Abstract
Background
The runt domain transcription factors are key regulators of developmental processes in bilaterians, involved both in cell proliferation and differentiation, and their disruption usually leads to disease. Three runt domain genes have been described in each vertebrate genome (the RUNX gene family), but only one in other chordates. Therefore, the common ancestor of vertebrates has been thought to have had a single runt domain gene.
Results
Analysis of the genome draft of the fugu pufferfish (Takifugu rubripes) reveals the existence of a fourth runt domain gene, FrRUNT, in addition to the orthologs of human RUNX1, RUNX2 and RUNX3. The tiny FrRUNT packs six exons and two putative promoters in just 3 kb of genomic sequence. The first exon is located within an intron of FrSUPT3H, the ortholog of human SUPT3H, and the first exon of FrSUPT3H resides within the first intron of FrRUNT. The two gene structures are therefore "interlocked". In the human genome, SUPT3H is instead interlocked with RUNX2. FrRUNT has no detectable ortholog in the genomes of mammals, birds or amphibians. We consider alternative explanations for an apparent contradiction between the phylogenetic data and the comparison of the genomic neighborhoods of human and fugu runt domain genes. We hypothesize that an ancient RUNT locus was lost in the tetrapod lineage, together with FrFSTL6, a member of a novel family of follistatin-like genes.
Conclusions
Our results suggest that the runt domain family may have started expanding in chordates much earlier than previously thought, and exemplify the importance of detailed analysis of whole-genome draft sequence to provide new insights into gene evolution.
Background
Since the initial description of the Drosophila segmentation gene runt over a decade ago [1], a small family of runt domain (RD) genes has been described and extensively analyzed in several species. The 130 amino acid long runt domain is very highly conserved and is readily identifiable computationally. RD transcription factors are developmental regulators involved both in cell proliferation and differentiation, and their disruption usually leads to disease [2].
In humans, three different RD genes were identified [3] and named according to various schemes, currently standardized by the human gene symbols RUNX1, RUNX2 and RUNX3. RUNX genes have two promoters (P1 and P2, also called distal and proximal, respectively) [4-7] separated by a long intron; the proximal promoter (P2) is always located within a large CpG island [8]. Extensive alternative splicing giving rise to many isoforms has been described for all RUNX genes [9-11].
Orthologs of all three human RUNX genes were identified in mouse. A single RD gene was described in Xenopus, presumed to be orthologous to RUNX1 [12]. An experimental search for RD genes in fugu showed the existence of a fugu ortholog of human RUNX2, and suggested the existence of a single additional RD gene in fugu [13], while a computational search of the fugu genomic sequence revealed three RUNX genes [14]. Four RD genes were identified in Drosophila [14,15], while a single RD gene exists in C. elegans [16], sea urchin and amphioxus [17]. Based on these data, current thought on the evolution of the RD gene family posits that a single RD gene was present in the common ancestor of chordates [17], and this ancestral gene triplicated during early vertebrate evolution, giving rise to the modern RUNX gene complement. The proposed mechanism of expansion involved large-scale genomic duplications, identifiable today as large paralogous segments [18]. The proper identification of true orthology relationships is often helpful for inferring gene function and translating knowledge between model organisms and more complex species. Under the current model, simple orthology relationships should be expected among vertebrate RUNX genes, but their functional relationship to the ancestral RD gene is unknown. The single known RD gene in C. elegans has been shown to be required for the formation of a functional gut; this role has been claimed to be conserved with mouse Runx3 [19].
The current availability of genome drafts for several vertebrate species, including Homo sapiens, Mus musculus, Rattus norvegicus, Canis familiaris, Gallus gallus, Takifugu rubripes, Tetraodon nigroviridis, Danio rerio and Xenopus tropicalis, allows us to explore a comprehensive set of vertebrate RD genes and characterize their genomic environments, shedding light on the structure and evolution of this important gene family.
Results
The fugu genome has at least four runt domain genes
Our search for RD genes in the fugu draft yielded four distinct genomic scaffolds (Fig. 1 and Table 1), each containing a single, complete RD gene. Each scaffold had one or more sequence gaps, some within the RD genes, others between them and their neighbors. We employed a directed sequencing approach to obtain the additional sequence needed to close the gaps in these four scaffolds and to improve sequence quality.
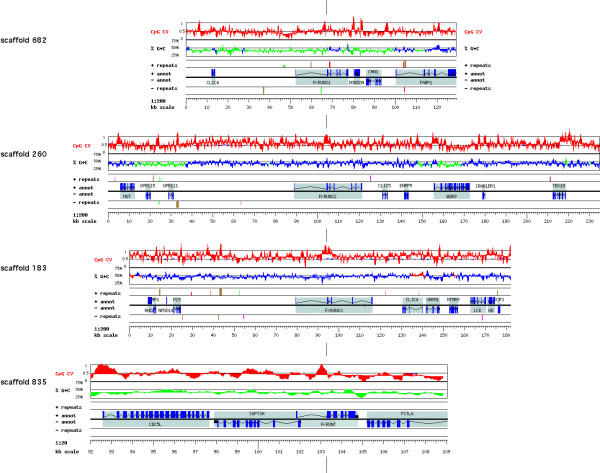
Figure 1.
The four fugu scaffolds analyzed and visualized using the GESTALT Workbench. The RD genes are aligned by the position of the proximal promoter. For each scaffold, graphs are shown of the CpG contrast values (observed/expected, typically ~0.5 for the fugu genome), G+C percentage (green – below 43%, blue – between 43% and 50%, red – above 50%), interspersed repeats (SINEs in red, LINEs in green, DNA and LTR elements in brown, other repeats in purple), and gene annotations. Genes and repeats displayed above each black midline are in the forward strand, while those displayed under the midline are in the reverse strand of the sequence. All scaffolds are shown at a resolution of 200 bp/pixel, except for scaffold 835, which includes the tiny FrRUNT gene. For clarity, only the last 17 kb of this scaffold are shown at 20 bp/pixel. The much longer scaffold scaffold 183 is also shown truncated at 182 kb; the LCK locus has been studied in detail elsewhere [42].
Table 1.
The four fugu RD genes and their human orthologs.
| Fugu gene | Human ortholog | |||||
| Name | Location | Size | %G+C | location | Gene size | %G+C |
| FrRUNT | scaffold_835 | 3.0 | 41.2% | N.O. | N.O. | N.O. |
| FrRUNX1 | scaffold_682 | 25.2 | 42.5% | chr21 | 262 | 43.6% |
| FrRUNX2 | scaffold_260 | 32.6 | 45.7% | chr6 | 219 | 39.9% |
| FrRUNX3 | scaffold_183 | 36.8 | 47.4% | chr1 | 66 | 54.8% |
Sizes are expressed in kb. N.O.: No ortholog.
