Abstract
p73 is a recently described member of the p53 family, and, like p53, it undergoes a number of posttranslational modifications. Here we show, by yeast two-hybrid screening, pull-down assays, and coimmunoprecipitation, that p73α, -β, and -γ bind to the protein inhibitor of activated STAT-1 (PIAS-1) and that this binding stabilizes p73. PIAS-1 also sumoylates p73α, although not the C-terminally truncated isoforms p73β and -γ, and this requires the RING finger domain of PIAS-1. The ΔNp73α isoform can also bind, and be sumoylated by, PIAS-1. PIAS-1-mediated sumoylation decreases p73 transcriptional activity on several target promoters, such as Bax. p73 is colocalized in the nucleus with PIAS-1, and sumoylated p73 is located exclusively in the nuclear matrix. PIAS-1 is expressed predominantly during S phase, and PIAS-1 overexpression reduces p73-mediated transcription of p21, with a reduction of cells in G1 and cell cycle reentry. Inhibition of endogenous PIAS-1 by RNA interference reduces the proportion of cells in S phase and induces G2 arrest. These data suggest that PIAS-1, acting partly through binding and sumoylation of p73, is an important component of the cell cycle machinery.
Signal transducer and activator of transcription (STAT) proteins are cytoplasmic transcription factors which, after phosphorylation, translocate to the nucleus, where they regulate expression of STAT-responsive genes (reviewed in references 4 and 14). STAT activity is partly regulated by a family of at least six related proteins, the protein inhibitors of activated STATs (PIAS). Thus, PIAS-1 inhibits DNA binding and gene activation by activated, tyrosine-phosphorylated STAT-1 (24). Another protein, PIASy, also inhibits the transcriptional activity of activated STAT-1, without, however, affecting STAT-1 DNA binding (25). A region located at the carboxy terminus of PIAS-1 directly interacts with STAT-1 dimers, though its interaction with phosphorylated and nonphosphorylated monomers is inhibited by the N-terminal domain (23). Recently, it has become clear that PIAS proteins can also regulate the activity of other transcription factors such as Smads (25), the mineralocorticoid and glucocorticoid receptors (40), and p53 (38).
At least part of the mechanism of transcriptional regulation by PIAS proteins depends on their E3 SUMO ligase activity, and PIAS-1 and SUMO-1 colocalize in nuclear granules (19). Thus, PIAS proteins sumoylate activated STAT-1, resulting in reduced STAT-1-mediated transcription (42). Moreover, the RING finger domain of PIAS-1 binds to the C terminus of the tumor suppressor p53 (33) and catalyzes its sumoylation (17), a modification which represses p53 activity on a reporter plasmid containing consensus p53 DNA binding sites (38). However, other studies using the p21 promoter have shown enhanced p53 transcriptional activity after interaction with PIAS-1, although this is independent of the RING finger domain and therefore of p53 sumoylation (26). Two homologues of p53, p63 and p73, have been described. The three proteins share a significant degree of sequence homology, particularly in the central sequence-specific DNA binding domain (DBD), the amino-terminal transactivation domain (TA), and the carboxyl-terminal oligomerization domain (OD) (for reviews, see references 15, 21, 22, 27, and 43). These structural similarities are paralleled by a certain degree of functional overlap; all three family members can regulate expression of the same genes through direct binding to p53 binding sites, although they also have other specific transcriptional targets. Transcriptional activation of the shared target genes leads to the induction of cell cycle arrest and apoptosis, cell growth-inhibitory responses that are thought to be critical for the tumor suppressor activities of p53. p73 is more complex than p53, since it is expressed as a number of alternatively spliced C-terminal isoforms whose expression can be initiated from two alternative promoters. Most of the splicing occurs at the 3′ end, in a part of the sequence that is not present in p53, and creates at least six proteins (α to η) that have different C termini (16, 5, 6, 41). In addition to these C-terminal variants, p73 is also expressed as N-terminally truncated ΔNp73 isoforms that lack the TA domain and which are derived from the use of an alternative promoter (P2) located in intron 3 (13). These ΔNp73 isoforms perform an important regulatory role, since they exert a dominant negative effect on p53 and TAp73 (12). Since PIAS-1 has been shown to sumoylate p53 and affect its transcriptional activity, we have studied here whether PIAS-1, similarly, binds to and sumoylates p73 and affects its stability, localization, and function.
MATERIALS AND METHODS
Cell cultures and transfections.
