Abstract
Arteriovenous malformation (AVM) is a fast-flow, congenital vascular anomaly that may arise anywhere in the body. AVMs typically progress, causing destruction of surrounding tissue and, sometimes, cardiac overload. AVMs are difficult to control; they often re-expand after embolization or resection, and pharmacologic therapy is unavailable. We studied extracranial AVMs in order to identify their biological basis. We performed whole-exome sequencing (WES) and whole-genome sequencing (WGS) on AVM tissue from affected individuals. Endothelial cells were separated from non-endothelial cells by immune-affinity purification. We used droplet digital PCR (ddPCR) to confirm mutations found by WES and WGS, to determine whether mutant alleles were enriched in endothelial or non-endothelial cells, and to screen additional AVM specimens. In seven of ten specimens, WES and WGS detected and ddPCR confirmed somatic mutations in mitogen activated protein kinase kinase 1 (MAP2K1), the gene that encodes MAP-extracellular signal-regulated kinase 1 (MEK1). Mutant alleles were enriched in endothelial cells and were not present in blood or saliva. 9 of 15 additional AVM specimens contained mutant MAP2K1 alleles. Mutations were missense or small in-frame deletions that affect amino acid residues within or adjacent to the protein’s negative regulatory domain. Several of these mutations have been found in cancers and shown to increase MEK1 activity. In summary, somatic mutations in MAP2K1 are a common cause of extracranial AVM. The likely mechanism is endothelial cell dysfunction due to increased MEK1 activity. MEK1 inhibitors, which are approved to treat several forms of cancer, are potential therapeutic agents for individuals with extracranial AVM.
Keywords: arteriovenous malformation, MEK1, MAP2K1, vascular anomaly
Main Text
Arteriovenous malformation (AVM) is a congenital vascular anomaly, comprised of abnormal connections between arteries and veins that are missing normal high-resistance capillary beds (Figure 1).1 Sporadic extracranial AVMs are solitary and may be localized or regional. Rapid blood flow is demonstrable by Doppler ultrasonography. Magnetic resonance imaging reveals signal voids consistent with fast-flow, while angiography shows the early filling of draining veins (Figure 1). With time, arterial to venous shunting causes tissue ischemia that leads to pain, ulceration, bleeding, and destruction of adjacent tissues. Treatment for AVM has been discouraging. Embolization and/or resection are often followed by expansion; there are no drug treatments.2 The purpose of this study was to identify the genetic basis for sporadic, extracranial AVM in an effort to devise a new therapeutic strategy.
Figure 1.
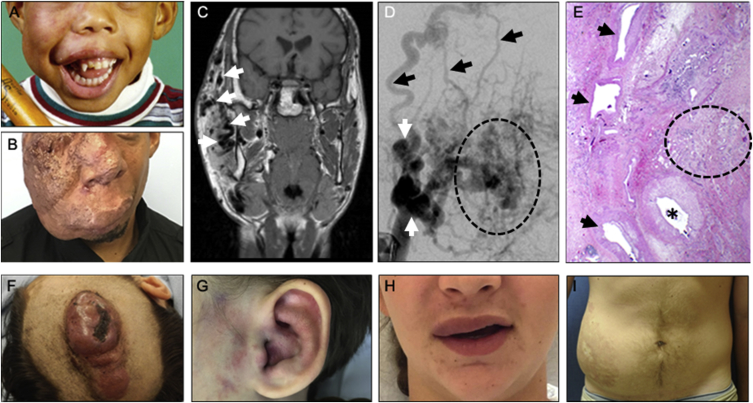
Solitary Extracranial AVMs
(A and B) Photograph of participant 23 during (A) childhood (stage I) and (B) adulthood (stage III). Note progressive growth of the facial AVM.
(C) Coronal magnetic resonance image illustrates the extent of the lesion with multiple signal voids consistent with fast-flow (white arrows).
(D) Angiogram showing tortuous arteries (white arrows) that feed the AVM, the “nidus” (dotted oval) where there are direct communications between numerous small arteries and veins, and early filling of draining veins (black arrows).
(E) Hematoxylin and eosin-stained section of participant 23’s affected tissue, obtained after initial resection. Note the large feeder artery (asterisk), hyper-muscularized veins (arrows), and an area where arteries and veins connect in the absence of a normal capillary bed (dotted oval).
(F) Participant 2, scalp AVM (stage III).
(G) Participant 6, ear AVM (stage I).
(H) Participant 12, upper lip AVM (stage I).
(I) Participant 19, abdominal AVM (stage II).
