Significance
The immune system defends against pathogens in part via a diverse population of T cells that display different surface receptor proteins [T-cell receptors (TCRs)] designed to recognize MHC-presented foreign peptides. Receptor diversity is produced by an initial random gene recombination process, followed by selection for proteins that fold correctly and bind weakly to self-peptides. Using data from mice of different ages, from embryo to young adult, we quantify the changes with time in the way receptors are generated and selected for function. We find a strong increase in repertoire diversity, occurring shortly after birth, due to a sharp increase in the number of random nucleotide insertions in the primitive TCR gene recombination process. Differences between thymic and blood TCR sequence distributions allow us to infer subtle details of this “turning on” of the mouse immune system.
Keywords: immunology, VDJ recombination, T cells, sequencing, mouse
Abstract
The ability of the adaptive immune system to respond to arbitrary pathogens stems from the broad diversity of immune cell surface receptors. This diversity originates in a stochastic DNA editing process (VDJ recombination) that acts on the surface receptor gene each time a new immune cell is created from a stem cell. By analyzing T-cell receptor (TCR) sequence repertoires taken from the blood and thymus of mice of different ages, we quantify the changes in the VDJ recombination process that occur from embryo to young adult. We find a rapid increase with age in the number of random insertions and a dramatic increase in diversity. Because the blood accumulates thymic output over time, blood repertoires are mixtures of different statistical recombination processes, and we unravel the mixture statistics to obtain a picture of the time evolution of the early immune system. Sequence repertoire analysis also allows us to detect the statistical impact of selection on the output of the VDJ recombination process. The effects we find are nearly identical between thymus and blood, suggesting that our analysis mainly detects selection for proper folding of the TCR receptor protein. We further find that selection is weaker in laboratory mice than in humans and it does not affect the diversity of the repertoire.
The adaptive immune system relies on cell surface receptors to recognize an unpredictable array of foreign pathogens. T cells perform their surveillance function through a highly diverse repertoire of T-cell receptors (TCRs): Any individual TCR recognizes only a small subset of the foreign peptides that it may encounter, and the system defends against a broad range of pathogens by having TCRs of many different specificities. This receptor diversity is created by a stochastic DNA editing process (VDJ recombination) that acts on the TCR gene each time a new immune cell is created from a stem cell and acts independently on each of the two chains (alpha and beta) that compose the receptor.
The resulting repertoire diversity can now be studied in great detail, using high-throughput sequencing of lymphocyte receptor repertoires (1–8). In previous work, we used human sequence repertoires to develop methods for inferring the details of the stochastic process of VDJ recombination (9, 38) and for characterizing the statistical effects of thymic selection (10). In this paper, we apply these methods to T-cell -chain sequence data collected from mice at several stages of development, from embryos to young adults, to study the T-cell repertoire maturation process. We exploit the flexibility of the mouse model to study aspects of immune system development and function that are not readily accessible in humans.
It is known that B- and T-cell receptors formed in embryonic or neonatal individuals are less diverse than in adults: They have fewer nontemplated insertions (11) due to the absence of terminal deoxynucleotidyl transferase (TdT) expression, the enzyme responsible for these insertions (12–14). Whereas this observation has been confirmed by deep sequencing of human TCR repertoires (15, 16), the precise form and time-resolved dynamics of VDJ recombination have not been assessed.
We analyze mouse T cells taken simultaneously from thymus and blood in the same individual to gain insights into the dynamics of repertoire development. First, because the periphery accumulates T cells produced at different times, whereas new T cells pass quickly through the thymus and reflect conditions at a single time point, the statistical structure of the blood T-cell repertoire can be very different from that of the thymus. This difference poses a challenge for statistical inference, and in this paper we develop a method for describing such repertoires derived from a mixture of different conditions. Second, the ability to compare thymus with blood sequence repertoires allows for a more refined view of the stages of receptor selection for functionality, from initial receptor generation to eventual passage into the periphery, and leads to a qualitatively different picture for mice than previously reported for humans.
Results
Inferring the Statistics of VDJ Recombination.
A new TCR gene is created from germline DNA by a series of stochastic events: choosing gene segments, deleting bases from the ends of the chosen gene segments, and inserting nucleotides between the modified gene segments. Because the same sequence can be generated by distinct recombination events, standard tools that assign a unique recombination scenario to each sequence (17–20) give biased estimates that would limit our ability to detect the developmental changes that interest us here. In previous work (9, 10) we showed how to overcome this problem, using an approach that assigns probabilities to different ways of generating a sequence (see Materials and Methods for details). This approach allows us to accurately quantify and track diversity as a function of developmental age, in both the thymus and the periphery.
Here, we apply these methods to thymic and peripheral sequence repertoires of TCR beta-chain (TRB) genes of mice of varying ages: 17 d after conception and 4 d, 21 d, and 42 d after birth (see Materials and Methods and Table S1 for a complete summary of data). A distinct set of generation parameters was inferred for each of these repertoires. Only out-of-frame sequences, which are nonproductive and thus thought to be free of selection effects, were included in the inference.
Table S1.
Sequence datasets used in analyses reported in main text
| Age | Tissue | Read length, bp | Productive | Nonproductive |
| 17 -d embryo | Thymus 2 | 87 | 11,199 | 15,368 |
| 17 -d embryo | Thymus 3 | 87 | 9,589 | 11,485 |
| 4 d | Thymus 1 | 87 | 9,165 | 4,687 |
| 4 d | Thymus 2 | 87 | 27,794 | 14,417 |
| 4 d | Spleen 1 | 87 | 447 | 325 |
| 4 d | Spleen 2 | 87 | 213 | 112 |
| 4 d | Spleen 3 | 87 | 1,356 | 668 |
| 21 d | Thymus | 60 | 95,164 | 43,418 |
| 21 d | Blood | 60 | 14,469 | 7,510 |
| 42 d | Thymus 1 | 87 | 33,028 | 16,159 |
| 42 d | Thymus 2 | 87 | 24,292 | 10,864 |
| 42 d | Thymus 3 | 87 | 24,846 | 12,006 |
| 42 d | Thymus 1 library replicate | 87 | 137,233 | 67,232 |
| 42 d | Thymus 2 library replicate | 87 | 61,990 | 30,390 |
| 42 d | Thymus 3 library replicate | 87 | 83,591 | 40,425 |
| 42 d | Blood 1 | 87 | 16,642 | 7,550 |
| 42 d | Blood 2 | 87 | 3,256 | 1,235 |
| 42 d | Blood 3 | 87 | 16,858 | 7,306 |
Thymic Repertoires Reveal Mechanism of Diversity Maturation.
