Abstract
Latent membrane protein 1 (LMP1), the Epstein-Barr virus oncoprotein, activates NF-κB, phosphatidylinositol 3-kinase, mitogen-activated protein kinase, and c-Jun N-terminal kinase signaling. To determine global transcriptional changes induced by LMP1 in epithelial cells, genomic analysis of C33A cells stably expressing LMP1 was performed. Relatively few genes were induced by LMP1. Expression of two members of the Id (inhibitor of differentiation) family of proteins, Id1 and Id3, was induced in the presence of LMP1 and confirmed by mRNA and protein in C33A and Rat-1 cells. In Rat-1 foci transformed by LMP1, Id1 protein was also increased. Id proteins are known negative regulators of E-box proteins that positively regulate p16 and potentially other cyclin-dependent kinase inhibitors (cdki's). In LMP1-expressing Rat-1 cells, cdki p27 was specifically downregulated. Decreased p27 was correlated with increased levels of Cdk2 and increased levels of phosphorylated retinoblastoma protein. This study describes new properties of LMP1 that likely contribute to transformation and oncogenesis.
Epstein-Barr virus (EBV) is a ubiquitous human pathogen that is associated with several malignancies (16, 28). Latent membrane protein 1 (LMP1) is considered the EBV oncoprotein and is expressed in many of the cancers associated with EBV. LMP1 transforms rodent fibroblasts by conferring anchorage-independent growth and loss of contact inhibition (32). Fibroblasts expressing LMP1 form tumors in nude mice and can grow under reduced-serum conditions. LMP1 is also essential for EBV-mediated transformation of B lymphocytes (13).
LMP1 is an integral membrane protein with a short amino-terminal cytoplasmic tail, six membrane-spanning domains, and a cytoplasmic carboxy-terminal domain. LMP1 functions as a constitutively active tumor necrosis factor (TNF) receptor, as oligomerization of LMP1 molecules via interactions of the transmembrane domains brings the carboxyl-terminal domains in close proximity to induce ligand-independent signaling (6, 8, 9). The carboxyl-terminal domain contains two signaling domains, C-terminal activation regions (CTARs). CTAR1 binds TNF receptor-associated factors (TRAFs), and CTAR2 binds the TNF receptor-associated death domain protein (designated TRADD) that recruits other signaling molecules. Signaling from the C-terminal domain activates NF-κB and leads to activation of several important signaling pathways, including the mitogen-activated protein kinase (MAPK), c-Jun N-terminal kinase, and phosphatidylinositol 3-kinase (PI3K) pathways (6).
LMP1 alters the cellular environment by inducing the expression of a number of genes. Genomic analyses of EBV-infected and LMP1-expressing lymphocytes indicate that most of the genes that are induced during EBV infection are a result of LMP1 and NF-κB signaling (3, 4). LMP1 induces the expression of cell surface receptors, epidermal growth factor receptor (EGFR) (22), CD40, CD54, and CD95. LMP1 also induces antiapoptotic proteins such as A20 (8), Bcl-2, cIAP, and Bfl-1, as well as invasion and angiogenic factors such as vascular endothelial growth factor, cyclooxygenase-2, and matrix metalloproteinase 9 (24, 34).
To gain greater understanding into the mechanism of transformation of epithelial cells by LMP1, genomic analysis of C33A cells expressing LMP1 was performed. Two members of the inhibitor of DNA binding or inhibitor of differentiation (Id) family of proteins, Id1 and Id3, were upregulated by LMP1. Induction of Id1 and Id3 mRNA was confirmed by quantitative PCR and correlated with increased proteins levels. Increased Id1 protein was detected by immunofluorescence in Rat-1 foci induced by transformation with LMP1, and Id1 and Id3 protein levels were increased in Rat-1 LMP1 stable cell lines. The Id proteins are potent regulators of cellular differentiation and cell cycle progression, and in the Rat-1 stable cell lines cyclin-dependent kinase inhibitor (cdki) p27 protein levels were reduced, while levels of cyclin-dependent kinase 2 (Cdk2) and phosphorylated retinoblastoma (Rb) protein were increased. The LMP1 mediated effects upon Id proteins, and cell cycle proteins were mapped to CTAR1. The data presented in this study identify key properties of LMP1 that affect cell cycle progression and likely contribute to transformation and oncogenesis.
MATERIALS AND METHODS
Plasmids.
The initial cloning of wild-type LMP1 and construction of LMP1 deletion mutants, 1-231 (previously, 231-STOP) and Δ187-351 has been described previously (23). Full-length LMP1 and LMP1 deletion mutants were subcloned by PCR with Platinum Pfx DNA polymerase (Invitrogen) according to the manufacturer's directions. Cloning into the myc-tagged expression vector was accomplished by amplification with LMP1myc5′ (CGACGGATCCATATGGAACACGACCTTGAGAGG) and LMP1-3′ (ATCACGAGGAATTCAATGTGGCTTTTCAGCCTAGAC), restriction enzyme digestion, and insertion into the BamHI and EcoRI sites of M3-pcDNA3 (18). Wild-type LMP1, 1-231, and Δ187-351 cloned into the expression vector resulted in plasmids pM3-LMP1, pM3-1-231, pM3-Δ187-351, respectively, expressing three N-terminal myc epitope tags on LMP1 molecules. Hemagglutinin (HA)-tagged LMP1 was cloned by amplification with LMP1-HA5′ containing the coding sequence of the HA epitope (GCCGGATCCATGGCTTACCCATACGATGTTCCAGATTACGCTAGCTTGGGTGGTCATATGGAACACGACCTTGAGAGG) and LMP1-3′, restriction enzyme digestion, and insertion into the BamHI and EcoRI sites of pBABE. Cloning into pBABE resulted in recombinant retrovirus vectors expressing N-terminal HA-tagged LMP1, 1-231, and Δ187-351 in pBABE-HA-LMP1, pBABE-HA-1-231, and pBABE-HA-Δ187-351, respectively.
Id1 promoter reporter constructs, kindly provided by Takenobu Katagiri, in pGL3-Basic (Promega) were described previously (12). Full-length reporter plasmid ID1-2.1 and truncated reporters ID1-985 and ID1-0.8 contain upstream promoter regions from −2,100, −985, and −800 bp, respectively. ID1-mutA and ID1-mutB are derived from ID1-985, which contains the BMP-2 serum-responsive region, and each contains the triple-point mutation (TTT) that alters putative Egr-1 and Sp1 binding sites, respectively.
Transfection and retrovirus transduction.
Cells were transfected with FuGENE 6 (Boehringer Mannheim) according to the manufacturer's directions. Recombinant retroviruses were generated as previously described (30) by transfection of 293T cells with pBABE, pBABE-HA-LMP1, pBABE-HA-1-231, or pBABE-HA-Δ187-351, and VSV-G (pG1-VSV-G)- and gag-pol (pGPZ9)-expressing plasmids. After 24 h, the transfection medium was removed and replaced with fresh medium, and cells were incubated at 33°C overnight. The following day, culture supernatants were harvested and centrifuged 1,000 × g for 10 min to remove any cells or cellular debris. Rat-1 cells were transduced with clarified 293T culture supernatants overnight with 8 μg of Polybrene/ml.
