Abstract
Hexaploid wheat consists of three subgenomes, namely, A, B, and D. These well-characterized ancestral genomes also exist at the diploid and tetraploid levels, thereby rendering wheat as a good model species for studying polyploidization. Here, we performed intra- and inter-species comparative analyses of wheat and its relatives to dissect polymorphism and differentiation of the TaGW2 genes. Our results showed that genetic diversity of TaGW2 decreased with progression from the diploids to tetraploids and hexaploids. The strongest selection occurred in the promoter regions of TaGW2-6A and TaGW2-6B. Phylogenetic trees clearly indicated that Triticum urartu and Ae. speltoides were the donors of the A and B genomes in tetraploid and hexaploid wheats. Haplotypes detected among hexaploid genotypes traced back to the tetraploid level. Fst and π values revealed that the strongest selection on TaGW2 occurred at the tetraploid level rather than in hexaploid wheat. This infers that grain size enlargement, especially increased kernel width, mainly occurred in tetraploid genotypes. In addition, relative expression levels of TaGW2s significantly declined from the diploid level to tetraploids and hexaploids, further indicating that these genes negatively regulate kernel size. Our results also revealed that the polyploidization events possibly caused much stronger differentiation than domestication and breeding.
Keywords: TaGW2, genetic differentiation, grain size, nucleotide polymorphism, Triticum aestivum
Introduction
Polyploidization has played an important role in the evolution of plant eukaryotes. Polyploids arise by chromosome doubling of an individual genome (autoploidy) or by chromosome doubling of hybrids between species whose chromosomes normally do not pair (allopolyploidy). Common or bread wheat (Triticum aestivum L., 2n = 6x = 42), which represents one of the best-characterized examples of polyploidization, evolved through two hybridization events (Marcussen et al., 2014). Common wheat consists of three sets (or genomes) of homologous chromosomes, named A, B, and D, with each composed of 7 chromosomes. Bread wheat evolved by two spontaneous hybridization events (Feuillet et al., 2008). Natural hybridization between diploid species T. urartu (2n = 2x = 14, AA) and an unknown B genome species, giving rise to a tetraploid species (T. dicoccoides L., 2n = 28, AABB), occurred about 500,000 years ago (Dvorák et al., 1993; Mori et al., 1995; Huang et al., 2002; Dvorak and Akhunov, 2005). The origin of the B genome has been a discussion topic for many years, and differences in viewpoint have hindered its elucidation (Sarkar and Stebbins, 1956; Kimber and Athwal, 1972; Dvorák and Zhang, 1990; Wang et al., 1997; Maestra and Naranjo, 1998; Huang et al., 2002). More recent studies generally support the view that Aegilops speltoides (2n = 2x = 14, SS) is the donor, or major contributor of the B genome (Petersen et al., 2006; Kilian et al., 2007). The second step hybridization took place 7,000–10,000 years ago between a tetraploid species and diploid Ae. tauschii (2n = 2x = 14, DD) (Kihara, 1944; McFadden and Sears, 1944), resulting in the bread wheat (2n = 6x = 42, AABBDD) (Kihara, 1944; Huang et al., 2002). Compared to other allopolyploids wheat is considered to be a relatively young polyploid. For this reason and its importance as a major food crop wheat has long been employed as a classical model for studying the process of allopolyploidization in crop plants.
Grain weight is an important domestication and breeding trait. Rice is an important crop model diploid plant and its yield genetics have been studied extensively (Xing and Zhang, 2010; Bai et al., 2012; Zuo and Li, 2014). The cloned GW2 on rice chromosome 2S encodes a ubiquitin E3 ligase, whose deletion leads to increased grain width and weight, thereby improving yield (Song et al., 2007), but it was not strongly selected during domestication or in breeding (Lu et al., 2013). Research on wheat GW2 homologs has been extensive, and includes gene cloning, functional marker development and elucidation of the genetic effects of each homolog (Su et al., 2011; Qin et al., 2014; Jaiswal et al., 2015). Expression analysis suggested that the TaGW2 genes were constitutively expressed in different tissues (Su et al., 2011). Yang et al. (2012) identified a single-base insertion in the eighth exon of TaGW2-6A in the landrace Lankaodali. This caused premature termination and led to increased grain width and weight. However, RNAi results showed that the patterns of TaGW2 regulation on grain development might be more complex (Bednarek et al., 2012; Hong et al., 2014). Simmonds et al. (2016) screened an EMS TILLING population of a tetraploid wheat cultivar “Kronos” and found that a GW2-A1 mutant allele significantly increased thousand grain weight, grain width and grain length in both durum and bread wheats. These studies mainly focused on gene cloning, marker development, and expression analysis, whereas the evolution of TaGW2s during wheat polyploidization was not examined yet.
Nucleotide polymorphism and genetic differentiation of three TaGW2 homologs in wheat and its ancestors and relatives were investigated in the present study, the aims of which were to: (1) determine the TaGW2 nucleotide diversity at the genomic level in 164 accessions of wheat and related species; (2) assess the genetic differentiation and interspecific relationships among diploids, tetraploids, and hexaploids based on Fst values; (3) analyze the diversity and genetic differentiation in wheat and related species in order to understand the evolutionary pattern of TaGW2 genes; (4) construct a TaGW2 haplotype network that developed during polyploidization and track the haplotypes of TaGW2-6A, and -6B present in common wheat and known to have undergone strong selection; and 5) elucidate the relationship between TaGW2 expression levels and grain weight in relation to polyploidization by real-time quantitative PCR. Finally, we also wished to compare the genetic diversity (π) and genetic differentiation of TaGW2 promoters and coding regions among diploids, tetraploids, and hexaploids for a better understanding of wheat evolution using a key gene.
Materials and methods
Plant materials
The TaGW2 sequences in 164 accessions of wheat and related species were generated. The accessions comprised 79 diploids, 55 tetraploids and 30 hexaploids (Table S1) and included 12 T. urartu (AA), 8 T. boeoticum (AA), 15 T. monococcum (AA), 12 Ae. speltoides (SS), 6 Ae. longissima (SS), 4 Ae. sharonensis (SS), 2 Ae. searsii (SS), 20 Ae. tauschii (DD), 8 T. dicoccoides (AABB), 14 T. dicoccum (AABB), 16 T. durum (AABB), 8 T. turgidum L. (AABB), 3 T. carthlicum (AABB), 2 T. polonicum (AABB), 2 T. turanicum (AABB), 2 T. araraticum (AAGG), and 30 T. aestivum (16 landraces and 14 modern cultivars, AABBDD) accessions. All were obtained from Chinese Crop Germplasm Resources Information System (http://www.cgris.net/zhongzhidinggou/index.php). Detailed information for each accession is given in Table S2.
Phenotypic traits
Cultivars used in this study were planted at the CAAS-Shunyi Experiment Station in Beijing (116.3°E, 40.0°N) during the wheat-growing season. Each cultivar was planted in 2 m double rows spaced 25 cm apart, with 20 seeds planted in each row. Field management followed local practices. Mean widths (mm) of 20 kernels, and 100-grain weights of two samples for each accession converted to 1,000-kernel weight were obtained for analysis. Detailed information is provided in Table S3.
DNA and RNA extraction
Genomic DNA was extracted from leaves of 15-day-old seedlings using the CTAB method (Chen and Ronald, 1999). Mature grains were ground to a powder in liquid nitrogen and total RNA was extracted using a TIANGEN RNAplant plus Reagent (Tiangen, Beijing) following instructions given with the kit. The cDNA was synthesized using the SuperScript II system (Invitrogen, Madison, WI, USA) according to the manufacturer's instructions, and then diluted 10-fold for subsequent quantitative real-time PCR (qRT-PCR) analysis.
