Abstract
Cyclin-dependent kinases (CDKs) play key roles in eukaryotic DNA replication and cell cycle progression. Phosphorylation of components of the preinitiation complex activates replication and prevents reinitiation. One mechanism is mediated by nuclear export of critical proteins. Human papillomavirus (HPV) DNA replication requires cellular machinery in addition to the viral replicative DNA helicase E1 and origin recognition protein E2. E1 phosphorylation by cyclin/CDK is critical for efficient viral DNA replication. We now show that E1 is phosphorylated by CDKs in vivo and that phosphorylation regulates its nucleocytoplasmic localization. We identified a conserved regulatory region for localization which contains a dominant leucine-rich nuclear export sequence (NES), the previously defined cyclin binding motif, three serine residues that are CDK substrates, and a putative bipartite nuclear localization sequence. We show that E1 is exported from the nucleus by a CRM1-dependent mechanism unless the NES is inactivated by CDK phosphorylation. Replication activities of E1 phosphorylation site mutations are reduced and correlate inversely with their increased cytoplasmic localization. Nuclear localization and replication activities of most of these mutations are enhanced or restored by mutations in the NES. Collectively, our data demonstrate that CDK phosphorylation controls E1 nuclear localization to support viral DNA amplification. Thus, HPV adopts and adapts the cellular regulatory mechanism to complete its reproductive program.
Precise and timely subcellular localization of proteins is essential for their biological functions, and conversely, the control of protein localization provides the cells with a convenient way to regulate their functions. Posttranslational modifications of proteins play critical roles in these aspects. For instance, three of the most important outcomes after protein phosphorylation are typically the changes in protein localization, stability, or activity (32, 38). Eukaryotic DNA replication is strictly controlled by mechanisms that regulate the cell cycle to ensure both genetic inheritance and stability. DNA replication is initiated at a precise time when the cellular replication machinery is ready, and then the genome is replicated for a single round in each cell cycle. Cell cycle entry and progression are regulated by cyclin/cyclin-dependent kinases (CDKs) that phosphorylate key regulatory proteins (5, 59, 65, 74). At the G1/S-phase transition, cyclin/CDK complexes phosphorylate the components of the cellular DNA prereplication complex (pre-RC), including origin recognition complex, Cdc6, Cdt1, and MCM2-7, enabling the initiation of replication and subsequently preventing reduplication in the same cell cycle (5, 6, 54). The specific mechanisms vary for different components in different organisms. For example, for the replicative DNA helicase complex composed of the MCM2-7 subunits (71), phosphorylation inhibits its activity in humans and mice (29, 30). In budding yeast, phosphorylation leads to nuclear export (17, 37, 51, 52, 68, 77), whereas in Xenopus laevis, phosphorylation prevents its association with chromatin (15, 27, 57). In addition, vertebrate Cdc6 protein, the loading factor for the MCM2-7 helicase, is exported out of the nucleus upon phosphorylation (20, 33, 56, 58).
Proteins that shuttle between the cytoplasm and nucleus often possess short peptide sequences that are recognized by transporting factors. Proteins with a nuclear localization sequence (NLS) can be transported into the nucleus by a family of importin α/β heterodimers, while those with a nuclear export sequence (NES) can form a complex with export receptors (exportins) and RanGTP to be transported from the nucleus (31, 34, 50, 72). The activities of the NLS and NES are often regulated by posttranslational modifications, including phosphorylation, that can control the affinity for and accessibility by the protein transporters (31, 32). As an example, CDK phosphorylation of the human Cdc6 protein is thought to expose an NES and promote its export from the nucleus after initiation of replication (20, 33). Translocation is mediated by the nuclear export receptor CRM1 (24, 66). In contrast, cyclin B1, which is essential for the cell cycle to progress to mitosis, is shuttled out of the nucleus during interphase, attributable to the activity of a potent NES (7, 22, 75). Phosphorylation of a single serine residue within the NES at G2/M phase promotes its retention in the nucleus by disrupting the specific cyclin B1-CRM1 interaction (78, 79). In the present work, we describe our findings that the replicative DNA helicase E1 of the human papillomavirus (HPV) possesses a potent NES and that its nucleocytoplasmic localization is controlled by cyclin/CDK complexes, thereby regulating viral DNA replication.
HPVs comprise a family of small DNA tumor viruses specific for squamous mucosal or cutaneous epithelia. Infections can trigger hyperproliferation of epithelial keratinocytes and cause benign warts or condylomata. Lesions associated with certain higher-risk HPV genotypes can progress to neoplastic dysplasias and malignant cancers (84). Most infections, however, are latent, and the viral DNA persists as extrachromosomal plasmids in low copy numbers in the basal germinal stratum as a result of minimal expression of the viral genes. Viral transcription is activated in differentiated keratinocytes, especially during episodes of temporary or long-term immunosuppression. Viral DNA replication requires the cellular DNA machinery, including DNA polymerase α/primase, DNA polymerase δ, replication protein A (RPA), PCNA, and topoisomerases (36, 80). The virus encodes oncoprotein E7. Its expression in differentiated keratinocytes reestablishes an S-phase environment to support viral DNA amplification in a subset of these cells in the mid- to upper strata (11, 23).
Initiation of viral DNA replication requires the HPV origin recognition protein E2 and the replicative helicase E1 (12, 13, 25, 28, 36, 42, 61-64, 73, 80, 81). The dimeric E2 protein (40 kDa per subunit) binds to three recognition sites in the ori sequence and recruits the E1 protein (70 kDa). Facilitated by heat shock-chaperone proteins, the HPV type 11 (HPV-11) E1 protein assembles into a dihexameric, bidirectional DNA helicase (42). In the presence of RPA and topoisomerase I, the E1 protein unwinds the supercoiled ori DNA efficiently (42). E1 also recruits RPA (45) and the DNA polymerase α/primase to the replication fork to initiate replication (2, 8, 14, 47, 55). E1 is essential throughout initiation and elongation whereas E2 is required only for the assembly of the pre-RC (43).
In addition, the E1 protein interacts with cyclin E (16, 46). In vitro, E1 binds multiple cyclins and is phosphorylated by the corresponding cyclin/CDK complexes, with the exception of cyclin D/CDK4. This interaction requires the consensus RxL cyclin binding motif, which is located in the amino-terminal domain of the E1 protein (46). There are four candidate CDK phosphorylation sites in HPV-11 E1, namely, S89, S93, S107, and T468, each followed by a proline (Fig. 1A). Mutation of all four residues or of the RxL motif severely compromises transient replication of viral origin-containing plasmids in transfected cells (46), suggesting that E1 phosphorylation by cyclin/CDK is critical for initiation of viral DNA replication. The mechanism was not understood. It remained a formal possibility that one or more of the mutations may have crippled E1 helicase assembly or activity. Since HPV replication depends on S-phase entry, it was not entirely unexpected that the E1 DNA helicase could be subject to regulation by cyclin/CDK, as occurs with the host cell MCM2-7 helicase complex. However, the purpose of the virus is to produce progeny virions. If phosphorylation of E1 also prevents reinitiation as is observed with the cellular replication proteins, how could the viral DNA amplify in the nondividing keratinocytes to productive copy numbers?
FIG. 1.
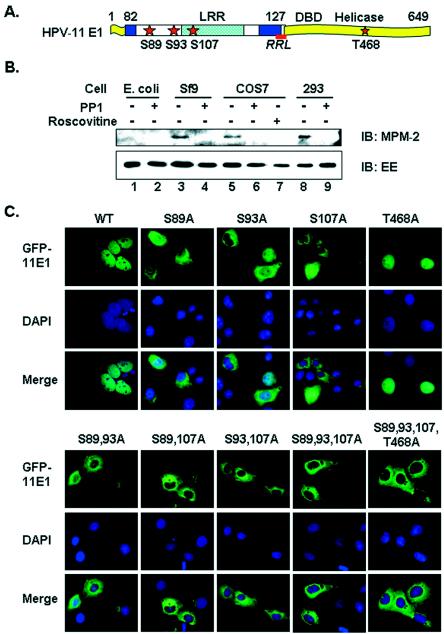
In vivo phosphorylation and subcellular localization of wild-type and mutant forms of the HPV-11 E1 protein. (A) Functional domains and critical sequences regulating HPV-11 E1 nucleocytoplasmic localization. The sketch is not to scale. LRR, residues 82 to 127, was defined inthis work. DBD (DNA binding domain) and the helicase domain were defined previously. Stars, candidate CDK phosphorylation sites. Crosshatched box, a leucine-rich NES. Underlined segment, the consensus cyclin binding motif (RRL). Filled boxes, bipartite NLS. (B) In vivo phosphorylation of EE-E1. EE-E1 protein purified from different sources with (+) or without (−) prior treatment with PP1 was blotted with MPM-2 antibody, which recognizes phospho-S/T-P. The membrane was stripped and reprobed with anti-EE antibody. IB, immunoblot. (C) Subcellular localization of GFP-11E1 and phosphorylation site mutations in transfected COS7 cells. Upper panels show the green fluorescent signals for GFP-11E1, middle panels show the DAPI-stained nuclei, and the lower panels show the merged images. WT, wild type.