The Committee on Clinical Investigation at Boston Children’s Hospital approved this study and informed consent was obtained from study participants. Ten AVM specimens that had been collected during a clinically indicated procedure, including matched unaffected tissue specimens from three of the study participants, had DNA extracted using the DNeasy Blood & Tissue Kit (QIAGEN); saliva DNA was extracted using the prepIT-L2P extraction kit (DNA Genotek). Five affected tissue samples underwent deep WES and another five had WES and WGS on Illumina platforms at Macrogen. WES and WGS used 101 basepair (bp) or 150 paired-end reads, respectively. Average on-target WES depths were between 224-fold and 281-fold for the AVM samples and between 135-fold and 176-fold for the unaffected tissues. Coverage ≥30-fold and ≥390-fold were obtained for 90% to 93% and for 81% to 92% of the exomes, respectively (Figure 2). The five AVM specimens and the three matched unaffected tissue samples subjected to WGS yielded average depths between 108-fold and 145-fold and between 37-fold and 50-fold, respectively.
Figure 2.
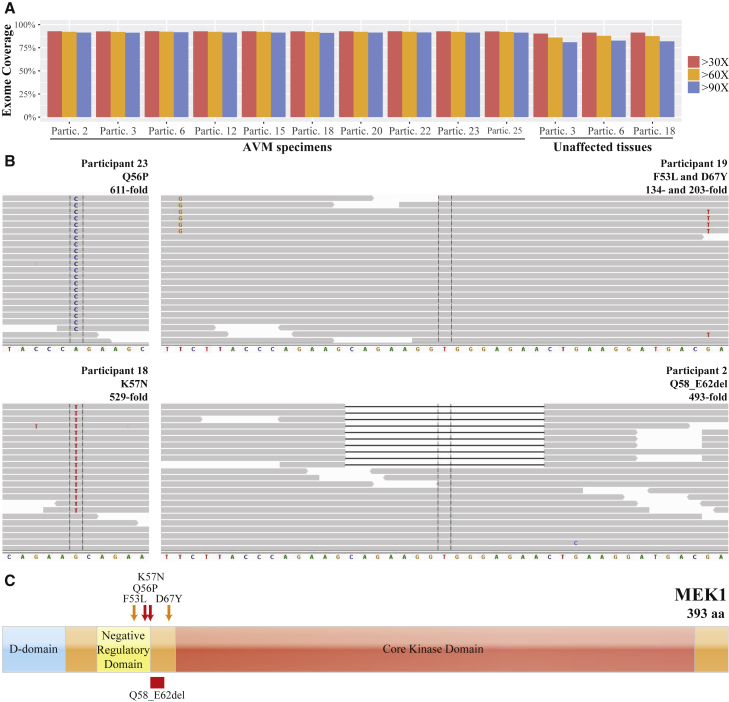
Somatic Mutation Detection in AVMs after Whole-Exome Sequencing
(A) Graph depicts the depth-of-coverage across the exome for ten affected and three unaffected tissue samples. Note ≥90-fold coverage for 90% of the exome obtained for each AVM sample.
(B) Integrative Genomic Viewer screenshots showing reads containing variant and reference alleles for four AVM samples. Total read depth at the site of the somatic mutation is indicated as “-fold” coverage. Note 4 reads in participant 19 indicate that the (c.159T>G) p.Phe53Leu and c.199G>T (p.Asp67Tyr) somatic mutations are in cis.
(C) Schematic diagram of MEK1 with approximate locations of the D, negative regulatory, and core kinase domains indicated. Note AVM somatic mutations cluster near the negative regulatory domain. The orange arrows indicate that p.Phe53Leu and p.Asp67Tyr variant were found in cis in a single individual. All other variants were found in two or more study participants.
Raw sequencing reads were aligned to the GRCh37 human reference genome using the Burrows-Wheeler Aligner.3 The reads were processed by Picard and the Genome Analysis Tool Kit4 for the removal of duplicated and error-prone reads, indel realignment, and base-quality recalibration. Candidate somatic mutations were identified using the single-sample (when only AVM DNA was available) and paired-sample (when AVM and blood/saliva DNA were available) modes of MosaicHunter.5 Somatic mutations with at least 2% mutant allele fraction and at least five reads supporting the variant allele were considered in subsequent analyses. We excluded common variants that were annotated in the Single Nucleotide Polymorphism,6 1000 Genomes Project,7 Exome Sequencing Project,8 or Exome Aggregation Consortium9 databases. Among the protein-altering mutations, only those predicted to be deleterious by Polymorphism Phenotyping (PolyPhen-2) or Sorting Intolerant From Tolerant (SIFT) algorithms10, 11 were analyzed further.
AVMs had between zero and four genes with putative protein-altering somatic mutations, but only MAP2K1 (GenBank: NM_002755; MIM: 176872), encoding the dual specificity mitogen-activated protein kinase MEK1 (GenBank: NP_002746.1), contained somatic mutations (c.171G>T [p.Lys57Asn] or c.167A>C [p.Gln56Pro]) in multiple samples (Table 1). Each AVM sample had ≥105-fold coverage over the entire MAP2K1 coding sequence. Because several specimens contained somatic MAP2K1 missense mutations, we reanalyzed samples in which we failed to detect a mutation at reduced stringency by lowering the detection threshold to ≥1% mutant allele fraction and ≥3 reads supporting the variant allele, and we also looked for short indels. This led to the identification of mutations in three additional deep WES samples (p.Lys57Asn [n = 2], c.173_187del [p.Gln58_Glu62del] [n = 1]) (Table 1) and in two of five AVM samples from other participants that had standard12 WES (Table 2). The specimen from participant 19 harbored two somatic MAP2K1 mutations (c.[159T>G;199G>T]; p.[Phe53Leu;Asp67Tyr]) in cis (Table 2).
