A cadmium-induced PvERF15 transcription factor binds to an AC-rich element to enhance the expression of transcription factor PvMTF-1 in bean, thus forming a PvERF15/PvMTF-1 transcriptional pathway.
Abstract
In plants, cadmium (Cd)-responsive transcription factors are key downstream effectors of Cd stress transcriptional pathways, which are capable of converging Cd stress signals through triggering the expression of Cd detoxification genes. However, the upstream transcriptional regulatory pathways that modulate their responses to Cd are less clear. Previously, we identified the bean (Phaseolus vulgaris) METAL RESPONSE ELEMENT-BINDING TRANSCRIPTION FACTOR1 (PvMTF-1) that responds to Cd and confers Cd tolerance in planta. Here, we demonstrate an upstream transcriptional regulation of the PvMTF-1 response to Cd. Using a yeast one-hybrid system, we cloned the bean ETHYLENE RESPONSE FACTOR15 (PvERF15) that binds to the PvMTF-1 promoter. PvERF15 was strongly induced by Cd stress, and its overexpression resulted in the up-regulation of PvMTF-1. DNA-protein interaction assays further revealed that PvERF15 binds directly to a 19-bp AC-rich element in the PvMTF-1 promoter. The AC-rich element serves as a positive element bound by PvERF15 to activate gene expression. More importantly, knockdown of PvERF15 by RNA interference resulted in reduced Cd-induced expression of PvMTF-1. PvERF15 seems to be involved in Cd tolerance, since knockdown of PvERF15 by RNA interference in bean leaf discs decreased Cd tolerance in a transient assay. Since PvERF15 is a component of the Cd stress transcriptional pathway in beans and PvMTF-1 is one of its downstream targets, our findings provide a PvERF15/PvMTF-1 transcriptional pathway and thereby contribute to the understanding of Cd stress transcriptional regulatory pathways in plants.
Cadmium (Cd) is highly toxic to plants. Genome-wide gene expression analyses in different plant species have revealed that plants alter transcriptomic expression profiles in response to Cd stress (Weber et al., 2006; Gao et al., 2013; He et al., 2013; Liu et al., 2015; Xu et al., 2015; Oono et al., 2016). These findings indicate that transcriptional regulation of Cd tolerance-related genes is a conserved strategy for plant response to this heavy metal. Cd-responsive transcription is often modulated by multiple transcription factors, some of which confer Cd tolerance through controlling the expression of Cd detoxification genes. Previous studies have characterized a few Cd-responsive transcription factors, including wheat (Triticum aestivum) heat shock transcription factor A4a (Shim et al., 2009), Indian mustard (Brassica juncea) BjCdR15 (Farinati et al., 2010), Arabidopsis (Arabidopsis thaliana) basic helix-loop-helix transcription factors bHLH29, bHLH38, and bHLH39 (Wu et al., 2012), maize (Zea mays) zinc finger protein OXIDATIVE STRESS2 (He et al., 2016), and Arabidopsis zinc finger transcription factor ZAT6 (Chen et al., 2016). In fact, these Cd-responsive transcription factors are the key downstream effectors of Cd stress transcriptional pathways. They trigger the expression of Cd detoxification genes and, thus, converge Cd stress signals (DalCorso et al., 2010; Chmielowska-Bąk et al., 2014). Although the downstream targets of these transcription factors have been identified extensively (Shim et al., 2009; Farinati et al., 2010; Wu et al., 2012; Sun et al., 2015; Chen et al., 2016; He et al., 2016), the upstream transcriptional regulatory pathways that modulate their responses to Cd remain unclear.
We previously identified bean (Phaseolus vulgaris) METAL RESPONSE ELEMENT-BINDING TRANSCRIPTION FACTOR1 (PvMTF-1) as a Cd-responsive transcription factor (Sun et al., 2015). PvMTF-1 contains a zinc finger-like domain and confers Cd tolerance of tobacco (Nicotiana tabacum) plants through activating Trp biosynthesis (Sun et al., 2015). The expression of PvMTF-1 is driven by an intronic promoter located in the bean STRESS-RELATED GENE2 (PvSR2) locus (Qi et al., 2007; Sun et al., 2015; Yang et al., 2015). This intronic promoter (hereafter named PvMTF-1 promoter) is composed of a 397-bp PvSR2 genomic sequence between the upstream transcription start site and the intronic transcription start site (Supplemental Fig. S1). In bean, PvSR2 confers Cd tolerance of tobacco plants (Chai et al., 2003) and is one of the PvMTF-1 downstream target genes (Yang et al., 2015).
Interestingly, the expression of PvMTF-1 is induced by Cd stress (Sun et al., 2015), suggesting that it should be activated by putative upstream transcription factors in the Cd response pathways. In this study, we identified and functionally characterized a bean ethylene response factor, PvERF15, involved in Cd-induced PvMTF-1 activation and Cd tolerance. It binds to an AC-rich element (ACE) in the promoter of the PvMTF-1 gene and then activates its expression. These findings provide new insights into understanding the Cd stress transcriptional pathways in plants.
RESULTS
PvERF15 Is a Cd-Responsive Transcription Factor That Binds to the PvMTF-1 Promoter
To identify putative transcription factors regulating PvMTF-1, we constructed a cDNA library from Cd-treated bean seedlings and performed the yeast one-hybrid (Y1H) assay with the PvMTF-1 promoter as bait. A total of 3 × 104 clones were screened for growth on His-free plates in the presence of 2 mm 3-amino-1,2,4-triazole, and 51 colonies tested positive. By sequencing cDNA from these clones, we isolated seven independent clones of a transcription factor with 255 amino acids that belongs to the bean ERF transcription factor family. The AP2/ERF domain of this bean ERF displays high similarity (62%) with that of Arabidopsis ERF15 (AtERF15; At2g31230); therefore, we designated this transcription factor as PvERF15 (PHAVU_007G193300g; Supplemental Fig. S2). We selected it as a potential transcriptional regulator of PvMTF-1 for further study. The Y1H assay was conducted to further confirm the screening results (Fig. 1A).
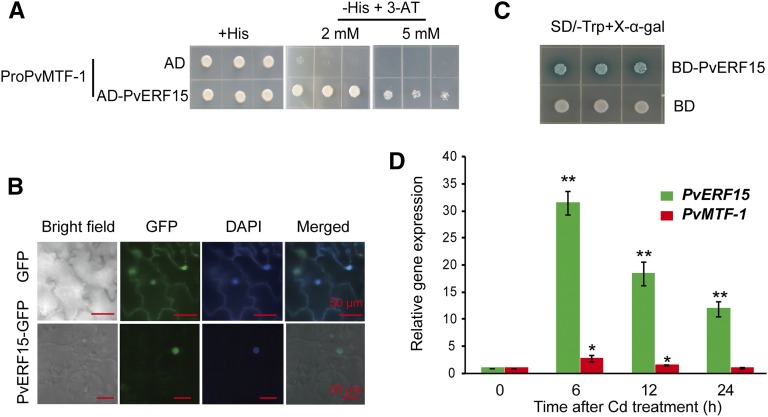
Figure 1.