Primers and PCR amplification
Primers (Table S4) designed using Primer Premier 5.0 software (http://www.premierbiosoft.com/) was synthesized by Shanghai Sangon Biological Technology Co., Ltd (http://www.sangon.com/). PCR were performed in total volumes of 15 μL comprising 50 ng of genomic DNA, 1 μL of 10 mM forward and reverse primers, 0.24 μL of 25 mM dNTPs, 7.5 μL of GC Buffer I, and 0.15 μL of LA Taq Polymerase (Takara, Dalian). Samples for PCR were incubated at 94°C for 4 min, followed by 35 cycles of 94°C for 45 s, annealing for 45 s, and extension at 72°C for 30 s to 3 min, with a final extension for 10 min. The annealing temperature and extension time varied according to the primer set and size of PCR product (Table S4).
Sequencing
Two pairs of primers (TaGW2-P-1 and TaGW2-P-2) for promoter amplification, and four pairs of primers (TaGW2-1, TaGW2-2, TaGW2-3, and TaGW2-4) for the coding sequences were designed for amplification (Table S4). PCR were as mentioned above. PCR products were separated by electrophoresis in agarose gels, and the target bands were extracted and cloned into pEASY-T1 simple vectors and transformed to DH5α-competent Escherichia coli by the heat shock method (Beijing Trans Gen Biotech Co., Ltd, Product Code: CT111). Positive clones were selected for sequencing by an ABI 3730XI DNA Analyzer (Applied Biosystems). PCR and DNA sequencing were repeated at least three times to ensure sequence accuracy. Promoter and coding sequences of TaGW2s for diploids, tetraploids, and hexaploids were submitted to GenBank (Accession numbers: BankIt1971968 KY264756-KY264772).
Expression analysis
Genome-specific primers were designed according to cDNA sequence differences of the three homologous genes and used to evaluate the correlation of gene expression levels of TaGW2-6A/6B/6D and grain weight. The primer sets for TaGW2 (TaGW2-6A-RT, TaGW2-6B-RT, and TaGW2-6D-RT) and Actin (Table S4) were used for amplification of TaGW2 and actin genes, respectively. The qRT-PCR was conducted using mRNA extracted from mature seeds of diploids, tetraploids and hexaploids, with SYBR Premix Ex-Taq (Takara, Dalian) on a 7,500 Real-time PCR system (Applied Biosystems, Foster City, CA). qRT-PCR were performed in total volumes of 20 μL, containing 2 μL of cDNA, 1 μL of 2 mM gene-specific primers, 0.4 μL of ROX Reference Dye (50×), and 10 μL of 2×SYBR Premix Ex-Taq. The relative expression values of TaGW2s were calculated by the 2−ΔΔCt method using actin gene as endogenous control, which was not variable in different tissues and developmental stages of wheat under our experiment (Livak and Schmittgen, 2001; Bednarek et al., 2012). Each measurement was determined on at least two independent biological samples, with three replicates for each sample.
Data analyses
The full length sequence alignments of TaGW2-6A/6B/6D genes were conducted using DNAStar (http://www.dnastar.com/). The Fst test was performed using Arlequin 3.5.1.2 (http://cmpg.unibe.ch/software/arlequin3/) based on the data from the software DNAStar. Diversity analyses, Tajima's D-tests (Tajima, 1989), and synonymous substitution tests were conducted using DnaSP 5.10 (http://www.ub.es/dnasp). Phylogenetic trees were drawn using MEGA 6.0 (http://www.megasoftware.net/). Haplotype networks based on the TaGW2 DNA sequences were constructed using TCS 1.21 (Clement et al., 2000). Variance analyses and significance tests were performed using SPSS System for Windows Version 12.0 (http://www-01.ibm.com/software/analytics/spss/). Tukey's test was used to determine statistical differences by one-way ANOVA.
Results
Nucleotide polymorphism of TaGW2s in diploids, tetraploids, and hexaploids
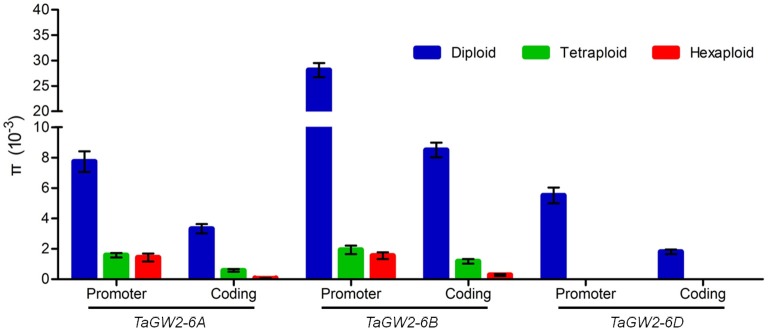
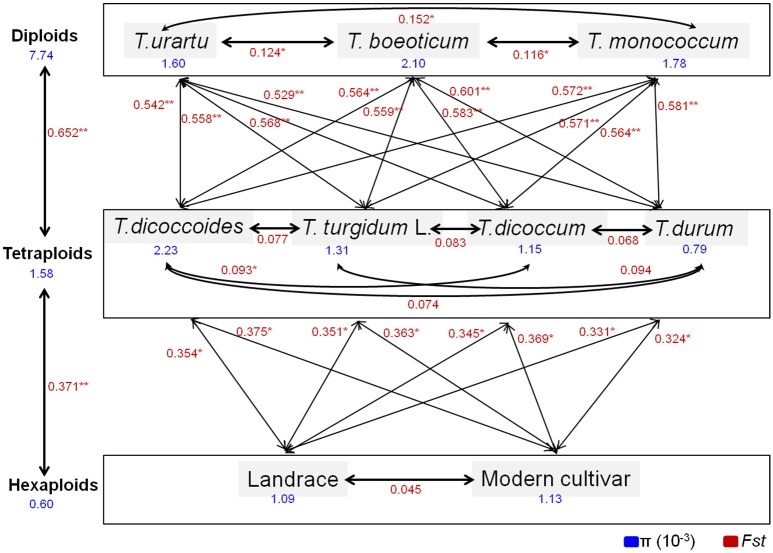
Nucleotide polymorphisms of the three TaGW2 homoeologs were obtained from ~2.9 kb of promoter regions and ~8.8 kb of coding regions. Genetic diversity decreased significantly (P < 0.01) with progression from diploids to hexaploids (Table 1). For example, there were 119 polymorphisms (SNPs + InDels) in diploids (π = 4.64 × 10−3), 34 in tetraploids (π = 1.25 × 10−3), and only 10 in hexaploids (π = 0.60 × 10−3) at TaGW2-6A. At TaGW2-6B, the comparable frequencies were 284, 36 and 15 with π values of 15.51 × 10−3, 1.45 × 10−3, and 0.81 × 10−3, respectively. At TaGW2-6D the number of polymorphic sites was 93 in Ae. tauschii with none being identified in hexaploid wheat. Diversity in the promoter regions of TaGW2s was significantly higher than in the coding regions (P < 0.05), indicating a high level of conservation in the gene-coding regions (Table 1 and Figure 1). In both promoters and coding regions TaGW2-6B had the highest genetic diversity followed by TaGW2-6A and TaGW2-6D (Figure 1).
Table 1.
