The regulation of seed germination by dormancy is controlled by the degradation of specific subsets of mRNA during imbibition through the involvement of a 5′ to 3′ decay machinery.
Abstract
The regulation of plant gene expression, necessary for development and adaptive responses, relies not only on RNA transcription but also on messenger RNA (mRNA) fate. To understand whether seed germination relies on the degradation of specific subsets of mRNA, we investigated whether the 5′ to 3′ RNA decay machinery participated in the regulation of this process. Arabidopsis (Arabidopsis thaliana) seeds of exoribonuclease4 (xrn4) and varicose (vcs) mutants displayed distinct dormancy phenotypes. Transcriptome analysis of xrn4-5 and vcs-8 mutant seeds allowed us to identify genes that are likely to play a role in the control of germination. Study of 5′ untranslated region features of these transcripts revealed that specific motifs, secondary energy, and GC content could play a role in their degradation by XRN4 and VCS, and Gene Ontology clustering revealed novel actors of seed dormancy and germination. Several specific transcripts identified as being putative targets of XRN4 and VCS in seeds (PECTIN LYASE-LIKE, ASPARTYL PROTEASE, DWD-HYPERSENSITIVE-TO-ABA3, and YELLOW STRIPE-LIKE5) were further studied by reverse genetics, and their functional roles in the germination process were confirmed by mutant analysis. These findings suggest that completion of germination and its regulation by dormancy also depend on the degradation of specific subsets of mRNA.
The presence of mRNAs in dried orthodox seeds was discovered 50 years ago (Dure and Waters, 1965), and since this pioneering work, they have been detected in all seed species studied so far (Nakabayashi et al., 2005; Kimura and Nambara, 2010; Bazin et al., 2011). They accumulate during seed development and are called stored mRNA or long-lived mRNA because they retain their function as long as the seed remains alive. Although they were initially thought to contribute to protein synthesis during the very early stages of seed germination (Dure and Waters, 1965), the absence of a requirement of active transcription for the completion of this process has shown that they can have a more important role than expected (Rajjou et al., 2004). Thus, it has been proposed that they may serve as templates for protein synthesis throughout the whole germination process and that the balance between their degradation, their sequestration by cytoplasmic granules, their association with polysomes, and the neosynthesis of novel subsets of mRNA governs germination completion and dormancy alleviation (Bazin et al., 2011, Galland et al., 2014; Basbouss-Serhal et al., 2015; Galland and Rajjou, 2015).
Seed dormancy is defined as the inability to germinate in apparently favorable conditions (Bewley, 1997) and allows timely appropriate germination, thus permitting suitable seedling development and subsequent population establishment. Seed dormancy is regulated by the environmental conditions prevailing at the time of seed dispersal and by the plant hormones abscisic acid (ABA) and GA, mainly. Major molecular regulators of seed germination have been identified using transcriptomic approaches (Holdsworth et al., 2008), which ultimately led to the buildup of integrative gene regulatory networks (Bassel et al., 2011). Gene coexpression networks have helped to identify novel molecular actors involved in transcriptome reprogramming during seed maturation (Righetti et al., 2015) and in germination (Dekkers et al., 2013). However, recent data also suggest that posttranscriptional events are likely to be more appropriate to explain the germination process. For example, it has been demonstrated that there is no apparent correlation between the transcriptome and the translatome (i.e. the polysome-associated fraction of mRNAs) in germinating seeds. Indeed, only a subset of the mRNAs stored during seed maturation or neosynthesized during seed imbibition are effectively targeted to translation, as shown in sunflower (Helianthus annuus; Layat et al., 2014) or in Arabidopsis (Arabidopsis thaliana; Basbouss-Serhal et al., 2015) seeds. In addition, Bazin et al. (2011) have shown that specific mRNAs could become oxidized during seed dormancy alleviation, thus impairing their translation during seed imbibition, although they remain present in the transcriptome. These data clearly raise the question of the molecular basis of seed germination and challenge the actual dogma of the transcriptional regulation of this process.
The stability and decay of mRNA has progressively emerged as a fundamental step in the regulation of eukaryotic gene expression. The 5′ to 3′ and 3′ to 5′ pathways of mRNA decay were first identified in yeast (Saccharomyces cerevisiae; Mitchell and Tollervey, 2000; Parker and Song, 2004), but most components of the RNA turnover machinery are conserved in multicellular eukaryotes, thus suggesting similar basic mechanisms (Chiba and Green, 2009). The first step of mRNA decay is the deadenylation process, which shortens the poly(A) tail through the activity of the CCR4-Not complex (Chen et al., 2002). The main 3′ to 5′ RNA degradation machinery of eukaryotic cells is a multisubunit complex called the exosome, found in both cytoplasm and nuclear compartments (Januszyk and Lima, 2014). In the nucleus, the exosome is involved in the processing or quality control of different types of RNA, such as mRNAs, tRNAs, ribosomal RNAs, and small nuclear RNAs (Lange et al., 2014). In the cytoplasm, the exosome functions in minor degradation pathways and the mRNA surveillance system, such as nonsense-mediated decay (Chlebowski et al., 2013). In the case of the 5′ to 3′ degradation, deadenylation is followed by decapping, which is catalyzed by the conserved eukaryotic decapping complex VARICOSE (VCS)-DCP1-DCP2 (Dunckley and Parker, 1999; She et al., 2004, 2006; Xu et al., 2006). DCP2 is the catalytic subunit, and its activity can be highly regulated (Xu et al., 2006; Ling et al., 2011). DCP1 interacts directly with DCP2 and is an essential activator of DCP2 activity (She et al., 2004; Xu et al., 2006). DCP1 phosphorylation promotes mRNA decapping, presumably through ribonucleic complex formation (Borja et al., 2011; Xu and Chua, 2012). VCS is the Arabidopsis ortholog of human Heldls/Ge1 and is required for the interaction of DCP1 with DCP2 (Fenger-Gron et al., 2005). For example, its interaction with DCP1 and DCP2 is involved in the postembryonic development in Arabidopsis (Xu et al., 2006). The activity of the decapping complex is regulated by environmental signals such as drought, which controls DCP1 phosphorylation by the MPK6 kinase (Xu and Chua, 2012). An important function of the decapping complex is to limit the accumulation of aberrant mRNAs, thereby preventing their entry into the small RNA pathway and silencing process (Martinez de Alba et al., 2015). The decapped transcripts are then digested by 5′ to 3′ exoribonucleases (Nagarajan et al., 2013). The RNA decay is performed in the cytosol by an exoribonuclease called XRN1 in yeast and mammals (PACMAN in Drosophila melanogaster), while in the nucleus, a second enzyme, called XRN2 in animals and RAT1 in yeast, is active.
In Arabidopsis, three genes have been identified as homologous to XRN2, while no sequence homology could be found for XRN1. AtXRN2 and AtXRN3 function in the nucleus, similar to yeast XRN2, while AtXRN4 is localized in the cytoplasm and is the functional homolog of yeast XRN1 (Kastenmayer and Green, 2000). XRN4 is a component of the signaling cascade for ethylene perception and is also called EIN5 (Potuschak et al., 2006; Olmedo et al., 2006). AtXRN4 degrades a limited set of polyadenylated transcripts and 3′ cleavage products of microRNA (miRNA)-mediated cleavage (Souret et al., 2004; Rymarquis et al., 2011) and is involved in the adaptation to heat stress (Merret et al., 2013). Decapping components DCP1, DCP2, and VCS and the 5′ to 3′ exoribonuclease XRN4 are present in cytoplasmic foci called processing bodies (Xu and Chua, 2011). Arabidopsis DCP5 also is considered an additional processing body component that associates with DCP1 and DCP2 and is required for mRNA decapping (Xu and Chua, 2009). Processing bodies constitute specialized cellular compartments of mRNA turnover (Fillman and Lykke-Andersen, 2005), and these dynamic complexes were first identified in yeast and human (Sheth and Parker, 2003; Cougot et al., 2004) and later in plants (Weber et al., 2008). The phenotypes of processing body mutants in Arabidopsis suggest that their function is more related to development or response to the environment than to housekeeping (Xu et al., 2006; Merret et al., 2013; Nguyen et al., 2015). The degradation of a specific mRNA is a tightly regulated process that relies on the changes of its specific interaction with mRNA-binding proteins to form ribonucleic complexes (Lee and Lykke-Andersen, 2013). It also depends on its innate characteristics, since the features of the 5′ and 3′ untranslated regions (UTRs), the cap structure at the 5′ end, the poly(A) tail at the 3′ end, and specific sequence elements located within a transcript are known to play a role in mRNA stability (Mignone et al., 2002; Geisberg et al., 2014). Additionally, miRNAs and small interfering RNAs also cause endonucleolytic cleavage, thus leading to gene silencing, a major mechanism affecting plant development (Gregory et al., 2008).
Seed germination being a developmental process that requires major timely regulated changes in the gene expression program, it is highly plausible that it could involve mRNA decay. However, the function of mRNA decay in seed germination and dormancy is largely unknown to date; therefore, we have investigated the role of 5′ to 3′ decay in this process. The objective of this work was to determine whether Arabidopsis seed germination, as modulated by dormancy, might rely on the 5′ to 3′ selective decay of transcripts. We have phenotyped the seed germination of mutants vcs and xrn4, which allows us to propose a possible role of 5′ to 3′ mRNA decay in seed germination and dormancy. The transcriptomes of seeds of xrn4-5 and vcs-8 mutants were analyzed during their imbibition at 25°C in darkness, a condition preventing the germination of dormant seeds only (Leymarie et al., 2012). This led to the identification and characterization of putative targets of XRN4 and VCS having a role in the regulation of dormancy. We conclude that the decapping complex and XRN4-mediated decay play critical roles in the regulation of Arabidopsis seed dormancy.
RESULTS
RNA Decay Mutants Have a Dormancy Phenotype
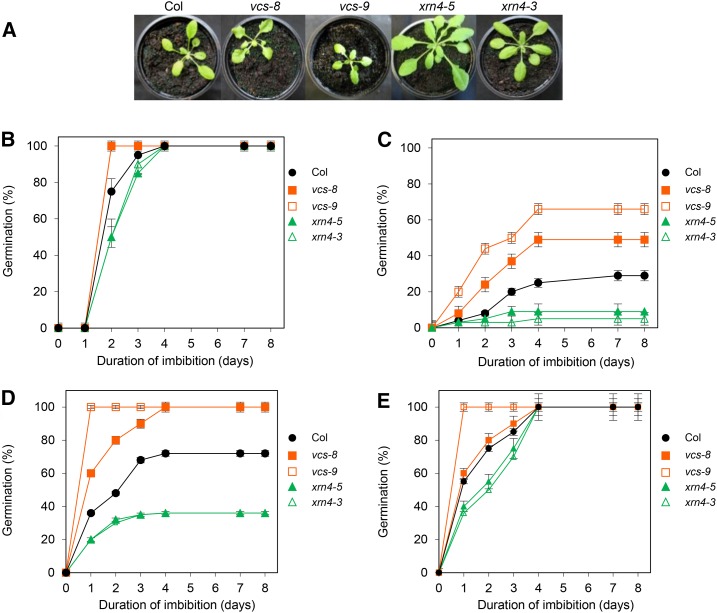
We have studied the phenotype of dormancy of seeds of mutants altered in the 5′ to 3′ mRNA decay machinery (i.e. the xrn4 [xrn4-3 and xrn4-5] and vcs [vcs-8 and vcs-9] mutants; Supplemental Table S1). Five weeks after sowing, xrn4 mutant plants presented a faster development, while vcs mutant plants were smaller when compared with wild-type plants (Fig. 1A). To confirm the phenotypes observed, we quantified some developmental traits for each genotype (Supplemental Fig. S1). The length of the floral stem of xrn4 mutant plants was similar to that of Columbia-0 (Col-0), while it was smaller in vcs mutant plants (Supplemental Fig. S1A). vcs mutant plants produced fewer seeds per silique and per plant and smaller seeds than wild-type plants (Supplemental Fig. S1, B–D). Seeds of xrn4 were more abundant per silique and per plant than in Col-0, but the mutation did not alter the seed size (Supplemental Fig. S1, B–D). Whatever the mutant, the plants produced morphologically normal seeds and in sufficient amounts for the rest of the study.