PvERF15 is a Cd-responsive transactivator that binds to the PvMTF-1 promoter. A, Y1H binding assay of PvERF15 to the PvMTF-1 promoter (ProPvMTF-1). A yeast strain with the HIS3 gene driven by ProPvMTF-1 was transformed with a plasmid encoding the GAL4 activation domain (AD) alone or with a PvERF15 fusion (AD-PvERF15). Interaction is indicated by the ability of cells to grow on His-deficient medium (−His) with or without 3-amino-1,-2,-4-triazole (3-AT). Three independent yeast clones are shown. B, Subcellular localization of PvERF15-GFP and GFP transiently expressed from the 35S promoter in N. benthamiana epidermal cells. DAPI, 4,6-Diamidino-2-phenylindole (nuclei staining). C, The transcriptional activation activity of PvERF15 was analyzed in yeast cells. LacZ reporter gene expression is indicated by blue color on synthetic defined (SD) medium lacking Trp (SD/-Trp) containing 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside (X-α-gal). The Gal4 DNA-binding domain (BD) alone was used as a negative control. Three independent yeast clones are shown. D, Time-course analysis of Cd-inducible gene expression in bean leaf discs. Bean leave discs were treated with 200 μm CdCl2 for 0, 6, 12, and 24 h. RNAs were then extracted and subjected to qRT-PCR. PvERF15 and PvMTF-1 mRNA abundance was expressed as a ratio relative to the pretreatment level (0 h), which was set to a value of 1. Data shown are averages of three independent qRT-PCR experiments for each time point. Error bars represent sd. Significance between experimental values was assessed using Student’s t test (*, P < 0.05; and **, P < 0.01).
Next, we checked whether PvERF15 acts as a transcription factor. PvERF15 was localized in the nucleus of Nicotiana benthamiana cells (Fig. 1B). The GAL4-binding domain (BD)-PvERF15 fusion protein (BD-PvERF15) but not the BD alone (negative control) was able to activate the expression of the GAL4 upstream activating sequence-driven LacZ reporter gene in yeast (Saccharomyces cerevisiae) cells (Fig. 1C). These results suggested that PvERF15 is a transcriptional activator in the nuclei. Since we sought to determine the transcriptional regulation of PvMTF-1 by Cd in this study, we checked whether PvERF15 also is regulated by Cd. A time-course analysis of Cd-inducible gene expression in bean leaf discs was performed using real-time quantitative reverse transcription (qRT)-PCR over 24 h. The mRNAs of these two genes were increased during a 6-h period (31.5- and 2.85-fold induction for PvERF15 and PvMTF-1, respectively) and gradually decreased thereafter (Fig. 1D). These results suggest that both PvMTF-1 and PvERF1 respond to Cd stress, but PvMTF-1 is more weakly induced by Cd than PvERF15. Taken together, we concluded that PvERF15 is a Cd-responsive transcription factor that binds to the PvMTF-1 promoter.
PvERF15 Binds to an ACE in the PvMTF-1 Promoter
To demonstrate the in vivo binding of PvERF15 on the PvMTF-1 promoter, we performed chromatin immunoprecipitation (ChIP) using transgenic bean leaf discs transiently expressing GFP-fused PvERF15 under the control of the cauliflower mosaic virus (CaMV) 35S promoter (35S:PERF15-GFP; Fig. 2A). The quantitative PCR (qPCR) analysis revealed significant enrichment (3.4-fold enrichment compared with that of the 3′ untranslated region [UTR]; P < 0.05) of PvERF15 at the PvMTF-1 promoter region 1 (−397/−175; P1) but not at the overlapping region 2 (−201/−4; P2). However, no significant enrichment of PvERF15 at P1 and P2 was observed in the wild-type control samples (Fig. 2B). These results suggest that PvERF15 binds directly to the 197-bp PvMTF-1 promoter region (−397/−201) in vivo.
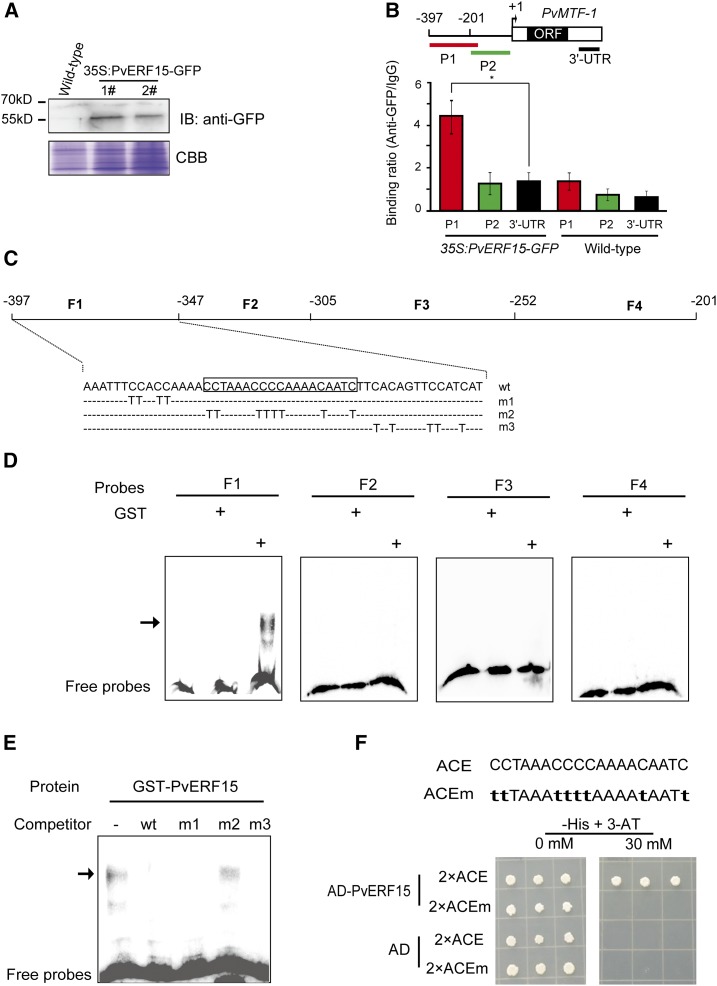
Figure 2.