We studied the four scaffold sequences using the GESTALT Workbench [20] and constructed hypothetical gene structures for the fugu RD genes by maximizing similarity to known vertebrate RD proteins. Three of the four RD genes found in the fugu genome have clear one-to-one similarity relationships with the three mammalian RUNX genes (see phylogenetic analysis below). They have been assumed to be their orthologs [14,17]; we call them FrRUNX1, FrRUNX2 and FrRUNX3 (Fig. 1). Their genomic structures are similar to those of their human counterparts, but their sizes have evolved differently. RUNX3 is the smallest of the three human RUNX genes, while in fugu FrRUNX3 is the largest (Table 1), and FrRUNX2 is significantly larger than FrRUNX1. FrRUNX1 has acquired an additional intron [17] that is not present in human RUNX1 or in any other RD gene. This intron is just 65 bp long, has canonical splice signals, and is in phase 0 with respect to the protein reading frame, at the beginning of the runt domain. An additional intron has been described at the 5' end of the coding region, yielding a short form that would be locally non-homologous to the other RD genes [14]. A detailed comparison of human RUNX2 and FrRUNX2 has been published [13]. In both human and in fugu, RUNX3 has the highest G+C content of the RD genes, while the G+C content of RUNX2 differs significantly between the two species (Table 1).
The fugu RUNT gene
In addition to the three RUNX genes, the fugu genome has a fourth and more divergent runt-domain gene, that we named FrRUNT. FrRUNT is an extremely compact gene, spanning just 3 kb of genomic sequence (Fig. 2). Based on sequence analysis only, FrRUNT appears to have two promoters, with an intron separating the hypothetical distal promoter (P1) and first exon from the main body of the gene. This intron is usually very long in RUNX genes. It is indeed the longest intron observed in FrRUNT, but it is nevertheless very short, spanning just 1372 bp. There is a local concentration of CpG dinucleotides 200–300 bp upstream of exon 2 (Figs. 1, 2), suggesting that an incipient CpG island might function as a proximal promoter (P2). The G+C content is not elevated in this area, in similarity to the CpG islands of the fugu RUNX genes (Fig. 1). The main body of the gene is split into five exons, separated by much shorter introns (69–190 bp long), all of which have canonical splice signals. The longest predicted FrRUNT product is 294 amino acids long, in contrast with the 496 aa, 463 aa and 421 aa observed for FrRUNX1, FrRUNX2 and FrRUNX3, respectively. The small number of exons in FrRUNT leaves little room for alternative splicing by exon skipping, without compromising functionally important domains of the protein. The overall compactness of the gene makes the incorporation of yet undetected exons improbable. Several cryptic splice sites within the exons could enable splicing variants altering exon length.
Figure 2.
FrRUNT sequence detail, showing its compact organization and its interlocking with the SUPT3H gene. The two genes are disposed in opposite orientations. The inverted lettering for SUPT3H denotes translation from the reverse strand. The two putative promoters and the extent of the runt domain (RD) are indicated; x1–x6 denote the six exons. The black triangle points at the asparagine residue absent from all previously known runt domain proteins.
FrRUNT is exceptional in that the length of the runt domain (131 residues) varies from the universally conserved 130 amino acids, due to the introduction of an asparagine residue after position 47 in the RD (Fig. 2). This appears to be the first report of such a mutation within this highly conserved domain. We also noted that the RD sequence of the tunicate Oikopleura dioica (AAS21356 in GenBank) has an insertion at the same position (of two amino acids, a proline and an isoleucine). Comparison to the published structure of this domain [21] shows that this variable region is located in loop L4, opposite the DNA-binding region, i.e. in the location least likely to disrupt the structure of the protein.
A surprising observation is that FrRUNT is "interlocked" with FrSUPT3H (Fig. 2), the gene orthologous to the human transcription factor SUPT3H [22]: The first exon of FrRUNT is located within the first intron of FrSUPT3H, and vice-versa. In the human genome, though, SUPT3H is interlocked with RUNX2, as shown in [13]. We discuss the puzzles posed by these differences in genomic organization below.
Four RD genes in Tetraodon and in zebrafish
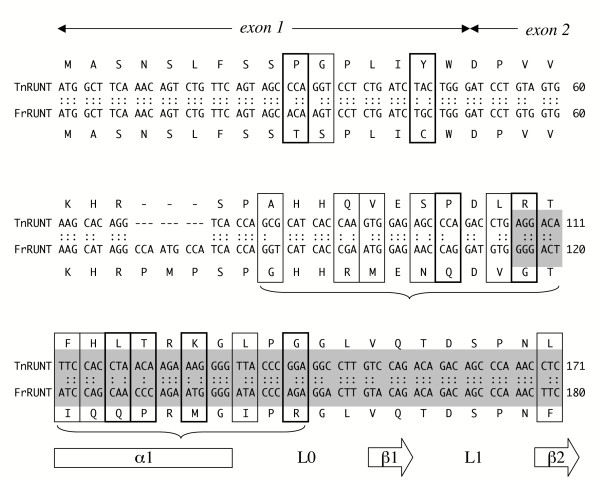
A genome draft of another pufferfish species, Tetraodon nigroviridis, has been released [23]. A computational search into this draft reveals four RD genes with clear orthologous relationships with the four fugu RD genes. We call them TnRUNX1, TnRUNX2 and TnRUNX3, and TnRUNT (Table 2). The RD portion of TnRUNT has been deposited in the EMBL database (accession CAG00330); in this work we report the complete gene structure of TnRUNT, which is similar to that of FrRUNT. In further similarity with the fugu genomic organization, TnRUNT is "interlocked" with the Tetraodon ortholog of SUPT3H (not shown). The three RUNX gene pairs are conserved between fugu and Tetraodon (Table 2) and display a larger percentage of protein identity than nucleotide identity, indicating a prevalence of conservative substitutions. In contrast, TnRUNT is only 83.3% identical to FrRUNT at the protein level, less than their nucleotide identity. Indeed, a striking series of non-synonymous mutations has created a highly divergent segment (only eight identical amino acids out of twenty-two) including the N-terminus of the runt domain (Fig. 3). The strong and unexpected divergence is not the result of local low sequence quality, as tested by examining the relevant Tetraodon entries from the NCBI Trace Archive, and by resequencing the fugu gene. The differences between TnRUNT and FrRUNT do not modify the Ig-like β-sandwich core of the runt domain [21], which is highly conserved as expected. Therefore, the structural integrity of the runt domain appears not to be compromised. We can only speculate that the extensive variation in its N-terminus may reflect species-specific constraints.
Table 2.