Figure 1.
Seeds of Arabidopsis mutants affected in 5′ to 3′ mRNA decay have a dormancy phenotype. A, Plant development 5 weeks after sowing. B to E, Germination of freshly harvested seeds of Col-0 (black circles), vcs-8 (closed orange squares), vcs-9 (open orange squares), xrn4-5 (closed green triangles), and xrn4-3 (open green triangles) in darkness at 15°C (B), at 25°C (C), and at 25°C after 2 weeks (D) and 3 weeks (E) of storage at 20°C and 56% relative humidity (after-ripening). Means ± sd of triplicate experiments are shown.
At harvest, Col-0 seeds were dormant, since 100% germinated at 15°C (Fig. 1B) but only 25% germinated at 25°C in darkness (Fig. 1C). After-ripening treatment (at 20°C and 56% relative humidity) increased the germination of Col-0 seeds at 25°C to 70% after 2 weeks and to 100% after 3 weeks (Fig. 1, D and E). At 15°C, all mutant lines germinated after 5 d, but xrn4 seeds germinated more slowly (Fig. 1B). Germination at 25°C revealed that xrn4 seeds were more dormant than Col-0 seeds, since only 5% germinated after 8 d, while vcs seeds were less dormant, since around 50% of seeds from this mutant germinated at this temperature (Fig. 1C). Two weeks of after-ripening were enough to totally alleviate the seed dormancy of vcs mutants, whereas only ∼40% of xrn4 seeds germinated at 25°C after this duration of dry storage (Fig. 1D). After 3 weeks of after-ripening, all genotypes became nondormant, since 100% germination was attained after 7 d at 25°C (Fig. 1E).
5′ to 3′ mRNA Decay Affects ABA/GA Metabolism and Signaling
The potential for germination of freshly harvested seeds of Col-0 and mutants was studied in the presence of the major hormonal regulators of dormancy, ABA and GA. ABA treatment (10−6 m) slowed germination at 15°C of seeds of all genotypes, since it increased by 50% to 70% the time to reach 50% germination for Col-0 and vcs and had an even stronger effect on the germination of xrn4 mutant seeds (Table I). At 25°C, GA3 (10−3 m) promoted the germination of all genotypes in a similar way. However, these treatments still discriminated the high-dormancy (xrn4) and low-dormancy (vcs) phenotypes, since seeds of vcs mutants germinated faster than seeds of xrn4 mutants (Table I).
Table I. Effects of ABA and GA3 on time to reach 50% termination (T50) of seeds of vcs-8, vcs-9, xrn4-3, and xrn4-5 mutants compared with Col-0 seeds at 15°C and 25°C in darkness.
Means ± sd of triplicate experiments are shown. T50 values are in hours. ND, Not determined (50% germination was not reached).
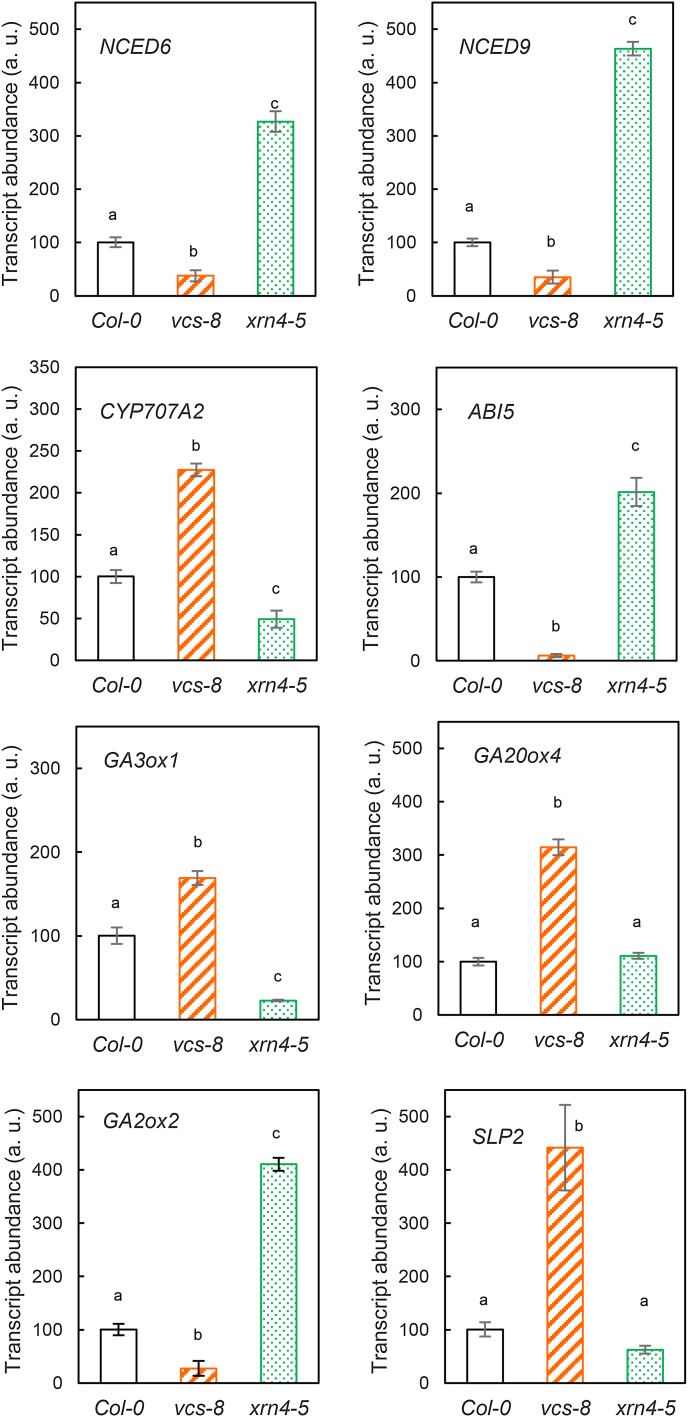
In addition, we determined the abundance of transcripts involved in ABA and GA metabolism and signaling using quantitative real-time reverse transcription (qRT)-PCR in seeds of Col-0, vcs-8 (its phenotype was roughly similar to that of vcs-9), and xrn4-5 after 24 h of imbibition in water in the dark at 25°C (i.e. before radicle protrusion; Fig. 2). We performed this study with the xrn4-5 mutant because RNA degradation data were already published for this allele (Souret et al., 2004; Rymarquis et al., 2011). In seeds of the vcs-8 mutant, the low-dormancy genotype, the abundance of transcripts related to ABA synthesis (NCED6 and NCED9), ABA signaling (ABA-INSENSITIVE5 [ABI5]), and GA catabolism (GA2ox2) was reduced compared with seeds of Col-0, while transcripts related to ABA catabolism (CYP707A2), GA activation (GA3ox1 and GA20ox4), and GA signaling (SLP2) were more abundant (Fig. 2). In contrast, in seeds of the xrn4-5 mutant, the abundance of transcripts related to ABA synthesis (NCED6 and NCED9), ABA signaling (ABI5), and GA catabolism (GA2ox2) was higher than in Col-0, whereas the abundance of transcripts of CYP707A2, related to ABA catabolism, and Ga3ox1, involved in GA activation, decreased. The abundance of SLP2 and GA20ox4 was not altered by the xrn4-5 mutation (Fig. 2).
Figure 2.
The transcript abundance of genes involved in ABA and GA pathways is affected in mRNA decay mutants. The quantification of transcript abundance is shown for NCED6, NCED9, CYP707A2, ABI5, SLP2, Ga3ox1, Ga20ox4, and Ga2ox2 in seeds of Col-0 (white bars), vcs-8 (striped bars), and xrn4-5 (dotted bars) after 24 h of imbibition at 25°C. a.u., Arbitrary unit (100 value was attributed to Col-0). Means ± sd of triplicate experiments are shown. Letters indicate homogenous groups in a corresponding class (ANOVA test and Newman-Keuls tests, P = 0.05).
Transcriptome Analysis Reveals Features of Transcripts Putatively Degraded by XRN4 and VCS during Seed Imbibition
A transcriptomic analysis was performed with seeds of vcs-8 and xrn4-5 mutants and Col-0 in order to identify the targets of XRN4 and VCS. We analyzed the transcripts in seeds after 24 h of imbibition at 25°C in the dark using CATMAv7 microarrays to compare each mutant versus the wild type. This time point was chosen based on previous work (Basbouss-Serhal et al., 2015) and because it corresponded to 75% of the amount of time required for the initiation of germination of nondormant seeds.
MA plots representing the distribution of expression ratios of all transcripts showed that major variations were included between log2 ratio values of −1.5 and +1.5 (Supplemental Fig. S2). Gene expression was considered to be different after Benjamini and Hochberg correction (P = 0.05) and for a log2 ratio above 0.5 or under −0.5, at least for one comparison. The microarray data were validated by performing qRT-PCR with 17 genes. They showed a high correlation between qRT-PCR and microarray transcript abundance, with r2 values ranging from 0.71 to 0.95 (Supplemental Fig. S3). A full list of differentially abundant transcripts in xrn4-5 and vcs-8 mutants is given in Supplemental Data Set S1; however, we focused the functional analysis on the genes that were specifically more abundant in the mutants. Indeed, transcripts being putative targets of mRNA decay mediated either by XRN4 or VCS in wild-type seeds should be more abundant in the corresponding mutants than in Col-0.