Table 1.
MAP2K1 Variant Identification and Filtering for Ten AVM Samples Subjected to Deep Whole-Exome Sequencing
|
Participant |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 3 | 6 | 12 | 15 | 18 | 20 | 22 | 23 | 25 | |
| Somatic mosaic sites identified by MosaicHunter with ≥2% variant allele fraction and ≥5 variant reads | 34 | 32 | 24 | 14 | 21 | 15 | 22 | 33 | 38 | 63 |
| Considering only variants not present in the general population | 7 | 6 | 9 | 3 | 3 | 3 | 2 | 8 | 2 | 43 |
| Considering only non-synonymous mutations | 2 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 2 | 7 |
| Considering only mutations predicted to be deleterious | 2 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 2 | 4 |
| Genes harboring variants found with the above criteria | PALM2-AKAP2, HAUS7 | ND | MAP2K1 | MAP2K1 | ND | MAP2K1 | ND | ND | MAP2K1, EPPK1 | ZBTB41, OR8B8, FREM2, ACADVL |
| MAP2K1 mutation containing samples found using initial or less stringent criteria (variant/total WES reads) | c.173_187del (p.Gln58_Glu62del) (10/493) | ND | c.171G>T (p.Lys57Asn) (13/646) | c.167A>C (p.Gln56Pro) 45/625 | ND | c.171G>T (p.Lys57Asn) (17/529) | c.171G>T (p.Lys57Asn) (9/584) | c.171G>T (p.Lys57Asn) (6/583) | c.167A>C (p.Gln56Pro) (21/611) | ND |
Abbreviation: ND indicates no mutant alleles were detected.
Table 2.
MAP2K1Variant Detection in 25 Participants with AVM
| Participant | Age | Sex | Location, Stage | DNA Sourcea | Variantb | WESc | WGSc | ddPCRd,e |
|---|---|---|---|---|---|---|---|---|
| 1 | 5 y | M | lip, stage II | frozen | p.Gln56Pro | 7% (3/45) | – | 11% (231/2,628) |
| 6 y | frozen | p.Gln56Pro | – | – | 2% (49/2,764) | |||
| saliva | ND | – | – | 0% (0/931) | ||||
| 2 | 7 y | M | scalp, stage III | frozen | p.Gln58_Glu62del | 2% (10/493) | 3% (2/58) | 6%f (954/15,592) |
| saliva | ND | – | – | 0% (0/1,745) | ||||
| 3 | 9 y | F | thigh, stage III | frozen | ND | 0% (0/633) | 0% (0/68) | 0% (0/4,828) |
| blood | ND | 0% (1/348)g | 0% (0/31) | 0% (0/5,006) | ||||
| 4 | 9 y | M | forehead, stage II | frozen | p.Lys57Asn | – | – | 6% (138/2,654) |
| frozen | p.Lys57Asn | – | – | 8% (105/1,379) | ||||
| CD31+ cells,a p3 | p.Lys57Asn | – | – | 31% (187/437) | ||||
| CD31− cells,a p3 | ND | – | – | 0% (0/624) | ||||
| blood | ND | – | – | 0% (0/2,062) | ||||
| 5 | 11 y | F | cheek, stage II | frozen | p.Lys57Asn | – | – | 5% (124/3,445) |
| saliva | ND | – | – | 0% (0/1,274) | ||||
| 6 | 11 y | M | ear, stage I | frozen | p.Lys57Asn | 2% (13/646) | – | 8% (157/2,208) |
| blood | ND | 0% (0/271) | 0% (0/35) | 0% (0/6,913) | ||||
| 7 | 12 y | F | cheek, stage II | frozen | ND | 0% (0/37) | – | 0% (0/3,207) |
| saliva | ND | – | – | 0% (0/4,085) | ||||
| 8 | 12 y | F | ear, stage I | frozen | p.Lys57Asn | 4% (2/52) | – | 13% (414/3,498) |
| saliva | ND | – | – | 0% (0/965) | ||||
| 9 | 13 y | M | foot, stage III | frozen | ND | – | – | 0% (0/2,461) |
| saliva | ND | – | – | 0% (0/1,926) | ||||
| 10 | 13 y | F | lip, stage II | frozen | p.Lys57Asn | – | – | 7% (172/3,117) |
| saliva | ND | – | – | 0% (0/533) | ||||
| 11 | 13 y | M | forehead, stage II | frozen | p.Lys57Asn | – | – | 4% (68/2,066) |
| blood | ND | – | – | 0% (0/3,698) | ||||
| 12 | 14 y | F | lip, stage I | frozen | p.Gln56Pro | 7% (45/625) | – | 9% (210/2,334) |
| 13 | 14 y | F | lip, stage II | frozen | p.Gln56Pro | – | – | 8% (172/2,980) |
| frozen | p.Gln56Pro | – | – | 6% (102/1,821) | ||||
| frozen | p.Gln56Pro | – | – | 10% (209/2,655) | ||||
| CD31+ cells,a p5 | p.Gln56Pro | – | – | 35% (175/514) | ||||
| CD31− cells,a p2 | ND | – | – | 0% (0/563) | ||||
| blood | ND | – | – | 0% (0/2,396) | ||||
| 14 | 15 y | M | knee, stage I | frozen | p.Gln58_Glu62del | – | – | 10%f (1,336/12,887) |
| 15 | 15 yo | M | cheek, stage III | frozen | ND | 0% (0/675) | 0% (0/143) | 0% (0/3,708) |
| 16 | 17 y | M | forehead, stage II | frozen | ND | – | – | 0% (0/4,025) |