Human small lung adenocarcinoma cells (H1299) and human osteosarcoma cells (Saos-2) were grown in RPMI medium (GibcoBRL, Gaithersburg, Md.), and Hek293 cells were grown in Dulbecco's modified Eagle's medium (GibcoBRL). All media were supplemented with 10% (vol/vol) fetal bovine serum (GibcoBRL), and cells were cultured at 37°C in a humidified atmosphere of 5% (vol/vol) CO2 in air.
Transient transfections were performed with Lipofectamine 2000 reagents according to the protocol of the manufacturer (GibcoBRL). For protein expression, H1299 cells were transfected with the indicated amounts of plasmids and were harvested after 36 h. For luciferase assays, H1299 cells were transfected with the indicated amounts of the different plasmids and harvested after 24 h.
Plasmids.
Hemagglutinin (HA)-tagged versions of p73α, -β, -γ, and -ΔN have been previously described (5). To generate a p73-glutathione S-transferase (GST) fusion protein, p73 was amplified and subcloned into the BamHI site of the pGEX-6P1 expression vector (Amersham Pharmacia Biotech) in frame with the GST element. The p73 K627R mutant was generated by site-directed mutagenesis with the Quickchange Multi-Site Direct Mutagenesis kit (Stratagene) according to the manufacturer's protocol. The primers used for the mutagenesis were 5′-GCAAGCAGCCCATCAGGGAGGAGTTTCA-3′ and 5′-TCCTGCAAGTCCTTCGTCATGGGCTGCTTG-3′. p73 deletion mutants were generated by PCR and cloned into pcDNA-HA by using NheI and NotI sites. The primers used for amplification were as follows: for p73(1-319), 5′-ATGGCCCAGTCCACC-3′ sense primer and 5′-TCAGGAGCTCTCGTT-3′ antisense primer; for p73(318-363), 5′-ATGAGCTCCGCCAAGAAC-3′ sense primer and 5′-TCAGTGGATCTCG-3′ antisense primer; and for p73(318-444), 5′-ATGAGCTCCGCCAAGAAC-3′ sense primer and TCAGTTGGGTGTAGCTGCCG-3′ antisense primer.
The C350S PIAS-1 mutant, containing an inactive RING finger domain, was generated from the Flag-tagged PIAS-1 plasmid (pCMV5-Flag) by using the same kit described above. The primers used were 5′-CTGTCGGGCACTTACCAGTCCCCACCTTCA-3′ and 5′-ACTGAAGGTGGGAGTGGTAAGTGCCCGAC-3′. The truncated PIAS-1 forms PIAS-1/401-651 and PIAS-1/300-401 were cloned by PCR by using the following primers derived from the published PIAS-1 cDNA sequence (sequence data are available from EMBL/GenBank/DDBJ under accession no. NM 019663, GeneInfo identifier [GI] 9790154): 5′-GACTGTGATGAAATACAA-3′ for PIAS-1/401-651, 5′-GCACTGCAGAAATTGGA-3′ for PIAS-1/300-401, and a common antisense primer, 5′-TCAGTCCAATGA-3′ The isolated PCR fragments were then cloned in frame with a Flag tag into pcDNA-Flag by using the NheI and NotI unique restriction sites.
SUMO-1 was amplified by using the following primers derived from the published SUMO-1 cDNA sequence (sequence data are available from EMBL/GenBank/DDBJ under accession no. U67122, GI 1762972): 5′-GAACACCACATTACA-3′ for reverse transcription (RT), 5′-ATGTCTGACCAGGA 3′, and 5′-CTAAACTGTTGAATGACC-3′. The fragments were cloned into pcDNA-Flag by using the NheI and NotI unique restriction sites and (RT)-PCR.
Yeast two-hybrid screening.
Yeast two-hybrid screening of the Jurkat library in the Y190 yeast strain (Clontech, Palo Alto, Calif.) was performed with the Matchmaker Two-Hybrid System 2 (Clontech) according to the manufacturer's protocol. The complexity of the library was 3 × 106, and we screened the complexity of the library once. The lithium acetate method, described by Gietz and coworkers (8) was used to introduce DNA into yeast cells. The construct used for the screening contains a p73α fragment from amino acid (aa) 345 to 636 fused to the DNA binding domain of GAL4. The construct was generated by amplifying the cDNA of the above-mentioned plasmid by using the primers 5-′CGGATTTCAAGAAGCGCGG-3′ and 5-′AGCGGATCCTCAGTCGATCTCG-3′ and cloning into the EcoRI and BamHI sites of the pAS2-1 vector (Clontech).