Nucleotide polymorphisms and π values of TaGW2 genes in diploids, tetraploids, and hexaploids.
| Gene | Region | Length (bp) | Diploids | Tetraploids | Hexaploids | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNPs | InDels | π ± S.E (10−3) | P-value | SNPs | InDels | π ± S.E (10−3) | P-value | SNPs | InDels | π ± S.E (10−3) | P-value | |||
| TaGW2-6A | Promoter | 2400 | 37 | 3 | 7.74 ± 0.68a (A) | 0.000 | 20 | 6 | 1.58 ± 0.15b (B) | 0.016 | 7 | 2 | 1.43 ± 0.26c (C) | 0.000 |
| Coding | 8816 | 70 | 9 | 3.32 ± 0.31a (A) | 7 | 1 | 0.58 ± 0.09b (B) | 1 | 0 | 0.05 ± 0.02c (C) | ||||
| Total | 11216 | 107 | 12 | 4.64 ± 0.52a (A) | 27 | 7 | 1.25 ± 0.24b (B) | 8 | 2 | 0.60 ± 0.45c (C) | ||||
| TaGW2-6B | Promoter | 2350 | 122 | 4 | 28.12 ± 1.39a (A) | 0.000 | 15 | 5 | 1.93 ± 0.28b (B) | 0.003 | 9 | 0 | 1.55 ± 0.22b (B) | 0.000 |
| Coding | 8888 | 150 | 8 | 8.50 ± 0.48a (A) | 14 | 2 | 1.18 ± 0.15b (B) | 4 | 2 | 0.28 ± 0.06c (C) | ||||
| Total | 11238 | 272 | 12 | 15.51 ± 0.82a (A) | 29 | 7 | 1.45 ± 0.14b (B) | 13 | 2 | 0.81 ± 0.11c (C) | ||||
| TaGW2-6D | Promoter | 2400 | 30 | 5 | 5.51 ± 0.52a (A) | 0.000 | – | – | – | 0 | 0 | 0 b (B) | ||
| Coding | 8825 | 46 | 12 | 1.81 ± 0.15a (A) | – | – | – | 0 | 0 | 0 b (B) | ||||
| Total | 11225 | 76 | 17 | 2.61 ± 0.34a (A) | – | – | – | 0 | 0 | 0 b (B) | ||||
π, average number of nucleotide differences between two sequences. Capital and small letters indicate significance levels at P < 0.01 and P < 0.05 in comparison between ploidy levels for each region.
Figure 1.
Comparison of gene diversities (π) of TaGW2s between the promoter and coding regions in diploids (blue), tetraploids (green), and hexaploids (red). Bars represent the standard errors.
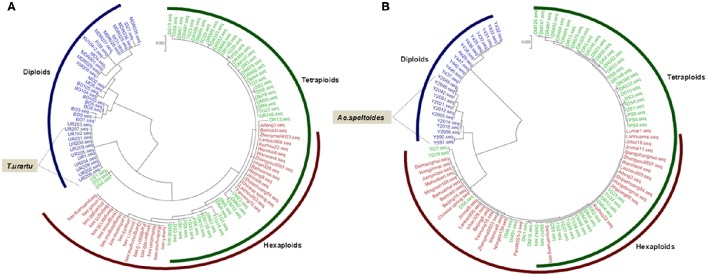
Phylogenetic analysis of TaGW2-6A in wheat and its relatives showed that diploid Triticum species clustered into a single subgroup, but the boundary between tetraploid and hexaploid species was not distinct. Interestingly, T. urartu was more closely related to the hexaploids than T. boeoticum and T. monococcum indicating that it may be the direct donor of the A genome (Figure 2A). At TaGW2-6B, Aegilops speltoides accessions clustered into a single major subgroup, with tetraploid and hexaploid accessions placed in another major subgroup. The closest phylogenetic relationship involved Ae. speltoides leading us to hypothesize that this species is the direct donor, or a major contributor, of TaGW2-6B (Figure 2B). As predicted, common wheat and Ae. tauschii clustered into subgroups based on TaGW2-6D (Figure S1).
Figure 2.
Phylogenic analysis of TaGW2-6A (A) and TaGW2-6B (B) in diploids (blue), tetraploids (green), and hexaploids (red).
The strongest genetic differentiation of TaGW2s occurred at polyploidization rather than during domestication or breeding
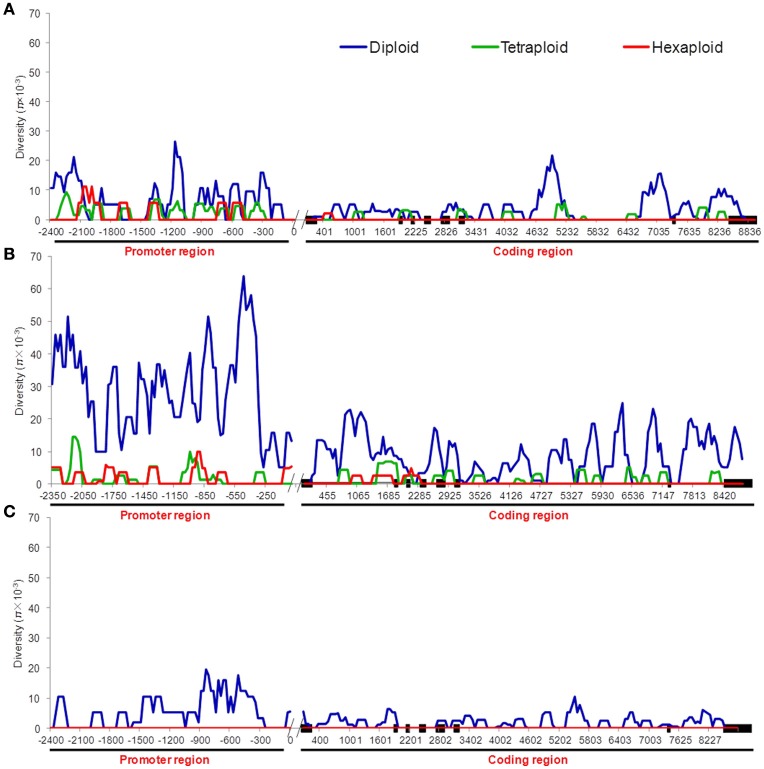
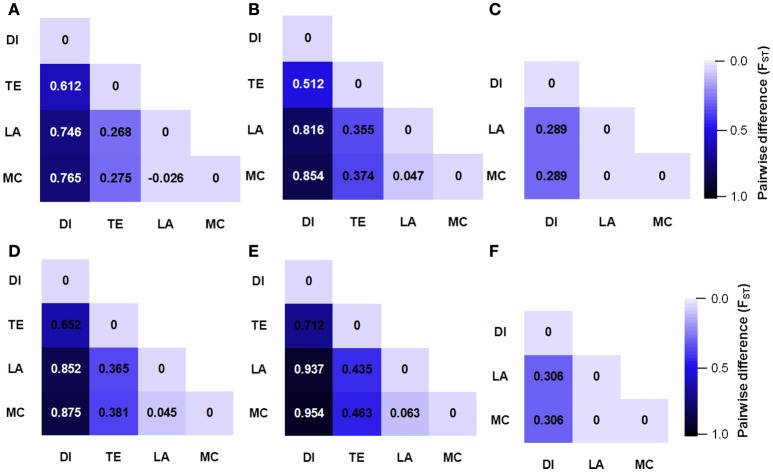
We compared the diversity and genetic differentiation of TaGW2s among hexaploids (modern cultivars and landraces), diploids, and tetraploids. A clear reduction in diversity occurred with progression from diploids to tetraploids (Figures 3, 4 and Table S5). Fst values for the coding and promoter regions of TaGW2-6A were 0.612 and 0.652 (P < 0.01) between diploids and tetraploids, 0.268 and 0.365 (P < 0.01) between tetraploids and landraces, and −0.026 and 0.045 between common wheat landraces and modern cultivars, respectively (Figure 4). In the coding region of TaGW2-6B, Fst was 0.512 (P < 0.01) between diploids and tetraploids, 0.355 and 0.374 (P < 0.01) between tetraploids and landraces and modern cultivars, and only 0.047 (P < 0.05) between landraces and modern cultivars. However, the Fst of the promoter region of TaGW2-6B was slightly higher than that of the coding region. In the coding region of TaGW2-6D the Fst between Ae. tauschii and common wheat was 0.289 (P < 0.01), and 0.306 in the promoter region (P < 0.01). There was no difference between landraces and modern cultivars in the coding and promoter regions of TaGW2-6D (Figures 3, 4). Therefore, the genetic differentiation between diploids and tetraploids was stronger than that between tetraploids and hexaploids in both the promoter and coding regions of TaGW2s. The Fst and π values (Figures 3, 4) also revealed stronger selection on TaGW2 during tetraploidization than during hexaploidization. This was further supported by Tajima's tests. A significant deviation from the value of zero (P < 0.05) between the promoter and coding regions was detected in both diploids and hexaploids, thereby indicating that TaGW2-6A underwent strong selection at the regions of this locus (Table S6). Only the promoter region of TaGW2-6B in both diploids and hexaploids underwent selection, and TaGW2-6D underwent selection in hexaploids (Table S6).