In this work, we show that the HPV DNA helicase E1 contains a potent leucine-rich NES, which dominates over NLS and promotes E1 nuclear export in a CRM1-dependent manner. The E1 protein is shuttled out of the nucleus unless the NES is inactivated by CDK phosphorylation on one or more of the candidate substrate sites. Thus, one of the mechanisms by which CDK controls the timing and extent of viral DNA replication is through the regulation on E1 subcellular localization. These results reveal that HPV has devised a clever adaptation of the cellular regulatory mechanism to support its own DNA replication.
MATERIALS AND METHODS
Plasmids and antibodies.
pMTX-EE-E1 and pMT2-E2 are mammalian expression vectors encoding an EE-epitope tagged HPV-11 E1 and native E2 proteins. p7730-7933/1-99 is an HPV-11 origin-containing plasmid (13, 36). pGFP-11E2 expresses a green fluorescent protein (GFP) fusion to the HPV-11 E2 protein (83). The NES mutation, the phosphorylation site mutations, and the cyclin binding motif mutations were each constructed in the context of pGFP-11E1dm, which encodes a GFP fused to the HPV-11 E1 protein. This vector is mutated at the dominant RNA splice donor site at nucleotide 847 without affecting the encoded amino acids (21). pMTX-EE-E1dm was derived from pMTX-EE-E1 by incorporating the E1dm mutation just described. All the E1 and E2 proteins expressed from the above vectors are replication competent. pMTX EE-E1 S107A and pMTX EE-E1 S107A-NEm were then prepared in pMTX-EE-E1dm. Oligonucleotide-directed mutagenesis was performed using standard PCR methods. All PCR-amplified fragments were confirmed by DNA sequencing.
The anti-EE monoclonal antibody has been described elsewhere (13, 26, 36). For immunoprecipitation, the anti-EE antibody was cross-linked to CNBr-activated Sepharose 4 FastFlow beads (Amersham Biosciences Corp., Piscataway, N.J.) according to the manufacturer's instructions. Phosphoserine/threonine-proline-specific monoclonal antibody MPM-2 was purchased from Upstate USA (Charlottesville, Va.), the anti-GFP monoclonal antibody JL-8 and polyclonal antibody were from Clontech Laboratories (Palo Alto, Calif.), and anti-mouse immunoglobulin G-fluorescein isothiocyanate (FITC) conjugate was from Molecular Probes (Eugene, Oreg.).
Cell culture, transfection, and drug treatment.
COS7, 293, and p21-9 cells (9) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum at 37°C and 5% CO2. Cells were transfected by electroporation as described previously (13, 83). Cells were treated with roscovitine (Sigma, St. Louis, Mo.) at 20 μM 4 h posttransfection and cultured for another 20 h. For p21cip induction, isopropyl-β-thiogalactosidase (IPTG) (Invitrogen Life Technologies, Carlsbad, Calif.) was added to the culture medium to a final concentration of 50 μM 4 h posttransfection, and the cells were grown for an additional 20 h. Leptomycin B (LMB; Sigma) at 5 μg/ml was added to the culture medium to a final concentration of 20 ng/ml at 24 h posttransfection, and the cells were cultured for 6 more h.
Fluorescence microscopy.
After electroporation, 105 cells were grown on two-well chamber slides (Nalge Nunc International, Naperville, Ill.). Some were treated with the various chemicals, as described above. Cells were fixed with 4% paraformaldehyde at the indicated times. For GFP-11E1 and GFP-11E2 observations, slides were directly overlaid with coverslips by using mounting media with DAPI (4′,6′-diamidino-2-phenylindole) (Vector Laboratories, Burlingame, Calif.). Immunostaining of EE-E1 was performed using monoclonal anti-EE antibody followed by anti-mouse immunoglobulin G-FITC conjugate. These slides were mounted with DAPI-containing media. Images were captured and processed with an Olympus AX70 fluorescence microscope with FITC-DAPI filters by using an AxioCam charge-coupled device digital camera (Carl Zeiss, Oberkochen, Germany).
Protein purification, immunoprecipitation, and protein phosphatase treatment.
Purification of Esherichia coli-expressed EE-E1 and Sf9-expressed EE-E1 protein was described previously (36, 42, 46). For EE-E1 immunoprecipitation, 5 × 107 COS7 cells or 1.5 × 108 293 cells transfected with pMTX-EE-E1dm plasmid were lysed in 5 ml of NETN buffer (20 mM Tris-Cl [pH 8.0], 100 mM NaCl, 1 mM Na-EDTA, 5 mM NaF, 0.5% NP-40) containing 1 mM phenylmethylsulfonyl fluoride and 1% protease inhibitor cocktail (Sigma). Lysates were mixed with 200 μl of Sepharose 4 beads to which anti-EE antibody was coupled at 4°C overnight and then extensively washed five times with NETN buffer. Proteins were eluted with 400 μl of 0.1 M glycine buffer (pH 3.0) and neutralized with 4 μl of 1.5 M Tris-Cl buffer (pH 9.0). The final solution was concentrated using Nanosep Centrifugal columns (Pall Co., Ann Arbor, Mich.). The concentrations of EE-E1 proteins recovered were determined by Western blotting with anti-EE antibody with known concentrations of purified EE-E1 as a standard. Fifty nanograms each was used for subsequent analysis. For phosphatase 1 treatment, 2 μl of purified or immunoprecipitated EE-E1 (50 ng) was mixed with 1 U of protein phosphatase 1 (PP1) catalytic subunit (Sigma) in reaction buffer (50 mM Tris-Cl [pH 7.5], 0.1 mM Na-EDTA, 1 mM MnCl2) to a final volume of 10 μl. The mixture was incubated at 30°C for an hour. All samples were resolved by electrophoresis through sodium dodecyl sulfate (SDS)-8% polyacrylamide gels, followed by immunoblotting with individual antibodies and detection by enhanced chemiluminescence (ECL) reagents (Amersham). The MPM-2 antibody was used to detect phosphorylated proteins. The membranes were stripped and reprobed with monoclonal anti-EE antibody. GFP-11E1 immunoprecipitation from transfected COS7 cells was performed in similar fashion, except that a polyclonal anti-GFP antibody was used to recover the fusion protein, and bound protein was eluted from protein A-Sepharose beads (Amersham) with 2× SDS-polyacrylamide gel electrophoresis sample buffer and subjected to immunoblotting. MPM-2 was used to detect phosphorylated protein. The membranes were stripped and reprobed with monoclonal anti-GFP antibody (JL-8).
Transient-replication assays.
Transient-replication assays were performed using 293 cells as described elsewhere (13, 21). In each assay, 50 to 500 ng of the pGFP-11E1 plasmid, 5 μg of the pMT2-E2 plasmid, and 0.5 μg of HPV-11 origin-containing plasmid p7730-99 were cotransfected into 5 × 106 cells by electroporation. Cells were harvested 48 h posttransfection, and low-molecular-weight DNA was isolated. One half of the recovered DNA was digested with HindIII restriction endonuclease to linearize the ori plasmid. The other half was double digested with HindIII together with DpnI, which cut the unreplicated input plasmids into small fragments. Standard Southern blotting was then performed using [α-32P]dCTP-labeled probes prepared from the ori plasmid p7730-99. The results were analyzed with a PhosphorImager instrument (Molecular Dynamics, Sunnyvale, Calif.).