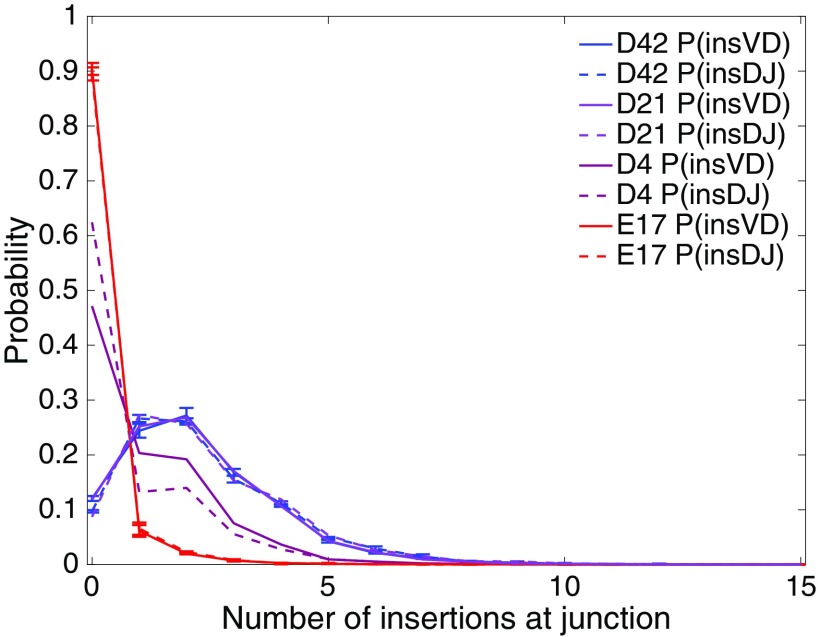
The best-documented element of VDJ recombination known to change between fetal and adult life is the number of nontemplated (N) insertions at the junctions. In Fig. 1 we plot the marginal distributions of the number of N insertions at the VD and DJ junctions inferred from out-of-frame thymic sequences of mice at a sequence of ages. During the passage from fetal to mature animal, this distribution changes dramatically and rapidly. In the embryonic mouse, 90% of the sequences have no insertions, whereas this fraction drops to 10% in adult mice. This trend is consistent with previous observations in neonates (11) and is explained by the low level of expression of TdT before birth (12, 14). TdT is turned on after birth, and Fig. 1 indicates that the asymptotic level of TdT in adults must be reached before 21 d, as there are no noticeable differences between 21 d and 42 d. The distribution at 4 d shows an intermediate situation, roughly halfway between embryonic and adult. Whereas Fig. 1 shows that the inferred distribution of insertions is identical at the VD and DJ junctions in the thymus of fetal or adult mice, it is different at 4 d. This difference could be due to the temporal ordering of VDJ recombination: DJ recombination occurs before VD recombination, with a short time delay between the two, and the rise of TdT expression during this delay could explain the increased mean number of VD insertions relative to that of DJ insertions.
Fig. 1.
Age-dependent insertion length distributions. Shown are distributions of the number of N insertions at the VD junction, , and at the DJ junction, , inferred from individual mouse thymus datasets at different ages: embryo day 17 (E17) and 4 d, 21 d, and 42 d postbirth (D4, D21, and D42). The error bars indicate the variation across individuals (Table S1).
We asked whether features of the recombination process other than the number of insertions changed between embryonic and mature mice. We found that D and J gene choice did not change significantly (, t test corrected for multiple testing), whereas a few V genes had their use significantly () increase (V4, V12-1, V26) or decrease (V14, V16, V17, V20, V22) with age (Figs. S1 and S2). The profiles of deletion showed no significant changes with age (Figs. S3 and S4) and neither did the frequencies of inserted N nucleotides (Fig. S5). These results confirm quantitatively that an increase in the number of untemplated insertions is the primary driver of diversity expansion in early life.
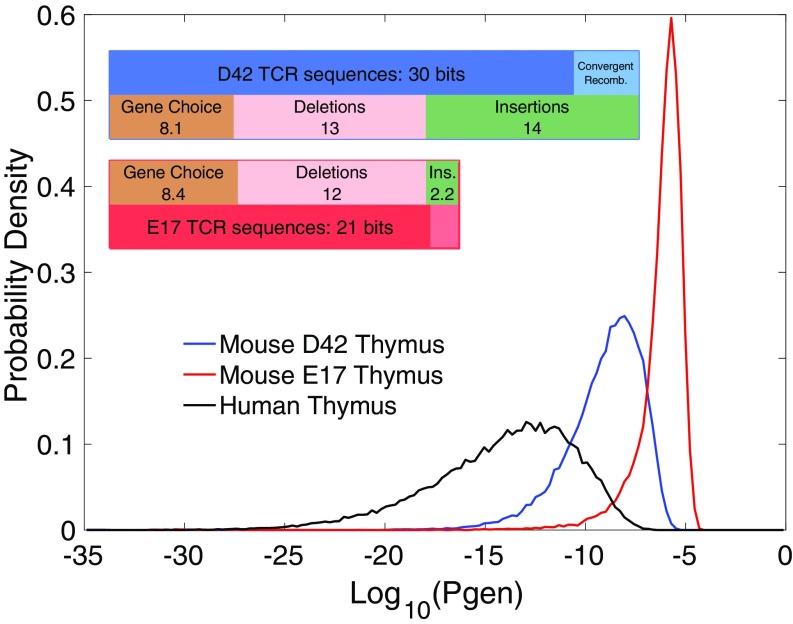
To quantify the overall change in diversity between TRB sequence repertoires at different ages, we calculated the Shannon entropy of their distributions. Entropy can be decomposed as a sum of contributions from gene choices, deletions, and insertions, from which a correction for convergent recombination must be subtracted (9). We find that the diversity of generated nucleotide sequences increased from 21 bits in fetal mice to 30 bits in adult mice. The change in repertoire diversity during this transition is almost entirely due to the change in the insertion profile, as can be seen in Fig. 2, Inset, where the different contributions to the sequence entropies of the fetal and mature sequence repertoires are compared.