Figure 3.
π value comparison of TaGW2-6A (A), TaGW2-6B (B), and TaGW2-6D (C) between promoter and coding regions in diploids (blue), tetraploids (green), and hexaploids (red). Black solid block in the horizontal axis indicates the exon, double slash indicates the boundary between coding and promoter regions, and numbers show the physical position of sequences.
Figure 4.
Genetic differentiation (Fst) at coding and promoter regions of TaGW2-6A (A,D), TaGW2-6B (B,E), and TaGW2-6D (C,F) between pairs of populations. The color gradient represents changes in Fst value from dark (1.0) to light blue (0.0). DI, diploids; TE, tetraploids; LA, landraces; MC, modern cultivars.
Further comparison of polymorphisms and Tajima's D-values of TaGW2s in wheat and its relatives (Table 2) showed that the π values of the promoter and coding regions at TaGW2-6A were the highest in T. boeoticum (2.1 × 10−3 and 1.6 × 10−3). Tajima's D-values indicated that the promoter of TaGW2-6A underwent selection in both landraces and modern cultivars (P < 0.05), whereas in the coding region, selection occurred in T. urartu and T. boeoticum. The π values of the promoter and coding regions at TaGW2-6B were the highest in Ae. speltoides (4.82 × 10−3 and 1.98 × 10−3), and the coding region underwent strong selection (Table 2). According to Tajima's D-values, the strongest selection occurred in the promoter regions of both TaGW2-6A and TaGW2-6B.
Table 2.
Nucleotide polymorphisms and Tajima's D of TaGW2 genes in wheat and related species.
| Gene | Ploidy | Species | No. of accessions | Promoter | Gene | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Length (bp) | SNPs | π (10−3) | θ (10−3) | Tajima's D | Length (bp) | SNPs | π (10−3) | θ (10−3) | Tajima's D | ||||
| TaGW2-6A | Diploid | T. urartu | 12 | 2400 | 12 | 1.60 | 1.20 | −1.45138 | 8816 | 30 | 1.16 | 1.14 | 0.07126* |
| T. boeoticum | 8 | 11 | 2.10 | 1.75 | 1.01793 | 41 | 1.60 | 1.78 | −0.52090 | ||||
| T. monococcum | 14 | 13 | 1.78 | 1.65 | 0.32036 | 20 | 0.82 | 0.91 | −0.40491 | ||||
| Tetraploid | T. dicoccoides | 8 | 12 | 2.23 | 1.89 | 0.90906 | 18 | 1.12 | 1.89 | 0.65742 | |||
| T. dicoccum | 14 | 7 | 1.15 | 0.90 | 1.06282 | 12 | 0.58 | 0.90 | 0.73268 | ||||
| T. durum | 16 | 12 | 0.79 | 1.47 | −1.77781 | 13 | 0.80 | 0.97 | 0.47695 | ||||
| T. turgidum | 8 | 8 | 1.31 | 1.25 | 0.20201 | 11 | 0.66 | 0.85 | 0.39853 | ||||
| Hexaploid | Landraces | 16 | 7 | 1.42 | 0.82 | 2.61475** | 1 | 0.05 | 0.04 | 0.67135 | |||
| Modern cultivar | 14 | 7 | 1.37 | 0.83 | 2.42088* | 1 | 0.04 | 0.04 | 0.84865 | ||||
| TaGW2-6B | Diploid | Ae. speltoides | 12 | 2350 | 30 | 4.82 | 4.21 | 0.64836 | 8888 | 39 | 1.98 | 1.82 | 1.38834* |
| Ae. longissima | 6 | 12 | 2.34 | 1.35 | −0.05002 | 23 | 1.13 | 0.98 | −0.43045 | ||||
| Ae. sharonensis | 4 | 15 | 2.66 | 2.87 | −1.33698 | 28 | 1.07 | 1.00 | 0.31789 | ||||
| Tetraploid | T. dicoccoides | 8 | 9 | 1.62 | 1.39 | 0.79684 | 19 | 0.81 | 0.99 | 0.46725 | |||
| T. dicoccum | 14 | 16 | 1.85 | 2.02 | −0.33910 | 16 | 0.93 | 1.02 | 0.73653 | ||||
| T. durum | 16 | 14 | 1.86 | 1.70 | 0.38178 | 14 | 0.93 | 0.91 | 0.56473 | ||||
| T. turgidum | 8 | 8 | 1.38 | 1.24 | 0.53786 | 11 | 0.69 | 1.24 | 0.46378 | ||||
| Hexaploid | Landraces | 16 | 8 | 1.50 | 1.14 | 1.07627 | 5 | 0.27 | 0.19 | 1.71112 | |||
| Modern cultivar | 14 | 9 | 1.60 | 1.02 | 1.44838 | 5 | 0.31 | 0.22 | 1.85358 | ||||
| TaGW2-6D | Diploid | Ae. tauschii | 20 | 2400 | 30 | 5.46 | 3.39 | 0.86383 | 8825 | 46 | 1.81 | 1.50 | 0.86383 |
| Hexaploid | Landraces | 16 | 0 | 0 | 0 | — | 0 | 0 | — | — | |||
| Modern cultivar | 14 | 0 | 0 | 0 | — | 0 | 0 | — | — | ||||
Significant at P < 0.05;
Significant at P < 0.01.
Comparisons of nucleotide polymorphisms (π) and genetic differentiation (Fst) within and between ploidy levels in both the promoter and coding regions of TaGW2-6A (Figure 5 and Figure S2) showed that the π value in diploids was the highest, followed by the tetraploids, and lastly hexaploids. Genetic differentiation (Fst) in the diploids (T. urartu, T. boeoticum, and T. monococcum) varied from 0.10 to 0.16, in tetraploids (T. dicoccoides, T. turgidum, T. dicoccum, and T. durum) from 0.05 to 0.10, and in hexaploids (landrace and modern cultivar groups) less than 0.05. Compared to TaGW2-6A the π and Fst values for TaGW2-6B showed similar patterns of variation (Figures S3, S4). The Fst values among related wheat species also showed that stronger differentiation occurred at polyploidization rather than during domestication or breeding, and further indicated that the strongest selection occurred in the promoter regions of TaGW2-6A and TaGW2-6B.