PvERF15 binds an ACE within the PvMTF-1 promoter. A, Immunoblot (IB) confirming the expression of PvERF15-GFP (56.75 kD) in bean leaf discs. Untransformed bean leaf discs (Wild-type) were used as a negative control. Two independent 35S:PvERF15-GFP lines (1# and 2#) were included. A Coomassie Brilliant Blue (CBB)-stained gel served as a loading control. B, ChIP experiments were used with GFP antibody and mouse IgG (mock control). The diagram of the PvMTF-1 gene indicates the amplicons (P1, P2, and 3′ UTR) used for subsequent qPCR analysis. Relative enrichment was calculated by comparing GFP antibody-immunoprecipitated DNA with those immunoprecipitated with the IgG control (binding ratio of GFP antibody to IgG). 3′ UTR was used as a negative control. Error bars represent sd. Significance between experimental values was assessed using Student’s t test (*, P < 0.05). ORF, Open Reading Frame. C, Diagram of the PvMTF-1 promoter subfragments as probes in EMSA. F1 sequences (wt) and mutations introduced into F1 (m1, m2, and m3) are shown at bottom. ACE is boxed. D, EMSA was performed using biotin-labeled probes with the affinity-purified recombinant GST-PvERF15 and GST (mock proteins). The bound complex is indicated by the arrow. E, Competition experiments using a 1,000-fold excess of unlabeled competitors (wt, m1, m2, and m3). The bound complex is indicated by the arrow. F, Y1H binding assay of PvERF15 to wild-type ACE or a mutant version (ACEm; mutations shown in lowercase letters) bait. Other details are given in the legend to Figure 1.
However, we were more interested in the PvMTF-1 promoter region, which contains unknown ERF-binding cis-elements such as the GCC box and drought-responsive element (DRE). Therefore, we further delineated the PvERF15-binding element into fragments by conducting an electrophoretic mobility shift assay (EMSA). The promoter fragment was partitioned into four ∼50-bp regions (F1–F4; Fig. 2C). The glutathione S-transferase (GST)-PvERF15 fusion protein (GST-PvERF15), but not GST alone, specifically recognized the biotin-labeled F1 probes but not the F2 to F4 probes (Fig. 2D).
Because F1 contained AC-rich sequences (78% AC content), we mutated C to T in the F1 and named these as m1, m2, and m3 (Fig. 2C). The binding ability of PvERF15 with these mutated probes was examined by the competition assay (Fig. 2E). Like wild-type F1, m1 and m3 were able to compete the interaction of PvERF15 with F1 probe. In contrast, m2 fails in inhibiting its binding to PvERF15. These results indicate that the 19-bp F1 sequence (−382/−364; 5′-CCTAAACCCCAAAACAATC-3′) is essential for the binding of PvERF15. We thereby named this sequence as the ACE. To confirm the binding of PvERF15 to this element, we performed a Y1H assay using two copies of ACE as bait. The results showed that PvERF15 was capable of binding to ACE but not to the mutated ACE (Fig. 2F).
ACE Is a Non-Cd Response Element But Acts as a Positive Element
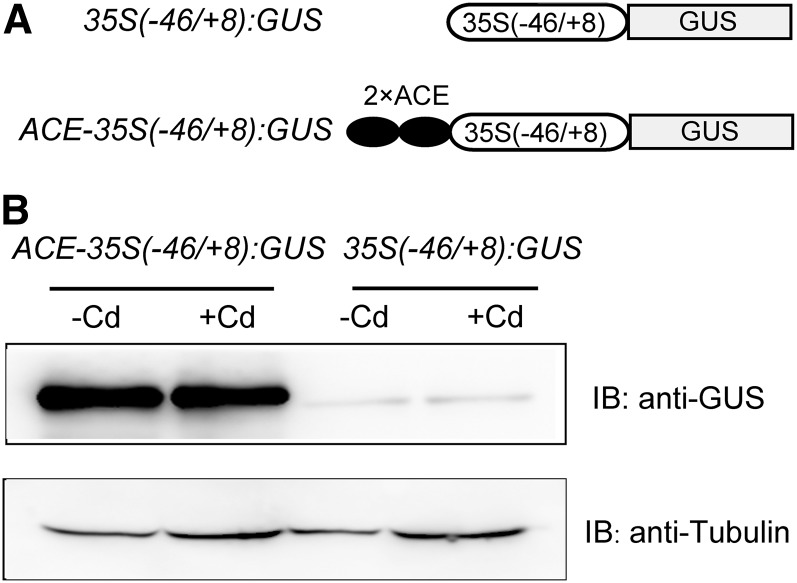
To investigate whether ACE acts as a Cd response element, we performed transient GUS assays in bean leaf discs as described by Peng et al. (2015). A GUS reporter gene controlled by two copies of ACE and the minimal (−46/+8) cauliflower mosaic virus 35S promoter [ACE-35S(−46/+8):GUS; Fig. 3A] was used to quantify the activity of ACE. In transient GUS assays, we used GUS under the control of a minimal 35S promoter [35S(−46/+8):GUS; Fig. 3A] as a vector control. The bean leaf discs transiently expressing each of these constructs were treated without or with Cd and then assayed for GUS protein accumulation by western blot (Fig. 3B). Low GUS accumulation was found in 35S(−46/+8):GUS transgenic control plants (Fig. 3B, lanes 3 and 4), reflecting a low activity of 35S(−46/+8). However, plants expressing ACE-35S(−46/+8):GUS exhibited higher GUS accumulation than control plants (Fig. 3B, lane 1). These results suggest that ACE acts as a positive element, since it confers gene expression when fused to a minimal promoter. ACE failed to confer Cd-responsive expression of the GUS gene, because the GUS accumulation level was not altered significantly by Cd treatment in the plants expressing ACE-35S(−46/+8):GUS (Fig. 3B, lanes 1 and 2). These results suggest that ACE is a non-Cd response element but acts as a positive element.
Figure 3.
Transient assays in bean leaf discs confirm ACE as a positive element. A, Schematic diagrams of the test constructs in the transient assays. B, Immunoblot analysis of plants expressing GUS. The bean leaf discs transiently transformed with each of the constructs were treated without (−Cd) and with 200 μm CdCl2 (+Cd) for 24 h. GUS expression was determined by immunoblot (IB) assays using anti-GUS and anti-tubulin (as a loading control) antibodies. The experiments were repeated independently two times with similar results.
