Significance
Induced pluripotent stem cells (iPSCs) open new possibilities for generating personalized disease models and drug testing. However, iPSC differentiation to a specific cell type can take weeks to complete, delaying the optimization process (maximize yield of desired cell types) for each patient’s iPSC. This task can be accelerated if the destination cell type can be determined early during cell lineage trajectory before cells manifest the desired phenotype. Our results indicate such a possibility: by quantifying the cell population structure during a critical state transition, we identified key regulators of lineages commitment and predicted the percentage of desired cell types for several protocol variations 2 wk in advance.
Keywords: single-cell analysis, critical state transitions, iPSC to cardiomyocyte differentiation, differentiation efficiency, prediction
Abstract
Steering the differentiation of induced pluripotent stem cells (iPSCs) toward specific cell types is crucial for patient-specific disease modeling and drug testing. This effort requires the capacity to predict and control when and how multipotent progenitor cells commit to the desired cell fate. Cell fate commitment represents a critical state transition or “tipping point” at which complex systems undergo a sudden qualitative shift. To characterize such transitions during iPSC to cardiomyocyte differentiation, we analyzed the gene expression patterns of 96 developmental genes at single-cell resolution. We identified a bifurcation event early in the trajectory when a primitive streak-like cell population segregated into the mesodermal and endodermal lineages. Before this branching point, we could detect the signature of an imminent critical transition: increase in cell heterogeneity and coordination of gene expression. Correlation analysis of gene expression profiles at the tipping point indicates transcription factors that drive the state transition toward each alternative cell fate and their relationships with specific phenotypic readouts. The latter helps us to facilitate small molecule screening for differentiation efficiency. To this end, we set up an analysis of cell population structure at the tipping point after systematic variation of the protocol to bias the differentiation toward mesodermal or endodermal cell lineage. We were able to predict the proportion of cardiomyocytes many days before cells manifest the differentiated phenotype. The analysis of cell populations undergoing a critical state transition thus affords a tool to forecast cell fate outcomes and can be used to optimize differentiation protocols to obtain desired cell populations.
The availability of human induced pluripotent stem cells (iPSCs) with their potential to differentiate into virtually any cell type creates unprecedented possibilities, not only to study human development and disease but also to generate patient-specific cells to determine personalized drug response (1, 2). However, steering iPSCs efficiently into pure populations of a specific cell type, such as cardiomyocytes, remains a challenge, because the binary nature of cell fate decisions often causes the “leakage” of cells into undesired lineages at each such decision point. Additionally, optimizing established differentiation protocols for a specific genetic background (i.e., patient-specific iPSC lines) to maximize differentiation efficiency is time-consuming because of the long time period (up to weeks) until cells display the differentiated phenotype that informs about the success of a differentiation protocol. Thus, it is paramount to develop tools that not only reveal the critical regulators that govern lineage-specific decision-making but at the same time, also facilitate and shorten the optimization procedures for iPSC differentiation protocols.
Toward this aim, longitudinal single-cell gene expression analysis provides a new avenue to understand lineage commitment in mouse and human pluripotent cells (3–5). Reconstructing lineage trajectories at single-cell resolution captures cell fate transitions in a large statistical ensemble of identical systems as each individual progenitor cell in a differentiating population, allowing us to dissect molecular and cellular patterns driving lineage commitment. For instance, single-cell resolution analyses have shown that cell types form discrete clusters when gene expression patterns are visualized in a low-dimensional space using, for example, principle component analysis or t-distributed stochastic neighbor-embedding plots (3, 6) (Fig. 1 A and B). This pattern is consistent with the concept of attractors [i.e., stable cell states of the gene regulatory network (GRN)], which correspond to the valleys in Waddington’s “epigenetic landscape” (7). A cell fate transition then corresponds to a switching between distinct attractors via transient unstable states and can be analyzed as coordinated shift of gene expression in a low-dimensional cell-state space (8) (Fig. 1 A and B). This formalism enables us to study universal patterns that underlie major transitions of GRN states (hence transitions of cell states) independent of specific molecular mechanisms, such as the specific structure of the regulatory network that drives the transition. Such phenomenological analysis of major state shifts in complex systems has been successfully used for ecosystems and social systems (9, 10).
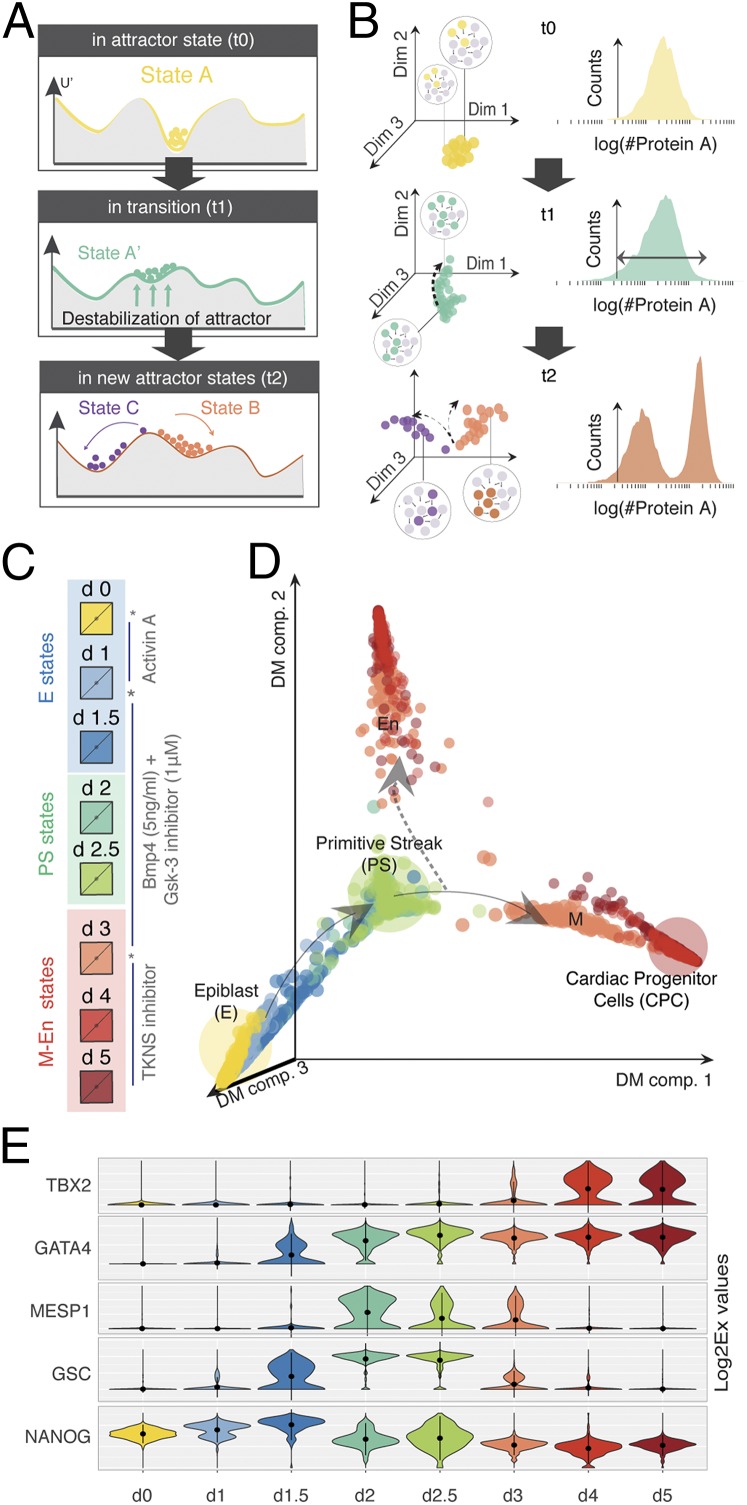
Fig. 1.
Directed differentiation at single-cell resolution. (A) Theoretical and (B) experimental framework to study cell differentiation as a transition between attractors. Before transition (time t0): Cells in state A (local minima in a quasipotential landscape) are defined by a distinct GRN state (expressed and nonexpressed genes are colored and gray, respectively). The state A attractor manifests as either a dense cloud of points in a high-dimensional cell-state space (as measured using single-cell qPCR) or a tight, uniform distribution of a single gene/dimension (as measured by flow cytometry) as shown in B. The tipping point (time t1): The attractor destabilizes (via changes in the quasipotential landscape), and cells become primed toward a future attractor state(s). Cells in the poised state A' exhibit increased cell diversity, which manifests as a shift in the high-dimensional state space or a wider distribution in a single dimension. Posttransition (time t2): Stable states B and C emerge through the stabilization of mutually exclusive GRN states that can be observed as two clouds occupying distinct positions in the high-dimensional cell-state space or bimodal distribution of the marker gene as shown in B. (C) Snapshot of the iPSC to iCM differentiation protocol used in this study. Asterisks mark the perturbation time points (days 0, 1, and 3) that also correspond to cell culture media exchanges. (D) Diffusion map (DM) of the iPSC to iCM differentiation based on 1,934 single-cell gene expression vectors of 96 genes. Color of each dot represents the day of collection during differentiation. Arrows indicate the direction of cell-state trajectories. The dashed arrow points toward the undesired cell state. (E) Dynamics of state-specific transcription factors. The violin plots show the variability of gene expression (log2Ex) across each cell population for five transcription factors: NANOG (stem cell marker), GSC (PS marker), MESP1 (posterior PS/cardiac mesoderm marker), GATA4 (mesoderm and endoderm marker), and TBX2 (cardiac marker).
