Abstract
The genetic nature and biological effects of recombination between porcine endogenous retroviruses (PERV) were studied. An infectious molecular clone was generated from a high-titer, human-tropic PERV isolate, PERV-A 14/220 (B. A. Oldmixon, et al. J. Virol. 76:3045-3048, 2002; T. A. Ericsson et al. Proc. Natl. Acad. Sci. USA 100:6759-6764, 2003). To analyze this sequence and 15 available full-length PERV nucleotide sequences, we developed a sequence comparison program, LOHATM to calculate local sequence homology between two sequences. This analysis determined that PERV-A 14/220 arose by homologous recombination of a PERV-C genome replacing an 850-bp region around the pol-env junction with that of a PERV-A sequence. This 850-bp PERV-A sequence encompasses the env receptor binding domain, thereby conferring a wide host range including human cells. In addition, we determined that multiple regions derived from PERV-C are responsible for the increased infectious titer of PERV-A 14/220. Thus, a single recombination event may be a fast and effective way to generate high-titer, potentially harmful PERV. Further, local homology and phylogenetic analyses between 16 full-length sequences revealed evidence for other recombination events in the past that give rise to other PERV genomes that possess the PERV-A, but not the PERV-B, env gene. These results indicate that PERV-A env is more prone to recombination with heterogeneous backbone genomes than PERV-B env. Such recombination events that generate more active PERV-A appear to occur in pigs rather frequently, which increases the potential risk of zoonotic PERV transmission. In this context, pigs lacking non-human-tropic PERV-C would be more suitable as donor animals for clinical xenotransplantation.
Pig-to-human xenotransplantation may alleviate the shortage of human donor organs and may benefit the treatment of a wide range of health problems, such as diabetes, neurological disorders, and organ failure. However, the potential risk of zoonotic pathogen transmission remains a major obstacle (23). While barrier derivation technologies allow the breeding of pigs free of exogenous pathogens, porcine endogenous retroviruses (PERV) are not eliminated due to their presence in germ line DNA. Moreover, certain PERV have been shown to infect human cells in vitro (25, 34, 54) although no in vivo transmission to human xenograft recipients has been observed as yet (5, 7, 8, 13, 22, 30, 32, 37, 52). Three replication-competent, gammaretrovirus subgroups of PERV (PERV-A, -B, and -C) have been identified in the genomic DNA of pigs (21, 33, 34, 48). While they share homologous gag and pol genes (>90% identity at the nucleotide level), significant differences within the env gene determine different receptor usage. PERV-A and PERV-B infect cells derived from human, pig, and some other species, whereas PERV-C infection is mainly restricted to pig cells (48).
Genetic recombination has important implications in the risk of PERV zoonosis (42, 43, 46, 49). In this regard, spontaneously occurring PERV recombinants may be more harmful than the endogenous forms of PERV present in pig genomes. For example, recombinant PERV, which were isolated from porcine primary cultures and display hybrid sequences within the env gene, replicate efficiently in human cells (28, 55). However, studies on PERV recombination in terms of its genetic nature, biological effects, and implication for xenotransplantation have been limited. Notably, data from partial sequencing of the PERV genome revealed evidence for possible recombination events (28, 40, 55) but not a genetic characterization of the whole genome of recombinant PERV. In contrast, studies analyzing longer genome sequences (17-19) have not directly linked genetic recombination with its effect on biological activity of PERV.
A recombinant isolate, PERV-A 14/220, was originally isolated by transmission from activated peripheral blood mononuclear cells (PBMC) from a miniature pig to human embryonic kidney 293 cells (28). PERV-A 14/220 infects human cells with a significantly higher titer than previous PERV-A and -B isolates (9). Its genome consists of the gag, pol, and env transmembrane (TM) sequences most related to PERV-C and the env receptor-binding domain (RBD) of PERV-A (28). Here we generated an infectious molecular clone of this isolate and further characterized its full-length sequence. Comparison of this sequence with 15 full-length PERV database sequences confirmed the likely origins of the parental sequences and identified the precise recombination crossover points. Furthermore, we obtained evidence for other recombination events, which led to formation of different forms of germ line- as well as cell line-derived PERV-A. Our study of the infectivity of the PERV-A 14/220 clone, a previously described prototype PERV-A clone (2), and chimeras derived from these two viruses demonstrated that there are multiple genetic determinants for the high infectivity of the recombinant PERV-A 14/220. The implications and consequences of PERV recombination for zoonotic transmission are discussed.
MATERIALS AND METHODS
PCR cloning and sequence determination of infectious PERV-A 14/220.
Genomic DNA from 293 cells infected with PERV-A 14/220 was extracted with a QIAamp DNA blood mini kit (QIAGEN) according to the manufacturer's instructions and used as a template in PCRs. A PCR-based method for the isolation of PERV proviruses was applied as previously described (2). It was expected that PERV-A 14/220 would contain more PERV-C-related than PERV-A-related sequence in the long terminal repeat (LTR) as in most other parts of its genome. We therefore designed a new primer, (+)5′- CTAATGAGAAGCTTAAAATTGTTCTGAATTC, matching with PERV-C LTR U3 sequences in the database (GenBank accession numbers AF038599 and AF038600) as the upstream primer of 5′ halves. While 3′ halves were obtained by using the previous reverse transcriptase (RT)-U3 primer pairs (2), 5′ halves were successfully amplified and cloned by using the new U3 primer together with the RT reverse primer. PCR products encoding the 5′ and 3′ halves were subcloned by using a Zero Blunt TOPO PCR cloning kit (Invitrogen) according to the manufacturer's instructions. Two clones (each) for the 3′ and 5′ halves were used to construct plasmids containing infectious proviruses by joining two halves at an NheI site in the RT region, resulting in four proviruses with different 3′ and 5′ combinations. DNA sequences were obtained by dye terminator cycle sequencing with a CEQ 2000 instrument and accessories (Beckman).
Sequence analyses. (i) Sequence retrieval.
Fifteen full-length PERV γ1 subgroup sequences were retrieved from GenBank after screening by BLAST searches of nonredundant (default, nr) and patent (pat) databases at the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov:80/BLAST) and the default database at the DNA Data Bank of Japan (www.ddbj.nig.ac.jp). The default database at the DNA Data Bank of Japan but not at the National Center for Biotechnology Information included the sequences deposited for patent purposes. A full-length sequence, A66552, was excluded from this study but was represented by another sequence from the same laboratory, A66553 (named BQOne in this study) (Table 1). While A66553 has all intact open reading frames (ORF) for gag, pol, and env, A66552 has a number of ORF nonsense errors and six internal deletions and is 11 nucleotides (nt) shorter at the 5′ end of the cDNA compared to the sequence of A66553. The following sequences (sized over 7 kbp) were also excluded as they do not display the full-length genomes: AF038601/AR130474 (7,333 nt), AX052638 (7,873 nts), AX052634 (7,362 nt), and Y17013 (7,808 nt).
TABLE 1.