Cell culture and stable cell lines.
Cell lines were maintained in Dulbecco modified Eagle medium (Gibco) supplemented with an antibiotic-antimycotic mixture and 10% (vol/vol) heat-inactivated fetal bovine serum. Stable cell lines were established in the human cervical carcinoma cell line C33A by transfection with vector control pcDNA3 (Invitrogen) or the LMP1-expressing plasmids M3-LMP1, M3-1-231, and M3-Δ187-351, followed by selection with G418 (0.6 mg/ml; Gibco). Rat-1 rodent fibroblast stable cell lines were established by transduction with vector control pBABE or LMP1-expressing retroviruses and selection with puromycin (1 μg/ml; Sigma). For serum starvation, selected cells were grown to confluence and then changed to medium with 0 to 0.2% serum for 24 to 48 h after being washed once with phosphate-buffered saline (PBS) to remove serum.
RNA isolation and microarray probe synthesis.
C33A cells were grown in 100-mm tissue culture plates, and total RNA was isolated with RNeasy Midi purification kits (QIAGEN) according to the manufacturer's directions. Eluted RNA was precipitated with sodium acetate and ethanol and stored at −80°C. Precipitated RNA was centrifuged, washed with 70% ethanol, and dried. Pelleted RNA was dissolved in water, quantitated, and electrophoresed on agarose gels to assess sample integrity. One quarter of the RNA was aliquoted and mixed with Direct Protect RPA lysis buffer (Ambion) and frozen at −20°C for future analysis. Remaining RNA, about 60 μg, was mixed with 4.5 μg of anchored oligo(dT) primer [(T)20VN] in a final volume of 40 μl, denatured for 10 min at 70°C, and cooled immediately in ice water. cDNA was synthesized with SuperScript II (Invitrogen) according to the manufacturer's directions by the addition of 18 μl of 5× buffer, 9 μl of deoxynucleoside triphosphates (final concentration, 0.5 mM dCTP, 0.5 mM dATP, 0.5 mM dGTP, and 0.15 mM dTTP), 9 μl of 5-(3-aminoallyl)-dUTP (final concentration, 0.3 mM; Molecular Probes), 9 μl of dithiothreitol (0.1 M stock), and 5.1 μl of SuperScript II (200 U/μl) for 1 h at 42°C. Reaction mixtures were spiked with an additional 3 μl of enzyme and incubated for an additional hour. The reverse transcription reaction was stopped and RNA was hydrolyzed by the addition of 39 μl of 1 M NaOH-2 mM EDTA and incubation at 65°C for 15 min, followed by neutralization with 39 μl of 1 M HCl. cDNA was concentrated and desalted in Microcon YM-30 (Amicon) according to the manufacturer's directions by the addition of 400 μl of nuclease-free water and centrifugation carried out three times. Probes were labeled with amine-reactive fluorescent dye with the ARES Alexa Fluor DNA Labeling Kit (Molecular Probes) according to the manufacturer's directions. Vector control pcDNA3 RNA was labeled with 647-nm dye for the Cy5 channel, while LMP1 stable cell line RNA was labeled with 555-nm dye for the Cy3 channel. Dye-labeled probes were purified with the QIAquick PCR purification kit (QIAGEN) according to the manufacturer's directions. cDNA yield and dye labeling efficiency were determined by spectrophotometric measurements according to the ARES kit directions. Between 2 to 4 μg of high-specific-activity probe with a dye-to-base ratio of 1:18 to 1:25 per reaction mixture was typical.
Probe preparation and array hybridization.
Equal amounts of control and experimental probes, 1 to 1.5 μg each, were combined with 35 μg of human COT-1 DNA (Invitrogen) and were concentrated to ≤25 μl with MicroconYM-30 (Amicon) columns according to the manufacturer's directions. To the probe-COT-1 mixture was added 10 μg of yeast tRNA (Invitrogen), 20 μg of poly(A) DNA (Amersham Pharmacia Biotech), 5.3 μl of 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.7 μl of 10% sodium dodecyl sulfate (SDS), and water to a final volume of 35 μl. Samples were placed in a boiling-water bath for 2 min and then incubated for 15 min at 42°C.
Human microarray slides were obtained from the UNC Lineberger Genomics Core Facility containing the Compugen oligo series representing more than 18,000 unique genes. Arrays were cross-linked with 600 mJ of UV irradiation, followed by incubation in prehybridization buffer (3× SSC, 0.1% SDS, and 0.2% bovine serum albumin) at 65°C for 45 min. Slides were then washed five times with nuclease-free water, washed one time with 95% ethanol, and dried. Probe mixtures, adjusted to 35 μl, were applied to slides, covered with a coverslip, and hybridized overnight in hybridization chambers at 65°C. Hybridized arrays were washed once with room temperature 2× SSC-0.1% SDS to remove the coverslip, once with room temperature 1× SSC for 5 min, and twice with 0.2× SSC at 65°C for 5 min. Slides were dried by centrifugation and scanned with an Axon 4000 scanner at 532 and 635 nm. Images were then processed with GenePix software (Axon) to generate the raw data files that were uploaded to the University of North Carolina Microarray Database for normalization. Clustering analysis was performed within the database, and data sets were combined in Microsoft Excel for manual sorting.
Real-time PCR.
Aliquots of RNA used for microarrays were treated with RNase-Free DNase (QIAGEN) and purified with the RNeasy Mini kit (QIAGEN) according to the manufacturer's directions. RNA concentration and purity were assessed by spectrophotometric measurements. Primer pairs (Table 1) were designed using Primer Express (Applied Biosystems). Quantitative reverse transcription-PCR was performed on DNase-treated RNA using the Quantitect SYBR Green RT-PCR Kit (QIAGEN) according to the manufacturer's directions. Amplification of target sequences was detected with an ABI 7900HT sequence detection system (Applied Biosystems) and analyzed with SDS 2.0 software (Applied Biosystems). The cycle threshold (Ct) was determined as the number of PCR cycles required for a given reaction to reach an arbitrary fluorescence value within the linear amplification range. The change in Ct (ΔCt) was determined between the same gene primer sets and different samples and the change in ΔCt (ΔΔCt) was determined by adjusting for the difference in the number of cycles required for actin to reach the Ct. Since each PCRcycle results in a twofold amplification of each product, the difference was determined as 2ΔΔCt.
TABLE 1.