Fig. 2.
Sequence entropy for thymic repertoires. Shown is distribution of the log generation probability for mouse thymic repertoires derived from 17 d embryo (E17) and 42 d postbirth animals (D42). The human generative probability distribution (9) is plotted for comparison. Shannon entropy is minus the mean over these distributions. Inset shows Shannon entropy at ages 17 d embryo and 42 d postbirth, decomposed into the components of the recombination scenario. Sequence entropy is recombination scenario entropy minus a correction for convergent recombination. Note that only the insertion component of scenario entropy changes significantly between embryonic and mature.
The entropy is mathematically equal to the negative of the mean over the sequence repertoire of the logarithm of the generation probability. A plot of the distribution of generation probabilities, , over a typical repertoire (Fig. 2) shows that the generation probability of individual sequences ranges from a few parts per million to less than 1 part in .
Peripheral Repertoires Reflect Past History of Thymic Diversity.
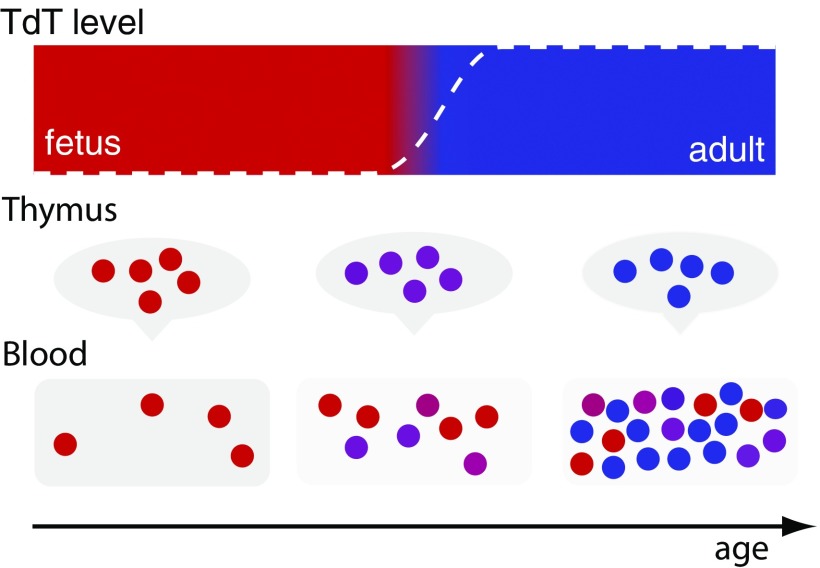
Our analysis of thymic repertoires has shown that the generative probability distribution for VDJ recombination changes dramatically in the days and weeks after birth. To understand how the evolution of VDJ recombination impacts repertoires, we need to account for the fact that peripheral compartments accumulate cells generated across earlier times, as sketched in Fig. 3. As a result, sequence repertoires must be described by a mixture of generative models with varying parameters that reflect past states of the generation process.
Fig. 3.
Age-dependent recombination affects repertoire statistics. T cells recombine according to an insertion profile that depends on a time-dependent effective TdT level (Top, dashed white line). Thymic repertoires have a unique statistical profile at any time and cells’ output to the periphery at different times are described by different values of (indicated by colors). The accumulating peripheral T-cell repertoire is described by a mixture model that accounts for the different numbers of T cells emitted at different times.
To use our inference procedure to quantify the state of mixing of repertoires, we must make some simplifying assumptions. First, given our observation that other features vary rather little with age (Figs. S1–S4), we assume that only the statistics of the untemplated insertions change with time. Second, we assume that the instantaneous insertion distribution function, , interpolates linearly between the embryonic and adult distributions by setting , where is the number of insertions at a junction, is the distribution for the 17-d embryo, and is the adult distribution at 42 d, and is an effective level of TdT measured by its impact on the number of insertions. This interpolation describes the data at day 4 (Fig. 2) accurately: The Kullback–Leibler divergence between (for the optimal choice of ) and the directly inferred distribution is bits, much less than the -bit entropy of the distribution. A more thorough validation would require data that more densely cover the early life period when the recombination machinery is changing.
The distribution describes the TRB generation process at a fixed TdT level . As explained above, repertoires in general represent the accumulated output of recombination events at earlier times and must be described by a mixture of processes at various values. The generic mixture model for insertions at the VD and DJ junctions can thus be written as
| [1] |
where is the distribution of in the repertoire reflecting the distribution of the past developmental ages at which its receptors were produced, and are its mean and variance, and . Conveniently, per the second line of Eq. 1, the mixture distribution depends only on the mean and variance of . The variance is constrained by and gives a measure of the level of mixing in the repertoire. Zero variance means no mixing; i.e., all cells were created at a single effective TdT level . Maximal variance and mixing are attained when a fraction of cells fully expresses TdT (), whereas the remaining fraction does not express TdT at all ().
