Abstract
Porcine endogenous retroviruses (PERVs) pose a potential stumbling block for therapeutic xenotransplantation, with the greatest threat coming from viruses generated by recombination between members of the PERV subgroup A (PERV-A) and PERV-C families (PERV-A/C recombinants). PERV-A and PERV-B have been shown to infect human cells in culture, albeit with low titers. PERV-C has a more restricted host range and cannot infect human cells. A recombinant PERV-A/C virus (PERV-A14/220) contains the PERV-A sequence between the end of pol and the middle of the SU region in env. The remaining sequence is derived from PERV-C. PERV-A14/220 is approximately 500-fold more infectious than PERV-A. To determine the molecular basis for the increased infectivity of PERV-A14/220, we have made a series of vector constructs. The primary determinant for the enhanced replicative potential of the recombinant virus appeared to be the env gene. Using a series of chimeric env genes, we could identify two determinants of high infectivity; one was an isoleucine to valine substitution at position 140 between variable regions A and B, and the other lies within the proline rich region. Taken together, these results show that the novel juxtaposition of env gene sequences enhanced the infectivity of PERV-A14/220 for human cells, perhaps by stabilization of the envelope glycoprotein or increased receptor binding.
Pig to human xenotransplantation is a potential solution to the shortage of human organs available for allotransplantation (19). However, the theoretical risk of a zoonotic, cross-species transmission of an infectious pathogen remains a concern (27, 36). Whereas exogenous pathogens can be removed by specific-pathogen-free breeding, these techniques cannot exclude inherited agents such as endogenous retroviruses that are passed down via the germ line (6, 34, 35).
PCR amplification studies with redundant primers directed to a conserved region of the pol gene have identified seven classes of endogenous provirus in pig (9, 14, 20). However, only one class of sequence, the so-called porcine endogenous retroviruses (PERVs), encodes infectious viruses. The PERVs have been divided into three subclasses, PERV-A, PERV-B, and PERV-C, by virtue of their host range properties (31). PERV-A and PERV-B have been shown to infect both human and pig cells, whereas PERV-C is more restricted in host range and can only infect pig cells (17, 21, 31). Most porcine endogenous retroviruses studied to date grow poorly if at all (10, 15, 39).
Partially inbred herds of miniature swine represent one potential source of organs and cells for xenotransplantation (7, 24). Extensive in vitro PERV transmission assays on these animals have identified certain animals that either do or do not transmit human-tropic replication-competent (HTRC) PERVs in vitro (18, 22, 40). When the HTRC viruses produced from transmitting miniature swine were analyzed, they were all found to be recombinants between PERV-A and PERV-C (18). The env gene of all of the recombinant viruses contained the PERV-A env receptor-binding domain (RBD) on a PERV-C background (10, 18, 39).
Direct analyses of the genomic DNA from miniature swine as well as screening of genomic libraries have found no evidence of inherited recombinations between PERV-A and PERV-C (PERV-A/C recombinants) and, therefore, no genomic HTRC PERVs (25). At the same time no functional PERV-A envelopes were identified in DNA from either the transmitting or nontransmitting miniature swine (25). Analysis of RNA from nonstimulated porcine lymphocytes found evidence of PERV-A/C recombinants, thereby excluding the possibility that PERV-A/C recombinants are generated only during the course of the in vitro transmission assay (40). Further, a correlation between the ability to produce A/C recombinants and the level of PERV-C replication was noted (40). Together, these observations suggest that replicating PERV-C recombines with the envelope sequence of endogenous PERV-A in pig cells, allowing the infection of human cells (40).
We have now compared the properties of one such recombinant virus, PERV-A14/220 (1, 10) (Fig. 1), to replication-competent PERV-A and PERV-B produced by PK15 cells (PERV-A PK and PERV-B PK, respectively). We have identified specific genetic determinants that specify high levels of infectivity in the recombinant virus. These data shed light on the increased infectivity of some of the PERV-A/C recombinants and the potential that these high-titer HTRC PERVs may be produced de novo in xenograft recipients after transplantation.
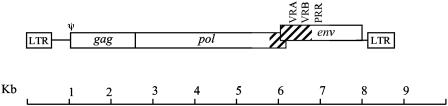
FIG. 1.
Structure of the PERV-A14/220 genome. PERV-A14/220 is the product of a recombination between PERV-A (striped) and PERV-C (open) that retains the tropism of PERV-A. The PERV-A-derived sequence consists of a maximum of 913 bp corresponding to nucleotides 5374 to 6280 in the PERV-A14/220 sequence. This region encompasses the 3′ end of pol as well as variable regions A and B (VRA and VRB, respectively) but not the PRR of env. Approximate locations of variable regions A and B, the PRR, and the packaging signal (ψ) are indicated.
MATERIALS AND METHODS
Cells and viruses.
All cell lines used have been previously described (32, 33). All were cultivated in Dulbecco's modified Eagle medium containing 10% fetal calf serum and antibiotics. Plasmids encoding full-length, replication-competent amphotropic murine leukemia virus (MLV) 4070A, PERV-A (clone Ap60), PERV-B (clone Bp17), and PERV-A14/220 (clone A14/220) proviruses have been described elsewhere (1, 2, 5). Virus was recovered by CaPO4-mediated transfection of 293T cells followed by repeated passage. These cell lines were subsequently transduced with a retroviral vector, p5G2, encoding enhanced green fluorescent protein (eGFP), and neomycin resistance genes (M. Yap, unpublished data) at a high multiplicity of infection. Virus produced from the chronically infected cell lines could be titrated by enumeration of G418-resistant colonies.
Viral vectors.
Two or three plasmids, providing genome vector, Gag-Pol, and Env functions, were introduced into 293T cells by CaPO4-mediated transient transfection (26). At 24 h after transfection, cells were grown for 8 to 10 h with 10 mM sodium butyrate to stimulate cytomegalovirus promoter-driven expression. At 48 h after transfection, the virus-containing supernatants were harvested and passed through a 0.45-μm-pore-size filter (Millipore).