| saliva | ND | – | – | 0% (0/1,396) | ||||
| 17 | 18 y | F | lip, stage II | frozen | ND | 0% (0/30) | – | 0% (0/3,757) |
| saliva | ND | – | – | 0% (0/2,458) | ||||
| 18 | 21 y | M | ear, stage II | frozen | p.Lys57Asn | 3% (17/529) | 4% (3/69) | 7% (155/2,559) |
| CD31+ cells,a p4 | p.Lys57Asn | – | – | 53% (468/946) | ||||
| CD31− cells,a p4 | ND | – | – | 0% (0/765) | ||||
| blood | ND | 0% (0/250) | 0% (0/25) | 0% (0/3,681) | ||||
| 19 | 22 y | M | abdomen, stage II | frozen | p.Phe53Leu | 5% (6/134) | – | 1% (9/1,318) |
| p.Asp67Tyr | 5% (10/203) | – | 1% (7/1,336) | |||||
| blood | ND | 0% (0/15) | – | – | ||||
| 20 | 22 y | M | scalp, stage I | frozen | p.Lys57Asn | 2% (9/584) | – | 4% (63/2,660) |
| blood | ND | – | – | 0% (0/3,171) | ||||
| 21 | 23 y | M | face, stage III | frozen | ND | – | – | 0% (0/4,061) |
| 22 | 24 y | M | cheek, stage I | frozen | p.Lys57Asn | 1% (6/583) | – | 5% (100/2,531) |
| saliva | ND | – | – | 0% (0/807) | ||||
| 23 | 32 y | M | cheek, stage III | frozen | p.Gln56Pro | 3% (21/611) | 4% (2/47) | 7% (128/2,103) |
| saliva | ND | – | – | 0% (0/1,494) | ||||
| 24 | 39 y | F | scalp, stage III | frozen | ND | – | – | 0% (0/3,640) |
| 25 | 65 y | F | nose, stage II | frozen | ND | 0% (0/656) | – | 0% (0/2,642) |
| saliva | ND | – | – | 0% (0/1,325) |
Abbreviations are as follows: ND indicates no mutant alleles were detected; dash (–) indicates no study was performed.
For cultured CD31+ and CD31− cells, the passage number (p) at the time DNA was extracted in indicated.
The effect of the mutation at the protein level is indicated.
Mutant allele percentages are provided as whole numbers and are calculated from the number of mutant reads/total reads at that locus.
Mutant allele percentages are provided as whole numbers and are calculated by counting droplets that contain mutant, mutant+wild type, and wild-type amplimers. For simplicity, the ratio of mutant amplimer containing droplets/all amplimer containing droplets is shown.
When no MAP2K1 mutation was detected in affected tissue, the denominator for the ddPCR assay is the sum of wild-type droplets for the four individual assays.
Because no ddPCR assay was developed for this mutation, the mutant allele percentages were determined using pseudocluster counts, which were observed with each of the other missense mutant ddPCR probes.
We cannot preclude this variant (p.Asp67Tyr) being a true positive somatic mutation; however, this variant was not detected in 3,867 DNA containing ddPCR droplets from affected tissue, nor in ddPCR droplets from the same blood sample. Therefore, we suspect the WES result is a false positive finding.
We developed ddPCR assays13 for each of the missense mutations that were identified by WES and WGS (Table S1). We used ddPCR to confirm the MAP2K1 mutations found in deep WES (n = 7) and standard WES (n = 2). We also used ddPCR to screen for somatic mutations in 16 additional individuals: 3 without mutations detected by deep WES, 3 without mutations detected by standard WES, and 10 whom we had not previously examined. Out of these 16 individuals, 7 had MAP2K1 mutations detected by ddPCR (Table 2). One person had a ddPCR pseudocluster that upon subcloning and sequencing was found to represent the same 15-bp deletion (c.173_187del [p.Gln58_Glu62del]) previously detected by WES in another individual. In 13 participants who had affected tissue with MAP2K1 mutations and a paired unaffected tissue sample, none of the unaffected tissue samples contained mutant MAP2K1 alleles (Table 2). We also did not identify mutant MAP2K1 alleles in affected tissue from individuals with other types of vascular anomalies, including infantile hemangioma, congenital hemangioma, capillary malformation, lymphatic malformation, venous malformation, and verrucous venous malformation (data not shown).