Figure 5.
π and Fst values for the promoter region of TaGW2-6A in wheat-related species. Blue font indicates the value of genetic diversity (π), and red font shows the value of genetic differentiation (Fst). *Significant at P < 0.05; **Significant at P < 0.01.
TaGW2 haplotypes in common wheat can be traced back to tetraploid wheat groups
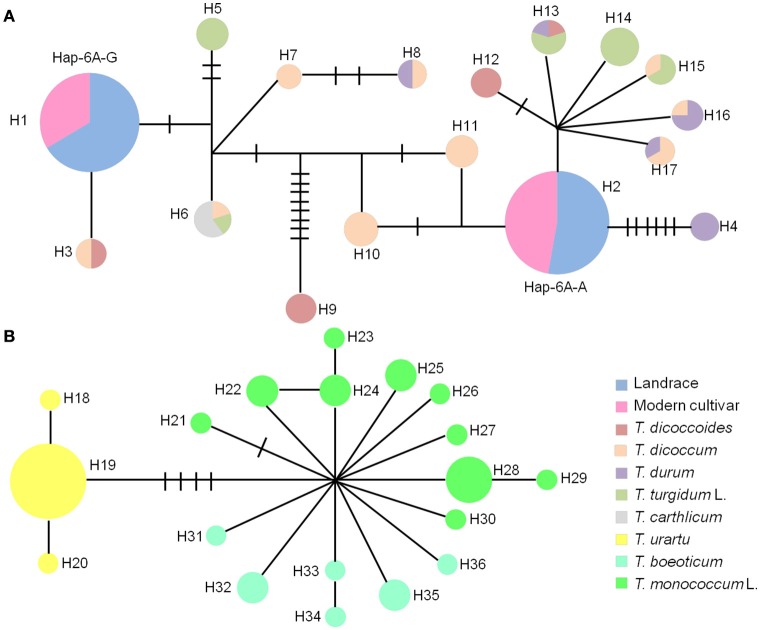
The haplotype network in wheat relatives showed that the A genomes of TaGW2 were clustered into two unconnected sub-networks. The hexaploids and tetraploids clustered in the same group, whereas diploids formed a distinct set (Figure 6). Fifteen TaGW2-6A haplotypes were detected in tetraploids and two in hexaploids, whereas there were 19 in diploids. The favorable (greater kernel width and weight) Hap-6A-A haplotype in common wheat was located close to that of T. durum, whereas Hap-6A-G was close to T. dicoccoides and T. dicoccum. The haplotype network of the B genomes also clustered into two sub-networks (Figure S5). The hexaploids and tetraploids clustered into the same sub-network, and diploids, including Ae. speltoides, Ae. Longissima, and Ae. sharonensis, were in a separate sub-network. Eleven haplotypes of TaGW2-6B were detected in tetraploids, whereas there were four in hexaploids, and 13 in the diploids. The favorable haplotype Hap-6B-1 detected in common wheat was located close to that of T. dicoccoides and T. dicoccum, whereas the origin of Hap-6B-2 was uncertain and could have been from any of the tetraploid species. The unfavorable haplotypes Hap-6B-3 and Hap-6B-4 formed a separate branch. The networks also showed the dramatic reduction in numbers of haplotypes at TaGW2s during polyploidization. A relative consistency of haplotypes existed between tetraploids and hexaploids.
Figure 6.
Haplotype networks of TaGW2-6A based on promoter sequences in diploids, tetraploids and hexaploids. (A) Haplotype networks of tetraploids and hexaploids. (B) Haplotype networks of diploids. Colored circles represent various subspecies.
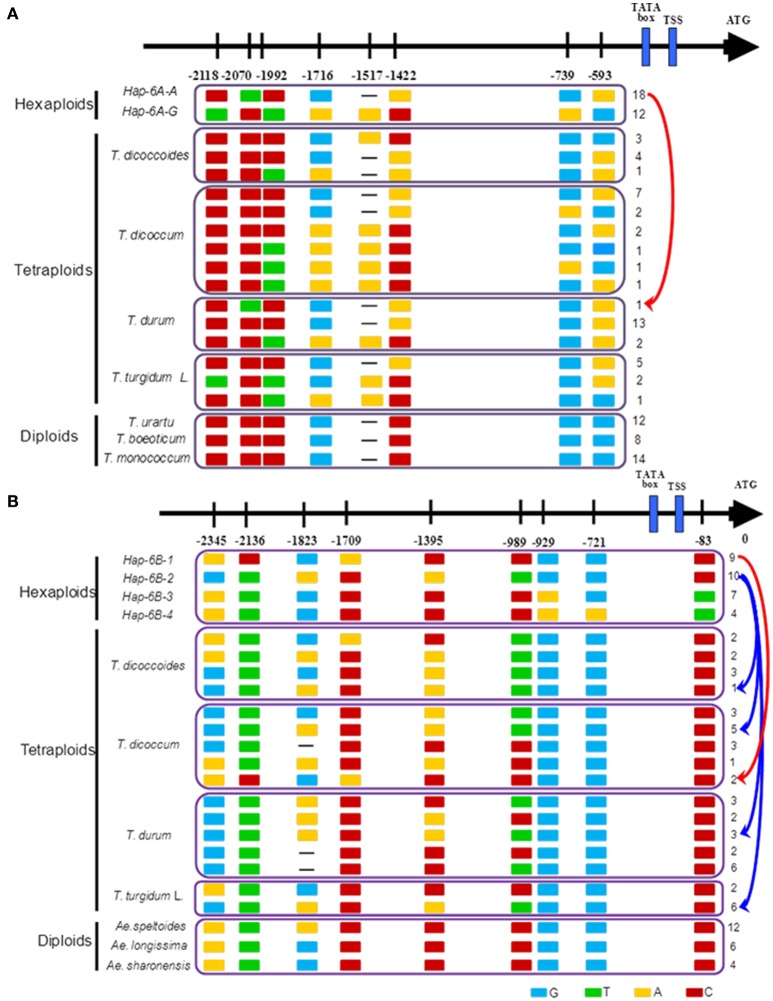
Previous studies (Su et al., 2011; Qin et al., 2014) and haplotype network analysis in this study (Figure 7) detected two haplotypes in the promoter region of TaGW2-6A, and four haplotypes in TaGW2-6B. After tracking them through wheat polyploidization, we found that the favored haplotypes in TaGW2-6A and TaGW2-6B were all from tetraploids, i.e., Hap-6A-A was present in T. durum (DR13), Hap-6B-1 in T. dicoccum (DM6 and DM51), and Hap-6B-2 in T. dicoccoides (DS10), T. dicoccum (DM4, DM44, DM46, DM135, and DM147), T. durum (DR13, DR487, and DR492), and T. turgidum L. (TG2, TG23, TG27, TG29, TG33, and TG39). More importantly, SNPs discovered in hexaploids were almost all monomorphic in diploids, but polymorphic in tetraploids.
Figure 7.
Favored haplotypes of TaGW2-6A (A) and TaGW2-6B (B) in hexaploids tracked to diploids and tetraploids. Rectangles colored brown, red, light blue and green represent nucleotide bases A, C, G, and T, respectively. Numbers indicate positions of polymorphism in promoters relative to the coding start codon.
