Significance
Point-of-care diagnostics in the developing world and resource-limited areas require numerous special design considerations to provide effective early detection of disease. Of particular need for these contexts are diagnostic technologies featuring low costs, ease of use, and broad applicability. Here we present a nanoparticle-inkjet-printable microfluidics-based platform that fulfills these criteria and that we expect to significantly reduce the footprint, complexity, and cost of clinical diagnostics. This reusable $0.01 platform is miniaturized to handle small sample volumes and can perform numerous analyses. It can perform complex, minimally invasive analyses of single cells without specialized equipment and personnel. This inexpensive, accessible platform has broad applications in precision diagnostics and is a step toward the democratization of medical technologies.
Keywords: lab on a chip, point of care, diagnostics, nanoparticles, microfluidics
Abstract
Isolation and characterization of rare cells and molecules from a heterogeneous population is of critical importance in diagnosis of common lethal diseases such as malaria, tuberculosis, HIV, and cancer. For the developing world, point-of-care (POC) diagnostics design must account for limited funds, modest public health infrastructure, and low power availability. To address these challenges, here we integrate microfluidics, electronics, and inkjet printing to build an ultra–low-cost, rapid, and miniaturized lab-on-a-chip (LOC) platform. This platform can perform label-free and rapid single-cell capture, efficient cellular manipulation, rare-cell isolation, selective analytical separation of biological species, sorting, concentration, positioning, enumeration, and characterization. The miniaturized format allows for small sample and reagent volumes. By keeping the electronics separate from microfluidic chips, the former can be reused and device lifetime is extended. Perhaps most notably, the device manufacturing is significantly less expensive, time-consuming, and complex than traditional LOC platforms, requiring only an inkjet printer rather than skilled personnel and clean-room facilities. Production only takes 20 min (vs. up to weeks) and $0.01—an unprecedented cost in clinical diagnostics. The platform works based on intrinsic physical characteristics of biomolecules (e.g., size and polarizability). We demonstrate biomedical applications and verify cell viability in our platform, whose multiplexing and integration of numerous steps and external analyses enhance its application in the clinic, including by nonspecialists. Through its massive cost reduction and usability we anticipate that our platform will enable greater access to diagnostic facilities in developed countries as well as POC diagnostics in resource-poor and developing countries.
Enabling early detection of diseases is one of the greatest opportunities we have for developing treatments and prevention strategies as well as reducing the costs of healthcare. In particular, the detection of rare or low-abundance cells and molecules of interest from a heterogeneous population is of critical importance in the diagnosis of several lethal diseases, such as malaria (1), tuberculosis, cancer (2), and HIV (3). This procedure requires rapid and accurate separation and sorting of target cell types toward an analyzer platform, which are traditionally performed through analyses such as membrane filtration (4), centrifugation method (5), FACS (6), and magnetic activated cell sorting (MACS) (7). Although all these methods are powerful techniques, most require bulky equipment, high costs, and highly trained personnel, and cannot simultaneously achieve high throughput and resolution, speed, and low cost (8). One of the key challenges for most of these techniques is the requirement for labeling, the attachment of any foreign molecule to the molecule of interest, to increase detection sensitivity. Labeling strategies are usually low-yield and may include lengthy synthesis and purification steps that can potentially alter intrinsic properties of cells. To address these limitations, lab-on-a-chip (LOC) devices have been widely developed over the last 10–15 y (9, 10); these devices integrate and automate multiple laboratory protocols into a miniature device that employs microfluidic technology to handle small sample volumes on the order of microliters to nanoliters (11). The majority of LOC devices can therefore be used for clinical assays (9) to perform a variety of tasks, including single-cell analysis (12), rare-cell isolation (13), sample preparation, pretreatment or preconcentration (14, 15, 16), purification (17), fractionation (18), drug screening (19, 20), enrichment of rare cells and molecules (21, 22), target cells of interest manipulation (23–26), positioning (27), counting and characterizing (28–30), DNA analysis (14), protein detection (31), environmental monitoring (32), and the detection of biohazards (33). Among different techniques, the family of electrokinetic phenomena (e.g., electrophoresis, electroosmosis, diffusiophoresis, and capillary osmosis) (34), is perfectly suitable and the most widely used concept in LOC devices for the applications mentioned above. This is due to the multipurpose nature of the electric field and the precise controllability of its direction, amplitude, and frequency for specific analyte response in fluids (35, 36). Analytes (bioparticles) of interest range from cancer cells, white blood cells, red blood cells, and platelets to bacteria, microorganisms, proteins, and DNA molecules. In these LOC devices interaction of an applied nonuniform electric field with the induced effective dipole moment of the bioparticle enables selective and label-free manipulation as well as analytical separation, sorting, and capturing of bioparticles of interest within a medium based on their intrinsic physical characteristics, including size, shape, deformability, density, polarizability, and more (6, 37). These devices are often subsets of microelectromechanical systems devices that are constructed by nanotechnology/ microtechnology, often referred to as “micro total analysis systems,” or μTAS. Electronic apparatus integration of such μTAS platforms often requires access to a clean room, sophisticated equipment, and highly trained personnel to perform several time-consuming and relatively expensive micro/nano manufacturing procedures (e.g. lithography, oxidation, etching, and thin film depositions). This entire procedure can take several days or weeks (38). These limitations add significant time, cost, and complexity to such devices, which hinders not only their mass production but also their point-of-care (POC) diagnostic applications. In addition, LOC-based POC devices are usually not fully integrated (i.e., they are incapable of performing all processes for assays such as cell manipulation, analytical separation, capturing, and analysis), which would be a very useful feature for improving the accessibility of miniaturized, low-cost clinical assays. According to the National Institutes of Health (39), almost 90% of people with infectious diseases, such as HIV, live in the developing world. Given the limited resources and access to medical technologies, the potential impact that inexpensive, multifunctional LOC devices could have on public health is perhaps the greatest in the developing world (40). For instance, according to the World Health Organization, breast cancer mortality (the most prevalent cancer in women worldwide) is increasing, particularly in developing countries; 58% of the deaths from this cancer occur in developing countries due to the lack of early detection programs. As another consequence of inferior access to diagnostics, the survival rates of breast cancer patients are as high as 80% in developed nations but are only 40% in low-income nations (41). These circumstances underscore the need for affordable POC diagnostics in developing and low-income nations to facilitate early diagnosis and improve survival rates. Consequently, the design of diagnostic platforms should incorporate considerations of limited funding, modest public health infrastructure, low power consumption, and challenging environmental conditions for transportation and storage (42). In other words, for LOC-based diagnostics to have an impact on global health the platforms must be rapid, sensitive, specific, robust, user-friendly, and, most importantly, affordable with minimal equipment and deliverable to end users (43, 44). To achieve the necessary cost and equipment reductions for a widely accessible POC test it is important to achieve full integration of all steps in a diagnostic assay while eliminating much of the manufacturing costs.
To remove some of the barriers described above, we combined microfluidics, electronics, and inkjet printing technologies to develop an ultra–low-cost microfluidic-based miniaturized LOC platform. This rapid, high-throughput, and label-free platform is made of reusable and flexible inkjet-nanoparticle-printed (FINP) electronic apparatuses and disposable microfluidic biochips, called FINP biochips. A unit of ready-to-use platform costs about $0.01 and its production time is approximately 20 min.
Results and Discussion
Platform Capabilities.