Full-length PERV sequences
| Name | Accession no. (patient)a | Source or origin | Cloning method | Genomic loci | ORF damage | Infectivity | Reference or source |
|---|---|---|---|---|---|---|---|
| Receptor group A | |||||||
| A14/220 | AY570980 | Inbred c/c minipig | PCR halves | NAc | None | Yes | This study |
| A130 | AJ279056 (AX546209) | Large White | BAC libraryb | Yesd | TM truncation | Yes | 26 |
| A151 | AF435967 | Large White | BAC library | Yesd | TM truncation | Yes | 26 |
| A463 | AF435966 | Large White | BAC library | Yesd | Stop in Pro | Yes | 26 |
| A58 | AJ293656 (AX546207) | PK 15 | Lambda library | Yesd | TM truncation | Yes | 19 |
| A42 | AJ133817 | 293/PK15 | Lambda library | NA | None | Yes | 19 |
| Ap60 | AY099323 | 293/PK15 | PCR halves | NA | None | Yes | 2 |
| Receptor group B | |||||||
| B192 | AJ279057 (AX546210) | Large White | BAC library | Yesd | Two stops in pol | Negative | 26 |
| Bimut (78) | AY056035 | Large White | Cosmid library | Yese | Frameshift in gag | Negative | 14 |
| B213 | AJ293657 (AX546208) | PK15 | Lambda library | Yesd | Unknown nucleotide in SU, but, frame potentially ok | Yes | 19 |
| B33 | AJ133816 | 293/PK15 | Lambda library | NA | Mutation at env initiation | Yes after correction | 6 |
| B43 | AJ133818 | 293/PK15 | Lambda library | NA | None | Yes | 6 |
| Bp17 | AY099324 | 293/PK15 | PCR halves | NA | None | Yes | 2 |
| BQOne | (A66553) | PK15 | cDNA library/PCR | NA | None | NA (cDNA) | 10 |
| Receptor group C | |||||||
| Cmsl | AF038600 (AR130475) | Minipig PBMC | cDNA library | NA | None | NA | 1 |
| Ctsukuba | AF038599 (AR130473) | Shimozuma cell line | cDNA library | NA | Two frameshifts in pol | NA | 1 |
(ii) Local homology analysis.
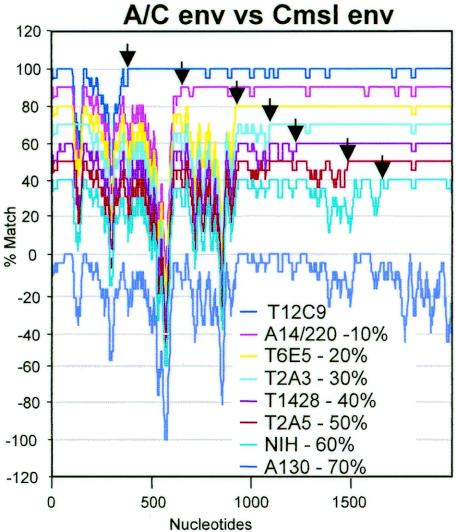
A program to align a set of sequences and calculate local homology along the alignment for all possible pairs of sequences was created (LOHATM: local homology analysis, Y. Takeuchi and R. Myers; available from the authors on request). LOHATM employs Clustal X (50) to align a number of sequences. All possible pairs of aligned sequences with gaps were analyzed to calculate the percent match for 41 positions (between 20 each 5′ and 3′ to a given position, but available positions decrease to 21 at 3′ and 5′ termini). Positions with gaps for both sequences were counted as matched, while positions with a gap for only one sequence were counted as mismatched. Analyses with different sizes of window, 21 and 81 instead of 41 bp, produced similar results albeit with different resolutions. A set of nine env genes of seven recombinants between PERV-A and PERV-C (A/C recombinants) derived from miniature swine and one each of PERV-A and PERV-C was also analyzed by LOHATM with a window size of 21 (see Fig. 4). The percent match (y axis) was plotted against the nucleotide positions (x axis) by using Microsoft Excel.
FIG. 4.
Recombination crossover points in PERV-A/C hybrid env genes. Six nucleotide sequences of env genes (NIH, accession no. AF130444 [55]; T2A3, AF417230; T2A5, AF417231; T6E5, AF417232; T12C9, AF417229; T1428, AY364236 [28]) and three env coding sequences extracted from A14/220, Cmsl, and A130 (their lengths are between 1,917 nt of Cmsl and 1,983 nt of A130), were analyzed by LOHATM with a window size of 21 nt. Homology plots of Cmsl against seven A/C recombinant sequences are shown (1,984 nt positions in the alignment [x axis]; 0, 10, 20, 30, 40, 50, or 60% was subtracted from percent match values as indicated [y axis]). A plot of Cmsl versus A130 is also shown (percent match, 100 [y axis]). The apparent crossover points from PERV-A- to PERV-C-like sequence are indicated by arrows.
(iii) Phylogenetic trees.
Phylogenetic neighbor-joining (N-J) trees (36) were created by using Clustal X for the PERV genome regions corresponding to the following nucleotide numbers in the A14/220 sequence (Table 1): leader (5′-R to gag initiation), 1 to 604; Gag, 605 to 2179; Pro, 2180 to 2536; RT, 2537 to 4425; integrase (IN), 4426 to 5761; surface (SU), 5637 to 7004; TM, 7005 to 7598; U3-R, 7648 to 8188 (putative junctions for Pro-RT, RT-IN, and SU-TM). The robustness of the N-J tree topology was assessed by bootstrap analysis, with 1,000 rounds of replication, by using the program PAUP version 4.0 (44). Phylogram trees were rooted against the A151 germ line sequence (Table 1) and edited by using TreeView (29).
Construction of chimeric proviruses.
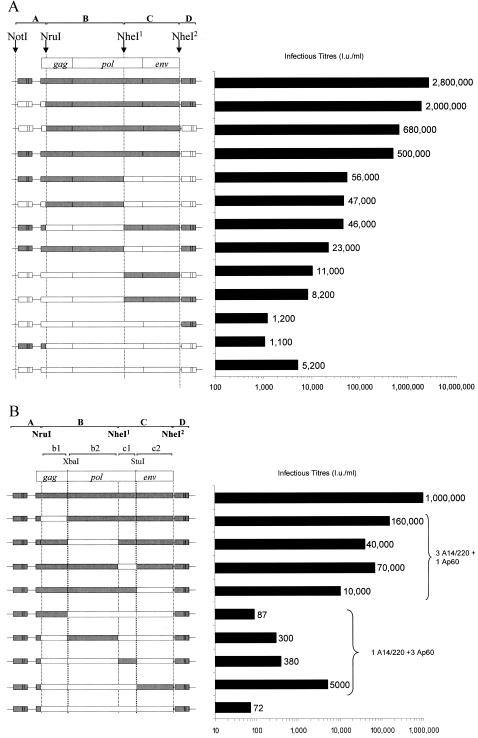
The indicated restriction sites NruI, NheI1, and NheI2 (see Fig. 5B) were used to construct chimeric proviruses by four-fragment mix-and-match cloning. For this purpose an NheI2 site was introduced into Ap60 immediately after the stop codon of env by PCR-mediated mutagenesis by using the primers 5′-GAGCTAGCGGTCAGTTTC-3′ and 5′-ACTGACCGCTAGCTTTCCCAGTTCT-3′; the PCR product sequence was verified. For the construction of further chimeras the XbaI and StuI sites, which are conserved between Ap60 and A14/220 (see Fig. 5B), were used. Because the A14/220 plasmid contains additional XbaI and StuI sites outside of the proviral genome, partial restriction digests were performed. All chimera were verified by restriction digests and partial sequencing.
FIG. 5.
Infectious titers of A14/220 and Ap60 chimeras. (A) Full-length proviral PERV chimeras consisting of A14/220 (gray bars) or Ap60 (white bars) sequences were constructed by using four restriction sites: two conserved sites (NruI and NheI1 sites), an NheI2 site (created in Ap60 by mutagenesis), and an NotI site outside of the provirus genome. Infectious titer (infectious units per milliliter) of replicating PERV following establishment of persistently infected cultures by DNA transfection was measured on human 293 cells by an immunocytological focus assay (2). A data set of a representative of two independent experiments is shown by bars as well as numbers. (B) Chimeric PERV containing a mix-and-match set of protein coding region (fragments B and C in panel A) between A14/220 (gray bars) and Ap60 (white bars) sequences as well as the rest of the genome derived from A14/220 were constructed. Fragments B were divided to subfragments b1 and b2 by XbaI and fragments C were divided into to c1 and c2 by StuI. Results for chimeras containing three subfragments from A14/220 (three A14/220 plus one Ap60) as well as one subfragment from A14/220 (one A14/220 plus three Ap60) are shown. Infectious titer (infectious units per milliliter) of replicating PERV harvested 3 days after DNA transfection was measured on human 293 cells by an immunocytological focus assay. A data set of a representative of two independent experiments (transfection followed by titration) is shown.