TaGW2s negatively regulate seed size
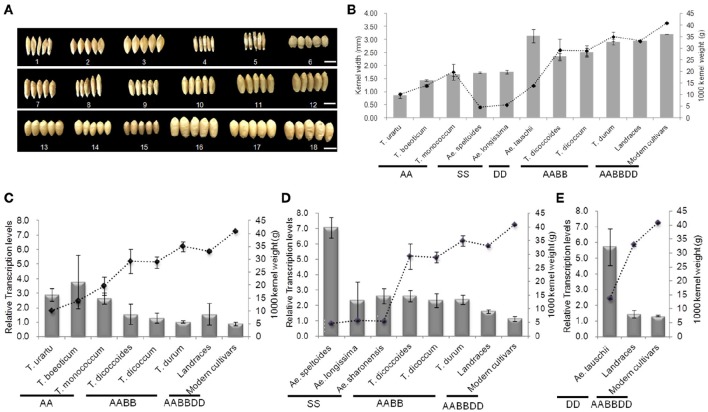
Genome-specific primers were designed according to cDNA sequence differences in the GW2 homologs on chromosomes 6A, 6B, and 6D, in order to evaluate the correlation between respective gene expression levels and grain width/weight during wheat polyploidization (Figure 8 and Table S7). Grain width and grain weight obviously increased following polyploidization (Figures 8A,B). The average relative expression of TaGW2-6A decreased from 3.128 in diploids to 1.281 in tetraploids, and 1.148 in hexaploids, whereas the average grain weights and widths increased from 14.560 g and 1.552 mm to 31.824 g and 2.603 mm in tetraploid, and 35.846 g and 3.155 mm in common wheat, respectively (Figure 8C and Table S7). The differences in expression levels were not significant. The average relative expression of TaGW2-6B in diploids was 5.168, in tetraploids 2.426, and in hexaploids 1.434 (Figure 8D and Table S7). The decreased expression levels were significant (P < 0.05), as were the increases in grain width and weight (P < 0.01). Similar results were obtained for TaGW2-6D (P < 0.05) (Figure 8E and Table S7). In addition, we measured overall relative expression values of TaGW2s in diploids, tetraploids and hexaploids (Table S8). The overall transcription levels of TaGW2-6A/6B/6D were 3.128, 5.168, and 5.734 in diploids, respectively. The overall relative expressions of TaGW2-6B/6D were significantly higher than that in tetraploids (3.426, P < 0.05) and in hexaploids (3.530, P < 0.05), as grain width and weight increased, which reflected that expression level for each genome was dramatically declined in wheat polyploidization. This further demonstrated the negative regulatory roles of TaGW2s on grain size, and strong selection of these yield-related genes during wheat evolution.
Figure 8.
Relationships between mean relative expression levels of TaGW2s and 1000-kernel weight in diploids, tetraploids, and hexaploids. (A) Kernel shape, 1–6: diploids; 7-12: tetraploids; 13-18: hexaploids; bars, 5 mm. (A) 1: UR207 (AA); 2: BO8 (AA); 3: MO1 (AA); 4: Y590 (SS); 5: Y435 (SS); 6: Y2280 (DD); 7: DS4; 8: DS8; 9: DM12; 10: DM147; 11: DR3; 12: DR146; 13: Baihuamai; 14: Chinese Spring; 15: Nongda 139; 16: Zhongyou 9507; 17: Wenmai 8; 18: Zhengmai 9023. (B) Kernel weight and width. Bars represent standard errors. (C–E) Mean relative expression levels of TaGW2-6A, TaGW2-6B, and TaGW2-6D.
Discussion
TaGW2s underwent stronger differentiation during wheat polyploidization than domestication and breeding
Common wheat has undergone ~8,000 years of artificial selection (Doebley et al., 2006; Feldman et al., 2012; Marcussen et al., 2014). The process of polyploidization of wheat involved a strong differentiation compared to the wild ancestral species, and genetic diversity significantly decreased, especially genes controlling important agronomic traits (Haudry et al., 2007).
In the present study, we compared sequence differences in TaGW2 homologs in diploids, tetraploids and hexaploid wheat species and various relatives. Dramatic declines in nucleotide diversity (π) and Fst values (Figures 1, 3, 4 and Table 1) occurred with each round of polyploidization. As shown in Figure 5 and Figure S2 π values for the promoter and coding regions of TaGW2-6A decreased 4.8- and 5.7-fold, respectively, from diploids to tetraploids, and further decreased 2.6- and 11.6-fold from tetraploids to hexaploids. For TaGW2-6B comparable 14- and 1.23-fold reductions occurred at the promoter and coding regions with tetraploidy, and further reductions of 7.2 and 4.2 times occurred with hexaploidy (Figures S3, S4). However, π value differences among accessions within ploidy levels varied by less than 1-fold. Moreover, Fst values of TaGW2-6A, -6B, and -6D in both promoter and coding regions between diploids and tetraploids were higher than between tetraploids and hexaploids (Figure 4). In addition, the haplotype numbers of TaGW2-6A and TaGW2-6B decreased from diploids, to tetraploids and hexaploids (Figure 6 and Figure S5). Dramatic reductions in diversity in other genes, such as TaSUS1-7A, TaGS5-5A, and TaCWI, following polyploidization reported in other species (Hou et al., 2014; Jiang et al., 2015; Ma et al., 2016). All of these reports indicate that strong differentiation of important yield-related genes occurred during polyploidization and domestication of both tetraploid and hexaploid wheats.
T. urartu and Ae. speltoides confirmed as the direct donors of the wheat A and B genomes
Common wheat arose following chromosome doubling of a natural hybrid of tetraploid T. dicoccum and diploid Ae. tauschii. This event that may have occurred as few as once or twice times caused an “evolutionary bottleneck”, and consequently much of the genetic variation present in diploid species and tetraploids with common genomes is not present in the hexaploid (Ogbonnaya et al., 2005; Ozkan et al., 2005).
In this study, the genetic relationships of common wheat and related species (Figure 2 and Figure S1) were evaluated by phylogenetic analysis of TaGW2 polymorphisms. In regard to TaGW2-6A and TaGW2-6B tetraploids and hexaploids were in a single subgroup, with diploid species individually clustered into different subgroups, consistent with earlier results of Buckler et al. (2001). During the evolution of common wheat, T. urartu and a B genome donor (herein suggested to be Ae. speltoides) hybridized to form tetraploid wheat, which later hybridized with the D genome donor Ae. tauschii to form common wheat. Many studies have focused on the prospective A, B, D genome donors of wheat (Kihara, 1944; McFadden and Sears, 1944; Dvorák et al., 1993; Kilian et al., 2007). T. urartu (Dvorák et al., 1993), Ae. speltoides (Petersen et al., 2006; Kilian et al., 2007) and Ae. tauschii (Kihara, 1944) may be the direct or main donors of the A, B and D genomes, respectively. In this study, phylogenetic analysis of TaGW2s further verified T. urartu as the direct donor of the A genome, Ae. speltoides was the likely donor or main donor of the B genome, and Ae. tauschii was the D genome donor of common wheat (Figure 2 and Figure S1).