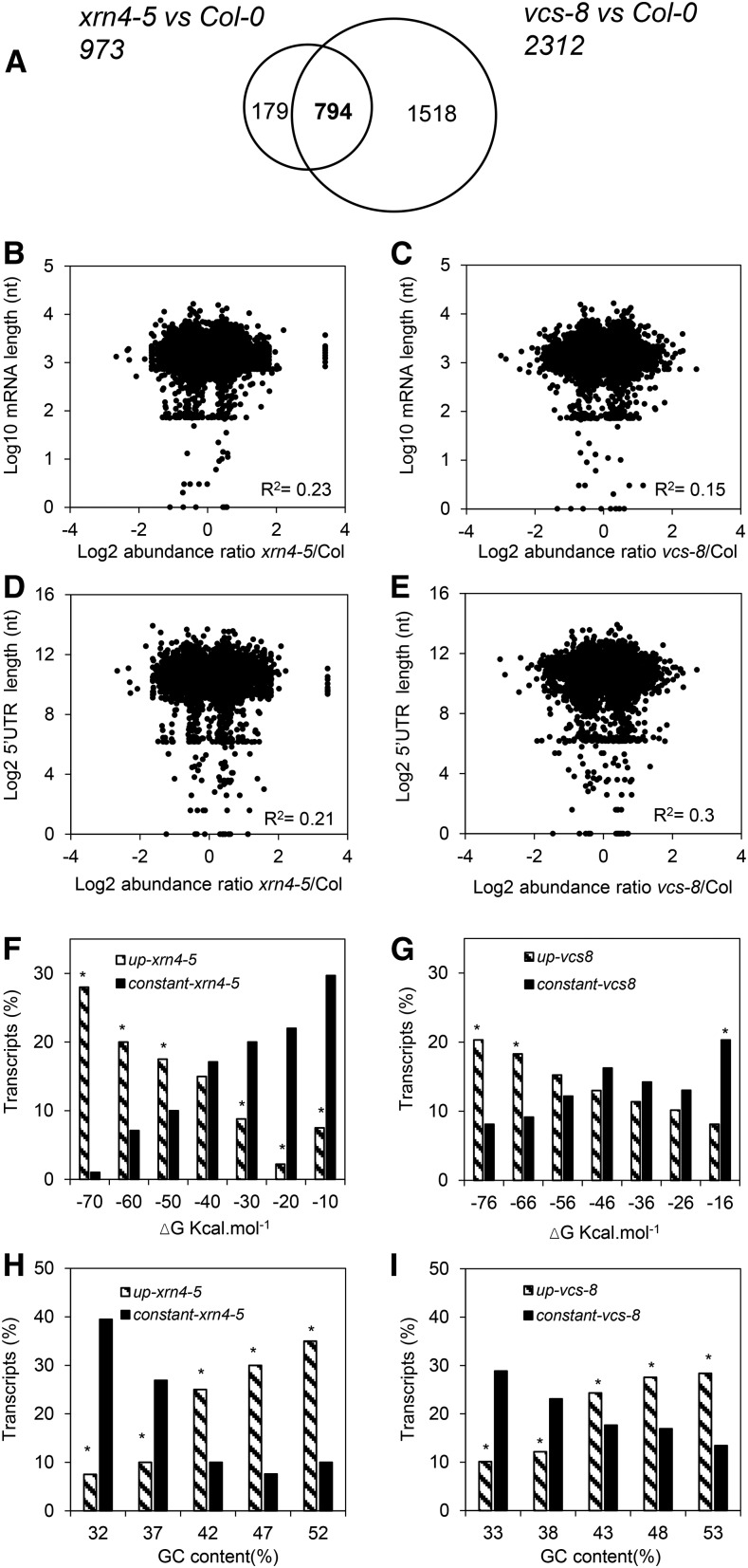
A Venn diagram (Fig. 3A) shows that 973 transcripts were more abundant in seeds of xrn4-5 and that 2,312 transcripts were more abundant in seeds of vcs-8 compared with seeds of Col-0 at 24 h of imbibition at 25°C. Among these differentially abundant transcripts, 794 transcripts were common between the two mutants, but we considered that they could not be related to dormancy since the phenotypes of the two mutants were opposite (Fig. 3A). Therefore, attention was paid to the transcripts specifically more abundant in each mutant, representing 179 transcripts in xrn4-5 (Supplemental Data Set S2) and 1,518 transcripts in vcs-8 (Supplemental Data Set S3). Genes known to be targets of some miRNAs represented 55 genes out of 179 more abundant in xrn4-5 but only 135 genes out of 1,518 up-regulated in vcs-8 (Supplemental Data Set S4).
Figure 3.
mRNA decay modifies the seed transcriptome and is related to transcript features. A, Venn diagram showing the number of transcripts displaying significant increases in abundance (after Benjamini and Hochberg correction; ratio > 0.5) in xrn4-5 and vcs-8 mutants compared with wild-type Col-0 after 24 h of imbibition at 25°C. Totals of 973 and 2,312 transcripts more abundant in xrn4-5 and vcs-8 mutants, respectively, were identified. B and C, Relationship between mRNA total length and mRNA abundance in xrn4-5 (B) and vcs-8 (C) mutants. Each point of the scatterplots represents log10 (mRNA total length) on the x axis versus the ratio of change in mRNA log2 expression (mutant to Col-0) on the y axis for transcripts more abundant in mutant seeds. nt, Nucleotides. D and E, Relationship between the length of mRNA 5′ UTRs and mRNA abundance in xrn4-5 (D) and vcs-8 (E) mutants. Each point of the scatterplots represents log2 (5′ UTR length) on the x axis versus the ratio of change in mRNA log2 expression (mutant to Col-0) for transcripts more abundant in mutants on the y axis. F and G, Distribution of transcripts specifically more abundant in xrn4-5 (F) and vcs-8 (G) mutants (striped bars) and of transcripts displaying no significant change in abundance (black bars), compared with Col-0, as a function of the minimal free energy (ΔG) of secondary structure. mRNA classes were clustered based on the free energy of their secondary structure using a 10 kcal mol−1 step. The distribution of up-accumulated or constant transcripts in xrn4-5 and vcs-8 seeds was statistically different, as determined by χ2 test (P < 0.05). H and I, Distribution of transcripts specifically more abundant in xrn4-5 (H) and vcs-8 (I) mutants (striped bars) and of transcripts displaying no significant change in abundance (black bars), compared with Col-0, as a function of their GC content. mRNAs classes were clustered based on the free energy of their secondary structure using a 5% GC content step. The distribution of up-accumulated or constant transcripts in xrn4-5 and vcs-8 seeds was statistically different, as determined by χ2 test (P < 0.05).
To determine the characteristics of the transcripts whose decay by XRN4 or VCS might play a role in dormancy, we explored the structural properties of mRNAs specifically more abundant in xrn4-5 or vcs-8 mutants. For both mutants, the total length of mRNAs and of the 5′ UTR (downloaded from The Arabidopsis Information Resource [TAIR]) of all transcripts was not correlated with their abundance (Fig. 3, B–E). Free secondary energy (ΔG) and GC content of the 5′ UTR were calculated for the transcripts specifically more abundant in each mutant but also for the transcripts whose abundance was constant between Col-0 and the mutants. To allow a valid comparison of constant and up-regulated genes, we first established a list of constant genes for each mutant (Supplemental Data Set S5). It corresponded to a range of ratios between −0.2 and +0.2 for xrn4-5 or between −0.3 and +0.3 for vcs-8. The major part of the transcripts more abundant in xrn4-5 and vcs-8 was characterized by a low ΔG and a high GC content (Fig. 3, F–I), while this was not the case for constant transcripts. We also searched motifs overrepresented in the 5′ UTR of transcripts specifically more abundant in xrn4-5 and vcs-8 mutants using TAIR and the Motif Analysis tool (Table II). In Table II, ME represents the factor of enrichment of the motif in the 5′ UTR of the transcript in comparison with its occurrence in the Arabidopsis genome and E represents the expect value corresponding to the significance of the motif. We identified 17 overrepresented motifs in xrn4-5 mutants characterized by ME values higher than 9.1 and E values lower than 2.22e−10 (Table II). For vcs-8, 10 overrepresented motifs were found with ME values between 6.1 and 11.12 and E values lower than 7.3e−3 (Table II). Three overrepresented common motifs were found in motifs of transcripts more abundant in xrn4-5 and vcs-8 mutants (Table II).
Table II. Common cis-elements identified in the 5′ UTR of the transcripts more abundant in seeds of xrn4-5 and vcs-8 mutants after 24 h of imbibition at 25°C.
Analysis was performed on subsets of 973 and 2,312 transcripts that were more abundant in xrn4-5 and vcs-8 mutants, respectively. ME represents the factor of enrichment of the motif in the 5′ UTR (up to −500 bp) of the transcript analyzed in comparison with its occurrence in the Arabidopsis genome. E represents the expect value corresponding to the significance of the motif. The motifs in boldface are common to both mutants.
| Mutant | Motif | ME | E |
|---|---|---|---|
| xrn4-5 | GCTCAG | 13.15 | 7.10e-21 |
| TTGACT | 22.14 | 8.84e-73 | |
| TGGTTA | 21.19 | 1.41e-82 | |
| CTCAGT | 18.61 | 2.74e-12 | |
| CGTCAA | 17.14 | 5.11e-22 | |
| AGCTGG | 15.16 | 2e-40 | |
| TTAGAG | 15.15 | 5.82e-51 | |
| TCTCCG | 15.41 | 2.22e-10 | |
| TGGAGC | 13.03 | 4.50e-20 | |
| CAGACG | 9.55 | 4.36e-99 | |
| GCGCTG | 9.55 | 4.36e-99 | |
| AGCGCT | 9.40 | 4.44e-63 | |
| CCGCAG | 9.34 | 3.20e-10 | |
| CTGCGG | 9.35 | 1.45e-20 | |
| CAGCTG | 9.22 | 3.20e-19 | |
| CCCCAG | 9.10 | 2.70e-20 | |
| ATGTAT | 9.10 | 4.10e-15 | |
| vcs-8 | TTTTTC | 11.12 | 2.88e-05 |
| ATTGCA | 11.12 | 2.88e-05 | |
| TGCTTG | 10.88 | 7.30e-03 | |
| CTCTCT | 10.51 | 1.28e-04 | |
| AGAGAG | 10.51 | 1.28e-04 | |
| GTCCTG | 9.58 | 2.45e-09 | |
| CAGACG | 9.21 | 2.16e-59 | |
| GCGCTG | 9.15 | 2.46e-29 | |
| AGCGCT | 9.10 | 1.34e-23 | |
| GGCCGC | 6.10 | 3.42e-12 |
Specific Targets of XRN4 and VCS Regulate Seed Germination
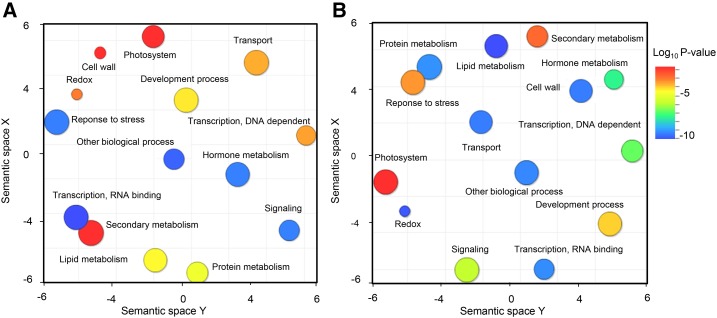
Putative functions were assigned to the transcripts more abundant in xrn4-5 and vcs-8 mutants using the Gene Ontology (GO) TAIR categorization, and they were visualized as functional clusters after redundancy reduction via REVIGO (Fig. 4). On this representation, colors indicate the user-provided P value and the size of the circles is relative to the frequency of GO terms in the underlying GO database. Interestingly, GO analysis of transcripts specifically abundant in the xrn4-5 mutant revealed that the biological functions response to stress, hormone metabolism, transcription, RNA binding, and signaling were the most represented (Fig. 4A). The transcripts more abundant in the vcs-8 mutant belonged to the lipid metabolism, protein metabolism, cell wall, transport, transcription, RNA binding, and redox categories (Fig. 4B). The more relevant GO categories are detailed in “Discussion.”
