Abstract
A novel coronavirus (SCoV) is the etiological agent of severe acute respiratory syndrome. Site-specific proteolysis plays a critical role in regulating a number of cellular and viral processes. Since the main protease of SCoV, also termed 3C-like protease, is an attractive target for drug therapy, we have developed a safe, simple, and rapid genetic screen assay to monitor the activity of the SCoV 3C-like protease. This genetic system is based on the bacteriophage lambda regulatory circuit, in which the viral repressor cI is specifically cleaved to initiate the lysogenic-to-lytic switch. A specific target for the SCoV 3C-like protease, P1/P2 (SAVLQ/SGFRK), was inserted into the lambda phage cI repressor. The target specificity of the SCoV P1/P2 repressor was evaluated by coexpression of this repressor with a chemically synthesized SCoV 3C-like protease gene construct. Upon infection of Escherichia coli cells containing the two plasmids encoding the cI. SCoV P1/P2-cro and the β-galactosidase-SCoV 3C-like protease constructs, lambda phage replicated up to 2,000-fold more efficiently than in cells that did not express the SCoV 3C-like protease. This simple and highly specific assay can be used to monitor the activity of the SCoV 3C-like protease, and it has the potential to be used for screening specific inhibitors.
The recently identified severe acute respiratory syndrome (SARS) coronavirus (CoV) (SCoV) (5, 9, 12, 19) causes a life-threatening highly contagious pneumonia and is the most pathogenic human CoV identified so far. This disease was first recognized in southern China in November 2002. By August 2003, 8,422 cases had occurred in 29 countries and 908 individuals had died from the disease (http://www.who.int/csr/sars/country/en/country2003_08_15.pdf). Its rapid transmission and the high mortality (10%) make SARS a potential global threat. Recent reports of several SARS cases show that new SARS outbreaks are possible in the near future (http://www.who.int/csr/don/en/). To date, neither a vaccine nor an effective therapy is available.
The activity of specific proteases is essential in many fundamental cellular and viral processes. Viral polyprotein processing is indispensable in the replication and maturation of many viruses (6). Consequently, site-specific proteolysis has been an attractive target for the development of antiviral therapies based on potent and selective viral inhibitors. The generation of such therapies based on the inhibition of site-specific proteolysis has been clearly illustrated in the development of effective inhibitors of human immunodeficiency virus type 1 (HIV-1) (10, 30) and hepatitis C virus (HCV) (13).
CoVs are large, enveloped, plus-strand RNA viruses, which have the largest genomes of all RNA viruses (11). The SCoV genomic RNA is nearly 30 kb and is capped and polyadenylated (14, 21, 22). The primary translation product of the viral RNA is largely processed into multiple proteins by the viral main protease, also called 3C-like protease (Fig. 1) to indicate the similarity of its cleavage site specificity to that observed for picornavirus 3C protease (1). The SCoV 3C-like protease has a molecular mass of nearly 35 kDa (7, 24, 31) and, like other CoV 3C-like proteases, has specificity for Gln at the P1 position (2). Recently, the crystal structure of the SCoV 3C-like protease has revealed that the protein fold can be described as a serine protease, but with a Cys-His at the active site (31).
FIG. 1.
Amino acid sequence of the SCoV 3C-like protease engineered in the present study. The autocleavage sites of the protease are marked with vertical arrows above the sequences. The cleavage site used as a target site in the genetic screen described here is shaded. Underlined are the catalytic-site residues Cys145 and His41.
It has been demonstrated that a bacteriophage lambda-based genetic screen can be used to isolate and characterize site-specific proteases (25). We have previously adapted this system, illustrated in Fig. 2, to study the HIV-1 and HCV proteases (3, 15, 16). This genetic screen system is based on the bacteriophage lambda cI-cro regulatory circuit, where the λ-encoded repressor cI is specifically cleaved to initiate the lysogenic-to-lytic switch (20). The inherent difficulties and safety requirements for the ex vivo propagation of SCoV prompted us to explore this genetic system as a simple alternative approach for the characterization of SCoV 3C-like protease activity. In this report, we demonstrate that the lambda-based genetic screen system can be used to monitor the activity of the SCoV 3C-like protease.