Quantitative PCR primers
| Gene name (accesssion no.) | Sense primer | Antisense primer |
|---|---|---|
| EGFR (X00588) | TGCGTCTCTTGCCGGAAT | GGCTCACCCTCCAGAAGC |
| H-ras (AF493916) | TACGGCATCCCCTACATCGA | ACCAACGTGTAGAAGGCATCCT |
| Rel A (M62399) | CCTGGAGCAGGCTATCAGTCA | CCCACGCTGCTCTTCTATAGGA |
| c-Src (NM_004383) | CGTGTTTGCGCTTGACCAT | AAATCCAAGCCCCTGACAGA |
| Actin (NM_001101) | TCACCCACACTGTGCCCATCTACGA | CAGCGGAACCGCTCATTGCCAATGG |
| Annexin A2 (NM_004039) | GATGAGGTCACCATTGTCAACATT | GGCGAAGGCAATATCCTGTCT |
| Cycin E1 (T54121) | AGGGTCAAGTAGCACCTTCCATAG | CCAGCCACCTCCCAGACA |
| DDX21 (U41387) | CGGACAGGAACTGGGAAGAC | TCTTGCAGTTCCCCATGAAGT |
| Id3 (NM_002167) | AGGGATGGGCCCCAACT | AGGTTTAGTCTCCAGGAAGGGATT |
| LZ16 (NM_013275) | AGTTTCTCTAACCAAGACCCCAAA | CTCGCTCCCTCACCTCCTT |
| Vimentin (NM_003380) | ACACCCTGCAATCTTTCAGACA | GATTCCACTTTGCGTTCAAGGT |
| Id1 (NM_002165) | AGAACCGCAAGGTGAGCAA | CCAACTGAAGGTCCCTGATGTAG |
| CSDA (NM_003651) | TCCAAACCAGCCGTCTGTTC | GGCGACGCCGGTAATTG |
| STIPI (NM_006819) | ACCGCTGCCCTCGAGTTC | AGAGCAGCCGAGACAGCAA |
| DNAJ A1 (NM_001539) | GCACTGTGTGGCTTCCAGAA | TGACCTGGATGAGAGGTGATGA |
Cell harvesting and Western blotting.
Cell lines were grown in 100-mm tissue culture plates to 70 to 95% confluency and harvested. Cells were washed with ice-cold PBS (Gibco) and lysed with 100 to 250 μl of RIPA buffer (10 mM Tris-HCl [pH 8.0], 140 mM NaCl, 1% Triton X-100, 0.1% SDS, 1% deoxycholic acid, and protease and phosphatase inhibitors σ). Cell lysates were clarified by centrifugation and quantitated by the Bio-Rad DC protein assay system. Samples were then boiled in SDS sample buffer, and indicated amounts of proteins were separated by SDS-polyacrylamide gel electrophoresis with 10 or 15% acrylamide and transferred to Immobilon P membranes (Millipore) for Western blotting analysis. Primary antibodies used included β-actin (I-19), HA-probe (Y-11), Id1 (C-20 and Z-8) and Id3 (H-70) (Santa Cruz), p27 (Calbiochem), Cdk2 (BD Biosciences), Phospho-Rb (Ab-1) (Oncogene), and CS1-4 (anti-LMP1) (Dako). Bound proteins were detected with horseradish peroxidase-conjugated secondary antibodies (Amersham Pharmacia and Dako) and the Supersignal West Pico system (Pierce), followed by exposure to film.
Rat-1 foci and immunofluorescence.
Rat-1 cells were plated at a 1:5 ratio in 6- or 12-well plates and infected the following day with pBABE or pBABE-HA-LMP1 retroviruses with 8 μg of Polybrene/ml. The medium was changed the following day and every 2 to 3 days thereafter, and foci were allowed to form for 10 to 15 days. Wells were washed once with PBS and fixed with methanol for 5 min at room temperature. Cells were blocked for 20 min with PBS containing 20% normal goat serum (NGS) and then incubated with primary antibodies in PBS containing 20% NGS overnight at 4°C. Cells were washed three times with PBS, incubated with fluorescein isothiocyanate-conjugated secondary antibody (Jackson Laboratories) in PBS with 20% NGS for 1 h at room temperature, and then washed three times with PBS. Stained wells were examined by phase-contrast and fluorescence microscopy for foci and immunofluorescence.
Id1 promoter luciferase assays.
Cells were plated at a 1:5 ratio into six-well plates 1 day prior to transfection. Cells were transfected with 0.75 μg of pRL-SV40 (Promega), 0.75 μg of pGL3-Basic or Id1 promoter plasmids, and 0.75 μg of pcDNA3 or pM3-LMP1. The following day, the medium was changed, and 40 h posttransfection cells were harvested and luciferase activity was assayed with the Dual-Luciferase reporter assay system (Promega) according to the manufacturer's directions. Relative luciferase activity was determined by dividing the firefly luciferase activity of the Id1 promoter constructs by the internal control Renilla luciferase activity. Each condition was done in triplicate and replicated in different experiments.
RESULTS
Genomic analysis of C33A cells expressing LMP1.
LMP1 induces NF-κB transcriptional activity, PI3K activity, and MAPK activity. To determine global transcriptional regulation by LMP1 in epithelial cells, microarray analysis was performed. C33A cells were stably transfected with pcDNA3 or M3-LMP1, and RNA was isolated. Microarray probes were synthesized, and two-color microarray analysis was performed on chips with 18,000 human genes. Comparisons between the vector control and LMP1-expressing C33A cells were performed in triplicate. Raw data files were normalized in the University of North Carolina Microarray Database, and clustering analysis was performed. Clustering analysis of the triplicate LMP1 arrays did not reveal linkage between different groups of genes (data not shown).
To determine individual genes consistently up- or downregulated by LMP1, normalized data files were loaded into Microsoft Excel and sorted manually by log2 ratio of the means and medians. Using the criteria of log2 ratio of the means or medians >2 or less than −2, which corresponds to a >4-fold change in expression, few genes were induced by LMP1 and none were repressed. Genes induced by LMP1 are listed in Table 2. Annexin A2 was the most highly induced gene at nearly sevenfold. Next at >5-fold were two separate spots corresponding to Id3. Interestingly, also scoring just above and below the fourfold cutoff were spots corresponding to Id4 and Id1, respectively. Vimentin was also upregulated, as has been previously reported (20). Several other proteins of interest that were induced slightly less than fourfold were also noted.
TABLE 2.
Genes induced by LMP1 on microarrays
| Gene | Description | Accession no. | Scorea |
|---|---|---|---|
| ANXA2 | Annexin A2 | NM_004039 | −2.75 |
| ID3 | Inhibitor of DNA binding 3, dominant negative helix-loop-helix protein | X69111 | −2.46 |
| ID3 | Inhibitor of DNA binding 3, dominant negative helix-loop-helix protein | AA482119 | −2.40 |
| GAGE1 | G antigen 1 | NM_001468 | −2.35 |
| DNAJA1 | DnaJ (Hsp40) homolog, subfamily A, member 1 | NM_001539 | −2.32 |
| CSDA | Cold shock domain protein A | NM_003651 | −2.32 |
| WDR3 | WD repeat domain 3 | NM_006784 | −2.22 |
| IL1RL1LG | Putative T1/ST2 receptor binding protein | NM_006858 | −2.18 |
| NDRG1 | N-myc downstream regulated gene 1 | NM_006096 | −2.15 |
| DDX21 | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 21 | U41387 | −2.13 |
| VIM | Vimentin | NM_003380 | −2.08 |
| SLC6A7 | Solute carrier family 6 (neurotransmitter transporter, l-proline), member 7 | NM_014228 | −2.08 |
| CNN3 | Calponin 3; acidic | NM_001839 | −2.06 |
| CCNE1 | Cyclin E1 | T54121 | −2.05 |
| ID4 | Inhibitor of DNA binding 4, dominant negative helix-loop-helix protein | AA453341 | −2.05 |
| STIP1 | Stress-induced-phosphoprotein 1 (Hsp70/Hsp90-organizing protein) | NM_006819 | −2.03 |
| BTF3 | Basic transcription factor 3 | NM_001207 | −2.01 |
| CCT5 | Chaperonin containing TCP1, subunit 5 (epsilon) | AF275798 | −2.00 |
| ID1 | Inhibitor of DNA binding 1, dominant negative helix-loop-helix protein | NM_002165 | −1.81 |
| LZ16 | Nasopharyngeal carcinoma susceptibility protein | NM_013275 | −1.43 |
| RELA | v-rel reticuloendotheliosis viral oncogene homolog A, nuclear factor of kappa light polypeptide gene enhancer in B-cells 3, p65 (avian) | M62399 | −1.3 |
| HRAS | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | NM_005343 | −1.27 |
| CSK | c-src tyrosine kinase | NM_004383 | −1.21 |
Score, average of the log2(mean red/green) and log2(median red/green) from three experiments. red, vector control; green, LMP1 signal.