PvERF15 Activates Gene Expression via ACE
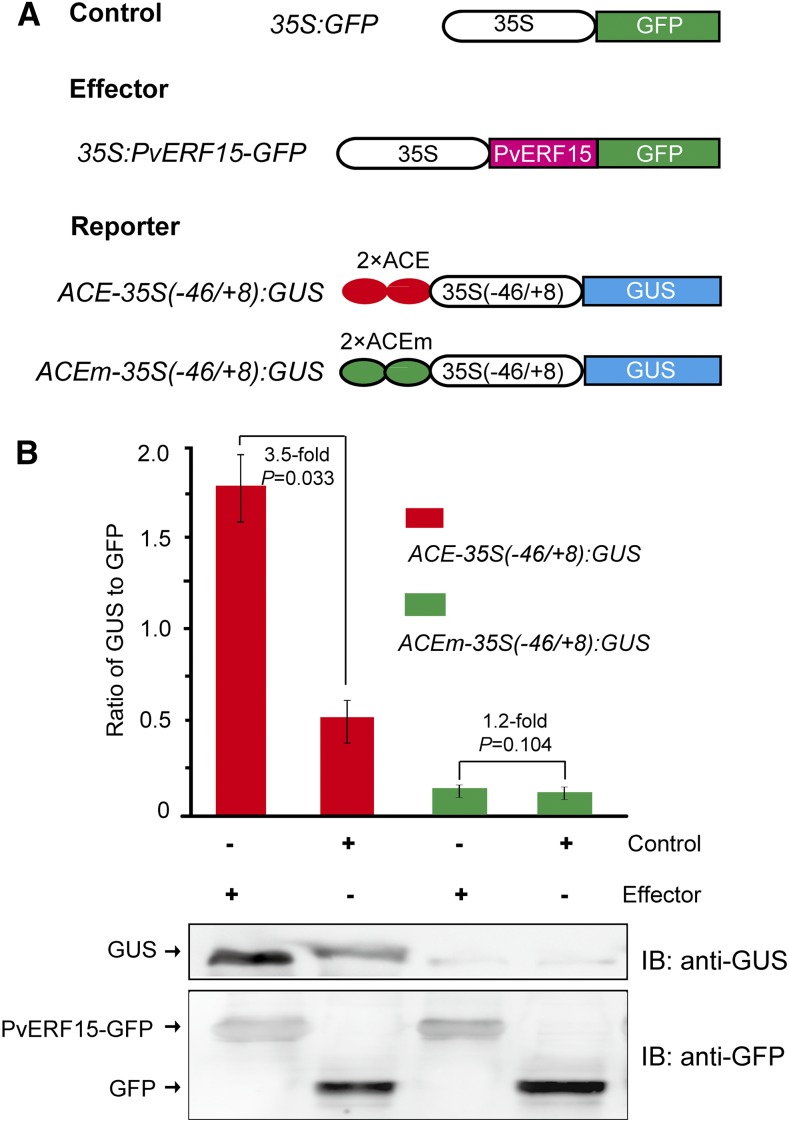
To test whether PvERF15 depends on ACE to activate gene expression in planta, we performed transient GUS assays in N. benthamiana as described by Shim et al. (2013). ACE-35S(−46/+8):GUS or its ACE mutant (C-to-T) version [ACEm-35S(−46/+8):GUS; Fig. 4A] was used as a reporter. 35S:PvERF15-GFP and 35S:GFP were used as effector and control, respectively (Fig. 4A). The reporter and effector were coinfiltrated into N. benthamiana leaf discs and then subjected to GUS activity analysis (Fig. 4B). In the ACE-35S(−46/+8):GUS transgenic reporter plants, the expression of PvERF15-GFP resulted in a more than 3.5-fold increase of GUS expression compared with the GFP-alone control, providing evidence for a positive transactivation activity of PvERF15. In the ACEm-35S(−46/+8):GUS transgenic reporter plants, however, PvERF15-GFP showed a GUS expression level similar to that of the GFP control, suggesting that mutations in the ACE completely abolished the transactivation of PvERF15. These data suggest that PvERF15 activates gene expression in an ACE-dependent manner.
Figure 4.
GUS transient assays of PvERF15 transcriptional activity in N. benthamiana leaf discs. A, Schematic diagram of constructs used in the experiments. B, N. benthamiana leaf discs were cotransformed with combinations of these constructs, as indicated. The expression of GUS, PvERF15-GFP, or GFP was determined by immunoblot (IB) using anti-GUS and anti-GFP antibodies. Transcriptional activity is measured as the relative immunoblotting signal intensity of GUS to GFP and represents an average from three independent experiments. Error bars represent sd. Significance between experimental values was assessed using Student’s t test, and P values are provided.
PvERF15 Is Essential for Both Basal and Cd-Inducible Expression of PvMTF-1
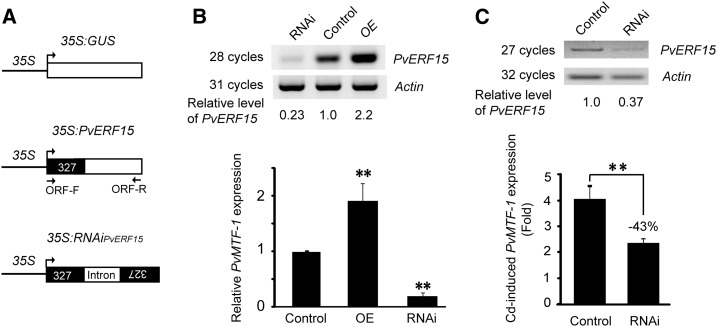
PvERF15 is able to bind directly to the ACE-containing PvMTF-1 promoter in vivo (Fig. 2), and PvERF15 activates gene expression via ACE (Fig. 4). These results imply that PvERF15 possibly acts as a transcriptional regulator of PvMTF-1. To demonstrate this, we examined the effects of overexpression and underexpression of PvERF15 on the PvMTF-1 expression in bean plants. 35S:PvERF15 (Fig. 5A) was introduced into bean leaf discs to generate PvERF15-overexpressing plants (OE). PvERF15 RNA interference (RNAi)-mediated knockdown plants were generated by transfecting leaf discs with the RNAi construct (35S:RNAiPvERF15; Fig. 5A). This RNAi construct contains a 327-bp PvERF15-specific DNA sequence (+1/+327, relative to the translational start codon). In transient assays, the 35S:GUS transgenic leaf discs were used as a control. RNAi plants and overexpression plants had less than 25% and more than 200% of the control PvERF15 mRNA level, respectively (Fig. 5B, top). This overexpression and underexpression of PvERF15 increased and suppressed the PvMTF-1 expression, respectively (Fig. 5B, bottom). These findings suggest that PvERF15 is required for the basal expression of PvMTF-1.
Figure 5.