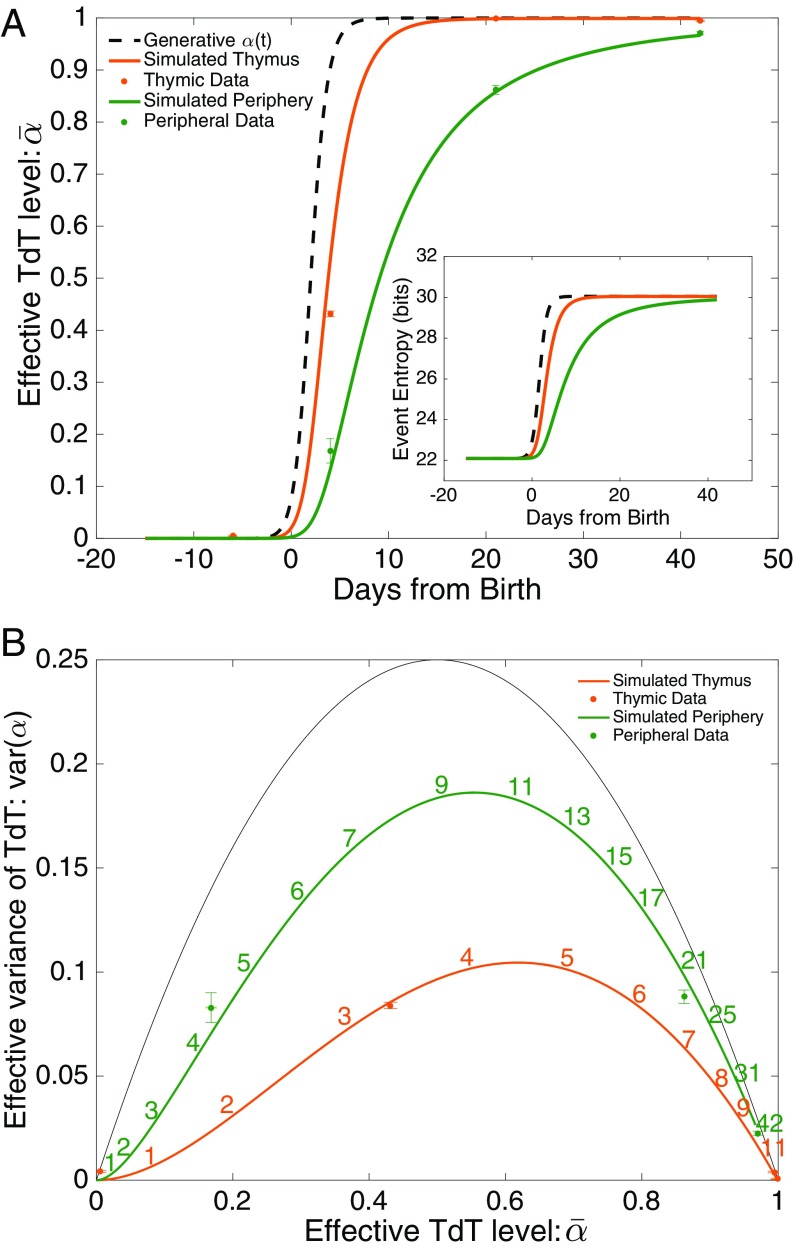
We estimate and for our datasets by first inferring the joint distribution of Eq. 2 in Materials and Methods, using our inference technique, and then adjusting and to obtain the best fit to Eq. 1. The data points in Fig. 4A show the mean effective TdT level as a function of age for thymus or blood datasets, whereas the data points in Fig. 4B report the associated values of var(). Fig. 4A shows that the blood repertoire transitions from embryonic () to mature () with a time delay relative to the thymic repertoire. This result is expected because blood T cells are first produced in the thymus. The rise in TdT level results in an increase of diversity, measured by the entropy of recombination events (Fig. 4A, Inset). Fig. 4B shows that, although embryonic and adult repertoires have no mixing, , all intermediate repertoires are mixed, with significantly larger than 0. Although the thymus does not accumulate cells, T cells do spend a finite time in the thymus and, when TdT levels are changing fast, thymic repertoires are described by mixtures. Still, blood repertoires are substantially more mixed than thymic repertoires, as expected because the thymus contains cells that have recombined over a narrow range of TdT levels , whereas blood contains cells with a greater range of ages and of values of .
Fig. 4.
Age dependence of the recombination process. The effective TdT level is estimated as an interpolation parameter between the recombination statistics of embryonic and mature animals. (A) Mean effective TdT at various ages (17 d embryo and 4 d, 21 d, and 42 d postbirth), from different tissues: thymus and periphery. Periphery is taken from blood, except for day 4 for which it is taken from spleen. The data points are compared with a minimal model of thymic entry, residence, and output, with a sharply increasing effective TdT level represented by the dashed line (Materials and Methods). A, Inset shows recombination entropy as a function of age, as predicted by the model. (B) The variance of , which measures the level of mixing in the repertoires, is shown as a function of its mean for both data (symbols) and the prediction of the minimal model (lines). The black line shows the maximal possible variance . Numbers represent age from birth in days.
The behavior of the data displayed in Fig. 4 can be better understood by comparison with a simple model (Materials and Methods and Mixture Model). In this model the effective TdT level in the thymus is given by a sharply rising Hill function (Fig. 4A, dashed curve), recombined cells are created at a rate that increases rapidly with time, and cells reside in the thymus for 3 d on average, after which they are released into the periphery. Although model parameters were chosen to reproduce the observed behavior quantitatively, we did not attempt a formal fit to the data, because of the paucity of data points. Results for and are displayed in Fig. 4 (orange and green curves). The model recapitulates the delay in maturation between thymus and blood (Fig. 4A) and also accounts for the observed level of mixing as a function of time in blood and thymus (Fig. 4B). The model curves in Fig. 4B are parametric in time (time stamps added for clarity) and it is significant that the data points lie close to points on the model curves at the right age.
Selection Shapes the In-Frame Repertoire.
Our discussion so far has focused on the evolution of the generative model for VDJ recombination, a model inferred from nonproductive, out-of-frame sequences. We now discuss what can be learned from in-frame sequences. Because they can code for functional surface receptor proteins, their statistics will be modified, relative to the generative model statistics, by selection effects. To quantify selection, we focus on the complementarity-determining region 3 (CDR3) of the beta chain, the region thought to encode most of the functional diversity of the T-cell repertoire. We define the CDR3 as the amino acid sequence running from a conserved cysteine in the V segment to a conserved phenylalanine in the J segment. We associate to each possible CDR3 amino acid sequence a selection factor , defined as the ratio of its probability of being observed in the data to its probability of having been generated. For computability, is taken to be a product of factors reflecting the selection effect of each amino acid at each position in a CDR3 of length . The collection of these subfactors defines a selection motif. The algorithms for inferring the subfactors from the data were developed in previous work on human TRB sequences (21) (see Materials and Methods for details).