Immunoblotting and in situ staining.
Cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred onto nitrocellulose membrane by using a semidry blotting system (Hoefer), and probed with the previously described rabbit anti-PERV capsid (CA) antibody (2) by chemiluminescence-based detection (Amersham), according to the manufacturer's instructions. The in situ detection of replicating PERV with the anti-PERV CA antibody and the detection of lacZ pseudotypes was performed as previously described (2, 47).
Viruses and cell culture.
The murine leukemia virus (MLV) lacZ vector and lacZ pseudotypes as well as the human embryonic kidney 293 cells have been described previously (48). Cell-free infection was carried out essentially as described previously (2, 47). Briefly, cells were seeded at 104 cells/well in 96-well plates. After 24 h virus dilutions were added in the presence of 4 μg of polybrene per ml and then incubated for 3 days before fixation and staining. Transfection into 293 cells was performed by using Lipofectamine (GIBCO) or FuGENE (Roche) according to the manufacturer's instructions. A similar transfection efficiency was achieved between different proviral constructs as estimated by cotransfection of a green fluorescent protein expression plasmid, pCNCG (41), followed by fluorescence-activated cell sorting analysis. Cross-contamination of chimeric PERV cultures was controlled by performing PCRs with PERV-A- and PERV-C-specific primers on genomic DNA of viral cell cultures based on the protocol described above in the cloning section. RT activity was measured by using a C-type RT enzyme-linked immunosorbent assay (Cavidi Tech) according to the manufacturer's instructions.
Nucleotide sequence accession number.
The sequence of the PERV-A 14/220 proviral clone A14/220 was determined and has been deposited in the GenBank database under accession number AY570980.
RESULTS
A high-titer PERV isolate from miniature swine.
A biological PERV isolate, PERV-A 14/220, was obtained from an animal of swine leukocyte antigen haplotype c/c during screening of inbred herds of miniature swine for production of infectious PERV in a previous study (28). This isolate was established by in vitro viral transmission from activated PBMC to human 293 cells. PERV-A 14/220 has been shown to possess a genome consisting of the gag, pol, and env TM subunit sequences most related to PERV-C and the RBD of the env SU protein, gp70 of PERV-A (28). Hence, this isolate uses the same receptors as PERV-A for cell entry (9). Nucleotide sequences of the env gene of PCR clones representing PERV-A 14/220 have been reported as GenBank AF417227 and AF417228 (28). In preliminary experiments, PERV-A 14/220 replicated in human 293 cells with faster kinetics than prototype PERV-A (PERV-A PK) isolated from porcine PK15 cells (9, 28, 34). The culture supernatant of 293 cells persistently infected with PERV-A 14/220 contained up to 100-fold more infectious virus than that of PERV-A PK (Table 2).
TABLE 2.
Titration of infection by biological and molecular PERV isolatesa
| Cell line (species) | Titer (IU/ml)
|
|||
|---|---|---|---|---|
| PERV-A 14/220 | A14/220 | PERV-A PK | Ap60 | |
| Replicating PERVb | ||||
| 293T (human) | 2,600,000 | 1,800,000 | 42,120 | 37,000 |
| Mv-1-Lu (mink) | 130,000 | 180,000 | 5,900 | 4,500 |
| CRFK (cat) | 410,000 | 170,000 | 17,000 | 12,000 |
| LacZ pseudotype | ||||
| 293T (human) | 15,000 | 21,000 | 1,200 | 420 |
| TE671 (human) | 11,000 | 77,000 | 1,400 | 930 |
| Pindak (squirrel monkey) | 1,500 | 1,300 | <10 | <10 |
| Mv-1-Lu (mink) | 17,000 | 76,000 | 1,000 | 1,000 |
| CRFK (cat) | 7,700 | 7,600 | 120 | 450 |
| ST-IOWA (pig) | 26,000 | 37,000 | 2,500 | 2,500 |
Biological isolates, PERV-A 14/220 and PK, and viruses derived from molecular clones, A14/220 and Ap60, were titrated. A data set representative of two independent experiments is shown.
Titration for replicating PERV was not possible because of high background in the immunostaining assay in TE671, Pindak, and ST-IOWA cells.
PCR cloning of infectious PERV-A 14/220.
To further characterize PERV-A 14/220, we constructed an infectious molecular clone of this isolate by using a PCR-based technique previously described for prototype PERV-A PK (Ap60, formerly called PERV-60) (Table 1) and PERV-B PK (Bp17, formerly called PERV-17) (2). Four plasmid constructs containing PERV-A 14/220 proviruses as well as the prototype PERV-A PK clone Ap60 were transfected into 293 cells, and cell lysates were analyzed for Gag-Pol expression with an anti-PERV CA antibody (2). All four PERV-A 14/220 proviruses produced detectable CA as early as 4 days after transfection, while it took 14 days for Ap60 CA to appear (data not shown). A preliminary virus transmission experiment on 293 cells showed that these molecular clones produced PERV as infectious as the biological isolate of PERV-A 14/220 (Table 2 and data not shown). The provirus clone that gave the highest titer was selected for use and was named A14/220 in further studies.
Sequence analyses. (i) Genetic make-up of PERV-A 14/220.
Previous partial sequence data of the biological isolate indicated that PERV-A 14/220 is a recombinant virus between PERV-C and PERV-A. To elucidate the origin of PERV-A 14/220 and identify recombination crossover points, we compared the A14/220 sequence with other full-length genomes of the PERV γ1 group (33) which encompasses the PERV-A, -B, and -C subgroups. In total, 12 proviral sequences and three cDNA clone sequences (Cmsl, Ctsukuba, and BQOne) were retrieved; these are listed in Table 1. These PERV genome sequences have been deposited following either screening for their infectivity (2, 6, 14, 19, 26), screening for the presence of ORF by partial sequencing (1, 28), or screening for their ability to produce full-length viral proteins (14, 26). Importantly, some of the germ line sequences, A130, A151, A463 and B192, were obtained from the same bacterial artificial chromosome library by the same, extensive screening (26). These clones are, therefore, likely to represent PERV that are currently infectious or were infectious until more recently than other PERV with more genetic defects in the pig genome.