We re-examined, at reduced stringency, the WES and WGS data for the AVM specimens which did not have detectable MAP2K1 mutations for mutations in other RAS/MAPK signaling pathway components (HRAS [MIM: 190020], KRAS [MIM: 190070], NRAS [MIM: 164790], ARAF [MIM: 311010], BRAF [MIM: 164757], RAF1 [MIM: 164760], MAP2K2 [MIM: 601263], MAPK1 [MIM: 176948], and MAPK3 [MIM: 601795]) as well as for previously reported AVM-associated genes (RASA [MIM: 139150], PTEN [MIM: 601728], ENG [MIM: 131195], ACVRL1 [MIM: 601284], SMAD4 [MIM: 600993], and GDF2 [MIM: 605120]). No suspicious somatic mutations were found.
We next examined whether somatic MAP2K1 mutations in AVMs were enriched in a specific cell type by separating endothelial cells from other cell types14 in three AVM specimens that harbored MAP2K1 mutations. In brief, specimens were digested with collagenase A (Roche) and dispase (BD Biosciences), filtered through a 70-μm strainer, incubated with anti-cluster of differentiation 31 protein (anti-CD31) antibody conjugated to magnetic Dynabeads (Invitrogen), and cultured. CD31+ cells were grown in endothelial cell growth medium and CD31− cells were cultured in mesenchymal stem cell growth medium (both from Lonza). We observed mutant MAP2K1 alleles only in the DNA we extracted from the endothelial cells (Figure 3).
Figure 3.
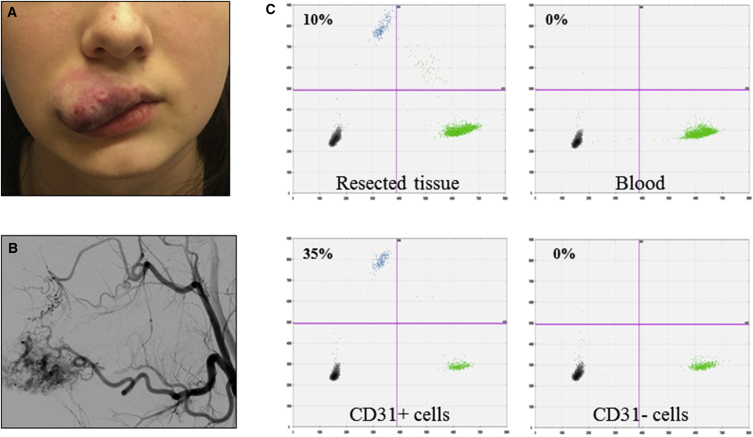
AVM Endothelial Cells Enriched for the MAP2K1 Mutation
(A) Photograph of an upper labial AVM (participant 13) from which CD31+ and CD31− cells were separated.
(B) Angiogram prior to resection shows arteriovenous shunting.
(C) ddPCR assay results performed on DNA extracted from the AVM (resected tissue), endothelial (CD31+), and non-endothelial (CD31−) cells, and peripheral blood. Droplets containing mutant only, mutant and wild-type, or wild-type only alleles appear in left upper, right upper, and right lower quadrants, respectively (empty droplets are in the left lower quadrant). Percentages of mutant alleles in each sample are indicated (droplet counts are provided in Table 2).
The most common type of AVM occurs sporadically and is solitary. The prevalence of AVMs is unknown. The Vascular Anomalies Center at Boston Children’s Hospital annually evaluates ∼50 new individuals with extracranial AVM.15 Solitary, extracranial AVM is not heritable, has similar incidence in males and females, and exhibits variable severity. These features are typical of disorders caused by a somatic mutation16 and are consistent with the finding of somatic MAP2K1 mutations in 16 of 25 tissue specimens. Solitary AVMs progress through four clinical stages: stage I lesions are typically small, warm to the touch, and exhibit arteriovenous shunting by Doppler ultrasonography; stage II lesions enlarge significantly, become pulsatile, and develop venous dilatation; stage III AVMs exhibit pain, ulceration, and/or bleeding; and stage IV lesions are characterized by cardiac failure.2, 17 We cannot exclude the possibility that somatic MAP2K1 mutations arise as a consequence of AVM expansion, rather than being the cause of AVM. Arguing against the former interpretation is the presence of mutations in stage I lesions (Figure 1 and Table 2) and the absence of mutations in other enlarging vascular malformations. Finding MAP2K1 mutations solely within the endothelial cell compartment of an AVM strongly suggests that mutant endothelial cells are responsible for the malformation process, perhaps by preventing capillary networks from forming between developing arteries and veins.
