthy human whole blood (Research Blood Components) was spiked with GFP-expressing PANC-1 cells for a final concentration of 200 or 2000 cells per mL of blood. The spiked whole blood was depleted of RBC with the addition of RBC lysis buffer (00-4300-54, eBioscience) at a 1:10 volume ratio of cell solution to lysis buffer, and placed on a tube rotator for 10 min. The remaining cells (mostly leukocytes and PANC-1 cells) were centrifuged at 212 RCF for 5 min and resuspended in 1200 µL of PBS. The cells were centrifuged again at 846 RCF for 5 min, and resuspend in 500 µL of PBS. Tetramethylrhodamine-conjugated 10 kDa dextran fluorescent polymer (D1816, ThermoFisher Scientific) was added to the suspended cells to a final concentration of 0.4 µg µL−1. 150 µL of the cell-dye solution was loaded into the reservoir inlet and passed through a microfluidic chip optimized for PANC-1 delivery at 50 psi on ice. The cells were collected from the reservoir outlet, and placed on ice. After a 5 min incubation, 500 µL of DMEM with 10% FBS and 1% Penicillin–Streptomycin was added to the cells. The cells were centrifuged at 846 RCF for 5 min and resuspended in 200 µL of PBS containing 0.2% bovine serum albumin (A9418, Sigma-Aldrich). 10 µL of Alexa Fluor 647 anti-human CD45 Antibody (clone HI30, Biolegend) was added to the cell solution to stain the leukocytes, and cells were incubated on ice for 30 min. The cells were then centrifuged at 846 RCF for 5 min, washed with PBS, and centrifuged again at 846 RCF for 5 min and prepared for FACS as described below. Two negative controls were employed for these experiments: (1) RBC-depleted whole blood that was not spiked with PANC-1 cells was incubated with the dextran dye and processed through the microfluidic delivery platform to establish delivery of dye into leukocytes and (2) GFP-expressing PANC-1 cells incubated with the dextran dye, but not processed through the microfluidic delivery platform, to establish the basal levels of surface receptor-mediated binding and non-specific uptake of dye.
Patient Sample
Whole blood was obtained from a patient (HTB1760) undergoing a Whipple resection of a moderately differentiated pancreatic ductal adenocarcinoma after obtaining informed consent in accordance with the Massachusetts General Hospital institutional review board. Blood was processed as described above in the “CTC cell spike” section. Cells from a formalin fixed paraffin embedded sample of the resected tumor were isolated by laser capture microdissection at the Advanced Tissue Resources Core at the Massachusetts General Hospital.
FACS
FACS buffer was prepared with 3% FBS and 1% F-68 Pluronics (P1300, Sigma-Aldrich) in PBS. 10 µL of 0.1 mg mL−1 concentration propidium iodide (P4864, Sigma-Aldrich) viability dye was added to 1 mL of FACS buffer. Cells were resuspended in the resulting solution of propidium iodide (PI) and FACS buffer and analyzed by FACS. Viable cells were gated in the low PI region. The cells with the intracellular cascade blue dextran polymer were gated in the high Pacific-Blue region. The cells with the intracellular rhodamine dextran polymer were gated in the high PE-TxRed region. Two flow cytometers were used for cell analysis, LSR HTS-2 and LSR Fortessa HTS, and one flow cytometer was used for cell sorting, Aria. All flow cytometers were from BD Biosciences.