Vector genomes included p5G2; pHIT111, a LacZ vector (26), and PERV-Ap60, PERV-Bp17, and PERV-A14/220 vectors encoding Gag-Pol and eGFP. The PERV plasmids were derived from the full-length plasmids described above by the following methods: (i) introducing a SalI site just downstream of pol by using the primers IH1 and IH2 (PERV-Ap60 and PERV-A14/220) or IH3 and IH4 (PERV-Bp17) by QuikChange site-directed mutagenesis (Stratagene), (ii) creating a second SalI site just upstream of the end of env with primers IH5 and IH6, (iii) deletion of the env gene by SalI digestion and religation, and (iv) introduction of a novel SalI fragment derived from pIRES2eGFP (Clontech) carrying an internal ribosome entry site (IRES) plus eGFP. Primers used are listed in Table 1; mutagenesis reactions were done with 15 ng of PERV DNA with 18 cycles of a 30-min extension time at 68°C. The MLV Gag-Pol expression plasmid (pHIT60) encoding NB-tropic capsid (CA) from Moloney MLV and Env expression constructs for the vesicular stomatitis virus (VSV) G protein (pczVSV-G) and amphotropic MLV Env (pHIT456) have been described before (3, 4, 26).
TABLE 1.
Primers used in this study
| Primer | Sequence |
|---|---|
| IH1 | TAACTCCTCAAGTTAATGGTCGACGCCTTGTGGACAGTCCGAAC |
| IH2 | GTTCGGACTGTCCACAAGGCGTCGACCATTAACTTGAGGAGTTA |
| IH3 | AACTCCCCAGGCCAGTAGTCGACGCCTTATAGACAGCTCG |
| IH4 | CGAGCTGTCTATAAGGCGTCGACTACTGGCCTGGGGAGTT |
| IH5 | GCAGTCCAGATCATGGTACGTCGACAACAGTACCAARGCC |
| IH6 | GGCT/CTTGGTACTGTTGTCGACGTACCATGATCTGGACTGC |
| IH7 | TCTAGACCACCATGCATCCCACGT |
| IH8 | TATTTTCCTGTTTTCCTTTTTCAG |
| IH9 | ATCGATAAGCTTGATATCGAATTC |
| IH10 | CTGAAAAAGGAAAACAGGAAAATA |
| IH11 | CCTCATCCAGGGAGCTTTTCAAGC |
| IH12 | GCTTGAAAAGCTCCCTGGATGAGG |
| IH13 | GAGCCCATATCCCTGACACTAGC |
| IH14 | GCTAGTGTCAGGGATATGGGCTC |
| IH15 | GGTAAACGCCTTGTGAACAGTCCGAACTCCC |
| IH16 | GGGAGTTCGGACTGTTCACAAGGCGTTTACC |
| IH17 | GCAAGCAATGGAGCTGCGTAACTTCTAATGATGGG |
| IH18 | CCCATCATTAGAAGTTACGCAGCTCCATTGCTTGC |
PERV Env expression constructs FBPERV-A and FBPERV-B have been described previously (31). The env gene of A14/220 was subcloned in the same expression plasmid (B. Bartosch, unpublished data). These constructs were modified as follows. Chimeric env constructs were produced by a joining PCR on two previously produced PCR products. Each product was produced by using either the 5′ or 3′ flanking primer and a primer at the recombination point. Amino acids 211 to 216 are the region in the PERV-A14/220 Env where PERV-A and -C recombined. To produce the chimeric PERV-A14/220 envelopes, PCR product 1, corresponding to amino acids 1 to 216, was synthesized by using the 5′ flanking primer IH7 and a reverse primer at the recombination site IH8. PCR product 2, corresponding to amino acids 211 to 653, was produced by using the 3′ flanking primer IH9 and a forward primer at the recombination site IH10. The 20-bp region of overlap enabled the two products to be joined in a third PCR. The RBD and surface and transmembrane (SU/TM) chimeras were produced by using the 5′, IH7, and 3′, IH9, flanking primers with IH11 and IH12 (RBD) or IH13 and IH14 (SU/TM). Site-directed mutagenesis was used to mutate positions 48 and 140 in the chimeric envelopes. The D48N mutation utilized the primers IH15 and IH16; I140V was synthesized by using the primers IH17 and IH18. Primers were designed with the same flanking sequence to produce the reciprocal mutations, N48D and V140I. For the PRR constructs the forward primer for the A14/220 recombination site, IH10, and the reverse primer for the RBD, IH12, were used to amplify by PCR the ≅300-bp section by using pfuTurbo (Stratagene). This section was then used as a site-directed mutagenesis primer. A total of 10 ng of the PERV-A and PERV-A14/220 Env constructs and approximately 750 ng of the PCR product were mutated in an 18-cycle PCR with a 12-min extension time. All changes introduced in env plasmids were confirmed by DNA sequencing.
Virus titration.
The titer of viral preparations was determined by three means (1): fluorescence-activated cell sorting (FACS), (2) antibiotic staining, or (3) LacZ staining.
(i) FACS.
A total of 4 × 104 to 1.8 × 105 target cells were plated per 12-well dish. Sixteen hours later, cells were transduced with virus in the absence of polybrene. At 24 to 36 h posttransduction, cells were harvested for FACS analysis. All FACS experiments were performed by using a FACS Calibur (Becton Dickinson). The data were analyzed with the FCSPress 1.3c software. Titers are given as infectious units per milliliter (IU/ml), calculated by multiplication of the number of cells infected with the dilution factor.
(ii) Antibiotic selection.
To determine the titer of retroviral vectors carrying neomycin resistance, target cells were plated at a density of 2 × 104 to 1 × 105 cells in 6-well plates. Sixteen hours later the cells were transduced with serial dilutions of virus containing 8 μg of polybrene per ml. At 24 to 48 h posttransduction, antibiotic selection was commenced with medium containing G418 at a final concentration of 1 mg/ml. Antibiotic-containing medium was replaced every 2 days for 10 to 15 days until colonies had formed. Colonies were then washed with phosphate-buffered saline (PBS) and fixed and stained by using a solution of 0.3% (wt/vol) crystal violet and 70% (vol/vol) methanol. Colonies were then counted and titers were calculated as infectious units per milliliter.
(iii) LacZ staining.