The four Tetraodon nigroviridis RD genes, and their identity levels to their respective fugu orthologs.
| Tetraodon gene | Identity | ||||
| Name | Location | Size | %G+C | Nucleotide | Protein |
| TnRUNT | SCAF14597 | 2.5 | 42.3% | 87.5% | 83.3% |
| TnRUNX1 | SCAF15084 | (*) 6.0 | 44.1% | 93.3% | 97.0% |
| TnRUNX2 | SCAF14590 | (*) 31.8 | 44.4% | 96.4% | 99.4% |
| TnRUNX3 | SCAF15009 | (*) 36.6 | 45.3% | 94.6% | 98.1% |
Sizes are expressed in kb. Asterisks denote minimal sizes due to the presence of gaps within the genes; the gene structure for TnRUNX1 is incomplete at its 3' end.
Figure 3.
Localized high divergence between TnRUNT and FrRUNT. This alignment between the first 171 bp of the predicted TnRUNT coding sequence, with the first 180 bp of FrRUNT, shows a cluster of mutations (curly brackets) around the N-terminus of the runt domain (gray background). Non-conservative mutations (negative BLOSUM scores) are surrounded with thick black frames; thin frames enclose conservative mutations, and synonymous mutations are not indicated. The first sixteen codons derive from exon 1, the rest of the alignment from exon 2. The secondary structures indicated (alpha helix, beta strands and loops) are after [21].
We also searched for RD genes in the genome draft of the zebrafish, Danio rerio [24]. Orthologs of RUNX1-3 are present, but no ortholog of FrRUNT could be found. This could be due to the incompleteness of the draft sequence. On the other hand, there are two copies of RUNX2, RUNX2A and RUNX2B, which have been shown to have somewhat different patterns of expression [25]. We next analyze the phylogenetic relationships between all the observed RD genes.
Phylogenetic distribution of runt domain genes
We performed exhaustive computational searches for RD genes, and in particular for potential orthologs of FrRUNT, using the available drafts of the human, chimp, mouse, rat, dog, chicken and frog genomes. In all cases, we identified three clear matches corresponding to orthologs of the three RUNX genes. None of these genomes included a potential ortholog of the fugu/Tetraodon RUNT gene. In principle, such orthologs might be found in the future within current sequencing gaps or heterochromatic regions, but considering the virtually finished human genome, and the combined coverage of all the genome drafts, we can infer that the RUNT gene is absent in mammals, and probably in all tetrapods.
Using representative protein sequences of the RUNX and RUNT genes (see Methods), we reconstructed a molecular tree (Fig. 4, top left) showing the relationship between the three RUNX proteins, FrRUNT and TnRUNT, and the runt proteins of Ciona intestinalis and Branchiostoma floridae (amphioxus). In this analysis, the FrRUNT protein is nearly equidistant from the three RUNX proteins and amphioxus RUNT (~72% identical, see Table 3). In comparison, the identity level between the RUNX proteins is 90%–98% in the same region (Table 3), and they are 90%–95% identical to amphioxus RUNT. Therefore, while the amphioxus RUNT protein is very closely related to the vertebrate RUNX, the pufferfish RUNT proteins are significantly more divergent.
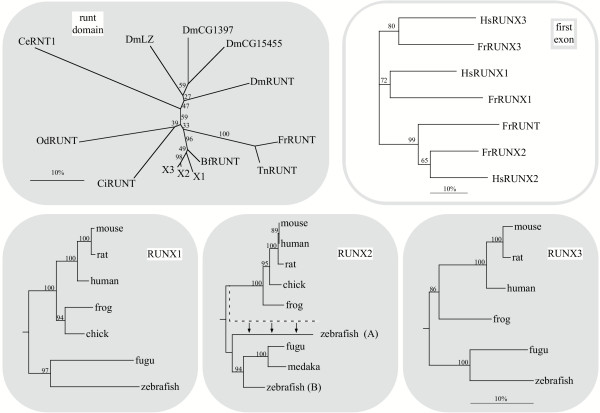
Figure 4.
Phylogenetic reconstruction of runt domain evolution. Top left: unrooted tree of RD proteins based on the runt domain; X1, X2 and X3 denote the RUNX clades, BfRUNT, FrRUNT, TnRUNT, CiRUNT, OdRUNT, SpRUNT and CeRNT1 represent RUNT proteins from amphioxus, fugu, Tetraodon, Ciona, Oikopleura dioica, sea urchin and C. elegans, respectively, while taxa named Dm- represent D. melanogaster RD proteins. Top right: unrooted tree of first exons of human and fugu runt domain genes. Bottom: expanded species trees for the three RUNX orthologous groups. For all trees, numbers indicate percent bootstrap support; the horizontal bars indicate 10% divergence along each branch. White and gray backgrounds indicate comparisons at the nucleotide and amino acid level, respectively. The dashed branch in the RUNX2 panel represents the position of zebrafish (A) prior to AsaturA analysis. The arrows indicate the change effected by this correction.
Table 3.
Identity matrix.
| FrRUNX2 | FrRUNX3 | HsRUNX1 | HsRUNX2 | HsRUNX3 | FrRUNT | |
| FrRUNX1 | 89.7% | 91.4% | 95.7% | 89.7% | 89.7% | 72.7% |
| FrRUNX2 | 95.7% | 92.3% | 96.6% | 96.6% | 71.2% | |
| FrRUNX3 | 94.8% | 97.4% | 97.4% | 72.7% | ||
| HsRUNX1 | 94.0% | 94.0% | 72.7% | |||
| HsRUNX2 | 98.3% | 72.7% | ||||
| HsRUNX3 | 72.7% |
Percentages of identity between the human and fugu runt domains.
A difficulty has been documented in phylogenetic reconstruction of gene families with anciently duplicated genes [26], in which saturation of frequently-mutating amino acids leads to erroneous "outgroup topologies". In these incorrect topologies, the duplication event appears to be more ancient than supported by the data, which in turn suggest the existence of lineage-specific gene loss events. We tested our phylogenetic reconstruction using the program ASaturA [26], which identifies and suppresses saturated amino acids, thereby correcting the affected tree topology. This analysis did not modify the location of FrRUNT and TnRUNT in our reconstruction, suggesting that mutational saturation is not causing the observed divergence age of the pufferfish RUNT genes.
In this protein-level comparison of the conserved runt domains, the pufferfish RUNT proteins appear to be surprisingly ancient, predating the divergence between craniates (including vertebrates) and cephalochordates (including amphioxus). On the other hand, when comparing the nucleotide sequences of the first exon from each human and fugu runt domain gene, we observed that FrRUNT is more closely related to RUNX2 (Fig. 4, top right), suggesting the possibility of a recombination event between these genes (see discussion below).