Whole Genome Amplification
3 µL of lysis buffer (1.83 m KOH and 0.42 m DTT, Sigma-Aldrich) was added to the wells of a 96-well PCR plate (Eppendorf) containing 30 µL of sorted cells. The plate was incubated for 10 min at 50 °C and 3 µL of 2 m HCl (Fluka Analytical) was added. Multiple displacement amplification was performed on a thermal cycler (Eppendorf) for 2 h in 106 µL total volume, containing 36 µL of DNA post lysis, 2 µL RepliPHI (Epicentre), 2.5 µL of 10 × 10−3 m random hexamers (IDT), 1 µL of 10 mg mL−1 BSA (NEB), 0.4 µL of 1 m DTT (Sigma-Aldrich), 10 µL of 10× reaction buffer (Epicentre), 1.6 µL of 25 × 10−3 m dNTPs (Epicentre), and 52.5 µL of sterile water (Invitrogen) per reaction. DNA was cleaned up using AmpureXP beads (A63880, Beckman Coulter) by splitting each reaction into two 53 µL wells and adding 100 µL of Ampure XP to each reaction, incubating for 5 min on a magnetic plate, removing supernatant and replacing with fresh 100 µL of 70% ethanol twice, then air drying on the magnet for 10 min. The beads were resuspended in 50 µL of Tris-EDTA buffer, pH 8 (T0224, Teknova), incubated 5 min off of the magnet, then placed back on the magnet for another 5 min. The supernatant was removed for quantification using the Quant-IT PicoGreen dsDNA assay (P11496, ThermoFisher Scientific).
Targeted Sequencing
DNA was adjusted to 4 ng µL−1 and used in the GeneRead DNAseq Breast Cancer Gene Panel (NGHS-001X, Qiagen). Since a pancreatic cancer-specific panel was not available, we chose to use the breast cancer panel. Amplicons were generated via this method for a panel of 20 genes, and library preparation was performed using the NEBNext DNA Library Prep Master Mix Set for Illumina (E6040L, New England Biolabs). Libraries were quantitated using the Kapa Library Quantification Kit (Kapa Biosystems) and loaded onto the Illumina MiSeq for sequencing. Data analysis was performed using the GeneRead data analysis portal (Qiagen).
Supplementary Material
Acknowledgments
M.T.S., A.S., and V.K.A. contributed equally to this work. This work was supported by National Institutes of Health under Grant Nos. R01GM101420-01A1 and P01CA117969, the Kathy and Curt Marble Cancer Research Fund, the Ragon Institute of Harvard/MIT/ MGH, and the Koch Institute Support (core) GrantP30-CA14051 from the National Cancer Institute.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Dr. May Tun Saung, Department of Chemical Engineering, Koch Institute for Integrative Cancer Research at MIT, Cambridge, MA 02139, USA
Dr. Armon Sharei, Department of Chemical Engineering, Koch Institute for Integrative Cancer Research at MIT, Cambridge, MA 02139, USA
Dr. Viktor A. Adalsteinsson, Department of Chemical Engineering, Koch Institute for Integrative Cancer Research at MIT, Cambridge, MA 02139, USA
Nahyun Cho, Department of Chemical Engineering, Koch Institute for Integrative Cancer Research at MIT, Cambridge, MA 02139, USA.
Tushar Kamath, Department of Chemical Engineering, Koch Institute for Integrative Cancer Research at MIT, Cambridge, MA 02139, USA.
Camilo Ruiz, Department of Chemical Engineering, Koch Institute for Integrative Cancer Research at MIT, Cambridge, MA 02139, USA.
Jesse Kirkpatrick, Department of Chemical Engineering, Koch Institute for Integrative Cancer Research at MIT, Cambridge, MA 02139, USA.
Nehal Patel, Advanced Tissue Resources Core, Massachusetts General Hospital, Charlestown Navy Yard, Charlestown, MA 02129, USA.
Dr. Mari Mino-Kenudson, Department of Pathology, Massachusetts General Hospital, Boston, MA 02114, USA
Dr. Sarah P. Thayer, Department of Surgery, Massachusetts General Hospital, Boston, MA 02114, USA
Dr. Robert Langer, Department of Chemical Engineering, Koch Institute for Integrative Cancer Research at MIT, Cambridge, MA 02139, USA
Dr. Klavs F. Jensen, Department of Chemical Engineering, Koch Institute for Integrative Cancer Research at MIT, Cambridge, MA 02139, USA
Dr. Andrew S. Liss, Department of Surgery, Massachusetts General Hospital, Boston, MA 02114, USA
Dr. J. Christopher Love, Department of Chemical Engineering, Koch Institute for Integrative Cancer Research at MIT, Cambridge, MA 02139, USA The Broad Institute of Harvard and MIT, Cambridge, MA 02142, USA.