Western blotting.
Samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride (PVDF) membranes by means of a semidry blotter. The membranes were blocked with 5% nonfat dry milk powder in Tris-buffered saline-0.05% Tween 20 for 1 h. Immunodetection was performed by incubating the membranes with the different primary antibodies diluted in blocking buffer for 2 h at room temperature or overnight at 4°C. After four washes with Tris-buffered saline-0.05% Tween 20, the membranes were incubated with secondary antibody conjugated with horseradish peroxidase for 1 h. After four washes, blots were developed with a ECL Plus detection kit (Amersham), and membranes were exposed to Kodak MS film.
The following antibodies were used: anti-HA polyclonal (Y-11; Santa Cruz), anti-Flag monoclonal (M2; Sigma), anti-PIAS-1 (C-20; Santa Cruz), antiactin (C-11; Santa Cruz), anti-β-tubulin (H-235; Santa Cruz), anti-lamin B (M-20; Santa Cruz), anti-p21 (H167; Santa Cruz), and anti-Sumo-1 (FL-101; Santa Cruz).
GST fusion proteins and pull-down assays.
GST and GST-p73 were transformed into Escherichia coli strain BL21(DE3). Cultures were grown overnight and used to inoculate fresh medium (1/100, vol/vol); the new cultures were subsequently grown for 2.5 h, induced with 1 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG), and grown for an additional 2 h. Cells were collected by centrifugation and resuspended in 2% of the original volume with 50 mM Tris-HCl (pH 8 at 4°C)-1 mM EDTA-5 mM 2-mercaptoethanol. The lysates were clarified by centrifugation, and GST fusion proteins were purified on glutathione-Sepharose beads (Amersham Biosciences, Inc.).
For pull-down assays, H1299 cells transiently transfected (as described above) with Flag-PIAS-1 or empty vector were lysed in lysis buffer (50 mM Tris HCl [pH 8], 150 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 10% glycerol, 100 mM NaF, and 1% Triton X-100) containing a protease and phosphatase inhibitor cocktail (Sigma) and centrifuged to precipitate cellular debris. A 1.5-mg quantity of total cellular proteins was first precleared with glutathione-Sepharose beads and then incubated for 2 h at 4°C with immobilized GST fusion proteins (25 μg). Unbound proteins were removed by washing four times with 0.1% Tween 20 in phosphate-buffered saline (PBS), and the precipitates were resolved by SDS-PAGE (8% polyacrylamide). The immunoblots were probed with antibodies as described above.
Immunoprecipitation.
H1299 cells were transiently transfected with 20 μg of total DNA of the indicated mammalian expression plasmids and harvested 48 h after transfection (11). The cells were then lysed in lysis buffer as described above. After preclearing for 1 h at 4°C, immunoprecipitation was performed by incubating 1.5 mg of whole-cell protein extracts with an anti-HA (Y-11) polyclonal antibody (Santa Cruz) or an anti-Flag (M2) monoclonal antibody (Sigma) with rocking at 4°C for 1 h. The immune complexes were collected by incubation with protein G-agarose (KPL, Guilford, Calif.) for 1 h and washed with Net-gel buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.25% gelatin, 0.1% NP-40). The beads were then resuspended in 5× SDS Laemmli sample buffer, subjected to SDS-PAGE (8% polyacrylamide) analysis, and electrotransferred onto PVDF membranes. The membranes were probed with primary antibodies as described above.
For endogenous immunoprecipitation experiments, Hek293 cells were lysed as described above and 5 mg of total protein was immunoprecipitated with PIAS-1 antibody (C20; Santa Cruz). As a control, the same amount of protein was immunoprecipitated with an antiactin antibody (C11; Santa Cruz).
Sumoylation assay.
H1299 cells were transiently transfected with 5 μg of total DNA of the indicated mammalian expression plasmids and harvested at 36 h after transfection. The cells were then lysed in 400 μl of 5× SDS Laemmli buffer and sonicated for 10 s, and the lysates were clarified by centrifugation and either used immediately or stored frozen at −80°C. Portions (25 μl) of the lysates were separated by SDS-PAGE (8% polyacrylamide) and electrotransferred onto PVDF membranes. The membranes were probed with primary antibodies as described above.