Specifically, we postulate that exit from pluripotency is not simply a jump between attractors but instead, is initiated by the gradual destabilization of the pluripotent stem cell attractor triggered by exogenous signals (i.e., growth factors and modulators of signaling pathways). This response is akin to flattening of the valley in the landscape, which facilitates exit from the attractor state until, at a critical point, the pluripotency attractor suddenly vanishes, providing access to two alternative cell fate attractors (Fig. 1 A and B). A destabilization of an attractor until it vanishes formally represents a bifurcation event (11, 12). The associated sudden qualitative system changes are known to produce the phenomenological signatures of a critical state transition (“tipping point”) (9, 10). As the cell population approaches a tipping point, we expect to observe two changes that can only be revealed by analyzing gene expression patterns at single-cell resolution: (i) increased cell population diversity because of the destabilization of the attractor and diminished “attracting force” and simultaneously, (ii) a higher coordination of gene expression across the cells as they move on a trajectory along which the attractor transition takes place (11, 12).
This framework of attractor destabilization can help us formalize how exogenous signals relate to the differentiation efficiency that is usually measured as the percentage of the desired cell type in the differentiated cell population. We hypothesized that the signals conveyed by the treatment to cause iPSC differentiation not only destabilize the attractor but also, bias the destabilization toward a specific lineage. This bias would be manifest before fate commitment. Thus, we examined by single-cell analysis the population structure after the treatment but before cell lineage commitment to determine if it can inform about the future course of the differentiation trajectory. To validate our hypotheses, we systematically varied the levels of differentiation cues for cardiomyocyte differentiation and investigated how a range of signals affected differentiation efficiency. We show that our analysis of the cell population structure at the tipping point can help us forecast the preference by differentiating iPSC for cardiac over other fate options (hence, to predict the efficiency of the desired differentiation).
Results
We first monitored changes in transcript levels at single-cell resolution during the first 6 days as cells exit pluripotency and move toward the cardiomyocyte cell fate (Fig. S1). Extensive prior knowledge guided us to identify (i) intermediate cell states at branch points of development, (ii) key transcriptional regulators that control cell fate decisions, and (iii) instructive signals that guide the differentiation process (2, 13–17). We used this knowledge to select 96 gene markers for our study (Dataset S1, Table S1). A standard method for induced pluripotent stem cell-derived cardiomyocytes (iCMs) differentiation (Fig. 1C) consisting of the sequential treatment of iPSCs with cytokines and other molecules that induce cardiac mesoderm in vivo was used: activin A (day 0), BMP4 (bone morphogenetic protein 4) combined with a Wnt pathway activator (day 1), and a Wnt antagonist (day 3) mimicking, at least partially, the differentiation signals that epiblast (E) cells are exposed to during heart development in vivo (13, 17, 18). This widely used protocol yields ∼70% cardiomyocytes within 2 wk in culture (Materials and Methods), although there is considerable variability depending on the initial conditions (i.e., iPSC density plating) as well as genetic background of the ES cell/iPSC line used (16).
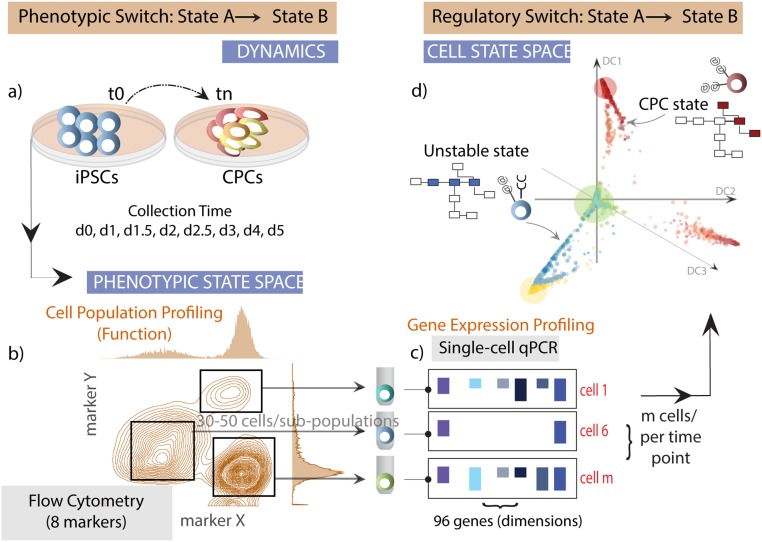
Fig. S1.
An integrative systems biology approach to study cell differentiation. We integrated single-cell qPCR measurements with flow cytometry to connect the phenotypic states (cell surface marker profile) to the regulatory states (transcription factor expression profile). (A) Single cells are collected using flow cytometry at specific time points as iPSCs differentiate to cardiac progenitors cells (CPCs) as described in the methods for downstream single-cell and population analyses. (B) We monitor the dynamics for eight specific cell surface markers, and individual cells from distinct cell populations are sorted into wells of a 96-well plate containing a buffer optimized for single-cell cDNA synthesis. (C) We performed qPCR on cDNA generated from sorted single cells using Fluidigm’s microfluidics-based platform (Biomark). The expression level of 96 genes (composed of transcription factors, signal transducers, and cell surface proteins) was measured in ∼1,900 individual cells. (D) Finally, we performed a rigorous computational analysis of single-cell gene expression data based on clustering, dimensionality reduction, and correlation analysis to map gene regulatory states and identify the differentiation trajectories and branch points.
To reconstruct the iPSC to iCM differentiation trajectory and identify lineage branch points, we measured transcript expression of the selected genes in ∼1,900 individual differentiating cells obtained during the first 6 days of differentiation (Fig. 1D, Fig. S2, and Dataset S2). A major lineage branching took place at day 3 when individual cells transitioned from a multipotent, primitive streak (PS)-like progenitor state to either a more differentiated mesodermal (M) state or an endodermal (En) state as indicated by lineage-specific transcripts (Fig. 1E). This abrupt disappearance of the progenitor state and its split into two gene expression programs suggest a bifurcation in the dynamics of the underlying GRN (12)—a critical state transition (9, 10).
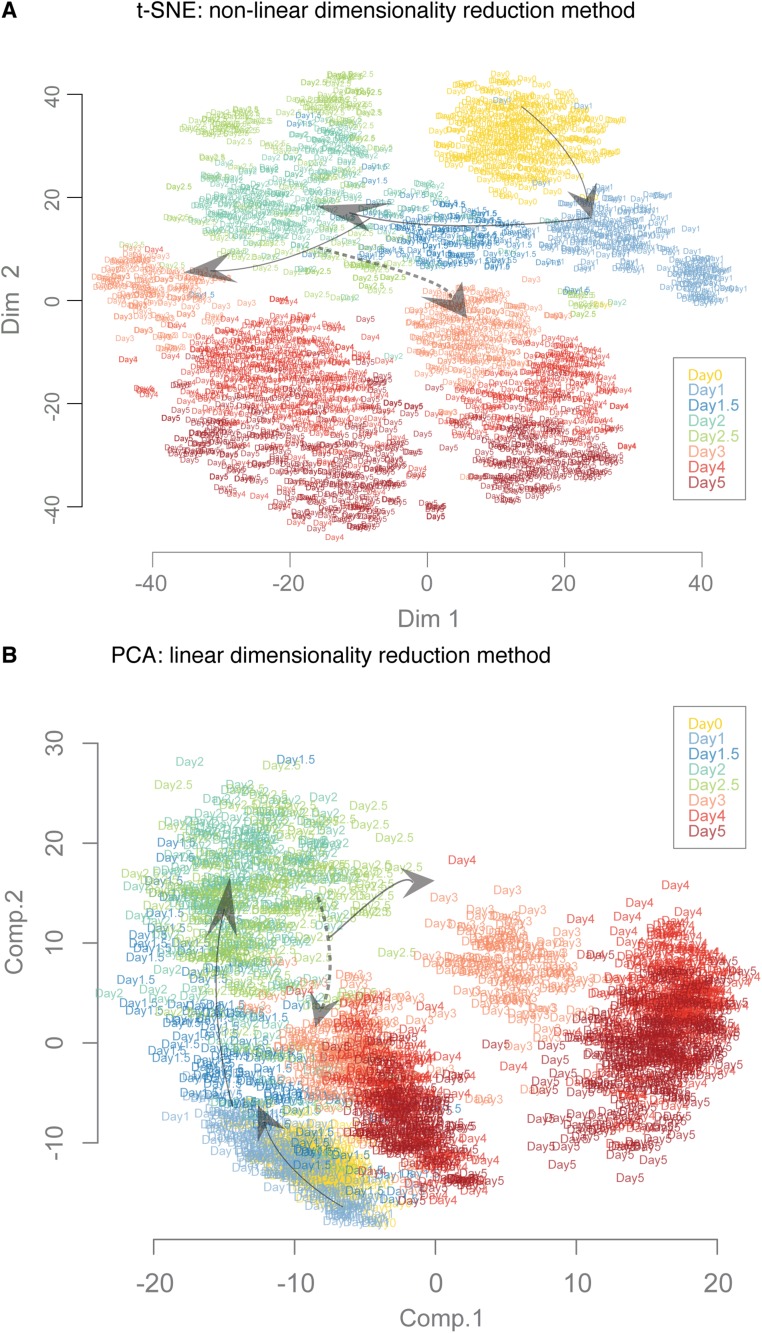
Fig. S2.