References
- 1.Fuchs E, Horsley V. Nat. Cell Biol. 2011;13:513. doi: 10.1038/ncb0511-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang L, Ridgway LD, Wetzel MD, Ngo J, Yin W, Kumar D, Goodman JC, Groves MD, Marchetti D. Sci. Transl. Med. 2013;5:180ra48. doi: 10.1126/scitranslmed.3005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gorges TM, Tinhofer I, Drosch M, Röse L, Zollner TM, Krahn T, von Ahsen O. BMC Cancer. 2012;12:178. doi: 10.1186/1471-2407-12-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vona G, Sabile A, Louha M, Sitruk V, Romana S, Schütze K, Capron F, Franco D, Pazzagli M, Vekemans M, Lacour B, Bréchot C, Paterlini-Bréchot P. Am. J. Pathol. 2000;156:57. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng S, Lin HK, Lu B, Williams A, Datar R, Cote RJ, Tai Y-C. Biomed. Microdevices. 2011;13:203. doi: 10.1007/s10544-010-9485-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan SJ, Lakshmi RL, Chen P, Lim W-T, Yobas L, Lim CT. Biosens. Bioelectron. 2010;26:1701. doi: 10.1016/j.bios.2010.07.054. [DOI] [PubMed] [Google Scholar]
- 7.Juratli MA, Sarimollaoglu M, Siegel ER, Nedosekin DA, Galanzha EI, Suen JY, Zharov VP. Head Neck. 2014;36:1207. doi: 10.1002/hed.23439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torchilin VP. Annu. Rev. Biomed. Eng. 2006;8:343. doi: 10.1146/annurev.bioeng.8.061505.095735. [DOI] [PubMed] [Google Scholar]
- 9.Birkholz K, Schwenkert M, Kellner C, Gross S, Fey G, Schuler-Thurner B, Schuler G, Schaft N, Dörrie J. Blood. 2010;116:2277. doi: 10.1182/blood-2010-02-268425. [DOI] [PubMed] [Google Scholar]
- 10.Bae Y, Jang W-D, Nishiyama N, Fukushima S, Kataoka K. Mol. Biosyst. 2005;1:242. doi: 10.1039/b500266d. [DOI] [PubMed] [Google Scholar]
- 11.Wolff JA, Rozema DB. Mol. Ther. 2008;16:8. doi: 10.1038/sj.mt.6300326. [DOI] [PubMed] [Google Scholar]
- 12.Kiss AL, Botos E. J. Cell. Mol. Med. 2009;13:1228. doi: 10.1111/j.1582-4934.2009.00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J, Sharei A, Sim WY, Adamo A, Langer R, Jensen KF, Bawendi MG. Nano Lett. 2012;12:6322. doi: 10.1021/nl303421h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharei A, Trifonova R, Jhunjhunwala S, Hartoularos GC, Eyerman AT, Lytton-Jean A, Angin M, Sharma S, Poceviciute R, Mao S, Heimann M, Liu S, Talkar T, Khan OF, Addo M, von Andrian UH, Anderson DG, Langer R, Lieberman J, Jensen KF. PLoS One. 2015;10:e0118803. doi: 10.1371/journal.pone.0118803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharei A, Zoldan J, Adamo A, Sim WY, Cho N, Jackson E, Mao S, Schneider S, Han M-J, Lytton-Jean A, Basto PA, Jhunjhunwala S, Lee J, Heller DA, Kang JW, Hartoularos GC, Kim K-S, Anderson DG, Langer R, Jensen KF. Proc. Natl. Acad. Sci. USA. 2013;110:2082. doi: 10.1073/pnas.1218705110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharei A, Poceviciute R, Jackson EL, Cho N, Mao