A total of 4 × 104 to 1.8 × 105 cells were plated in 12-well plates; 16 h later the cells were transduced with a serial dilution of virus containing 8 μg of polybrene per ml. At 48 h posttransduction cells were washed in PBS and fixed at room temperature for 5 min (2% formaldehyde, 0.2% glutaraldehyde in PBS). Cells were then left overnight at 37°C in staining solution (4 mM potassium ferricyanide, 4 mM potassium ferrocyanide, 2 mM magnesium chloride, and 0.4 mg of X-Gal in dimethylformamide per ml, made up in PBS). The following day, stained colonies were counted and titers were calculated as infectious units per milliliter.
RT assay.
Reverse transcriptase (RT) assays were performed by using a modified version of the protocol described by S. Goff et al. (11). Viral supernatants were filtered through 0.45-μm-pore-size filters (Millipore) and spun down at 55,000 rpm for 30 min in a TLA 100.1 rotor (Beckman). The supernatant was removed, and pellets were resuspended in 10 μl of 1× RT buffer [50 mM Tris-hydrochloride (pH 8.3), 0.05% (vol/vol), NP-40, 5 μg of oligodeoxythymidylic acid per ml, 10 μg of poly(A) per ml, 20 mM dithiothreitol, 0.6 mM manganese chloride, 60 mM sodium chloride] containing 37 Bq of [32P]dTTP (Amersham). Samples were incubated for 2 h at 37°C before 5 μl was spotted onto DEAE paper (DE-81; Whatman) in duplicate and air dried. The paper was washed twice with 0.6 M sodium chloride-0.06 M sodium citrate for 15 min, followed by 95% ethanol for 15 min, and then air dried. The incorporation of 32P was measured by scintillation counting.
RESULTS
Replication-competent PERVs.
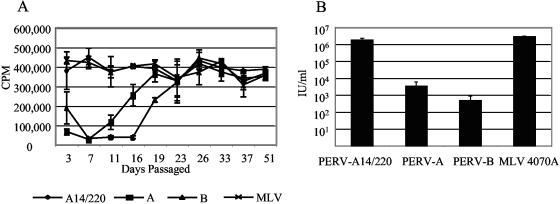
To enable comparison of the three PERVs (PERV-A14/220, PERV-A, and PERV-B) and to confirm previous reports on their relative infectivity, stable producer lines were established. PERV proviral constructs (1, 2) and the amphotropic MLV 4070A control (5) were transfected into 293T cells and passaged for 50 days. Virus-containing supernatants were harvested on each passage. The level of pelletable RT in each sample was measured to follow virus release (Fig. 2A). PERV-A14/220 and amphotropic MLV quickly became established, with the level of RT remaining relatively constant from the initial transfection. PERV-A showed a lag phase of 10 to 15 days before the virus could be detected in the culture, with PERV-B following one passage behind. Once infected, the PERV-A and -B cell lines released similar levels of RT compared to the PERV-A14/220 and MLV lines. Similar levels of Gag protein were expressed in these cells (data not shown). One possible explanation for these data is that PERV-A and PERV-B have reduced particle infectivity compared to PERV-A14/220 and MLV.
FIG. 2.
Chronically infected PERV cell lines. (A) Level (cpm) of RT in the supernatant of 293T cells transfected with the following proviral constructs and passaged at 3- to 5-day intervals for approximately 50 days: PERV-A14/220 (A14/220) (1), PERV-Ap60 (A) and PERV-Bp17 (B) (2), and MLV 4070A (5). (B) Infectious titers of chronically infected cell lines, transduced with the p5G2 vector at a multiplicity of infection of >7 at day 40 of culture. Virus-containing supernatant was harvested 2 days later and titrated on human 293 cells. Levels of virus packaging the neomycin resistance gene were measured by G418 selection and represented as infectious per milliliter.
To examine this question, cells were transduced with a retroviral vector, p5G2, encoding selectable marker neomycin under conditions where essentially all cells should become infected (multiplicity of infection of >7). Particles released from these cells can package the transduced vector and can be titrated on 293 cells by G418 selection (Fig. 2B). PERV-A14/220-producing cells release virus encoding G418 resistance at a level comparable to that of the amphotropic MLV control, both reaching approximately 106 IU/ml. Although releasing as much pelletable, i.e., particle-associated, RT as PERV-A14/220 or amphotropic virus, the PERV-A and -B show a much lower level of infectivity, releasing approximately 5 × 103 and 5 × 102 IU/ml, respectively. Although PERV-A14/220 and PERV-A use the same receptor (10), their titers differ by approximately 500-fold. Two possible reasons can be suggested for this increase in infectivity. The recombination event in the env gene is at the edge of the RBD, and such alterations have been shown to increase receptor binding by other gammaretroviruses (30). Alternatively, PERV-C, which contributes the majority of the PERV-A14/220 sequence, contains cis- or trans-acting determinants that render it more infectious, perhaps by increasing Env expression.
Mapping of PERV high-titer determinants.
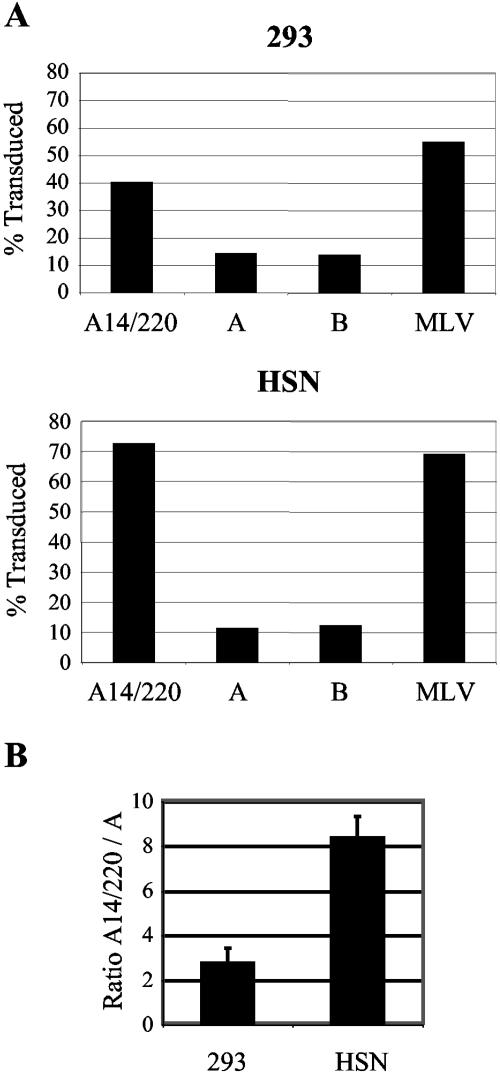
To investigate the reasons for the difference between PERV-A and PERV-A14/220, the Gag-Pol and Env constituent parts were studied independently. Removal of the env gene from the proviral constructs enabled the comparison of the Gag-Pol and other cis-acting sequences. Two SalI sites were engineered into the env gene of three PERVs, the first just after the pol gene and the second just before the stop codon in env. The majority of the env sequence was then removed and replaced with IRES eGFP. Use of these constructs, pseudotyped with VSV-G, enables the direct comparison of PERV-A and PERV-A14/220 Gag-Pol and long terminal repeat (LTR) functions.