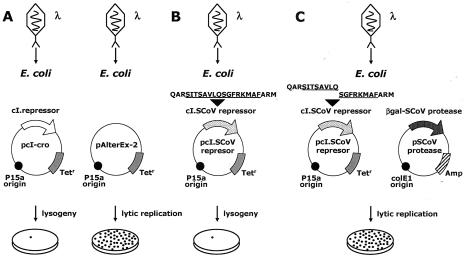
FIG. 2.
Lambda-based genetic screen to monitor the activity of SCoV 3C-like protease. This genetic screen system is based on the bacteriophage lambda cI-cro regulatory circuit, where the viral repressor cI is specifically cleaved to initiate the lysogenic-to-lytic switch. (A) Expression of the phage-encoded repressor (cI) results in repression of the bacteriophage's lytic functions (lysogeny). (B) SCoV target repressor containing the P1/P2 cleavage site; as illustrated in Fig. 3 and 4, this repressor efficiently represses the infecting phage (lysogeny). (C) When phages infect E. coli cells that express recombinant cI.SCoV repressor and β-Gal-SCoV 3C-like protease, infection results in lytic replication.
We chemically synthesized the SCoV 3C-like protease gene (Fig. 1) from synthetic oligonucleotides as chemical building blocks without employing any viral component formed in vivo or ex vivo (4, 27). The strategy of synthesizing the SCoV 3C-like protease was as follows. Three overlapping DNA fragments of 340, 340, and 268 bp were combined by PCR, using the Overlap Extension protocol (23), to obtain the full-length SCoV 3C-like protease. Each of the former DNA fragments was synthesized by assembling eight purified oligonucleotides (average length, 60 nucleotides [nt]) of plus and minus polarities with an overlapping complementary sequence of 20 nt at their termini (Table 1). Synthetic oligonucleotides were assembled in an asymmetric PCR assay previously described (18) and were designed to synthesize the SCoV 3C-like protease gene reported by Anand et al. (2). Next, the full-length SCoV 3C-like protease gene was reamplified by PCR with oligonucleotides SProL (sense, 5′GGGTTGAATTCTGCTGTTCTGCAGAGT 3′; underlining indicates an EcoRI restriction site) and SProR (antisense; Table 1), digested with EcoRI and XhoI, and cloned into pBluescript SK(−) (pBSK−; Stratagene) to generate the β-galactosidase (β-Gal)-SCoV 3C-like protease fusion (Fig. 2). Sequence analysis of several full-length β-Gal-SCoV 3C-like protease clones verified the accuracy of these synthetic genes. One of the former clones had exactly the intended sequence (Fig. 1).
TABLE 1.
Synthetic oligonucleotides for engineering the full-length SCoV 3C-like protease used in this work
| Name | Sequence (5′-3′) | nta | Orientation |
|---|---|---|---|
| S1 | TCTGCTGTTCTGCAGAGTGGTTTTAGGAAAATGGCATTCCCGTCAGGCAAAGTTGAAGGG | −15-45 | Sense |
| S2 | CATTAAGAGTTGTAGTTCCACAGGTTACTTGTACCATGCACCCTTCAACTTTGCCTGACG | 26-85 | Antisense |
| S3 | TGGAACTACAACTCTTAATGGATTGTGGTTGGATGACACAGTATACTGTCCAAGACATGT | 66-125 | Sense |