Comparison of dimensionality reduction algorithms to visualize the trajectories and transitions. We compared the data projections based on 1,934 single-cell gene expression profiles using three different dimensionality reduction methods. Results from diffusion maps are described and discussed in the text: (A) t-SNE—a nonlinear method—and (B) principle component analysis (PCA)—a linear method. The cells are represented by labels according to the collection day. The arrows indicate the general direction of progression of differentiation. The broken line arrows indicate the branching of En lineage from PS-like population.
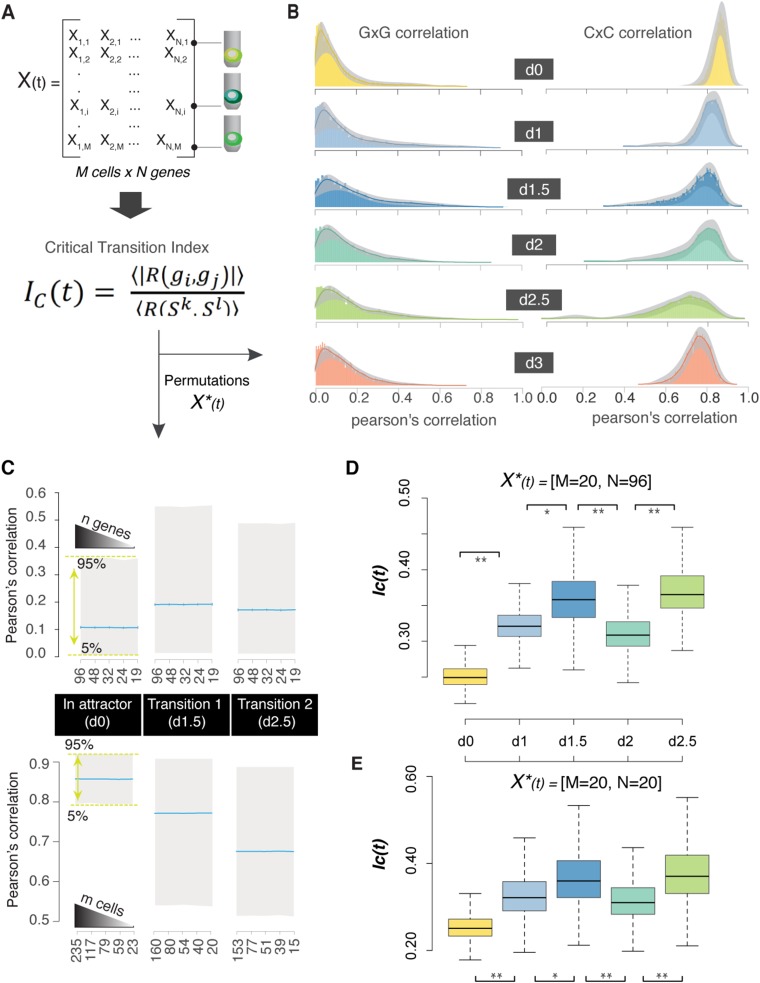
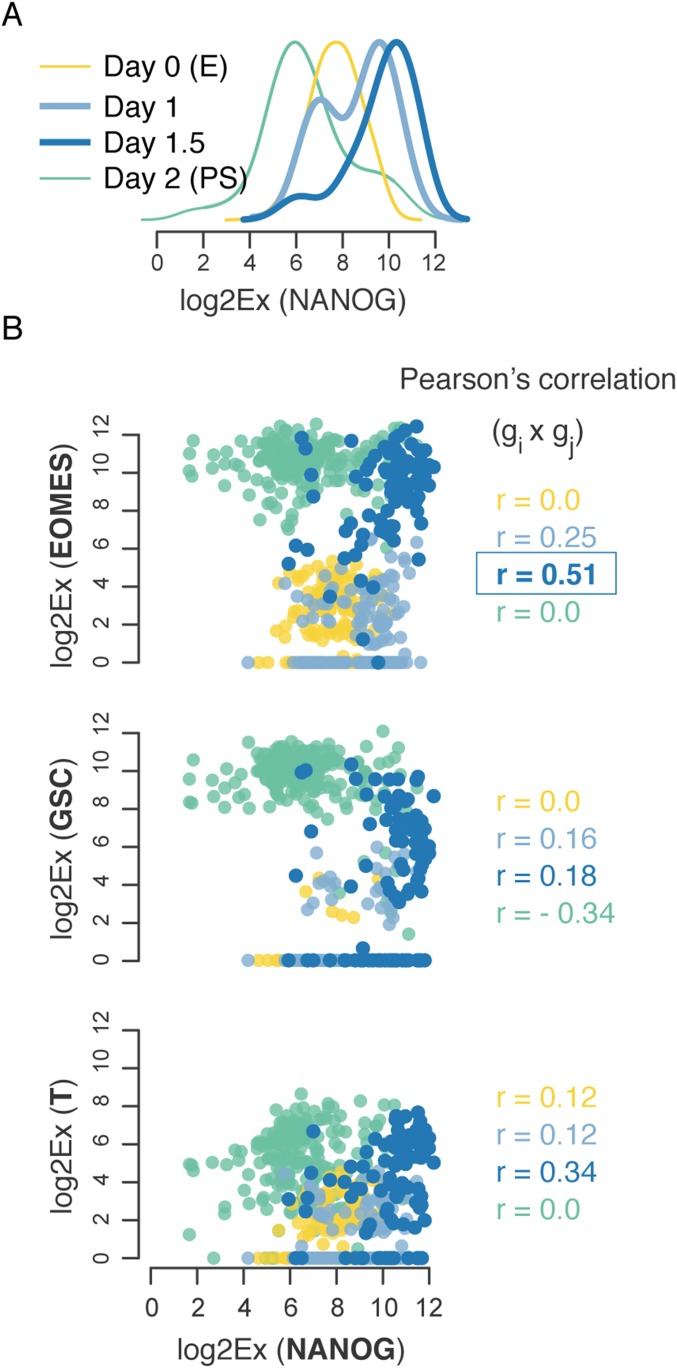
The signature of a critical state transition that can be identified by single-cell resolution analysis of cell populations consists of (i) a decrease of overall cell to cell correlation with respect to gene expression and (ii) a concomitant increase in overall gene to gene correlation across the cells (11). The first one manifests an increase in cell diversity as the attractor destabilizing and allowing access to new GRN states (lineage priming). However, the counterintuitive increase in overall gene to gene correlation reveals a tight coordination of gene expression before the transition (Fig. 2 and Fig. S3) (11). These changes can be summarized by the critical transition index IC(t) computed for each measured time point t (Fig. 2A and Fig. S3), which is defined as the ratio of the average of all pairs of gene to gene correlation coefficients to the average of all pairs of cell to cell correlation coefficients. Computing the IC(t) values (from day 0 to day 3) revealed a significant increase as the differentiating cell population approached the M–En branch point, indicating a bifurcation (Fig. 2A and Fig. S3). To show that the observed trend was independent of the quality and quantity of genes or cells, we calculated the Ic for randomly selected subsets of our dataset (Fig. S3). Cells were indistinguishable at day 0 (E state), and there was no apparent correlation between pluripotency and lineage-specific transcripts (Fig. 2B and Dataset S1, Table S2)—consistent with the theory that, in an attractor state, cell population diversity is mainly caused by symmetric fluctuations around the “set point” caused by gene expression noise (7, 19). Specifically, on destabilization of the E state triggered by the first differentiation signal (activin A), cells diversified, and gene to gene correlation between NANOG, an E state-specific marker, and PS state-specific markers increased (Fig. 2C and Dataset S1, Table S3). Our data confirm previously reported interactions between NANOG and the transcription factor EOMES (20, 21) as a major regulatory interaction that drives exit from pluripotency toward PS state (Fig. S4). At day 2, when the PS-like cells are still uniform with respect to lineage-specific markers, we observed a temporary decrease, still significant, in the critical transition index (Fig. 2A), consistent with the PS state being a distinct and observable, although transient, stabilized state. By day 2.5, the value of IC increased again driven by the emergence of correlations and anticorrelations in the expression of lineage-specific transcription factors (Fig. 2D and Dataset S1, Table S4). After cells were committed to a specific lineage, cell-state variability (within each new subpopulation) decreased, thus lowering Ic for each individual day 3 cell subpopulation (Fig. 2A).
Fig. 2.