Diversity differences in TaGW2s mainly occurred in the promoter regions during polyploidization of wheat
Natural diversity influencing gene expression levels of some yield-related genes in graminaceous crops often occurs in the promoter regions. Examples include OsGS5, ZmGS3, ZmGW2-CHR4, TaTEF-7A, and TaCWI-4A (Li et al., 2010a,b, 2011; Zheng et al., 2014; Jiang et al., 2015). Previous studies (Su et al., 2011; Qin et al., 2014) also showed that genetic diversity mainly occurred in the promoter regions of TaGW2-6A and TaGW2-6B in common wheat. Expression levels of TaGW2 genes in developing seeds were negatively correlated with grain width and grain weight.
In the present study diversity (π) in the promoter regions was significantly higher than in the coding regions of TaGW2 genes in various species (Table 1 and Figure 1). Genetic differentiation (Fst) in the promoter regions was higher than in the coding regions among diploids, tetraploids, and hexaploid groups (Figure 4). In addition, deviations of Tajima's D from zero for TaGW2-6A and -6B in the promoter regions of diploids and hexaploids further demonstrated that the promoter regions underwent selection. Compared to the conserved coding regions, the extensive variation that occurred in the promoter regions regulated grain size through variation in expression level. Correlation of gene expression levels of TaGW2-6A, -6B and -6D with grain width/weight during wheat polyploidization showed that grain width/weight increased with progression from diploids to hexaploids, but the relative expression levels of the genes significantly decreased (Figure 8 and Table S7).
TaGW2s are conserved in function but have different fates in rice and wheat
OsGW2, first cloned in rice following genetic analysis of an induced mutant, encodes a ubiquitin E3 ligase (Song et al., 2007). There was no significant variation between landraces and modern cultivars, indicating that the locus had not been subjected to selection during domestication and breeding. Moreover, indica and japonica sub-populations showed different patterns of variation, suggesting that OsGW2 might have undergone long-term purifying selection during evolution and improvement of rice (Lu et al., 2013). Huang et al. (2012) performed a genome-wide association study (GWAS) of flowering time and grain yield traits in a panel of 950 worldwide rice varieties and did not detect an association of OsGW2 and yield. TaGW2s in wheat are functional RING-type E3 ligases (Bednarek et al., 2012), and gene expression analysis and RNAi demonstrated that variation in them was negatively correlated with grain weight, a function that was similar to OsGW2 in rice (Su et al., 2011; Yang et al., 2012; Hong et al., 2014; Qin et al., 2014). Strong selection of certain TaGW2-6A and TaGW2-6B haplotypes occurred in global wheat breeding (Su et al., 2011; Qin et al., 2014). In this study, we conducted a systematic analysis of the TaGW2 genes during polyploidization of wheat. Haplotype networks and haplotype analyses (Figures 6, 7 and Figure S5) showed that favorable haplotypes of TaGW2-6A and TaGW2-6B in common wheat were also found in tetraploids, but were not detected in diploids. Strong selection of favorable variants of TaGW2-6A and TaGW2-6B apparently occurred in both tetraploid and hexaploid wheats. Clearly the agronomic effects of variation in TaGW2 genes in polyploid wheat and rice were different. This work demonstrates the value of comparative gene homology studies in grass species.
Author contributions
LQ, CH, and XZ designed research. LQ, JZ, and CH performed research. LQ, TL, and JH contributed new reagents or analytical tools. LQ and CH analyzed data. LQ, XZ, and CH drafted the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge help from Prof. Robert A McIntosh, University of Sydney, with English editing. This work was supported by the China Natural Science Foundation (31270036, 30900898), National Key Research and Development Program of China (2016YFD0100302) and CAAS-Innovation Team Project.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00318/full#supplementary-material
References
- Bai X. F., Wu B., Xing Y. Z. (2012). Yield-related QTLs and their applications in rice genetic improvement. J. Integr. Plant Biol. 54, 300–311. 10.1111/j.1744-7909.2012.01117.x [DOI] [PubMed] [Google Scholar]
- Bednarek J., Boulaflous A., Girousse C., Ravel C., Tassy C., Barret P., et al. (2012). Down-regulation of the TaGW2 gene by RNA interference results in decreased grain size and weight in wheat. J. Exp. Bot. 63, 5945–5955. 10.1093/jxb/ers249 [DOI] [PubMed] [Google Scholar]
- Buckler E. S., Thornsberry J. M., Kresovich S. (2001). Molecular diversity, structure and domestication of grasses. Genet. Res. 77, 213–218. 10.1017/S0016672301005158 [DOI] [PubMed] [Google Scholar]
- Chen D. H., Ronald P. C. (1999). A rapid DNA minipreparation method suitable for AFLP and other PCR applications. Plant Mol. Biol. Rep. 17, 53–57. 10.1023/A:1007585532036 [DOI] [Google Scholar]
- Clement M., Posada D., Crandall K. A. (2000). TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1659. 10.1046/j.1365-294x.2000.01020.x [DOI] [PubMed] [Google Scholar]
- Doebley J. F., Gaut B. S., Smith B. D. (2006). The molecular genetics of crop domestication. Cell 127, 1309–1321. 10.1016/j.cell.2006.12.006 [DOI] [PubMed] [Google Scholar]
- Dvorak J., Akhunov E. D. (2005). Tempos of gene locus deletions and duplications and their relationship to recombination rate during diploid and polyploid evolution in the Aegilops-Triticum alliance. Genetics 171, 323–332. 10.1534/genetics.105.041632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorák J., Terlizzi P. D., Zhang H. B., Resta P. (1993). The evolution of polyploid wheats: identification of the A genome donor species. Genome 36, 21–31. 10.1139/g93-004 [DOI] [PubMed] [Google Scholar]
- Dvorák J., Zhang H. B. (1990). Variation in repeated nucleotide sequences sheds light on the phylogeny of the wheat B and G genomes. Proc. Natl. Acad. Sci. U.S.A. 87, 9640–9644. 10.1073/pnas.87.24.9640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M., Levy A. A., Fahima T., Korol A. (2012). Genomic asymmetry in allopolyploid plants: wheat as a model. J. Exp. Bot. 63, 5045–5059. 10.1093/jxb/ers192 [DOI] [PubMed] [Google Scholar]
- Feuillet C., Langridge P., Waugh R. (2008). Cereal breeding takes a walk on the wild side. Trends Genet. 24, 24–32. 10.1016/j.tig.2007.11.001 [DOI] [PubMed] [Google Scholar]
- Haudry A., Cenci A., Ravel C., Bataillon T., Brunel D., Poncet C., et al. (2007). Grinding up wheat: a massive loss of nucleotide diversity since domestication. Mol. Biol. Evol. 24, 1506–1517. 10.1093/molbev/msm077 [DOI] [PubMed] [Google Scholar]
- Hong Y. T., Chen L. F., Du L. P., Su Z. Q., Wang J. F., Ye X. G., et al. (2014). Transcript suppression of TaGW2 increased grain width and weight in bread wheat. Funct. Integr. Genomics 14, 341–349. 10.1007/s10142-014-0380-5 [DOI] [PubMed] [Google Scholar]