Quantitative PCR in C33A Cells.
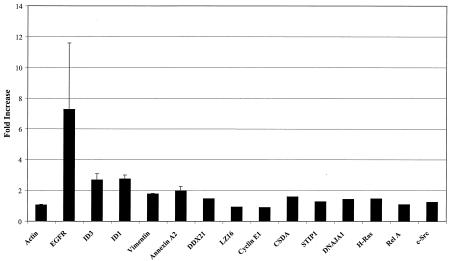
Genes observed to be increased by LMP1 in the microarrays (Table 2) were tested by real-time reverse transcription-PCR. Genes whose expression level was altered less than twofold (ΔΔCt ≤ 1) were judged to be unchanged. Genes whose expression was changed more than twofold (ΔΔCt ≥ 1) were judged to be regulated by LMP1, and analysis was repeated three or more times. EGFR, although not induced on the arrays, is known to be upregulated by LMP1 in C33A cells (22) and was used as a positive control. EGFR was induced by LMP1 by quantitative PCR (Fig. 1). Id1, Id3, vimentin, and annexin A2 upregulation was also confirmed by quantitative PCR. The remaining genes that scored as significant by microarray analysis were unchanged by quantitative PCR analysis. These discrepancies in RNA increases between quantitative PCR and microarray analyses reflect likely differences in the relative abundance of the mRNAs.
FIG. 1.
Quantitative PCR analysis of gene expression in the presence of LMP1. RNA from LMP1 and control C33A cells was purified, and quantitative reverse transcription-PCR was performed. The number of cycles required to reach a given fluorescence intensity threshold (Ct) was determined, and different conditions were compared using ΔCt and ΔΔCt relative to actin control amplification. Differences were calculated as 2ΔΔCt. Each reaction and condition were performed in triplicate, and genes judged to be significantly altered (ΔΔCt ≥ 1) were repeated. Error bars represent the standard deviation of three different experiments.
Id1 promoter reporter assays.
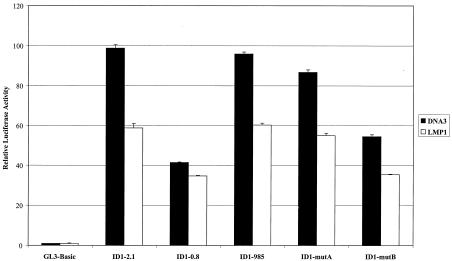
As LMP1 clearly alters epithelial cell growth, the induction of proteins of the Id family of proteins was of particular interest. Id proteins bind to helix-loop-helix transcription factor family members, inhibit differentiation, and promote proliferation. To characterize induction of the Id1 promoter by LMP1, reporter assays were performed with Id1 promoter plasmids. Full-length Id1 promoter reporter plasmid contains 2.1 kb of upstream sequences (ID1-2.1), and promoters truncated to −985 and −800 (ID1-985 and ID1-0.8) differ by the presence or absence, respectively, of serum-responsive elements between −985 and −947 bases. Mutants ID-mutA and ID1-mutB are derived from ID1-985 but contain triple-point mutations that alter putative Egr-1 and Sp1 binding sites, respectively, that have previously been shown to be responsible for activation of the promoter by serum components. Promoter reporter constructs were assayed with vector control or LMP1 under various conditions and cell lines, including C33A or Rat-1 cells, transient or stable cell lines, and presence or absence of serum. Activation of Id1 promoter reporter plasmids by LMP1 was not detected under any conditions. As represented in Fig. 2 by C33A cells transiently expressing LMP1 in the presence of serum, the activity of each promoter was slightly decreased in the presence of LMP1. These data indicate either that the LMP1 responsive sequences are not contained within these constructs or that the promoter is activated by LMP1 by mechanisms not sensitive to the experimental conditions tested.
FIG. 2.
Id1 promoter reporter assays. C33A cells were transfected with control pRL-SV40, pGL3-Basic, or Id1 promoter reporter plasmids and vector control or pM3-LMP1. Full-length reporter plasmid ID1-2.1 and truncated reporters ID1-985 and ID1-0.8 contain upstream promoter regions from −2,100, −985, and −800 bp, respectively. ID1-mutA and ID1-mutB are derived from ID1-985, which contains the BMP-2 serum-responsive region, and each contains the triple-point mutations (TTT) that alter putative Egr-1 and Sp1 binding sites, respectively. Forty hours posttransfection, cells were harvested, and dual-luciferase assays were performed. Relative luciferase activity was determined by the firefly luciferase activity of the Id1 promoter constructs relative to control Renilla luciferase activity.
Id proteins in C33A cells.
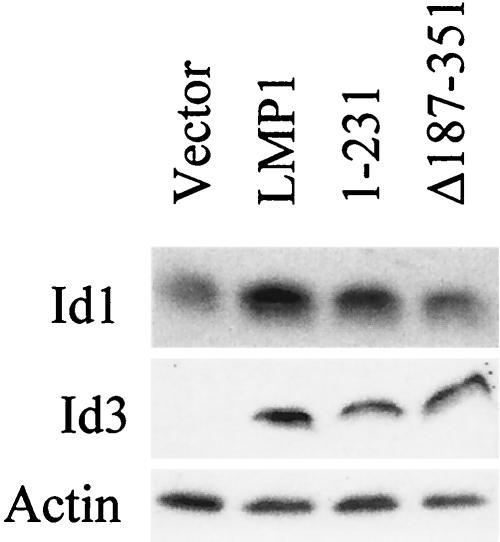
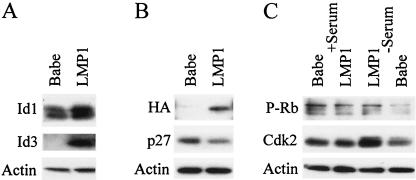
To determine whether increased RNA levels led to increased Id proteins levels, Western blotting was performed. C33A cells stably expressing LMP1 or LMP1 mutants 1-231 (CTAR2 deleted) or Δ187-351 (CTAR1 deleted) were examined for Id1 and Id3 protein levels relative to vector control cells. As with the mRNA levels, both Id1 and Id3 proteins were increased in the presence of LMP1 (Fig. 3). Id1 was detected in vector control cells and dramatically increased in the LMP1 expressing cells (Fig. 3, top). Id3 was not detected in the absence of LMP1 but clearly induced when LMP1 was expressed. The 1-231 mutant also induced the expression of both Id1 and Id3, but the Δ187-351 mutant only induced Id3. These data indicate that LMP1 containing CTAR1 is sufficient to induce expression of both Id1 and Id3, while LMP1 lacking CTAR1 can increase Id3 but cannot induce Id1. Densitometry quantification of the Id1 bands indicated that LMP1 and 1-231 induce Id1 protein three- and twofold, respectively. Id1 is similar to other genes that have been identified that are uniquely activated through CTAR1, including the EGFR and TRAF1.