Figure 4.
mRNA decay targets molecular regulators of seed germination and dormancy. GO classification of transcripts specifically more abundant in xrn4-5 (A) and vcs-8 (B) mutants after 24 h of imbibition at 25°C was obtained using REVIGO (http://revigo.irb.hr; Supek et al., 2011), which allows a two-dimensional space representation according to the semantic similarity of GO terms (semantically similar GO terms remain close together in the plot). The size of the circles indicates the frequency of the GO term in the GO database (more general terms are represented by larger circles), and the color of the circles indicates the P value (scale at right; blue and green circles represent GO terms with more significant P values than orange and red circles).
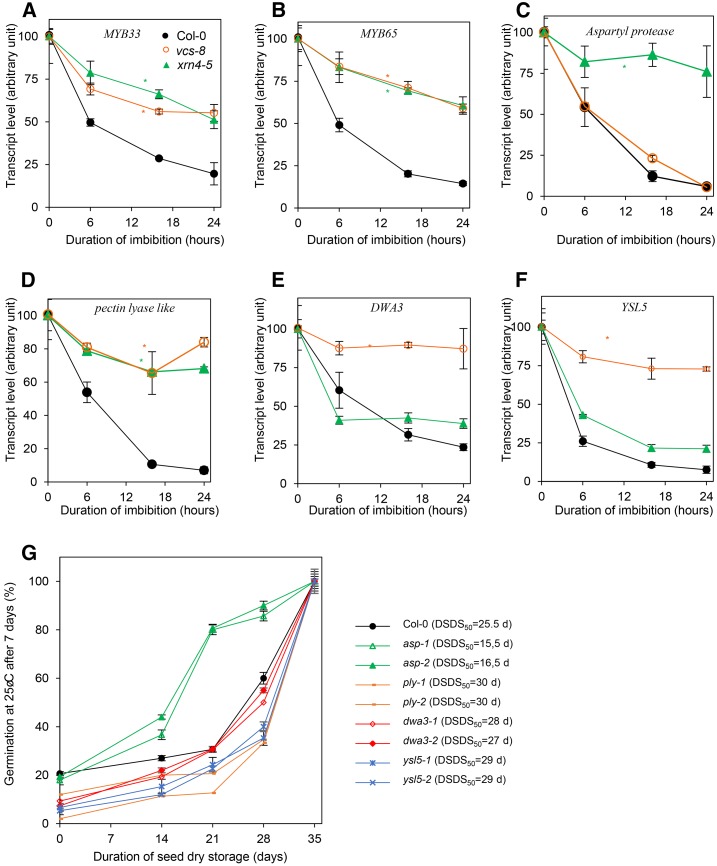
Given that XRN4 and VCS are known to be involved in the 5′ to 3′ decay process, we postulated that the transcripts up-regulated in each mutant were degraded through this pathway in the wild type, even though the participation of other decay mechanisms cannot be totally excluded. To assess that point, we followed the stability or the degradation of some of these transcripts in seeds of the wild type and the xrn4-5 and vcs-8 mutants. This was achieved by determining the abundance of these transcripts at different time points during seed imbibition in the presence of cordycepin, an inhibitor of transcription (Gutierrez et al., 2002). We chose four genes presenting high ratio changes in our transcriptomic analysis (Supplemental Data Sets S2 and S3): putative ASPARTYL PROTEASE (ASP; At1g66180), PECTIN LYASE-LIKE (PLY; At3g62110; Cao, 2012), DWD-HYPERSENSITIVE-TO-ABA3 (DWA3; At1g61210), and YELLOW STRIPE-LIKE5 (YSL5; At3g17650). As controls, we also analyzed the change in abundance of transcripts known to be targeted by XRN4 (i.e. MYB33 [At5g06100] and MYB65 [At3g114401]; Souret et al., 2004). In the Col-0 background, the level of all transcripts tested decreased during seed imbibition in the presence of cordycepin (Fig. 5). The decay of control genes (MYB33 and MYB65) depended on XRN4 but also on VCS, since their abundance was not decreased markedly in seeds of xrn4-5 and vcs-8 mutants during imbibition in the presence of cordycepin (Fig. 5, A and B). ASP degradation was XRN4 dependent, since the abundance of this transcript did not vary in seeds of xrn4-5 imbibed in the presence of cordycepin (Fig. 5C). Degradation of DWA3 and YSL5 was VCS dependent (Fig. 5, E and F), while PLY degradation depended on both genes (Fig. 5D).
Figure 5.
The 5′ to 3′ mRNA decay of specific transcripts is associated with dormancy alleviation. A to F, mRNA stability analyzed by the quantification of transcript abundance during seed imbibition with cordycepin (a transcription inhibitor) in Col-0 (black circles), vcs-8 (open orange circles), and xrn4-5 (closed green triangles) for transcripts of MYB33 (At5g06100; A), MYB65 (At3g11440; B), ASP (At1g66180; C), PLY (At3g62110; D), DWA3 (At1g61210; E), and YSL5 (At3g17650; F). Means ± sd of triplicate experiments are shown. Asterisks indicate differences between mutant and Col-0 (ANOVA and Newman-Keuls tests, P = 0.05). G, Effect of the duration of after-ripening treatment at 20°C at 56% relative humidity on subsequent germination percentage after 7 d at 25°C of Arabidopsis seeds. Col-0, asp-1 (SALK_025595), asp-2 (SALK_041567), ply-1 (SALK_017839), ply-2 (SALK_031921), dwa3-1 (SALK_092993), dwa3-2 (SALK_058391), ysl5-1 (SALK_105596), and ysl5-2 (SALK_058656) mutants were assayed during dry storage. The after-ripening requirement for each genotype was estimated by calculating the number of days of after-ripening required to reach 50% germination (DSDS50) and is indicated at right. Values are means of three biological replicates ± sd.
In order to gain functional insights on the role of mRNA decay in seed dormancy, we characterized the phenotype of dormancy alleviation of two mutant alleles corresponding to each of the previously studied genes: i.e. asp, ply, dwa3, and ysl5 (for mutant accessions, see Supplemental Table S1). The germination percentages, measured after 7 d at 25°C in the dark, of seeds from these mutants was evaluated after different durations of after-ripening (Fig. 5G). The number of days of after-ripening required to reach 50% germination is a criterion that is related to dormancy intensity. It corresponded to 25.5 d in Col-0 but only 16 d for asp. In contrast, it increased to 27.5 d for dwa3, 29 d for ysl5, and 30 d for ply mutants (Fig. 5G). This suggested that seeds from asp mutants were less dormant while seeds of the other analyzed mutants were slightly more dormant than Col-0 seeds (Fig. 5G). Since seeds of asp mutants displayed a clear phenotype of low dormancy (Fig. 5G), we investigated their responses to ABA and GA treatments (Supplemental Fig. S4). ABA (10−6 m) similarly delayed the germination of freshly harvested seeds of the wild type and asp mutants at 15°C, whereas the beneficial effect of GA3 (5 × 10−4 m) on germination at 25°C was magnified in seeds of both asp mutants (Supplemental Fig. S4).
DISCUSSION
Although mRNA decay was historically less studied than transcription, there is increasing evidence that it can play a major role in developmental and environmental responses (Belostotsky and Sieburth, 2009; Maldonado-Bonilla, 2014). The degradation of RNA could indeed be more efficient and rapid for fine-tuning gene expression than mobilizing the transcription machinery, and we hypothesized that it could play a major role in germination.
To address this hypothesis, we studied mutants affected in two major components of the 5′ to 3′ mRNA degradation pathway in eukaryotes (i.e. deadenylation-dependent decapping and nucleotic cleavage by exoribonuclease), VCS and XRN4, respectively (Chiba and Green, 2009). The effect of the mutations on plant development was assessed in order to exclude any strong phenotype that might have an indirect effect on seed germination. The effects were most important for vcs mutants (Deyholos et al., 2003), although vcs-8 and vcs-9 appeared as leaky mutant alleles compared with vcs-7 (Goeres et al., 2007). The effects of xrn4 mutations, already described elsewhere (Olmedo et al., 2006; Potuschak et al., 2006; Gregory et al., 2008), were less drastic in our growth conditions. For example, we did not observe late flowering, as mentioned by Olmedo et al. (2006). The absence of a strong pleiotropic effect of mutations on plant development and seed production pattern, therefore, allowed comparisons of germination phenotypes. As shown previously (Leymarie et al., 2012), freshly harvested Arabidopsis Col-0 seeds fully germinated at 15°C in the dark, when dormancy is not expressed, but seeds of xrn4-5 mutants germinated slowly (Fig. 1B). At 25°C, only 25% of freshly harvested Col-0 seeds germinated, confirming that they were dormant (Fig. 1C). After-ripening of Col-0 seeds for 4 weeks at 20°C and 56% relative humidity allowed dormancy alleviation, as shown by Basbouss-Serhal et al. (2016; Fig. 2E). Seeds of vcs mutants were less dormant than Col-0 seeds, whereas seeds of xrn4-5 were more dormant, as shown with freshly harvested seeds and during after-ripening (Fig. 2, C–E). These results indicate that VCS and XRN4 proteins are likely to play a role in the regulation of dormancy, which was confirmed by the effect of hormone application (ABA or GA3) during the germination of freshly harvested seeds, since it allowed us to similarly rank the mutants according to their level of dormancy (Table I). The cross talk between the dormancy phenotypes of mutants and hormones also was investigated by analyzing the transcript abundance of genes related to Arabidopsis seed dormancy in xrn4-5 and vcs-8 mutants. We focused on ABA and GA metabolism and responses because the balance of ABA and GA is the major regulator of Arabidopsis seed dormancy (Finch-Savage and Leubner-Metzger, 2006; Finkelstein et al., 2008; Nambara et al., 2010). As expected, seeds of the dormant mutant xrn4-5 displayed a higher abundance of transcripts related to dormancy (ABA anabolism and response, GA inactivation) and lower levels of genes related to germination (ABA catabolism, GA activation and response), while seeds of nondormant vcs mutants displayed the opposite pattern of gene expression (Fig. 2). Thus, the alteration of germination in 5′ to 3′ mRNA decay mutants used here is likely to be related to the canonical hormonal pathway of dormancy regulation.