The FINP platform described herein is designed to perform multiple functions, including label-free single bioparticle trapping; label-free single-cell quantification and enumeration through impedance microcytometry; and high-speed and label-free cellular manipulation, concentration, sorting, patterning, and selective analytical separation and isolation of the cells of interest. The final cost for a unit of ready-to-use platform is lower than that of separate individual exciting platforms developed for each of the above applications. It includes both conventional nonmicrofluidics-based techniques (e.g., FACS) (6) and MACS (7), as well as micro/nanofabricated LOC platforms. Our approach to fabricate electronic apertures of this LOC platform not only fully eliminates the traditional need of using clean-room facilities, as well as costly and time-consuming manufacturing procedures, but also only involves two steps: (i) drawing any required electronic aperture configurations (e.g., circular configuration) using any open-source vector-drawing software and (ii) integrating the drawn electronic patterns with a desired resolution (e.g., ∼20 μm) around conductive nanoparticles on a flexible substrate through a low-cost and rapid inkjet printing technology easily applied by any end user. In addition, a variety of electronic aperture configurations can be easily redesigned and integrated ad infinitum at no additional cost. This is extremely useful and important in many fields [e.g., tissue engineering (45)] and is not an easy option in traditional microfabrication procedures. As another important advantage, the time to produce such devices is cut from weeks/days to approximately 20 min; production is achievable by end users, which allows high-throughput mass production. In contrast to the reusable electronic apertures, the microfluidic blocks of this platform are designed as disposable microfluidic biochips to remove the need for storage or transport to a medical facility for sterilization. These noninvasive microfluidic biochips are made of inexpensive polymers by rapid, direct prototyping through a simple and fast procedure. Taking overall costs into consideration for the proposed LOC platform, a unit of ready-to-use FINP device can be manufactured for the cost of about one cent. As another important feature of this platform, all processed samples through our device can be recollected from each distinct chamber for further studies of individual cells, which is essential for complete end-to-end diagnostics. The platform is designed as a contactless platform, where the electronic apparatuses are not in contact with the biological samples. This eliminates the possibility of bubble formation, fouling, and cross-contamination of the chip, all of which are common issues in most current microfluidics-based LOC cell manipulators (3, 46). Lifetime, as another important feature of any POC platform, is improved in our device by designing the flexible electronic apparatus as detachable components from the disposable microfluidic biochips. It enables unlimited reuse of the chips simply by replacing the disposable microfluidic biochips. This is in contrast to most developed microfluidics-based cell manipulators (47) and single-cell impedance microcytometers in which any residue left from previous tests is detrimental to device lifetime, accuracy, and reliability. The only solution for these devices is to replace the chips or to require elaborate costly and time-consuming cleaning procedures that may not be possible on the consumer end. Finally, we have successfully implemented our FINP platform and demonstrated its utility to electrically and label-free perform (no labeling or using any external tags) a variety of individual functions including single-particle capture, cell manipulation, rare-cell isolation, selective analytical separation, sorting, concentrating, aligning, positioning, and patterning as well as quantification, enumeration, and characterization. We also conducted viability and proliferation tests to verify that our device is entirely safe for cells by regrowing the cells and analyzing their growth rate. We believe this multifunctional, rapid, ultra–low-cost, miniaturized, label-free, efficient, and high-throughput LOC platform that we introduce here is expected to effectively minimize the footprint, complexity, and cost of clinical diagnostics by implementing a solution for facilitating consumer-end diagnostic facilities in developed countries as well as POC diagnostic applications in resource poor and developing countries.
Platform Design and Integration.
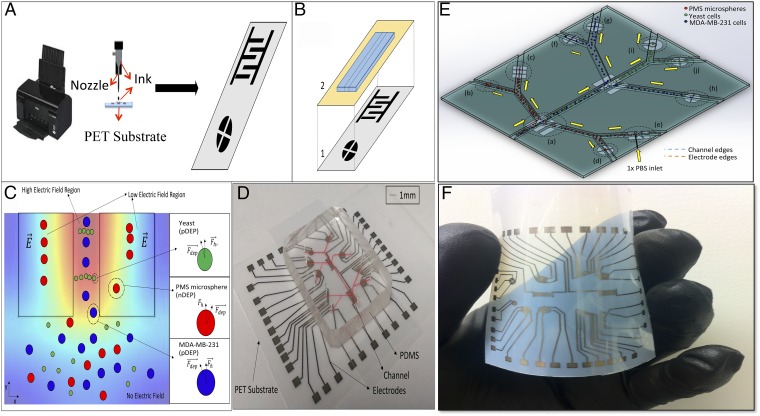
Our FINP chip is designed as a layered stack of (i) multiple-outlines inkjet-printed reusable electronic apparatuses made of conductive nanoparticles on a flexible microporous polyethylene terephthalate (PET) substrate, (ii) disposable microfluidic biochips made of polydimethylsiloxane (PDMS), and (iii) a disposable thin insulating barrier to insulate disposable microfluidic biochip from the electronic apparatuses (Fig. 1 A, B, D, and F). The chip has individual contactless cell micromanipulator chambers for label-free selective analytical cell separation, label-free and rapid cell concentration, sorting, patterning, electroorientation cell immobilization, and multiplex single-cell trapping functions (Fig. 1E). In addition, miniaturized and contactless single-cell impedance microcytometer chambers are implemented as sensing stages of our platform to perform label-free, real-time, and rapid single-cell morphology discrimination, detection, enumeration, and analysis. Although all these chambers might not be necessary for a certain application, we designed and validated all of the chambers individually and nonsimultaneously to demonstrate the utility of our platform. Manipulating the motion of bioparticles in micromanipulator chambers of our platform is achieved by polarizing and forming dipole moments within a medium in response to an induced nonuniform electric field (37). This results a force (Fig. 1C) on the bioparticles known as the dielectrophoresis (DEP) force (FDEP). DEP is extensively studied in the literature (48, 49) and briefly is modeled using three main equations (SI Materials and Methods). In these equations, the Clausius–Mossotti (CM) factor shows the effective polarizability of a bioparticle, where its magnitude determines the strength of the polarization effect and its sign determines the direction of the particle motion, which changes at the particle’s cross-over frequency. A positive CM factor indicates a positive force that pulls the particle toward the global or local regions with the highest electric field gradient, whereas a negative CM factor or negative force pulls particles toward the regions with the lowest gradient. The DEP force directly depends on the nonuniform electric field gradient (Eq. S1). Consequently, the effectiveness of DEP-based devices strongly depends on the geometry and configuration of electronic apertures producing the electric field distribution. For instance, interdigitated configurations are popular for bioparticle separation as well as single-cell capture (50), because they provide multiple field maxima and minima with a range of magnitudes, ideal for particle manipulation. Quadrupole configurations are popular for cell patterning in tissue engineering to form an inner defined area (27). As a result, according to the function assigned to each chamber, different electronic aperture configurations are used in our FINP platform. In our sensing stages, impedance microcytometer extensions, enumeration, and identification of individual bioparticles is on the basis of differences in size and dielectric properties by constant monitoring of the modulated impedance across a focusing micropore located in the microfluidic biochip. Passage of individual cells modulates the alternating electric current, giving impedance-based characterization of cells.
Fig. 1.
Principle of the FINP platform. (A) The fabrication procedure showing schematic illustration of a high-resolution inkjet printing system to print reusable electronic apertures made of conductive nanoparticles on flexible microporous PET substrates. (B) FINP chip expanded view, schematic stack-up: (1) multioutline inkjet-printed reusable electronic apparatuses on flexible PET substrate and (2) disposable microfluidic biochips made of PDMS bound on disposable thin insulating barrier. (C) Conceptual view of dielectrophoretic separation of bioparticles under influence of DEP force (FDEP) used in selective analytical separation and isolation chambers; bioparticle movement direction is altered in the presence of a nonuniform electric field. (D) Image of an LOC device in which a disposable microfluidic biochip is integrated with the printed electronic block containing a variety of electronic aperture configurations used in different device chambers, assigned to perform multiple functions of single-cell trapping, cellular manipulation, concentrating, sorting, patterning, selective analytical separation, and single-cell quantification. (E) Schematic of an LOC platform in which (a) is a microseparator chamber for selective analytical separation of bioparticles, (b) is a bioparticle patterning chamber, (c) is a multiplex dielectrophoretic single bioparticle trapping array, (d) is a contactless concentrating microchamber, (e) is a miniaturized and label-free impedance microcytometer, (f), (g), (h), and (j) are concentrating microchambers, and (i) is an electroorientation bioparticle immobilizing chamber. (F) Flexible reusable electronic block containing a variety of electronic aperture configurations used in different device chambers.