We inferred selection motifs for a variety of thymic and blood repertoires (Fig. S6). These motifs are very consistent between thymus and blood of mice of the same age, with weaker consistency between mice of different ages (Fig. S7). Similarity between blood and thymus may seem surprising, as we could have expected a significant fraction of TRBs from thymic cells to have been sequenced before any selection effect, making them statistically closer to out-of-frame sequences. These observations suggest that our selection factors primarily capture selection for the ability of the coded protein to fold into a displayable receptor and may not capture more subtle effects such as negative selection against self-recognition. In Fig. S6, we also display patterns of correlation between mature mouse selection factors and quantitative amino acid biochemical properties; significant, but hard-to-interpret, patterns are apparent.
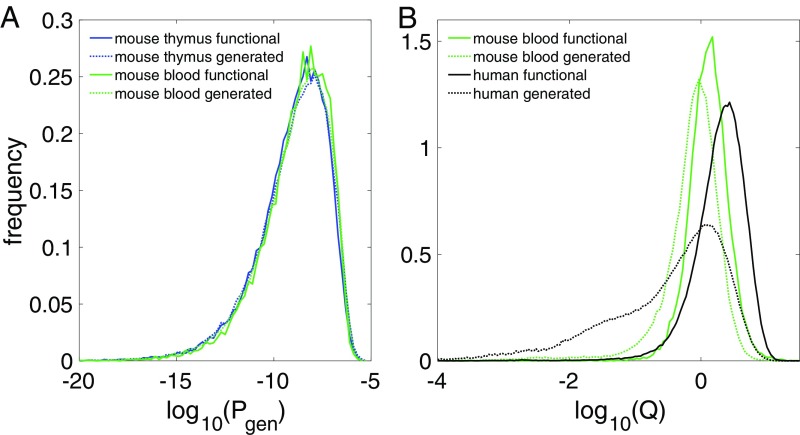
Our method attaches two hidden variables to each in-frame sequence: its probability of being generated in a VDJ recombination event and the selection factor governing its probability of then appearing in a thymic or peripheral sequence repertoire. Distributions of sequence repertoires over these variables give interesting insights into selection. We recall that, for humans, we found that the distribution of in-frame sequences was strongly skewed to higher : If a sequence was more likely to be created, it was more likely to be selected (21). Fig. 5A shows that this correlation does not hold for mice: The distribution for in-frame repertoires is virtually the same as that created by VDJ recombination. The difference between humans and mice is even more apparent in the distribution of sequence repertoires over the selection factor (Fig. 5B). For mice, selection is a weak effect: The distribution over is narrow, nearly centered about (no selection), and moves to only slightly higher values of in going from generated to selected repertoires. For humans, the primitively generated repertoire has a large fraction of sequences with a low probability of being selected (Fig. 5B). Consequently, selection purges a large fraction of sequences and substantially modifies the repertoire statistics.
Fig. 5.
Distributions with respect to generation probability and selection factor. (A) The distribution of mature mouse in-frame sequences with respect to primitive generation probability. The distributions for thymus and blood are essentially the same; the predicted distribution using Q factors on primitively generated sequences agrees perfectly with data. (B) Distribution over selection factor Q of primitively generated sequences vs. in-frame data sequences. For mice, both distributions are concentrated around ; for humans, one-third of primitively generated sequences have (i.e., are strongly selected against).
Mixture Model
In this model, T cells are introduced into the thymus with rate , with an effective TdT level . Cells leave the thymus into the periphery (blood and spleen) with constant rate . Under these assumptions, the total number of cells in the thymus, , and the periphery, , read
| [S2] |
| [S3] |
The distributions of in these compartments are given by
| [S4] |
The mean and variance of , plotted in Fig. 5 of the main text, are calculated from these expressions. They are sufficient to calculate the joint distribution of insertions at the two junctions, per Eq. 2 of the main text.
Discussion
VDJ recombination is a stochastic process that produces the initial diversity on which the adaptive immune system relies to develop a functional and diverse repertoire of receptor specificities. Previous studies have shown that this diversity is limited in neonates compared with adults, either by biasing the choice of gene segments (22–25) or by having a small number of N insertions (11, 26). Combining high-throughput sequencing with statistical analysis of murine T-cell receptor beta chains, we analyzed the dynamics of maturation of VDJ recombination. This analysis allowed us to precisely quantify, in bits, how diversity increases with age, from embryo to adult. We found that the most significant change in the recombination statistics was the number of untemplated N insertions, which sharply increases around the age of 4 d, from almost no insertions to the amount found in adults. Low numbers of insertions in neonates and during embryonic development are common to both B- and T-cell receptors (27) and are attributed to low TdT expression (12–14). Diversity can be further reduced in embryos by concentrating gene use on only a few combinations, as was shown for Ig in mice (22, 23), humans (24), and more recently zebrafish, using high-throughput sequencing (25). Similar observations were made on human TCR beta chains (28, 29). By contrast, we found only minor differences in TRB gene use between embryonic and adult mice (Fig. S1), meaning that the reduced number of N insertions is the only factor limiting diversity in the embryo relative to the adult.
One can only speculate about the biological function of the lack of N insertions in embryos and very young individuals. Rearrangements with no insertions may encode particular specificities that are effectively innate. The invariant TCRs of mucosal-associated invariant T cells (MAIT) and natural killer T cells (NKT) are specific examples of such genetically encoded receptors (30). These TCRs, which lack N insertions, are formed with high probability by VDJ recombination (31) and are further selected to be very conserved. Receptors lacking N insertions may provide neonates with a minimal set of innate-like specificities, ensuring basic immunity (32), which is later completed by the full diversity of receptors endowed with N insertions.
Our analysis highlights the importance of focusing on the underlying statistical ensembles from which repertoires are drawn, rather than looking for significance in the sequences themselves. Although sequence repertoires are contingent and noisy, with little to no overlap between individuals, their statistical properties are consistent between individuals, as was already noted for humans (9, 21). Crucially, a statistical treatment is essential for tracking the precise dynamics of N insertions with age, as deterministic assignments give systematically biased estimates of these numbers (Fig. S8). In our study of the development of repertoire diversity, we avoided the confounding factor of possibly time-dependent selection by analyzing out-of-frame rearrangements.