S, Hartoularos GC, Jang DY, Jhunjhunwala S, Eyerman A, Schoettle T, Langer R, Jensen KF. Integr. Biol. (Camb) 2014;6:470. doi: 10.1039/c3ib40215k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szeto GL, Van Egeren D, Worku H, Sharei A, Alejandro B, Park C, Frew K, Brefo M, Mao S, Heimann M, Langer R, Jensen K, Irvine DJ. Sci. Rep. 2015;5:10276. doi: 10.1038/srep10276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaffer CL, Weinberg RA. Science. 2011;331:1559. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 19.Butz J, Wickstrom E, Edwards J. BMC Biotechnol. 2003;3:11. doi: 10.1186/1472-6750-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hou Y, Song L, Zhu P, Zhang B, Tao Y, Xu X, Li F, Wu K, Liang J, Shao D, Wu H, Ye X, Ye C, Wu R, Jian M, Chen Y, Xie W, Zhang R, Chen L, Liu X, Yao X, Zheng H, Yu C, Li Q, Gong Z, Mao M, Yang X, Yang L, Li J, Wang W, Lu Z, Gu N, Laurie G, Bolund L, Kristiansen K, Wang J, Yang H, Li Y, Zhang X, Wang J. Cell. 2012;148:873. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 21.Smerage JB, Barlow WE, Hortobagyi GN, Winer EP, Leyland-Jones B, Srkalovic G, Tejwani S, Schott AF, O’Rourke MA, Lew DL, Doyle GV, Gralow JR, Livingston RB, Hayes DF. J. Clin. Oncol. 2014;32:3483. doi: 10.1200/JCO.2014.56.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lohr JG, Adalsteinsson VA, Cibulskis K, Choudhury AD, Rosenberg M, Cruz-Gordillo P, Francis JM, Zhang C-Z, Shalek AK, Satija R, Trombetta JJ, Lu D, Tallapragada N, Tahirova N, Kim S, Blumenstiel B, Sougnez C, Lowe A, Wong B, Auclair D, Van Allen EM, Nakabayashi M, Lis RT, Lee G-SM, Li T, Chabot MS, Ly A, Taplin M-E, Clancy TE, Loda M, Regev A, Meyerson M, Hahn WC, Kantoff PW, Golub TR, Getz G, Boehm JS, Love JC. Nat. Biotechnol. 2014;32:479. doi: 10.1038/nbt.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kollmannsperger A, Sharei A, Raulf A, Heilemann M, Langer R, Jensen KF, Wieneke R, Tampé R. Nat. Commun. 2016;7:10372. doi: 10.1038/ncomms10372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raab M, Gentili M, de Belly H, Thiam HR, Vargas P, Jimenez AJ, Lautenschlaeger F, Voituriez R, Lennon-Duménil AM, Manel N, Piel M, Lee CP, Liu PT, Kung HN, Su MT, Chua HH, Chang YH, Chang CW, Tsai CH, Liu FT, Chen MR, Hatch E, Hetzer M, Fidziańska A, Bilińska ZT, Tesson F, Wagner T, Walski M, Grzybowski J, Ruzyłło W, Hausmanowa-Petrusewicz I, Harada T, Swift J, Irianto J, Shin JW, Spinler KR, Athirasala A, Diegmiller R, Dingal PC, Ivanovska IL, Discher DE, Rowat AC, Jaalouk DE, Zwerger M, Ung WL, Eydelnant IA, Olins DE, Olins AL, Herrmann H, Weitz DA, Lammerding J, Davidson PM, Denais C, Bakshi MC, Lammerding J, Sixt M, Wolf K, Te Lindert M, Krause M, Alexander S, Te Riet J, Willis AL, Hoffman RM, Figdor CG, Weiss SJ, Friedl P, Pflicke H, Sixt M, Heuzé ML, Collin O, Terriac E, Lennon-Duménil AM, Piel M, Krause M, Wolf K, Le Berre