Platform Validation.
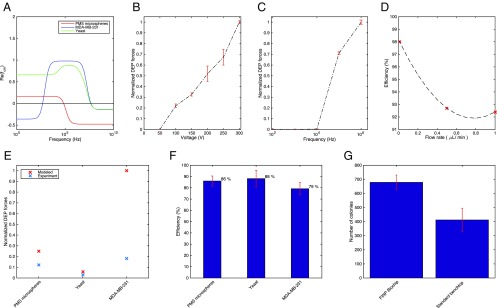
To implement the multiple intended functions of our LOC device we designed and tested a variety of electronic aperture configurations for each chamber (e.g., interdigitated configuration and focusing configuration). Numerical simulations were also performed for individual configurations using finite element analysis software (COMSOL Multiphysics; COMSOL Inc.). As an initial validation of our platform design and performance, we used different-sized (5-μm and 10-μm) polystyrene microspheres coated with streptavidin (PMS microspheres) as particle models (Fig. 2). These particles were chosen due to their spherical shape and size similarity to the cells used in later experiments. We optimized the chip configuration parameters (e.g., electronic apertures and microfluidic channels dimensions). A series of experiments were performed using 10-μm PMS microspheres to better understand the effect of influential factors on different assigned functions (Fig. 2). As shown in Fig 2 B and C, the effects of various input signal voltages and various excitation input signal frequencies on the velocity and experienced DEP force by the PMS microspheres were experimentally computed (SI Materials and Methods). Furthermore, the influence of higher flow rates (i.e., 0.5 μL⋅min−1 and 1 μL⋅min−1) on the device capturing efficiency, defined as the ratio of captured target particles at objective locations to their corresponding number in the initial sample, is shown in Fig. 2D. An approximate 5–10% drop in capturing efficiency was observed due to the higher domination of hydrodynamic force under higher flow rates. In addition, expected DEP forces on cells predicted by our models were experimentally validated (Fig. 2E). Our validation samples included the DAPI-stained breast adenocarcinoma (MDA-MB-231) cell line, representing advanced stages of the disease (12.4 μm diameter) (51) and BY4741 Saccharomyces cerevisiae yeast cells (5-μm diameter) (52). The cell line MDA-MB-231 was chosen for these studies because breast cancer is the most prevalent cancer in women worldwide and the ability to distinguish rare circulating tumor cells (CTCs) can advance the understanding of cancer metastasis to better treat cancer patients, especially in developing countries. Yeast cells were chosen because of their frequent use as a model organism for studying cell responses, due to their simple and representative structure.
Fig. 2.
FINP platform characterization. (A) The interfacial polarization factor as a function of electric field frequency for the cancer cell line MDA-MB-231 breast adenocarcinoma cells, yeast cells, and streptavidin-coated PMS microspheres, with medium conductivity of σ = 3 μS/m, using a two-shell model (53). (B) Normalized DEP force on 10-μm streptavidin-coated microsphere particles along the length of the microfluidic channel as a function of input voltages computed from the experiments, excitation at 1 MHz. (C) Normalized DEP spectra of 10-μm PMS microspheres in aqueous medium solution at input voltage Vrms = 106 V, frequency range from 100 Hz to 1 MHz. (D) Capture rates of 10-μm PMS microspheres in our chip under flow rate of suspension. (E) A comparison between the modeled and computed from the experiments DEP force on the three different bioparticles, MDA-MB-231 breast adenocarcinoma cells, yeast cells, and PMS microspheres at their cross-over frequencies, normalized to the maximum value. (F) The collection efficiency and purity of isolated bioparticles at outlets indicates separation efficiency of 79, 88, and 86% for the breast adenocarcinoma cells, yeast cells, and PMS microspheres, respectively. (G) Quantification of the viability of the yeast cells exposed to the electric field in the FINP platform compared with standard bench-top methods, where the transformation efficiency improved by about 1.5-fold.
Multiplex Single-Bioparticle Trapping Chambers.
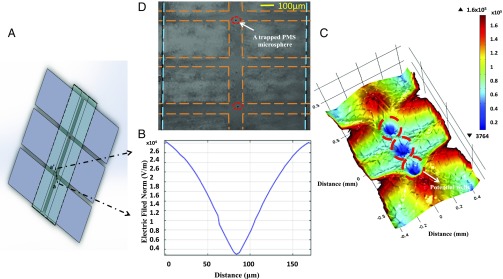
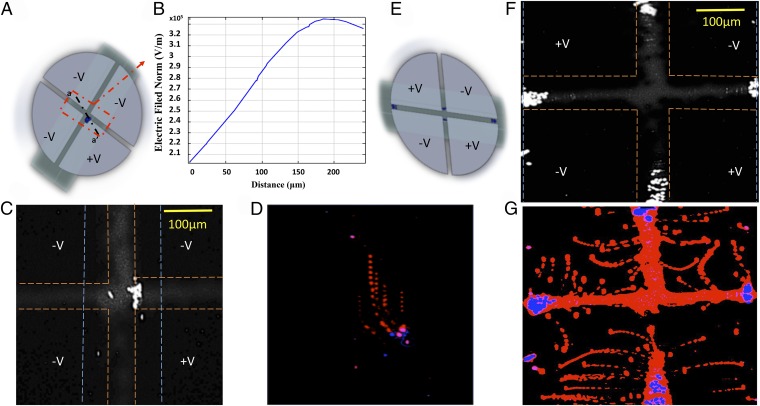
Selective trapping of individual cells (e.g., CTCs) at specific locations is essential for single-cell studies, especially those requiring downstream analysis of cellular content. To add this capability to our platform we designed an array of contactless single-bioparticle dielectrophoretic-traps. These arrays use dielectrophoretic forces to specified bioparticles (e.g., cells) and hold them in well-defined microregions. Such a platform has wide applications in cell engineering (e.g., genome editing), evaluation of chemotherapeutic reagents for cancer therapy, analysis of apoptosis, drug testing, and other phenotypic assays at the single-cell level (53). The designed multiplex single-bioparticle trapping chambers consist of an array of facing electrodes configuration (Fig. 3), with the dimensions of 150-μm width and 100-μm gaps along the arms between electrodes, for efficient and reliable bioparticle trapping. Intact individual single particles, experiencing negative DEP (nDEP), are trapped in defined multiplexed electrical filed (potential well) cages at the center of the electrode array, without the need for surface modification, labeling, or robotic equipment. Electric fields were modeled for our designed structure using finite element analysis software (COMSOL Multiphysics; COMSOL Inc.), in which the designed configuration has multiple well-defined regions of electrical field minima at the centers of the traps ( V/m) that isolate and trap single bioparticles experiencing nDEP. Fig. 3B shows a plot of electric field across the center of the electrodes at a height of 1 μm above the electrodes (at Vrms = 106 V) showing the field variation in the x–y plane (a–a′), and Fig. 3C shows a 3D plot of the electric field gradient of this configuration. The approximate minimum particle radius (r) for stable trapping in our designed array was also calculated to be ∼291 nm using the equation below (54):
| [1] |
where Δd is a small region over which force (namely the field gradient) is constant, is the Boltzmann’s constant, is the electric field gradient, and T is the temperature.