Using an analysis tailored to the productive repertoire, we were able to assign to any CDR3 sequence a statistical measure of selection, quantifying the probability that an amino acid sequence, once generated, goes on to become a functional receptor. The inferred selection motifs were very similar in thymic and blood repertoires. This lack of difference suggests that our measure of selection is mainly sensitive to basic selection against misfolding of the encoded receptor protein (which would have the same effect on thymic and blood repertoires) and is relatively insensitive to thymic selection against self-recognizing receptors (which would be present in blood but not in thymus). Negative selection in the thymus of course exists, but previous work on statistically characterizing its nature has shown that strong effects are localized in the few residues that actually contact presented peptides (33–35). Because we are statistically characterizing selection across the more extensive CDR3 region (11 aa long on average), it is perhaps not surprising that localized negative selection effects are washed out. It would clearly be productive to incorporate such insights into our approach.
The study of mice raises interesting and puzzling questions about sequence diversity. We found that the mature mouse repertoire is 9 bits (or -fold) more diverse than the embryonic repertoire. This wide diversity of sequences is accompanied by a wide diversity of generation probabilities: In the mature repertoire, typical generation probabilities vary from more than to less than (Fig. 3). TCRs with very low generation probabilities should be private, i.e., not likely to be generated independently in two mice, whereas TCRs with the highest generation probabilities can be public, i.e., frequently found in different mice (36). The estimate of generation probabilities afforded by our model could therefore be useful for studying the origin of public TCR repertoires in mice (37).
The mature mouse repertoire is 14 bits (or -fold) less diverse than the human T-cell repertoire, owing to a lower number of N insertions (typically per junction in mice vs. in humans). Humans and mice have to deal with presumably equally complex pathogen environments, and it would be natural to expect their immune systems to have similar levels of sequence diversity. It is intriguing that this -fold difference in potential diversity closely reflects the difference in the number of T cells in the two species ( in mice vs. in humans). Another difference with humans is the timing of the transition. The number of TCR N insertions increases as early as the first semester of gestation in humans (28) and from the second semester for B-cell receptors (BCRs) (27). By contrast, our results for mice show a sharp transition soon after birth. Finally, we found that the inferred selection factors are weaker in mice than in humans. They are also not correlated with generation probability. As a result, selection does not affect the entropy of the mouse repertoire, as it does to the human repertoire (21). The reason for this stark difference is not clear, and it would be interesting to see whether wild mice, as opposed to the inbred laboratory mice we have studied, show the same effect.
Materials and Methods
Datasets.
The data used in our analyses are 87-bp (and 60-bp) nucleotide sequences covering the variable region of the rearranged mouse TRB gene. The sequences were obtained by Adaptive Biosciences, using their TRB DNA sequencing protocol (including error correction on the basis of multiple reads of each unique DNA sequence) applied to biological samples provided by two of the authors (A.J.L. and C.S.D.). The samples comprised blood, spleen, and thymus samples from mice sacrificed at four different ages: 17-d embryo and 4 d, 21 d, and 42 d postbirth (the library preparation and sequencing for day 42 thymic samples were replicated). The mice were Black 6 laboratory mice (Jackson Laboratories) raised in standard laboratory conditions. The animal care committee of the New Jersey Medical School provided approvals for the experiments carried out with mice in this publication. The numbers of unique sequences in the various datasets were a few tens of thousands on average (with a few datasets providing more than unique sequences). The sequencing of the mature (D42) thymus samples was replicated once. Detailed statistics on the datasets are provided in Table S1. The full sequence datasets are available, along with an explanatory README file, at princeton.edu/∼ccallan/MousePaper/data/.
Stochastic Model for VDJ Recombination.
Out-of-frame data sequences were used to infer the statistical ensemble of sequences produced directly by the VDJ recombination process. We assume that the probability distribution for the generative events involved in VDJ recombination of TRB has the form (9, 38)
| [2] |
where is a recombination scenario (defined by gene choice, numbers of deletions, and number and identity of insertions) and where each factor in the equation is a distribution over the possible elements of the scenario, is the distribution of choices of the three kinds of gene segments (note that a correlation between the two D genes and the two clusters of J genes is imposed by genome topology), and is the distribution of numbers of deletions from the end of a particular gene V (and likewise for D and J). A scenario includes specific N nucleotide insertions and at the VD and DJ junctions, and is the distribution of the total numbers of such insertions, whereas , etc. describes the probability of inserting particular N nucleotides. Note that Eq. 2 gives the probability of recombination scenarios, not sequences. To obtain the probability of generating a specific sequence, one must sum the expression in Eq. 2 over all of the recombination scenarios that result in that sequence. We determine the component probability distributions in Eq. 2, , , , and so forth, directly from the data using the principle of maximum likelihood. The likelihood of a whole dataset is given by the product, over all of the unique out-of-frame sequences in the dataset, of the generation probabilities of those sequences according to the model. In practice, likelihood maximization is performed using an expectation–maximization algorithm, as explained in ref. 9.
The main assumption underlying Eq. 2 is its simple product structure, reflecting the independence of the enzymes that carry out different steps of the process. Another assumption is that the probability of inserting a given N nucleotide depends only on the identity of the nucleotide that precedes it (Markov assumption). We self-consistently checked the validity of these assumptions by verifying a posteriori that almost no unaccounted correlations between the recombination events were left in the data that were not explicitly assumed (Validation of the Structure of the Sequence Generation Model and Fig. S9) and by showing that the statistics of triplets of N insertions were well predicted by the Markov model (Fig. S5). We also compared our probabilistically inferred distributions of recombination scenario variables with distributions assembled from assignments made by a standard VDJ alignment software package (17). We found that these nonprobabilistic alignment methods greatly overestimate the fraction of sequences with no N nucleotide insertions and significantly violate the D-J pairing rule imposed by genome topology, whereas the probabilistic method does not (Fig. S8). This discrepancy is what motivates our use of a probabilistic approach. The inferred model features were very reproducible across individuals of the same age (Figs. S2 and S4).
Selection Model.
Following previous work on human TRB sequences (21), we associate to each possible CDR3 amino acid sequence of the TRB repertoire a selection factor defined as the ratio between the probability of generation of in VDJ recombination and its probability of occurrence in unique in-frame data sequences. The selection factor is assumed to be a product of subfactors related to the V and J gene choice (), CDR3 length (), and amino acid identity at each position of the CDR3 ():
| [3] |
The are normalized such that their sum over amino acids at each and is unity and Z enforces an overall normalization. The set of all these subfactors defines a motif of selection across all possible TRB sequences, and a likelihood maximization procedure allows us to infer the best selection factors from the data.