We then analyzed the transcriptomes of freshly harvested seeds of xrn4-5 and vcs-8 mutants (after 24 h of imbibition at 25°C) and compared them with those of the wild-type Col-0. This work was focused on the transcripts that were more abundant in the mutants, since they are likely to be direct targets of decay in the wild type (Souret et al., 2004; Gregory et al., 2008), while transcripts less abundant in mutants would correspond to secondary effects of the mutations (Rymarquis et al., 2011). We identified 973 transcripts more abundant in the xrn4-5 mutant (i.e. putative targets of XRN4 in wild-type seeds) and 2,312 transcripts more abundant in the vcs-8 mutant (i.e. putative targets of VCS; Fig. 4A). This discrepancy in the number of transcripts has to be considered with regard to the stronger phenotype of the vcs-8 mutant for other development aspects (Fig. 1; Supplemental Fig. S1; Deyholos et al., 2003) and is in agreement with previous work, since XRN4 appears to target a limited set of transcripts in standard growth conditions (Merret et al., 2013). We identified 794 common transcripts between the two mutants, and they represent the major part of XRN4 targets (Fig. 3A), which is probably related to the fact that the exoribonuclease activity of XRN4 is directed against mRNA decapped by the decapping complex involving VCS. This overlap was not observed when comparing mRNA decay in dcp2 and xrn4 mutants, leading to the hypothesis that other exoribonucleases than XRN4 might exist (Goeres et al., 2007) and also could explain why the two-thirds of the transcripts more abundant in vcs mutants were not more abundant in xrn4.
In addition, the transcriptomic analysis of xrn4-5 and vcs-8 mutants revealed an increase of numerous miRNA targets compared with the wild type (Supplemental Data Set S4). This is particularly relevant because XRN4 and VCS have been shown to be involved in either the degradation of 3′ products of miRNA-mediated cleavage or in miRNA-guided translational repression, respectively (Souret et al., 2004; Motomura et al., 2012). The proportion of miRNA targets was particularly high for transcripts accumulated in xrn4 (55 of 79; Supplemental Data Set S4), in accordance with the contribution of XRN4 to the miRNA-mediated decay already demonstrated by Souret et al. (2004). For example, Souret et al. (2004) and Rymarquis et al. (2011) also identified the following targets of miRNA as being regulated by XRN4 in Arabidopsis: the cleavage of SCARECROW-like HAM1 (At2g45160) RNA is mediated by miR171 (Llave et al., 2002), ARF8 (At5g37020) is a target of miR167, AP2-like (At4g36920) is a target of miR172, AGO1 (At1g48410) is a target of miR168, and MYB33/65 (At5g06100 and At3g11440) is a target of miR159 (Rhoades et al., 2002; Kasschau et al., 2003). For vcs-8 mutants, we identified only 135 miRNA targets among the 1,518 up-regulated transcripts (Supplemental Data Set S4). CUC2 (At5g53950), a target of miR164 (Laufs et al., 2004), was identified previously by Motomura et al. (2012) as being not degraded in the vcs-6 mutant. The increase of miRNA targets in the exoribonuclease and decapping mutants suggests that the miRNA-dependent degradation process might play a role in seed dormancy, as suggested by Nonogaki (2014).
Our attempt to identify mRNA properties associated with the abundance of transcripts in xrn4-5 and vcs-8 mutants highlighted the role of the 5′ UTR, while the analysis of the 3′ UTR did not reveal any conclusion (data not shown). Our analysis indicates that total mRNA length and 5′ UTR length were not implicated in the selectivity of XRN4 and VCS (Fig. 4, B–E), but we show that a strong secondary structure and high GC content of the 5′ UTR made the transcripts more sensitive to degradation (Fig. 4, F–I), as reported previously in yeast (Muhlrad et al., 1995; Doma and Parker, 2006) and in mammals (Zhang et al., 2011). It already has been suggested that secondary structures or upstream open reading frames in the 5′ UTR can play a role in mRNA degradation (Oliveira and McCarthy, 1995). The GC content of the 5′ UTR and 5′ UTR length can influence mRNA structure near the 5′ cap, thus regulating posttranscriptional processes (Gu et al., 2014). In particular, the competition between the formation of the translation initiation complex and the degradation machinery at the 5′ UTR has been shown to regulate the stability of various transcripts (de la Cruz et al., 2002; Dvir et al., 2013). However, the role of the RNA structure of the 5′ UTR in RNA degradation is still poorly documented in plants. To determine the role of the 5′ UTR on mRNA decay, we identified 17 hexamer motifs overrepresented in the 5′ UTR of transcripts more abundant in xrn4-5 mutants (Table II). Nine motifs (GCTCAG, TTGACT, CTCAGT, CGTCAA, AGCTGG, TTAGAG, TCTCCG, TGGAGC, and GCGCTG) were identified previously as overrepresented in targets of XRN4 in Arabidopsis plants (Rymarquis et al., 2011). For the vcs-8 mutant, we identified 10 overrepresented motifs (Table I). Some of them (TTTTTC, ATTGCA, and TGCTTG) were identified as specific for uncapped transcripts in the Arabidopsis flower system (Jiao et al., 2008). Three motifs were common between transcripts up-regulated in xrn4-5 and vcs-8 mutants (CAGACG, GCGCTG, and AGCGCTT) and could directly interact with the decapping complex and XRN4.
With regard to the opposite dormancy phenotypes of vcs-8 and xrn4-5 seeds (Fig. 1C), VCS is likely to be involved in decapping transcripts that can positively regulate germination (1,518) or that are not related to this process (794), whereas the activity of XRN4 targets 179 transcripts involved in the maintenance of dormancy. The clusters of transcripts found specifically more abundant in xrn4-5 or vcs-8 mutants were classified according to GO functions and visualized as REVIGO scatterplots (Fig. 4). The GO category transcription, RNA binding was common to both mutants and contained many transcription factors, which demonstrates that both germination and dormancy maintenance require specific transcription, although this process is not strictly necessary for sensu stricto germination (Rajjou et al., 2004). For example, several WRKY transcription factors (Tripathi et al., 2014) were identified only in the nondormant vcs-8 mutants (Supplemental Data Set S3). WRKY60 (At2g25000) and WRKY63 (At1g66600) have already been shown as being negative regulators of ABA signaling (Liu et al., 2012; Ren et al., 2010), and WRKY22 (At4g01250) is induced significantly by hydrogen peroxide treatment (Zhou et al., 2011), which could be related to the role of reactive oxygen species (ROS) in germination or dormancy alleviation (Bailly et al., 2008; Leymarie et al., 2012). Auxin-inducible transcription factors also were more represented in vcs-8 (ARF5 [At1g19850], IAA4 [At5g43700], IAA2 [At3g23030], IAA11 [At4g28640], IAA18 [At1g51950], and AXR5 [At4g14560]; Guilfoyle and Hagen, 2007). They could be related to the growth process occurring after germination sensu stricto. Among the transcription factors regulated by XRN4 (Supplemental Data Set S2), several transcripts related to ABA signaling were identified, such as ABI5 (At2g36270; Lopez-Molina et al., 2002; Dekkers et al., 2016) or MYB33 (At5g06100; Kim et al., 2008).
Clustering analysis showed that the transcripts that were specifically more abundant in the xrn4-5 mutant (i.e. that should be degraded in wild-type seeds to permit dormancy alleviation) fell mainly into the three categories: hormone metabolism, response to stress, and signaling. In the GO category hormone metabolism, we found regulators of hormonal pathways related to dormancy (Supplemental Data Set S2). For example, the ABA response element-binding factors ABF3 and ABF4 (At4g34000 and At3g19290) are involved in ABA signaling (Muñiz Garcia et al., 2014), and At3g02480 encodes a protein of the Late Embryogenesis Abundant family that is known to be often ABA induced (Parcy et al., 1994). PHYTOCHROME-INTERACTING FACTOR6 (At3g62090) already has been suggested to play a role in Arabidopsis primary dormancy (Penfield et al., 2009). Inhibition of germination in the wild type also might be related to the presence of many transcripts of heat shock proteins, such as HSP17 (At5g12030), HSP20 (At1g52560, At1g53540, At1g59860, and At1g07400), HSP70 (At3g12580), or HSP90 (At5g52640), degraded by XRN4. The involvement of heat shock proteins in the inhibition of germination already has been shown in sunflower (Layat et al., 2014) and Arabidopsis (Basbouss-Serhal et al., 2015), suggesting that their chaperone activity can play a role in the maintenance of dormancy.
Transcripts accumulated in the vcs-8 mutant, whose degradation would prevent seed germination of the wild type, were enriched in the GO categories hormone, redox, cell wall, and transport (Fig. 4C; Supplemental Data Set S3). In the GO category hormone metabolism, we found regulators of the hormonal balance that drives the germination process (Finch-Savage and Leubner-Metzger, 2006). The ABA hydroxylase CYP707A1 (At4g19230), for example, is involved in ABA degradation and is considered a key point of the control of germination (Millar et al., 2006; Okamoto et al., 2006). The 1-aminocyclopropane-1-carboxylic acid oxidase ACO1 (At2g19590) represents a major step of ethylene synthesis (Corbineau et al., 2014), and At2g14900 is involved in the GA response. The involvement of ROS signaling in dormancy alleviation was confirmed here by the overrepresentation of the GO category redox in the vcs-8 mutant. Our data suggest that the increase in ROS content occurring during seed germination could be related to a decay in transcripts related to the enzymatic ROS scavenging, such as the monodehydroascorbate reductases MDAR1 (At3g52880) and MDAR4 (At3g27820; Bailly et al., 2004) and several thioredoxins (At3g51030, At5g39950, At2g32920, and At1g77510), a family of proteins known to regulate seed germination (Marx et al., 2003; Montrichard et al., 2003; El-Maarouf-Bouteau et al., 2013). Glutathione involvement also was suggested with the presence of the glutathione-disulfide reductase GR1 (At3g24170), GLUTATHIONE SYNTHETASE2 (At5g27380), and a glutaredoxin (At3g57070). Cell wall modifications (extensibility, degradation, and synthesis) are critical to allow cell expansion and radicle elongation (Bewley, 1997; Nonogaki et al., 2007; Wang et al., 2015; Sechet et al., 2016). Here, in seeds of the low-dormancy mutant vcs-8, transcripts of expansins such as EXPA20 (At4g38210), extensins (At1g76930 and At2g43150), several pectin lyases (At3g06770, At3g16850, At5g48900, At1g67750, and At3g62110), the galacturonosyltransferase GAUT1 (At3g61130), cellulose synthases (At5g22740, At4g07960, and At3g07330), and the Xyl transferase XGD1 (At5g33290) appeared to be more abundant than in seeds of the wild type (Supplemental Data Set S3). Cytoskeleton elements also were overrepresented in this category with tubulin (At4g20890 and At1g75780) and actin (At5g07740, At5g55400, and At1g52080). Myosins (At3g19960, At3g56480, At2g34730, At5g07890, and At2g20290) and dynein (At1g23220) accumulation could be related to the stimulation of intracellular trafficking (Diaz-Camino et al., 2005; Wang et al., 2015). The GO category transport also was overrepresented in seeds of the vcs-8 mutant, including sugar or oligopeptide transporters that may be associated with the germination process (Aluri and Büttner, 2007). For example, the peptide transporter PTR1 (At3g54140) already has been identified in germinating seeds (Dietrich et al., 2004; Stacey et al., 2006; Layat et al., 2014), such as the metal transporter YSL5 (At3g17650; Waters et al., 2006).