Such viral particles were transduced onto human 293 and rat HSN cells; the percentage of infected cells was determined 2 days later by FACS. Figure 3A shows a representative experiment, where PERV-A and -B particles share a similar titer of approximately 8 × 104 IU/ml on HSN cells and 4 × 105 IU/ml on 293 cells. PERV-A14/220 has a titer of 5 × 105 IU/ml on HSN cells and 1 × 106 IU/ml on 293 cells. The PERV-A14/220 particle had a level of infectivity similar to that of MLV. Figure 3B shows the ratio of infected cells between PERV-A14/220 and PERV-A particles, summarizing data from three independent experiments. Thus, the 500-fold increase in the titer of PERV-A14/220 over PERV-A or -B is reduced to a three- to eightfold difference in the absence of the envelope gene.
FIG. 3.
Comparison of PERV Gag-Pols. (A) VSV-G pseudotyped particles containing the Gag-Pol cores from PERV-A14/220, PERV-A, PERV-B, or amphotropic MLV were titrated onto human 293 or rat HSN cells. The percentage of transduction was measured by FACS analysis. A representative experiment is shown reflecting transduction with 135 μl of each virus. (B) Ratio of the level of transduction of PERV-A14/220 Gag-Pol to PERV-A Gag-Pol. Results are the means of three independent experiments for 293 and HSN cells.
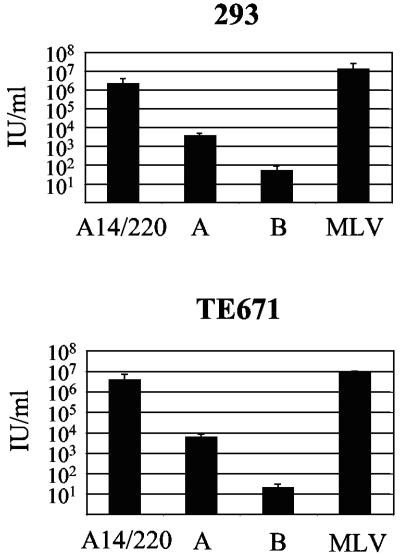
PERV envelopes.
To assess the effects of the env recombination on the titer of PERV-A14/220, the PERV Env proteins were compared. Pseudotyped virions containing MLV cores carrying the gene for neomycin resistance were titrated on human 293 and TE671 cells. Comparison of the levels of CA released from transient transfections showed no significant difference in particle release (data not shown). Results for PERV-A and -B Env proteins reflect those described by Takeuchi et al. (31) and mimic the results seen with the chronically infected cell lines (Fig. 2B). PERV-A pseudotyped particles yielded an infectious titer of approximately 103 IU/ml, while PERV-B pseudotypes were at least 10-fold less infectious. Both PERV-A14/220 and the amphotropic MLV control Env yielded a titer of greater than 106 IU/ml (Fig. 4).
FIG. 4.
Comparison of PERV Env proteins. Moloney MLV cores were pseudotyped with PERV and amphotropic MLV envelopes. Viruses were titrated on 293 and TE671 cells by G418 selection.
PERV-A14/220 and PERV-A have been shown to use the same receptor (10); however, PERV-A14/220 Env is approximately 500-fold more infectious than the PERV-A pseudotype. There are two possibilities that could account for the increase in infectivity of the A14/220 Env. The TM section of the A14/220 Env is derived from PERV-C. This TM region may enable increased Env incorporation or allow for increased budding of the virus particle. Alternatively, the recombination event, on the edge of the RBD, may have generated an Env protein with enhanced infectivity. This phenomenon has previously been described with the feline leukemia virus subgroup B (FeLV-B) Env protein (30). These data suggest that the Env recombination event accounts for the major part of the difference in infectivity of PERV-A14/220 compared to PERV-A. PERV-A and PERV-B both appear to possess relatively inefficient Env proteins, at least as assessed on human cells.
Chimeric PERV envelopes.
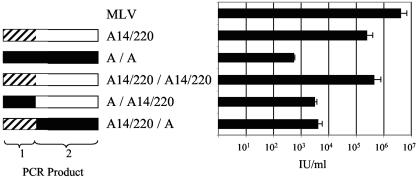
To investigate the increased titer of the A14/220 Env, we made a number of chimeric env genes. A 20-bp region at the site of the recombination in A14/220 was conserved in PERV-A. This enabled the production of two PCR products for each env gene that overlapped at this 20-bp site (Fig. 5, products 1 and 2). These two halves were then joined to produce an env that mimicked the recombination in A14/220 (Fig. 5, left). MLV cores were pseudotyped with these chimeric envelopes and titrated on 293 cells (Fig. 5, right). The (A14/220)/(A14/220) and A/A constructs are controls in which the original env genes were reengineered from the two PCR products. Both (A14/220)/(A14/220) and A/A gave levels of infection corresponding to their respective starting plasmids. The (A)/(A14/220) construct should contain the same Env sequence as A14/220 and, therefore, a high titer would be expected. However, the titer seen with this construct, 3 × 103 IU/ml, although above that observed with A/A, was at least 100-fold lower than that with A14/220. Further, introduction of PERV-A-derived fragment 2 into the A14/220 Env also resulted in a 100-fold reduction in titer. These results imply that sequences both upstream and downstream of the recombination point are important for enhanced Env function in PERV-A14/220.