Fig. 3.
Multiplexed single bioparticle trapping. (A) Schematic of contactless microfluidics-based single-cell trap array. (B) A plot of the variation in electric field in a potential cage, perpendicular to the microfluidic channel flow direction, where particles are trapped by nDEP in regions of low–field-strength, dielectrophoretic traps (potential well traps). (C) Three-dimensional simulated electric fields gradient at z-elevation within the microfluidics channel; there are three strong dielectrophoretic traps (potential well traps) in the centers of the microfluidic channel. (D) Microscopic images taken showing the individual trapping and holding of single 10-μm PMS microspheres in well-defined potential traps from a larger array of up to 20 traps. The dashed lines indicate the edges of the microfluidics channel (blue) and electrodes (orange).
These simulation data were validated experimentally using 10-μm streptavidin-coated PMS microspheres as a model study. A suspension of PMS microspheres suspension was injected into the microfluidic channel and single bioparticles trapped at defined electrical field cages, where an input signal of 106 Vrms at 1 MHz was applied to the electrodes array. Fig. 3D shows an optical image of two of PMS microspheres trapped in electrical field cages at the center of the electrodes array under nDEP forces. Single trapped particles were confined into well defined microregions of electric filed cages. In addition, once trapped, a particle was isolated in the electric filed minimum and no further particles were observed to collect. One should note that the designed traps are individually controllable and suitable for arrayed operation.
Cell Viability and Genetic Transformation.
Next, we sought to verify the viability of cells in our platform. We investigated this by reculturing yeast cells (BY4741) after transformation of exogenous DNA into the cells in our platform or using standard benchtop transformation as a control (Materials and Methods). Yeast cells were first concentrated and patterned among the gaps in an interdigitated shape array of electronic aperture before the addition of exogenous DNA and transformation buffers. After several incubation steps of the mixture at the required temperatures we counted the number of colonies after 2 d. Our platform yielded ∼1.5-fold greater transformation efficiency than the traditional benchtop protocol (Fig. 2G; 679 colonies vs. 412). This increased efficiency may be due to an increased interacting interface (surface area) between the cells and DNA facilitated by the device’s cell-patterning function (55). These results thus establish not only the viability of cells upon exposure to an a.c. field in our platform but also the potential for using our platform for high-throughput, high-efficiency, miniaturized genetic transformation of cells.
Label-Free Selective Analytical Sorting and Isolation of Cells.
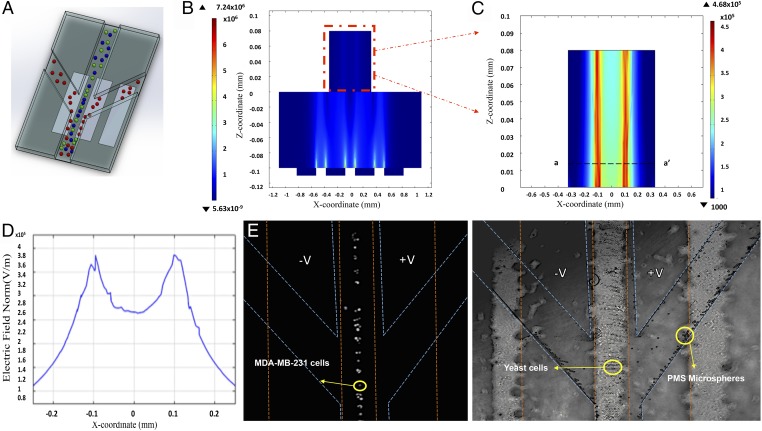
Early diagnosis of diseases involving rare cells requires rapid and accurate selective analytical separation and isolation of target cell types. We tested the capacity of our platform to isolate cells of interest using a mixture of live breast adenocarcinoma cells (MDA-MB-231), yeast cells, and streptavidin-coated PMS microspheres (10 μm). These bioparticles have different polarization properties and their CM factor numerical modeling (Fig. 2A) shows that each bioparticle type has its own unique cross-over frequency between 1 kHz and 1 MHz. These indicate a good separation resolution, defined as the minimum size difference in bioparticles that are still separable with high efficiency. In these experiments, our contactless disposable microfluidic biochip has two microseparator chambers. The structure of an individual microseparator chamber is shown in Fig. 4A, which consists of three distinct arms (100-μm width and 80-μm height), including a main channel and two side branches. As shown in Fig. 4A, optimized electronic apertures used in these microseparator chambers were interdigitated electrodes (150-μm width and 100-μm gap). These electrodes were placed in the side branches of the first separator chamber and in the main channel of the second microseparator chamber, under the microfluidic biochip, aligned with the main flow channel. The electronic apertures were physically separated from the microfluidic biochip and biological sample to prevent the possibility of bubble formation, fouling, and the cross-contamination of the chip. The integrated configuration allowed separated target bioparticles to flow toward the side arms, as a result of DEP force, whereas the rest of mixture flows through the main middle channel of microseparators. Sine waves of alternating voltage (a.c.) signals (Vrms = 106 V at f = 1 MHz, at the first microseparator chamber, and at f = 10 kHz at the second microseparator chamber, chosen according to the CM factor modeling) were applied across the electrodes. Applied voltage to the electrodes induced electric field gradient minima ( V/m) at the side arms of the first microseparator chamber (middle arm of the second separator chamber) and electric field maxima ( V/m) at the middle arm of the first microseparator chamber (side arms of the second microseparator chamber), according to our numerical finite element modeling (Fig. 4 B–D). This configuration exerts negative DEP force on the PMS microspheres flowing over the electrodes in microchannel of the first microseparator chamber, which directs them toward the side branches. Fig. 4E shows an image of the particle mixture in the first microseparator chamber, where the PMS microspheres experiencing nDEP were deviated toward the side branches. Meanwhile, the breast adenocarcinoma (MDA-MB-231) cell line and the yeast cells experiencing positive DEP (pDEP) maintained their path in the main channel and were then transported into the second microseparator chamber. In the second microseparator chamber, breast adenocarcinoma cells (MDA-MB-231) were pushed toward the side channels, indicating a strong pDEP response, whereas yeast cells were pulled toward the middle channel, indicating a strong nDEP response at a new frequency. For these experiments, the input signal amplitude was chosen to provide sufficient DEP force against the hydrodynamic force (h), deriving to have the resulting net force on the particles (net = h+DEP) in the desired direction. It should be noted that because our nanoparticles-based, inkjet-printed electrodes have a porous and coarse surface they provide an even stronger nonuniform electric field than smooth metal electrodes. This improves the DEP forces imposed on particles and results in enhanced separation efficiency. To quantitatively evaluate separating performance, the collection efficiency and purity of isolated bioparticles at outlets were calculated. We calculated separation efficiency (enrichment factor) as (nc/nt) × 100%, where nc is the number of correctly separated target bioparticles and nt is the initial number of bioparticles in starting samples. The results indicate separation efficiency of 79, 88, and 86% for the breast adenocarcinoma cell, yeast cells, and PMS microspheres, respectively (Fig. 2F).
Fig. 4.
Label-free selective analytical sorting and isolation of cells. (A) Conceptual view of the hydrodynamic DEP-based separation process, in which different cell types are indicated in different colors. (B) Numerical simulation of the electric field distribution over the electrodes at the cross-section. (C) Inset from B. (D) Variation in electric field at the first microseparator chamber, showing the magnitude of electric field maxima and minima. Bioparticles experiencing nDEP deviated toward the side branches, whereas the other bioparticles experiencing pDEP maintained their path in the main channel. (E) Fluorescent (Left) and bright-field (Right) microscopic images of mixture of live breast adenocarcinoma cells (MDA-MB-231), yeast cells, and streptavidin-coated polystyrene beads in the first microseparator chamber, in which streptavidin-coated PMSs were separated and directed to the side channels as a result of negative DEP force. The dashed lines indicate the edges of the microfluidics channel (blue) and electrodes (orange).