Measurement of p73 half-life. (i) Cycloheximide block.
Cycloheximide (20 μg/ml) was added to H1299 cells at 24 h after transfection with a total of 3 μg of the indicated plasmids at a p73/PIAS ratio of 1:2. p73 or tubulin protein levels were determined by collecting cells at the indicated time points and performing immunoblotting as described above. The relative amount of p73 protein was evaluated by densitometry and normalized to tubulin.
(ii) 35S pulse-chase.
H1299 cells were transfected as described above. At 48 h posttransfection, cells were starved for 30 min in Dulbecco's modified Eagle's medium with dialyzed serum and then labeled with 250μCi of Redivue PRO-MIX (l-35S in vitro cell labeling mix) per ml for 60 min. Unlabeled methionine and cysteine (1 mg/ml) were added, and cells were collected in radioimmunoprecipitation assay buffer (200 mM Tris [pH 8], 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 1% NP-40, and 0.2 mM EDTA) at the indicated times. Immunoprecipitation were performed with 150 μg of total protein lysate and 4 μl of anti-HA (Y-11) polyclonal antibody (Santa Cruz). Immunoprecipitates were washed six times in radioimmunoprecipitation assay buffer and six times in NET-gel, run in an 8% polyacrylamide gel, and detected by autoradiography.
Indirect immunofluorescence.
Saos-2 cells, grown on glass coverslips, were transiently transfected as described above with 1 μg of expression plasmid encoding Flag-tagged PIAS-1 and 1 μg of a plasmid encoding HA-p73α. The cells were washed twice with cold PBS, fixed in 4% (wt/vol) paraformaldehyde in PBS, and permeabilized with 0.1% Triton X-100 in PBS for 2 min. After permeabilization, cells were incubated for 30 min in 5% goat serum-PBS, incubated for 1 h with anti-HA (Y-11) polyclonal antibody (1:100 dilution), washed, and then incubated with the secondary antibody (goat anti-rabbit-Alexa 488; Molecular Probes) for 30 min. The cells were then incubated with anti-Flag (M2) monoclonal antibody, followed by the secondary antibody (goat anti-mouse-Alexa 568; Molecular Probes).
Confocal imaging was performed by using a 480-nm ion argon laser and a 542-nm helium-neon laser connected to a Nikon C1 microscope with a 60× numerical aperture 1.4 lens and analyzed with EZC1 software from Nikon.
Luciferase assay.
H1299 cells were transiently transfected in 96-well plates with the indicated combinations of plasmids (20 ng/well) together with a Bax-luciferase and p21-luciferase reporter plasmid (60 ng/well) and Renilla luciferase reporter (1.2 ng/well). The total amount of transfected DNA in each dish was kept constant by the addition of different amounts of empty vector. Thirty-six hours later, the luciferase activity was quantified by using a commercially available kit (dual-luciferase reporter assay system; Promega) with a Perkin-Elmer Victor2 luminometer.
Cell cycle detection.
The cell cycle was analyzed by flow cytometric evaluation of DNA content by the method of Nicoletti et al. (31). Cells were collected by trypsinization, pelleted at 800 × g for 10 min, and fixed in 70% ethanol. DNA content was evaluated by flow cytometry with propidium iodide (PI) staining. Twenty thousand events were evaluated by using the Cell Quest program (Becton Dickinson, San Jose, Calif.). Electronic gating (forward scatter area versus forward scatter height) was used, when appropriate, to eliminate cell aggregates and to identify the transfected population. The same method was used to evaluate the cell cycle by using ModFit LT software (Verity Software).
ChIP.
Saos-2 cells (1.5 × 106) were transfected with pcDNA 3.1 HA-p73TA vector alone or in association with Flag-PIAS-1 or with both Flag-PIAS-1 and Flag-SUMO-1. Cells were cross-linked, using 1% formaldehyde-5 mM HEPES-KOH (pH 8)-0.1 mM EDTA-10 mM NaCl, for 40 min at 24 h after transfection. The cross-linking reaction was stopped by incubating the cells with 0.125 M glycine for 5 min at room temperature. After being washed with ice-cold PBS, the cells were harvested in lysis buffer (50 mM HEPES-KOH [pH 8], 1 mM EDTA, 140 mM NaCl, 25% glycerol, 0.5% NP-40, 0.25% Triton X-100) containing protease inhibitors. Nuclei were collected by centrifugation and were resuspended in wash buffer (10 mM Tris-HCl [pH 8], 1 mM EDTA, 200 mM NaCl) containing protease inhibitors. After centrifugation, the nuclei were resuspended in 1.8 ml of chromatin immunoprecipitation (ChIP) dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 0.0167 M Tris-HCl, 0.167 M NaCl) with 200 μl of SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl [pH 8.1]).