PvERF15 is a transcriptional regulator of PvMTF-1. A, Schematic diagram of plasmids used in the transient assay. 35S:RNAiPvERF15 contains a 327-bp coding sequence (black box) of PvERF15 in the sense and antisense orientation interspersed by intron 1 (intron) of the potato GA20 OXIDASE gene. Arrows indicate the primers used in the RT-PCR assay for PvERF15. B and C, Each of the plasmids was transformed into bean leaf discs to generate PvERF15 overexpression (OE), PvERF15 RNAi (RNAi), and 35S:GUS (Control) plants. PvERF15 overexpression and knockdown were confirmed by RT-PCR using a pair of primers, ORF-F and ORF-R. The ACTIN gene was used as a loading control. PvERF15 relative abundance is shown at bottom, and the control was set as 1. B, PvMTF-1 expression levels in overexpression and RNAi plants relative to the control (set as 1) determined by qRT-PCR analysis. C, Control and RNAi plants treated with Cd for 6 h and then subjected to qRT-PCR analysis for Cd-inducible PvMTF-1 expression relative to pretreatment (0 h; set as 1). sd values are from three technical replicates of one biological experiment. The experiments were repeated independently two times with similar results. Significance between experimental values was assessed using Student’s t test (**, P < 0.01).
Furthermore, we determined whether knockdown of PvERF15 affects Cd-induced PvMTF-1 expression. We generated RNAi plants that had less than 40% of control PvERF15 transcript level (Fig. 5C, top). Because PvMTF-1 was strongly induced at 6 h of Cd stress (Fig. 1D), we selected this time point in this experiment. Approximately 4.1- and 2.35-fold induction of PvMTF-1 were detected in the control and RNAi plants, respectively (Fig. 5C, bottom). The result showed that knockdown of PvERF15 decreased Cd-inducible PvMTF-1 expression level by approximately 43% compared with the control, indicating an important role of PvERF15 in Cd-induced expression of PvMTF-1.
Knockdown of PvERF15 Decreases Cd Tolerance in a Transient Assay
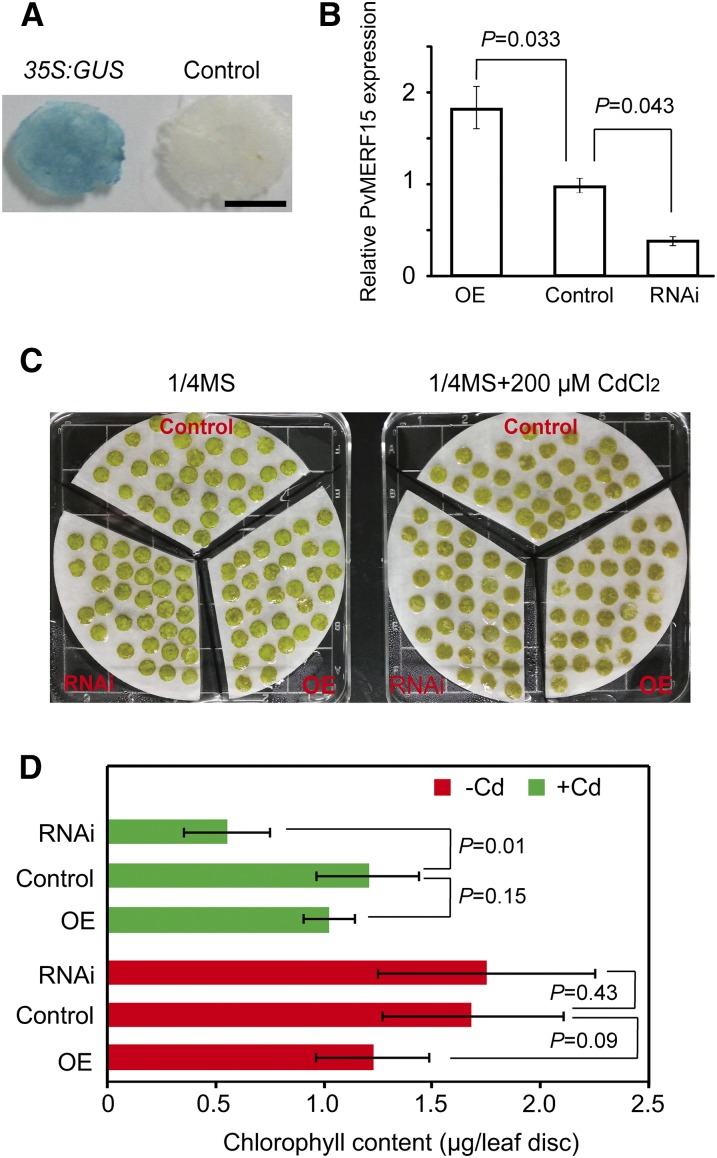
To assess the functional relevance of PvERF15 in bean against Cd stress, we used a transient assay to check the capability of PvERF15-OE and PvERF15-RNAi bean leaf discs to tolerate Cd stress. In the transient assays, the bean leaf discs transiently expressing 35S:GUS were used as a control. The effectiveness of transformation and the level of PvERF15 transcript accumulation were detected 24 h after transformation. Detectable GUS activity in the 35S:GUS control leaf discs confirmed the validity of transformation (Fig. 6A). Further qRT-PCR analysis showed that RNAi plants and overexpression plants had less than 45% and more than 180% of the control PvERF15 mRNA accumulation level, respectively (Fig. 6B). Compared with untreated leaf discs, the leaf discs showed chlorosis 48 h after Cd treatment (Fig. 6C), indicative of a symptom of Cd toxicity (Laspina et al., 2005). No distinguishable difference was observed among these Cd-treated leaf discs. To gain further insight into the toxicity of Cd, we determined the changes in the chlorophyll level of leaf discs in response to Cd (Fig. 6D). Under both Cd stress and non-Cd stress conditions, overexpression plants showed no substantial difference (P > 0.05, Student’s t test) compared with the control plants. Under the condition without Cd stress, there was no substantial difference (P > 0.05, Student’s t test) between the RNAi plants and the control plants. After Cd treatment, however, chlorophyll content in the RNAi plants was much lower (P < 0.05, Student’s t test) than that of the control plants. This finding suggested that knockdown of PvERF15 resulted in a decreased Cd tolerance, supporting a possible role of PvERF15 in bean leaf tissues against Cd stress.
Figure 6.
Transient expression assays in bean leaf discs for Cd tolerance. PvERF15 overexpression (OE), PvERF15 RNAi (RNAi), and 35S:GUS (Control) bean leaf discs were generated by A. tumefaciens-mediated plant transformation. A, Histochemical staining in the 35S:GUS leaf discs but not in untransformed control to monitor the validity of transformation. At least six leaf discs were examined, and a typical disc is presented. Bar = 2.5 mm. B, PvERF15 accumulation levels in overexpression and RNAi plants relative to the control (set as 1) determined by qRT-PCR analysis. C and D, Phenotype (C) and chlorophyll content (D) from overexpression, RNAi, and control bean leaf discs treated with 200 μm CdCl2 (+Cd) or without (−Cd) for 48 h. Chlorophyll content (μg per leaf disc) is given as means ± sd of three independent experiments (at least 30 leaf discs were analyzed for each transgenic line in an experiment). Significance between experimental values was assessed using Student’s t test, and P values are provided.