| S4 | TTCATAGTTAGGATTAAGCATGTCTTCTGCTGTGCAAATGACATGTCTTGGACAGTATAC | 106-165 | Antisense |
| S5 | TGCTTAATCCTAACTATGAAGATCTGCTCATTCGCAAATCCAACCATAGCTTTCTTGTTC | 146-205 | Sense |
| S6 | ATAGAATGGCCAATAACACGAAGTTGAACATTGCCAGCCTGAACAAGAAAGCTATGGTTG | 186-245 | Antisense |
| S7 | CGTGTTATTGGCCATTCTATGCAAAATTGTCTGCTTAGGCTTAAAGTTGATACTTCTAAC | 226-285 | Sense |
| S8 | CAGGTTGGATACGGACAAATTTATACTTGGGTGTCTTAGGGTTAGAAGTATCAACTTTAA | 266-325 | Antisense |
| S9 | ATTTGTCCGTATCCAACCTGGTCAAACATTTTCAGTTCTAGCATGCTACAATGGTTCACC | 306-365 | Sense |
| S10 | GGTATGATTAGGTCTCATGGCACACTGATAAACACCAGATGGTGAACCATTGTAGCATGC | 346-405 | Antisense |
| S11 | CCATGAGACCTAATCATACCATTAAAGGTTCTTTCCTTAATGGATCATGTGGTAGTGTTG | 386-445 | Sense |
| S12 | ATATAGCAGAAAGACACGCAATCATAATCAATGTTAAAACCAACACTACCACATGATCCA | 426-485 | Antisense |
| S13 | TGCGTGTCTTTCTGCTATATGCATCATATGGAGCTTCCAACAGGAGTACACGCTGGTACT | 466-525 | Sense |
| S14 | GTCTGTCAACAAATGGACCATAGAATTTACCTTCTAAGTCAGTACCAGCGTGTACTCCTG | 506-565 | Antisense |
| S15 | TGGTCCATTTGTTGACAGACAAACTGCACAGGCTGCAGGTACAGACACAACCATAACATT | 546-605 | Sense |
| S16 | ACCATTGATAACAGCAGCATACAGCCATGCCAAAACATTTAATGTTATGGTTGTGTCTGT | 586-645 | Antisense |
| S17 | ATGCTGCTGTTATCAATGGTGATAGGTGGTTTCTTAATAGATTCACCACTACTTTGAATG | 626-685 | Sense |
| S18 | AAAGGTTCATAGTTGTACTTCATTGCCACAAGGTTAAAGTCATTCAAAGTAGTGGTGAAT | 666-725 | Antisense |
| S19 | AAGTACAACTATGAACCTTTGACACAAGATCATGTTGACATATTGGGACCTCTTTCTGCT | 706-766 | Sense |
| S20 | TCAAAGCAGCACACATATCTAAGACGGCAATTCCTGTTTGAGCAGAAAGAGGTCCCAATA | 746-805 | Antisense |
| S21 | AGATATGTGTGCTGCTTTGAAAGAGCTGCTGCAGAATGGTATGAATGGTCGTACTATCCT | 786-845 | Sense |
| S22 | ATCAAATGGTGTAAACTCATCTTCTAAAATAGTGCTACCAAGGATAGTACGACCATTCAT | 826-885 | Antisense |
| S23 | ATGAGTTTACACCATTTGATGTTGTTAGACAATGCTCTGGTGTTACCTTCCAAGGTAAGTTCAAGAAA | 866-948 | Sense |
| SProR | GGGAGGGGGCTCGAGTCATTTCTTGAACTTACCTTGb | 916-948 | Antisense |
Numerical position on the SCoV 3C-like protease.
Underlining indicates an XhoI restriction site.
By using a unique restriction site (BssH2 site) located in the coding sequence of the linker domain of cI (25), the SCoV 3C-like protease P1/P2 (SAVLQ/SGFRK) cleavage site was inserted into the λ cI repressor (cI.SCoV) (Fig. 2B). The oligonucleotides encoding the SCoV proteolytic P1/P2 cleavage site were 5′GTTCAGGCGCGCGCTTCAATCACTTCTGCTGTTCTGCAGAGTGGTTTTAGGAAAATGGCATTCGCGCGCATGTTC3′ (sense) and 5′GAACATGCGCGCGAATGCCATTTTCCTAAAACCACTCTGCAGAACAGCAGAAGTGATTGAAGCGCGCGCCTGAAC3′ (antisense). A control mutant site (SAVLA/SGFRK) was also inserted in the λ cI repressor (cI.SCoVmt). As illustrated (Fig. 3), Escherichia coli JM109 cells expressing these two repressors efficiently repressed the infecting phage.
FIG. 3.