We found MAP2K1 mutations in 64% of the specimens; the remaining samples may contain mutations at levels below our detection limit, in regions of the gene not interrogated by ddPCR, or in other genes. AVMs also occur in individuals segregating three Mendelian genetic diseases; however, these AVMs are clinically, radiologically, and/or histologically distinct from AVMs with MAP2K1 mutations. AVMs associated with hereditary hemorrhagic telangiectasia (HHT [MIM: 187300, 600376, 175050, 615506]), caused by mutations in ENG, ACVRL1, SMAD4, or GDF2, are multifocal, small, and affect lungs, gastrointestinal system, and brain (see McDonald and Pyeritz, GeneReviews, in Web Resources). Families with RASA1 mutations have capillary malformation-arteriovenous malformation (CM-AVM [MIM: 608354]). This syndrome is characterized by multiple, small, cutaneous fast-flow lesions; some affected individuals also have intracranial or extracranial AVMs (see Beyrak-Toydemir and Stevenson, GeneReviews, in Web Resources). Multiple, intramuscular AVMs occur in individuals with phosphatase and tensin homolog (PTEN) hamartoma-tumor syndrome (MIM: 601728) (see Eng, GeneReviews, in Web Resources).
The MAP2K1 protein product MEK1 plays an important role in the RAS/MAPK signaling pathway that controls numerous cellular and developmental processes.18, 19 The somatic mutations we identified cluster within or adjacent to the protein’s negative regulatory domain (Figure 2); they have been observed in neoplasms, including melanoma, lung cancer, and hematopoietic malignancies,20, 21, 22, 23, 24, 25 and have been shown to constitutively increase MEK1 activity.26, 27, 28 MEK1 and its paralog MEK2 phosphorylate ERK1 and ERK2. MAPK signaling is activated by receptor tyrosine kinases, integrins, and G protein coupled receptors and is modulated by cross-talk with several other signaling pathways including the AKT-mTOR pathway.19 Somatic mutations affecting proteins upstream of MEK1 occur in cancer and other types of vascular malformations.12, 19, 29, 30, 31, 32, 33, 34 The MAP2K1 mutations we detected in AVMs likely alter the function of MEK1 by producing a hypermorphic or neomorphic effect, since these mutations have previously been shown to increase ERK1 and ERK2 phosphorylation in tumors and in cultured cells. Consistent with this hypothesis, mice that are heterozygous for Mek1 knockout alleles do not exhibit a phenotype, whereas homozygous knockout alleles cause embryonic lethality and placental defects.35 Germline mutations in MAP2K1 have been found in persons with Noonan syndrome (NS [MIM: 163950])36 and cardio-facio-cutaneous syndrome (CFC [MIM: 115150]).37 Individuals with NS or CFC have cardiac malformations, including pulmonic stenosis and atrial septal defect, but not AVM (see Allanson and Roberts, GeneReviews, and Rauen, GeneReviews, in Web Resources). The MAP2K1 mutations we detected in AVMs differ from those found in NS and CFC.
The AVMs in which MAP2K1 mutations were found do not metastasize, but they enlarge over time.38 Incompletely resected AVMs often re-expand and attempts to embolize or ligate feeding arteries result in increased flow from collateral vessels.2 Delineating similarities and differences between MAP2K1 mutation-containing AVMs and cancers will provide insights regarding cell-type-specific functions for MEK1 and broader roles for MEK1 in responding to environmental stress such as tissue injury or hypoxia. MEK1 inhibitors currently are in use against various cancers.18 Further study is required to determine whether these drugs have efficacy for AVM. Current agents are cytostatic rather than cytotoxic, but might benefit individuals with AVM if they cause mutant endothelial cells to terminally differentiate into capillary beds or prevent the regrowth of AVMs after resection or embolization.
Acknowledgments
The authors thank Mr. Matthew P. Vivero and Dr. Steven Hann for their scientific expertise, Dr. Katherine A. Rauen for sharing unpublished data regarding MAP2K1 mutations in CFC, the study participants, and the members of the Vascular Anomalies Center at Boston Children’s Hospital. Research reported in this manuscript was supported by the NIH awards NICHD-81004 (A.K.G.), NICHD-82606 (A.K.G.), NHLBI-12703 (A.K.G.), and NIAMS AR-64231 (M.L.W.), the Translational Research Program Career Award Boston Children’s Hospital (A.K.G.), and the American Society of Plastic Surgeons/Plastic Surgery Foundation (grant PSF-71190 to A.K.G.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Published: February 9, 2017
Footnotes
Supplemental Data include one table and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.01.018.