Cell lysates were sonicated to obtain chromatin fragments of ∼700 bp, and, following centrifugation at 12,000 × g for 20 min to remove cell debris, 100 μg of total protein was precleared with 100 μl of protein A-agarose-salmon sperm DNA (Upstate; catalog no. 16-157) and 2 μg of mouse immunoglobulin G1 κ (BD) for 2 h at 4°C. The precleared extracts were incubated with 2 μg of mouse anti-HA antibody (Babco). The control was incubated with 2 μg of mouse anti-K5 antibody (Santa Cruz) overnight at 4°C, followed by incubation with protein A-agarose-salmon sperm DNA (60 μl) for 1 h 30 min at 4°C. The immunocomplexes were washed twice with low-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, 0.15 M NaCl), five times with high-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, 0.5 M NaCl), once with LiCl salt wash buffer (1 mM EDTA, 10 mM Tris-HCl, 0.25 M LiCl, 1% NP-40, 1% deoxycholate), and twice with Tris-EDTA buffer. The precipitates were extracted twice with 250 μl of immunoprecipitation elution buffer (1% SDS, 0.1 M NaHCO3). The total eluates (500 μl) were pooled by adding 20 μl of 5 M NaCl and incubated at 65°C for at least 6 h to reverse the formaldehyde cross-linking. DNA fragments were purified by phenol-chloroform extraction. In order to compare the levels of expression of the transfected cDNA, parallel green fluorescent protein (GFP) transfections were performed and expression was normalized by Western blotting. DNA samples were then analyzed with 38 cycles of PCR to amplify p21 promoter sequences. The primers used were 5′-CCTCTTCGGCCGGTGGAC-3′ (forward) and 5′-CCGTTTTCGACCCTGAGAG-3′ (reverse).
Colony suppression assays.
H1299 cells were plated in 60-mm-diameter plates and cotransfected with the indicated pcDNA-HA- and pcDNA-Flag-derived vectors together with a pBabe-Pure vector by using the calcium phosphate method (calcium phosphate transfection kit; Invitrogen) according to the manufacturer's protocol. The ratio between the pcDNA-HA p73 expression plasmid (1 μg of DNA/transfection) and pcDNA-Flag PIAS-1 and pcDNA-Flag SUMO-1 (2 μg of DNA/transfection) was 1:2:2. Equal amounts of pBabe-Puro (0.5 μg/transfection) were added to each transfection mixture. At 48 h after transfection, cells were placed in selection medium containing 2 μg of puromycin (Sigma) per ml. Two weeks later, colonies were fixed in methanol, stained with crystal violet, washed with water, counted, and photographed.
Cell cycle synchronization.
To generate synchronized populations, Hek293 cells were arrested at the G1/S boundary by a double thymidine block. The first thymidine block was imposed by treatment with fresh medium containing 2 mM thymidine (Sigma) for 16 h. Cells were washed in PBS, and fresh medium containing 24 μM deoxycytidine (Sigma) was added and left for 9 h. Following the 9-h release period, a second thymidine block was imposed by addition of thymidine to a final concentration of 2 mM. After 16 h, cells were released from the second thymidine block by removal of the thymidine-containing medium. Cells were collected at the indicated time points after the second thymidine block.
To generate a population synchronized in G2, nocodazole (Sigma) was added to a final concentration of 0.2 μg/ml in Hek293 cell cultures and left for 16 h. The cells were then washed, grown in normal medium, and harvested and analyzed at the times indicated.
Growth arrest was obtained in Saos-2 cells by serum starvation for 48 h, after which the medium was replaced with serum-containing medium and cells were harvested at 0, 3, 6, and 24 h.
RT-PCR.