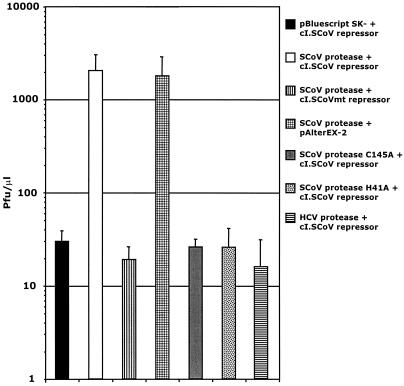
Selective growth of λ in E. coli cells coexpressing the β-Gal-SCoV 3C-like protease construct and the cI.SCoV repressor. Expression of the protease was induced with IPTG for 1 h, and the cells were infected with λ for three additional hours. The graph illustrates the resulting phage titer per microliter. Plasmids pBSK− and pAlterEX-2 were used as controls for the β-Gal-SCoV 3C-like protease construct and the cI.SCoV repressor, respectively. cI.SCoVmt was also used as a negative control for the cI.SCoV repressor. As shown, selection in cells coexpressing the β-Gal-SCoV 3C-like protease construct and the cI.SCoV3 repressor resulted in λ replication, whereas the replication of λ was severely compromised in cells expressing the mutant cI.SCoVmt repressor. Lack of phage replication was also observed in cells expressing mutated forms of β-Gal-SCoV 3C-like protease that included catalytic-site residue substitutions C145A and H41A. Similarly, expression of another protease (HCV serine protease) also prevented phage replication. Values are the means ± standard deviations (error bars) of at least four experiments.
We next tested the target specificity of the SCoV repressors by coexpressing these repressors with a β-Gal-SCoV 3C-like protease fusion construct. E. coli JM109 cells were then cotransformed with plasmids encoding the cI.SCoV repressor and the β-Gal-SCoV 3C-like protease constructs (Fig. 2C). The resulting cells were grown overnight at 30°C in the presence of 0.2% maltose, harvested by centrifugation, and resuspended to an optical density at 600 nm (OD600) of 2.0 per ml in 10 mM MgSO4. To induce the expression of SCoV 3C-like protease, cells (200 μl) were incubated in 1 ml of Luria-Bertani (LB) medium containing 12.5 μg of tetracycline, 20 μg of ampicillin, 0.2% maltose, 10 mM MgSO4, and 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 1 h. Thereafter, cell cultures were infected with 107 PFU of λ phage. After 3 h at 37°C, the titer of the resulting phage was determined by coplating the cultures with 200 μl of E. coli XL-1 Blue cells (OD600 = 2.0/ml in 10 mM MgSO4) on LB plates using 3 ml of top agar containing 12.5 μg of tetracycline per ml, 0.2% maltose, and 0.1 mM IPTG. After incubation at 37°C for 6 h, the resulting phage plaques were counted for growth scores. As shown in Fig. 3, λ phage replicated up to 2,000-fold more efficiently in cells expressing the cI.SCoV repressor and the β-Gal-SCoV 3C-like protease than in cells that did not express the β-Gal-SCoV 3C-like protease (Fig. 3).
The specificity of this trans-cleavage reaction was further demonstrated by the lack of phage replication in cells expressing mutated forms of the SCoV 3C-like protease that included catalytic-site residue substitutions C145A and H41A (Fig. 3). To demonstrate that the engineered cI.SCoV repressor is highly specific for the SCoV 3C-like protease, cells expressing the cI.SCoV repressor were transformed with an HCV serine protease construct. As shown in Fig. 3, the expression of the HCV serine protease did not allow phage replication. Likewise, phage replication was also abolished in cells expressing the control cI.SCoVmt repressor, arguing that cI.SCoV degradation was specifically mediated by the SCoV 3C-like protease.
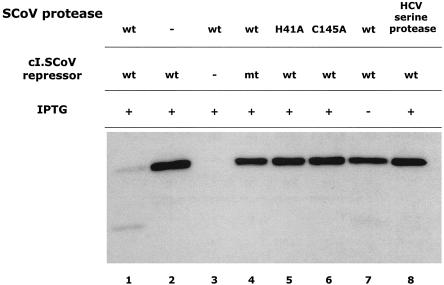
Finally, a Western blot also demonstrated (Fig. 4) that the expression of the SCoV 3C-like protease resulted in nearly complete cleavage of the cI.SCoV repressor (Fig. 4, lane 1) but had no effect on the control cI.SCoVmt (Fig. 4, lane 4). Expression of proteases that included catalytic-site residue substitutions C145A and H41A (Fig. 4, lanes 6 and 5, respectively) completely abolished the cleavage of cI.SCoV. Furthermore, expression of another protease as the HCV serine protease also abolished wild-type cI.SCoV repressor cleavage (Fig. 4, lane 8).