We also studied the relationships between the RUNX1, RUNX2 and RUNX3 orthologs in several vertebrates and found the species trees to be largely as expected (Fig. 4, bottom row). One of the two zebrafish RUNX2 protein sequences (RUNX2A) appeared to be slightly more closely related to tetrapod RUNX2 genes than to the other RUNX2 genes in fish species, including zebrafish RUNX2B (dashed branch in Fig. 4, RUNX2 panel). We tested this result using ASaturA [26], and found it to be an artifact of mutational saturation: in the corrected tree (Fig. 4), RUNX2A is more closely related to the other fish RUNX2 genes.
Comparative genomics of runt domain genes
Four RD genes have been identified in Drosophila [14], more similar to each other than to the vertebrate RUNX genes: they represent an independent family expansion in insects. The four Drosophila RD genes are all linked on chromosome X. Moreover, three of these genes are clustered within a 150 kb region. There is no linkage of RD genes, however, in the human genome: each gene is on a different chromosome. Their genomic environments usually show some conservation: the three human RUNX genes are followed by CLIC genes in the complementary strand (Fig. 5), and linked to members of the DSCR1 family. The RD genes appear not to be clustered in the fugu genome, though this conclusion is limited by the fragmentary nature of the current genome draft. All four fugu RD genes are flanked by at least one non-RD gene on each side. Fugu RUNX genes are followed by CLIC genes except for FrRUNX1, but a CLIC gene is located ~55 kb upstream of FrRUNX1 and in the same orientation. This organization could have arisen by an inversion event in the fugu lineage. To ensure a misassembly did not cause this apparent inversion, we performed a 3x shotgun sampling of the BAC clone OML73850, which spans the range 1–97413 of scaffold 682. This quality control step failed to uncover any misassemblies, and confirmed the genomic organization observed in this fugu scaffold. FrRUNT does not appear to be linked to any CLIC gene, and no DSCR1 family members can be discerned near any of the fugu RD genes.
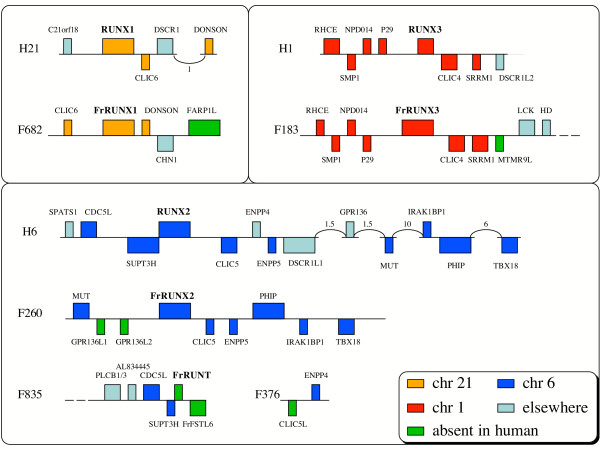
Figure 5.
Comparative genomic organization. The genomic neighborhoods of the human and fugu RD genes are compared and contrasted, highlighting synteny conservation (same color between the two species) and gene loss (green features). Feature widths represent rough gene size but are not exactly proportional to gene lengths; likewise intergenic distances are not meant to be precise. Arcs indicate larger genomic distances with one or more intervening genes (not displayed). The numbers associated with the arcs represent the distance in Mb.
When studying the wider genomic environments of fugu and human RD genes, we observed significant synteny conservation, in agreement with the observations reported for chromosome X genes [27]. Indeed, when comparing each of the genes neighboring fugu RUNX genes to the human genome, we find that their orthologs tend to be located in the corresponding human chromosome, e.g. most of the genes linked to FrRUNX3 have orthologs on human chromosome 1, where human RUNX3 resides (Fig. 5). Some inversion events can be inferred, e.g. one involving the genes PHIP and IRAK1BP1 and another involving MUT. Gene order and orientation has changed, and intergenic distances have changed drastically, but the overall gene synteny is largely preserved, lending support to the assignments of orthology between the human and fugu RUNX gene pairs.
An exception to the conservation of synteny involves the genes CDC5L and SUPT3H, which in the human genome are found immediately upstream of RUNX2, but in fugu are instead located upstream of FrRUNT, including the first-exon interlocking with SUPT3H mentioned earlier. Evidence points at a larger duplication in fishes, encompassing at least CDC5L, SUPT3H, RUNX2, CLIC5, ENPP4 and ENPP5, followed by differential gene loss. One duplicate copy would have retained FrRUNX2, CLIC5 and ENPP5 (see Fig. 5), while the other copy (currently represented by scaffolds 835 and 376) would have retained CDC5L, SUPT3H, a second RD gene, a second copy of CLIC5 (CLIC5L) and ENPP4. The presence of remnants of SUPT3H upstream of FrRUNX2 [13], and of two copies of RUNX2 in zebrafish, lends further support to this hypothesis. Thus, comparative genomic analysis of human, fugu and zebrafish suggests that FrRUNT may be a derivative form of a duplicated RUNX2 gene. However, this contradicts the conclusions from phylogenetic analysis or the protein sequences; we discuss this contradiction below.
A new family of follistatin-like genes
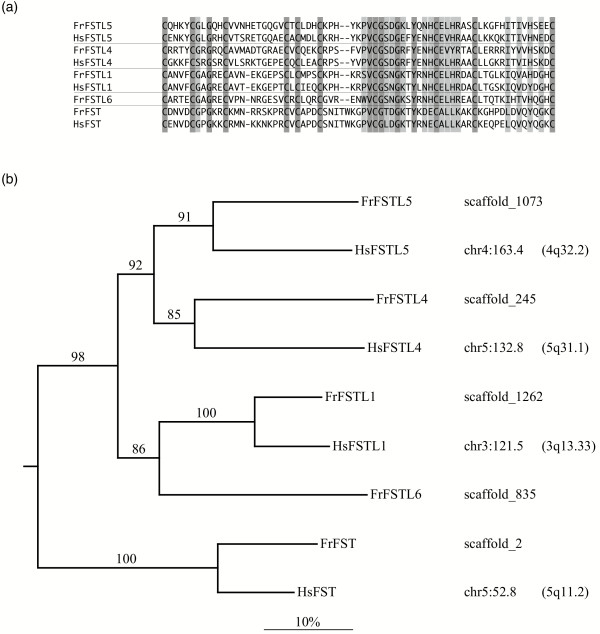
We find several fugu genes for which no human ortholog can be discerned (green features in Fig. 5), among them FrRUNT. Immediately downstream to FrRUNT we identified a novel gene (FrFSTL6) from the follistatin family, most closely related to FSTL1. A detailed computational search for additional sequences of this family identified two novel, large human follistatin-like genes, which we called FSTL4 and FSTL5. We then found clear orthologs for both in the fugu genome (FrFSTL4 and FrFSTL5, respectively). The genes in this family share a Kazal-type cysteine-rich domain (Fig. 6a) and a calcium-binding EF-hand domain (not shown).