The functional role of VCS and XRN4 in the regulation of germination was finally confirmed. Using some selected genes, we showed that the transcripts more abundant in xrn4-5 and vcs-8 mutants were indeed targets of XRN4 and VCS by assessing their stability using the transcriptional inhibitor cordycepin. Our data confirmed the known pathways of decay of MYB33 and MYB65 by XRN4, and we showed that they also were targets of VCS. The decay of ASP was XRN4 dependent, while the degradation of PLY, DWA3, and YSL5 was VCS dependent (Fig. 5, D–F). We phenotyped seeds of mutant plants corresponding to these transcripts (two alleles for each) in order to demonstrate their role in the regulation of dormancy. While asp seeds were less dormant than those of Col-0, seeds of ply, ysl5, and dwa3 were more dormant (Fig. 5G). This suggests that ASP protein might be involved in dormancy maintenance and that the degradation of the related mRNA is made by XRN4, thus releasing dormancy. ASP is a predicted A1 family aspartyl protease that is responsive to ascorbic acid and light, but its role in plants is not yet elucidated (Gao et al., 2011). Interestingly, we show here that seeds of asp mutants were more sensitive to GA3, which suggests that ASP might be involved in the repression of GA signaling, whereas it would not interact with ABA signaling (Supplemental Fig. S4). PLY, YSL5, and DWA3, whose seeds of the mutants were more dormant, could play a role in the completion of the germination process. Enzymes involved in cell wall degradation, such as pectin lyases, have been shown to play important roles in germination (Bewley, 1997). No data are available about YSL5, but YSL1 and YSL3 are involved in iron transport at the seed level (Waters et al., 2006). DWA3 is involved in the negative regulation of ABA responses (Lee et al., 2008, 2010). Interestingly, comparison of the translatome between dormant and nondormant seeds shows that PLY, YSL5, and DWA3 transcripts were more associated with polysomes in germinating nondormant seeds after 24 h of imbibition at 25°C (Basbouss-Serhal et al., 2015). Therefore, these transcripts must be degraded to maintain dormancy.
CONCLUSION
In conclusion, we show here that the regulation of germination by dormancy in Arabidopsis seeds relies on the specific decay of positive and negative regulators of this complex process. We demonstrate the role of 5′ to 3′ mRNA degradation and the involvement of mRNA secondary structure and specific motifs in the selectivity of the decay. This work, which also allowed us to reveal several new regulators of seed dormancy and germination, opens novel fields of investigation in seed biology that should lead to other pathways of mRNA fate in seeds.
MATERIALS AND METHODS
Plant Material and Germination Assays
All mutant lines from Arabidopsis (Arabidopsis thaliana) were in the Col-0 background and are listed in Supplemental Table S1. T-DNA insertions in homozygous mutant plants were checked by PCR using two sets of primers designed on the Salk database (http://signal.salk.edu/cgi-bin/tdnaexpress). Seeds were stratified at 4°C for 2 to 4 d before sowing, and plants were grown at 20°C to 22°C under long days (16 h of light/8 h of dark). Seeds were harvested dormant and were either stored at harvest at −20°C to maintain dormancy or dormancy was alleviated by after-ripening consisting of storage in 56% relative humidity at 20°C for 3 weeks (Basbouss-Serhal et al., 2016). Germination assays were performed at 15°C or 25°C in darkness in three biological replicates of 50 seeds for each genotype in 9-cm petri dishes on a layer of cotton wool covered by a filter paper sheet soaked with water or with ABA (10−6 m) or GA3 (10−3 m). A seed was considered germinated when the radicle had protruded through the testa. Germination was scored daily for 8 d, and the results presented correspond to the mean of the germination percentages obtained for three biological replicates.
RNA Extraction
RNA was extracted from dormant seeds after 24 h of imbibition at 25°C in three biological replicates. For each extraction, 30 mg of seeds was ground in liquid nitrogen, and total RNA was extracted by a hot phenol and polyvinylpyrrolidone procedure according to Verwoerd et al. (1989) and purified with Macherey-Nagel nucleospin XS columns according to the manufacturer’s instructions. RNA extract was analyzed by a spectrophotometer (Nanovue; GE Healthcare) to determine the RNA concentration at 260 nm, and RNA quality (230–400 nm) also was checked on agarose gels.
qRT-PCR
Total RNA (1 µg) was treated with DNase I (Sigma) and then was reverse transcribed with Revertaid reverse transcriptase (Fermentas). The cDNAs obtained were checked by agarose gel electrophoresis and with PCR amplification of the actin gene (26 cycles). Amplification was then performed with real-time PCR in Mastercycler epgradients Realplex2 (Eppendorf) using 5 µL of 30× diluted cDNA solution. Real-time PCR was performed with the Maxima SYBR Green/qPCR Master Mix (Fermentas) in a 15-µL reaction volume with 0.23 µm of each primer (Eurogentec). The program for running quantitative PCR was initiated at 95°C for 10 min followed by 30 cycles at 95°C for 30 s, 56°C for 30 s, and 72°C for 30 s. As reference genes, fragments of UBIQUITIN5 and At4g12590 were amplified. The sequences of the primers used are presented in Supplemental Table S2. Critical thresholds were calculated using Realplex2 software (Eppendorf), and relative expression was calculated according to Hellemans et al. (2007). An arbitrary value of 100 was assigned to the Col-0 seeds, which were used as a control sample for normalization (Pfaffl, 2001). Results presented are means ± sd of three biological replicates. Statistical analyses were performed with StatBox 6.40 software (Grimmer Logiciel). Data were normally distributed and with equal variance, and Newman-Keuls tests (α = 0.05) were performed after ANOVA.
DNA Microarray Hybridization Studies and Analysis
Microarray analysis was carried out at the Institute of Plant Sciences Paris-Saclay using the CATMAv7 array based on Agilent technology. The CATMAv7 design of the Arabidopsis genome was made with gene annotations included in FLAGdb++, an integrative database around the plant genome (http://urgv.evry.inra.fr/FLAGdb; Dèrozier et al., 2011). The single high-density CATMAv7 microarray slide contains four chambers, each containing 149,916 primers. Each 60-bp primer is triplicate in each chamber for robust analysis and in both strands. As part of all probes, 35,754 triplicates correspond to genes in TAIR 8 (among which 476 probes correspond to mitochondrial and chloroplast genes), 1,289 probes correspond to EUGENE software predictions + 658 probes for miRNA/MIR, and finally there are 240 controls. Three independent biological replicates were produced. For each comparison, one technical replicate with fluorochrome reversal was performed for each biological replicate (i.e. four hybridizations per comparison). The labeling of complementary RNAs with Cy3-dUTP or Cy5-dUTP was performed as described in the Two-Color Microarray-Based Gene Expression Analysis Low Input Quick Amp Labeling manual (Agilent Technologies). The hybridization and washing were performed according to Agilent Microarray Hybridization Chamber User Guide instructions (Agilent Technologies). Two-micrometer scanning was performed with InnoScan900 scanner (Innopsys), and raw data were extracted using Mapix software (Innopsys). For each array, the raw data comprised the logarithm of median feature pixel intensity at wavelengths 635 nm (red) and 532 nm (green). For each array, a global intensity-dependent normalization using the loess procedure (Yang et al., 2002) was performed to correct the dye bias. The differential analysis is based on log ratios averaging over the duplicate probes and over the technical replicates. Hence, the number of available data points for each gene equals the number of biological replicates and is used to calculate the moderated Student’s t test (Smyth, 2004). Under the null hypothesis, no evidence that the specific variances vary between probes is highlighted by limma; consequently, the moderated t statistic is assumed to follow a standard normal distribution. To control the false discovery rate, adjusted P values found using the optimized false discovery rate approach of Storey and Tibshirani (2003) were calculated. We considered as differentially expressed probes with an adjusted P ≤ 0.05. Analysis was done with R software. The function SqueezeVar of the library limma was used to smooth the specific variances by computing empirical Bayes posterior means. The library kerfdr was used to calculate the adjusted P values.
Microarray data from this article were deposited at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/; accession no. GSE68268) and at CATdb (http://urgv.evry.inra.fr/CATdb/; project no. AU14-08_xrn_vcs) according to Minimum Information about a Microarray Experiment standards.
GO Analysis
Subsets of genes were classified according to GO categories with the Classification SuperViewer Tool in the Bio-Analytic Resource for Plant Biology (http://bar.utoronto.ca). GO analysis results are presented using the software REVIGO (http://revigo.irb.hr; Supek et al., 2011), which allows one to plot the GO data in two-dimensional space. REVIGO summarizes long, unintelligible lists of GO terms by finding a representative subset of the terms using a simple clustering algorithm that relies on semantic similarity measures. REVIGO allows one to visualize the clusters in scatterplots in two-dimensional space, assigning x and y coordinates to each term so that more semantically similar GO terms also are closer in the plot. The disc color gradient (blue to red colors) indicates the degree of GO enrichment corresponding to P values, while the disc size is proportional to the frequency of the GO term in the underlying database. The allowed similarity used was small (0.5), and the semantic similarity measure was SimRel.
Characterization of the 5′ UTR
A database of the 5′ UTR of transcripts that are differentially abundant in xrn4-5 and vcs-8 mutants was generated from TAIR (http://www.arabidopsis.org using the tool Bulk data retrieval with the Arabidopsis Genome Initiative numbers). Since this analysis took into account the 5′ UTR sequences up to −500 bp, we identified 2,298 5′ UTR sequences from the 2,312 transcripts differentially abundant in vcs-8 and 826 5′ UTR sequences from the 973 transcripts differentially abundant in xrn4-5. The minimal energy for secondary structures was calculated using RNAfold. The nucleotide composition of UTRs was determined using MFOLD (http://mfold.rna.albany.edu/). Overrepresented motifs in the 5′ UTR were identified on UTRs of transcripts more abundant in mutants with TAIR. Motif enrichment represents the ratio of the frequency of the motif overrepresented in the coexpressed genes to the frequency of this motif in the 5′ UTR in the whole genome of Arabidopsis (calculated with TAIR). Annotations of miRNA targets were according to the Arabidopsis miRNA target database (http://ppi.bioinfo.asia.edu.tw/At_miRNA/index.php).
mRNA Stability
To assess mRNA stability, we analyzed the level of several transcripts in Col-0, xrn4-5, and vcs-8 dry seeds and seeds incubated for 6, 16, and 24 h at 25°C in darkness with 50 µm cordycepin, which inhibits transcription. Total RNA was extracted, and mRNA levels were assessed by qRT-PCR analyses as described above. The percentages of the remaining mRNAs calculated relative to dry seeds for each genotype were plotted against time, and a regression curve was obtained.
Accession Numbers
Gene data from this article can be found in Supplemental Table S1 and correspond to Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) accession number GSE68268.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phenotypic characterization of the growth and seed production of Col-0, xrn4-3, xrn4-5, vcs-8, and vcs-9 plants grown at 22°C.
Supplemental Figure S2. MA plots of transcripts in xrn4-5 and vcs-8 mutants.
Supplemental Figure S3. Validation of microarray results by qRT-PCR.
Supplemental Figure S4. Effects of ABA and GA on the germination of seeds of asp mutants.
Supplemental Table S1. Characterization of the T-DNA mutant lines studied.
Supplemental Table S2. Sequences of primers used for qRT-PCR experiments.