FIG. 4.
The SCoV 3C-like protease reduces the expression levels of cI.CoV. The cI.SCoV and cI.SCoVmt (lane 4) repressors were coexpressed with the SCoV 3C-like protease. Expression of the protease was induced with IPTG for 3 h. The ODs of the cultures after 3 h (in the presence of IPTG) were measured to assure that equivalent amounts of total cell protein were blotted. No significant differences were observed when the ODs of the different cultures were compared, suggesting that the expression of the SCoV 3C-like protease did not affect the growth of the bacteria. Control proteases with catalytic residue substitutions C145A and H41A and another protease (HCV serine protease) were also included in this experiment (lanes 5, 6, and 8, respectively). Lane 7 cells were grown in the absence of IPTG. Reduced signal and cleavage products were observed only when the wild-type (wt) SCoV 3C-like protease was expressed (lane 1); cleavage products were also observed in the absence of IPTG, suggesting residual expression of the wild-type SCoV 3C-like protease (lane 7). E. coli JM109 cells were cotransformed with pAlterEx-2 repressor plasmids and the pBSK− plasmid containing wild-type or mutated SCoV 3C-like proteases. Cultures were lysed in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis sample buffer, resolved in 18% gradient SDS-polyacrylamide gels, transferred to nitrocellulose membranes, and blocked in phosphate-buffered saline-0.1% Tween 20-10% nonfat dry milk. For immunochemical detection of the cI.SCoV repressor, membranes were subsequently incubated with rabbit serum containing polyclonal anti-cI antibodies (anti-cI sera; Invitrogen). Bound antibodies were visualized with peroxidase-linked anti-rabbit immunoglobulin G (Pierce) and the ECL Plus kit (Amersham Biosciences).
The genetic screen system used here to monitor the activity of the SCoV 3C-like protease is based on the well-characterized bacteriophage λ lytic-lysogenic cycle (20). When the cI repressor is functional, lytic gene products are silenced and the phage enters a lysogenic phase. Endogenous bacterial protease RecA cleaves the cI repressor at a specific region. cI repressor cleavage allows the expression of cro and progression into the lytic replication cycle. This lysogenic-to-lytic switch was previously adapted to develop a genetic screen system for the characterization of the HIV-1 and HCV proteases (3, 15, 16, 25, 26). The simplicity and specificity of this system prompted us to explore this genetic system as a new approach for the characterization of SCoV 3C-like protease activity. Moreover, the different biological properties of the HIV-1, HCV, and SCoV proteases offered us the opportunity to explore whether this system can be used to characterize proteases with different mechanisms of action. Thus, the cI repressor was modified to replace the normal site of RecA-mediated cleavage with a SCoV 3C-like protease target cleavage site (Fig. 2B). The coexpression of the cI.SCoV repressor and the SCoV 3C-like protease resulted in λ phage replication (Fig. 2C, 3, and 4). In contrast, phage replication was efficiently repressed in control cells that did not express the SCoV 3C-like protease (Fig. 3 and 4). Therefore, we demonstrate here that this lambda-based system can be used to monitor the catalytic activity of the SCoV 3C-like protease. This simple assay can augment biochemical approaches to the analysis of this protease.
As mentioned in the beginning of this report, proteases have been an attractive target for developing effective HIV-1 and HCV therapeutics, and this seems to be the case for SCoV. SCoV can be detected at 14 days postexposure in 97% of patients (19); with influenza virus or rhinoviruses, nearly every patient will test negative for the virus at day 5 (29). This protracted period of SCoV replication means that there is a window of opportunity for intervention in SARS with an antiviral. Another reason for the development of in vitro cell-free enzymatic methods for the characterization of the different SCoV proteins is the safety procedures (biosafety level 3 [BSL-3]) required for SCoV ex vivo propagation. Even BSL-3 equipment did not prevent the infection of laboratory researchers with this virus (17). It is important to emphasize that last winter the reported cases of SARS, after the outbreak was contained in July 2003, were due to laboratory contamination of researchers working with SCoV (17). The simplicity of our system can be seen as a complement to the classical biochemical approach for monitoring SCoV 3C-like proteolytic activity. As we and others have previously demonstrated for the HIV-1 protease (3, 15, 26), this system allows the characterization of enzymes with different proteolytic activities. Coupling mutant sequence libraries with this positive genetic selection system will allow the study of a huge number of functional mutants. Mutant proteases may be of interest for characterizing the catalytic properties of the enzyme in the absence or presence of specific inhibitors as well as for predicting the protease inhibitor resistance profile. To perform these experiments using classical biochemical approaches would be difficult and time-consuming.