DISCUSSION
PvERF15 and PvMTF-1 Form a Cd Stress Transcriptional Regulatory Pathway
PvMTF-1 is a key component of the Cd stress transcriptional pathway in plants. In this study, we demonstrate that PvERF15 is located upstream of PvMTF-1 and acts as a transcriptional regulator of PvMTF-1 expression via the ACE of its promoter. A working model of PvERF15 on the PvMTF-1 response to Cd is presented. Upon Cd stress, PvERF15 expression is induced to bind a positive element ACE and enhance PvMTF-1 expression. Several lines of evidence support this model: (1) PvERF15 is strongly induced by Cd stress, and its overexpression results in the up-regulation of PvMTF-1; (2) knockdown of PvERF15 by RNAi resulted in reduced Cd-induced expression of PvMTF-1; (3) ACE acts as a positive element through which PvERF15 activates gene expression; and (4) PvERF15 seems to be involved in Cd tolerance, since knockdown of PvERF15 in bean leaf discs decreased Cd tolerance in a transient assay. Although it is unknown how PvERF15 senses stress signals and regulates the bean response to Cd stress, the results presented here strongly support that PvERF15 is a component of the Cd stress transcriptional pathway in beans and PvMTF-1 is one of its downstream targets. Our findings provide a PvERF15/PvMTF-1 transcriptional pathway and thereby contribute to the understanding of Cd stress transcriptional pathways in plants. In this study, genetic evidence for the biological relevance of PvERF15 is based mainly on RNAi-mediated knockdown of PvERF15 in bean leaf discs. To have a more complete understanding of PvERF15 function in the bean Cd response, stable PvERF15 knockout bean lines will be needed for future study.
Cd-Induced Transcription Can Be Modulated by a Non-Cd Response Element
The working models of Cd-induced transcription remain largely unclear. We previously revealed a Cd-responsive transcription factor (such as PvMTF-1)/Cd response element (such as the metal response element) working model (Sun et al., 2015). However, unlike the metal response element, ACE is a positive element and interacts with a Cd-responsive PvERF15 transcription factor to regulate Cd-induced transcription. Our findings, therefore, suggest a non-Cd response element/Cd-responsive transcription factor working model. Our results thereby extend our knowledge of Cd-induced transcriptional regulation.
ACE Is a New ERF-Binding Element
Interestingly, ACE is different from the known ERF transcription factor-binding sites such as the GCC box and DRE, indicating that it is a new ERF-binding element. It is well established that ERFs are involved in different biological processes through binding a diverse set of cis-elements present in downstream targets (Sasaki et al., 2007). In the future, experiments assessing the core sequences of ACE and its role in the ethylene response will be necessary.
MATERIALS AND METHODS
Plant Materials and Treatments
The leaves of 4- to 5-week-old Nicotiana benthamiana plants were used for transient expression assays and subcellular localization of PvERF15-GFP protein. The seeds of bean (Phaseolus vulgaris ‘Saxa’) were sown on a soil mix of commercial potting soil:perlite (3:1) at 22°C with a 16-h/8-h light/dark cycle. The detached leaves (8 –10 cm long and 4–5 cm wide) from bean seedlings were used for transient expression assays and Cd stress. For Cd treatment, bean leaf discs (5 mm diameter) were transferred to a filter paper soaked with one-quarter-strength Murashige and Skoog (MS) liquid medium supplemented with 200 μm CdCl2 in the greenhouse. The samples were removed from the cultures and frozen in liquid nitrogen at 0, 6, 12, and 24 h.
Y1H Library Screening
Two-week-old bean seedlings were grown on MS liquid medium (control) with 200 μm CdCl2 for 24 h and then used to construct a GAL4 AD-cDNA library using the yeast SfiI-digested pGADT7 vector and the Creator SMART cDNA Library Construction Kit (Clontech). The resulting PvMTF-1 promoter bait plasmid and AD-cDNA library were introduced into the yeast (Saccharomyces cerevisiae) strain Y187 (Clontech). Because the bait plasmid did not undergo self-activation in Y187 yeast cells, positive clones were isolated by growth on His-free SD medium containing 1 mm 3-amino-1,2,4-triazole. Plasmid DNA was isolated from yeast cells and sequenced. The Y1H assays were performed as described previously (Sun et al., 2015).
Primers
Oligonucleotide primers used for PCR and the construction of expression vectors are listed in Supplemental Table S1.
Plasmid Construction
35S:PvERF15 and 35S:PvERF15-GFP were generated using the ClonExpress II One Step Cloning Kit (Vazyme Biotech). The coding region of PvERF15 was PCR amplified using the primers 121-PvERF15-F and 121-PvERF15-R and then cloned into the BamHI/SacI sites of pBI121 (Clontech). The coding region of PvERF15 was PCR amplified using the primers 1302-PvERF15-F and 1302-PvERF15-R and then cloned into the BglII/SpeI sites of pCAMIBA1302 (Clontech). The 35S:RNAiPvERF15 plasmid was constructed by fusion PCR of three amplified overlapping PCR fragments. Using AD-PvERF15 as a template, the sense RNAiPvERF15 fragment was amplified using the primers ERF15-sF and ERF15-sR and the antisense RNAiPvERF15 fragment was amplified using the primers ERF15-asF and ERF15-asR. The intron of the potato (Solanum tuberosum) GA20 OXIDASE gene was obtained from a pUCCRNAi plasmid by PCR using the primers GA20in-F and GA20in-R. The final PCR was performed using mixed PCR products (33 ng of each PCR product) using ERF15-sF and ERF15-asR. The PCR products were cloned into the BamHI/SacI sites of pBI121 (Clontech). For bait construction, the PvMTF-1 promoter was amplified from bean genomic DNA using the primers ProPvMTF-1-F and ProPvMTF-1-R. 2×ACE and 2×ACEm DNA were synthesized by Shanghai Sangon Biotechnology. These baits were then cloned into the EcoRI/SpeI sites of pHIS2.1 (Clontech) to generate bait constructs. All of the constructs were verified by sequencing using the corresponding sequencing primers as described by Sun et al. (2015).
We used an asymmetric overlap extension PCR method to construct ACE-35S(−46/+8):GUS and ACEm-35S(−46/+8):GUS plasmids using AC-35S-F and ACEm-35S-F and a common reverse primer, AC-35S-R. The PCR products were then cloned into the HindIII/XbaI sites of pBI121 instead of the 35S promoter and sequenced using GUS-seqR (5′-CCCACACTTTGCCGTAATGA-3′).
Subcellular Localization
The 35S:PvERF15-GFP and pCAMBIA1302 empty vector (35S:GFP) constructs were transformed into N. benthamiana leaves by Agrobacterium tumefaciens infiltration. The fluorescence signal was observed with a Zeiss 5 Live laser scanning confocal microscope at 42 h after transformation.