FIG. 5.
Structure and function of chimeric Env proteins. Envelope constructs that mimic the recombination event that occurred in PERV-A14/220 were tested for biological activity by pseudotyping an MLV-based LacZ vector and by titration on 293T cells. Open boxes, PERV-A14/220-derived sequences of PERV-C origin; hashed boxes, PERV-A14/220-derived sequences of PERV-A origin; solid boxes, PERV-A-derived sequences. Constructs are named to identify their plasmids of origin; thus, (A14/220)/(A) env consists of 5′ sequences from a PERV-A14/220-derived sequence and 3′ sequences from PERV-A.
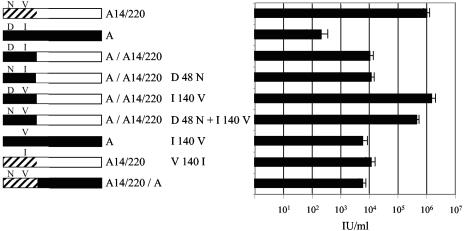
Although amino acids 1 to 216 of the A14/220 clone are derived from a member of the PERV-A class of endogenous elements (Fig. 5, product 1), sequence comparisons revealed two amino acid differences in this region compared to the prototypic PERV-A. In the A14/220 Env, D at position 48 has been replaced with N, and I at position 140 has been replaced by V. To examine the role played by N48D and V140I, the residues were independently mutated (Fig. 6, left). Pseudotyped MLV particles were titrated onto 293T cells (Fig. 6, right). The (A)/(A14/220) construct gave a titer of 104 IU/ml, which was 100-fold lower than that of the A14/220 Env. Mutation of D48N in (A)/(A14/220) had no effect on the infectivity of the Env, whereas the I140V mutation increased the infectivity of (A)/(A14/220) to that of A14/220. The double mutant Env also reached the same titer as the A14/220 Env. Introduction of the I140V change into PERV-A gave a 10-fold increase in infectivity in the PERV-A envelope; the reciprocal V140I change in PERV-A14/220 led to a 100-fold reduction in titer. Taken together, these results show that residue 140 plays a key role in determining the infectivity of PERV-A14/220. However, the extent of this effect is modulated by the nature of sequences on the C-terminal side of the recombination breakpoint.
FIG. 6.
Effect of site-directed mutagenesis on residues 48 (D48N) and 140 (I140V) of the chimeric envelopes of PERV-A14/220 and PERV-A. Env function was assayed by LacZ titration. Open boxes, PERV-A14/220-derived sequences of PERV-C origin; hashed boxes, PERV-A14/220-derived sequences of PERV-A origin; solid boxes, PERV-A-derived sequences. Constructs are named as described in the legend of Fig. 5.
C-terminal region in PERV-A14/220.
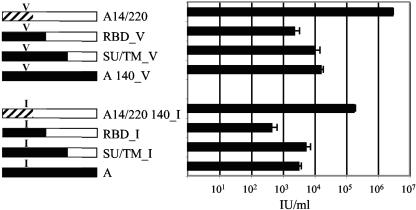
To identify the region in the C terminus of A14/220 Env that contributes to increased infectivity, additional chimeric envelopes were produced. A chimera joined at the boundary of SU/TM and a second where the recombination was modeled just after the proline-rich region (PRR) (RBD chimera) were produced (Fig. 7, left). Envelopes with both V and I at position 140 were tested. Pseudotyped particles were titrated on 293 cells (Fig. 7, right). Regardless of the residue at position 140, the SU/TM chimeric envelopes showed no higher infectivity than the PERV-A envelopes. The RBD chimeric envelopes were actually less infectious than those of PERV-A. These findings suggest that the PRR plays an important role in the increased infectivity of PERV-A14/220.
FIG. 7.
Analysis of the role of PERV-C-derived sequences in the C-terminal portion of the PERV-A14/220 envelope. Chimeric envelopes were engineered to mimic a recombination event between PERV-A and PERV-C that occurred at the SU/TM boundary or just after the RBD and were tested for function as described in the legend of Fig. 5. The presence of a valine or isoleucine at position 140 is indicated. Open boxes, PERV-A14/220-derived sequences of PERV-C origin; hashed boxes, PERV-A14/220-derived sequences of PERV-A origin; solid boxes, PERV-A-derived sequences. Constructs are named as described in the legend of Fig. 5.
PRR chimeras.
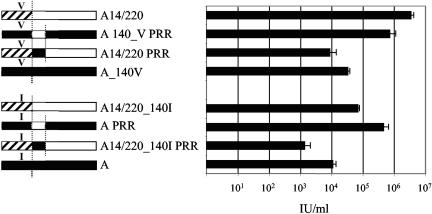
To test the role of the PRR, the 300-bp section between the site of recombination in the A14/220 and RBD envelope was introduced into a variety of backgrounds (Fig. 8, left). Pseudotyped particles were titrated onto 293 cells (Fig. 8, right). The addition of the PRR from PERV-A into the A14/220 envelope reduced its infectivity to below that of PERV-A I140V, whereas the PRR from A14/220 mutated into PERV-A I140V increased its titer of infection to approximately 106 IU/ml. Thus, the I-to-V substitution plus the PRR from A14/220 have increased the infectivity of the PERV-A envelope almost to that of A14/220. Interestingly, the A14/220 PRR introduced into the PERV-A envelope also gave a large increase in titer. This envelope construct was nearly 10-fold more infectious than the A14/220 V140I construct and nearly as infectious as the A I140V plus A14/220 PRR construct. It thus appears that the sequence responsible for the increase in infectivity of PERV-A14/220 maps mainly to env, with the change at residue 140 and the PPR substitution making equal contributions to the enhanced infectivity.
FIG. 8.