A critical transition signature for differentiation branch points. (A) Time point-specific boxplots represent the distribution of IC(t) values from 1,000 permutations of 25 randomly selected genes. After bifurcation, we used cells that cluster as M lineage for day 3. The mean value corresponds to the IC(t) value [X(t) = 96 genes × M cells]. **P value < 2e-10 for comparison between the time points (Kolmogorov–Smirnov test and Wilcoxon rank sum test). (B) Gene to gene (GxG) correlation plots for six lineage-specific transcription factors at day 0 (“in attractor”). The shade corresponds to the Pearson’s correlation across all of the cells for each pairwise comparison, whereas the shape of the data cloud shows the distribution of Pearson’s correlation across all of the cells for each gene pair. (C and D) GxG correlation plots for six lineage-specific transcription factors during two state transitions. We can observe distinct patterns for individual genes, such as EOMES (important during E → PS but not PS → M transition), or small regulatory circuits (i.e., day 2.5 shows anticorrelated networks that are related with lineage segregation). (E) Early iPSC to iCM differentiation model. Each cell state (stable or transitional) can be marked by specific transcription factors.
Fig. S3.
Robustness of the IC values using bootstrap analysis. (A) Time point-specific matrices—X(t)—were generated based on the Log2Ex gene expression values (columns correspond to genes, and rows correspond to cells). After bifurcation, we generated time- and lineage-specific matrices (i.e., cells that cluster as M lineage on day 3). For each matrix, we calculated the critical transition index. (B) Dynamics of gene to gene (GxG) and cell to cell (CxC) correlations. Solid lines correspond to the density plot based on the correlation values of the X(t), whereas the gray regions show the confidence intervals (5 and 95%) computed from 1,000 permutations of 50 randomly selected cells for each population [X*(t) = 50 cells × 96 genes]. (C) Boxplots present the distribution of Pearson’s correlation values from 1,000 permutations of n randomly selected genes [X*(t) = n genes × M cells] or m randomly selected cells [X*(t) = N genes × m cells] for each time point. Blue lines indicate the mean values that are used for the IC(t) calculations. The graph shows that mean values for both GxG and CxC correlations, as a component of the IC index, are not affected by the number of selected variables. (D and E) Boxplots present the distribution of IC(t) values from 1,000 permutations with specific matrix sizes: X*(t) = 96 genes × 20 cells or X*(t) = 20 genes × 20 cells. As expected, the size of the vector (n = 96 vs. n = 20) influences the possible index value, although the trend (increasing during transitions) remains constant; the relative trend is there. *P value < 5e-10 for comparison between the time points (Kolmogorov–Smirnov test and Wilcoxon rank sum test); **P value < 2e-10 for comparison between the time points (Kolmogorov–Smirnov test and Wilcoxon rank sum test).
Fig. S4.
Transcription factor EOMES drives the E to PS transition. (A) Dynamics of the NANOG gene expression. Density plots of NANOG gene expression—a pluripotency marker for day 0 (pluripotency state) to 2 (PS state). (B) Dynamics of the regulatory relationships between NANOG and PS specifiers. Plots based on Log2Ex values for each pairwise comparison (i.e., NANOG vs. EOMES) illustrate the molecular noise during the transition (colored by cell population across time). Pearson’s correlations were calculated separately for each time point.
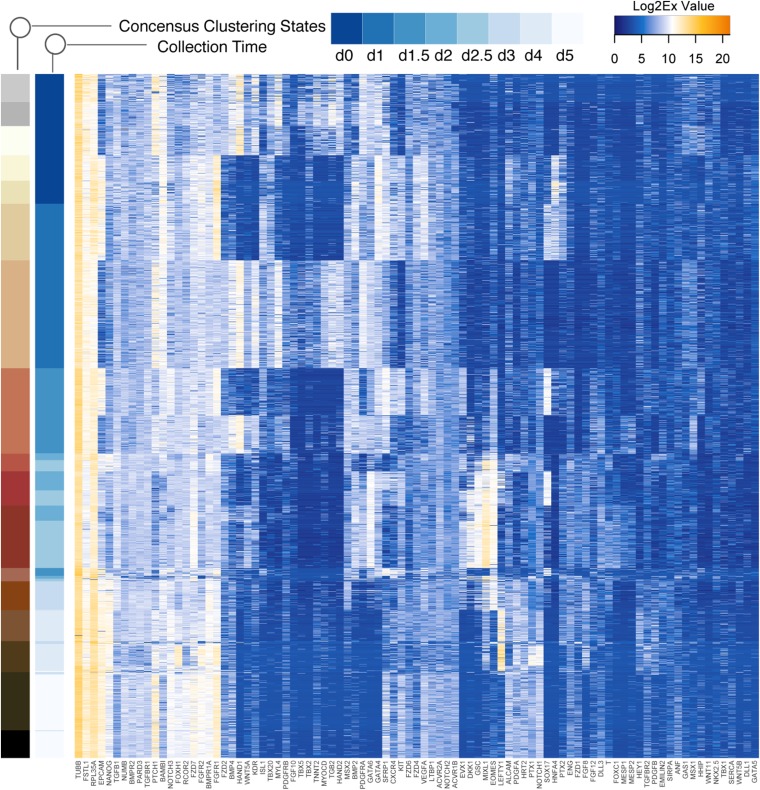
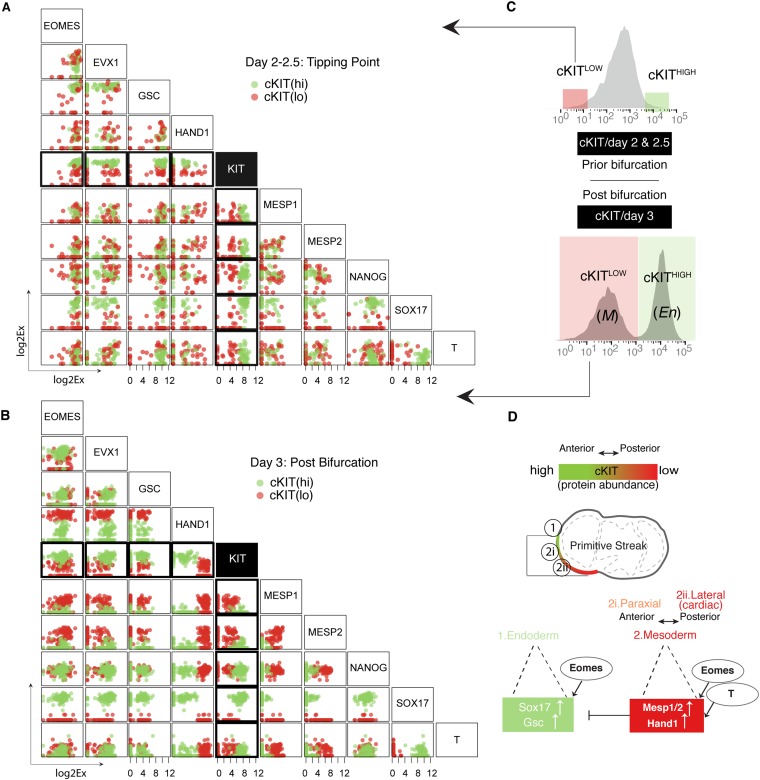
Combining the above findings with consensus clustering and correlation analysis allowed us to build a comprehensive model of early iPSC to iCM differentiation (Fig. 2E). Our data support two distinct cell (sub)states after day 2 (Fig. S5), which were evident in the mutually exclusive expression of the fate-determining transcription factors indicative of binary lineage branching (22). The identified heterogeneity at this stage can be correlated with distinct in vivo states during the anterior–posterior patterning of the PS (Figs. S6 and S7). In particular, SOX17 (23) and HAND1 (24) appeared to display the familiar toggle switch-like binary behavior that segregates the PS-like cells into two distinctly primed populations: if HAND1 >> SOX17, cells were primed toward M fates (posterior PS); if HAND1 << SOX17, they were primed to the En fate (anterior PS). Similar observations have been reported by other single-cell studies for mesoderm differentiation (3, 4). However, our analysis additionally revealed that the expression of the cell surface marker, cKIT, correlated with this anterior vs. posterior PS specification (Fig. S6). Thus, we decided to investigate the distribution of the cKIT protein expression phenotype and its association with mesoderm–endoderm branching.
Fig. S5.
Consensus clustering to identify regulatory states and state specifiers. Heat map showing regulatory states based on consensus clustering of the gene expression data (Log2Ex values) for individual cells for all time points. Color bars indicate 17 robust clusters (>80% accuracy) and day of collection.
Fig. S6.