RESULTS
We have previously shown that a predominant intragenic splice occurs within the RNA derived from the HPV-11 E1 expression vector (21). The vector pGFP-11E1 was initially designed to encode the GFP fused to the N terminus of the wild-type HPV-11 E1 sequence. This vector supports transient replication. However, it primarily expresses a pancellular GFP variant of slightly higher molecular mass, resulting from fusion to a truncated peptide generated from the E1 intragenic splice. This predominant splice also takes place in transcripts derived from an expression vector of the native E1 protein, but the resulting peptide was too short to detect previously. A mutation at the donor splice site at nucleotide 847 (E1dm), which does not alter the encoded E1 amino acids, eliminates this splice, and the pGFP-11E1dm vector produces an abundant, replication-competent GFP-11E1 protein (21). Thus, this mutation increases the quantity of the E1 protein expressed and allows us to monitor the subcellular localization of the GFP-11E1 protein. In experiments described in this report, all E1 expression vectors contain this donor site mutation. However, for simplicity, dm is not indicated in the clones.
E1 is phosphorylated by CDKs in vivo.
Our previous report showed that the HPV-11 E1 protein is phosphorylated by multiple cyclin/CDK complexes in vitro (46). We asked whether the E1 protein is also phosphorylated in vivo. HPV-11 E1 tagged at its amino terminus with a Glu-rich (EE) epitope was expressed in E. coli, insect Sf9 cells, monkey COS7 cells, and human 293 cells. The EE-E1 protein functions as a potent DNA helicase in vitro and is replication competent in transient-replication and cell-free replication assays (36, 42, 46). The protein was purified from E. coli and Sf9 cells as described previously (36, 42-44) or was immunoprecipitated from transfected mammalian cells by using Sepharose beads conjugated to monoclonal antibody to the EE epitope. The recovered protein was then analyzed by SDS-polyacrylamide gel electrophoresis and Western blotted with an antibody specific for phospho-Ser/Thr-Pro resulting from phosphorylation by CDKs. Of several such antibodies tested, only MPM-2 (18) specifically recognizes phosphorylated EE-E1. Figure 1B shows that this antibody detected EE-E1 protein expressed in insect and mammalian cells (lanes 3, 5, and 8). However, if the EE-E1 was first treated with PP1, a serine/threonine phosphatase, reactivity with the MPM-2 antibody was abolished (Fig. 1B, lanes 4, 6, and 9), while reactivity with the anti-EE antibody was unchanged (Fig. 1B, lanes 3 to 6, 8, and 9). EE-E1 purified from E. coli was not phosphorylated and was detected only by the anti-EE antibody, not by MPM-2 antibody, with or without PP1 treatment (Fig. 1B, lanes 1 and 2). Collectively, these results demonstrate the specificity of the MPM-2 antibody and the fact that the HPV-11 E1 is indeed phosphorylated in eukaryotic cells.
To substantiate this conclusion, we treated COS7 cells expressing EE-E1 for 20 h with 20 μM roscovitine. Roscovitine is a purine derivative that reversibly competes for ATP binding and specifically inhibits the activity of CDK1 and CDK2 but has no effect on CDK4 and CDK6 (19, 49), nor on the mitogen-activated protein kinases (4), which have the same consensus substrate sites as CDKs do. Upon treatment with roscovitine, the ability to detect EE-E1 with the MPM-2 antibody was abolished (Fig. 1B, lane 7). Thus, we conclude that the HPV E1 protein is phosphorylated in vivo by one or more CDK complexes on the consensus CDK substrate site(s). Mutational analyses described below strongly suggest that CDK phosphorylation substrates included S89, S93, and S107.
Single mutations in S89, S93, or S107 significantly reduce nuclear localization.
We have previously shown that mutant forms of E1, in which the consensus cyclin binding motif or all four CDK phosphorylation sites were mutated, exhibited very low transient replication activities (46). We have also shown that, in the majority of the transfected cells, wild-type GFP-11E1 is restricted to the nucleus, whereas a small percentage of the cells exhibit cytoplasmic or pancellular signals (21). To understand the mechanism by which CDK phosphorylation regulates the E1 replication activity, we asked whether phosphorylation could affect its subcellular localization. Thus, we tracked the subcellular localization of GFP-11E1 fusion protein after the incorporation of single or multiple Ser-to-Ala mutations at the candidate CDK phosphorylation sites. Direct fluorescence microscopy of GFP-11E1 or mutations was conducted in the monkey kidney epithelial cell line COS7 and in the human kidney epithelial cell line 293. The results were identical. Only images from COS7 are presented, as these cells are larger, more adherent, and more suitable for such investigations.
Unlike the wild-type GFP-11E1, the majority of cells expressing the S89A, S93A, or S107A single mutations exhibited significant cytoplasmic signals (Fig. 1C). S107A was most defective in nuclear localization. More than 80% of the cells with GFP-11E1 S107A contained only cytoplasmic signals, whereas in S89A- and S93A-expressing cells about 60 to 70% had exclusive cytoplasmic signals. In the remaining cells for all three mutations, there was some nuclear signal in addition to a dominant cytoplasmic signal. In contrast, the GFP-11E1 T468A protein was detected predominantly in the nucleus, as with the wild-type GFP-11E1 protein (Fig. 1C). Since the T468A mutation did not affect the nucleocytoplasmic localization of E1, the subsequent experiments were mainly focused on the first three serine residues.
With the three double mutations S89,93A, S89,107A, and S93,107A, more than 90% of the transfected cells had exclusively cytoplasmic GFP-11E1 protein (Fig. 1C). When all three serine residues or all four CDK substrates were mutated, as in S89,93,107A and S89,93,107,T468A, all the GFP-11E1 protein was restricted to the cytoplasm (Fig. 1C). These observations strongly suggest that CDK phosphorylations on S89, S93, and S107 are all required for effective retention of E1 in the nucleus, with S107 being the most critical residue for this function.
Lack of phosphorylation by cyclin/CDK leads to cytoplasmic E1.
A prediction from the above interpretation is that E1 mutations unable to bind cyclins and no longer phosphorylated by CDK complexes should exhibit a phenotype similar to that for the phosphorylation site mutations. Thus, we constructed and tested GFP-11E1 mutated in the consensus cyclin binding motif RxL (46). The RRL tripeptide was mutated to RRA, KRA, or ARA (see Fig. 3A). The ARA mutation has previously been shown to disrupt the binding to cyclins (46). Our recent experiments showed that HPV-11 E1 protein had a bipartite NLS, which partially overlaps the cyclin binding motif, and that mutation of the RR residues decreased E1 nuclear localization (J. H. Yu, W. Deng, B. Y. Lin, T. R. Broker, and L. T. Chow, unpublished observation) (see Fig. 3A). Without alteration of the basic residues, the RRA and perhaps KRA mutations are not expected to compromise the NLS activity. This supposition is demonstrated for the RRA mutation, as will be described below (see Fig. 3B for the localization of RRA-NEm). Interestingly, we observed that all three mutant proteins were localized to the cytoplasm (Fig. 2A, left panel). Western blotting with the phospho-S/TP-specific antibody MPM-2 failed to detect the GFP-11E1 RRA, although the protein was observed at a level similar to that of the wild-type protein by using an anti-GFP antibody (Fig. 2A, right panel, compare lanes 2 and 3). In contrast, the wild-type GFP-11E1, which is primarily nuclear (Fig. 1C), was detected by the MPM-2 and GFP antibodies, consistent with the observation made with the EE-E1 protein (Fig. 1B). In addition, the GFP-11E1 S89,93,107,T468A protein, which is exclusively cytoplasmic (Fig. 1C), did not produce a detectable signal when probed with the MPM-2 antibody (Fig. 2A, right panel, lane 1). Collectively, these results support our interpretation that E1 phosphorylation at S89, S93, and S107 is required for efficient nuclear localization.
FIG. 3.
Identification of the leucine-rich NES necessary and sufficient for HPV-11 E1 export. All images shown were taken from transfected COS7 cells. (A) The E1 proteins of HPVs contain a conserved LRR, which consists of the CDK phosphorylation sites (red letters), the cyclin binding motif (RRL, purple letters), a putative bipartite NLS (letters in italics), and a leucine-rich NES (green box). The consensus NES core sequence and the mutations in the consensus cyclin binding motif are illustrated above, and the NEm mutation is shown below. Underlined sequence, the leucine-rich NES core of 10 amino acids (residues 106 to 115). Green letters indicate the conserved large hydrophobic amino acids in the NES. Alignments with E1 sequences of other papillomaviruses are presented at the bottom. Note that the BPV-1 E1 lacks the NES. (B) Subcellular localization of GFP-11E1 mutated in the NES (NEm). WT, wild type. (C) Subcellular localization of EE-E1 and mutations, as revealed by indirect immunofluorescence (FITC) with anti-EE antibody. (D) Delineation of the functional NES in HPV-11 E1. E1 peptides were fused downstream of GFP. The positions of the appended peptides are indicated in parentheses.