M, Aubertin J, Piel M, Jimenez AJ, Maiuri P, Lafaurie-Janvore J, Divoux S, Piel M, Perez F, Olmos Y, Hodgson L, Mantell J, Verkade P, Carlton JG, Vietri M, Schink KO, Campsteijn C, Wegner CS, Schultz SW, Christ L, Thoresen SB, Brech A, Raiborg C, Stenmark H, Poser I, Sarov M, Hutchins JR, Hériché JK, Toyoda Y, Pozniakovsky A, Weigl D, Nitzsche A, Hegemann B, Bird AW, Pelletier L, Kittler R, Hua S, Naumann R, Augsburg M, Sykora MM, Hofemeister H, Zhang Y, Nasmyth K, White KP, Dietzel S, Mechtler K, Durbin R, Stewart AF, Peters JM, Buchholz F, Hyman AA, Scheffer LL, Sreetama SC, Sharma N, Medikayala S, Brown KJ, Defour A, Jaiswal JK, De Vos WH, Houben F, Kamps M, Malhas A, Verheyen F, Cox J, Manders EM, Verstraeten VL, van Steensel MA, Marcelis CL, van den Wijngaard A, Vaux DJ, Ramaekers FC, Broers JL, Vargas JD, Hatch EM, Anderson DJ, Hetzer MW, Gonzalo S, Kreienkamp R, Janssen A, van der Burg M, Szuhai K, Kops GJ, Medema RH, Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC, Maciejowski J, Li Y, Bosco N, Campbell PJ, de Lange T, Sarbajna S, Davies D, West SC, Gritenaite D, Princz LN, Szakal B, Bantele SC, Wendeler L, Schilbach S, Habermann BH, Matos J, Lisby M, Branzei D, Pfander B, Chan YW, West SC, Denais CM, Gilbert RM, Isermann P, McGregor AL, te Lindert M, Weigelin B, Davidson P, Friedl P, Wolf K, Lammerding J, Shimi T, Goldman RD, Hatch EM, Fischer AH, Deerinck TJ, Hetzer MW, Soria-Valles C, Osorio FG, Gutiérrez-Fernández A, Angeles ADL, Bueno C, Menéndez P, Martín-Subero JI, Daley GQ, Freije JM, López-Otín C, Liu YJ, Le Berre M, Lautenschlaeger F, Maiuri P, Callan-Jones A, Heuzé M, Takaki T, Voituriez R, Piel M, Faure-André G, Vargas P, Yuseff MI, Heuzé M, Diaz J, Lankar D, Steri V, Manry J, Hugues S, Vascotto F, Boulanger J, Raposo G, Bono MR, Rosemblatt M, Piel M, Lennon-Duménil AM, Riedl J, Flynn KC, Raducanu A, Gärtner F, Beck G, Bösl M, Bradke F, Massberg S, Aszodi A, Sixt M, Wedlich-Söldner R, Holmer L, Worman HJ, Manel N, Hogstad B, Wang Y, Levy DE, Unutmaz D, Littman DR, Satoh T, Manel N, Nègre D, Mangeot PE, Duisit G, Blanchard S, Vidalain PO, Leissner P, Winter AJ, Rabourdin-Combe C, Mehtali M, Moullier P, Darlix JL, Cosset FL, Vargas P, Maiuri P, Bretou M, Sáez PJ, Pierobon P, Maurin M, Chabaud M, Lankar D, Obino D, Terriac E, Raab M, Thiam HR, Brocker T, Kitchen-Goosen SM, Alberts AS, Sunareni P, Xia S, Li R, Voituriez R, Piel M, Lennon-Duménil AM, Lahaye X, Satoh T, Gentili M, Cerboni S, Conrad C, Hurbain I, El Marjou A, Lacabaratz C, Lelièvre JD, Manel N. Science. 2016;352:359. doi: 10.1126/science.aad7611. [DOI] [PubMed] [Google Scholar]