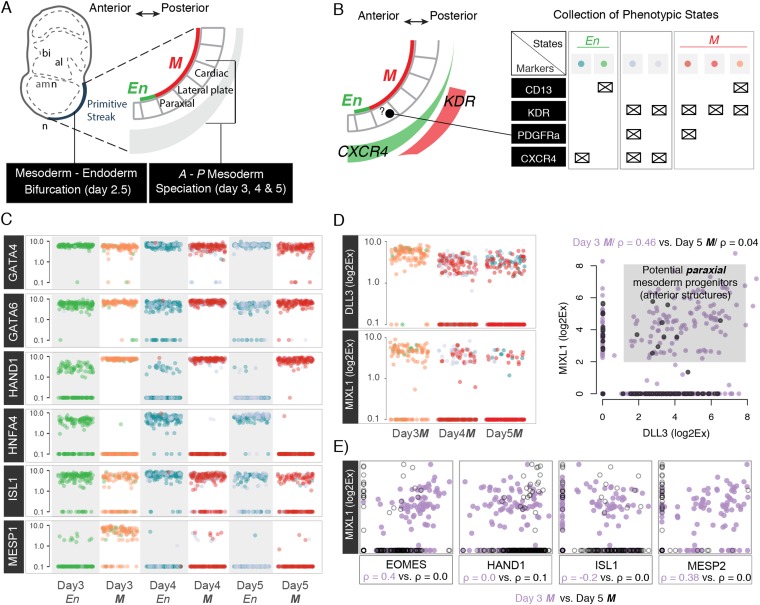
cKIT protein abundance correlates with anterior–posterior patterning factors. (A) Correlation plots for nine transcription factors and c-KIT for days 2 and 2.5 (prior bifurcation). The color corresponds to the phenotypic properties of the cells (more specifically, the protein abundance of cell surface marker cKIT; C). (B) Correlation plots for nine transcription factors and c-KIT for day 3 (postbifurcation). The color corresponds to the phenotypic properties of the cells (more specifically, the protein abundance of cell surface marker cKIT; C). (C) Flow cytometry analysis for cKIT protein. Boxes illustrate the fraction of populations that was analyzed as cKITLOW–cKIT(lo) or cKITHIGH—cKIT(hi). (D) A model for anterior–posterior patterning of PS. cKITHIGH correlates with En cell fates that are located in the most anterior part of PS and express high levels of SOX17 and GSC. The rest of the PS correlates with cKITLOW and associates with M cell fates.
Fig. S7.
Cell states during anterior–posterior mesoderm specification. (A) Model of the anterior–posterior patterning of PS. (B) Distribution of cell surface molecules in PS and collected phenotypic states that are related to En or M cell fates. For instance, KDR+ cells are primarily related to lateral mesoderm fate, whereas PDGFRa is mainly a paraxial mesoderm marker. The color scheme is used below to indicate how different transcription factors are distributed between each phenotype. (C) Plots of Log2Ex values for six transcriptions factors that define M or En cell fates: GATA4 and GATA6 indicate both cell fates; HAND1, ISL1, and MESP1 indicate cardiac cell fates/midposterior PS; and HNFA4 indicates En cell fates/anterior PS. (D) MIXL1 (a transcription factor) and DLL3 (a Notch ligand) have been recently reported (4) as paraxial mesoderm markers. They can be detected between days 3 and 5, but they correlate only during day 3 (correlation plot), suggesting that there might be a paraxial mesoderm population early during mesoderm specification process. (E) Correlation plots for days 3 and 5 gene expression between MIXL1 (a paraxial mesoderm regulator) and four other transcription factors. Notice that EOMES that is associated with anterior PS specification has a positive correlation with MIXL1, whereas ISL1 that is expressed in the most posterior parts is slightly anticorrelated with MIXL1. However, all correlations are lost by day 5.
We found that, at the tipping point, cKIT protein expression varied among individual cells, displaying the widest spread, consistent with maximal cell to cell variability (Fig. 3A). Additionally, around the tipping point, the heterogeneous cell population transiently exhibited an M–En continuum, in which individual cells expressed the molecular signature, indicating priming toward either the desired cardiac (cKIT− and HAND1+/SOX17− cells) or the undesired noncardiac (cKIT+ and HAND1−/SOX17+ cells) fate. Single-cell gene expression profiling of the extreme tails of the cKIT distribution (outliers), cKITlow and cKIThigh cells, mapped them to cell states primed for the M and En lineages, respectively. Thus, information on prospective fate is hidden in the bulk population distributions and seems most pronounced in the outlier subpopulations (25) as evident at days 2 and 2.5. Although at this point, the population is still unimodal with respect to cKIT expression, the cKIThigh cells expressed higher levels of SOX17, whereas the cKITlow cells expressed higher levels of HAND1, and both displayed decreased expression of NANOG (Fig. 3B). Therefore, the population distribution of cKIT protein expression can act as surrogate marker for the position in the HAND1–SOX17 expression axis of cells poised to differentiate to M or En cell lineages, respectively.
Fig. 3.
cKIT distribution at the tipping point hides M–En primed states. (A) Dynamics of cKIT distribution from day 1 (activin A-induced cells) to 3 (postbranching event). The unimodal distribution before (day 1) and during the tipping point (days 2–2.5) slowly changes to the characteristic bimodal distribution after the branching event (day 3). The cKIT−/HAND1+ cells will eventually commit to the cardiac cell fate. (B) Heat map of cell to cell correlations based on the cKITHigh, cKITMedium, and cKITLow single-cell gene expression vectors (30–50 cells per phenotypic state per time point). Cells have been sorted based on the phenotypic state [low (L) → average (M) → high (H)] and time collected (day 0 → day 3). By day 2.5, cKITHigh and cKITLow cells are presented as two distinct states that expressed SOX17 or HAND1, respectively (side bar). These results are supported by consensus clustering analysis (Fig. S5).
Accordingly, this behavior can be exploited to identify primed states and predict lineage commitment using a single surface marker—cKIT (12, 19). We reasoned that the cell population structure at the tipping point (day 2), which reflects attractor destabilization that can be biased in either direction by the instructive signal, may determine the M vs. En decision and thus, influence the ultimate efficiency of differentiation into iCMs as observed on day 28. In other words, the final percentage of cells in the desired state (iCM) may already be destined at a critical fork in the road (trajectory) far earlier in the developmental journey. Thus, the transient PS-like state may be sensitive to tuning that “tilts” the cells toward either fate.
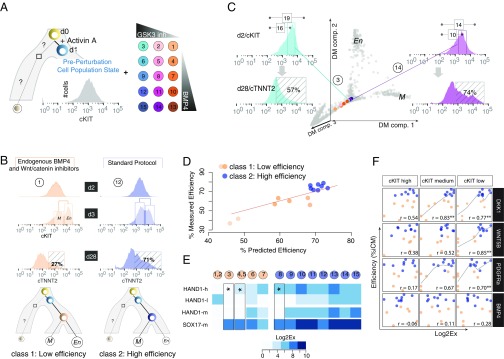
To test the idea that the cell population structure at this tipping point sets the course of the long-term trajectory and can predict the efficiency of differentiation into future states, we varied the protocol for differentiation by gradually modulating the concentration for the two inducers, BMP4 and Wnt pathway activator (glycogen synthase kinase 3 (GSK3) inhibitor) (Materials and Methods and Fig. 4A). We then monitored the structure of the differentiating cell populations with respect to five state-specific surface markers and the final efficiency of iCM differentiation (Fig. 4 A and B and Fig. S8). Indeed, the features of the cKIT distribution (i.e., mean and dispersion) correlate with the low- vs. high-efficiency protocols. The M–En branching at day 3 took place in the absence of any exogenous signals (no BMP4/no GSK3 inhibitor), suggesting that Activin A-induced iPSCs are in a transient, unstable state driven by the endogenous TGF-β/BMP and Wnt pathways to undergo a critical state transition (Fig. 4 B and C). It seems, however, that Activin A alone induced a biased destabilization of the E attractor toward the En states and resulted in low iCM efficiency (<30%); this observation is consistent with use of Activin A in definitive endoderm (hepatocytes and pancreatic) differentiation protocols (26).
Fig. 4.
Cell-state diversity at the tipping point predicts differentiation efficiency. (A) Perturbation scheme to influence the M–En branching event on day 3. In total, we tested 15 BMP4/GSK3 inhibitor combinations highlighted with a unique color or number in a circle across all panels herein. (B) Dynamics of the cKIT distribution during early differentiation reflect the effect of each small molecule combination. We observed different cKIT distributions on days 2 and 3 for each treatment (flow cytometry data shown in Fig. S8), which inform predictions about protocol efficiency as measured by the proportion of cTNNT2+ cells—the most commonly used molecules to show the cardiac fate—on day 28 (shown in striped boxes). (C) Projections of the 100-cell pool cKITHigh, cKITMedium, and cKITLow gene expression profiles onto the diffusion map (DM). The single-cell gene expression profiles are shown in gray, and combination-specific, population-specific pooled gene expression vectors are represented with the corresponding colors from A. The position in the trajectory at day 2 as well as the variable distribution of cKIT outlier fractions (shown as numbers in boxes above histograms) can explain huge variation in the efficiencies of the protocols (measured as cTNNT2+ cells at day 28). (D) Random Forest analysis to identify transcripts or cell subpopulations that predict high- vs. low-efficiency protocols. We classified every combination that produces >60% iCMs as high efficiency (dark blue), whereas any combination with efficiency <60% was classified as low efficiency (orange). (E) The cKITHigh, cKITMedium, and cKITLow concatenated gene expression vectors outperform the cKITMedium vector classification. HAND1 gene expression in cKITHigh and cKITLow populations discriminates between three different combinations with different efficiencies (marked by asterisks), whereas SOX17 and HAND1 gene expression in cKITMedium population is the same. (F) Correlation plots for signaling molecules and combination class (low vs. high efficiency). Low-efficiency protocols are marked by lower dosages of exogenous BMP4 (<2.5 μM), but it seems that there is no correlation between exogenous administration and endogenous BMP4 levels (measured as gene expression; no correlation). However, higher dosages of exogenous BMP4 are highly correlated with the expression levels of DKK1 and Wnt5B in the cKITLow population that will commit to the cardiac cell fate. Again, cKITHigh or cKITLow gene expression vectors perform better than cKITMedium.