FIG. 2.
In vivo phosphorylation and subcellular localization of GFP-11E1 and mutations in the presence of inhibitors of CDKs or CRM1. Images were captured from transfected COS7 cells except those in panel C. (A) GFP-11E1 phosphorylation was blocked by mutations in the cyclin binding motif (RRL to RRA, KRA, or ARA). GFP-11E1 wild-type or mutated proteins were immunoprecipitated using polyclonal GFP antibody, blotted with MPM-2, and reprobed with monoclonal anti-GFP after stripping. (B) GFP-11E1 was localized to the cytoplasm (left panel) after CDK activity was inhibited by roscovitine (middle panels). In contrast, the nuclear localization of GFP-11E2 was not affected by roscovitine (right panels). Immunoprecipitation and Western blotting were performed as described above. (C) GFP-11E1 was localized to the cytoplasm after CDK activity was inhibited in p21-9 cells upon p21cip1 induction by IPTG (9) (left panels). The nuclear localization of GFP-11E2 was not affected by p21cip1 induction (right panels). (D) Inhibition of CRM1 with LMB (33, 53) restored nuclear localization of GFP-11E1 phosphorylation mutations. Twenty nanograms of LMB per milliliter was added to culture medium for 6 h. IB, immunoblot; WT, wild type; IP, immunoprecipitation.
Inhibition of CDK activities results in cytoplasmic E1 protein.
To verify the above interpretation further, we treated the COS7 cells transfected with GFP-11E1 with 20 μM roscovitine for 20 h. The specific inhibition of CDK activities resulted in a GFP-11E1 which was no longer detectable with the MPM-2 antibody (Fig. 2B, middle panel). On the cellular level, the GFP-11E1 protein was observed exclusively in the cytoplasm (Fig. 2B, left panel, compared with Fig. 1C). As a control, roscovitine was also used to treat cells expressing another HPV-11 protein, GFP-11E2. E2 is a nuclear protein with a known NLS and is also a substrate of CDKs (46, 83). Treatment with roscovitine did not alter the localization of GFP-11E2 (Fig. 2B, right panel). Thus, the effect of roscovitine on the localization of GFF-11E1 cannot be due to any general cytotoxicity attributable to this CDK inhibitor.
In another experiment, we used elevated p21cip1 expression to interfere with CDK activity. p21cip1 is a potent inhibitor of cyclin E/CDK2 and cyclin A/CDK2 complexes that regulate the G1/S-phase transition and also plays a key role in G1/S or G2 checkpoints following DNA damage (1, 65, 69). We transfected the GFP-11E1 expression vector into the p21-9 cell line. This cell line is derived from HT1080 human fibrosarcoma cells with an inducible p21cip1 under the control of the bacterial lacI repressor (9). Maximal induction of the p21cip1 protein is achieved by adding IPTG to the medium at 50 μM for 14 to 24 h (9). In the absence of IPTG, the GFP-11E1 protein was primarily nuclear, as in COS7 or 293 cells. Upon induction with IPTG, GFP-11E1 was observed in the cytoplasm in the majority of the cells (Fig. 2C, left panel). In contrast, induction of p21cip1 expression did not change the nuclear localization of GFP-11E2 (Fig. 2C, right panel). Collectively, these results with two different CDK inhibitors support the conclusion that phosphorylation of E1 by cyclin/CDK complexes, particularly CDK2, is required for E1 nuclear retention. These results also suggest that HPV-11 E2 is not subjected to the same regulation as HPV-11 E1 in its subcellular localization.
Mutations in E1 phosphorylation sites promote CRM1-mediated nuclear export.
To determine whether phosphorylation affected E1 nuclear import or nuclear export, we treated the cells with LMB. LMB is a natural product which binds to the nuclear export receptor CRM1 in a highly specific manner and efficiently inhibits nuclear export (35, 53). We treated COS7 cells expressing wild-type GFP-11E1 proteins with 20 ng of LMB/ml for 6 h (Fig. 2D). Virtually all cells were observed to have the wild-type GFP-11E1 protein in the nucleus. In untreated cells, about 20% contained some cytoplasmic signal, as described previously (21). With GFP-11E1 S107A, GFP-11E1 S89,93,107A, and GFP-11E1 RRA, exposure to LMB led to exclusive or significant nuclear signals, in contrast to their original, primarily cytoplasmic distribution in the absence of LMB (compare Fig. 2D with Fig. 1C and 2A). Prolonged treatment with LMB increased nuclear localization, and all the protein was localized to the nucleus at 11 h (data not shown). Similarly, all other E1 phosphorylation site mutations were also found in the nucleus upon LMB treatment (data not shown). Collectively, these results demonstrated that cytoplasmic distribution for each of these E1 mutations was caused by the rapid, continuous export from the nucleus mediated by CRM1. These observations also indicate that neither the loss of phosphorylation by CDK nor the mutation in the consensus cyclin binding motif (RRL) was able to abolish its nuclear entry completely.
A conserved, putative leucine-rich NES controls E1 export.
CRM1 binds specifically to proteins with a leucine-rich NES and, in a complex with RanGTP, transports the cargo out of the nucleus (66). The leucine-rich NES is a short peptide sequence sufficient to promote rapid protein export from the nucleus (22, 75). It has a core sequence of about 10 residues that are highly enriched with large hydrophobic residues such as leucine, isoleucine, and valine. A consensus sequence for the core peptide has been proposed to be L-X (2-3)-L/I/V/F/M-X (2-3)-L-X-L/I (7). Upon inspecting the entire E1 sequence with special attention to the region close to the three CDK substrates, we identified only one region (residues 82 to 126) which is rich in leucine, isoleucine, or valine residues (Fig. 3A), and contains a putative NES core consensus sequence, ISPRLDAIKL (residues 106 to 115, green underlining).
To test whether this region might be an NES, we mutated two of the hydrophobic resides (L110 and I113) in this putative core sequence to alanine (designated NEm) in the wild-type GFP-11E1 and in a number of CDK phosphorylation site mutations. Expression vectors for these mutations were separately transfected into COS7 cells. The nuclear signals of S89A-NEm, S93A-NEm, and S89,93,107A-NEm were increased to differing extents. In contrast, the wild-type E1-NEm and S107A-NEm were both exclusively nuclear (Fig. 3B, compare with Fig. 1C). The dramatic difference between S107A and S107A-NEm, compared with the other phosphorylation site mutations and the corresponding NEm, suggests that the primary function of phosphorylation of S107 is to inactivate the NES.
CDK phosphorylation inactivates the NES of the E1 protein.
To substantiate our hypothesis that one of the critical roles of CDK phosphorylation is to inactivate the NES, we also introduced the NEm into the GFP-11E1 RRA mutated in the consensus cyclin binding motif. GFP-11E1 RRA was cytoplasmic and was not detected by the MPM-2 antibody in Western blot assays (Fig. 2A). The introduction of the NEm did not restore CDK phosphorylation, as the protein did not react with the MPM-2 antibody (Fig. 2A, right panel, lanes 3 and 4). Nevertheless, the GFP-11E1 RRA-NEm protein was primarily located in the nucleus (Fig. 3B). Thus, this observation supports our hypotheses that the L110A,I113A mutation inactivates the NES and that the NES mutation can at least partially substitute for the requirement for CDK phosphorylation for E1 nuclear retention. We also treated cells transfected with the NEm mutations with roscovitine. This CDK inhibitor caused the wild-type E1 protein to redistribute to the cytoplasm (Fig. 2B), but it had no effect on the subcellular localization of any of the NES mutations (data not shown). Taken together, these results demonstrate that the NES is effectively inactivated by the L110A,I113A mutation and hence that CDK phosphorylation is no longer necessary for its inactivation. Moreover, these results also support our hypothesis that the leucine-rich peptide spanning residues 106 to 115 is indeed an NES core sequence and that one of the functions of CDK complexes is to inactivate it by phosphorylation, primarily on S107.