Cell pellets were collected and RNA was extracted by using Trizol. First-strand cDNA was synthesized by using poly(dT) primers. PCR was performed with the PIAS-1 primers 5′-CCACGCCTTCCTGCTGTAGA-3′ (sense primer) and 5′-TATCACACAGGCAGTCTTAGAT-3′ (antisense primer) or the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) primers 5′-GGCTGAGAACGGGAAGCTTGTCTAT-3′ (sense primer) and 5′-CAGCCTTCTCCATGGTGGTGAAGA-3′ (antisense primer), using SYBR green fluorescence for real-time PCR analysis on an Opticon (MJ Research) thermocycler. The program used was 40 cycles of 95°C for 5 min, 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s. An aliquot of each mixture was also run on a 1.5% agarose gel.
PIAS-1 small interfering RNA (siRNA).
Saos-2 cells were electroporated with 20 μl of a 20 μM solution of 21-nucleotide RNA, synthesized by Qiagen, using a Bio-Rad electroporation apparatus. The PIAS-1 target sequence was AAGGTCATTCTAGAGCTTTA, and the scrambled sequence was AATTCTCCGAACGTGTCACGT. Cells were collected after 48 h, fixed, stained with PI, and analyzed for DNA content by flow cytometry as described above. An aliquot of the cells was lysed and subjected to Western blotting for PIAS-1 detection as described above.
RESULTS
PIAS-1 binds to p73.
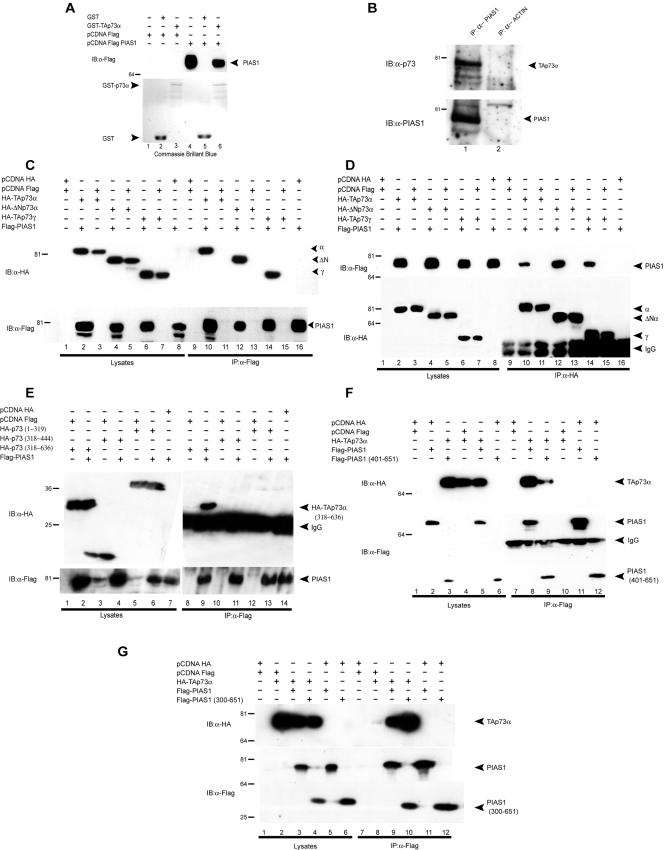
In order to identify proteins that are able to bind p73 and regulate p73 function, we performed a yeast two-hybrid screening with a C-terminal fragment of p73α as bait (Fig. 1A). The fragment from aa 345 to 636 contained the OD and the sterile alpha motif (SAM) domain (Fig. 1A). Seven independent partially overlapping clones contained the 3′ end sequence of PIAS-1 (Fig. 1B).
FIG. 1.
p73 colocalizes with PIAS-1. (A) Schematic representation of the p73 constructs used in this study. Boxes indicate the different functional domains: transactivation domain (TA) in red, DNA binding domain (DBD) in yellow, oligomerization domain (OD) in green, and sterile alpha motif (SAM) in pink. The fragment of p73 used as bait in the yeast two-hybrid screening is also shown. The asterisk indicates the position of lysine 627, where sumoylation occurs. (B) Schematic representation of the PIAS-1 constructs used in this study. Boxes indicate the different functional domains: SAF-AB Acinus and Pias domain in orange, PINIT domain (essential for nuclear retention) (7) in blue, zinc finger-like RING domain in green, and serine/threonine rich region (34) in purple. The asterisk indicates the position of the catalytic cysteine at position 350. (C) p73 and PIAS-1 colocalize in the nucleus when overexpressed in Saos-2 cells. An HA-TAp73α expression vector and a Flag-PIAS-1 expression vector were transiently transfected in Saos-2 cells alone or in combination. At 24 h after transfection, cells were fixed and stained with HA polyclonal (Y-11) and anti-Flag monoclonal (M2) antibodies.