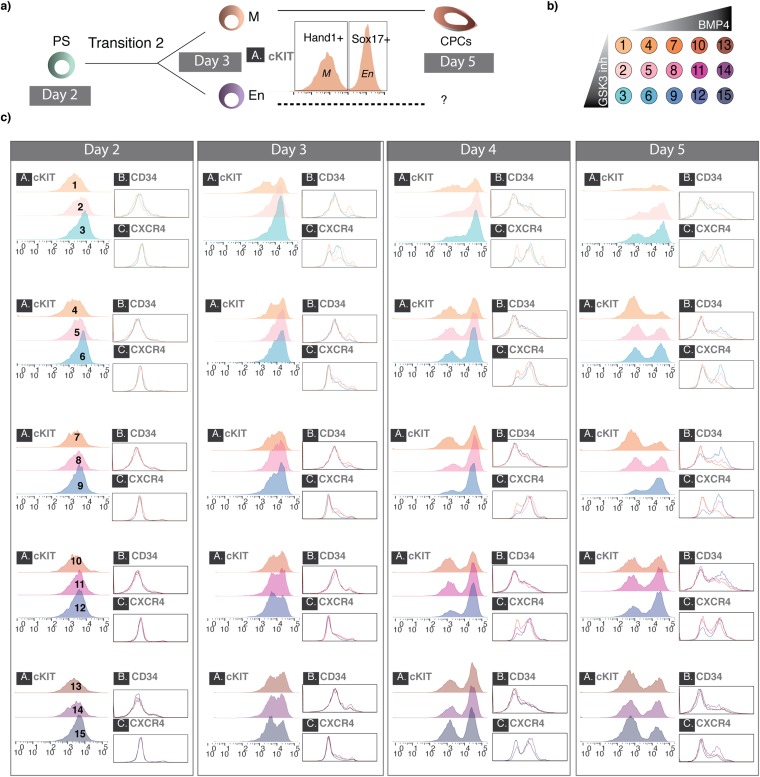
Fig. S8.
Cell population dynamics during early iCM differentiation based on individual protein markers. (A) A model of M–En bifurcation after the PS-like state on day 2 helps to navigate the cell population dynamics (i.e., bimodality of certain markers after day 3). Cell state abbreviations: CPC, cardiac progenitor cell; En, endoderm; M, mesoderm; PS, primitive streak. (B) Representation of the perturbation matrix used to explore the effect of exogenous cues during early cardiomyocyte differentiation. Each color represents a unique BMP4–GSK3 inhibitor combination (GSK3 inhibitor: 0, 0.5, and 1 μM; BMP4, 0, 1.25, 2.5, 5, and 10 ng/mL). In total, there are 15 combinations, and each combination is assigned to a number (1–15) that is used in C. (C) Individual columns represent days during differentiation postinduction with Activin. Each panel in the columns represents three concentrations of GSK3 inhibitor for each BMP4 concentration. FACS plots are marked as in B for each BMP4–GSK3 inhibitor combination and represent expression for (A) cKIT, (B) CD34, and (C) CXCR4.
Because the distribution of cKIT marks the priming at the M–En branch point, we also profiled the abundance of the same 96 transcripts in cKITHigh, cKITMedium, and cKITLow population fractions (100-cell pools) in the day 2 cells for each of the tested protocol modifications to determine if the gene expression profiles also predicted the terminal iCM differentiation efficiency. Using a Random Forest classification to extract genes that are associated with low- or high-efficiency treatments, we identify a molecular signature at the tipping point that predicted whether the efficiency of cardiomyocyte differentiation at day 28 was high (>70% iCMs) or low (<70% iCMs) (Fig. 4D). Importantly, we achieved a better classification when we used the information afforded by the gene expression of the populations fractions cKITHigh-cKITMedium-cKITLow separately (concatenated gene expression vectors) compared with just the gene expression of the cKITMedium fraction (Fig. 4 D–F and Dataset S1, Table S5), corroborating the informative potential of the cKIT outliers. Finally, we investigated how the expression of important endogenous signaling molecules, including BMP4 and DKK1, a canonical Wnt pathway inhibitor that promotes the cardiac cell fate, is influenced by the levels of exogenous BMP4 and GSK3 inhibitor (Dataset S1, Table S7). We could confirm that poor cardiomyocyte differentiation efficiency correlated with low levels of noncanonical Wnt or PDGF pathway activation, implicating a role for cell–cell communication in cell fate commitment. Similar to the classification analysis, the “average cell profile,” indicated by the expression profiles of the cKITMedium population, can distinguish between poor and high efficiency. However, we could identify cases where signaling interactions with predictive power were either exclusively seen (Wnt5B) or better detected (i.e., PDGFRa) in the outlier fractions (Fig. 4F).
Discussion
Understanding the hierarchy of cell fate decisions in mammals has enormous translational applications beyond insights in the biology of embryonic development and how pluripotent cells commit to diversity of adult cell types. Although separated from the tissue context, iPSCs hold great promise for unraveling the principles of cell fate determination on a dish because of ease and potential for massively parallel studies. Such endeavor will be critical for deciphering the idiosyncratic cell lineage trajectories that may vary between individuals and are altered in iPSC lines derived from patients (1, 27). Here, we harnessed the information inherent in cell population structure afforded by single-cell resolution analysis and showed that such integrative analysis of single-cell expression data contains clues about the propensity of multipotential cells toward a particular fate, even before apparent phenotypic fate decisions.
Specifically, we have analyzed the expression of 96 genes in ∼1,900 individual cells over 6 days in a way that represents a major departure from all recent single-cell transcript studies. Current computational analysis of single-cell gene expression profiles focuses on the descriptive identification of cell clusters and differentially expressed genes. Here, we take a dynamical systems approach that considers the governing principles that generate these patterns in the first place (28). Our analysis is grounded in first principles of nonlinear dynamical systems and provides strong evidence for the notion that cell types, such as iPSCs, are stable states (attractors) in the epigenetic landscape. As a consequence, fate commitment and differentiation are quasidiscrete, involving stepwise switching between distinct stable states. Indeed, we detected a critical cell-state transition (tipping point) during early cardiac differentiation (day 2.5). As predicted by theory, the initial (PS-like cells) and final (M or En cells) states were discrete populations within the developmental trajectory from iPSC to cardiomyocyte.
At the molecular level, a cell fate transition does not only involve abrupt shifts in the transcriptional state of a cell population—the main pattern on which traditional gene expression analysis relies to identify cell fate-specific transcripts. Now, we observe that, concomitant to increased cell heterogeneity, which itself is a predicted manifestation of critical dynamics, a cell population at a tipping point is characterized by increased gene to gene correlations—suggesting a coordination between the functionally related nodes of the GRN as recently shown (11). Thus, sampling longitudinally, even before the bifurcation, can reveal the pair of genes that increases their coordination and could serve as markers for the upcoming cell fate transition. Integrating the molecular profiles and cell population structure proved a powerful analysis tool to dissect the regulatory molecules that drive the cell fate transitions: E–PS–M or En. Analyzing the gene expression activity and how it shifts between days 2, 2.5, and 3 (prior- and postbifurcation), we could identify and quantify the cells that are primed (day 2) and committed (day 2.5/3) to M or En cell fate. However, this result takes a step beyond the descriptive analysis and has enormous practical implications: we show that it is possible to exploit knowledge of dynamical trajectories of a gene regulatory circuit (HAND1-SOX17), when measured at single-cell resolution, to predict the ultimate course of a biological process. In other words, we show how we can exploit a phenomenological signature to gain mechanistic insights about a cell fate specification event. The key element of our approach is to identify the tipping points in the trajectory using single-cell transcriptomics, which reveal primed states and their phenotypic markers. This framework can be applied to study the directionality and dynamics of cell lineage trajectories for well-established or newly defined differentiation protocols.
After the desired high-dimensional cell fate decision can be reduced to a transcriptional circuit (HAND1-SOX17) and a phenotypic readout (cKIT distribution), we could study how the exogenous differentiation cues bias the destabilization of the pluripotency attractor, channeling the cells toward a specific lineage. This observation shows that the population structure of heterogeneous differentiating cells at the tipping point can serve as an early readout for the proportion of cells that will commit to a given fate. Thus, such analyses can help to predict the efficiency of protocols for directing differentiation to a desired cell type many days before cells enter terminal differentiation (predict percentage of cardiac cells at day 2). These predictions have an enormous potential for translational applications, because iPSCs are being used to study diseases with developmental components, such as neurodegeneration. Thus, the most concrete utility of our approach is the optimization of directed differentiation protocols for patient-specific iPSC lines, which we show herein can be evaluated and optimized through a high-throughput screening procedure.
To conclude, the dynamics of cell population structure with respect to high-dimensional gene expression states are an important “biological observable.” Our study shows that single-cell resolution analysis in combination with dynamical systems theory is an invaluable tool for predicting the trajectory of cellular and tissue responses and potentially, predicting impending transitions between health and disease (10).