Transcriptional Activation Activity Assay in Yeast Cells
The coding sequence of PvERF15 was PCR amplified and cloned into pGBKT7 (Clontech) to generate BD-PvERF15. Its transcriptional activation activity was determined in SD medium lacking Trp and containing 40 µg mL−1 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside (Sigma-Aldrich).
RNA Isolation and cDNA Synthesis
Total RNA was extracted from plant material using the RNAprep pure plant kit with on-column DNase digestion (Tiangen Biotech). RNA (2 μg) was used to synthesize the first-strand cDNA with oligo(dT) primers with the PrimeScript first-strand cDNA synthesis kit (Takara).
A. tumefaciens-Mediated Gene Transfer by Infiltration of Plant Leaf Discs
Various constructs tested were introduced into A. tumefaciens GV3101. The leaf discs of bean or N. benthamiana were vacuum infiltrated with A. tumefaciens as described previously by Marion et al. (2008). Briefly, A. tumefaciens cells were collected and resuspended at an optical density at 600 nm of 2 in one-quarter-strength MS liquid medium containing 5% (w/v) Suc, 200 μm acetosyringone, and 0.01% (v/v) Silwet. After vacuum infiltration, transformed bean leaf discs were transferred to a filter paper soaked with one-quarter-strength MS solution medium.
ChIP Assay
Bean leaf discs expressing PvERF15-GFP fusion proteins and nontransformed leaf discs (wild-type control) were subjected to ChIP using an anti-GFP antibody (ab290; Abcam) following the instructions of the EpiQuik Plant ChIP Kit (Epigentek). Western blot analysis of the PvERF15-GFP protein was performed using the anti-GFP antibody (ab290; Abcam). For ChIP-qPCR analysis, 1 μL of 20-fold diluted immunoprecipitated DNA was analyzed by qPCR on a CFX96 Real-Time System (Bio-Rad) with the SYBR Premix Ex-Taq Kit (Takara). Enrichment of the ChIP target was expressed as a binding ratio between GFP antibody-immunoprecipitated samples and those immunoprecipitated with the IgG control. The binding ratio was calculated using the 2−ΔΔCt method as described previously (Mukhopadhyay et al., 2008). The following primers were used: P1-F and P1-R for P1, P2-F and P2-R for P2, and 3′-UTR-F and 3′-UTR-R for 3′ UTR.
Fusion Protein Preparation and EMSA
A GST-PvERF15 fusion protein was prepared by cloning the PvERF15 coding region into the BamHI/SalI sites of the pGEX 4T-1 plasmid (GE Healthcare Bio-Sciences). GST-PvERF15 fusion proteins were isolated according to the manufacturer’s protocol (GE Healthcare Bio-Sciences) and subsequently subjected to an EMSA using the LightShift Chemiluminescent EMSA Kit (Pierce). The 5′-biotin-labeled DNA probes or completed DNA were synthesized by Shanghai Sangon Biotechnology.
Transient Gene Expression Analysis in Bean and N. benthamiana Leaf Discs
For ACE functional analysis, transformed N. benthamiana leaf discs were treated without (−Cd) and with 200 μm CdCl2 (+Cd) for 24 h. GUS expression was determined by western blot using anti-GUS and anti-tubulin (as a loading control) antibodies. In GUS transient assays of PvERF15 transcriptional activity, transformed N. benthamiana leaf discs were grown on one-quarter-strength MS for 2 d. GUS and GFP expression was determined by western blot using anti-GUS or anti-GFP antibodies. To determine the expression of PvMTF-1 regulated by PvERF15, transformed bean leaf discs were grown on one-quarter-strength MS solution medium for 1 d. One-half of the samples were removed from the cultures and frozen in liquid nitrogen at 0 h, and the other half were transferred to a filter paper soaked with fresh one-quarter-strength MS solution with 200 μm CdCl2 for 6 h.
Total RNA was isolated and then subjected to cDNA synthesis. The expression of PvERF15 was confirmed by semiquantitative reverse transcription-PCR using the primers ORF-F and ORF-R. qRT-PCR analysis was performed on the CFX96 Real-Time System (Bio-Rad) with the SYBR Premix Ex-Taq Kit (Takara). Real-time qRT-PCR analysis of PvERF15 or PvMTF-1 transcripts was performed using PvERF15-rF and PvERF15-rR for PvERF15 and PvMTF-1-rF and PvMTF-1-rR for PvMTF-1. The bean ACTIN gene was used as an internal control using the primers Actin-F and Actin-R.
Leaf Disc Assay for Cd Tolerance
Transformed bean leaf discs were grown on one-quarter-strength MS solution medium for 1 d. GUS staining of bean leaf discs transiently expressing 35S:GUS was monitored for transformation efficiency. Real-time qRT-PCR analysis of PvERF15 transcripts was performed using PvERF15-rF and PvERF15-rR. The bean ACTIN gene was used as an internal control using the primers Actin-F and Actin-R. These bean leaf discs were then transferred to a filter paper soaked with fresh one-quarter-strength MS solution without (control) or with 200 μm CdCl2 for another 2 d. Chlorophyll was extracted in 80% acetone and quantified according to Arnon (1949). The concentration was calculated using the following equation: total chlorophyll (μg mL−1) = 20.2 × OD663 + 8.02 × OD645, where OD represents optical density at the given values.
Total Protein Extraction and Western Blot
Total protein from the plant samples was extracted using extraction buffer (50 mm Tris-HCl [pH 7.5], 150 mm NaCl, 1 mm EDTA,10% [v/v] glycerol, 5 mm dithiothreitol, 0.5% [v/v] Triton X-100, 1 mm phenylmethylsulfonyl fluoride, and 1% [v/v] Nonidet P-40). Proteins separated on a gel were electrophoretically transferred to a pure nitrocellulose blotting membrane (Pall Life Sciences). The membrane was cut across the molecular mass region of corresponding proteins and separately probed with anti-GUS antibody (Clontech), anti-GFP antibody (Abcam), or anti-tubulin antibody (Sigma-Aldrich). Protein blots were developed with an ECL kit (Amersham Pharmacia Biotech), and images were obtained using the LAS3000 image-capture system (Fujifilm).
Statistical Analysis
Statistical analysis (sd and P values) was performed using Microsoft Excel 2007. Significance (P < 0.05 and P < 0.01) was assessed using Student’s t test.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers XM_007144842 (PvERF15), AB067722 (ACTIN), and DQ109993 and U54704 (PvMTF-1).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Schematic representation of the PvMTF-1 gene located within the PvSR2 locus.