Marker-Free Concentration and Enrichment of Rare Cells.
Increasing cell concentration and reducing the sample volume in a preconcentration stage can noticeably improve natural limitations (e.g., analytical parameters such as sensitivity and the detection limit of biosensors) and the performance of biosensing systems, which are crucial factors for reliable detection of particular cells or metabolites (56) in complex samples (57). For example, the concentration of CTCs in peripheral blood is prognostic of metastatic disease in breast (54), prostate, colon (58), and other tumor types (59, 60). We used marker-free human breast adenocarcinoma cells (MDA-MB-231 cells) as a model of CTCs in which they were rapidly concentrated and collected in contactless DEP-based concentrating microchambers of the device. At the microconcentrator chambers, the polarities of the applied voltages and the microconcentrator electronic aperture configurations (i.e., focusing electrodes) precisely define the locations where the target cells are to be relocated and concentrated (Fig. 5 A and E). Numerical finite element modeling of inner electric field gradient of Fig. 5A is shown in Fig. 5B, in which the electric field maxima is the defined concentrating location. Experimentally, the onset of cell concentration from breast cancer adenocarcinoma MDA-MB-231 cells occurred at voltages of 106 Vrms when a frequency of 1 MHz was applied. Breast adenocarcinoma cells (MDA-MB-231), displaying a strong positive DEP response at this frequency, were rapidly (<6 s) pulled toward the defined electric field gradient maxima ( V/m) and concentrated effectively (Fig. 5 C and F). To visualize these effects, we superimposed the cell trajectories in two different trap models onto color images (Fig. 5 D and G), in which cell trajectories (red) toward the defined concentrating positions (blue) are shown. These concentrated cells can be ultimately recollected from each chamber independently for further downstream analysis, including nucleic acid extraction, purification, and sequencing (48).
Fig. 5.
Marker-free concentration and enrichment of rare cells. (A) Schematic of the designed electronic apertures in a contactless DEP-based concentrating microchamber, where the concentrating locations were precisely defined by the polarities of the applied voltages. (B) A plot of the variation in electric field in a concentrating microchamber, showing the magnitude of electric field maxima along the a–a′ line of A. (C) On-chip label-free selective concentration of breast adenocarcinoma cells (MDA-MB-231) at the defined concentrating traps of A. (D) The superimposed cell trajectories of MDAMB-231 alternating AC field of 106 Vrms at 1 MHz for the defined concentrating positions in A. (E) Changing the polarities of the applied voltages will change the concentrating locations. (F) Concentration of breast adenocarcinoma cells (MDA-MB-231) at the new defined concentrating location of E. (G) The superimposed cell trajectories of MDA-MB-231 for the new defined concentrating location in E. The dashed lines in F and C indicate the edges of the microfluidics channel (blue) and electrodes (orange).
Label-Free and Real-Time Single-Cell Quantification and Enumeration in Impedance Microcytometer.
Diagnosing infectious diseases in the developing world, such as tuberculosis and malaria, is often performed with tests that count lymphocytes (1) and monocytes (61), respectively. These efforts would benefit greatly from a lower-cost and disposable diagnostic platform capable of rapidly and accurately counting cell types of interest. The conventional method using flow cytometry, using an instrument such as the Beckman-Coulter FC500, requires an up-front cost of $100,000 as well as operator training and expertise in a clinical setting.
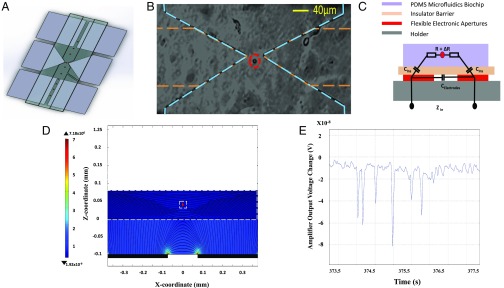
This detection and characterization of single cells by our microcytometer is performed on the basis of differences in size and dielectric properties using impedance spectroscopy as a real-time and label-free electrical technique. Other published designs of impedance cytometers (1, 61–64) have not endeavored to reduce costs or to integrate them with other LOC analysis modules, which has limited their use for POC diagnostics in limited resource settings. Our high-throughput, ultra–low-cost, and label-free version of the classic Coulter counter constantly monitors and measures the a.c. electrical properties of particles in suspension passing through a focusing micropore (40 μm) designed in our disposable microfluidic biochip. The focusing micropore of this microfluidic biochip is precisely aligned above two pairs of nanoparticle inkjet-printed coplanar electrodes, in a contactless format. The measured impedance level (ΔR) is modulated by the passing individual bioparticles (e.g., cells) that temporary block the focusing micropore and create a barrier to the current flow (Fig. 6 A, C, and D). This results in a modulation of the alternating electric current, ΔI, expressed as
| [2] |
where ΔI is the alternating electric current, R is the series resistance of microfluidic channel and pore, ΔR is impedance variation, Cins is the thin film insulator layer capacitance between each electrode and carrying fluids, and ω is the angular frequency of the applied a.c. field.
Fig. 6.
Label-free and real-time single-cell quantification and enumeration. (A) Schematic of the microcytometer, two pairs of rectangular focusing electrodes and a pair of triangular measurement electrodes at the middle. (B) A microscopic image of a single PMS microsphere flowing in the microfluidic channel during the sensing, at the center of the focusing-shaped electrodes toward the micropore. (C) Equivalent circuit model of the proposed a.c. impedance microcytometer with a bioparticle in the aperture. R is the impedance of carrying fluids, series resistance of microfluidic channel, and pore; Celectrods is the parasitic capacitance between two electrodes; Cins is the equivalent capacitance between electrode and carrying fluids due to the insulating thin film; and ΔR is the modulated impedance due to the temporary blockage by an individual bioparticle. (D) Simulation of the electric field in the DEP focusing section performed using COMSOL Multiphysics. The electric field is simulated at an applied potential of 12 V a.c. and frequency of 900 kHz. It simulates a system with a domain consisting of a pair of focusing triangle electrodes, 40 μm apart from each other, which are placed at the bottom of lateral channels. The bioparticle passes over electrodes and the impedance of the detection volume is measured (side view). (E) The measured electric output voltage modulation, induced by the passage of six individual PMS microspheres through the micropore (peaks) detected by the impedance microcytometer. The dashed lines in B indicate the edges of the microfluidics channel (blue) and electrodes (orange).
The sensitivity and accuracy of our miniaturized microcytometer is further improved by maintaining the same position and the same orientation for each bioparticle while passing through the sensing region (i.e., focusing micropore). We achieved this by designing a pair of dielectrophoretic focusing/electro-orientating electrodes (150-μm width and 80-μm gap) to focus the bioparticles toward the midline of the flow direction (Fig. 6A) before they reach the sensing zone. To examine the utility of our impedance microcytometer chamber, we quantified and enumerated streptavidin-coated PMS microspheres suspended in PBS. According to our experiments, we determined the operational conditions to be at f = 900 kH (coplanar electrodes excited with a 12-V a.c. signal), where all interfacial parasitic capacitance at the surface of the electrodes (Fig. 6C) are bypassed, and the measured signal depends only on the channel resistance term (SI Materials and Methods) of Eq. 2, (). At these running conditions, current signals were simultaneously recorded in real time while PMS microspheres passed through the sensing position. Using an optical microscope, particle passage was simultaneously monitored and verified optically (Fig. 6B).