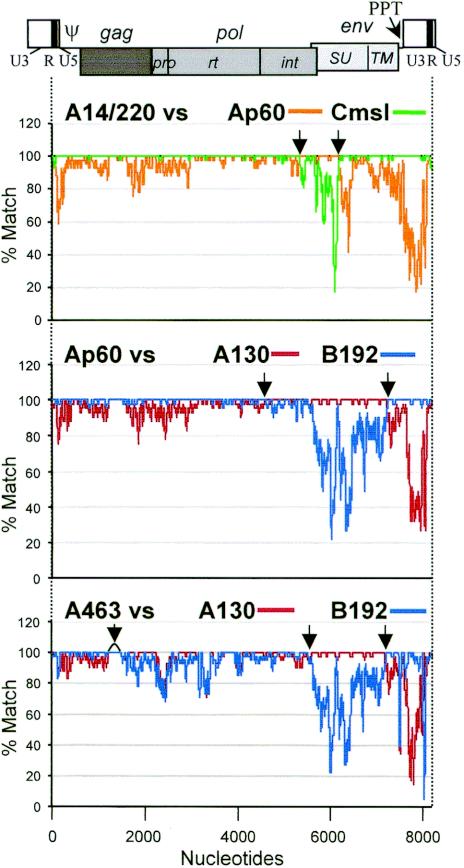
The full-length sequences corresponding to the RNA genome (5′-R to 3′-R) were extracted from proviral sequences including A14/220, while those for cDNA clone sequences were generated by restoring the 5′-R sequence with the corresponding 3′-R sequence. The lengths of these sequences range from 8,143 to 8,299 nt, due mainly to size differences in their LTR and SU sequences (Fig. 1 and 22). Sets of two to four sequences were aligned and then analyzed for local sequence homologies. Nucleotide positions of the alignment (x axis) were plotted against percent match for a window of 41 nt (y axis) between a pair of aligned sequences by using the program LOHATM (Fig. 1 and 2). First, to reveal the genetic nature of A14/220 as a recombinant, we aligned A14/220 with Cmsl (representing PERV-C), Ap60 (PK15 cell-derived prototype PERV-A, also compared in biological studies; see below) and A130 (representing germ line PERV-A). Homology plots of A14/220 against Cmsl and Ap60 based on this alignment are shown in Fig. 1 (top panel). The plot of A14/220 versus Cmsl shows that these two sequences are highly related through the whole genome, apart from approximately 850 bp spanning the region of the pol-env junction. In this 850-bp region, A14/220 is highly related to Ap60 (Fig. 1, top) and A130 (data not shown). Most likely, the recombination crossover points (Fig. 1, arrows in top panel) where these sequences are conserved are nt 5374 to 5426 and 6260 to 6280 based on the nucleotide numbering of A14/220. These results strongly indicate that homologous recombination of a PERV-C sequence replacing its pol-env 850-bp region with that of a PERV-A sequence gave rise to A14/220, a PERV using PERV-A receptors.
FIG. 1.
PERV recombination profiles shown by local homology analysis. (Top) Four full-length nucleotide sequences, A14/220 (8,188 nt), Cmsl (8,144 nt), Ap60 (8,261 nt), and A130 (8,299 nt), were analyzed by LOHA with 41-nt windows. The alignment length was 8,325 nt positions and a percent match plot for A14/220 versus Cmsl was overlaid on that for A14/220 versus Ap60. (Middle) Ap60, A130, and B192 (8,255 nt) were analyzed, and a percent match plot for Ap60 versus B192 was overlaid on that for Ap60 versus A130. The alignment size was 8,372 positions. (Bottom) A463 (8,204 nt), A130, and B192 were aligned: A463 versus B192 overlaid on A463 versus A130 for 8453 positions. A schema of a PERV provirus is displayed on top to indicate the positions of the viral genes and domains. The apparent recombination crossover points are indicated by arrows.
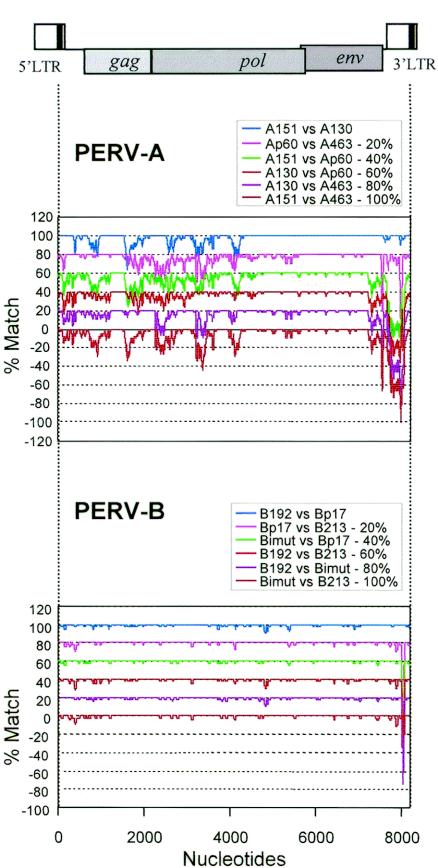
FIG. 2.
Genetic variability of PERV-A and -B proviruses. (Top) The three germ line proviral sequences A130, A151 (8,291 nt), and A463 as well as the prototype cell line-derived PERV-A sequence Ap60 were analyzed by LOHA (aligned into 8,356 positions). (Bottom) The three germ line proviral sequences B192, B213 (8,215 nt), and Bimut (8,217 nt) as well as the prototype cell line-derived PERV-B sequence Bp17 (8,256 nt) were analyzed (aligned into 8,253 nt positions). All six possible homology plots (0, 20, 40, 60, 80, or 100% was subtracted from percent match values as indicated [y axis]) for each set of sequences are shown.
(ii) Evidence for other recombination events.
In the above study on A14/220, we noticed that two PERV-A sequences, Ap60 and A130, were considerably different from each other outside of their env gene, suggesting that other recombination events may have resulted in the variability of PERV genomes. By applying the same local homology analysis on selected sets of sequences, evidence for two other prominent events of past recombination was discovered. Homology plots of Ap60 against A130 and B192 are shown in Fig. 1 (middle panel). Ap60, an infectious PERV-A transmitted from a pig cell line PK15 to human 293 cells, appeared to contain a PERV-B-like sequence for the major part of the genome but with a PERV-A sequence for parts of pol and env. Similar to the relationship between A14/220 and PERV-C, Ap60 likely arose by recombination of PERV-B acquiring a PERV-A env sequence. Although it is unknown when such a recombination event occurred, there is no evidence that the Ap60 type of PERV-A sequence is present in pig germ line chromosomes. Likewise, another potential recombinant PERV is A463 (Fig. 1, bottom panel). Compared with A130 and B192, A463 appeared to contain an LTR and its proximal 5′ and 3′ regions from PERV-B, parts of pol and env from PERV-A, and a unique sequence for gag and the 5′ part of pol (see Fig. 3). A463 is likely to be a genuinely endogenous PERV present in germ line DNA as it has been obtained from a pig genomic library, and PERV proviruses have been found in the same integration locus in different individual pigs (26, 27). These results suggest that recombination between various PERV sequences has been instrumental in the formation of both germ line PERV sequences as well as pre-germ line fixation forms of exogenous or replicating PERV.
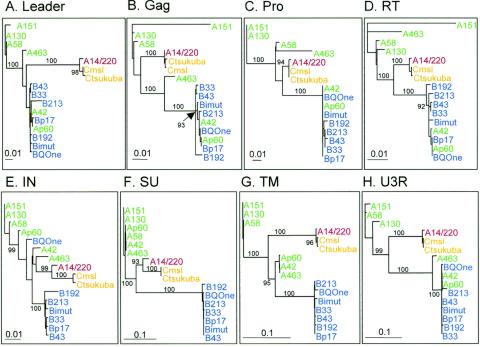
FIG. 3.
Phylogenetic trees for PERV genome regions of leader, Gag, PRO, RT, IN, SU, TM, and U3R. Phylogram trees were obtained for 16 different full-length PERV nucleotide sequences rooted against sequence A151. Scales show substitution(s) per site. Bootstrap values higher than >90% are shown.
Our analyses uncovered evidence for recombination that generates PERV containing the PERV-A RBD and, therefore, is able to use PERV-A receptors (9) but not the other receptor groups, PERV-B and -C. These results suggested that the PERV-A env gene may be more prone to recombination and bound to more heterogeneous sequences than the PERV-B env. To investigate this possibility, four sequences each for PERV-A and PERV-B (three germ line and one cell line PERV sequence each) were aligned and all six possible pairwise homology plots for each set were obtained (Fig. 2). Each of the four PERV-A sequences appear to be variable from each other in the 5′ half of the genome, as all six plots show a match of less than 90% in many nucleotide positions. Conversely, the 3′ end (end of env to U3) of these sequences can be classified into two types (the A130/A151 group and Ap60/A436 group) (Fig. 2, top panel). These results demonstrate that the PERV-A receptor group of viruses has heterogeneous sequences outside env (in the case of A14/220 env, the RBD). Homology in the 3′ half of pol and env between A130 and A151 is remarkably high, despite the fact that they are different in the other parts of the genome. This suggests that either sequence or both may also have been generated by recent recombination events. In contrast, PERV-B sequences are conserved all along the genome, apart from some differences shown as troughs in the U3 region due to different numbers of enhancer repeats (Fig. 2, bottom panel) (38, 51, 53). There is no variability bias toward specific genes or genetic elements, indicating no evidence of recombination in recently active PERV using PERV-B receptors.