Figure 6.
Comparison of selected follistatin-like proteins. FST: follistatin. (a) Multiple alignment of the cysteine-rich domain. Darker shading indicates perfect conservation, lighter shading indicates positions that can be explained assuming one or two mutations. (b) Neighbor-joining tree rooted using human osteonectin/SPARC (NP_003109) as outgroup. Numbers on branches indicate the percent support in 1000 bootstrap replicates. The horizontal bar indicates 10% divergence along each branch. The column on the right indicates the scaffold in which each gene is located (fugu) or its genomic location in Mb coordinates and cytogenetic band (human).
Having characterized the complete gene family in both human and fugu, we performed a phylogenetic reconstruction based on the conserved Kazal-type domain (Fig. 6b). FrFSTL6 appears to have no ortholog in the human genome, nor could we identify a potential ortholog in the mouse and frog genomes. This suggests that FrFSTL6 was also lost in the tetrapod lineage. It is reasonable to hypothesize that the neighboring FrRUNT and FrFSTL6 genes were lost in a single deletion event.
Discussion
We have taken advantage of the availability of genomic drafts for several vertebrate species, including the finished human genome, to identify the orthologs of all currently known runt-domain (RD) genes, as well as a novel member of this small gene family. Both pufferfish species (Takifugu rubripes and Tetraodon nigroviridis) have four RD genes; since these genomes are only available as draft assemblies, additional RD genes might be found when the finished genomic sequences are made available.
The function of the novel FrRUNT/TnRUNT gene is currently unknown. Based on the phylogenetic analysis, this novel gene appears to represent an ancestral form of the RD family in vertebrates, subsequently lost in the tetrapod lineage. It is therefore surprising that its gene structure, and not that of RUNX2 as in humans, is interlocked with the SUPT3H gene. Based on the comparative genomics analysis alone, one could hypothesize that FrRUNT is simply a derivative form of RUNX2, i.e. the ortholog of zebrafish RUNX2A. In this case, though, one would expect FrRUNT to be more similar to FrRUNX2 than it is to either FrRUNX1 or FrRUNX3, but in terms of amino acid sequence similarity, it appears to be equidistant from the three RUNX genes. This discrepancy might be explained by invoking accelerated evolution of the pufferfish RUNT genes, perhaps as a lineage-specific adaptation. Typically, nucleotide sequences diverge much faster than amino acid sequences, and the first exons of RD genes are significantly less conserved than the runt domain itself, on which we based our phylogenetic analysis. Therefore, we find it hard to sustain that, while the first exon of FrRUNT maintains its nucleotide similarity to the first exon of RUNX2, the amino acid sequence of the (normally highly conserved) runt domain itself has diverged at such an accelerated pace. Furthermore, we found this not to be an artifact of mutational saturation [26]. A similar situation is observed for the neighboring FSTL6 gene: parsimony considerations could lead one to assume that FSTL6 is a fish-specific duplicate of FSTL1, though contradicting the phylogenetic reconstruction of the evolution of this gene family (Fig. 6). This situation could again be explained by assuming accelerated evolution of FSTL6, but we consider this to be a remarkable coincidence.
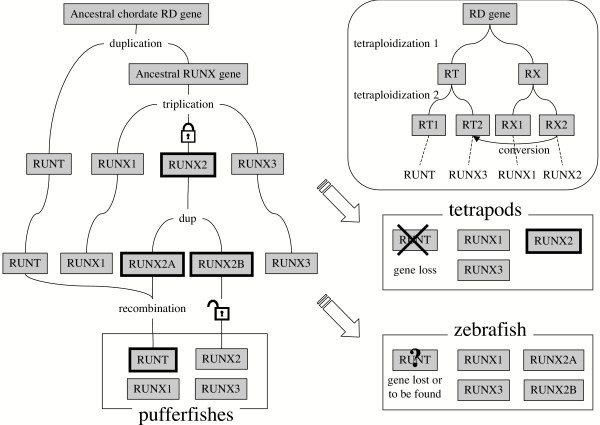
The conundrum is whether FrRUNT is an ancestral form, or is derived from RUNX2. Both hypotheses contradict part of the available data. We propose here a third hypothesis, in the form of an evolutionary history (see Fig. 7): An ancestral RD gene duplicated in chordates, after divergence from sea urchin, which has a single RD gene [17]. One of the two resulting RD genes became the RUNX family founder, which expanded by triplication, and one of the three RUNX genes (namely RUNX2) became interlocked with SUPT3H. After the teleost/tetrapod divergence, a regional duplication in teleosts created a second copy of RUNX2 and its neighboring genes. In the tetrapod lineage, the ancestral RUNT gene was lost, in conjunction with FSTL6. In the pufferfish lineage, the RUNT gene replaced most of RUNX2A, perhaps by recombination (Fig. 8). This is supported by the clear similarity between the first exons of FrRUNT and RUNX2. The copy of SUPT3H interlocked with RUNX2B, apparently superfluous, is being lost by gradual degradation, and only small fragments of it remain [13]. This scenario is compatible with all the data observed. While it posits a small number of additional evolutionary events, it does not involve highly improbable events like the accelerated evolution of a normally highly conserved protein structural domain. Interestingly, the first duplication event could correspond to the first round of vertebrate genome tetraploidization [28]. A second round of tetraploidization in the ancestral vertebrate could have produced a set of four paralogous runt domain genes, and a hypothetical gene conversion event may have led to the current complement of three RUNX genes (Fig. 7 inset). Conversion between paralogous copies of genes derived from tetraploidization events has been demonstrated [29].
Figure 7.
Hypothesis for the evolution of vertebrate RD genes. Features with thick edges represent RD genes interlocked with SUPT3H family members. The closed padlock icon represents the interlocking event between the RUNX2 and SUPT3H, and the open padlock represents the degradation of a copy of SUPT3H, releasing RUNX2 from interlocking. Not enough information is yet available to establish which of the zebrafish RUNX2 genes is interlocked with a SUPT3H gene. "dup": duplication. Top right inset: Hypothesis of how the ancestral RD duplication could map to the first vertebrate tetraploidization event. RT and RX represent the ancestral forms of RUNT and RUNX genes.
Figure 8.