FIG. 3.
Id1 and Id3 protein levels in C33A cells. C33A stable cell lines were analyzed by Western blotting. A total of 50 μg of total cell lysates from the vector control, LMP1, 1-231, and Δ187-351 was examined for expression of Id1, Id3, and actin.
Id1 expression in LMP1-transformed Rat-1 foci.
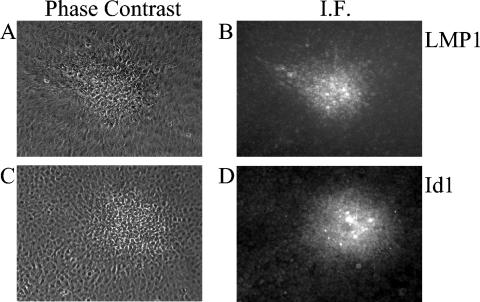
Because the Id proteins are known regulators of cell growth, it is possible that the Id proteins contribute to LMP1-mediated transformation of rodent fibroblasts. To determine if the induction of Id proteins by LMP1 occurs during rodent fibroblast transformation, Rat-1 foci from cells transduced with control and LMP1-expressing retroviruses were examined by phase-contrast and immunofluorescence microscopy. Foci were only observed in LMP1 transduced cells (Fig. 4A and C and data not shown). Immunofluorescence staining for LMP1 was restricted to the foci (Fig. 4B). These data reveal that LMP1 expression always induces focus formation. Furthermore, bright staining for Id1 was restricted to the foci and coincident with LMP1 expression (Fig. 4D). Under low contrast, low levels of Id1 expression were detected in control transduced cells and cells surrounding the LMP1 foci (data not shown). These data indicate that Id1 is increased in Rat-1 foci transformed by LMP1.
FIG. 4.
Detection of LMP1 and increased Id1 expression in transformed foci of Rat-1 cells. Rat-1 cells were transduced with control and LMP1 retroviruses, and foci were allowed to form for 10 days. Cells were fixed, and immunofluorescence staining was performed. Phase-contrast (A and C) and immunofluorescence (B and D) micrographs of LMP1 and Id1 are depicted. The same foci were examined in panels A and B and in panels C and D.
Id proteins and cdki's in Rat-1 cells.
To determine levels of Id proteins and potential effects on cell cycle control proteins, Rat-1 cells stably transduced with control or LMP1-expressing retroviruses were selected and analyzed by Western blotting. As with the C33A cells, Rat-1 cells expressing LMP1 had increased levels of Id1 and Id3 protein (Fig. 5A). Id1 has been shown to negatively regulate cdki p16 expression by binding to and sequestering Ets1 and Ets2, E-box proteins that regulate p16 expression (26). Potential E-box binding sites are located in the upstream regulatory regions of a number of cdki's, including p16, p15, p27, and p21 that are conserved between rat, mouse, and humans (data not shown). In agreement with previous studies (1, 29), the expression of cdki's p16, p19, and p21 was not detected in Rat-1 cells (data not shown). However, p27, which is expressed in Rat-1 cells, was strikingly downregulated in presence of LMP1 (Fig. 5B).
FIG. 5.
Id proteins and cell cycle control proteins in Rat-1 cells. Rat-1 cells transduced with Babe (control) or LMP1-expressing retroviruses were examined by Western blotting. Levels of Id1 and Id3 were determined in the presence of LMP1 (A), and levels of HA-tagged LMP1 and p27 expression were determined (B). Cell cycle control proteins Cdk2 and phosphorylated Rb were detected in the presence of serum or following serum starvation for 36 h (C). Actin was used as a control for equal loading, and 50 μg of protein was loaded in each lane.
Cdk levels and activity in Rat-1 cells.
cdki's, like p27, bind to and inhibit the activity of Cdk2 and Cdk4, thereby inhibiting cell cycle progression. Studies with v-src transformation in Rat-1 cells have shown that lower p27 levels result in increased Cdk2 levels and activity. To determine if the decrease in p27 affected levels of Cdks, Cdk2 and Cdk4 were analyzed. Serum starvation inhibits cell cycle progression; however, in the presence of LMP1, a large increase in Cdk2 levels was detected (Fig. 5C). In the presence of serum, Cdk2 levels were unchanged and Cdk4 was not detected in Rat-1 cells (data not shown). An important target of the Cdks in the regulation of the cell cycle is the Rb protein. To determine if LMP1 not only regulates the levels, but also the activity of Cdks, phosphorylated Rb protein was examined with a phosphospecific Rb antibody. Levels of Rb protein phosphorylated on T373 were greatly increased in serum-starved LMP1-expressing cells but not in cells transduced with control retroviruses (Fig. 5C). Total Rb was increased approximately threefold in LMP1-expressing Rat-1 cells under low-serum conditions (data not shown). Levels of phosphorylated Rb protein under high-serum conditions were high in both LMP1-expressing and control cells. Increased levels of Cdk2 and phosphorylation of Rb indicates activation of Cdk2 complexes under low-serum conditions. These data reveal that LMP1 expression abrogates the normal cell cycle regulation imposed by serum deprivation.
Effects of LMP1 functional domain mutants on Id1, Id3, p27, and Cdk2 in Rat-1 cells.
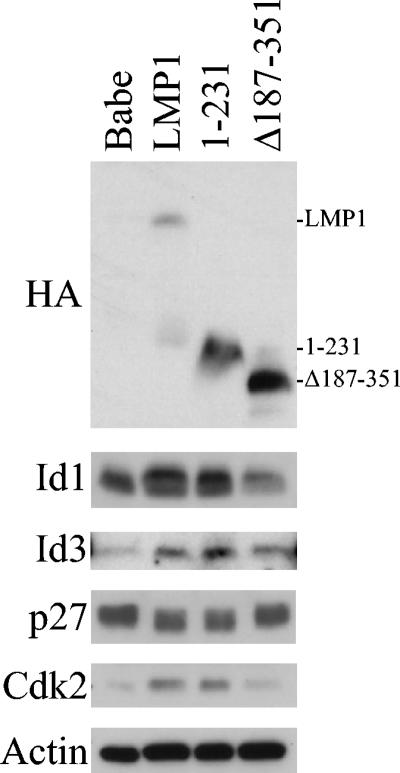
LMP1 CTAR1 and CTAR2 mutants were also tested in Rat-1 cells for effects on the Id proteins and cell cycle proteins. Rat-1 cells expressing LMP1 or 1-231 (CTAR2 deleted) increased Id1 and Id3 protein levels compared to control cells, whereas levels were unchanged in cells expressing LMP1 deleted for CTAR1 (Δ187-351), despite the high levels of Δ187-351 expression (Fig. 6). This is in contrast to C33A cells where Δ187-351 increased Id3 levels. Coordinate downregulation of p27 and increased Cdk2 were also detected in LMP1 and 1-231-expressing cells, but p27 and Cdk2 levels were unchanged in cells expressing the CTAR1 deletion mutant Δ187-351. These data indicate that in Rat-1 cells, LMP1 expression and signaling from CTAR1 are responsible for the induction of the Id proteins, downregulation of p27, and increased levels of Cdk2.