(iii) Phylogenetic relationships between PERV full-length sequences.
To confirm the above observations and to further study relationships between full-length PERV sequences, phylogenetic trees in different genome regions were produced (Fig. 3). For ease of comparison, all trees were rooted by using A151 as the outgroup. Overall, PERV-A group sequences are scattered in trees apart from that for the SU region (A14/220 in SU is an exception). In contrast, all of the PERV-B sequences are generally clustered together with the exception of the IN of BQOne (see below). These results confirm that PERV-A SU is combined with more heterogeneous elements than PERV-B SU. The SU tree shows that A14/220 is unique in that its recombination occurred in the middle of SU, resulting in an SU derived from PERV-A RBD with the 3′ half of SU from PERV-C (Fig. 3F). In the regions other than SU and IN, A14/220 clusters with two PERV-C sequences, consistent with the above results (Fig. 1, top). In contrast, Ap60 clusters with PERV-B from LTR to RT (Fig. 3A, B, C, D, and H), consistent with Fig. 1 (middle panel). A436 occurs in various places in different trees, confirming that it contains PERV-A, PERV-B, and unique elements (Fig. 1, bottom, and Fig. 3). Of the six PERV-A sequences, apart from A14/220, this analysis separated them into two groups of three in the TM tree (Fig. 3G). The positions of Ap60, A42, and A463 reflect that these are recombined with a PERV-B-related sequence near the end of TM, as shown for Ap60 and A436 in Fig. 1 (middle and bottom panels, respectively). It has been proposed that such recombination may rescue replication-defective endogenous PERV-A env, whose TM appears to be truncated (40). The U3-R tree, 3′ to this recombination point, demonstrates that A463, A42, and Ap60, unlike other PERV-A and -C genomes with so-called repeatless LTR, contain the PERV-B type LTR with enhancer repeats (51) (Fig. 3H). Finally, the significance of BQOne's separation from the rest of PERV-B in IN (Fig. 3E; also noted in a previous study [18]) is not clear, because this sequence was created by multiple PCR cloning from a cDNA library (39) and was, therefore, potentially more susceptible to technical artifacts. The IN region appears to be most conserved as indicated by the data shown in Fig. 1 and 2 and as reflected by a rather complicated tree with many branches of medium length dividing smaller clusters (Fig. 3E).
Subgroup classification of the PERV γ1 group has been based on receptor usage and/or env gene homology. Our results demonstrate that this classification applies only to the env gene or, more strictly, to its RBD but not to the other regions. This may cause some confusion as one could identify PERV-A 14/220 and PERV-A PK as PERV-C and -B, respectively, even though they contain parts of the PERV-A env sequence and use PERV-A receptors. While the limit of this env-based classification should be noted, we consider that it is still a valid, convenient classification.
Env crossover points in various A/C recombinants.
PERV transmission experiments by cocultivation of activated miniature swine PBMC with human 293 cells have led to the isolation of several PERV able to replicate in human 293 cells (28, 40, 54). All these isolates, including PERV-A 14/220, appear to have hybrid env genes consisting of PERV-A- and -C-like sequence (28, 55). To test if these env genes share recombination crossover points, A14/220 env and six env sequences (five from the Immerge laboratory and one from the National Institutes of Health [NIH]) were analyzed for local homology against Cmsl env (Fig. 4). These six env fragments were cloned and sequenced from productively PERV-infected 293 cells. Although their function for cell entry has not been tested, most if not all of them are likely to represent env genes of infectious PERV recombinants. Indeed, previous env sequences reported for PERV-A 14/220 (GenBank accession numbers AF417227 and AF417228 [28]) have the same crossover point as A14/220 and differ from the A14/220 sequence by only a few point mutations (data not shown). As for the nonrecombinant PERV-A reference, A130 env was also analyzed. Figure 4 shows that seven env sequences have different crossover points, scattered through the whole env gene. These results suggest that A-to-C recombination can occur in many places in the env gene, resulting in functional env genes with PERV-A receptors, although their levels of env activity may differ. No apparent recombination hot spots were found.
Difference in infectivity between A14/220 and Ap60 on different cell lines.
It has previously been reported that a PERV-A 14/220 biological isolate infects several human cell lines and a pig cell line more efficiently than the prototype PERV-A PK (9). It is noteworthy that PERV-A isolates and infectious molecular clones derived from PK15 cells have proved to be more infectious than any of the genomic clones derived from the germ line PERV (19, 26). This, together with the sequence analyses above, suggests that PERV-A PK, like PERV-A 14/220, may be a recombinant PERV-A that has higher infectivity compared to its PERV-A germ line ancestor. The infectivity of biological isolates of PERV-A 14/220 and PK and their molecular clones (A14/220 and Ap60, respectively) was studied to confirm differences in titers between these viruses on different cell lines and to test if the molecular clones maintain the phenotype of biological isolates. Virus supernatant from 293 cells persistently infected with PERV that harbor an MLV lacZ vector was titrated on five cell lines (two human and one each of pig, mink, and cat) for replicating PERV by immunostaining and MLV lacZ vector transfer as previously described (2). All virus producer cell lines persistently produced RT activity in their supernatants at a similar level (data not shown). Table 2 shows that in both assays, biological isolates and their corresponding molecular clones have similar infectivity on each cell line, indicating that the molecular clones represent the biological isolates in terms of infectivity. PERV-A 14/220 and A14/220 had up to 100-fold higher titer than PERV-A PK and Ap60 on all cell lines. In addition, the replicating virus titer on 293 cells was standardized against RT activity. A14/220 had 443 infectious units (IU) per mU of RT activity versus 3.3 IU/mU of RT for PERV-A PK, suggesting that an A14/220 PERV particle is about 100 times more infectious than a PERV-A PK particle. These results indicated that PERV-A 14/220 is more infectious on a wide range of cell lines than PERV-A PK and therefore more infectious than most PERV-A isolates or clones reported to date.
Genetic determinants for high-titer in A14/220.
To investigate the mechanism of high infectivity of PERV-A 14/220 and the genetic changes that can turn a relatively low-titer, prototype PERV-A into a high-titer virus, we attempted to map the genetic determinants for the titer difference between A14/220 and Ap60. First, chimeric constructs by mix-and-match of four fragments between A14/220 and Ap60 were produced and tested for infectivity after establishment of persistent infection in 293 cells. PERV replication was assayed by immunostaining (Fig. 5A). While the titer difference between the parental A14/220 and Ap60 constructs was about 1,000-fold in this series of experiments, most of the chimeric constructs showed titers intermediate between those of A14/220 and Ap60. The phenotypes of A14/220 and Ap60 were not attributable to a single region, although constructs with relatively higher titers appeared to contain certain protein coding regions (Fig. 5A, fragments B and C) from A14/220. In contrast, viral cis elements had no (5′-LTR, fragment A) or only little (3′-LTR, fragment D) effects. These results suggested that there are multiple determinants for the titer difference in the protein coding region (fragments B and C).
To further define the infectivity determinants, fragments B and C were further dissected into two fragments each (Fig. 5B, fragments b1, b2, c1, and c2). Constructs containing a mix and match of A14/220 and Ap60 for these fragments as well as fragments A and D from A 14/220 were tested. In this series, virus was harvested 3 days after transfection of 293T cells and then titrated on 293 cells. Again, the chimeric constructs had intermediate titers compared to those of A14/220 and Ap60 (Fig. 5B). Fragment b1 corresponding to gag had the least effect on the viral titer, whereas the other fragments, b2, c1, and c2, affected virus titer substantially. These results indicated that there are multiple determinants for a difference in titers between A14/220 and Ap60 in the pol and env coding regions.