The above localization experiments were conducted by direct observation of GFP-11E1 protein and mutant forms of this protein. The GFP-11E1 protein is fully functional in supporting transient replication (21). To verify that our observations are not confounded by the GFP moiety, we expressed wild-type and mutant forms of EE-E1 protein in COS7 cells. Indirect immunofluorescence staining of EE-E1, EE-E1 S107A, and EE-E1 S107A-NEm with antibody to the EE epitope showed that each of the proteins exhibited exactly the same pattern of subcellular localization as did the corresponding GFP-11E1 counterparts (Fig. 3C, compare with Fig. 1C and 3B). EE-E1 was observed in the nucleus in most cells. EE-E1 S107A was primarily in the cytoplasm, whereas EE-E1 S107A-NEm was localized entirely in the nucleus. Hence, through observations made on both GFP-tagged and EE-tagged E1, we demonstrated that E1 phosphorylation by CDKs determines its nuclear localization through the disruption of a leucine-rich NES by phosphorylation, primarily on S107.
Delineation of a bona fide leucine-rich NES in the E1 protein.
To define the NES of the E1 protein, we fused a series of short peptides spanning the 10-amino-acid NES core (residues 106 to 115) to the C terminus of the GFP. We limited the region between residues 86 and 119 purposefully to circumvent any complications which might result from the NLS or from phosphorylation by cyclin/CDK, the association of which depends on the consensus cyclin binding motif (residues 124 to 126). As shown in Fig. 3D, because of it small size, GFP alone was observed both in the nucleus and in the cytoplasm. The localization of the fusion proteins showed that all E1 peptides which contain residues 96 to 115 conferred an exclusive cytoplasmic localization on GFP. Each contains the 10-amino-acid-long core sequence (residues 106 to 115) and also includes two additional valine residues (Fig. 3A, green box). In contrast, the 10-amino-acid core sequence was not sufficient (Fig. 3D). Thus, we defined a bona fide leucine-rich NES in E1 between residues 96 and 115, and this sequence is necessary and sufficient for export from the nucleus.
E1 replication activity correlates with its subcellular localization.
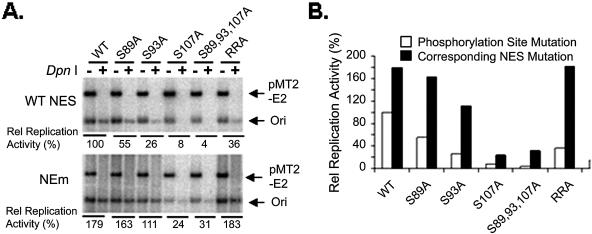
If one of the functions of E1 phosphorylation is to determine its nuclear localization, this would explain why the E1 S89,93,107,T468A mutation supported virus origin DNA replication in vivo extremely poorly, if at all (46). A prediction of this conclusion is that, if we could abolish nuclear export, we might observe a recovery of the replication activities of mutations that can no longer be phosphorylated by CDKs. We thus performed transient-replication assays to test this hypothesis (Fig. 4). 293 cells were cotransfected with 50 to 500 ng of GFP-11E1 expression plasmid, 5 μg of pMT2-E2 expression plasmid, and 0.5 μg of an HPV origin-containing plasmid. A subset of the GFP-11E1 phosphorylation mutations and their corresponding NES mutations (NEm) were tested. The replicated origin-containing plasmid (DpnI+ lanes) was revealed by Southern blotting 48 h posttransfection. The signals of replicated origin DNA were quantified, and their ratios to that of the wild-type GFP-11E1 were then calculated.
FIG. 4.
Transient replication of wild-type and mutant forms of the GFP-11E1 protein. (A) Activities of the wild-type GFP-11E1, the phosphorylation mutations, and the corresponding NES mutations (NEm) in transient replication assays. Linearized ori (arrow) in DpnI− lanes reflects the total amount of both the residual input ori and newly replicated ori plasmids, while DpnI+ lanes represent the newly replicated ori DNA. The signals from the pMT2-E2 plasmid served as a control for the complete DpnI digestion, equal ori plasmid input, transfection efficiency, and plasmid recovery. (B) Signals of replicated origin DNA (ori band from DpnI+ lane) were quantified, and the ratios to that obtained with wild-type E1 (100%) were presented as relative replication activities. WT, wild type.
In general, the absolute replication efficiency increased with the amounts of the E1 plasmid transfected, but a similar trend was observed among the various mutations. The results of one such experiment with 50 ng of E1 expression vectors are presented in Fig. 4. The data show that E1 proteins mutated at the phosphorylation site(s) or the consensus cyclin binding motif had much-reduced activities in initiating viral DNA replication. The extent of decrease correlated largely with the percentages of cells containing primarily cytoplasmic E1 protein (Fig. 1C and 2A). When the nuclear localizations of these mutations were restored through mutations in the NES (Fig. 3B), their abilities to support replication were greatly increased. In fact, the extents of replication by WT-NEm, S89A-NEm, S93A-NEm, and RRA-NEm were as high as or higher than that of the original wild-type GFP-11E1 protein. These results clearly demonstrate that cyclin/CDK phosphorylation inactivates the NES, promotes E1 nuclear retention, and consequently controls virus DNA replication. The observations also suggest that the S89A or the S93A mutation did not affect the ability of the E1 protein to function as a replicative helicase or to interact with the E2 protein and the host proteins to initiate replication. However, mutations involving S107 appeared to have additional functional impairment. The replication activities of S107A-NEm and S89,93,107A-NEm were the lowest among all the NES mutations, even though the nuclear localization of S107A-NEm and S89,93,107A-NEm was fully or largely restored, respectively (Fig. 3B).
DISCUSSION
In this work, we showed that the HPV-11 replicative DNA helicase E1 is phosphorylated by cyclin/CDK complexes in vivo (Fig. 1) and that phosphorylation on three serine residues in the amino-terminal portion of the protein determines its nuclear localization (Fig. 1). We also demonstrated that E1 has a CRM1-regulated nuclear export sequence which dominates over its NLS. Thus, unless the NES is inactivated by CDKs, E1 protein is rapidly shuttled out of the nucleus (Fig. 2 and 3). Only upon phosphorylation is E1 retained in the nucleus to support efficient virus DNA replication (Fig. 4). Our results in aggregate clearly demonstrate that, once expressed, E1 phosphorylation by CDK complexes, in particular CDK2, controls, at least in part, virus DNA replication through the regulation of its subcellular localization. We note that cyclin E/CDK2 is also required for cell-free replication where nuclear localization of E1 is irrelevant (41). The mechanism of this latter regulation remains to be elucidated.
The DNA binding domain and the helicase domain of E1 have previously been defined in HPV E1 proteins and in the bovine papillomavirus type 1 (BPV-1) E1 (Fig. 1A) (3, 10, 47, 67, 70, 76). Although some functionally significant modifications occur near the amino terminus of E1, such as phosphorylation by CDKs, casein kinase II, protein kinase A, or protein kinase C (16, 46, 48, 82), the mechanisms of regulation are not understood. The work described here has now identified a conserved region of 46 amino acids (residues 82 to 127) in the HPV E1 amino-terminal domain as a localization regulatory region (LRR) (Fig. 3A). The LRR contains the NES (residues 96 to 116) and spans the previously identified consensus cyclin binding motif RxL (residues 124 to 126) (46), the three CDK phosphorylation sites (S89, S93, and S107), and a bipartite NLS (residues 83 to 85, KRK, and residues 120 to 125, KKVKRR) (J. H. Yu, W. Deng, B. Y. Lin, T. R. Broker, and L. T. Chow, unpublished results) (Fig. 3A). These sequence elements are highly conserved among E1 proteins of many HPV genotypes, suggesting that there could exist a ubiquitous regulatory mechanism among E1 helicases of HPVs. The functions of this region are consistent with reports that the N-terminal 166 residues of HPV-11 E1 are dispensable for cell-free replication (2).
The E1 nuclear export is mediated by CRM1, and the E1 NES is inactivated by CDK phosphorylation on serine residues within or around the NES. Phosphorylation may have affected the accessibility or affinity of this sequence for CRM1. However, repeated attempts to isolate the E1/CRM1 complex from transfected cells were not successful, suggesting that the interaction is very transient. Interestingly, although the BPV-1 E1 also contains an analogous cyclin binding motif, one CDK phosphorylation site, and a bipartite NLS (16, 39, 40), it does not appear to harbor a consensus leucine-rich NES (Fig. 3A). Thus, BPV-1 E1 protein may have a different mechanism of regulation. For instance, sumoylation has been reported to be critical for the nuclear localization of BPV-1 E1 (60). It is not known whether sumoylation is also important for nuclear localization of the HPV E1 protein.