Fig. 6E shows the representative experimental data for the passage of six PMS microspheres, where all of the peaks measured by our cytometer corresponded to the actual passage of individual PMS microspheres through the pore. According to our measurements the passage of each detected single PMS microsphere through the pore resulted in a signal-to-noise-ratio of at least 2.5 dB, which indicates that our platform is capable of effective quantification of bioparticles (e.g., cells).
In summary, we have combined inkjet-printing technology with electronics and microfluidic technologies to develop an ultra–low-cost, miniaturized, contactless, and reusable LOC platform. To the best of our knowledge, such a platform with similar functionalities, cost and advantages has not yet been reported. Using label-free electrical detection, our platform is capable of integrating not only standard LOC protocols but also multiple analyses of biological samples, including the capture, sorting, and isolation of various bioparticles of interest. We demonstrate that the operation of our FINP platform is not detrimental to cell viability or proliferation. Notably, it is straightforward to adapt our platform for a variety of bioparticles and cells with differing sizes and properties, simply by varying the configuration of the electronic apertures and reoptimize it for the cells of interest. New tailored printable electronic apertures then can be accessible by an end user. In addition, manufacturing the device is significantly less expensive, time-consuming, and complex than traditional LOC platforms and does not require skilled personnel and clean-room facilities. Device production only takes 20 min and $0.01, which is an unprecedented cost in clinical diagnostics. Although our current version of the platform employs commercial syringe pumps to control the fluid flow, we are confident based on work from other groups (65) that our design can be adapted to a hand-held format with a disposable biochip and reusable electronic apertures. The miniaturized format allows for robust and rapid results from small sample volumes, addressing potential issues of sample abundance and urgency in POC applications. We believe this multifunctional and ultra–low-cost FINP platform that we introduce here is an important step toward minimizing the footprint, complexity, and cost of clinical diagnostics. In doing so, we are hopeful that this work will enable greater individual access to diagnostic facilities in developed countries as well as POC diagnostic applications in resource-poor and developing countries.
SI Materials and Methods
DEP Equations.
DEP can be modeled using the following three equations. These equations are valid for the case of a spherical lossless dielectric sphere immersed in a dielectric liquid and ignoring higher order effects of polarization:
| [S1] |
| [S2] |
| [S3] |
where is the bioparticle radius, is the suspending medium permittivity, is the gradient of the electric field, is CM factor, is the real part of the CM factor, are the complex permittivities of the bioparticles and suspending medium, n is the multipolar term (for the spherical case, we assumed a pure dipole, n = 1), is the angular frequency, is the permittivity, and σ is the electrical conductivity.
System Calibration with Polyethylene Beads.
Ten-micrometer and 5-μm polyethylene beads (Cospheric LLC) coated with streptavidin were used for the platform calibration. Experimental resulting plots of the force vs. applied voltage, velocity vs. applied voltage, CM factor values vs. frequency, and capturing efficiency vs. flow rate were obtained. Upon electric field initiation cells and bioparticles gained velocity v and FDEP is exerted on the bioparticles. FDEP is calculated for spherical object as follows:
| [S4] |
where is the radius of the bioparticles, η is the dynamic viscosity of the medium, and fd is the drag coefficient, which is equal to 1 when the cell is far away from the channel wall.
Impedance Microcytometer.
The resistance variation ΔR manifests itself in the form of change in the baseline current ΔI, for the applied a.c. voltage Vin. In our measurement setup, ΔI is amplified through a transimpedance stage with a fixed constant gain, and after down-conversion to base band, is low-pass filtered and measured in the form of ΔVout, the change in the output baseline voltage. The change in resistance, ΔR, resulting from a particle passing through the pore, can be theoretically calculated from
| [S5] |
where is the resistivity of electrolyte, is the diameter of a bioparticle, and is the dimension of the pore.
Materials and Methods
Cell Viability Examination and Transformation.
The commercially available Frozen-EZ Yeast Transformation II Kit from Zymo Research was used for transformation procedures. First, 20 μL of yeast cells (BY4741), suspended in a low-conductivity medium with a measured conductivity of σ = 3 μS/m, was injected into a disposable microfluidic well (used instead of microfluidic channels for ease of use), bound on a disposable thin insolating barrier (100 μm thick), was placed on an inkjet-printed electronic apparatus (interdigitated configuration). Then, 1 μL of plasmid DNA (pRS416) was added to the concentrated yeast cells in potential traps defined by the interdigitated configuration (at Vrms = 106 V, f = 1 MHz), which was followed by addition of 20 μL of buffer 3 from the kit to the system. After incubation of the mixture at 30 °C for 45 min, the transformation mixture was spread on synthetic complete -ura plates and incubated at 30 °C for 2 d. The control transformation experiment was performed using a standard polypropylene tubing method to compare the efficiencies. The same protocol and identical quantities of samples were used in both cases, but in the bench-top protocol the yeast cells and buffer 3 and plasmid DNA were mixed directly before transferring to the polypropylene tube. After 2 d, the efficiencies of the transformation methods were estimated by counting the number of grown yeast cell colonies.
Cell Culture.
MDA-MB-231 cells were cultured in DMEM supplemented with 10% (vol/vol) FBS and 100 units·mL−1 penicillin–streptomycin (Invitrogen Corp.). The cells were grown at 37 °C and 5% CO2 in a humidified atmosphere.
Fabrication of Electronic Apertures.
Silver nanoparticles ink was obtained from Mitsubishi Paper Mills. The ink is a colloidal solution of silver nanoparticles, ethylene glycol, ethanol, and isopropyl alcohol. The distribution of silver nanoparticle diameters is centered at 20 nm. The adherence and subsequent conductivity of the silver nanoparticle suspension is enhanced with a microporous coating on top of the desired print media (i.e., flexible microporous PET) substrate. Posts printing the electrodes were immediately conductive and ready for experimental use.
Microfluidic Biochips.
Disposable microfluidic biochips were fabricated by mixing PDMS prepolymer and curing agents at a ratio of 10:1 and then casting the mixture on premade molds. This was followed by degassing in a vacuum and curing in an oven for ∼2 h at 80 °C. PDMS channels were then cut and peeled off from the molds and reservoir holes were punched at the designed locations. The microfluidic channels then were bonded on this insulating barrier after 10 s of plasma treatment to form the final biochip.
Modeling and Simulation Results.
Numerical finite element modeling was performed using COMSOL Multiphysics (COMSOL Inc.), using similar or exact parameters used for each individual experiment. The three-shield model results were analyzed with in-house-developed MATLAB code, MATLAB (R2010b; The MathWorks Inc.).
Experimental Systems for Microfluidics.
Disposable microfluidic biochips are made of PDMS. The impedance microcytometer branches are 150 μm wide and taper down to a smaller 40-μm-wide-diameter pore.
Electronics.
A standard low-cost consumer-grade inkjet printer (Epson 300) was used to print electronic apertures. For the impedance microcytometer, an impedance spectroscope (ziControl software of the HF2IS; Zurich Instruments) and a transimpedance amplifier (Zurich Instruments HF2TA) were used to excite the coplanar electrodes (12 V a.c. voltage at f = 900 kHz) and monitor the real-time variation in output signal due to passage of the PMS microspheres. The function generator used was a 20-MHz Agilent Keysight 33220A, and the hf amplifier was a TREK model 2100HF piezo amplifier.
Thin Film Insulator.
Standard cover glasses with a thickness of 80–130 μm were used as thin isolative barriers. Cover glass bottom sides were coated with mineral oil for better contact with electronic apertures.
Syringe Pumps.
All buffer exchanges were performed by manual withdrawal, and flow was applied by controlled withdrawal using a Harvard Apparatus Pump 11 Elite syringe pump. Flow rates varied from 0.07 μL/min to 1 μL/min depending on the experiment.