Supplemental Data Set S1. mRNA abundance in xrn4-5 and vcs-8 mutants after 24 h of imbibition at 25°C.
Supplemental Data Set S2. GO clustering of transcripts more abundant in xrn4-5.
Supplemental Data Set S3. GO clustering of transcripts more abundant in vcs-8.
Supplemental Data Set S4. Target of miRNA up-regulated in xrn4-5 and vcs-8 mutants.
Supplemental Data Set S5. Constant genes chosen for the mRNA feature analysis (ΔG and GC content calculations).
Supplementary Material
Acknowledgments
We thank Hervé Vaucheret (IJPB) and Pascal Genschik (IBMP) for the gifts of Arabidopsis mutant seeds and Jean-Jacques Favory and Cécile Bousquet-Antonelli (LGBP) for useful discussions. We also thank Huifang Yan for her kind help with germination assays.
Glossary
- ABA
abscisic acid
- miRNA
microRNA
- UTR
untranslated region
- Col-0
Columbia-0
- qRT
quantitative real-time reverse transcription
- TAIR
The Arabidopsis Information Resource
- GO
Gene Ontology
- ROS
reactive oxygen species
Footnotes
This work was supported by the European Commission (Seventh Framework Program, Theme KBBE2012.1.1-01, EcoSeed project).
References
- Aluri S, Büttner M (2007) Identification and functional expression of the Arabidopsis thaliana vacuolar glucose transporter 1 and its role in seed germination and flowering. Proc Natl Acad Sci USA 104: 2537–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly C, El-Maarouf-Bouteau H, Corbineau F (2008) From intracellular signaling networks to cell death: the dual role of reactive oxygen species in seed physiology. C R Biol 331: 806–814 [DOI] [PubMed] [Google Scholar]
- Bailly C, Leymarie J, Lehner A, Rousseau S, Côme D, Corbineau F (2004) Catalase activity and expression in developing sunflower seeds as related to drying. J Exp Bot 55: 475–483 [DOI] [PubMed] [Google Scholar]
- Basbouss-Serhal I, Leymarie J, Bailly C (2016) Fluctuation of Arabidopsis seed dormancy with relative humidity and temperature during dry storage. J Exp Bot 67: 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basbouss-Serhal I, Soubigou-Taconnat L, Bailly C, Leymarie J (2015) Germination potential of dormant and non-dormant Arabidopsis seeds is driven by distinct recruitment of mRNAs to polysomes. Plant Physiol 168: 1049–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel GW, Lan H, Glaab E, Gibbs DJ, Gerjets T, Krasnogor N, Bonner AJ, Holdsworth MJ, Provart NJ (2011) Genome-wide network model capturing seed germination reveals coordinated regulation of plant cellular phase transitions. Proc Natl Acad Sci USA 108: 9709–9714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazin J, Langlade N, Vincourt P, Arribat S, Balzergue S, El-Maarouf-Bouteau H, Bailly C (2011) Targeted mRNA oxidation regulates sunflower seed dormancy alleviation during dry after-ripening. Plant Cell 23: 2196–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belostotsky DA, Sieburth LE (2009) Kill the messenger: mRNA decay and plant development. Curr Opin Plant Biol 12: 96–102 [DOI] [PubMed] [Google Scholar]
- Bewley JD. (1997) Seed germination and dormancy. Plant Cell 9: 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borja MS, Piotukh K, Freund C, Gross JD (2011) Dcp1 links coactivators of mRNA decapping to Dcp2 by proline recognition. RNA 17: 278–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J. (2012) The pectin lyases in Arabidopsis thaliana: evolution, selection and expression profiles. PLoS ONE 7: e46944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Chiang YC, Denis CL (2002) CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J 21: 1414–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba Y, Green PJ (2009) mRNA degradation machinery in plants. Plant Biol 52: 114–124 [Google Scholar]
- Chlebowski A, Lubas M, Jensen TH, Dziembowski A (2013) RNA decay machines: the exosome. Biochim Biophys Acta 1829: 552–560 [DOI] [PubMed] [Google Scholar]
- Corbineau F, Xia Q, Bailly C, El-Maarouf-Bouteau H (2014) Ethylene, a key factor in the regulation of seed dormancy. Front Plant Sci 5: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N, Babajko S, Seraphin B (2004) Cytoplasmic foci are sites of mRNA decay in human cells. Cell Biol 165: 31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers BJW, He H, Hanson J, Willems LAJ, Jamar DCL, Cueff G, Rajjou L, Hilhorst HWM, Bentsink L (2016) The Arabidopsis DELAY OF GERMINATION 1 gene affects ABSCISIC ACID INSENSITIVE 5 (ABI5) expression and genetically interacts with ABI3 during Arabidopsis seed development. Plant J 85: 451–465 [DOI] [PubMed] [Google Scholar]
- Dekkers BJW, Pearce S, van Bolderen-Veldkamp RP, Marshall A, Widera P, Gilbert J, Drost HG, Bassel GW, Mueller K, King JR, et al. (2013) Transcriptional dynamics of two seed compartments with opposing roles in Arabidopsis seed germination. Plant Physiol 163: 205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz BJ, Prieto S, Scheffler IE (2002) The role of the 5′ untranslated region (UTR) in glucose-dependent mRNA decay. Yeast 19: 887–902 [DOI] [PubMed] [Google Scholar]
- Dèrozier S, Samson F, Tamby JP, Guichard C, Brunaud V, Grevet P, Gagnot S, Label P, Leple JC, Lecharny A, et al. (2011) Exploration of plant genomes in the FLAGdb(++) environment. Plant Methods 7: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyholos MK, Cavaness GF, Hall B, King E, Punwani J, Van Norman JLE (2003) VARICOSE, a WD domain protein, is required for leaf blade development. Development 130: 6577–6588 [DOI] [PubMed] [Google Scholar]
- Diaz-Camino C, Conde R, Ovsenek N, Villanueva MA (2005) Actin expression is induced and three isoforms are differentially expressed during germination in Zea mays. J Exp Bot 56: 557–565 [DOI] [PubMed] [Google Scholar]
- Dietrich D, Hammes U, Thor K, Suter-Grotemeyer M, Fluckiger R, Slusarenko AJ, Ward JM, Rentsch D (2004) AtPTR1, a plasma membrane peptide transporter expressed during seed germination and in vascular tissue of Arabidopsis. Plant J 40: 488–499 [DOI] [PubMed] [Google Scholar]
- Doma MK, Parker R (2006) Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440: 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunckley T, Parker R (1999) The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J 18: 5411–5422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dure L, Waters L (1965) Long-lived messenger RNA: evidence from cotton seed germination. Science 147: 410–412 [DOI] [PubMed] [Google Scholar]
- Dvir S, Velten L, Sharon E, Zeevi D, Carey LB, Weinberger A, Segal E (2013) Deciphering the rules by which 5′-UTR sequences affect protein expression in yeast. Proc Natl Acad Sci USA 110: E2792–E2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Maarouf-Bouteau H, Meimoun P, Job C, Job D, Bailly C (2013) Role of protein and mRNA oxidation in seed dormancy and germination. Front Plant Sci 4: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J (2005) Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell 20: 905–915 [DOI] [PubMed] [Google Scholar]
- Fillman C, Lykke-Andersen J (2005) RNA decapping inside and outside of processing bodies. Curr Opin Cell Biol 17: 326–331 [DOI] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G (2006) Seed dormancy and the control of germination. New Phytol 171: 501–523 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Reeves W, Ariizumi T, Steber C (2008) Molecular aspects of seed dormancy. Annu Rev Plant Biol 59: 387–415 [DOI] [PubMed] [Google Scholar]
- Galland M, Huguet R, Arc E, Cueff G, Job D, Rajjou L (2014) Dynamic proteomics emphasizes the importance of selective mRNA translation and protein turnover during Arabidopsis seed germination. Mol Cell Proteomics 13: 252–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galland M, Rajjou L (2015) Regulation of mRNA translation controls seed germination and is critical for seedling vigor. Front Plant Sci 6: 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Nishikawa H, Badejo AA, Shibata H, Sawa Y, Nakagaw T, Maruta T, Shigeoka S, Smirnoff N, Ishikawa T (2011) Expression of aspartyl protease and C3HC4-type RING zinc finger genes are responsive to ascorbic acid in Arabidopsis thaliana. J Exp Bot 62: 3647–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisberg JV, Moqtaderi Z, Fan X, Ozsolak F, Struhl K (2014) Global analysis of mRNA isoform half-lives reveals stabilizing and destabilizing elements in yeast. Cell 156: 812–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeres DC, Van Norman JM, Zhang WP, Fauver NA, Spencer ML, Sieburth LE (2007) Components of the Arabidopsis mRNA decapping complex are required for early seedling development. Plant Cell 19: 1549–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory BD, O’Malley RC, Liste R, Urich MA, Tonti-Filippini J, Chen H, Millar AH, Ecker JR (2008) A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell 14: 854–866 [DOI] [PubMed] [Google Scholar]
- Gu W, Xu Y, Xie X, Wang T, Ko JH, Zhou T (2014) The role of RNA structure at 5′ untranslated region in microRNA-mediated gene regulation. RNA 20: 1369–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G (2007) Auxin response factors. Curr Opin Plant Biol 10: 453–460 [DOI] [PubMed] [Google Scholar]
- Gutierrez RA, Ewing RM, Cherry JM, Green PJ (2002) Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc Natl Acad Sci USA 99: 11513–11518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdsworth MJ, Bentsink L, Soppe WJJ (2008) Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol 179: 33–54 [DOI] [PubMed] [Google Scholar]
- Januszyk K, Lima CD (2014) The eukaryotic RNA exosome. Curr Opin Struct Biol 24: 132–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao YL, Riechmann JL, Meyerowitz EM (2008) Transcriptome-wide analysis of uncapped mRNAs in Arabidopsis reveals regulation of mRNA degradation. Plant Cell 20: 2571–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau KD, Xie Z, Allen E, Llave C, Chapmann EJ, Krizan KA, Carrington JC (2003) P1/HC Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA junction. Dev Cell 4: 205–217 [DOI] [PubMed] [Google Scholar]
- Kastenmayer JP, Green PJ (2000) Novel features of the XRN-family in Arabidopsis: evidence that AtXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc Natl Acad Sci USA 97: 13985–13990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Yang JY, Xu J, Jan IC, Prigge MJ, Chua NH (2008) Two cap-binding proteins CBP20 and CBP80 are involved in processing primary microRNAs. Plant Cell Physiol 49: 1634–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Nambara E (2010) Stored and neosynthesized mRNA in Arabidopsis seeds: effects of cycloheximide and controlled deterioration treatment on the resumption of transcription during imbibition. Plant Mol Biol 73: 119–129 [DOI] [PubMed] [Google Scholar]
- Lange H, Zuber H, Sement FM, Chicher J, Kuhn L, Hammann P, Brunaud V, Berard C, Bouteiller N, Balzergue S, et al. (2014) The RNA helicases AtMTR4 and HEN2 target specific subsets of nuclear transcripts for degradation by the nuclear exosome in Arabidopsis thaliana. PLoS Genet 10: e1004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs P, Peaucelle A, Morin H, Traas J (2004) MicroRNA regulation of the CUC genesis required for boundary size control in Arabidopsis meristems. Development 131: 4311–4322 [DOI] [PubMed] [Google Scholar]