Three serine residues in the LRR, S89, S93, and S107, are candidate CDK substrates. Our in vivo data show that GFP-11E1 and EE-E1 are substrates of CDK complexes, and phosphorylated E1 can be detected by the antibody MPM-2, which reacts with phospho-S/T-P (Fig. 1B and 2A and B). Of the three Ser residues, S107 is located within the NES and the E1 S107A mutation is most defective in its nuclear localization (Fig. 1C), but it was rescued very effectively by mutations in the NES (see GFP-11E1 S107A-NEm in Fig. 3B). Thus, the primary function of S107 phosphorylation by CDK is to inactivate the NES. In contrast, a small amount of the GFP-11E1 S89A-NEm, S93A-NEm, and S89,93,107A-NEm proteins remained in the cytoplasm (Fig. 3B). These data suggest two possible interpretations that are not mutually exclusive. First, phosphorylation of S89 and S93, in addition to S107, is necessary to inactivate the NES. Second, phosphorylation of S89 and S93 may play an additional role in enhancing the NLS activity. In support of this latter interpretation, in a time course experiment during LMB treatment, E1 S107A accumulated in the nucleus much more quickly than the E1 S89A and E1 S93A did (data not shown). In that case, a cytoplasmic kinase would be responsible for their phosphorylation prior to nuclear import. Once in the nucleus, regardless of the presence or absence of this other kinase, CDK alone would be able to maintain the protein in the phosphorylated states. These hypotheses would explain the nucleocytoplasmic localization of GFP-11E1 RRA mutated in the consensus cyclin binding motif and the corresponding double mutation GFP-11E1 RRA-NEm. Since the E1-RRA protein cannot be phosphorylated by CDKs to inactivate NES (Fig. 2A), the protein was localized to the cytoplasm (Fig. 2A). However, when the NES mutation was also introduced, the nuclear localization was restored (Fig. 3B). Thus, this double mutation is more effectively transported into the nucleus than the phosphorylation site mutations S89A-NEm and S93A-NEm (compare Fig. 1A, 2A, and 3B), consistent with a role of phosphorylation of S89 and S93 by kinases other than CDK in increasing the NLS activity.
We showed that the replication activities achieved by the wild-type or mutant forms of GFP-11E1 proteins correlated with their ability to localize to the nucleus (Fig. 1 to 4), except for E1 proteins that harbor the S107A mutation. Although the nuclear localization of GFP-11E1 S107A and that of GFP-11E1S89,93,107A were effectively or largely restored by the NES mutation, their replication activities remained extremely low relative to those of other NES mutations (Fig. 4). We propose that, in addition to being a CDK substrate to inactivate NES, S107 might be very important structurally for protein conformation and thus very sensitive to any mutations at the site. It is also possible that the S107A mutation may disrupt certain interactions with viral E2 or host proteins. More work will be needed to identify the molecular basis of the defects of the S107A mutation.
The regulation of NLS and NES of the E1 protein by CDK phosphorylation provides a stringent control for E1 nuclear localization. Only when all three sites are phosphorylated can E1 be effectively transported to and retained in the nucleus. Our previous (41, 46) and present data strongly suggest that the responsible kinases include cyclin E/CDK2 or cyclin A/CDK2 (Fig. 2B and C), kinases required for S-phase entry and progression. Recent in vitro experiments have shown that each of the three serine residues at the amino terminus was indeed phosphorylated by cyclin E/CDK2 in vitro (B.-Y. Lin, T. R. Broker, and L. T. Chow, unpublished results). This would explain why, in unsynchronized transfected cells, a small fraction of cells exhibited some cytoplasmic E1 signal (21; this study); these cells might be in G1 phase, with little or no CDK2 activities. We suggest that E1, a potent DNA helicase, which very efficiently unwinds supercoiled DNA to single strands (42), must be tightly regulated in its nucleocytoplasmic localization so that E1 unwinds viral DNA only when all the cellular DNA replication machinery and substrates are available to support viral DNA replication. This interpretation agrees with the observation that E1 does not bind the G1 cyclin D and is not a substrate of cyclin D/CDK4 in vitro (46). Even though we have no direct in vivo data on the mitotic cyclin B/CDK1, it is unlikely that this kinase is involved in phosphorylating E1 to promote its nuclear localization for the reason just discussed.
As an essential component of pre-RC assembly for virus DNA replication, HPV E1 appears to be regulated by CDK complexes in a different manner concerning its subcellular localization compared to the cellular pre-RC components. Instead of being transported to the cytoplasm after a round of replication, as are the phosphorylated forms of human Cdc6 and the yeast MCM complex to prevent reinitiation and endoreduplication, phosphorylation of HPV E1 ensures its nuclear retention as long as the appropriate CDKs are present. These outcomes are consistent with their respective functions. Eukaryotic cells must maintain genetic stability and permit DNA replication once per cell cycle, whereas the virus needs to amplify its genome for progeny production. In conclusion, HPV has provided an excellent example demonstrating how viruses adopt and adapt the host cell mechanisms to promote their own reproductive programs.
Acknowledgments
This research is supported by USPHS CA83679.
We thank Igor Roninson for the p21-9 cells.
REFERENCES
- 1.Abraham, R. T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15:2177-2196. [DOI] [PubMed] [Google Scholar]
- 2.Amin, A. A., S. Titolo, A. Pelletier, D. Fink, M. G. Cordingley, and J. Archambault. 2000. Identification of domains of the HPV11 E1 protein required for DNA replication in vitro. Virology 272:137-150. [DOI] [PubMed] [Google Scholar]
- 3.Auster, A. S., and L. Joshua-Tor. 2004. The DNA-binding domain of human papillomavirus type 18 E1: crystal structure, dimerization, and DNA binding. J. Biol. Chem. 279:3733-3742. [DOI] [PubMed] [Google Scholar]
- 4.Bain, J., H. McLauchlan, M. Elliott, and P. Cohen. 2003. The specificities of protein kinase inhibitors: an update. Biochem. J. 371:199-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333-374. [DOI] [PubMed] [Google Scholar]
- 6.Blow, J. J., and B. Hodgson. 2002. Replication licensing—origin licensing: defining the proliferative state? Trends Cell Biol. 12:72-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogerd, H., R. Fridell, R. Benson, J. Hua, and B. Cullen. 1996. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol. Cell. Biol. 16:4207-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonne-Andrea, C., S. Santucci, P. Clertant, and F. Tillier. 1995. Bovine papillomavirus E1 protein binds specifically DNA polymerase α but not replication protein A. J. Virol. 69:2341-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, B.-D., K. Watanabe, E. V. Broude, J. Fang, J. C. Poole, T. V. Kalinichenko, and I. B. Roninson. 2000. Effects of p21Waf1/Cip1/Sdi1 on cellular gene expression: implications for carcinogenesis, senescence, and age-related diseases. Proc. Natl. Acad. Sci. USA 97:4291-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, G., and A. Stenlund. 1998. Characterization of the DNA-binding domain of the bovine papillomavirus replication initiator E1. J. Virol. 72:2567-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng, S., D.-C. Schmidt-Grimminger, T. Murant, T. R. Broker, and L. T. Chow. 1995. Differentiation-dependent up-regulation of the human papillomavirus E7 gene reactivates cellular DNA replication in suprabasal differentiated keratinocytes. Genes Dev. 9:2335-2349. [DOI] [PubMed] [Google Scholar]