FIG. 6.
Regulation of Id proteins and cell cycle proteins by LMP1 mutants in Rat-1 cells. Rat-1 cells transduced with Babe (control), LMP1, or LMP1 mutant 1-231 (CTAR2 deleted) or Δ187-351 (CTAR1 deleted) retroviruses were examined by Western blotting. Expression of wild-type and mutant LMP1 proteins was detected with the HA epitope tag (top). Id proteins (Id1 and Id3) and cell cycle control proteins (Cdk2 and p27) were also detected (see the panels below). Lysates used for Cdk2 were from cells that were serum starved for 30 h. Actin was used as a loading control, and 25 μg of total cell lysates was loaded in each well.
DISCUSSION
In this study, genomic analysis of cells expressing LMP1 revealed induction of two genes in the family of inhibitors of differentiation, the Id1 and Id3 genes. Few other genes were induced by LMP1 as detected by microarrays. Id1 and Id3 protein levels were increased in C33A and Rat-1 cells, and Rat-1 foci transformed with LMP1 had increased Id1 protein. Id proteins are known to downregulate cdki's. In this study, p27 was decreased in LMP1-expressing Rat-1 cells, and Cdk2 levels and phosphorylation of Rb were increased. Importantly, LMP1 regulation of Id1, Id3, p27, and Cdk2 mapped to the CTAR1 signaling domain.
Microarray analyses are widely used to determine global transcriptional changes in cell lines and tumor samples and often reveal hundreds of genes regulated under different conditions. In the present study, the limited number of genes affected in the presence of LMP1 was surprising. LMP1 activates NF-κB, c-Jun N-terminal kinase, MAPK, and PI3K pathways and would be expected to alter the expression of a number of genes and gene families. Studies from our laboratory indicated that as stable cell lines are passaged, LMP1 expression declines. Although initial, low-passage, high-LMP1-expressing cells may dramatically regulate the expression of more genes, lower levels of LMP1 are more likely to be physiologically relevant to the levels of LMP1 expressed in EBV-infected cells.
The lack of sensitivity of microarrays may have limited the number of genes identified in the present study. In C33A cells, upregulation of EGFR in the presence of LMP1 was expected (22). Three separate spots for EGFR on the microarray did not score as significantly different in the presence of LMP1 despite Western blots and RNase protection assays for EGFR that indicated upregulation in these cells. The quantitative PCR analysis reveals that EGFR expression is low in C33A cells, taking about 25 cycles to reach the fluorescence detection level, while it only took actin 12 cycles and most other genes between 16 to 18 cycles (data not shown). Therefore, EGFR is expressed between at levels 100-fold to several thousandfold lower than other genes examined, and differences in gene expression levels may have been masked by the inherent noise in the microarray technique. Other genes, like the EGFR gene (whose expression is low), may be significantly regulated by LMP1 but not scored by microarray because of a lack of sensitivity. Alternatively, expression of genes important for growth may be enhanced by passage in tissue culture and obscure the effects of LMP1.
Two important genes induced by LMP1 are the Id1 and Id3 genes. These two genes are members of a family of genes named for their function as inhibitors of differentiation and inhibitors of DNA binding (31). The proteins expressed by these genes (Id1, Id2, Id3, and Id4) all contain helix-loop-helix domains that allow them to bind to helix-loop-helix transcription factors, called E-box proteins. However, the Id proteins lack DNA binding domains and act as dominant-negative transcription factors. Id proteins are highly expressed in proliferating cells and some cancers and downregulated in differentiating cells and during senescence. LMP1 has been reported to inhibit epithelial cell differentiation, and this effect likely reflects the increased Id expression (5).
In a genomic analysis of LMP1-transduced nasopharyngeal carcinoma (NPC) cells, the NP69 cell line revealed only 28 genes as being regulated more than twofold by LMP1 (20). These data and our findings suggest that LMP1 may not regulate the expression of a large number of genes in epithelial cells. Vimentin was upregulated in both C33A and NP69 cells as determined by arrays. Vascular endothelial growth factor induction by LMP1 that has been identified by other studies (24) was also indicated by array in NP69 cells. The Id proteins were not identified as being upregulated in NP69 cells, but several markers for differentiation, cytokeratin 14 and cytokeratin 19 and differentiation control molecules Jagged2 and Notch1m were downregulated. Although induction of Id proteins was not detected in NP69 cells, downregulation of differentiation markers would be consistent with the activity of Id proteins.
A recent report indicated that LMP1 induces Id1 in epithelial cells (19). Upregulation of Id1 was inhibited by NF-κB inhibitors and dominant-negative IκBα and was not inhibited by MAPK or SAPK inhibitors, or dominant-negative MAPK or SAPK mutants. This study identified two putative NF-κB sites at positions −1110 and −1947 within the Id1 promoter but did not test their function. While these sites were contained within the full-length reporter construct, ID-2.1, this construct was unresponsive to LMP1 in C33A and Rat-1 cells in the presence or absence of serum (Fig. 2). These sets of promoter reporter constructs respond appropriately to serum, as has been previously described (11, 12).
The roles of the different signaling domains of LMP1 in the upregulation of Id proteins are presently unclear. In the present study with Rat-1 cells, the CTAR2-deleted mutant, 1-231, induced Id1 and Id3 and downstream effector molecules, but the CTAR1-deleted mutant Δ187-351 did not induce either Id protein or alter cell cycle proteins. However, in C33A cells, although 1-231 was able to upregulate Id1 and Id3, Δ187-351 was only able to upregulate Id3. Finally, in work published by others using 293 cells (19), either CTAR1 or CTAR2 was able to induce Id1 expression, although Id3 and downstream molecules were not examined. Differing activities of the CTAR mutants in other cell lines likely reflects cell specific differences between 293, C33A, and Rat-1 cells.