Web Resources
GeneReviews, Allanson, J.E., and Roberts, A.E. (1993). Noonan syndrome. https://www.ncbi.nlm.nih.gov/books/NBK1124/
GeneReviews, Bayrak-Toydemir, P., and Stevenson, D. (1993). RASA1-related disorders. https://www.ncbi.nlm.nih.gov/books/NBK52764/
GeneReviews, Eng, C. (1993). PTEN hamartoma tumor syndrome. https://www.ncbi.nlm.nih.gov/books/NBK1488/
GeneReviews, McDonald, J., and Pyeritz, R.E. (1993). Hereditary hemorrhagic telangiectasia, https://www.ncbi.nlm.nih.gov/books/NBK1351/
GeneReviews, Rauen, K.A. (1993). Cardiofaciocutaneous syndrome. https://www.ncbi.nlm.nih.gov/books/NBK1186/
Mosaic Hunter, http://mosaichunter.cbi.pku.edu.cn/
OMIM, http://www.omim.org/
Supplemental Data
References
- 1.Mulliken J.B., Burrows P.E., Fishman S.J., Mulliken J.B. Second Edition. Oxford University Press; Oxford: 2013. Mulliken and Young’s Vascular Anomalies: Hemangiomas and Malformations. [Google Scholar]
- 2.Liu A.S., Mulliken J.B., Zurakowski D., Fishman S.J., Greene A.K. Extracranial arteriovenous malformations: natural progression and recurrence after treatment. Plast. Reconstr. Surg. 2010;125:1185–1194. doi: 10.1097/PRS.0b013e3181d18070. [DOI] [PubMed] [Google Scholar]
- 3.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang A.Y., Xu X., Ye A.Y., Wu Q., Yan L., Zhao B., Yang X., He Y., Wang S., Zhang Z. Postzygotic single-nucleotide mosaicisms in whole-genome sequences of clinically unremarkable individuals. Cell Res. 2014;24:1311–1327. doi: 10.1038/cr.2014.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tennessen J.A., Bigham A.W., O’Connor T.D., Fu W., Kenny E.E., Gravel S., McGee S., Do R., Liu X., Jun G., Broad GO. Seattle GO. NHLBI Exome Sequencing Project Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 12.Couto J.A., Vivero M.P., Kozakewich H.P., Taghinia A.H., Mulliken J.B., Warman M.L., Greene A.K. A somatic MAP3K3 mutation is associated with verrucous venous malformation. Am. J. Hum. Genet. 2015;96:480–486. doi: 10.1016/j.ajhg.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luks V.L., Kamitaki N., Vivero M.P., Uller W., Rab R., Bovée J.V., Rialon K.L., Guevara C.J., Alomari A.I., Greene A.K. Lymphatic and other vascular malformative/overgrowth disorders are caused by somatic mutations in PIK3CA. J. Pediatr. 2015;166 doi: 10.1016/j.jpeds.2014.12.069. 1048–54.e1, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin R.Z., Moreno-Luna R., Muñoz-Hernandez R., Li D., Jaminet S.C., Greene A.K., Melero-Martin J.M. Human white adipose tissue vasculature contains endothelial colony-forming cells with robust in vivo vasculogenic potential. Angiogenesis. 2013;16:735–744. doi: 10.1007/s10456-013-9350-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greene A.K., Liu A.S., Mulliken J.B., Chalache K., Fishman S.J. Vascular anomalies in 5,621 patients: guidelines for referral. J. Pediatr. Surg. 2011;46:1784–1789. doi: 10.1016/j.jpedsurg.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 16.Happle R. Lethal genes surviving by mosaicism: a possible explanation for sporadic birth defects involving the skin. J. Am. Acad. Dermatol. 1987;16:899–906. doi: 10.1016/s0190-9622(87)80249-9. [DOI] [PubMed] [Google Scholar]
- 17.Kohout M.P., Hansen M., Pribaz J.J., Mulliken J.B. Arteriovenous malformations of the head and neck: natural history and management. Plast. Reconstr. Surg. 1998;102:643–654. doi: 10.1097/00006534-199809030-00006. [DOI] [PubMed] [Google Scholar]
- 18.Caunt C.J., Sale M.J., Smith P.D., Cook S.J. MEK1 and MEK2 inhibitors and cancer therapy: the long and winding road. Nat. Rev. Cancer. 2015;15:577–592. doi: 10.1038/nrc4000. [DOI] [PubMed] [Google Scholar]
- 19.Wortzel I., Seger R. The ERK Cascade: Distinct Functions within Various Subcellular Organelles. Genes Cancer. 2011;2:195–209. doi: 10.1177/1947601911407328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arcila M.E., Drilon A., Sylvester B.E., Lovly C.M., Borsu L., Reva B., Kris M.G., Solit D.B., Ladanyi M. MAP2K1 (MEK1) Mutations Define a Distinct Subset of Lung Adenocarcinoma Associated with Smoking. Clin. Cancer Res. 2015;21:1935–1943. doi: 10.1158/1078-0432.CCR-14-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bansal A., Ramirez R.D., Minna J.D. Mutation analysis of the coding sequences of MEK-1 and MEK-2 genes in human lung cancer cell lines. Oncogene. 1997;14:1231–1234. doi: 10.1038/sj.onc.1200947. [DOI] [PubMed] [Google Scholar]