Materials and Methods
MHF2 (GM05387; Coriell Institute) cells were dedifferentiated to iPSCs using episomal iPSC reprogramming vectors (A14703; Thermo Fisher) according to the manufacturer’s instructions. We confirmed pluripotency and maintained the cells in feeder-free culture conditions. The iPSCs were differentiated in monolayer as described by Palpant et al. (18). For BMP4 and CHIR-99021 gradient experiment, we created a gradient of BMP4 and CHIR-99021, resulting in 15 different combinations used for culturing. All antibodies used for flow cytometry on BD FACSAria II are shown in Dataset S1, Table S6. We used Biomark (Fluidigm) to platform single-cell quantitative RT-PCR according to the manufacturer’s guidelines. The genes and primer sequences are given in Dataset S1, Table S1. The quantitative PCR (qPCR) data were processed and analyzed as described in SI Materials and Methods. Detailed methods are described in SI Materials and Methods.
SI Materials and Methods
Stem Cell Culture.
MHF2 (GM05387; Coriell Institute) cells at passage 14 were transfected with episomal iPSC reprogramming vectors (A14703; Thermo Fisher) according to the manufacturer’s instructions. These episomal vectors contain six reprogramming factors (Oct4, Sox2, Nanog, Lin28, Klf4, and lMyc). The transfected cells were cultured at different densities for 28 d, and several clones were picked for subculturing on mouse embryonic fibroblasts (MEFs) in standard stem cell culturing conditions. We randomly selected one of the clones that confirmed the alkaline phosphatase staining, exhibited stable stem cell morphology, and positively stained for Tra1-60 and SSEA-4 as measured by flow cytometry (Dataset S1, Table S6). Before differentiation, cells were acclimated to feeder-free conditions by growing them on Matrigel (1:30)-coated plates and in MEF-conditioned media for at least five passages. From here on, the iPSC line was maintained in MEF conditioned media (referred to here as conditioned media) unless otherwise mentioned.
Cardiac Differentiation.
Cells were differentiated in monolayer as described by Palpant et al. (18). Briefly, undifferentiated cells were plated as single cells (1.6 × 105 cells per 1 cm2) on Matrigel-coated 24-well plates. At ∼90–95% confluency, the conditioned media were supplemented with CHIR-99021 (1 μM; S2924; Selleck Chemicals) for 24 h (referred to as day −1). On day 0, the culture was shifted to normoxic conditions after the spent media were replaced with RPMI containing B27 without insulin (referred to here as differentiation media) supplemented with activin A (100 ng/mL; 338-AC-050; R&D SYSTEMS) and 1:30 Matrigel, creating a Matrigel sandwich. After 18 h, the media were replaced with fresh differentiation medium supplemented with BMP4 (5 ng/mL; 314-BP-050; R&D SYSTEMS) and CHIR-99021 (1 μM). On day 3, the media were replaced with fresh differentiation media containing XAV-939 (1 μM; S1180; Selleck Chemicals). On day 5, the media were replaced with differentiation media without supplements. After day 5, media were changed every 2 d with RPMI containing B27 with insulin medium (referred to here as maintenance medium). Beating cells were usually observed on days 9–11 after the initial induction with activin A.
BMP4 and CHIR-99021 Gradient Experiment.
We seeded cells for differentiation, and activin A was given as described above. On day 1, we created a gradient of BMP4 (0, 1.25, 2.5, 5, and 10 ng/mL) as well as three different concentrations for CHIR-99021 (0, 0.5, and 1 μM), resulting in 15 different combinations of BMP4 and CHIR-99021 concentrations used for culturing. Cells were treated as described above on and after day 3.
Flow Cytometry.
All antibodies used in this study are shown in Dataset S1, Table S6. Cells were washed once with PBS and trypsinized at 37 °C for up to 5 min to get a single-cell suspension. Live cells were stained in Iscove's Modified Dulbecco's Medium (IMDM) with 5% (vol/vol) FBS with the corresponding fluorophore-conjugated antibodies. Cells were prepared for flow cytometry by washing in PBS with 5% (vol/vol) FBS three times. For intracellular staining of cardiac troponin T, trypsinized cells were first fixed by resuspending the cells in 4% (wt/vol) paraformaldehyde for 10 min at room temperature. Cells were then washed and stained in 0.75% saponin for permeabilization. Cells were analyzed on the BD FACSAria II (Becton Dickinson) with FACSDiva software (BD Biosciences) according to the manufacturer’s guidelines, and data analysis was performed using FlowJo (v.10.0.8). Single cells and their representative 100-cell populations were sorted from the same subpopulations (i.e., gates) according to the manufacturer’s instructions. For cKITHigh, cKITMedium, and cKITLow fractions, we created gates that captured the top 5–8% of high cKIT-expressing cells, 48–52% around the mean cKIT expression, and 5–8% of low cKIT-expressing cells, respectively. Unstained cells were used as a control in all of the experiments to create negative gates, and appropriate compensation was performed for all multiantibody stains.
Single-Cell Quantitative RT-PCR.
For 76 human transcripts, we procured DELTAgene assays (Fluidigm) that were used for both gene-specific preamplification and detection. For 20 other transcripts, we procured two sets of nested DELTAgene assays. The outer primers were used for gene-specific preamplification, and the inner primers were used for detection. The genes and primer sequences are given in Dataset S1, Table S1. We used Fluidigm’s two-step, EvaGreen-based protocol (appendix C in PN 68000088 K1) on the Biomark (Fluidigm) platform for single-cell gene expression analysis (29). Briefly, we collected single cells or cell populations (100 cells) by directly sorting them in 5 μL lysis buffer containing VILO Buffer (PN 11754–250; Invitrogen), Nonidet P-40, and SUPERase-In (AM2696; Ambion). We included two empty wells as no template controls as well as 100-cell representative populations as positive controls. We used SuperScript Enzyme Mix (PN 11754–250; Invitrogen) to perform reverse transcription. We performed gene-specific preamplification using 500 nM each primer in DELTAgene assays and 2× TaqMan PreAmp Master Mix (PN 4391128; Invitrogen). Preamplification was performed for 15 and 16 cycles for 100 and single cells, respectively. The preamplification product was cleaned up to remove any unincorporated primers by Exonuclease (M0293L; New England BioLabs) treatment. The resulting reaction was diluted fivefold to be used for qPCR using 96.96 Dynamic Array Integrated Fluidic Circuits (IFCs) and the Biomark System from Fluidigm. Preprocessing of the IFCs was performed according to the manufacturer’s instructions. The samples were mixed with 2× SsoFast EvaGreen Supermix with Low ROX (PN 172–5211; Bio-Rad Laboratories) and 20× DNA Binding Dye Sample Loading Reagent (PN 100–3738; Fluidigm). Assay mix was prepared by adding 100 μM each forward and reverse gene detection DELTAgene assays with 2× Assay Loading Reagent (PN 85000736; Fluidigm); 5 μL each sample mix and assay mix were added to the respective inlets in the IFC and transferred to the IFC controller for loading. The loaded IFC was then transferred to the Biomark for qPCR using the GE Fast 96 × 96 PCR + Melt v2.pcl protocol. The data were analyzed using Fluidigm Real-Time PCR Analysis software, and we used the “Linear Derivative” method for baseline correction and the AutoGlobal method to set the cycle threshold (Ct).
Data Analysis.
Quality control and initial data processing.
We exported the Ct values for individual chips and used the R package—SINGuLAR (Fluidigm) for quality control and data processing. We used the SINGuLAR outlier identification method to remove apoptotic cells from subsequent analyses; briefly, outlier cells did not produce Ct values above the limit of detection (LOD) threshold (here, 25) for a set of tested genes that are expressed at least in 50% of the total cell population. Of all collected cells, 105 cells (105/2,024 = 0.05%) were removed based on these criteria. Raw Ct values were transformed into Log2Ex values as follows: Log2Ex = LOD–Ct − raw Ct. Expression values below the detection level were treated as zeros.
Dimensionality reduction methods and visualization.
To reveal the population structure and visualize the cell-state space based on gene expression vectors (1,934 individual cells, 96 genes—Log2Ex values), we used three different dimensionality reduction methods: (i) principle component analysis [data transformation is linear; stats R package (30)], (ii) t-distributed stochastic neighbor-embedding [t-SNE; data transformation is nonlinear; tsne R package (31)], and (iii) diffusion maps [data transformation is nonlinear; destiny R package (32)]. We used the optimal parameters as specified for the algorithms (i.e., for diffusion maps, nearest neighbor constant k = 50, and diffusion coefficient sigma = 12). Depending on the method, the first three major components can explain up to 70% of the total variability on the data. The first component reveals features related to pluripotency vs. differentiated states, whereas the second component separates the two sister lineages (M to En cell states).
Consensus clustering.
Cell-specific gene expression data were clustered using consensus clustering [R package: ConsensusClusterPlus (33)]. Final analyses were based on 1,000 permuted matrices subsampling 70% of the cells and 80% of the genes (method: k means; tested solutions: 2–20 clusters; distance metric: 1 − Pearson’s correlation). After calculating cluster consensus and item consensus, we identified 17 robust clusters, with >80% of their members being assigned with high accuracy.
Critical transition index (Ic).