Recently, a previously undescribed CoV associated with respiratory disease of unknown etiology in humans has been identified (8, 28). Easily, the system developed in this report can be extended to other CoV 3C-like proteases. Here we developed a safe, simple, and rapid genetic screen assay to monitor the activity of the CoV 3C-like protease. This system should be also useful for the development of a screening method to identify SCoV 3C-like protease inhibitors.
Nucleotide sequence accession number
The SCoV 3C-like protease nucleotide sequence constructed and used in the present work has been submitted to GenBank database under accession number AY609081.
Acknowledgments
This work was supported by Fundació irsiCaixa.
REFERENCES
- 1.Anand, K., G. J. Palm, J. R. Mesters, S. G. Siddell, J. Ziebuhr, and R. Hilgenfeld. 2002. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 21:3213-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anand, K., J. Ziebuhr, P. Wadhwani, J. R. Mesters, and R. Hilgenfeld. 2003. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300:1763-1767. [DOI] [PubMed] [Google Scholar]
- 3.Cabana, M., G. Fernandez, M. Parera, B. Clotet, and M. A. Martinez. 2002. Catalytic efficiency and phenotype of HIV-1 proteases encoding single critical resistance substitutions. Virology 300:71-78. [DOI] [PubMed] [Google Scholar]
- 4.Cello, J., A. V. Paul, and E. Wimmer. 2002. Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science 297:1016-1018. [DOI] [PubMed] [Google Scholar]
- 5.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 6.Dunn, B. M. (ed.) 1999. Proteases of infectious agents. Academic Press, San Diego, Calif.
- 7.Fan, K., P. Wei, Q. Feng, S. Chen, C. Huang, L. Ma, B. Lai, J. Pei, Y. Liu, J. Chen, and L. Lai. 2004. Biosynthesis, purification, and substrate specificity of severe acute respiratory syndrome coronavirus 3C-like proteinase. J. Biol. Chem. 279:1637-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fouchier, R. A., N. G. Hartwig, T. M. Bestebroer, B. Niemeyer, J. C. De Jong, J. H. Simon, and A. D. Osterhaus. 2004. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl. Acad. Sci. USA 101:6212-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fouchier, R. A., T. Kuiken, M. Schutten, G. van Amerongen, G. J. van Doornum, B. G. van den Hoogen, M. Peiris, W. Lim, K. Stohr, and A. D. Osterhaus. 2003. Aetiology: Koch's postulates fulfilled for SARS virus. Nature 423:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho, D. D., A. U. Neumann, A. S. Perelson, W. Chen, J. M. Leonard, and M. Markowitz. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373:123-126. [DOI] [PubMed] [Google Scholar]
- 11.Holmes, K. V., and M. M. C. Lai. 1996. Coronaviridae: the viruses and their replication, p. 1075-1094. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 1. Lippincott-Raven Publishers, Philadelphia, Pa. [Google Scholar]
- 12.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 13.Lamarre, D., P. C. Anderson, M. Bailey, P. Beaulieu, G. Bolger, P. Bonneau, M. Bos, D. R. Cameron, M. Cartier, M. G. Cordingley, A. M. Faucher, N. Goudreau, S. H. Kawai, G. Kukolj, L. Lagace, S. R. LaPlante, H. Narjes, M. A. Poupart, J. Rancourt, R. E. Sentjens, R. St George, B. Simoneau, G. Steinmann, D. Thibeault, Y. S. Tsantrizos, S. M. Weldon, C. L. Yong, and M. Llinas-Brunet. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186-189. [DOI] [PubMed] [Google Scholar]