- Layat E, Leymarie J, El-Maarouf-Bouteau H, Caius J, Langlade N, Bailly C (2014) Translatome profiling in dormant and nondormant sunflower (Helianthus annuus) seeds highlights post-transcriptional regulation of germination. New Phytol 4: 864–872 [DOI] [PubMed] [Google Scholar]
- Lee JH, Terzaghi W, Gusmaroli G, Charron JB, Yoon HJ, Chen H, He YJ, Xiong Y, Deng XW (2008) Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell 20: 152–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Yoon HJ, Terzaghi W, Martinez C, Dai M, Li J, Byun MO, Deng XW (2010) DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4-based E3 ligases, act together as negative regulators in ABA signal transduction. Plant Cell 22: 1716–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Lykke-Andersen J (2013) Emerging roles for ribonucleoprotein modification and remodeling in controlling RNA fate. Trends Cell Biol 23: 504–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leymarie J, Vitkauskaité G, Hoang HH, Gendreau E, Chazoule V, Meimoun P, Corbineau F, El-Maarouf-Bouteau H, Bailly C (2012) Role of reactive oxygen species in the regulation of Arabidopsis seed dormancy. Plant Cell Physiol 53: 96–106 [DOI] [PubMed] [Google Scholar]
- Ling SHM, Qamra R, Song HW (2011) Structural and functional insights into eukaryotic mRNA decapping. Wiley Interdiscip Rev RNA 2: 193–208 [DOI] [PubMed] [Google Scholar]
- Liu MJ, Wu SH, Chen HM, Wu SH (2012) Widespread translational control contributes to the regulation of Arabidopsis photomorphogenesis. Mol Syst Biol 8: 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C, Xie ZX, Kasschau KD, Carrington JC (2002) Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053–2056 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin D, Chait B, Chua NH (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 32: 317–328 [DOI] [PubMed] [Google Scholar]
- Maldonado-Bonilla LD. (2014) Composition and function of P bodies in Arabidopsis thaliana. Front Plant Sci 5: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez de Alba AE, Moreno AB, Gabriel M, Mallory AC, Christ A, Bounon R, Balzergue S, Aubourg S, Gautheret D, Crespi MD, et al. (2015) In plants, decapping prevents RDR6-dependent production of small interfering RNAs from endogenous mRNAs. Nucleic Acids Res 43: 2902–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx C, Wong J, Buchanan B (2003) Thioredoxin and germinating barley: targets and protein redox changes. Planta 216: 454–460 [DOI] [PubMed] [Google Scholar]
- Merret R, Descombin J, Juan YT, Favory JJ, Carpentier MC, Chaparro C, Charng YY, Deragon JM, Bousquet-Antonelli C (2013) XRN4 and LARP1 are required for a heat-triggered mRNA decay pathway involved in plant acclimation and survival during thermal stress. Cell Rep 5: 1279–1293 [DOI] [PubMed] [Google Scholar]
- Mignone F, Gissi C, Liuni S, Pesole G (2002) Untranslated regions of mRNAs. Genome Biol 3: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AA, Jacobsen JV, Ross JJ, Helliwell CA, Poole AT, Scofield G, Reid JB, Gubler F (2006) Seed dormancy and ABA metabolism in Arabidopsis and barley: the role of ABA 8′-hydroxylase. Plant J 45: 942–954 [DOI] [PubMed] [Google Scholar]
- Mitchell P, Tollervey D (2000) mRNA stability in eukaryotes. Curr Opin Genet Dev 10: 193–198 [DOI] [PubMed] [Google Scholar]
- Montrichard F, Renard M, Alkhalfioui F, Duval FD, Macherel D (2003) Identification and differential expression of two thioredoxin h isoforms in germinating seeds from pea. Plant Physiol 132: 1707–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motomura K, Le QTN, Kumakura N, Fukaya T, Takeda A, Watanabe Y (2012) The role of decapping proteins in the miRNA accumulation in Arabidopsis thaliana. RNA Biol 9: 644–652 [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Decker CJ, Parker R (1995) Turnover mechanisms of the stable yeast Pgk messenger RNA. Mol Cell Biol 15: 2145–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñiz Garcia MN, Stritzler M, Capiati DA (2014) Heterologous expression of Arabidopsis ABF4 gene in potato enhances tuberization through ABA-GA crosstalk regulation. Planta 239: 615–631 [DOI] [PubMed] [Google Scholar]
- Nagarajan VK, Jones CI, Newbury SF, Green PJ (2013) XRN 5′ → 3′ exoribonucleases: structure, mechanisms and functions. Biochim Biophys Acta 1829: 590–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J 41: 697–709 [DOI] [PubMed] [Google Scholar]
- Nambara E, Okamoto M, Tatematsu K, Yano R, Seo M, Kamiya Y (2010) Abscisic acid and the control of seed dormancy and germination. Seed Sci Res 20: 55–67 [Google Scholar]
- Nguyen AH, Matsui A, Tanaka M, Mizunashi K, Nakaminami K, Hayashi M, Iida K, Toyoda T, Nguyen DV, Seki M (2015) Loss of Arabidopsis 5′–3′ exoribonuclease AtXRN4 function enhances heat stress tolerance of plants subjected to severe heat stress. Plant Cell Physiol 56: 1762–1772 [DOI] [PubMed] [Google Scholar]
- Nonogaki H. (2014) Seed dormancy and germination emerging mechanisms and new hypotheses. Front Plant Sci 5: 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonogaki H, Chen F, Bradford K (2007) Mechanisms and genes involved in germination sensu stricto. In Bradford K, Nonogaki H, eds, Seed Development, Dormancy and Germination. Blackwell Publishing, Oxford, pp 264–304 [Google Scholar]
- Okamoto M, Kuwahara A, Seo M, Kushiro T, Asami T, Hirai N, Kamiya Y, Koshiba T, Nambara E (2006) CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol 141: 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira CC, McCarthy JEG (1995) The relationship between eukaryotic translation and mRNA stability: a short upstream open reading frame strongly inhibits translational initiation and greatly accelerates mRNA degradation in the yeast Saccharomyces cerevisiae. J Biol Chem 270: 8936–8943 [DOI] [PubMed] [Google Scholar]
- Olmedo G, Guo H, Gregory BD, Nourizadeh SD, Aguilar-Henonin L, Li H, An F, Guzman P, Ecker JR (2006) Ethylene-Insensitive 5 encodes a 5′-3′ exoribonuclease required for regulation of the EIN3-targeting F-box proteins EBF1/2. Proc Natl Acad Sci USA 103: 13286–13293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F, Valon C, Raynal M, Gaubier CP, Delseny M, Giraudat J (1994) Regulation of gene expression programs during Arabidopsis seed development: roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6: 1567–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Song HW (2004) The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol 11: 121–127 [DOI] [PubMed] [Google Scholar]
- Penfield A, Josse EM, Halliday KJ (2009) Role for an alternative splice variant of PIF6 in the control of Arabidopsis primary seed dormancy. Plant Mol Biol 73: 89–95 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T, Vansiri A, Binder BM, Lechner E, Vierstra RD, Genschik P (2006) The exoribonuclease XRN4 is a component of the ethylene response pathway in Arabidopsis. Plant Cell 18: 3047–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L, Gallardo K, Debeaujon I, Vandekerckhove J, Job C, Job D (2004) The effect of α-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol 134: 1598–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren XZ, Chen ZZ, Liu Y, Zhang HR, Zhang M, Liu QA, Hong XH, Zhu JK, Gong ZZ (2010) ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J 63: 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110: 513–520 [DOI] [PubMed] [Google Scholar]
- Righetti K, Vu JL, Pelletier S, Vu BL, Glaab E, Lalanne D, Pasha A, Patel RV, Provart NJ, Verdier J, et al. (2015) Inference of longevity-related genes from a robust coexpression network of seed maturation identifies regulators linking seed storability to biotic defense-related pathways. Plant Cell 27: 2692–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymarquis LA, Souret FF, Green PJ (2011) Evidence that XRN4, an Arabidopsis homolog of exoribonuclease XRN1, preferentially impacts transcripts with certain sequences or in particular functional categories. RNA 17: 501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sechet J, Frey A, Effroy-Cuzzi D, Berger A, Perreau F, Cueff G, Charif D, Rajjou L, Mouille G, North HM, et al. (2016) Xyloglucan metabolism differentially impacts the cell wall characteristics of the endosperm and embryo during Arabidopsis seed germination. Plant Physiol 170: 1367–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She M, Decker CJ, Chen N, Tumati S, Parker R, Song H (2006) Crystal structure and functional analysis of Dcp2p from Schizosaccharomyces pombe. Nat Struct Mol Biol 13: 63–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She MP, Decker CJ, Sundramurthy K, Liu YY, Chen N, Parker R, Song HW (2004) Crystal structure of Dcp1p and its functional implications in mRNA decapping. Nat Struct Mol Biol 11: 249–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R (2003) Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: 3. [DOI] [PubMed] [Google Scholar]
- Souret FF, Kastenmayer JP, Green PJ (2004) AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol Cell 15: 173–183 [DOI] [PubMed] [Google Scholar]
- Stacey MG, Osawa H, Patel A, Gassmann W, Stacey G (2006) Expression analyses of Arabidopsis oligopeptide transporters during seed germination, vegetative growth and reproduction. Planta 223: 291–305 [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bosnjak M, Skunca N, Smuc T (2011) REVIGO summarizes and visualizes long lists of Gene Ontology terms. PLoS ONE 6: e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi P, Rabara RC, Rushton PJ (2014) A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta 239: 255–266 [DOI] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BMM, Hoekema A (1989) A small scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res 17: 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WQ, Liu SJ, Song SQ, Møller IM (2015) Proteomics of seed development, desiccation tolerance, germination and vigor. Plant Physiol Biochem 86: 1–15 [DOI] [PubMed] [Google Scholar]
- Waters BM, Chu HH, Didonato RJ, Roberts LA, Eisley RB, Lahner B, Salt DE, Walker EL (2006) Mutations in Arabidopsis Yellow Stripe-Like1 and Yellow Stripe-Like3 reveal their roles in metal ion homeostasis and loading of metal ions in seeds. Plant Physiol 141: 1446–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber C, Nover L, Fauth M (2008) Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J 56: 517–530 [DOI] [PubMed] [Google Scholar]
- Xu J, Chua NH (2009) Arabidopsis Decapping 5 is required for mRNA decapping, P-body formation, and translational repression during postembryonic development. Plant Cell 21: 3270–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Chua NH (2011) Processing bodies and plant development. Curr Opin Plant Biol 14: 88–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Chua NH (2012) Dehydration stress activates Arabidopsis MPK6 to signal DCP1 phosphorylation. EMBO J 31: 1975–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Yang JY, Niu QW, Chua NH (2006) Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell 18: 3386–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kuo CCJ, Chen LA (2011) GC content around splice sites affects splicing through pre mRNA secondary structures. BMC Genomics 12: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Jiang YJ, Yu DQ (2011) WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol Cells 31: 303–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.