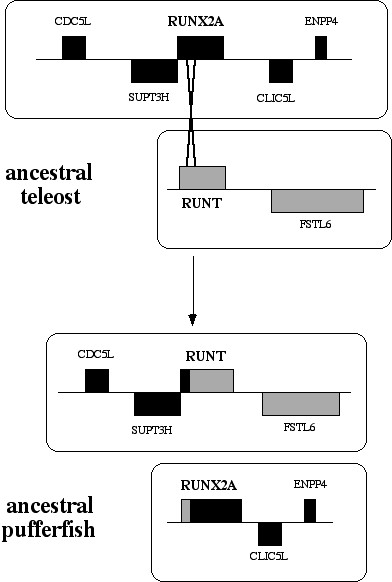
The hypothesized recombination event between the ancestral RUNX2A and RUNT genes. After the recombination, the RUNT gene is linked to SUPT3H and has a RUNX2-like first exon. The derivative RUNX2A gene has not yet been observed in pufferfishes, and may have been lost in evolution.
Under the proposed scenario, none of the three extant RUNX genes in mammals represents the ancestral vertebrate RD form. Rather, these derivative genes coexisted with an additional RUNT gene and still do so in teleost genomes. The single known RD gene in amphioxus is more similar to the vertebrate RUNX genes than it is to FrRUNT. It is possible, therefore, that cephalochordates (including amphioxus) have a second RD gene, short and divergent enough to escape experimental detection by DNA hybridization [17]. Why was one of the ancestral RD genes lost in the tetrapod lineage? The three RUNX genes bind to the same DNA motif and modify the expression of target genes through recruitment of transcriptional modulators, which are also shared [30]; functional differences between the three RUNX genes are attained by way of tightly regulated spatiotemporal expression patterns. We hypothesize that the ancestral RUNT gene became inessential to amniotes by functional reprogramming of the remaining three RUNX genes. Its loss would therefore represent an example of evolution by reduction in complexity. In pufferfishes, the hypothesized recombination event would have placed the RUNT gene under the regulatory control of the former RUNX2A promoter. The viability of such a sudden regulatory change would in turn suggest a significant level of functional redundancy among the RD genes.
Conclusions
We identified a fourth runt domain gene in the fugu genome, which appears to represent either a pufferfish-specific, fast-evolving derivative of RUNX2, or a direct descendant of the ancestral chordate RUNT gene. We find the latter hypothesis more reasonable. This novel gene evolved in parallel with the vertebrate RUNX genes, and while it has been preserved in pufferfishes, it appears to have been lost entirely in tetrapods. This suggests that the ancestral vertebrate was more complex than previously suspected.
By studying a very limited set of fugu genomic regions, namely the scaffolds related to RD genes, we have identified seven apparently functional fugu genes that are absent from the human genome (Fig. 5), and were probably lost early in tetrapod history. In the process of identifying relevant homologs for one of these genes (FrFSTL6), we have identified a new family of follistatin-like genes in the human genome. Phylogenetic analysis of the RD protein sequences led to results that contradict those derived from comparative genomics, but we showed that the two could be reconciled into a coherent evolutionary model. These results underscore the importance of obtaining complete genomic sequences of strongly divergent vertebrates, and the value to be derived by performing detailed and integrated analyses of their gene complements.
Methods
Search for RD genes
We used the human RUNX1, RUNX2 and RUNX3 proteins (SwissProt entries Q01196, Q13950 and Q13761, respectively) as queries in a TFASTY [31] search into the Takifugu rubripes "assembly3" genome draft [32] released after publication of the fugu genome [33]. These data have been provided freely by the Fugu Genome Consortium for use in this publication only. This search resulted in the unambiguous identification of four complete RD genes in scaffolds 183, 260, 682 and 835. Scaffold 25789 is nearly identical with range 115299–115845 of scaffold 183, partially overlapping the last exon of FrRUNX3. No further evidence was found for an additional RUNX3 gene: we conclude that scaffold 25789 is an assembly artifact. We similarly searched the genome drafts for Tetraodon nigroviridis produced by the Whitehead Institute and the Genoscope [23], and Danio rerio (Zv1/06 assembly, which was produced by the Zebrafish Sequencing Group at the Sanger Institute [24]. We analyzed, visualized and annotated all resulting genomic sequences using the GESTALT Workbench [20,34], and produced a detailed gene model for each RD gene. Lacking cDNA or EST data, we reconstructed the putative gene structures by maximizing similarity to known RD proteins. Genomic sequence data have been submitted to GenBank with accessions AY739093-AY739096; the predicted sequences for fugu RD proteins have accessions AAU14190-AAU14193.
In a second round of analysis, we used the newly identified RD genes as queries for renewed TFASTY searches of the genome drafts of human (July 03), mouse (February 03), Xenopus [35] (December 03 assembly) and Ciona intestinalis [36]. We also used BLAT to search into the updated "freezes" of human (May 04), chimp (November 03), mouse (May 04), rat (June 03), dog (July 04), and chicken (Feb 04).
Sequence finishing
Large insert clones spanning the gaps in scaffolds from the version 3 assembly were identified by BLAST searches against the BAC/cosmid database in the v.3.0 JGI website [37]. Cosmids (cloned in Lawrist4) were grown in LB media with kanamycin at 37 ° C for 14 hrs, and DNA was prepared on the Autogen 740 DNA Isolation system in accordance with the manufacturer's instructions. BACs (cloned in pBeloBAC 11) were similarly prepared by growing in media with chloramphenicol. Primers were designed in both directions, across all gaps. Oligonucleotide-directed sequencing from clones and Polymerase Chain Reaction (PCR) methods were used to fill the gaps. PCR amplification was performed on spanning BAC/cosmid or genomic DNA of Takifugu rubripes, generously provided by Dr. Greg Elgar. PCR products were purified with sephacryl (Amersham Pharmacia) and sequenced directly using Applied Biosystems Big dye terminator kit reagents. Whenever necessary, additional pairs of primers were designed for oligonucleotide-directed sequencing to close gaps. Shotgun sequencing data was obtained from the BAC clone OML73850 (b193C08) for part of scaffold 682. OML73850 was fragmented by sonication, end-repaired and electrophoresed to select insert size of 2–5 kb. Insert was ligated into pUC18 vector, transformed and plasmid DNA was made using Eppendorf – 5 Prime PERFECTprep robot and sequenced from both ends. Assembly was carried out using Phrap [38]. Analysis of the resulting sequence shows that OML73850 spans the first 97413 bp of scaffold 682, and links it to scaffold 4260. The additional sequence data generated in-house were combined with the consensus sequences of the scaffolds produced by the JGI WGS assembly v.3 for the purpose of producing a contiguous sequence for each scaffold.