- 25.Denais CM, Gilbert RM, Isermann P, McGregor AL, te Lindert M, Weigelin B, Davidson PM, Friedl P, Wolf K, Lammerding J, Burke B, Stewart CL, Hatch E, Hetzer M, Weigelin B, Bakker G-J, Friedl P, Harada T, Swift J, Irianto J, Shin JW, Spinler KR, Athirasala A, Diegmiller R, Dingal PC, Ivanovska IL, Discher DE, Thomas DG, Yenepalli A, Denais CM, Rape A, Beach JR, Wang YL, Schiemann WP, Baskaran H, Lammerding J, Egelhoff TT, Wolf K, Te Lindert M, Krause M, Alexander S, Te Riet J, Willis AL, Hoffman RM, Figdor CG, Weiss SJ, Friedl P, Friedl P, Wolf K, Lammerding J, Davidson PM, Denais C, Bakshi MC, Lammerding J, Rowat AC, Jaalouk DE, Zwerger M, Ung WL, Eydelnant IA, Olins DE, Olins AL, Herrmann H, Weitz DA, Lammerding J, Fu Y, Chin LK, Bourouina T, Liu AQ, VanDongen AM, Davidson PM, Sliz J, Isermann P, Denais C, Lammerding J, De Vos WH, Houben F, Kamps M, Malhas A, Verheyen F, Cox J, Manders EM, Verstraeten VL, van Steensel MA, Marcelis CL, van den Wijngaard A, Vaux DJ, Ramaekers FC, Broers JL, Vargas JD, Hatch EM, Anderson DJ, Hetzer MW, Hatch EM, Fischer AH, Deerinck TJ, Hetzer MW, Jamin A, Wiebe MS, Civril F, Deimling T, deO. Mann CC, Ablasser A, Moldt M, Witte G, Hornung V, Hopfner KP, Nakamura AJ, Rao VA, Pommier Y, Bonner WM, Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, Meyerson M, Pellman D, Bekker-Jensen S, Lukas C, Melander F, Bartek J, Lukas J, Loewer A, Karanam K, Mock C, Lahav G, Le Berre M, Aubertin J, Piel M, Broers JL, Peeters EA, Kuijpers HJ, Endert J, Bouten CV, Oomens CW, Baaijens FP, Ramaekers FC, Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT, Shimi T, Pfleghaar K, Kojima S, Pack CG, Solovei I, Goldman AE, Adam SA, Shumaker DK, Kinjo M, Cremer T, Goldman RD, Shimi T, Kittisopikul M, Tran J, Goldman AE, Adam SA, Zheng Y, Jaqaman K, Goldman RD, Petrie RJ, Koo H, Yamada KM, Neelam S, Chancellor TJ, Li Y, Nickerson JA, Roux KJ, Dickinson RB, Lele TP, Olmos Y, Hodgson L, Mantell J, Verkade P, Carlton JG, Vietri M, Schink KO, Campsteijn C, Wegner CS, Schultz SW, Christ L, Thoresen SB, Brech A, Raiborg C, Stenmark H, Hurley JH, Raab M, Gentili M, de Belly H, Thiam HR, Vargas P, Jimenez AJ, Lautenschlaeger F, Voituriez R, Lennon-Duménil AM, Manel N, Piel M, Hutchison CJ, Matsumoto A, Hieda M, Yokoyama Y, Nishioka Y, Yoshidome K, Tsujimoto M, Matsuura N, Nam HS, Benezra R, Hanson PI, Roth R, Lin Y, Heuser JE, Zwerger M, Jaalouk DE, Lombardi ML, Isermann P, Mauermann M, Dialynas G, Herrmann H, Wallrath LL, Lammerding J, Carey SP, Starchenko A, McGregor AL, Reinhart-King CA, Carey SP, Kraning-Rush CM, Williams RM, Reinhart-King CA, Alexander S, Koehl GE, Hirschberg M, Geissler EK, Friedl P, Jimenez AJ, Maiuri P, Lafaurie-Janvore J, Divoux S, Piel M, Perez F, Zipfel WR, Williams RM, Christie R, Nikitin AY, Hyman BT, Webb WW, Needham D, Nunn RS, Scheffer LL, Sreetama SC, Sharma N, Medikayala S, Brown KJ, Defour A, Jaiswal JK. Science. 2016;352:353. doi: 10.1126/science.aad7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McNeil PL, Kirchhausen T. Nat. Rev. Mol. Cell Biol. 2005;6:499. doi: 10.1038/nrm1665. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.