To estimate the Ic, we generated time point-specific Log2Ex matrices. For each of them (days 0, 1, 1.5, 2, 2.5, and 3/M—only mesoderm-specific cells), we generated two Pearson’s correlation matrices: (i) based on all of the gene pairs (96 × 96 correlation matrix) and (ii) based on all of the cell pairs (mxm; m = number of cells per time point). To calculate the index , we transformed the upper triangular of each matrix into a vector (unique values for each pair of comparisons), and for each vector, we estimated the absolute mean of the gene to gene correlation. To show that feature selection does not affect the calculation of the Ic, we subsampled either the gene vector (finally down to 20 genes, almost 1/5 of the initial 96 genes) or cells (finally down to the 1/10 of each cell population) for 1,000 permutations and repeated the Ic calculation (Fig. S3).
Random Forest classification.
We prepared two matrices based on the 96 Log2Ex gene expression values for each of the 100-cell pools: (i) “control” that contains only the cKITMedium values (15 schemes × 96 gene expression values) and (ii) “concatenated” that contains all cKIT fractions (15 schemes × 288 gene expression values). We filtered out genes that show no or very low (<15%) expression among samples and performed supervised Random Forest [randomForest R package (34)] for classification and to visualize regression trees. For the classification problem, we separated our 15 schemes into low- and high-efficiency classes based on their abundance of cTNNT2+ cells (schemes with >70% cTNNT2+ cells are high, and schemes with <70% cTNNT2+ cells are low). For the regression problem, we used the same percentage of cTNNT2+ cells thresholds described above. To decide if the cKITLow and cKITHigh vectors improve the classifier, we compared the output of these two runs using the R package importance, which provides a table of all of the response variables (a summary of both mean decrease in Gini and mean decrease in accuracy). The 10 more important variables based on the “concatenate” matrix are available in Dataset S1, Table S5.
Supplementary Material
Acknowledgments
We thank Mitra Mojtahedi, Danielle Yi, and Manisha Ray for advice on single-cell qPCR experiments; and Aymeric d’Herouel, Laleh Haghverdi, Carsten Marr, and Florian Buettner for advice on diffusion maps. We also thank William Longabaugh, Leah Rommereim, Lee Rowen, Cory Funk, and Gil Omenn for critically reading the manuscript and advice. We thank the Institute for Systems Biology’s Core Facilities for help with flow cytometry and single-cell qPCR and the Institute for Stem Cell & Regenerative Medicine at the University of Washington for providing access to stem cell culture facilities. This work was supported by Institute for Systems Biology-Luxembourg Center for Systems Biomedicine Strategic Partnership, National Institute of General Medical Sciences (NIGMS) Grant R01GM109964, and NIGMS National Centers for Systems Biology Grant 2P50GM076547-06A1. M.N.S. was supported, in part, by a United Negro College Fund (UNCF)–Merck Postdoctoral Science Research Fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1621412114/-/DCSupplemental.
References
- 1.Avior Y, Sagi I, Benvenisty N. Pluripotent stem cells in disease modelling and drug discovery. Nat Rev Mol Cell Biol. 2016;17(3):170–182. doi: 10.1038/nrm.2015.27. [DOI] [PubMed] [Google Scholar]
- 2.Nelson TJ, Martinez-Fernandez A, Terzic A. Induced pluripotent stem cells: Developmental biology to regenerative medicine. Nat Rev Cardiol. 2010;7(12):700–710. doi: 10.1038/nrcardio.2010.159. [DOI] [PubMed] [Google Scholar]
- 3.Scialdone A, et al. Resolving early mesoderm diversification through single-cell expression profiling. Nature. 2016;535(7611):289–293. doi: 10.1038/nature18633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loh KM, et al. Mapping the pairwise choices leading from pluripotency to human bone, heart, and other mesoderm cell types. Cell. 2016;166(2):451–467. doi: 10.1016/j.cell.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Semrau S, et al. 2016. Dynamics of lineage commitment revealed by single-cell transcriptomics of differentiating embryonic stem cells. bioRxiv:10.1101/068288.
- 6.Heath JR, Ribas A, Mischel PS. Single-cell analysis tools for drug discovery and development. Nat Rev Drug Discov. 2016;15(3):204–216. doi: 10.1038/nrd.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang S. The molecular and mathematical basis of Waddington’s epigenetic landscape: A framework for post-Darwinian biology? BioEssays. 2012;34(2):149–157. doi: 10.1002/bies.201100031. [DOI] [PubMed] [Google Scholar]
- 8.Moris N, Pina C, Arias AM. Transition states and cell fate decisions in epigenetic landscapes. Nat Rev Genet. 2016;17(11):693–703. doi: 10.1038/nrg.2016.98. [DOI] [PubMed] [Google Scholar]
- 9.Scheffer M, et al. Anticipating critical transitions. Science. 2012;338(6105):344–348. doi: 10.1126/science.1225244. [DOI] [PubMed] [Google Scholar]
- 10.Trefois C, Antony PMA, Goncalves J, Skupin A, Balling R. Critical transitions in chronic disease: Transferring concepts from ecology to systems medicine. Curr Opin Biotechnol. 2015;34:48–55. doi: 10.1016/j.copbio.2014.11.020. [DOI] [PubMed] [Google Scholar]
- 11.Mojtahedi M, et al. Cell fate decision as high-dimensional critical state transition. PLoS Biol. 2016;14(12):e2000640. doi: 10.1371/journal.pbio.2000640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou JX, Huang S. Understanding gene circuits at cell-fate branch points for rational cell reprogramming. Trends Genet. 2011;27(2):55–62. doi: 10.1016/j.tig.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: Lessons from embryonic development. Cell. 2008;132(4):661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 14.Drukker M, et al. Isolation of primitive endoderm, mesoderm, vascular endothelial and trophoblast progenitors from human pluripotent stem cells. Nat Biotechnol. 2012;30(6):531–542. doi: 10.1038/nbt.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brade T, Pane LS, Moretti A, Chien KR, Laugwitz KL. Embryonic heart progenitors and cardiogenesis. Cold Spring Harb Perspect Med. 2013;3(10):a013847. doi: 10.1101/cshperspect.a013847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mummery CL, et al. Differentiation of human embryonic stem cells and induced pluripotent stem cells to cardiomyocytes: A methods overview. Circ Res. 2012;111(3):344–358. doi: 10.1161/CIRCRESAHA.110.227512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burridge PW, Keller G, Gold JD, Wu JC. Production of de novo cardiomyocytes: Human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell. 2012;10(1):16–28. doi: 10.1016/j.stem.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palpant NJ, Hofsteen P, Pabon L, Reinecke H, Murry CE. Cardiac development in zebrafish and human embryonic stem cells is inhibited by exposure to tobacco cigarettes and e-cigarettes. PLoS One. 2015;10(5):e0126259. doi: 10.1371/journal.pone.0126259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hough SR, et al. Single-cell gene expression profiles define self-renewing, pluripotent, and lineage primed states of human pluripotent stem cells. Stem Cell Rep. 2014;2(6):881–895. doi: 10.1016/j.stemcr.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teo AKK, et al. Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev. 2011;25(3):238–250. doi: 10.1101/gad.607311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, Oron E, Nelson B, Razis S, Ivanova N. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell. 2012;10(4):440–454. doi: 10.1016/j.stem.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 22.Heinäniemi M, et al. Gene-pair expression signatures reveal lineage control. Nat Methods. 2013;10(6):577–583. doi: 10.1038/nmeth.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, et al. Sox17 is essential for the specification of cardiac mesoderm in embryonic stem cells. Proc Natl Acad Sci USA. 2007;104(10):3859–3864. doi: 10.1073/pnas.0609100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riley P, Anson-Cartwright L, Cross JC. The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nat Genet. 1998;18(3):271–275. doi: 10.1038/ng0398-271. [DOI] [PubMed] [Google Scholar]
- 25.Li Q, et al. Dynamics inside the cancer cell attractor reveal cell heterogeneity, limits of stability, and escape. Proc Natl Acad Sci USA. 2016;113(10):2672–2677. doi: 10.1073/pnas.1519210113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loh KM, et al. Efficient endoderm induction from human pluripotent stem cells by logically directing signals controlling lineage bifurcations. Cell Stem Cell. 2014;14(2):237–252. doi: 10.1016/j.stem.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J, Li S, He X-B, Cheng C, Le W. Induced pluripotent stem cells in Alzheimer’s disease: Applications for disease modeling and cell-replacement therapy. Mol Neurodegener. 2016;11(1):39. doi: 10.1186/s13024-016-0106-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marr C, Zhou JX, Huang S. Single-cell gene expression profiling and cell state dynamics: Collecting data, correlating data points and connecting the dots. Curr Opin Biotechnol. 2016;39:207–214. doi: 10.1016/j.copbio.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, et al. Methods for qPCR gene expression profiling applied to 1440 lymphoblastoid single cells. Methods. 2013;59(1):71–79. doi: 10.1016/j.ymeth.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.R Core Team 2016 R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna). Available at https://www.R-project.org/. Accessed January 19, 2017.
- 31.Van Der Maaten L, Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 32.Angerer P, et al. Destiny: Diffusion maps for large-scale single-cell data in R. Bioinformatics. 2016;32(8):1241–1243. doi: 10.1093/bioinformatics/btv715. [DOI] [PubMed] [Google Scholar]
- 33.Wilkerson MD, Hayes DN. ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liaw A, Wiener M. Classification and regression by randomForest. R News. 2002;2:18–22. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.