Phylogenetic reconstruction
The sequences were aligned using ClustalW [39]. Phylogenetic trees were built using the neighbor-joining algorithm [40] and tested with 1000 rounds of bootstrapping. Graphics were produced with TreeView [41]. Since full-length protein sequences cannot be reliably aligned for extremely divergent RD genes, we used only the runt domain to reconstruct the relationship between the pufferfish RUNT, human RUNX1, RUNX2 and RUNX3 (NP_00175, NP_033950 and NP_004341, respectively), ciona (C. intestinalis) RUNT, Oikopleura dioica RUNT (AAS21356), amphioxus (B. floridae) RUNT (AY146617), sea urchin S. purpuratus RUNT (NP_999779) and the four Drosophila melanogaster RD sequences (NP_523424, NP_511099, NP_572693 and NP_608398). The tree was rooted using the C. elegans RUN protein (AB027412) as outgroup. We further excluded the first thirteen amino acids of the runt domain, to avoid the topological distortion expected in this region from the highly divergent pufferfish RUNT sequences. For the separate phylogenetic trees of the three RUNX genes, we used the complete protein sequences, with gap opening and extension penalties of 5 and 0.1, respectively. ASaturA analyses were performed using PAM250, Kimura's correction and a cutoff value of 9, with 1000 rounds of bootstrap. The first exons of human and fugu runt domain genes were compared at the nucleotide level. For each exon, we selected the range from 30 nucleotides upstream of the ATG codon, to 15 downstream of the splicing donor site, i.e. 103 nucleotides for each RUNX gene and 94 nucleotides for FrRUNT.
Authors' contributions
GG conceived of the study, performed the bioinformatics analyses and prepared the manuscript. AK generated the sequence data required for finishing. LH, the laboratory director, provided guidance and contributed to the preparation of the manuscript. LR supervised the Institute for Systems Biology's contributions to the Pufferfish Finishing Consortium and contributed to the preparation of the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgments
We wish to thank Yoram Groner, Jared Roach, Irit Rubin, Arian Smit and Raafat El-Gewely for helpful discussions and support, Sydney Brenner for organizing the Pufferfish Finishing Consortium, the Joint Genome Institute for their public release of fugu assembly v.3 and for providing BAC and cosmid libraries, and the Institute for Molecular and Cell Biology at Singapore for funding fugu sequencing in our laboratory.
Contributor Information
Gustavo Glusman, Email: cgojp@systemsbiology.org.
Amardeep Kaur, Email: akaur@SystemsBiology.org.
Leroy Hood, Email: lhood@SystemsBiology.org.
Lee Rowen, Email: lrowen@SystemsBiology.org.
References
- Kania MA, Bonner AS, Duffy JB, Gergen JP. The Drosophila segmentation gene runt encodes a novel nuclear regulatory protein that is also expressed in the developing nervous system. Genes Dev. 1990;4:1701–1713. doi: 10.1101/gad.4.10.1701. [DOI] [PubMed] [Google Scholar]
- Coffman JA. Runx transcription factors and the developmental balance between cell proliferation and differentiation. Cell Biol Int. 2003;27:315–324. doi: 10.1016/S1065-6995(03)00018-0. [DOI] [PubMed] [Google Scholar]
- Levanon D, Negreanu V, Bernstein Y, Bar-Am I, Avivi L, Groner Y. AML1, AML2, and AML3, the human members of the runt domain gene-family: cDNA structure, expression, and chromosomal localization. Genomics. 1994;23:425–432. doi: 10.1006/geno.1994.1519. [DOI] [PubMed] [Google Scholar]
- Ogawa E, Maruyama M, Kagoshima H, Inuzuka M, Lu J, Satake M, Shigesada K, Ito Y. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc Natl Acad Sci U S A. 1993;90:6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghozi MC, Bernstein Y, Negreanu V, Levanon D, Groner Y. Expression of the human acute myeloid leukemia gene AML1 is regulated by two promoter regions. Proc Natl Acad Sci U S A. 1996;93:1935–1940. doi: 10.1073/pnas.93.5.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangsow C, Rubins N, Glusman G, Bernstein Y, Negreanu V, Goldenberg D, Lotem J, Ben-Asher E, Lancet D, Levanon D, Groner Y. The RUNX3 gene - sequence, structure and regulated expression. Gene. 2001;279:221–232. doi: 10.1016/S0378-1119(01)00760-0. [DOI] [PubMed] [Google Scholar]
- Rini D, Calabi F. Identification and comparative analysis of a second runx3 promoter. Gene. 2001;273:13–22. doi: 10.1016/S0378-1119(01)00579-0. [DOI] [PubMed] [Google Scholar]
- Levanon D, Glusman G, Bangsow T, Ben-Asher E, Male DA, Avidan N, Bangsow C, Hattori M, Taylor TD, Taudien S, Blechschmidt K, Shimizu N, Rosenthal A, Sakaki Y, Lancet D, Groner Y. Architecture and anatomy of the genomic locus encoding the human leukemia-associated transcription factor RUNX1/AML1. Gene. 2001;262:23–33. doi: 10.1016/S0378-1119(00)00532-1. [DOI] [PubMed] [Google Scholar]
- Miyoshi H, Ohira M, Shimizu K, Mitani K, Hirai H, Imai T, Yokoyama K, Soeda E, Ohki M. Alternative splicing and genomic structure of the AML1 gene involved in acute myeloid leukemia. Nucleic Acids Res. 1995;23:2762–2769. doi: 10.1093/nar/23.14.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon D, Bettoun D, Harris-Cerruti C, Woolf E, Negreanu V, Eilam R, Bernstein Y, Goldenberg D, Xiao C, Fliegauf M, Kremer E, Otto F, Brenner O, Lev-Tov A, Groner Y. The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. Embo J. 2002;21:3454–3463. doi: 10.1093/emboj/cdf370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M, Terry A, Hu M, O'Hara M, Blyth K, Baxter E, Cameron ER, Onions DE, Neil JC. Proviral insertions induce the expression of bone-specific isoforms of PEBP2alphaA (CBFA1): evidence for a new myc collaborating oncogene. Proc Natl Acad Sci U S A. 1997;94:8646–8651. doi: 10.1073/pnas.94.16.8646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracey WDJ, Pepling ME, Horb ME, Thomsen GH, Gergen JP. A Xenopus homologue of aml-1 reveals unexpected patterning mechanisms leading to the formation of embryonic blood. Development. 1998;125:1371–1380. doi: 10.1242/dev.125.8.1371. [DOI] [PubMed] [Google Scholar]