DISCUSSION
We conducted this study to gain insights into questions related to PERV recombination, such as how frequently PERV recombination occurs, what types of recombinant PERV can be formed, and what the consequence is in terms of zoonotic risk particularly for pig-to-human xenotransplantation.
An infectious molecular clone of high-titer, human-tropic PERV isolate, PERV-A 14/220, was generated. Most parts of the sequence of this clone were found to be almost identical to those of PERV-C. PERV-C infect porcine but not human cells, because human cells lack PERV-C receptors (48). PERV-A 14/220, however, contains a PERV-A sequence of about 850 bp around the pol-env junction including the RBD (Fig. 1 and 4), enabling this isolate to use PERV-A receptors and infect human cells (9). Similar recombined forms of the env gene, containing PERV-A and -C sequences at the 5′ and 3′ ends, respectively, have been obtained as parts of human-tropic PERV isolated by transmission to a human cell line from miniature swine PBMC (28, 55). Such recombinant env genes have not been detected in miniature swine genomic DNA by extensive screenings (40). Each of these recombined env genes had different A-to-C crossover points (Fig. 4), while at least three recombination patterns have been found for C-to-A recombination upstream of the RBD (18, 28). It is, therefore, most likely that these A/C recombinant PERV, which are genetically distinct from each other, have arisen in individual pigs, since such A/C recombinant env sequences have also been found in nonstimulated porcine PBMC (56). The possibility, however, remains that some of them were generated in the process of primary cell culture of porcine PBMC followed by their cocultivation with human cells. The relatively frequent isolation of recombinant, human-tropic PERV from the miniature swine indicates that these pigs are prone to produce such human-tropic, recombinant PERV.
PERV-A 14/220 infects several cell lines from different species at higher titers than the prototype PERV-A derived from PK15 cells (Table 2). As it has previously been reported that PERV-A PK infectivity was at least 100-fold lower than that of other standard gammaretroviruses, such as gibbon ape leukemia virus and MLV-A (2), the infectivity of PERV-A 14/220 appears to be close to the level of highly infectious gammaretrovirus isolates. In contrast, porcine genomic clones of germ line PERV-A have been reported to be even less infectious than PERV-A PK (19, 26). Another PERV isolate which is also an A/C recombinant and more infectious than PERV-A PK is the PERV NIH isolate (Fig. 4 and reference 55). In our preliminary experiments PERV-NIH (kindly provided by C. Wilson) was severalfold less infectious than PERV-A 14/220 (data not shown). The A/C recombinant PERV-A 14/220, therefore, appears to be the highest titer PERV with PERV-A receptors reported to date.
To investigate molecular mechanisms for the high infectivity of PERV-A 14/220 and genetic changes that can turn low-titer PERV-A to high-titer PERV-A, genetic determinants for a titer difference between A14/220 and Ap60 were mapped (Fig. 5). One may predict that the U3 region which is most divergent between A14/220 and Ap60 (Fig. 1) would be a major determinant. However, previous studies have shown that PERV-C LTR (found in A14/220 and PERV-NIH) has a similar or lesser level of activity in human cells compared to those of prototype PERV-A or PERV-B (found in Ap60) (38, 53). Our infectivity analyses showed that the LTR has only minor, if any, effects on the titer difference, consistent with previous LTR studies (Fig. 5). While the gag region had little effect, we identified three genome segments covering the pol and env region containing determinants for the titer difference, with the env segment having the most pronounced effect (Fig. 5B). In parallel with this study, the env gene has been further dissected and has been shown to have two separate determinants, making at least four determinants in total: one determinant was mapped to a single amino acid change in the RBD (a change within the PERV-A-derived sequence of A14/220), while the other lies in the PERV-C sequence (12). These consistent results suggest that genetic elements responsible for the high infectivity of PERV-A 14/220 are multiple and many; at least three of them reside in the PERV-C-derived sequence. Therefore, a single mutation is not sufficient to allow low-titer PERV-A to become as infectious as PERV-A 14/220 or other standard gammaretrovirus isolates. A single recombination event, rather than multiple mutations, may be a faster and more effective way to generate higher-titer, potentially more harmful PERV. Indeed, the prototype PERV-A PK represented by Ap60 and A42 clones, which is more infectious than known germ line PERV-A, appears to be a PERV-B/A recombinant (Fig. 1, middle panel) (40). Overall, recombinant, exogenous forms of PERV with PERV-A receptors are likely to pose a greater zoonotic risk than germ line forms of PERV-A.
Evidence for other types of recombination in the past that generated PERV using the PERV-A receptor were obtained (Fig. 1, middle and bottom panels). We also revealed that PERV-A but not PERV-B among PERV which are currently active, or were active until recently, are variable in the genome outside the env gene (Fig. 2 and 3). Such variability is likely to have arisen by recombination events where PERV-A env genes recombine with different genome elements, although sequence diversion by point mutations may have played some role. These results suggest that PERV-A env is more prone to recombination than PERV-B env and may have been propagating or surviving by hitchhiking on various types of PERV backbone. This is reminiscent of the env gene of the RD114 receptor group of beta- and gammaretroviruses, which have recombined to a wide range of viral backbone genomes and spread to a wide range of host species (3, 16, 20, 35, 45). Acquisition of endogenous env sequences is also regularly seen in lymphomagenesis by murine and feline gamma retroviruses (4). It is not clear why PERV-A, but not PERV-B, env sequences recombine frequently.
There are several possible explanations for the promiscuity of PERV-A env. First, germ line PERV-A proviruses have less competent sequences outside of env, and therefore their env genes require more extensive recombination in order to generate fully infectious, exogenous PERV. Second, it is known that some endogenous retrovirus env can be toxic or beneficial to the host under certain circumstances (3, 4, 15). A fine balance in the host-parasite relationship is required for an endogenous retrovirus to cohabit with the host. Thus, PERV-A env may be still adapting to the host environment by recombination, whereas PERV-B may have fully adapted to pigs and may at present be gradually decaying. Indeed, only one germ line clone of PERV-B, B213 has been reported to be weakly infectious (26), and the exact origin of the prototype infectious virus, PERV-B PK represented by the Bp17, B33, and B43 clones, is unclear.
Additionally, the recombination frequencies could be affected by proviral copy number and genome transcription in virus producer cells. In this regard, it is noteworthy that PERV-A proviruses are present at higher levels than PERV-B proviruses in most pig herds including miniature swine (21, 31) and that PERV-A RNA transcripts appear to be more abundant than those of PERV-B in miniature swine PBMC cultures (31). These facts may explain why replication-competent A/C, but not B/C, recombinants have been isolated from miniature swine PBMC (28). Currently, however, it is unknown where (in what species or breeds or tissues or cells) and when the other recombinant PERV occurred in the past, and, therefore, no data on proviral gene dosage and RNA level are available.
There is evidence that PERV-B is older than PERV-A or PERV-C in the pig genome. PERV-B-type LTR with enhancer repeats has been estimated to be twice as old as the repeatless LTR found in PERV-A germ line clones (51). Also, only the PERV-B, but not the PERV-A, env sequence was present in a warthog DNA sample, while samples from red river hog and bushpig, proposed to be closer relatives to the domestic pig than warthog (11), had both PERV-A and -B (33). In terms of PERV-A evolution, genetic recombination complicates age estimation and interpretation of phylogenetic relationships among PERV-A. It has been proposed that some germ line PERV-A sequences including A130 and A151 are linked to repeatless, younger LTR and, therefore, are youngest among PERV-A env genes (51). It is, however, possible that such germ line proviruses may be older than many others, such as PERV-A PK and germ line A436, that have arisen by recombination with PERV-B. The type of LTR linked to many env sequences analyzed in a previous study (51) is unknown.