Analysis of the role of the PRR. The PCR product between the recombination position in PERV-A14/220 to the end of the PRR was used to produce the PERV-A14/220 and PERV-A envelopes. The 300-bp product was then used as a site-directed mutagenesis primer to produce envelopes with the alternative PRR (indicated by the underscore). The presence of a valine or isoleucine at position 140 is indicated. The titers of the PRR envelopes were determined by LacZ staining on 293 cells. Open boxes, PERV-A14/220-derived sequences of PERV-C origin; hashed boxes, PERV-A14/220-derived sequences of PERV-A origin; solid boxes, PERV-A-derived sequences.
DISCUSSION
Inbred herds of miniature swine are potential organ sources for xenotransplantation. Extensive in vitro coculture transmission assays conducted on miniature swine of various haplotypes have identified animals that do not produce HTRC PERV-A or -B. However, PERV-A/C recombinant viruses were consistently identified in a subset of these animals (18, 22). One such PERV-A/C recombinant, PERV-A 14/220 and its molecular clone (A14/220), show considerably higher infectivity on human cells than PERV-A PK (1, 10). In this report the recombination event that occurred in PERV-A14/220 has been studied so that regions responsible for the increased infectivity could be identified. Chronically infected cell lines were produced from molecular PERV clones, Ap60 and A14/220. Although the amount of released RT reached the same levels, the infectivity of PERV-A14/220, as measured by transforming target cells with neomycin resistance, was at least 500-fold higher than that of PERV-A. To investigate where these high-titer determining regions lie, the Env gene was deleted from the molecular clones, and viral functions were assayed in a vector system. The Gag-Pol LTR from PERV-A14/220 is derived predominantly from the PERV-C sequence but showed only a three- to eightfold increase in infectivity compared to PERV-A. This difference cannot be attributed to differences in the LTR or other cis-acting sequences (1), implying that it lies within gag or pol coding sequences. By contrast, comparisons of the envelope genes found that PERV-A14/220 Env encodes a substantially increased infectivity compared to PERV-A Env. Studies with chimeric envelopes identified the PRR and the I140V substitution as causes for this increase. Others have reported increases in titer following duplications of potential transcription factor binding sites located within the proviral LTRs (8, 37). It remains to be seen whether recombining Env and LTR changes could generate a virus with a still higher titer.
One other PERV-A/C recombinant virus isolated from miniature swine, PERV-NIH (23, 38, 39), has been studied in some detail. This, too, has an enhanced infectivity compared to PERV-A but apparently not to the same extent. This can be explained by the position of the 3′ recombination event. PERV-NIH contains the I140A substitution but retains the PRR of PERV-A; thus, the enhanced infectivity can be attributed only to the RBD change.
Position 140 is in the RBD of the SU; however, structural models based on MLV predict that this residue lies on the opposite side of the envelope from the regions directly involved in receptor binding, variable regions A and B (R. Russell, personal communication). Therefore, a direct role for this relatively conserved mutation in altering receptor binding seems unlikely. Interestingly, in comparisons of residue 140 from replication-competent molecular clones, isoleucine is found in PK15 PERV-A (clone 42; accession no. AJ133817), PK15 PERV-A (clone 58; accession no. AJ293656), Bac-PERV-A (clone 463H12; accession no. AF435966), PERV-A (clone Ap60; accession no. AY099323), and FBSALF-A (accession no. Y12238), whereas Valine is found in PERV-A14/220 (accession no. AF417228), PERV-NIH (clone 1.15; accession no. AF130444), Bac-PERV-A (clone 130A12; accession no. AJ279056), and Bac-PERV-A (clone 151B10; accession no. AF435967). Therefore, both isoleucine and valine appear to be circulating in position 140 in the PERV-A population, with the valine resulting in approximately 10-fold higher infectivity.
The second region that was identified as increasing the infectivity of PERV-A14/220 was a section containing the PRR. Comparison of the sequence in the region of the PRR in PERV-A PK and PERV-A14/220 shows a small difference, with 10-amino-acid variations before the PRR, then 16 changes within the PRR, and an additional 5 amino acids in the PERV-A variable PRR (Fig. 9). The PRR has been described as a flexible hinge that allows the RBD of the envelope to flex during receptor binding and infection. It is predicted that the N-terminal constant region of the PRR is essential for enabling the conformational change in Env required for cell fusion. Consistent with this, mutations in this region of the PRR have been seen to increase SU shedding (12, 16, 41). The larger variable section of the PRR has a role in allowing flexibility during receptor binding. Therefore, mutations in this region have been shown to affect a virus's ability to infect cells that are expressing low levels of receptor (16). An alternative, but not mutually exclusive, theory for the role of the PRR is the accommodation of proximal protein domains in the trimeric envelope structure. According to this model, the variability in the PRR enables the accommodation of the slightly varying structures of the different RBDs (16). Indeed, changes in the PRR domain may compensate for changes at residue 140. Interestingly, one possible consequence of the substitution of isoleucine for valine at position 140 is to cause secondary movement in the envelope in the region predicted to be the trimerization interface.
FIG. 9.
Comparison of the PRR region of PERV-A and PERV-A14/220. ClustalW alignment of the section containing the PRR mutated as described in the legend of Fig. 8. Predicted conserved and variable regions of the PRR are shown in the first and second boxes, respectively. Symbols: asterisk, identical; colon, conserved; period, semiconserved.
Like PERV-A, PERV-B also shows a substantially lower titer than PERV-A14/220. Again, a relatively inefficient env gene appears to be responsible. No PERV-B/C recombinants have been recovered from miniature swine, and our initial attempts to model changes analogous to those seen with PERV-A on a PERV-B env have not been successful (data not shown). Therefore, these effects appear specific for PERV-A.
Two functional receptors for PERV-A have been cloned from human cells, HuPAR1 and HuPAR2 (10). PERV-A 14/220 is approximately 20-fold more infectious than PERV-A PK on HuPAR2, whereas on HuPAR1 PERV-A14/220 is about 1,000-fold more infectious. It will thus be of considerable interest to see whether the use of cells transfected with these receptors will allow the functional dissection of the roles of the RBD and PPR in virus infection. We note that the modified polytropic class of MLV contains a nine-amino-acid deletion compared to polytropic isolates; this is apparently associated with a significant decrease in the ability to interact with its receptor on mink but not Mus dunni cells (28).