- Eggers JH, Stock M, Fliegauf M, Vonderstrass B, Otto F. Genomic characterization of the RUNX2 gene of Fugu rubripes. Gene. 2002;291:159–167. doi: 10.1016/S0378-1119(02)00592-9. [DOI] [PubMed] [Google Scholar]
- Rennert J, Coffman JA, Mushegian AR, Robertson AJ. The evolution of runx genes I. A comparative study of sequences from phylogenetically diverse model organisms. BMC Evolutionary Biology. 2003;3:4. doi: 10.1186/1471-2148-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daga A, Karlovich CA, Dumstrei K, Banerjee U. Patterning of cells in the Drosophila eye by Lozenge, which shares homologous domains with AML1. Genes Dev. 1996;10:1194–1205. doi: 10.1101/gad.10.10.1194. [DOI] [PubMed] [Google Scholar]
- Bae SC, Lee J. cDNA cloning of run, a Caenorhabditis elegans Runt domain encoding gene. Gene. 2000;241:255–258. doi: 10.1016/S0378-1119(99)00488-6. [DOI] [PubMed] [Google Scholar]
- Stricker S, Poustka AJ, Wiecha U, Stiege A, Hecht J, Panopoulou G, Vilcinskas A, Mundlos S, Seitz V. A single amphioxus and sea urchin runt-gene suggests that runt-gene duplications occurred in early chordate evolution. Dev Comp Immunol. 2003;27:673–684. doi: 10.1016/S0145-305X(03)00037-5. [DOI] [PubMed] [Google Scholar]
- Strippoli P, D'Addabbo P, Lenzi L, Giannone S, Canaider S, Casadei R, Vitale L, Carinci P, Zannotti M. Segmental paralogy in the human genome: a large-scale triplication on 1p, 6p, and 21q. Mammalian Genome. 2002;13:456–462. doi: 10.1007/s00335-001-2157-0. [DOI] [PubMed] [Google Scholar]
- Nam S, Jin YH, Li QL, Lee KY, Jeong GB, Ito Y, Lee J, Bae SC. Expression pattern, regulation, and biological role of runt domain transcription factor, run, in Caenorhabditis elegans. Mol Cell Biol. 2002;22:547–554. doi: 10.1128/MCB.22.2.547-554.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glusman G, Lancet D. GESTALT: a workbench for automatic integration and visualization of large-scale genomic sequence analyses. Bioinformatics. 2000;16:482–483. doi: 10.1093/bioinformatics/16.5.482. [DOI] [PubMed] [Google Scholar]
- Backstrom S, Wolf-Watz M, Grundstrom C, Hard T, Grundstrom T, Sauer UH. The RUNX1 Runt domain at 1.25A resolution: a structural switch and specifically bound chloride ions modulate DNA binding. J Mol Biol. 2002;322:259–272. doi: 10.1016/S0022-2836(02)00702-7. [DOI] [PubMed] [Google Scholar]
- Yu J, Madison JM, Mundlos S, Winston F, Olsen BR. Characterization of a human homologue of the Saccharomyces cerevisiae transcription factor spt3 (SUPT3H) Genomics. 1998;53:90–96. doi: 10.1006/geno.1998.5500. [DOI] [PubMed] [Google Scholar]
- Tetraodon Genome Browser http://www.genoscope.cns.fr/externe/tetraodon
- Zebrafish assembly 6 download ftp://ftp.ensembl.org/pub/traces/zebrafish/assembly/assembly06
- Flores MV, Tsang VW, Hu W, Kalev-Zylinska M, Postlethwait J, Crosier P, Crosier K, Fisher S. Duplicate zebrafish runx2 orthologues are expressed in developing skeletal elements. Gene Expr Patterns. 2004;4:573–581. doi: 10.1016/j.modgep.2004.01.016. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, Frickey T, Taylor JS, Meyer A. Dealing with saturation at the amino acid level: a case study based on anciently duplicated zebrafish genes. Gene. 2002;295:205–211. doi: 10.1016/S0378-1119(02)00689-3. [DOI] [PubMed] [Google Scholar]
- Grutzner F, Crollius HR, Lutjens G, Jaillon O, Weissenbach J, Ropers HH, Haaf T. Four-hundred million years of conserved synteny of human Xp and Xq genes on three Tetraodon chromosomes. Genome Res. 2002;12:1316–1322. doi: 10.1101/gr.222402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin LG. Evolution of the vertebrate genome as reflected in paralogous chromosomal regions in man and the house mouse. Genomics. 1993;16:1–19. doi: 10.1006/geno.1993.1133. [DOI] [PubMed] [Google Scholar]
- Angers B, Gharbi K, Estoup A. Evidence of gene conversion events between paralogous sequences produced by tetraploidization in Salmoninae fish. J Mol Evol. 2002;54:501–510. doi: 10.1007/s00239-001-0041-x. [DOI] [PubMed] [Google Scholar]
- Levanon D, Groner Y. Structure and regulated expression of mammalian RUNX genes. Oncogene. 2004;23:4211–4219. doi: 10.1038/sj.onc.1207670. [DOI] [PubMed] [Google Scholar]
- Pearson WR, Wood T, Zhang Z, Miller W. Comparison of DNA sequences with protein sequences. Genomics. 1997;46:24–36. doi: 10.1006/geno.1997.4995. [DOI] [PubMed] [Google Scholar]
- JGI Fugu v3.0 Download http://genome.jgi-psf.org/fugu6/fugu6.download.ftp.html
- Aparicio S, Chapman J, Stupka E, Putnam N, Chia JM, Dehal PS, Christoffels A, Rash S, Hoon S, Smit AFA, Gelpke MD, Roach J, Oh T, Ho IY, Wong M, Detter C, Verhoef F, Predki P, Tay A, Lucas S, Richardson S, Smith S, Clark M, Edwards Y, Doggett N, Zharkikh A, Tavtigian SV, Pruss D, Barnstead M, Evans C, Baden H, Powell J, Glusman G, Rowen L, Hood L, Tan YH, Elgar G, Hawkins T, Venkatesh B, Rokhsar D, Brenner S. Whole genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- Glusman G, Lancet D. Visualizing large-scale genomic sequences. IEEE Eng Med Biol Mag. 2001;20:49–54. doi: 10.1109/51.940046. [DOI] [PubMed] [Google Scholar]
- JGI Xenopus download http://genome.jgi-psf.org/pub/JGI_data/Frog
- JGI Ciona v.1 http://genome.jgi-psf.org/ciona4/ciona4.download.ftp.html
- JGI Fugu v3.0 BLAST http://aluminum.jgi-psf.org/prod/bin/blast.fugu6.cgi
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Higgins DG, Thompson JD, Gibson TJ. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Brenner S, Venkatesh B, Yap WH, Chou CF, Tay A, Ponniah S, Wang Y, Tan YH. Conserved regulation of the lymphocyte-specific expression of lck in the Fugu and mammals. Proc Natl Acad Sci U S A. 2002;99:2936–2941. doi: 10.1073/pnas.032680599. [DOI] [PMC free article] [PubMed] [Google Scholar]