Media.
PBS with σ = 1.6 S⋅m−1 (εr = 80) and diluted PBS with a conductivity of 3 μS/m were used for the experiments.
Bioparticles.
Yeast cells (BY4741), breast adenocarcinoma (MDA-MB-231) cell lines, and 10-μm and 5-μm polyethylene beads (Cospheric LLC) coated with streptavidin were used for these experiments.
Acknowledgments
We thank Laurel Diane Crosby, Raeka Aiyar, Bob St. Onge, and Professor Hanlee P. Ji for useful comments, discussions, and support. This work was supported by NIH Grant P01 HG000205 (to R.W.D.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1621318114/-/DCSupplemental.
References
- 1.McKenzie FE, et al. White blood cell counts and malaria. J Infect Dis. 2005;192(2):323–330. doi: 10.1086/431152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao Y, Yuan Z. Nanotechnology for the detection and kill of circulating tumor cells. Nanoscale Res Lett. 2014;9(1):500. doi: 10.1186/1556-276X-9-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emaminejad S, Javanmard M, Dutton RW, Davis RW. Microfluidic diagnostic tool for the developing world: Contactless impedance flow cytometry. Lab Chip. 2012;12(21):4499–4507. doi: 10.1039/c2lc40759k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei H, et al. Particle sorting using a porous membrane in a microfluidic device. Lab Chip. 2011;11(2):238–245. doi: 10.1039/c0lc00121j. [DOI] [PubMed] [Google Scholar]
- 5.Pertoft H. Fractionation of cells and subcellular particles with Percoll. J Biochem Biophys Methods. 2000;44(1-2):1–30. doi: 10.1016/s0165-022x(00)00066-x. [DOI] [PubMed] [Google Scholar]
- 6.Kersaudy-Kerhoas M, Dhariwal R, Desmulliez MP. Recent advances in microparticle continuous separation. IET Nanobiotechnol. 2008;2(1):1–13. doi: 10.1049/iet-nbt:20070025. [DOI] [PubMed] [Google Scholar]
- 7.Henighan T, et al. Manipulation of magnetically labeled and unlabeled cells with mobile magnetic traps. Biophys J. 2010;98(3):412–417. doi: 10.1016/j.bpj.2009.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsiung S-K, et al. Microfluidic chip with microweir structure for continuous sample separating and collecting applications. Genomic Medicine, Biomarkers, and Health Sciences. 2012;4(1):70–75. [Google Scholar]
- 9.Chin CD, Linder V, Sia SK. Lab-on-a-chip devices for global health: Past studies and future opportunities. Lab Chip. 2007;7(1):41–57. doi: 10.1039/b611455e. [DOI] [PubMed] [Google Scholar]
- 10.Malic L, Brassard D, Veres T, Tabrizian M. Integration and detection of biochemical assays in digital microfluidic LOC devices. Lab Chip. 2010;10(4):418–431. doi: 10.1039/b917668c. [DOI] [PubMed] [Google Scholar]
- 11.Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442(7101):368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 12.Andersson H, van den Berg A. Microtechnologies and nanotechnologies for single-cell analysis. Curr Opin Biotechnol. 2004;15(1):44–49. doi: 10.1016/j.copbio.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, et al. Rare cell isolation and analysis in microfluidics. Lab Chip. 2014;14(4):626–645. doi: 10.1039/c3lc90136j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paegel BM, Blazej RG, Mathies RA. Microfluidic devices for DNA sequencing: Sample preparation and electrophoretic analysis. Curr Opin Biotechnol. 2003;14(1):42–50. doi: 10.1016/s0958-1669(02)00004-6. [DOI] [PubMed] [Google Scholar]
- 15.Breadmore MC, et al. Microchip-based purification of DNA from biological samples. Anal Chem. 2003;75(8):1880–1886. doi: 10.1021/ac0204855. [DOI] [PubMed] [Google Scholar]
- 16.Medoro G, et al. A lab-on-a-chip for cell detection and manipulation. IEEE Sens J. 2003;3(3):317–325. [Google Scholar]
- 17.Cheng I-F, Chang H-C, Hou D, Chang H-C. An integrated dielectrophoretic chip for continuous bioparticle filtering, focusing, sorting, trapping, and detecting. Biomicrofluidics. 2007;1(2):21503. doi: 10.1063/1.2723669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schilling EA, Kamholz AE, Yager P. Cell lysis and protein extraction in a microfluidic device with detection by a fluorogenic enzyme assay. Anal Chem. 2002;74(8):1798–1804. doi: 10.1021/ac015640e. [DOI] [PubMed] [Google Scholar]
- 19.Dittrich PS, Manz A. Lab-on-a-chip: Microfluidics in drug discovery. Nat Rev Drug Discov. 2006;5(3):210–218. doi: 10.1038/nrd1985. [DOI] [PubMed] [Google Scholar]
- 20.Kang L, Chung BG, Langer R, Khademhosseini A. Microfluidics for drug discovery and development: From target selection to product lifecycle management. Drug Discov Today. 2008;13(1-2):1–13. doi: 10.1016/j.drudis.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagrath S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450(7173):1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pratt ED, Huang C, Hawkins BG, Gleghorn JP, Kirby BJ. Rare cell capture in microfluidic devices. Chem Eng Sci. 2011;66(7):1508–1522. doi: 10.1016/j.ces.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jubery TZ, Srivastava SK, Dutta P. Dielectrophoretic separation of bioparticles in microdevices: A review. Electrophoresis. 2014;35(5):691–713. doi: 10.1002/elps.201300424. [DOI] [PubMed] [Google Scholar]
- 24.Bhagat AAS, et al. Microfluidics for cell separation. Med Biol Eng Comput. 2010;48(10):999–1014. doi: 10.1007/s11517-010-0611-4. [DOI] [PubMed] [Google Scholar]
- 25.Chiou PY, Ohta AT, Wu MC. Massively parallel manipulation of single cells and microparticles using optical images. Nature. 2005;436(7049):370–372. doi: 10.1038/nature03831. [DOI] [PubMed] [Google Scholar]
- 26.Miltenyi S, Müller W, Weichel W, Radbruch A. High gradient magnetic cell separation with MACS. Cytometry. 1990;11(2):231–238. doi: 10.1002/cyto.990110203. [DOI] [PubMed] [Google Scholar]
- 27.Alp B, et al. Building structured biomaterials using AC electrokinetics. IEEE Eng Med Biol Mag. 2003;22(6):91–97. doi: 10.1109/memb.2003.1266052. [DOI] [PubMed] [Google Scholar]
- 28.Cheng X, et al. A microfluidic device for practical label-free CD4(+) T cell counting of HIV-infected subjects. Lab Chip. 2007;7(2):170–178. doi: 10.1039/b612966h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huh D, Gu W, Kamotani Y, Grotberg JB, Takayama S. Microfluidics for flow cytometric analysis of cells and particles. Physiol Meas. 2005;26(3):R73–R98. doi: 10.1088/0967-3334/26/3/R02. [DOI] [PubMed] [Google Scholar]
- 30.Zhang S, et al. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 2008;68(11):4311–4320. doi: 10.1158/0008-5472.CAN-08-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oh S-H, Lee S-H, Kenrick SA, Daugherty PS, Soh HT. Microfluidic protein detection through genetically engineered bacterial cells. J Proteome Res. 2006;5(12):3433–3437. doi: 10.1021/pr060193a. [DOI] [PubMed] [Google Scholar]
- 32.Gardeniers JG, van den Berg A. Lab-on-a-chip systems for biomedical and environmental monitoring. Anal Bioanal Chem. 2004;378(7):1700–1703. doi: 10.1007/s00216-003-2435-7. [DOI] [PubMed] [Google Scholar]
- 33.Fatoyinbo HO, Hughes MP, Martin SP, Pashby P, Labeed FH. Dielectrophoretic separation of Bacillus subtilis spores from environmental diesel particles. J Environ Monit. 2007;9(1):87–90. doi: 10.1039/b614556f. [DOI] [PubMed] [Google Scholar]
- 34.Voldman J. Electrical forces for microscale cell manipulation. Annu Rev Biomed Eng. 2006;8:425–454. doi: 10.1146/annurev.bioeng.8.061505.095739. [DOI] [PubMed] [Google Scholar]
- 35.Ramos A, Morgan H, Green NG, Castellanos A. Ac electrokinetics: A review of forces in microelectrode structures. J Phys D Appl Phys. 1998;31(18):2338. [Google Scholar]
- 36.Hughes MP. AC electrokinetics: Applications for nanotechnology. Nanotechnology. 2000;11(2):124. [Google Scholar]
- 37.Çetin B, Li D. Dielectrophoresis in microfluidics technology. Electrophoresis. 2011;32(18):2410–2427. doi: 10.1002/elps.201100167. [DOI] [PubMed] [Google Scholar]
- 38.Martinez-Duarte R. Microfabrication technologies in dielectrophoresis applications--A review. Electrophoresis. 2012;33(21):3110–3132. doi: 10.1002/elps.201200242. [DOI] [PubMed] [Google Scholar]
- 39.Fonkwo PN. Pricing infectious disease. The economic and health implications of infectious diseases. EMBO Rep. 2008;9(Suppl 1):S13–S17. doi: 10.1038/embor.2008.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee WG, Kim Y-G, Chung BG, Demirci U, Khademhosseini A. Nano/Microfluidics for diagnosis of infectious diseases in developing countries. Adv Drug Deliv Rev. 2010;62(4-5):449–457. doi: 10.1016/j.addr.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.World Health Organization 2016 World Health Organization. Available at www.who.int/cancer/detection/breastcancer/en/index1.html. Accessed December 14, 2016.