The data presented in the present study suggest that LMP1-mediated Id induction and regulation of cell cycle proteins during Rat-1 cell transformation occurs primarily through signaling initiated by CTAR1. It will be important to determine the role of Id protein expression in the transformation of Rat-1 cells by LMP1 and LMP1 CTAR mutants. Studies of B-lymphocyte transformation by LMP1 have revealed that CTAR1 is required for transformation and that deletion of CTAR2 can be complemented by growth on fibroblast feeder layers (2, 7, 14, 15).
cdki's and the Rb pathway are important regulators of cell cycle G1/S entry and replicative senescence. The cdki p16 is often not expressed in NPC cells (21, 33). LMP1 stimulates the CRM1-dependent nuclear export of Ets2, leading to a decrease in p16 expression (25). Id1 and several E-box proteins, E47, Ets1, and Ets2, regulate the p16INK4 promoter; E-box binding sites within the promoter of p16 are conserved between mouse, human, and rat cells (26, 36). Another E-box protein, E2A, regulates cdki p21 in conjunction with Id1, controlling cell growth (27). Although regulation of cdki's by Id proteins in Rat-1 cells is unknown, examination of the promoters of several cdki's, including p16, p15, p27, and p21, revealed the presence of putative E-box binding sites contained within the upstream regulatory regions of all of these promoters that were conserved between rats, mice, and humans (data not shown). This indicates that all of these promoters might be regulated by Id proteins by a similar mechanism. Regulation of cdki p27 in Rat-1 cells and cdki p16 in NPC cells by LMP1 via induction of Id proteins may represent a common mechanism of transformation by LMP1 by altering cdki levels.
It is likely that the effects of LMP1 on Id proteins (downregulation of p27, increased Cdk2, and phosphorylation of Rb) are important for the transformation of rodent fibroblasts. The importance of these effects is supported by transformation of Rat-1 cells with v-src (10, 29). Transformation with v-src also leads to the downregulation of p27, activation of Cdk2, and phosphorylation of Rb. Interestingly, regulation of p27 by v-src was dependent upon PI3K signaling and led to increased degradation of p27. LMP1 also activates PI3K signaling (6) and could negatively regulate p27 at the level of proteolytic degradation induced by PI3K activation, as well as the transcriptional repression via the binding of Id proteins to E-box proteins. The activation of PI3K by LMP1 is a CTAR1-specific activity. CTAR1 was responsible for effects on p27 in the Rat-1 cells, supporting the importance of this signaling domain.
The importance of p27 regulation in EBV-infected cells is indicated by other studies. EBNA3C, which regulates LMP1 transcription during EBV latency (35), competes with p27 for binding to cyclin A in lymphocytes. EBNA3C interacts with cyclin A, stimulates cyclin A/Cdk2 kinase activity, and rescues p27 inhibition of cyclin A/Cdk2 (17). Phosphorylation of Rb was also stimulated in the presence of EBNA3C. In lymphocytes, activities of LMP1 and EBNA3C represent a two-pronged attack upon the Cdks and reveal two distinct mechanisms EBV has developed to deregulate the mammalian cell cycle. However, since EBNA3C is not expressed in most EBV-associated cancers, LMP1 may be responsible for effects on cell cycle proteins in these malignancies.
Many cancers have dysregulation of the cell cycle through mutations or deletions of the p53 or pRb genes, and DNA tumor viruses also target cell cycle regulatory and checkpoint molecules. Human papillomavirus (HPV) encodes two proteins that target cell cycle proteins for degradation, E6 and E7. HPV E6 induces the degradation of p53, and E7 binds and promotes the degradation of pRB and pRb family members p107 and p130. These activities of E6 and E7 lead to increased E2F-mediated gene transcription, unscheduled cyclin/Cdk2 activity, and cell cycle progression. Thus, in HPV-associated cancers, p53 and Rb are not mutated. EBV-associated cancers also have wild-type p53 and Rb, indicating that EBV also likely affects the regulation of the mammalian cell cycle.
The data presented in this study suggest that the EBV oncogene LMP1 affects the cell cycle by inducing the expression of cell cycle regulatory proteins, thereby eliminating the necessity for mutations. As opposed to inducing the degradation of p53 and pRB, LMP1 induces the expression of Id proteins that in turn regulate the expression of cell cycle proteins inducing transformation. This study identifies a new mechanism through which LMP1 and EBV affect cell growth.
Acknowledgments
We thank Jennifer A. Morrison, Natalie J. Thornburg, and Kathy Shair for critical reading of the manuscript.
This work was supported by NIH grants CA 32979 and CA 19014-26.
REFERENCES
- 1.Allan, L. A., T. Duhig, M. Read, and M. Fried. 2000. The p21WAF1/CIP1 promoter is methylated in Rat-1 cells: stable restoration of p53-dependent p2WAF1/CIP1 expression after transfection of a genomic clone containing the p21WAF1/CIP1 gene. Mol. Cell. Biol. 20:1291-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cahir-McFarland, E. D., K. M. Izumi, and G. Mosialos. 1999. Epstein-Barr virus transformation: involvement of latent membrane protein 1-mediated activation of NF-κB. Oncogene 18:6959-6964. [DOI] [PubMed] [Google Scholar]
- 3.Cahir-McFarland, E. D., K. Carter, A. Rosenwald, J. M. Giltnane, S. E. Henrickson, L. M. Staudt, and E. Kieff. 2004. Role of NF-κB in cell survival and transcription of latent membrane protein 1-expressing or Epstein-Barr virus latency III-infected cells. J. Virol. 78:4108-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cahir-McFarland, E. D., D. M. Davidson, S. L. Schauer, J. Duong, and E. Kieff. 2000. NF-κB inhibition causes spontaneous apoptosis in Epstein-Barr virus-transformed lymphoblastoid cells. Proc. Natl. Acad. Sci. USA 97:6055-6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawson, C. W., A. B. Rickinson, and L. S. Young. 1990. Epstein-Barr virus latent membrane protein inhibits human epithelial cell differentiation. Nature 344:777-780. [DOI] [PubMed] [Google Scholar]
- 6.Dawson, C. W., G. Tramountanis, A. G. Eliopoulos, and L. S. Young. 2003. Epstein-Barr virus latent membrane protein 1 (LMP1) activates the phosphatidylinositol 3-kinase/Akt pathway to promote cell survival and induce actin filament remodeling. J. Biol. Chem. 278:3694-3704. [DOI] [PubMed] [Google Scholar]
- 7.Dirmeier, U., B. Neuhierl, E. Kilger, G. Reisbach, M. L. Sandberg, and W. Hammerschmidt. 2003. Latent membrane protein 1 is critical for efficient growth transformation of human B cells by Epstein-Barr virus. Cancer Res. 63:2982-2989. [PubMed] [Google Scholar]