To test the consistency of the selection model, Eq. 3, we also learned a more general model where, instead of taking the to depend on amino acid identity, they were functions of the 62 codons. Using the same inference procedure, we found that selection factors for degenerate codons for the same amino acid were consistent (Fig. S10). This agreement justifies the assumption that selection depends only on amino acid composition.
Code Availability.
The Matlab software for implementing the inference procedures is available at princeton.edu/∼ccallan/MousePaper/software/. The results of the inference, along with instructions on how to use these files to recreate the figures in this paper, are available at princeton.edu/∼ccallan/MousePaper/results/.
Model of Repertoire Maturation.
TCRs are produced in the thymus with a time-dependent effective TdT level , with a production rate (arbitrary units). Time is in days, with birth at , , and . Cells reside in the thymus for an average of 3 d (exponentially distributed time), after which they are released into the periphery. The simulation is followed from (early embryo) to (age of oldest dataset).
Validation of the Structure of the Sequence Generation Model
Our inference procedure rests on a presumption of independence of the various factors in the generative model and a verification of that independence is an important aspect of our analysis. To address this issue, we compute the mutual information—a nonparametric measure of dependence between random variables—between the various recombination scenario variables and compare these numbers between the data and the generative model inferred from the same data.
The mutual information between two random variables and jointly distributed according to is defined as
| [S1] |
The mutual information between the variables defining recombination scenarios, as computed from the inferred model itself, can be calculated exactly and is shown in the below-diagonal halves of the matrices of Fig. S9. By construction, the generative model has zero mutual information between certain variable pairs, e.g., the number of VD insertions and the choice of J gene, and nonzero mutual information between variables that correlate with each other either directly or indirectly, for example, between D and J gene choice, or between choice and number of deletions.
On the other hand, the inference procedure assigns multiple scenarios, each with its own probability, to each data sequence. For any pair of scenario variables (e.g., insVD and delJ) we can use these assignments over all of the data sequences to populate a list of pairs of values, weighted by scenario probabilities. From this list we can then compute the mutual information between the two elements of the pair, using the Treves–Panzeri correction (39) to account for small sample sizes. The mutual information computed in this fashion is displayed in the above-diagonal halves of the matrices of Fig. S9. The model form is considered accurate if the obtained mutual information agrees with that predicted by the model, i.e., if the matrices of Fig. S9 are symmetric.
Selection Model
The selection factor is defined as the fold change between the probability of generation of a TRB sequence , , and its probability among productive in-frame sequences, :
| [S5] |
We assume that depends on only through the amino acid translation of its CDR3 and that it takes the factorized form
| [S6] |
where is the amino acid sequence of the CDR3, and is its length. The factors are length-specific factors, whereas are composition-specific factors. All factors are simultaneously inferred by maximizing the likelihood of the in-frame sequences using gradient ascent, as explained in detail in ref. 21. Fig. S6 shows the values of inferred from D42 mouse thymic sequences, and Fig. S7 compares the values of inferred from different datasets. The selection factors were normalized for these figures such that indicates a positive contribution to the overall selection, whereas a value below indicates a negative contribution.
Supplementary Material
Acknowledgments
The work of Y.E., T.M., and A.M.W. was supported in part by European Research Council Starting Grant 306312. The work of Y.E. was supported in part by The V Foundation for Cancer Research Grant D2015-032. The work of C.G.C. and Z.S. was supported in part by National Science Foundation Grant PHY-1305525.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700241114/-/DCSupplemental.
References
- 1.Weinstein JA, Jiang N, White RA, Fisher DS, Quake SR. High-throughput sequencing of the zebrafish antibody repertoire. Science. 2009;324:807–810. doi: 10.1126/science.1170020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyd SD, et al. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel {VDJ} pyrosequencing. Sci Transl Med. 2009;1:12ra23. doi: 10.1126/scitranslmed.3000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robins HS, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114:4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benichou J, Ben-Hamo R, Louzoun Y, Efroni S. Rep-Seq: Uncovering the immunological repertoire through next-generation sequencing. Immunology. 2012;135(3):183–191. doi: 10.1111/j.1365-2567.2011.03527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baum PD, Venturi V, Price DA. Wrestling with the repertoire: The promise and perils of next generation sequencing for antigen receptors. Eur J Immunol. 2012;42(11):2834–2839. doi: 10.1002/eji.201242999. [DOI] [PubMed] [Google Scholar]
- 6.Six A, et al. The past, present and future of immune repertoire biology - the rise of next-generation repertoire analysis. Front Immunol. 2013;4:413. doi: 10.3389/fimmu.2013.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Georgiou G, et al. The promise and challenge of high-throughput sequencing of the antibody repertoire. Nat Biotechnol. 2014;32:158–168. doi: 10.1038/nbt.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calis JJ, Rosenberg BR. Characterizing immune repertoires by high throughput sequencing: Strategies and applications. Trends Immunol. 2014;35(12):581–590. doi: 10.1016/j.it.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murugan A, Mora T, Walczak AM, Callan CG. Statistical inference of the generation probability of T-cell receptors from sequence repertoires. Proc Natl Acad Sci USA. 2012;109:16161–16166. doi: 10.1073/pnas.1212755109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elhanati Y, Marcou Q, Mora T, Walczak AM. repgenHMM: A dynamic programming tool to infer the rules of immune receptor generation from sequence data. Bioinformatics. 2016;32(13):1943–1951. doi: 10.1093/bioinformatics/btw112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feeney AJ. Lack of N regions in fetal and neonatal mouse immunoglobulin V-D-J junctional sequences. J Exp Med. 1990;172:1377–1390. doi: 10.1084/jem.172.5.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bogue M, Gilfillan S, Benoist C, Mathis D. Regulation of N-region diversity in antigen receptors through thymocyte differentiation and thymus ontogeny. Proc Natl Acad Sci USA. 1992;89:11011–11015. doi: 10.1073/pnas.89.22.11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komori T, Okada A, Stewart V, Alt FW. Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science. 1993;261:1171–1175. doi: 10.1126/science.8356451. [DOI] [PubMed] [Google Scholar]
- 14.Gilfillan S, Dierich A, Lemeur M, Benoist C, Mathis D. Mice lacking TdT: Mature animals with an immature lymphocyte repertoire. Science. 1993;261:1175–1178. doi: 10.1126/science.8356452. [DOI] [PubMed] [Google Scholar]
- 15.Britanova OV, et al. Dynamics of individual T cell repertoires: From cord blood to centenarians. J Immunol. 2016;196(12):5005–5013. doi: 10.4049/jimmunol.1600005. [DOI] [PubMed] [Google Scholar]
- 16.Pogorelyy MV, et al. 2016. Persisting fetal clonotypes influence the structure and overlap of adult human T cell receptor repertoires. arXiv qbio:1–21.