- 42.Gervais L, de Rooij N, Delamarche E. Microfluidic chips for point-of-care immunodiagnostics. Adv Mater. 2011;23(24):H151–H176. doi: 10.1002/adma.201100464. [DOI] [PubMed] [Google Scholar]
- 43.Pai NP, Vadnais C, Denkinger C, Engel N, Pai M. Point-of-care testing for infectious diseases: Diversity, complexity, and barriers in low- and middle-income countries. PLoS Med. 2012;9(9):e1001306. doi: 10.1371/journal.pmed.1001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yager P, et al. Microfluidic diagnostic technologies for global public health. Nature. 2006;442(7101):412–418. doi: 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- 45.Lin RZ, Ho CT, Liu CH, Chang HY. Dielectrophoresis based-cell patterning for tissue engineering. Biotechnol J. 2006;1(9):949–957. doi: 10.1002/biot.200600112. [DOI] [PubMed] [Google Scholar]
- 46.Shafiee H, Caldwell JL, Sano MB, Davalos RV. Contactless dielectrophoresis: A new technique for cell manipulation. Biomed Microdevices. 2009;11(5):997–1006. doi: 10.1007/s10544-009-9317-5. [DOI] [PubMed] [Google Scholar]
- 47.Park K, Suk H-J, Akin D, Bashir R. Dielectrophoresis-based cell manipulation using electrodes on a reusable printed circuit board. Lab Chip. 2009;9(15):2224–2229. doi: 10.1039/b904328d. [DOI] [PubMed] [Google Scholar]
- 48.Camarda M, et al. Theoretical and experimental study of the role of cell-cell dipole interaction in dielectrophoretic devices: Application to polynomial electrodes. Biomed Eng Online. 2014;13(1):71. doi: 10.1186/1475-925X-13-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pethig R. Review article-dielectrophoresis: Status of the theory, technology, and applications. Biomicrofluidics. 2010;4(2):022811. doi: 10.1063/1.3456626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Müller T, et al. High-frequency electric-field trap for micron and submicron particles. Il Nuovo Cimento D. 1995;17(4):425–432. [Google Scholar]
- 51.Henslee EA, Sano MB, Rojas AD, Schmelz EM, Davalos RV. Selective concentration of human cancer cells using contactless dielectrophoresis. Electrophoresis. 2011;32(18):2523–2529. doi: 10.1002/elps.201100081. [DOI] [PubMed] [Google Scholar]
- 52.Zhu H, Lin X, Su Y, Dong H, Wu J. Screen-printed microfluidic dielectrophoresis chip for cell separation. Biosens Bioelectron. 2015;63:371–378. doi: 10.1016/j.bios.2014.07.072. [DOI] [PubMed] [Google Scholar]
- 53.Alshareef M, et al. Separation of tumor cells with dielectrophoresis-based microfluidic chip. Biomicrofluidics. 2013;7(1):11803. doi: 10.1063/1.4774312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dawood S, et al. Circulating tumor cells in metastatic breast cancer: from prognostic stratification to modification of the staging system? Cancer. 2008;113(9):2422–2430. doi: 10.1002/cncr.23852. [DOI] [PubMed] [Google Scholar]
- 55.Zhan Y, et al. Low-frequency ac electroporation shows strong frequency dependence and yields comparable transfection results to dc electroporation. J Control Release. 2012;160(3):570–576. doi: 10.1016/j.jconrel.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 56.Lin C-C, Hsu J-L, Lee G-B. Sample preconcentration in microfluidic devices. Microfluid Nanofluidics. 2011;10(3):481–511. [Google Scholar]
- 57.Jokerst JC, Emory JM, Henry CS. Advances in microfluidics for environmental analysis. Analyst (Lond) 2012;137(1):24–34. doi: 10.1039/c1an15368d. [DOI] [PubMed] [Google Scholar]
- 58.de Bono JS, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14(19):6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 59.Xu W, et al. Isolation of circulating tumor cells in patients with hepatocellular carcinoma using a novel cell separation strategy. Clin Cancer Res. 2011;17(11):3783–3793. doi: 10.1158/1078-0432.CCR-10-0498. [DOI] [PubMed] [Google Scholar]
- 60.Koyanagi K, et al. Serial monitoring of circulating tumor cells predicts outcome of induction biochemotherapy plus maintenance biotherapy for metastatic melanoma. Clin Cancer Res. 2010;16(8):2402–2408. doi: 10.1158/1078-0432.CCR-10-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martin DJ, et al. CD4+ lymphocyte count in African patients co-infected with HIV and tuberculosis. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;8(4):386–391. [PubMed] [Google Scholar]
- 62.Cheung K, Gawad S, Renaud P. Impedance spectroscopy flow cytometry: On-chip label-free cell differentiation. Cytometry A. 2005;65(2):124–132. doi: 10.1002/cyto.a.20141. [DOI] [PubMed] [Google Scholar]
- 63.Kim KB, Chun H, Kim HC, Chung TD. Red blood cell quantification microfluidic chip using polyelectrolytic gel electrodes. Electrophoresis. 2009;30(9):1464–1469. doi: 10.1002/elps.200800448. [DOI] [PubMed] [Google Scholar]
- 64.Holmes D, Morgan H. Single cell impedance cytometry for identification and counting of CD4 T-cells in human blood using impedance labels. Anal Chem. 2010;82(4):1455–1461. doi: 10.1021/ac902568p. [DOI] [PubMed] [Google Scholar]
- 65.Hong J, Edel JB, deMello AJ. Micro- and nanofluidic systems for high-throughput biological screening. Drug Discov Today. 2009;14(3-4):134–146. doi: 10.1016/j.drudis.2008.10.001. [DOI] [PubMed] [Google Scholar]