- 12.Chiang, C.-M., G. Dong, T. R. Broker, and L. T. Chow. 1992. Control of human papillomavirus type 11 origin of replication by the E2 family of transcription regulatory proteins. J. Virol. 66:5224-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiang, C.-M., M. Ustav, A. Stenlund, T. F. Ho, T. R. Broker, and L. T. Chow. 1992. Viral E1 and E2 proteins support replication of homologous and heterologous papillomaviral origins. Proc. Natl. Acad. Sci. USA 89:5799-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conger, K. L., J.-S. Liu, S.-R. Kuo, L. T. Chow, and T. S.-F. Wang. 1999. Human papillomavirus DNA replication. Interactions between the viral E1 protein and two subunits of human DNA polymerase α/primase. J. Biol. Chem. 274:2696-2705. [DOI] [PubMed] [Google Scholar]
- 15.Coue, M., S. E. Kearsey, and M. Mechali. 1996. Chromatin binding, nuclear localization and phosphorylation of Xenopus cdc21 are cell-cycle dependent and associated with the control of initiation of DNA replication. EMBO J. 15:1085-1097. [PMC free article] [PubMed] [Google Scholar]
- 16.Cueille, N., R. Nougarede, F. Mechali, M. Philippe, and C. Bonne-Andrea. 1998. Functional interaction between the bovine papillomavirus virus type 1 replicative helicase E1 and cyclin E-Cdk2. J. Virol. 72:7255-7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalton, S., and L. Whitbread. 1995. Cell cycle-regulated nuclear import and export of Cdc47, a protein essential for initiation of DNA replication in budding yeast. Proc. Natl. Acad. Sci. USA 92:2514-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis, F. M., T. Y. Tsao, S. K. Fowler, and P. N. Rao. 1983. Monoclonal antibodies to mitotic cells. Proc. Natl. Acad. Sci. USA 80:2926-2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Azevedo, W. F., S. Leclerc, L. Meijer, L. Havlicek, M. Strnad, and S. H. Kim. 1997. Inhibition of cyclin-dependent kinases by purine analogues: crystal structure of human cdk2 complexed with roscovitine. Eur. J. Biochem. 243:518-526. [DOI] [PubMed] [Google Scholar]
- 20.Delmolino, L. M., P. Saha, and A. Dutta. 2001. Multiple mechanisms regulate subcellular localization of human CDC6. J. Biol. Chem. 276:26947-26954. [DOI] [PubMed] [Google Scholar]
- 21.Deng, W., G. Jin, B.-Y. Lin, B. A. Van Tine, T. R. Broker, and L. T. Chow. 2003. mRNA splicing regulates human papillomavirus type 11 E1 protein production and DNA replication. J. Virol. 77:10213-10226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fischer, U., J. Huber, W. C. Boelens, I. W. Mattaj, and R. Luhrmann. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82:475-483. [DOI] [PubMed] [Google Scholar]
- 23.Flores, E. R., B. L. Allen-Hoffmann, D. Lee, and P. F. Lambert. 2000. The human papillomavirus type 16 E7 oncogene is required for the productive stage of the viral life cycle. J. Virol. 74:6622-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fornerod, M., M. Ohno, M. Yoshida, and I. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 25.Fouts, E. T., X. Yu, E. H. Egelman, and M. R. Botchan. 1999. Biochemical and electron microscopic image analysis of the hexameric E1 helicase. J. Biol. Chem. 274:4447-4458. [DOI] [PubMed] [Google Scholar]
- 26.Grussenmeyer, T., K. H. Scheidtmann, M. A. Hutchinson, W. Eckhart, and G. Walter. 1985. Complexes of polyoma virus medium T antigen and cellular proteins. Proc. Natl. Acad. Sci. USA 82:7952-7954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hendrickson, M., M. Madine, S. Dalton, and J. Gautier. 1996. Phosphorylation of MCM4 by cdc2 protein kinase inhibits the activity of the minichromosome maintenance complex. Proc. Natl. Acad. Sci. USA 93:12223-12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes, F. J., and M. A. Romanos. 1993. E1 protein of human papillomavirus is a DNA helicase/ATPase. Nucleic Acids Res. 21:5817-5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishimi, Y., and Y. Komamura-Kohno. 2001. Phosphorylation of Mcm4 at specific sites by cyclin-dependent kinase leads to loss of Mcm4,6,7 helicase activity. J. Biol. Chem. 276:34428-34433. [DOI] [PubMed] [Google Scholar]
- 30.Ishimi, Y., Y. Komamura-Kohno, Z. You, A. Omori, and M. Kitagawa. 2000. Inhibition of Mcm4,6,7 helicase activity by phosphorylation with cyclin A/Cdk2. J. Biol. Chem. 275:16235-16241. [DOI] [PubMed] [Google Scholar]
- 31.Jans, D., C. Y. Xiao, and M. H. Lam. 2000. Nuclear targeting signal recognition: a key control point in nuclear transport? Bioessays 22:532-544. [DOI] [PubMed] [Google Scholar]
- 32.Jans, D. A., and S. Hubner. 1996. Regulation of protein transport to the nucleus: central role of phosphorylation. Physiol. Rev. 76:651-685. [DOI] [PubMed] [Google Scholar]
- 33.Jiang, W., N. J. Wells, and T. Hunter. 1999. Multistep regulation of DNA replication by Cdk phosphorylation of HsCdc6. Proc. Natl. Acad. Sci. USA 96:6193-6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Komeili, A., and E. K. O'Shea. 2001. New perspectives on nuclear transport. Annu. Rev. Genet. 35:341-364. [DOI] [PubMed] [Google Scholar]
- 35.Kudo, N., N. Matsumori, H. Taoka, D. Fujiwara, E. P. Schreiner, B. Wolff, M. Yoshida, and S. Horinouchi. 1999. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 96:9112-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuo, S.-R., J.-S. Liu, T. R. Broker, and L. T. Chow. 1994. Cell-free replication of the human papillomavirus DNA with homologous viral E1 and E2 proteins and human cell extracts. J. Biol. Chem. 269:24058-24065. [PubMed] [Google Scholar]
- 37.Labib, K., J. F. Diffley, and S. E. Kearsey. 1999. G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nat. Cell Biol. 1:415-422. [DOI] [PubMed] [Google Scholar]
- 38.Laney, J. D., and M. Hochstrasser. 1999. Substrate targeting in the ubiquitin system. Cell 97:427-430. [DOI] [PubMed] [Google Scholar]
- 39.Leng, X., V. G. Wilson, and X. L. Xiao. 1994. Genetically defined nuclear localization signal sequence of bovine papillomavirus E1 protein is necessary and sufficient for the nuclear localization of E1-beta-galactosidase fusion proteins. J. Gen. Virol. 75:2463-2467. [DOI] [PubMed] [Google Scholar]
- 40.Lentz, M. R., D. Pak, I. Mohr, and M. R. Botchan. 1993. The E1 replication protein of bovine papillomavirus type 1 contains an extended nuclear localization signal that includes a p34cdc2 phosphorylation site. J. Virol. 67:1414-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin, B. Y., T. Ma, J.-S. Liu, S.-R. Kuo, G. Jin, T. R. Broker, J. W. Harper, and L. T. Chow. 2000. HeLa cells are phenotypically limiting in cyclin E/CDK2 for efficient human papillomavirus DNA replication. J. Biol. Chem. 275:6167-6174. [DOI] [PubMed] [Google Scholar]
- 42.Lin, B. Y., A. M. Makhov, J. D. Griffith, T. R. Broker, and L. T. Chow. 2002. Chaperone proteins abrogate inhibition of the human papillomavirus (HPV) E1 replicative helicase by the HPV E2 protein. Mol. Cell. Biol. 22:6592-6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu, J.-S., S.-R. Kuo, T. R. Broker, and L. T. Chow. 1995. The functions of human papillomavirus type 11 E1, E2, and E2C proteins in cell-free DNA replication. J. Biol. Chem. 270:27283-27291. [DOI] [PubMed] [Google Scholar]
- 44.Liu, J.-S., S.-R. Kuo, A. M. Makhov, D. M. Cyr, J. D. Griffith, T. R. Broker, and L. T. Chow. 1998. Human Hsp70 and Hsp40 chaperone proteins facilitate human papillomavirus-11 E1 protein binding to the origin and stimulate cell-free DNA replication. J. Biol. Chem. 273:30704-30712. [DOI] [PubMed] [Google Scholar]
- 45.Loo, Y.-M., and T. Melendy. 2004. Recruitment of replication protein A by the papillomavirus E1 protein and modulation by single-stranded DNA. J. Virol. 78:1605-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma, T., N. Zou, B. Y. Lin, L. T. Chow, and J. W. Harper. 1999. Interaction between cyclin-dependent kinases and human papillomavirus replication-initiation protein E1 is required for efficient viral replication. Proc. Natl. Acad. Sci. USA 96:382-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Masterson, P. J., M. A. Stanley, A. P. Lewis, and M. A. Romanos. 1998. A C-terminal helicase domain of the human papillomavirus E1 protein binds E2 and the DNA polymerase α-primase p68 subunit. J. Virol. 72:7407-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McShan, G. D., and V. G. Wilson. 2000. Contribution of bovine papillomavirus type 1 E1 protein residue 48 to replication function. J. Gen. Virol. 81:1995-2004. [DOI] [PubMed] [Google Scholar]