- 14.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 15.Martinez, M. A., M. Cabana, M. Parera, A. Gutierrez, J. A. Este, and B. Clotet. 2000. A bacteriophage lambda-based genetic screen for characterization of the activity and phenotype of the human immunodeficiency virus type 1 protease. Antimicrob. Agents Chemother. 44:1132-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez, M. A., and B. Clotet. 2003. Genetic screen for monitoring hepatitis C virus NS3 serine protease activity. Antimicrob. Agents Chemother. 47:1760-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Normile, D. 2004. Second lab accident fuels fears about SARS. Science 303:26. [DOI] [PubMed] [Google Scholar]
- 18.Pan, W., E. Ravot, R. Tolle, R. Frank, R. Mosbach, I. Turbachova, and H. Bujard. 1999. Vaccine candidate MSP-1 from Plasmodium falciparum: a redesigned 4917 bp polynucleotide enables synthesis and isolation of full-length protein from Escherichia coli and mammalian cells. Nucleic Acids Res. 27:1094-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peiris, J. S., C. M. Chu, V. C. Cheng, K. S. Chan, I. F. Hung, L. L. Poon, K. I. Law, B. S. Tang, T. Y. Hon, C. S. Chan, K. H. Chan, J. S. Ng, B. J. Zheng, W. L. Ng, R. W. Lai, Y. Guan, and K. Y. Yuen. 2003. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 361:1767-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ptashne, M. 1986. A genetic switch. Cell Press, Cambridge, Mass.
- 21.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 22.Ruan, Y. J., C. L. Wei, A. L. Ee, V. B. Vega, H. Thoreau, S. T. Su, J. M. Chia, P. Ng, K. P. Chiu, L. Lim, T. Zhang, C. K. Peng, E. O. Lin, N. M. Lee, S. L. Yee, L. F. Ng, R. E. Chee, L. W. Stanton, P. M. Long, and E. T. Liu. 2003. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet 361:1779-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Shi, J., Z. Wei, and J. Song. 2004. Dissection study on the SARS 3C-like protease reveals the critical role of the extra domain in dimerization of the enzyme: defining the extra domain as a new target for design of highly specific protease inhibitors. J. Biol. Chem. 279:24765-24773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sices, H. J., and T. M. Kristie. 1998. A genetic screen for the isolation and characterization of site-specific proteases. Proc. Natl. Acad. Sci. USA 95:2828-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sices, H. J., M. D. Leusink, A. Pacheco, and T. M. Kristie. 2001. Rapid genetic selection of inhibitor-resistant protease mutants: clinically relevant and novel mutants of the HIV protease. AIDS Res. Hum. Retroviruses 17:1249-1255. [DOI] [PubMed] [Google Scholar]
- 27.Smith, H. O., C. A. Hutchison III, C. Pfannkoch, and J. C. Venter. 2003. Generating a synthetic genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides. Proc. Natl. Acad. Sci. USA 100:15440-15445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Der Hoek, L., K. Pyrc, M. F. Jebbink, W. Vermeulen-Oost, R. J. Berkhout, K. C. Wolthers, P. M. Wertheim-Van Dillen, J. Kaandorp, J. Spaargaren, and B. Berkhout. 2004. Identification of a new human coronavirus. Nat. Med. 10:368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vastag, B. 2003. Old drugs for a new bug: influenza, HIV drugs enlisted to fight SARS. JAMA 290:1695-1696. [DOI] [PubMed] [Google Scholar]
- 30.Wei, X., S. K. Ghosh, M. E. Taylor, V. A. Johnson, E. A. Emini, P. Deutsch, J. D. Lifson, S. Bonhoeffer, M. A. Nowak, B. H. Hahn, et al. 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373:117-122. [DOI] [PubMed] [Google Scholar]
- 31.Yang, H., M. Yang, Y. Ding, Y. Liu, Z. Lou, Z. Zhou, L. Sun, L. Mo, S. Ye, H. Pang, G. F. Gao, K. Anand, M. Bartlam, R. Hilgenfeld, and Z. Rao. 2003. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. USA 100:13190-13195. [DOI] [PMC free article] [PubMed] [Google Scholar]