- 8.Fries, K. L., W. E. Miller, and N. Raab-Traub. 1996. Epstein-Barr virus latent membrane protein 1 blocks p53-mediated apoptosis through the induction of the A20 gene. J. Virol. 70:8653-8659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatzivassiliou, E., W. E. Miller, N. Raab-Traub, E. Kieff, and G. Mosialos. 1998. A fusion of the EBV latent membrane protein-1 (LMP1) transmembrane domains to the CD40 cytoplasmic domain is similar to LMP1 in constitutive activation of epidermal growth factor receptor expression, nuclear factor-kappa B, and stress-activated protein kinase. J. Immunol. 160:1116-1121. [PubMed] [Google Scholar]
- 10.Johnson, D., M. C. Frame, and J. A. Wyke. 1998. Expression of the v-Src oncoprotein in fibroblasts disrupts normal regulation of the CDK inhibitor p27 and inhibits quiescence. Oncogene 16:2017-2028. [DOI] [PubMed] [Google Scholar]
- 11.Kang, Y., C. R. Chen, and J. Massague. 2003. A self-enabling TGFβ response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol. Cell 11:915-926. [DOI] [PubMed] [Google Scholar]
- 12.Katagiri, T., M. Imada, T. Yanai, T. Suda, N. Takahashi, and R. Kamijo. 2002. Identification of a BMP-responsive element in Id1, the gene for inhibition of myogenesis. Genes Cells 7:949-960. [DOI] [PubMed] [Google Scholar]
- 13.Kaye, K. M., K. M. Izumi, and E. Kieff. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc. Natl. Acad. Sci. USA 90:9150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaye, K. M., K. M. Izumi, H. Li, E. Johannsen, D. Davidson, R. Longnecker, and E. Kieff. 1999. An Epstein-Barr virus that expresses only the first 231 LMP1 amino acids efficiently initiates primary B-lymphocyte growth transformation. J. Virol. 73:10525-10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaye, K. M., K. M. Izumi, G. Mosialos, and E. Kieff. 1995. The Epstein-Barr virus LMP1 cytoplasmic carboxy terminus is essential for B-lymphocyte transformation; fibroblast cocultivation complements a critical function within the terminal 155 residues. J. Virol. 69:675-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kieff, E., and A. B. Rickinson. 2001. Epstein-Barr Virus and Its Replication, p. 2511-2573. In D. M. Knipe, P. M. Howley, D. E. Griffin, S. R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Strause (ed.), Field's Virology, 4th ed., vol. 2. Lippincott Williams & Wilkins Publishers, Philadelphia, Pa. [Google Scholar]
- 17.Knight, J. S., and E. S. Robertson. 2004. Epstein-Barr virus nuclear antigen 3C regulates cyclin A/p27 complexes and enhances cyclin A-dependent kinase activity. J. Virol. 78:1981-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kusano, S., and N. Raab-Traub. 2002. I-mfa domain proteins interact with Axin and affect its regulation of the Wnt and c-Jun N-terminal kinase signaling pathways. Mol. Cell. Biol. 22:6393-6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, H. M., Z. H. Zhuang, Q. Wang, J. C. Pang, X. H. Wang, H. L. Wong, H. C. Feng, D. Y. Jin, M. T. Ling, Y. C. Wong, A. G. Eliopoulos, L. S. Young, D. P. Huang, and S. W. Tsao. 2004. Epstein-Barr virus latent membrane protein 1 (LMP1) upregulates Id1 expression in nasopharyngeal epithelial cells. Oncogene 23:4488-4494. [DOI] [PubMed] [Google Scholar]
- 20.Lo, A. K., Y. Liu, X. H. Wang, D. P. Huang, P. W. Yuen, Y. C. Wong, and G. S. Tsao. 2003. Alterations of biologic properties and gene expression in nasopharyngeal epithelial cells by the Epstein-Barr virus-encoded latent membrane protein 1. Lab. Investig. 83:697-709. [DOI] [PubMed] [Google Scholar]
- 21.Lo, K. W., and D. P. Huang. 2002. Genetic and epigenetic changes in nasopharyngeal carcinoma. Semin. Cancer Biol. 12:451-462. [DOI] [PubMed] [Google Scholar]
- 22.Miller, W. E., H. S. Earp, and N. Raab-Traub. 1995. The Epstein-Barr virus latent membrane protein 1 induces expression of the epidermal growth factor receptor. J. Virol. 69:4390-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, W. E., G. Mosialos, E. Kieff, and N. Raab-Traub. 1997. Epstein-Barr virus LMP1 induction of the epidermal growth factor receptor is mediated through a TRAF signaling pathway distinct from NF-κB activation. J. Virol. 71:586-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murono, S., H. Inoue, T. Tanabe, I. Joab, T. Yoshizaki, M. Furukawa, and J. S. Pagano. 2001. Induction of cyclooxygenase-2 by Epstein-Barr virus latent membrane protein 1 is involved in vascular endothelial growth factor production in nasopharyngeal carcinoma cells. Proc. Natl. Acad. Sci. USA 98:6905-6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohtani, N., P. Brennan, S. Gaubatz, E. Sanij, P. Hertzog, E. Wolvetang, J. Ghysdael, M. Rowe, and E. Hara. 2003. Epstein-Barr virus LMP1 blocks p16INK4a-RB pathway by promoting nuclear export of E2F4/5. J. Cell Biol. 162:173-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohtani, N., Z. Zebedee, T. J. Huot, J. A. Stinson, M. Sugimoto, Y. Ohashi, A. D. Sharrocks, G. Peters, and E. Hara. 2001. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature 409:1067-1070. [DOI] [PubMed] [Google Scholar]
- 27.Prabhu, S., A. Ignatova, S. T. Park, and X. H. Sun. 1997. Regulation of the expression of cyclin-dependent kinase inhibitor p21 by E2A and Id proteins. Mol. Cell. Biol. 17:5888-5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr Virus, p. 2575-2627. In D. M. Knipe, P. M. Howley, D. E. Griffin, S. R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Strause (ed.), Field's Virology, 4th ed., vol. 2. Lippincott Williams & Wilkins Publishers, Philadelphia, Pa. [Google Scholar]
- 29.Riley, D., N. O. Carragher, M. C. Frame, and J. A. Wyke. 2001. The mechanism of cell cycle regulation by v-Src. Oncogene 20:5941-5950. [DOI] [PubMed] [Google Scholar]
- 30.Scholle, F., K. M. Bendt, and N. Raab-Traub. 2000. Epstein-Barr virus LMP2A transforms epithelial cells, inhibits cell differentiation, and activates Akt. J. Virol. 74:10681-10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sikder, H. A., M. K. Devlin, S. Dunlap, B. Ryu, and R. M. Alani. 2003. Id proteins in cell growth and tumorigenesis. Cancer Cell 3:525-530. [DOI] [PubMed] [Google Scholar]
- 32.Wang, D., D. Liebowitz, and E. Kieff. 1985. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell 43:831-840. [DOI] [PubMed] [Google Scholar]
- 33.Wong, T. S., D. L. Kwong, J. S. Sham, W. I. Wei, Y. L. Kwong, and A. P. Yuen. 2004. Quantitative plasma hypermethylated DNA markers of undifferentiated nasopharyngeal carcinoma. Clin. Cancer Res. 10:2401-2406. [DOI] [PubMed] [Google Scholar]
- 34.Yoshizaki, T., H. Sato, M. Furukawa, and J. S. Pagano. 1998. The expression of matrix metalloproteinase 9 is enhanced by Epstein-Barr virus latent membrane protein 1. Proc. Natl. Acad. Sci. USA 95:3621-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao, B., and C. E. Sample. 2000. Epstein-Barr virus nuclear antigen 3C activates the latent membrane protein 1 promoter in the presence of Epstein-Barr virus nuclear antigen 2 through sequences encompassing an Spi-1/Spi-B binding site. J. Virol. 74:5151-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng, W., H. Wang, L. Xue, Z. Zhang, and T. Tong. 2004. Regulation of cellular senescence and p16INK4a expression by Id1 and E47 proteins in human diploid fibroblast. J. Biol. Chem. 279:31524-31532. [DOI] [PubMed] [Google Scholar]