- 22.Chakraborty R., Hampton O.A., Shen X., Simko S.J., Shih A., Abhyankar H., Lim K.P., Covington K.R., Trevino L., Dewal N. Mutually exclusive recurrent somatic mutations in MAP2K1 and BRAF support a central role for ERK activation in LCH pathogenesis. Blood. 2014;124:3007–3015. doi: 10.1182/blood-2014-05-577825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi Y.L., Soda M., Ueno T., Hamada T., Haruta H., Yamato A., Fukumura K., Ando M., Kawazu M., Yamashita Y., Mano H. Oncogenic MAP2K1 mutations in human epithelial tumors. Carcinogenesis. 2012;33:956–961. doi: 10.1093/carcin/bgs099. [DOI] [PubMed] [Google Scholar]
- 24.Estep A.L., Palmer C., McCormick F., Rauen K.A. Mutation analysis of BRAF, MEK1 and MEK2 in 15 ovarian cancer cell lines: implications for therapy. PLoS ONE. 2007;2:e1279. doi: 10.1371/journal.pone.0001279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikolaev S.I., Rimoldi D., Iseli C., Valsesia A., Robyr D., Gehrig C., Harshman K., Guipponi M., Bukach O., Zoete V. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat. Genet. 2011;44:133–139. doi: 10.1038/ng.1026. [DOI] [PubMed] [Google Scholar]
- 26.Mansour S.J., Matten W.T., Hermann A.S., Candia J.M., Rong S., Fukasawa K., Vande Woude G.F., Ahn N.G. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- 27.Marks J.L., Gong Y., Chitale D., Golas B., McLellan M.D., Kasai Y., Ding L., Mardis E.R., Wilson R.K., Solit D. Novel MEK1 mutation identified by mutational analysis of epidermal growth factor receptor signaling pathway genes in lung adenocarcinoma. Cancer Res. 2008;68:5524–5528. doi: 10.1158/0008-5472.CAN-08-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherry S.T., Ward M., Sirotkin K. Use of molecular variation in the NCBI dbSNP database. Hum. Mutat. 2000;15:68–75. doi: 10.1002/(SICI)1098-1004(200001)15:1<68::AID-HUMU14>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 29.Aoki Y., Matsubara Y. Ras/MAPK syndromes and childhood hemato-oncological diseases. Int. J. Hematol. 2013;97:30–36. doi: 10.1007/s12185-012-1239-y. [DOI] [PubMed] [Google Scholar]
- 30.Knight T., Irving J.A. Ras/Raf/MEK/ERK Pathway Activation in Childhood Acute Lymphoblastic Leukemia and Its Therapeutic Targeting. Front. Oncol. 2014;4:160. doi: 10.3389/fonc.2014.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim Y.H., Bacchiocchi A., Qiu J., Straub R., Bruckner A., Bercovitch L., Narayan D., McNiff J., Ko C., Robinson-Bostom L., Yale Center for Mendelian Genomics GNA14 Somatic Mutation Causes Congenital and Sporadic Vascular Tumors by MAPK Activation. Am. J. Hum. Genet. 2016;99:443–450. doi: 10.1016/j.ajhg.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim Y.H., Douglas S.R., Ko C.J., Antaya R.J., McNiff J.M., Zhou J., Choate K.A., Narayan D. Somatic Activating RAS Mutations Cause Vascular Tumors Including Pyogenic Granuloma. J. Invest. Dermatol. 2015;135:1698–1700. doi: 10.1038/jid.2015.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Solus J.F., Kraft S. Ras, Raf, and MAP kinase in melanoma. Adv. Anat. Pathol. 2013;20:217–226. doi: 10.1097/PAP.0b013e3182976c94. [DOI] [PubMed] [Google Scholar]
- 34.Zenker M. Clinical manifestations of mutations in RAS and related intracellular signal transduction factors. Curr. Opin. Pediatr. 2011;23:443–451. doi: 10.1097/MOP.0b013e32834881dd. [DOI] [PubMed] [Google Scholar]
- 35.Giroux S., Tremblay M., Bernard D., Cardin-Girard J.F., Aubry S., Larouche L., Rousseau S., Huot J., Landry J., Jeannotte L., Charron J. Embryonic death of Mek1-deficient mice reveals a role for this kinase in angiogenesis in the labyrinthine region of the placenta. Curr. Biol. 1999;9:369–372. doi: 10.1016/s0960-9822(99)80164-x. [DOI] [PubMed] [Google Scholar]
- 36.Nava C., Hanna N., Michot C., Pereira S., Pouvreau N., Niihori T., Aoki Y., Matsubara Y., Arveiler B., Lacombe D. Cardio-facio-cutaneous and Noonan syndromes due to mutations in the RAS/MAPK signalling pathway: genotype-phenotype relationships and overlap with Costello syndrome. J. Med. Genet. 2007;44:763–771. doi: 10.1136/jmg.2007.050450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez-Viciana P., Tetsu O., Tidyman W.E., Estep A.L., Conger B.A., Cruz M.S., McCormick F., Rauen K.A. Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science. 2006;311:1287–1290. doi: 10.1126/science.1124642. [DOI] [PubMed] [Google Scholar]
- 38.Greene A.K., Orbach D.B. Management of arteriovenous malformations. Clin. Plast. Surg. 2011;38:95–106. doi: 10.1016/j.cps.2010.08.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.