Although a variety of PERV recombinants have been identified in the pig (1, 14), recent A/C recombinants appear absent from the genome of miniature swine (25, 40). Rather, the HTRC PERV-A/C recombinants that have been isolated appear to result from a series of de novo recombination events (40) between replication-competent PERV-C and nonfunctional endogenous PERV-A proviruses. Wood and colleagues used two analogies to describe the derivation of HTRC PERV-A. The first example considered was AKR mice in which recombination between the expressed form of a replication-competent ecotropic virus with a defective endogenous polytropic provirus results in the formation of a virus with the ability to replicate on a variety of cell types (13). De novo formation of recombinant virus rapidly occurs in essentially every mouse of the AKR strain (29); however, the host range of the novel virus is thought to reflect the properties of the endogenous polytropic RBD (28, 29). The second example involves recombination between an exogenous ecotropic virus, FeLV, with endogenous FeLV-like envelope sequence to produce the FeLV-B virus. As with PERV-A/C recombinants, the recombination site varies along the env gene. In this case the site of recombination apparently affects receptor usage by the recombinant. Studies with chimeric env genes have shown that sequences outside the RBD, near the C terminus of SU, permit usage of human Pit2 as receptor in addition to the Pit1 specified by the RBD (30). It will be interesting to compare receptor usage by PERV-A PK, PERV-A14/220, and the different chimeras generated during this study.
The single recombination event leading to the formation of PERV-A14/220 has not only generated a virus capable of replication on human cells but has also increased its infectivity on pig cells (10). Apparently, recombination has brought together multiple independent positive factors to produce a virus with increased infectivity for a wide range of cells. Clearly, such viruses would be a complication for xenotransplantation. However, the solution to this problem appears reasonably straightforward. It is likely that recombination is driven by replication of PERV-C; by selecting animals lacking PERV-C expression, one will minimize risks both of the germ line reinsertion of recombinant viruses as well as de novo generation of such viruses prior to or following transplantation.
Acknowledgments
This work was supported by the United Kingdom Medical Research Council.
We thank Robin Weiss, Clive Patience, Rupert Russell, and Melvyn Yap for helpful discussions.
REFERENCES
- 1.Bartosch, B., D. Stefanidis, R. Myers, R. Weiss, C. Patience, and Y. Takeuchi. 2004. Evidence and consequence of porcine endogenous retrovirus recombination. J. Virol. 78:13880-13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartosch, B., R. A. Weiss, and Y. Takeuchi. 2002. PCR-based cloning and immunocytological titration of infectious porcine endogenous retrovirus subgroup A and B. J. Gen. Virol. 83:2231-2240. [DOI] [PubMed] [Google Scholar]
- 3.Bock, M., K. N. Bishop, G. Towers, and J. P. Stoye. 2000. Use of a transient assay for studying the genetic determinants of Fv1 restriction. J. Virol. 74:7422-7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cannon, P. M., N. Kim, S. M. Kingsman, and A. J. Kingsman. 1996. Murine leukemia virus-based Tat-inducible long terminal repeat replacement vectors: a new system for anti-human immunodeficiency virus gene therapy. J. Virol. 70:8234-8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chattopadhyay, S. K., A. I. Oliff, D. L. Linemeyer, M. R. Lander, and D. R. Lowy. 1981. Genomes of murine leukemia viruses isolated from wild mice. J. Virol. 39:777-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark, D. A., J. F. Fryer, A. W. Tucker, P. D. McArdle, A. E. Hughes, V. C. Emery, and P. D. Griffiths. 2003. Porcine cytomegalovirus in pigs being bred for xenograft organs: progress towards control. Xenotransplantation 10:142-148. [DOI] [PubMed] [Google Scholar]
- 7.Cooper, D. K., B. Gollackner, and D. H. Sachs. 2002. Will the pig solve the transplantation backlog? Annu. Rev. Med. 53:133-147. [DOI] [PubMed] [Google Scholar]
- 8.Denner, J., V. Specke, U. Thiesen, A. Karlas, and R. Kurth. 2003. Genetic alterations of the long terminal repeat of an ecotropic porcine endogenous retrovirus during passage in human cells. Virology 314:125-133. [DOI] [PubMed] [Google Scholar]
- 9.Ericsson, T., B. Oldmixon, J. Blomberg, M. Rosa, C. Patience, and G. Andersson. 2001. Identification of novel porcine endogenous betaretrovirus sequences in miniature swine. J. Virol. 75:2765-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ericsson, T. A., Y. Takeuchi, C. Templin, G. Quinn, S. F. Farhadian, J. C. Wood, B. A. Oldmixon, K. M. Suling, J. K. Ishii, Y. Kitagawa, T. Miyazawa, D. R. Salomon, R. A. Weiss, and C. Patience. 2003. Identification of receptors for pig endogenous retrovirus. Proc. Natl. Acad. Sci. USA 100:6759-6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goff, S., P. Traktman, and D. Baltimore. 1981. Isolation and properties of Moloney murine leukemia virus mutants: use of a rapid assay for release of virion reverse transcriptase. J. Virol. 38:239-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray, K. D., and M. J. Roth. 1993. Mutational analysis of the envelope gene of Moloney murine leukemia virus. J. Virol. 67:3489-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartley, J. W., N. K. Wolford, L. J. Old, and W. P. Rowe. 1977. A new class of murine leukemia virus associated with development of spontaneous lymphomas. Proc. Natl. Acad. Sci. USA 74:789-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klymiuk, N., M. Muller, G. Brem, and B. Aigner. 2002. Characterization of porcine endogenous retrovirus γ pro-pol nucleotide sequences. J. Virol. 76:11738-11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krach, U., N. Fischer, F. Czauderna, and R. R. Tonjes. 2001. Comparison of replication-competent molecular clones of porcine endogenous retrovirus class A and class B derived from pig and human cells. J. Virol. 75:5465-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lavillette, D., M. Maurice, C. Roche, S. J. Russell, M. Sitbon, and F. L. Cosset. 1998. A proline-rich motif downstream of the receptor binding domain modulates conformation and fusogenicity of murine retroviral envelopes. J. Virol. 72:9955-9965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Tissier, P., J. P. Stoye, Y. Takeuchi, C. Patience, and R. A. Weiss. 1997. Two sets of human-tropic pig retrovirus. Nature 389:681-682. [DOI] [PubMed] [Google Scholar]
- 18.Oldmixon, B. A., J. C. Wood, T. A. Ericsson, C. A. Wilson, M. E. White-Scharf, G. Andersson, J. L. Greenstein, H. J. Schuurman, and C. Patience. 2002. Porcine endogenous retrovirus transmission characteristics of an inbred herd of miniature swine. J. Virol. 76:3045-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patience, C., and J. P. Stoye. Infectious risk of clinical xenotransplantation. Curr. Opin. Organ Transplant., in press.