- Hou J., Jiang Q. Y., Hao C. Y., Wang Y. Q., Zhang H. N., Zhang X. Y. (2014). Global selection on sucrose synthase haplotypes during a century of wheat breeding. Plant Physiol. 164, 1918–1929. 10.1104/pp.113.232454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. X., Sirikhachornkit A., Su X. J., Faris J., Gill B., Haselkorn R., et al. (2002). Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc. Natl. Acad. Sci. U.S.A. 99, 8133–8138. 10.1073/pnas.072223799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. H., Zhao Y., Wei X. H., Li C. Y., Wang A. H., Zhao Q., et al. (2012). Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat. Genet. 44, 32–39. 10.1038/ng.1018 [DOI] [PubMed] [Google Scholar]
- Jaiswal V., Gahlaut V., Mathur S., Agarwal P., Khandelwal M. K., Khurana J. P., et al. (2015). Identification of novel SNP in promoter sequence of TaGW2-6A associated with grain weight and other agronomic traits in wheat (Triticum aestivum L.). PLoS ONE 10:e0129400. 10.1371/journal.pone.0129400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y. M., Jiang Q. Y., Hao C. Y., Hou J., Wang L. F., Zhang H. N., et al. (2015). A yield-associated gene TaCWI, in wheat: its function, selection and evolution in global breeding revealed by haplotype analysis. Theor. Appl. Genet. 128, 131–143. 10.1007/s00122-014-2417-5 [DOI] [PubMed] [Google Scholar]
- Kihara H. (1944). Discovery of the DD-analyser, one of the ancestors of Triticum vulgare. Biol. Agric. Horticult. 19, 13–14. [Google Scholar]
- Kilian B., Ozkan H., Deusch O., Effgen S., Brandolini A., Kohl J., et al. (2007). Independent wheat B and G genome origins in outcrossing Aegilops progenitor haplotypes. Mol. Biol. Evol. 24, 217–227. 10.1093/molbev/msl151 [DOI] [PubMed] [Google Scholar]
- Kimber G., Athwal R. S. (1972). A reassessment of the course of evolution of wheat. Proc. Natl. Acad. Sci. U.S.A. 69, 912–915. 10.1073/pnas.69.4.912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Li L., Yang X. H., Warburton M. L., Bai G. H., Dai J. R., et al. (2010b). Relationship, evolutionary fate and function of two maize co-orthologs of rice GW2 associated with kernel size and weight. BMC Plant Biol. 10:143. 10.1186/1471-2229-10-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Yang X. H., Bai G. H., Warburton M. L., Mahuku G., Gore M., et al. (2010a). Cloning and characterization of a putative GS3 ortholog involved in maize kernel development. Theor. Appl. Genet. 120, 753–763. 10.1007/s00122-009-1196-x [DOI] [PubMed] [Google Scholar]
- Li Y. B., Fan C. C., Xing Y. Z., Jiang Y. H., Luo L. J., Sun L., et al. (2011). Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43, 1266–1269. 10.1038/ng.977 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lu L., Shao D., Qiu X. J., Sun L., Yan W. H., Zhou X. C., et al. (2013). Natural variation and artificial selection in four genes determine grain shape in rice. New Phytol. 200, 1269–1280. 10.1111/nph.12430 [DOI] [PubMed] [Google Scholar]
- Ma L., Li T., Hao C. Y., Wang Y. Q., Chen X. H., Zhang X. Y. (2016). TaGS5-3A, a grain size gene selected during wheat improvement for larger kernel and yield. Plant Biotechnol. J. 14, 269–1280. 10.1111/pbi.12492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestra B., Naranjo T. (1998). Homoeologous relationships of Aegilops speltoides chromosomes of bread wheat. Theor. Appl. Genet. 97, 181–186. 10.1007/s001220050883 [DOI] [Google Scholar]
- Marcussen T., Sandve S. R., Heier L., Spannagl M., Pfeifer M., International Wheat Genome Sequencing Consortium. et al. (2014). Ancient hybridizations among the ancestral genomes of bread wheat. Science 345:1250092. 10.1126/science.1250092 [DOI] [PubMed] [Google Scholar]
- McFadden E. S., Sears E. R. (1944). The artificial synthesis of Triticum spelta. Rec. Genet. Soc. Am. 13, 26–27. [Google Scholar]
- Mori N., Liu Y. G., Tsunewaki K. (1995). Wheat phylogeny determined by RFLP analysis of nuclear DNA. 2. wild tetraploid wheats. Theor. Appl. Genet. 90, 129–134. 10.1007/BF00221006 [DOI] [PubMed] [Google Scholar]
- Ogbonnaya F. C., Halloran G. M., Lagudah E. S. (2005). D genome of wheat: 60 years on from Kihara, Sears and McFadden. Wheat Inform. Serv. 100, 205–220. [Google Scholar]
- Ozkan H., Brandolini A., Pozzi C., Effgen S., Wunder J., Salamini F. (2005). A reconsideration of the domestication geography of tetraploid wheats. Theor. Appl. Genet. 110, 1052–1060. 10.1007/s00122-005-1925-8 [DOI] [PubMed] [Google Scholar]
- Petersen G., Seberg O., Yde M., Berthelsen K. (2006). Phylogenetic relationships of Triticum and Aegilops and evidence for the origin of the A, B, and D genomes of common wheat (Triticum aestivum). Mol. Phylogenet. Evol. 39, 70–82. 10.1016/j.ympev.2006.01.023 [DOI] [PubMed] [Google Scholar]
- Qin L., Hao C. Y., Hou J., Wang Y. Q., Li T., Wang L. F., et al. (2014). Homologous haplotypes, expression, genetic effects and geographic distribution of the wheat yield gene TaGW2. BMC Plant Biol. 14:107. 10.1186/1471-2229-14-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar P., Stebbins G. L. (1956). Morphological evidence concerning the origin of the B genome in wheat. Am. J. Bot. 43, 297–304. 10.2307/2438947 [DOI] [Google Scholar]
- Simmonds J., Scott P., Brinton J., Mestre T. C., Bush M., Del Blanco A., et al. (2016). A splice acceptor site mutation in TaGW2-A1 increases thousand grain weight in tetraploid and hexaploid wheat through wider and longer grains. Theor. Appl. Genet. 129, 1099–1112. 10.1007/s00122-016-2686-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X. J., Huang W., Shi M., Zhu M. Z., Lin H. X. (2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39, 623–630. 10.1038/ng2014 [DOI] [PubMed] [Google Scholar]
- Su Z. Q., Hao C. Y., Wang L. F., Dong Y. C., Zhang X. Y. (2011). Identification and development of a functional marker of TaGW2 associated with grain weight in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 122, 211–223. 10.1007/s00122-010-1437-z [DOI] [PubMed] [Google Scholar]
- Tajima F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. Z., Miyashita N. T., Tsunewaki K. (1997). Plasmon analysis of Triticum (wheat) and Aegilops: PCR-single stranded conformational polymorphism (PCR-SSCP) analysis of organellar DNAs. Proc. Natl. Acad. Sci. U.S.A. 94, 14570–14577. 10.1073/pnas.94.26.14570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y. Z., Zhang Q. F. (2010). Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 61, 421–442. 10.1146/annurev-arplant-042809-112209 [DOI] [PubMed] [Google Scholar]
- Yang Z. B., Bai Z. Y., Li X. L., Wang P., Wu Q. X., Yang L., et al. (2012). SNP identification and allelic-specific PCR markers development for TaGW2, a gene linked to wheat kernel weight. Theor. Appl. Genet. 125, 1057–1068. 10.1007/s00122-012-1895-6 [DOI] [PubMed] [Google Scholar]
- Zheng J., Liu H., Wang Y. Q., Wang L. F., Chang X. P., Jing R. L., et al. (2014). TEF-7A, a transcript elongation factor gene, influences yield-related traits in bread wheat (Triticum aestivum L.). J. Exp. Bot. 65, 5351–5365. 10.1093/jxb/eru306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J. R., Li J. Y. (2014). Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu. Rev. Genet. 48, 99–118. 10.1146/annurev-genet-120213-092138 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.