- 49.Meijer, L., A. Borgne, O. Mulner, J. Chong, J. Blow, N. Inagaki, M. Inagaki, J. Delcros, and J. Moulinoux. 1997. Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin-dependent kinases cdc2, cdk2 and cdk5. Eur. J. Biochem. 243:527-536. [DOI] [PubMed] [Google Scholar]
- 50.Nakielny, S., and G. Dreyfuss. 1997. Nuclear export of proteins and RNAs. Curr. Opin. Cell Biol. 9:420-429. [DOI] [PubMed] [Google Scholar]
- 51.Nguyen, V. Q., C. Co, K. Irie, and J. J. Li. 2000. Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2-7. Curr. Biol. 10:195-205. [DOI] [PubMed] [Google Scholar]
- 52.Nguyen, V. Q., C. Co, and J. J. Li. 2001. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature 411:1068-1073. [DOI] [PubMed] [Google Scholar]
- 53.Nishi, K., M. Yoshida, D. Fujiwara, M. Nishikawa, S. Horinouchi, and T. Beppu. 1994. Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J. Biol. Chem. 269:6320-6324. [PubMed] [Google Scholar]
- 54.Nishitani, H., and Z. Lygerou. 2002. Control of DNA replication licensing in a cell cycle. Genes Cells 7:523-534. [DOI] [PubMed] [Google Scholar]
- 55.Park, P., W. Copeland, L. Yang, T. Wang, M. R. Botchan, and I. J. Mohr. 1994. The cellular DNA polymerase alpha-primase is required for papillomavirus DNA replication and associates with the viral E1 helicase. Proc. Natl. Acad. Sci. USA 91:8700-8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pelizon, C., M. A. Madine, P. Romanowski, and R. A. Laskey. 2000. Unphosphorylatable mutants of Cdc6 disrupt its nuclear export but still support DNA replication once per cell cycle. Genes Dev. 14:2526-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pereverzeva, I., E. Whitmire, B. Khan, and M. Coue. 2000. Distinct phosphoisoforms of the Xenopus Mcm4 protein regulate the function of the Mcm complex. Mol. Cell. Biol. 20:3667-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petersen, B. O., J. Lukas, C. S. Sorensen, J. Bartek, and K. Helin. 1999. Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. EMBO J. 18:396-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pines, J. 1999. Four-dimensional control of the cell cycle. Nat. Cell Biol. 1:E73-E79. [DOI] [PubMed] [Google Scholar]
- 60.Rangasamy, D., K. Woytek, S. A. Khan, and V. G. Wilson. 2000. SUMO-1 modification of bovine papillomavirus E1 protein is required for intranuclear accumulation. J. Biol. Chem. 275:37999-38004. [DOI] [PubMed] [Google Scholar]
- 61.Remm, M., R. Brain, and J. R. Jenkins. 1992. The E2 binding sites determine the efficiency of replication for the origin of human papillomavirus type 18. Nucleic Acids Res. 20:6015-6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sedman, J., and A. Stenlund. 1998. The papillomavirus E1 protein forms a DNA-dependent hexameric complex with ATPase and DNA helicase activities. J. Virol. 72:6893-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seo, Y.-S., F. Müller, M. Lusky, E. Gibbs, H.-Y. Kim, B. Phillips, and J. Hurwitz. 1993. Bovine papilloma virus (BPV)-encoded E2 protein enhances binding of E1 protein to the BPV replication origin. Proc. Natl. Acad. Sci. USA 90:2865-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seo, Y.-S., F. Müller, M. Lusky, and J. Hurwitz. 1993. Bovine papilloma virus (BPV)-encoded E1 protein contains multiple activities required for BPV DNA replication. Proc. Natl. Acad. Sci. USA 90:702-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13:1501-1512. [DOI] [PubMed] [Google Scholar]
- 66.Stade, K., C. S. Ford, C. Guthrie, and K. Weis. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90:1041-1050. [DOI] [PubMed] [Google Scholar]
- 67.Sun, Y., H. Han, and D. McCance. 1998. Active domains of human papillomavirus type 11 E1 protein for origin replication. J. Gen. Virol. 79:1651-1658. [DOI] [PubMed] [Google Scholar]
- 68.Tanaka, S., and J. F. Diffley. 2002. Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2-7 during G1 phase. Nat. Cell Biol. 4:198-207. [DOI] [PubMed] [Google Scholar]
- 69.Taylor, W. R., and G. R. Stark. 2001. Regulation of the G2/M transition by p53. Oncogene 20:1803-1815. [DOI] [PubMed] [Google Scholar]
- 70.Titolo, S., K. Brault, J. Majewski, P. W. White, and J. Archambault. 2003. Characterization of the minimal DNA binding domain of the human papillomavirus E1 helicase: fluorescence anisotropy studies and characterization of a dimerization-defective mutant protein. J. Virol. 77:5178-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tye, B. K. 1999. MCM proteins in DNA replication. Annu. Rev. Biochem. 68:649-686. [DOI] [PubMed] [Google Scholar]
- 72.Ullman, K. S., M. A. Powers, and D. J. Forbes. 1997. Nuclear export receptors: from importin to exportin. Cell 90:967-970. [DOI] [PubMed] [Google Scholar]
- 73.Ustav, M., and A. Stenlund. 1991. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 10:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weis, K. 2003. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell 112:441-451. [DOI] [PubMed] [Google Scholar]
- 75.Wen, W., J. L. Meinkoth, R. Y. Tsien, and S. S. Taylor. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell 82:463-473. [DOI] [PubMed] [Google Scholar]
- 76.White, P. W., A. Pelletier, K. Brault, S. Titolo, E. Welchner, L. Thauvette, M. Fazekas, M. G. Cordingley, and J. Archambault. 2001. Characterization of recombinant HPV6 and 11 E1 helicases. Effect of ATP on the interaction of E1 with E2 and mapping of a minimal helicase domain. J. Biol. Chem. 276:22426-22438. [DOI] [PubMed] [Google Scholar]
- 77.Yan, H., A. M. Merchant, and B. K. Tye. 1993. Cell cycle-regulated nuclear localization of MCM2 and MCM3, which are required for the initiation of DNA synthesis at chromosomal replication origins in yeast. Genes Dev. 7:2149-2160. [DOI] [PubMed] [Google Scholar]
- 78.Yang, J., E. S. G. Bardes, J. D. Moore, J. Brennan, M. A. Powers, and S. Kornbluth. 1998. Control of cyclin B1 localization through regulated binding of the nuclear export factor CRM1. Genes Dev. 12:2131-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang, J., H. Song, S. Walsh, E. S. G. Bardes, and S. Kornbluth. 2001. Combinatorial control of cyclin B1 nuclear trafficking through phosphorylation at multiple sites. J. Biol. Chem. 276:3604-3609. [DOI] [PubMed] [Google Scholar]
- 80.Yang, L., R. Li, I. J. Mohr, R. Clark, and M. R. Botchan. 1991. Activation of BPV-1 replication in vitro by the transcription factor E2. Nature 353:628-632. [DOI] [PubMed] [Google Scholar]
- 81.Yang, L., I. Mohr, E. Fouts, D. A. Lim, M. Nohaile, and M. R. Botchan. 1993. The E1 protein of bovine papilloma virus 1 is an ATP-dependent DNA helicase. Proc. Natl. Acad. Sci. USA 90:5086-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zanardi, T. A., C. M. Stanley, B. M. Saville, S. M. Spacek, and M. R. Lentz. 1997. Modulation of bovine papillomavirus DNA replication by phosphorylation of the viral E1 protein. Virology 228:1-10. [DOI] [PubMed] [Google Scholar]
- 83.Zou, N., B. Y. Lin, F. Duan, K.-Y. Lee, G. Jin, R. Guan, G. Yao, E. J. Lefkowitz, T. R. Broker, and L. T. Chow. 2000. The hinge of the human papillomavirus type 11 E2 protein contains major determinants for nuclear localization and nuclear matrix association. J. Virol. 74:3761-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.zur Hausen, H. 2000. Papillomaviruses causing cancer: evasion from host-cell control in early events in carcinogenesis. J. Natl. Cancer Inst. 92:690-698. [DOI] [PubMed] [Google Scholar]