- 20.Patience, C., W. M. Switzer, Y. Takeuchi, D. J. Griffiths, M. E. Goward, W. Heneine, J. P. Stoye, and R. A. Weiss. 2001. Multiple groups of novel retroviral genomes in pigs and related species. J. Virol. 75:2771-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patience, C., Y. Takeuchi, and R. A. Weiss. 1997. Infection of human cells by an endogenous retrovirus of pigs. Nat. Med. 3:282-286. [DOI] [PubMed] [Google Scholar]
- 22.Quinn, G., J. Wood, K. Suling, S. Arn, D. H. Sachs, H. J. Schuurman, and C. Patience. 2004. Genotyping of porcine endogenous retroviruses from a family of miniature swine. J. Virol. 78:314-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ritzhaupt, A., L. J. Van Der Laan, D. R. Salomon, and C. A. Wilson. 2002. Porcine endogenous retrovirus infects but does not replicate in nonhuman primate primary cells and cell lines. J. Virol. 76:11312-11320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sachs, D. H., M. Sykes, S. C. Robson, and D. K. Cooper. 2001. Xenotransplantation. Adv. Immunol. 79:129-223. [DOI] [PubMed] [Google Scholar]
- 25.Scobie, L., S. Taylor, J. C. Wood, K. M. Suling, G. Quinn, S. Meikle, C. Patience, H. J. Schuurman, and D. E. Onions. 2004. Absence of replication-competent human-tropic porcine endogenous retroviruses in the germ line DNA of inbred miniature swine. J. Virol. 78:2502-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soneoka, Y., P. M. Cannon, E. E. Ramsdale, J. C. Griffiths, G. Romano, S. M. Kingsman, and A. J. Kingsman. 1995. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 23:628-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stoye, J. P., and J. M. Coffin. 1995. The dangers of xenotransplantation. Nat. Med. 1:1100. [DOI] [PubMed] [Google Scholar]
- 28.Stoye, J. P., and J. M. Coffin. 1987. The four classes of endogenous murine leukemia virus: structural relationships and potential for recombination. J. Virol. 61:2659-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoye, J. P., C. Moroni, and J. M. Coffin. 1991. Virological events leading to spontaneous AKR thymomas. J. Virol. 65:1273-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sugai, J., M. Eiden, M. M. Anderson, N. Van Hoeven, C. D. Meiering, and J. Overbaugh. 2001. Identification of envelope determinants of feline leukemia virus subgroup B that permit infection and gene transfer to cells expressing human Pit1 or Pit2. J. Virol. 75:6841-6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takeuchi, Y., C. Patience, S. Magre, R. A. Weiss, P. T. Banerjee, P. Le Tissier, and J. P. Stoye. 1998. Host range and interference studies of three classes of pig endogenous retrovirus. J. Virol. 72:9986-9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takeuchi, Y., G. Simpson, R. G. Vile, R. A. Weiss, and M. K. Collins. 1992. Retroviral pseudotypes produced by rescue of a Moloney murine leukemia virus vector by C-type, but not D-type, retroviruses. Virology 186:792-794. [DOI] [PubMed] [Google Scholar]
- 33.Thomson, D. M., S. Eccles, and P. Alexander. 1973. Antibodies and soluble tumour-specific antigens in blood and lymph of rats with chemically induced sarcomata. Br. J. Cancer. 28:6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tucker, A., C. Belcher, B. Moloo, J. Bell, T. Mazzulli, A. Humar, A. Hughes, P. McArdle, and A. Talbot. 2002. The production of transgenic pigs for potential use in clinical xenotransplantation: microbiological evaluation. Xenotransplantation 9:191-202. [DOI] [PubMed] [Google Scholar]
- 35.Tucker, A. W., F. McNeilly, B. Meehan, D. Galbraith, P. D. McArdle, G. Allan, and C. Patience. 2003. Methods for the exclusion of circoviruses and gammaherpesviruses from pigs. Xenotransplantation 10:343-348. [DOI] [PubMed] [Google Scholar]
- 36.Weiss, R. A., S. Magre, and Y. Takeuchi. 2000. Infection hazards of xenotransplantation. J. Infect. 40:21-25. [DOI] [PubMed] [Google Scholar]
- 37.Wilson, C. A., S. Laeeq, A. Ritzhaupt, W. Colon-Moran, and F. K. Yoshimura. 2003. Sequence analysis of porcine endogenous retrovirus long terminal repeats and identification of transcriptional regulatory regions. J. Virol. 77:142-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson, C. A., S. Wong, J. Muller, C. E. Davidson, T. M. Rose, and P. Burd. 1998. Type C retrovirus released from porcine primary peripheral blood mononuclear cells infects human cells. J. Virol. 72:3082-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson, C. A., S. Wong, M. VanBrocklin, and M. J. Federspiel. 2000. Extended analysis of the in vitro tropism of porcine endogenous retrovirus. J. Virol. 74:49-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wood, J. C., G. Quinn, K. M. Suling, B. A. Oldmixon, B. A. Van Tine, R. Cina, S. Arn, C. A. Huang, L. Scobie, D. E. Onions, D. H. Sachs, H. J. Schuurman, J. A. Fishman, and C. Patience. 2004. Identification of exogenous forms of human-tropic porcine endogenous retrovirus in miniature swine. J. Virol. 78:2494-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu, B. W., P. M. Cannon, E. M. Gordon, F. L. Hall, and W. F. Anderson. 1998. Characterization of the proline-rich region of murine leukemia virus envelope protein. J. Virol. 72:5383-5391. [DOI] [PMC free article] [PubMed] [Google Scholar]