Supplemental Figure S2. Nucleotide and deduced amino acid sequences of PvERF15 cDNA.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Ligeng Ma (College of Life Science, Capital Normal University) for providing pUCCRNAi plasmid, anti-GUS antibody, and anti-tubulin antibody and Weiwei Zhang (College of Life Science, Capital Normal University) for linguistic assistance during the preparation of this article.
Glossary
- Cd
cadmium
- Y1H
yeast one-hybrid
- qRT
quantitative reverse transcription
- ChIP
chromatin immunoprecipitation
- UTR
untranslated region
- EMSA
electrophoretic mobility shift assay
- ACE
AC-rich element
- RNAi
RNA interference
- MS
Murashige and Skoog
- SD
synthetic defined
- qPCR
quantitative PCR
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31271293 and 30770181 to X.Q.) and the Beijing Natural Science Foundation (grant no. 5112005 to X.Q.).
Articles can be viewed without a subscription.
References
- Arnon DI. (1949) Copper enzymes in isolate chloroplast: polyphenoloxidase in Beta vulgaris. Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai T, Chen Q, Zhang Y, Dong J, An C (2003) Cadmium resistance in transgenic tobacco plants enhanced by expressing bean heavy metal-responsive gene PvSR2. Sci China C Life Sci 46: 623–630 [DOI] [PubMed] [Google Scholar]
- Chen J, Yang L, Yan X, Liu Y, Wang R, Fan T, Ren Y, Tang X, Xiao F, Liu Y, et al. (2016) Zinc-finger transcription factor ZAT6 positively regulates cadmium tolerance through the glutathione-dependent pathway in Arabidopsis. Plant Physiol 171: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmielowska-Bąk J, Gzyl J, Rucińska-Sobkowiak R, Arasimowicz-Jelonek M, Deckert J (2014) The new insights into cadmium sensing. Front Plant Sci 5: 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DalCorso G, Farinati S, Furini A (2010) Regulatory networks of cadmium stress in plants. Plant Signal Behav 5: 663–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinati S, DalCorso G, Varotto S, Furini A (2010) The Brassica juncea BjCdR15, an ortholog of Arabidopsis TGA3, is a regulator of cadmium uptake, transport and accumulation in shoots and confers cadmium tolerance in transgenic plants. New Phytol 185: 964–978 [DOI] [PubMed] [Google Scholar]
- Gao J, Sun L, Yang X, Liu JX (2013) Transcriptomic analysis of cadmium stress response in the heavy metal hyperaccumulator Sedum alfredii Hance. PLoS ONE 8: e64643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Li H, Luo J, Ma C, Li S, Qu L, Gai Y, Jiang X, Janz D, Polle A, et al. (2013) A transcriptomic network underlies microstructural and physiological responses to cadmium in Populus × canescens. Plant Physiol 162: 424–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Ma X, Li Z, Jiao Z, Li Y, Ow DW (2016) Maize OXIDATIVE STRESS2 homologs enhance cadmium tolerance in Arabidopsis through activation of a putative SAM-dependent methyltransferase gene. Plant Physiol 171: 1675–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laspina NV, Groppa MD, Tomaro ML, Benavides MP (2005) Nitric oxide protects sunflower leaves against Cd-induced oxidative stress. Plant Sci 169: 323–330 [Google Scholar]
- Liu T, Zhu S, Tang Q, Tang S (2015) Genome-wide transcriptomic profiling of ramie (Boehmeria nivea L. Gaud) in response to cadmium stress. Gene 558: 131–137 [DOI] [PubMed] [Google Scholar]
- Marion J, Bach L, Bellec Y, Meyer C, Gissot L, Faure JD (2008) Systematic analysis of protein subcellular localization and interaction using high-throughput transient transformation of Arabidopsis seedlings. Plant J 56: 169–179 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A, Deplancke B, Walhout AJ, Tissenbaum HA (2008) Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real-time PCR to study transcription factor binding to DNA in Caenorhabditis elegans. Nat Protoc 3: 698–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oono Y, Yazawa T, Kanamori H, Sasaki H, Mori S, Handa H, Matsumoto T (2016) Genome-wide transcriptome analysis of cadmium stress in rice. Biomed Res Int 2016: 9739505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Chen L, Lu Y, Wu Y, Dumenil J, Zhu Z, Bevan MW, Li Y (2015) The ubiquitin receptors DA1, DAR1, and DAR2 redundantly regulate endoreduplication by modulating the stability of TCP14/15 in Arabidopsis. Plant Cell 27: 649–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi XT, Zhang YX, Chai TY (2007) The bean PvSR2 gene produces two transcripts by alternative promoter usage. Biochem Biophys Res Commun 356: 273–278 [DOI] [PubMed] [Google Scholar]
- Sasaki K, Mitsuhara I, Seo S, Ito H, Matsui H, Ohashi Y (2007) Two novel AP2/ERF domain proteins interact with cis-element VWRE for wound-induced expression of the tobacco tpoxN1 gene. Plant J 50: 1079–1092 [DOI] [PubMed] [Google Scholar]
- Shim D, Hwang JU, Lee J, Lee S, Choi Y, An G, Martinoia E, Lee Y (2009) Orthologs of the class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice. Plant Cell 21: 4031–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim JS, Jung C, Lee S, Min K, Lee YW, Choi Y, Lee JS, Song JT, Kim JK, Choi YD (2013) AtMYB44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling. Plant J 73: 483–495 [DOI] [PubMed] [Google Scholar]
- Sun N, Liu M, Zhang W, Yang W, Bei X, Ma H, Qiao F, Qi X (2015) Bean metal-responsive element-binding transcription factor confers cadmium resistance in tobacco. Plant Physiol 167: 1136–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Trampczynska A, Clemens S (2006) Comparative transcriptome analysis of toxic metal responses in Arabidopsis thaliana and the Cd2+-hypertolerant facultative metallophyte Arabidopsis halleri. Plant Cell Environ 29: 950–963 [DOI] [PubMed] [Google Scholar]
- Wu H, Chen C, Du J, Liu H, Cui Y, Zhang Y, He Y, Wang Y, Chu C, Feng Z, et al. (2012) Co-overexpression FIT with AtbHLH38 or AtbHLH39 in Arabidopsis-enhanced cadmium tolerance via increased cadmium sequestration in roots and improved iron homeostasis of shoots. Plant Physiol 158: 790–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Wang Y, Liu W, Wang J, Zhu X, Zhang K, Yu R, Wang R, Xie Y, Zhang W, et al. (2015) De novo sequencing of root transcriptome reveals complex cadmium-responsive regulatory networks in radish (Raphanus sativus L.). Plant Sci 236: 313–323 [DOI] [PubMed] [Google Scholar]
- Yang W, Bei X, Liu M, Qi X (2015) Intronic promoter-mediated feedback loop regulates bean PvSR2 gene expression. Biochem Biophys Res Commun 463: 1097–1101 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.