- 17.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: The highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas N, Heather J, Ndifon W, Shawe-Taylor J, Chain B. Decombinator: A tool for fast, efficient gene assignment in T-cell receptor sequences using a finite state machine. Bioinformatics. 2013;29:542–550. doi: 10.1093/bioinformatics/btt004. [DOI] [PubMed] [Google Scholar]
- 19.Bolotin DA, et al. MiXCR: Software for comprehensive adaptive immunity profiling. Nat Methods. 2015;12:380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- 20.Yu Y, Ceredig R, Seoighe C. LymAnalyzer: A tool for comprehensive analysis of next generation sequencing data of T cell receptors and immunoglobulins. Nucleic Acids Res. 2015;44(4):e31. doi: 10.1093/nar/gkv1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elhanati Y, Murugan A, Callan CG, Mora T, Walczak AM. Quantifying selection in immune receptor repertoires. Proc Natl Acad Sci USA. 2014;111:9875–9880. doi: 10.1073/pnas.1409572111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yancopoulos GD, et al. Preferential utilization of the most JH-proximal VH gene segments in pre-B-cell lines. Nature. 1984;311:727–733. doi: 10.1038/311727a0. [DOI] [PubMed] [Google Scholar]
- 23.Perlmutter RM, Kearney JF, Chang SP, Hood LE. Developmentally controlled expression of immunoglobulin VH genes. Science. 1985;227:1597–1601. doi: 10.1126/science.3975629. [DOI] [PubMed] [Google Scholar]
- 24.Schroeder HW, et al. Physical linkage of a human immunoglobulin heavy chain variable region gene segment to diversity and joining region elements. Proc Natl Acad Sci USA. 1988;85:8196–8200. doi: 10.1073/pnas.85.21.8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang N, et al. Determinism and stochasticity during maturation of the zebrafish antibody repertoire. Proc Natl Acad Sci USA. 2011;108:5348–5353. doi: 10.1073/pnas.1014277108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feeney AJ. Junctional sequences of fetal T cell receptor beta chains have few N regions. J Exp Med. 1991;174:115–124. doi: 10.1084/jem.174.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schroeder HW, Zhang L, Philips JB. Slow, programmed maturation of the immunoglobulin HCDR3 repertoire during the third trimester of fetal life. Blood. 2001;98:2745–2751. doi: 10.1182/blood.v98.9.2745. [DOI] [PubMed] [Google Scholar]
- 28.George JF, Schroeder HW. Developmental regulation of D beta reading frame and junctional diversity in T cell receptor-beta transcripts from human thymus. J Immunol. 1992;148:1230–1239. [PubMed] [Google Scholar]
- 29.Raaphorst FM, Kaijzel EL, van Tol MJ, Vossen JM, van den Elsen PJ. Non-random employment of V beta 6 and J beta gene elements and conserved amino acid usage profiles in CDR3 regions of human fetal and adult TCR beta chain rearrangements. Int Immunol. 1994;6:1–9. doi: 10.1093/intimm/6.1.1. [DOI] [PubMed] [Google Scholar]
- 30.Le Bourhis L, et al. Mucosal-associated invariant T cells: Unconventional development and function. Trends Immunol. 2011;32:212–218. doi: 10.1016/j.it.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 31.Greenaway HY, et al. NKT and MAIT invariant TCR sequences can be produced efficiently by VJ gene recombination. Immunobiology. 2013;218:213–224. doi: 10.1016/j.imbio.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 32.Gilfillan S, et al. Efficient immune responses in mice lacking N-region diversity. Eur J Immunol. 1995;25:3115–3122. doi: 10.1002/eji.1830251119. [DOI] [PubMed] [Google Scholar]
- 33.Kosmrlj A, Jha AK, Huseby ES, Kardar M, Chakraborty AK. How the thymus designs antigen-specific and self-tolerant T cell receptor sequences. Proc Natl Acad Sci USA. 2008;105:16671–16676. doi: 10.1073/pnas.0808081105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kosmrlj A, et al. Effects of thymic selection of the T-cell repertoire on HLA class I-associated control of HIV infection. Nature. 2010;465:350–354. doi: 10.1038/nature08997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stadinski BD, et al. Hydrophobic CDR3 residues promote the development of self-reactive T cells. Nat Immunol. 2016;17(8):946–955. doi: 10.1038/ni.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venturi V, Price DA, Douek DC, Davenport MP. The molecular basis for public T-cell responses? Nat Rev Immunol. 2008;8:231–238. doi: 10.1038/nri2260. [DOI] [PubMed] [Google Scholar]
- 37.Madi A, et al. T-cell receptor repertoires share a restricted set of public and abundant CDR3 sequences that are associated with self-related immunity. Genome Res. 2014;24:1603–1612. doi: 10.1101/gr.170753.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elhanati Y, et al. Inferring processes underlying B-cell repertoire diversity. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140243. doi: 10.1098/rstb.2014.0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Treves A, Panzeri S. The upward bias in measures of information derived from limited data samples. Neural Comput. 1995;7(2):399–407. [Google Scholar]
- 40.Monod MY, Giudicelli V, Chaume D, Lefranc MP. IMGT/JunctionAnalysis: The first tool for the analysis of the immunoglobulin and T cell receptor complex V-J and V-D-J JUNCTIONs. Bioinformatics. 2004;20:i379–i385. doi: 10.1093/bioinformatics/bth945. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.