Compared with PERV-A and -B, PERV-C appears to be even younger in the pig lineage as not all domestic pigs have PERV-C sequence (33). Miniature swine may be a unique pig breed as they possess more copies of PERV-C sequence (typically five to nine copies [31]) than many other pig herds (one to three copies [24]), suggesting that PERV-C is active in miniature swine. Although only limited studies have been performed on PERV transmission to human cells from primary cultures from other pig herds such as Large Whites, it is intriguing that A/C recombinant PERV have been reported from miniature swine but not from other herds. Miniature swine may be more prone to production of human-tropic, high-titer A/C recombinant PERV, like PERV A 14/220 or PERV-NIH, due to their possession of active PERV-C. Thus, miniature swine that possess replication-competent PERV-C may not represent an optimal source for xenotransplantation materials. However, miniature swine that do not produce replication-competent PERV-C have been identified (56). Further careful studies on PERV genetics and biology are required in the quest for an improved xenotransplantation source.
Acknowledgments
We thank Stephane Hugh for help in phylogenetic analyses, Rachel Frasier, Christelle Granier, Beth Oldmixon, and James Wood for technical assistance, and Francois-Loic Cosset, Paul Kellam, Jonathan Stoye, and Greg Towers for critical manuscript reading and discussion.
This study was supported by the United Kingdom Medical Research Council.
REFERENCES
- 1.Akiyoshi, D. E., M. Denaro, H. Zhu, J. L. Greenstein, P. Banerjee, and J. A. Fishman. 1998. Identification of a full-length cDNA for an endogenous retrovirus of miniature swine. J. Virol. 72:4503-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartosch, B., R. A. Weiss, and Y. Takeuchi. 2002. PCR-based cloning and immunocytological titration of infectious porcine endogenous retrovirus subgroup A and B. J. Gen. Virol. 83:2231-2240. [DOI] [PubMed] [Google Scholar]
- 3.Blond, J. L., D. Lavillette, V. Cheynet, O. Bouton, G. Oriol, S. Chapel-Fernandes, B. Mandrand, F. Mallet, and F. L. Cosset. 2000. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 74:3321-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coffin, J. M., S. H. Hughes, and H. E. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 5.Cunningham, D. A., C. Herring, X. M. Fernandez-Suarez, A. J. Whittam, K. Paradis, and G. A. Langford. 2001. Analysis of patients treated with living pig tissue for evidence of infection by porcine endogenous retroviruses. Trends Cardiovasc. Med. 11:190-196. [DOI] [PubMed] [Google Scholar]
- 6.Czauderna, F., N. Fischer, K. Boller, R. Kurth, and R. R. Tonjes. 2000. Establishment and characterization of molecular clones of porcine endogenous retroviruses replicating on human cells. J. Virol. 74:4028-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dinsmore, J. H., C. Manhart, R. Raineri, D. B. Jacoby, and A. Moore. 2000. No evidence for infection of human cells with porcine endogenous retrovirus (PERV) after exposure to porcine fetal neuronal cells. Transplantation 70:1382-1389. [DOI] [PubMed] [Google Scholar]
- 8.Elliott, R. B., L. Escobar, O. Garkavenko, M. C. Croxson, B. A. Schroeder, M. McGregor, G. Ferguson, N. Beckman, and S. Ferguson. 2000. No evidence of infection with porcine endogenous retrovirus in recipients of encapsulated porcine islet xenografts. Cell Transplant. 9:895-901. [DOI] [PubMed] [Google Scholar]
- 9.Ericsson, T. A., Y. Takeuchi, C. Templin, G. Quinn, S. F. Farhadian, J. C. Wood, B. A. Oldmixon, K. M. Suling, J. K. Ishii, Y. Kitagawa, T. Miyazawa, D. R. Salomon, R. A. Weiss, and C. Patience. 2003. Identification of receptors for pig endogenous retrovirus. Proc. Natl. Acad. Sci. USA 100:6759-6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galbraith, D. N., C. Haworth, G. M. Lees, and K. T. Smith. October 1997. Porcine retrovirus. Patent WO 97/40167.
- 11.Groves, C. P. 1981. Ancestors for the pigs: taxonomy and phylogeny of the genus Sus. Technical bulletin no. 3. Australian National University, Canberra, Australia.
- 12.Harrison, I., Y. Takeuchi, B. Bartosch, and J. P. Stoye. 2004. Determinants of high titer in recombinant porcine endogenous retroviruses. J. Virol. 78:13871-13879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heneine, W., A. Tibell, W. M. Switzer, P. Sandstrom, G. V. Rosales, A. Mathews, O. Korsgren, L. E. Chapman, T. M. Folks, and C. G. Groth. 1998. No evidence of infection with porcine endogenous retrovirus in recipients of porcine islet-cell xenografts. Lancet 352:695-699. [DOI] [PubMed] [Google Scholar]
- 14.Herring, C., G. Quinn, R. Bower, N. Parsons, N. A. Logan, A. Brawley, K. Elsome, A. Whittam, X. M. Fernandez-Suarez, D. Cunningham, D. Onions, G. Langford, and L. Scobie. 2001. Mapping full-length porcine endogenous retroviruses in a Large White pig. J. Virol. 75:12252-12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikeda, H., and H. Sugimura. 1989. Fv-4 resistance gene: a truncated endogenous murine leukemia virus with ecotropic interference properties. J. Virol. 63:5405-5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kewalramani, V. N., A. T. Panganiban, and M. Emerman. 1992. Spleen necrosis virus, an avian immunosuppressive retrovirus, shares a receptor with the type D simian retroviruses. J. Virol. 66:3026-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klymiuk, N., M. Muller, G. Brem, and B. Aigner. 2002. Characterization of porcine endogenous retrovirus γ pro-pol nucleotide sequences. J. Virol. 76:11738-11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klymiuk, N., M. Muller, G. Brem, and B. Aigner. 2003. Recombination analysis of human-tropic porcine endogenous retroviruses. J. Gen. Virol. 84:2729-2734. [DOI] [PubMed] [Google Scholar]
- 19.Krach, U., N. Fischer, F. Czauderna, and R. R. Tonjes. 2001. Comparison of replication-competent molecular clones of porcine endogenous retrovirus class A and class B derived from pig and human cells. J. Virol. 75:5465-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavillette, D., M. Marin, A. Ruggieri, F. Mallet, F. L. Cosset, and D. Kabat. 2002. The envelope glycoprotein of human endogenous retrovirus type W uses a divergent family of amino acid transporters/cell surface receptors. J. Virol. 76:6442-6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Tissier, P., J. P. Stoye, Y. Takeuchi, C. Patience, and R. A. Weiss. 1997. Two sets of human-tropic pig retrovirus. Nature 389:681-682. [DOI] [PubMed] [Google Scholar]
- 22.Levy, M. F., J. Crippin, S. Sutton, G. Netto, J. McCormack, T. Curiel, R. M. Goldstein, J. T. Newman, T. A. Gonwa, J. Banchereau, L. E. Diamond, G. Byrne, J. Logan, and G. B. Klintmalm. 2000. Liver allotransplantation after extracorporeal hepatic support with transgenic (hCD55/hCD59) porcine livers: clinical results and lack of pig-to-human transmission of the porcine endogenous retrovirus. Transplantation 69:272-280. [DOI] [PubMed] [Google Scholar]
- 23.Magre, S., Y. Takeuchi, and B. Bartosch. 2003. Xenotransplantation and pig endogenous retroviruses. Rev. Med. Virol. 13:311-329. [DOI] [PubMed] [Google Scholar]
- 24.Mang, R., J. Maas, X. Chen, J. Goudsmit, and A. C. van Der Kuyl. 2001. Identification of a novel type C porcine endogenous retrovirus: evidence that copy number of endogenous retroviruses increases during host inbreeding. J. Gen. Virol. 82:1829-1834. [DOI] [PubMed] [Google Scholar]
- 25.Martin, U., V. Kiessig, J. H. Blusch, A. Haverich, K. von der Helm, T. Herden, and G. Steinhoff. 1998. Expression of pig endogenous retrovirus by primary porcine endothelial cells and infection of human cells. Lancet 352:692-694. [DOI] [PubMed] [Google Scholar]
- 26.Niebert, M., C. Rogel-Gaillard, P. Chardon, and R. R. Tonjes. 2002. Characterization of chromosomally assigned replication-competent gamma porcine endogenous retroviruses derived from a Large White pig and expression in human cells. J. Virol. 76:2714-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niebert, M., and R. R. Tonjes. 2003. Analyses of prevalence and polymorphisms of six replication-competent and chromosomally assigned porcine endogenous retroviruses in individual pigs and pig subspecies. Virology 313:427-434. [DOI] [PubMed] [Google Scholar]
- 28.Oldmixon, B. A., J. C. Wood, T. A. Ericsson, C. A. Wilson, M. E. White-Scharf, G. Andersson, J. L. Greenstein, H. J. Schuurman, and C. Patience. 2002. Porcine endogenous retrovirus transmission characteristics of an inbred herd of miniature swine. J. Virol. 76:3045-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Page, R. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 30.Paradis, K., G. Langford, Z. Long, W. Heneine, P. Sandstrom, W. M. Switzer, L. E. Chapman, C. Lockey, D. Onions, E. Otto, et al. 1999. Search for cross-species transmission of porcine endogenous retrovirus in patients treated with living pig tissue. Science 285:1236-1241. [DOI] [PubMed] [Google Scholar]
- 31.Patience, C. Unpublished data.
- 32.Patience, C., G. S. Patton, Y. Takeuchi, R. A. Weiss, M. O. McClure, L. Rydberg, and M. E. Breimer. 1998. No evidence of pig DNA or retroviral infection in patients with short-term extracorporeal connection to pig kidneys. Lancet 352:699-701. [DOI] [PubMed] [Google Scholar]
- 33.Patience, C., W. M. Switzer, Y. Takeuchi, D. J. Griffiths, M. E. Goward, W. Heneine, J. P. Stoye, and R. A. Weiss. 2001. Multiple groups of novel retroviral genomes in pigs and related species. J. Virol. 75:2771-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patience, C., Y. Takeuchi, and R. A. Weiss. 1997. Infection of human cells by an endogenous retrovirus of pigs. Nat. Med. 3:282-286. [DOI] [PubMed] [Google Scholar]
- 35.Rasko, J. E., J. L. Battini, R. J. Gottschalk, I. Mazo, and A. D. Miller. 1999. The RD114/simian type D retrovirus receptor is a neutral amino acid transporter. Proc. Natl. Acad. Sci. USA 96:2129-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 37.Sauer, I. M., D. Kardassis, K. Zeillinger, A. Pascher, A. Gruenwald, G. Pless, M. Irgang, M. Kraemer, G. Puhl, J. Frank, A. R. Muller, T. Steinmuller, J. Denner, P. Neuhaus, and J. C. Gerlach. 2003. Clinical extracorporeal hybrid liver support: phase I study with primary porcine liver cells. Xenotransplantation 10:460-469. [DOI] [PubMed] [Google Scholar]
- 38.Scheef, G., N. Fischer, E. Flory, I. Schmitt, and R. R. Tonjes. 2002. Transcriptional regulation of porcine endogenous retroviruses released from porcine and infected human cells by heterotrimeric protein complex NF-Y and impact of immunosuppressive drugs. J. Virol. 76:12553-12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scobie, L. Personal communication.
- 40.Scobie, L., S. Taylor, J. C. Wood, K. M. Suling, G. Quinn, S. Meikle, C. Patience, H. J. Schuurman, and D. E. Onions. 2004. Absence of replication-competent human-tropic porcine endogenous retroviruses in the germ line DNA of inbred miniature swine. J. Virol. 78:2502-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soneoka, Y., P. M. Cannon, E. E. Ramsdale, J. C. Griffiths, G. Romano, S. M. Kingsman, and A. J. Kingsman. 1995. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 23:628-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stoye, J. P. 1999. The pathogenic potential of endogenous retroviruses: a sceptical view. Trends Microbiol. 7:430. [DOI] [PubMed] [Google Scholar]
- 43.Suling, K., G. Quinn, J. Wood, and C. Patience. 2003. Packaging of human endogenous retrovirus sequences is undetectable in porcine endogenous retrovirus particles produced from human cells. Virology 312:330-336. [DOI] [PubMed] [Google Scholar]
- 44.Swafford, D. 2001. PAUP v 4.0: phylogenetic analysis using parsimony (and other methods). Sinaur Associates, Sunderland, Mass.
- 45.Tailor, C. S., A. Nouri, Y. Zhao, Y. Takeuchi, and D. Kabat. 1999. A sodium-dependent neutral-amino-acid transporter mediates infections of feline and baboon endogenous retroviruses and simian type D retroviruses. J. Virol. 73:4470-4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takeuchi, Y. 1998. Retrovirus time travel. Trends Microbiol. 6:430. [DOI] [PubMed] [Google Scholar]
- 47.Takeuchi, Y., F. L. Cosset, P. J. Lachmann, H. Okada, R. A. Weiss, and M. K. Collins. 1994. Type C retrovirus inactivation by human complement is determined by both the viral genome and the producer cell. J. Virol. 68:8001-8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takeuchi, Y., C. Patience, S. Magre, R. A. Weiss, P. T. Banerjee, P. Le Tissier, and J. P. Stoye. 1998. Host range and interference studies of three classes of pig endogenous retrovirus. J. Virol. 72:9986-9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takeuchi, Y., and R. A. Weiss. 2000. Xenotransplantation: reappraising the risk of retroviral zoonosis. Curr. Opin. Immunol. 12:504-507. [DOI] [PubMed] [Google Scholar]
- 50.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tönjes, R. R., and M. Niebert. 2003. Relative age of proviral porcine endogenous retrovirus sequences in Sus scrofa based on the molecular clock hypothesis. J. Virol. 77:12363-12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van de Kerkhove, M. P., E. Di Florio, V. Scuderi, A. Mancini, A. Belli, A. Bracco, D. Scala, S. Scala, L. Zeuli, G. Di Nicuolo, P. Amoroso, F. Calise, and R. A. Chamuleau. 2003. Bridging a patient with acute liver failure to liver transplantation by the AMC-bioartificial liver. Cell Transplant. 12:563-568. [PubMed] [Google Scholar]
- 53.Wilson, C. A., S. Laeeq, A. Ritzhaupt, W. Colon-Moran, and F. K. Yoshimura. 2003. Sequence analysis of porcine endogenous retrovirus long terminal repeats and identification of transcriptional regulatory regions. J. Virol. 77:142-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson, C. A., S. Wong, J. Muller, C. E. Davidson, T. M. Rose, and P. Burd. 1998. Type C retrovirus released from porcine primary peripheral blood mononuclear cells infects human cells. J. Virol. 72:3082-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson, C. A., S. Wong, M. VanBrocklin, and M. J. Federspiel. 2000. Extended analysis of the in vitro tropism of porcine endogenous retrovirus. J. Virol. 74:49-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wood, J. C., G. Quinn, K. M. Suling, B. A. Oldmixon, B. A. Van Tine, R. Cina, S. Arn, C. A. Huang, L. Scobie, D. E. Onions, D. H. Sachs, H. J. Schuurman, J. A. Fishman, and C. Patience. 2004. Identification of exogenous forms of human-tropic porcine endogenous retrovirus in miniature swine. J. Virol. 78:2494-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]