The interaction between p73 and PIAS-1 was initially confirmed by yeast two-hybrid screening with PIAS-1 both as bait and as prey (data not shown). In addition, when overexpressed in Saos-2 cells, TAp73α and PIAS-1 colocalized in the nucleus (Fig. 1C). Moreover, addition of PIAS-1 changed the TAp73α distribution within the nucleus from diffuse to more punctate (Fig. 1C).
To further confirm direct interaction, we performed a GST pull-down experiment with recombinant GST-TAp73α and overexpressed Flag-PIAS-1. As shown in Fig. 2A, full-length TAp73α specifically interacted in vitro with PIAS-1. Figure 2B shows that endogenous TAp73α can also be coimmunoprecipitated with endogenous PIAS-1.
FIG. 2.
p73 binds PIAS-1. (A) GST pull-down assays showing p73 and PIAS-1 interaction. H1299 cells were transiently transfected with 5 μg of Flag-PIAS-1 or empty vector (pcDNA-Flag), and 1.5 mg of cell lysates was incubated with GST alone or with a GST fusion protein containing full-length p73 (GST-p73α). The retained proteins were immunoblotted (IB) with anti-Flag (M2) monoclonal antibody (upper panel). Lanes 1 and 4 contain aliquots of unprocessed lysates. A Coomassie brilliant blue-stained replica gel is also shown (lower panel). (B) Coimmunoprecipitation of endogenous p73 with endogenous PIAS-1. Hek293 cell extracts were precleared with protein G-agarose and immunoprecipitated (IP) with anti-PIAS-1 antibody (C20; Santa Cruz). The immune complexes were subjected to Western blot analysis with anti-rabbit polyclonal anti-p73 antibody (upper panel). The same blot was reprobed with anti-PIAS-1 antibody (C20; Santa Cruz) (lower panel). (C) Coimmunoprecipitation of PIAS-1 and p73. HA-p73α, HA-ΔNp73α, and HA-TAp73γ mammalian expression vectors were transiently transfected in H1299 cells together with the Flag-PIAS-1 expression vector. Cell extracts were immunoprecipitated with anti-Flag antibody and subjected to Western blot analysis (lanes 9 to 16) with anti-HA antibody (upper panel). The same blot was reprobed with anti-Flag antibody (lower panel). Aliquots of total cell extracts from unprocessed cells were also loaded on the gel (lanes 1 to 8). (D) Coimmunoprecipitation of p73 and PIAS-1. HA-p73α, HA-ΔNp73α, and HA-TAp73γ expression vectors were transiently transfected in H1299 cells together with the Flag-PIAS-1 expression vector. Cell extracts were immunoprecipitated with anti-HA antibody and subjected to Western blot analysis (lanes 9 to 16) with anti-Flag antibody (upper panel). The same blot was reprobed with anti-HA antibody (lower panel). Aliquots of total cell extracts from unprocessed cells were also loaded on the gel (lanes 1 to 8). (E) The p73(318-636) fragment is still capable of binding p73. HA-p73(1-319), HA-p73(318-444), and HA-p73(318-636) expression vectors were transiently transfected in H1299 cells together with the Flag-PIAS-1 expression vector. Cell extracts were immunoprecipitated with anti-Flag antibody and subjected to Western blot analysis with anti-HA antibody (upper panel). The same blot was reprobed with anti-Flag antibody (lower panel). Aliquots of total cell extracts from unprocessed cells were also loaded on the gel (lanes 1 to 7). (F) The PIAS-1(401-651) fragment is still capable of binding p73. Full-length Flag-PIAS-1 and Flag-PIAS-1(401-651) expression vectors were transfected in H1299 cells together with the HA-p73α expression vector. Cell extracts were immunoprecipitated with anti-Flag antibody and subjected to Western blot analysis with anti-HA antibody (upper panel). The same blot was reprobed with anti-Flag antibody (lower panel). Aliquots of total cell extracts from unprocessed cells (100 μg/lane) were also loaded on the gel (lanes 1 to 6). (G) The PIAS-1(300-651) fragment is still capable of binding p73. Full-length Flag-PIAS-1 and Flag-PIAS-1(300-651) expression vectors were transfected in H1299 cells together with the HA-p73α expression vector. Cell extracts were immunoprecipitated with anti-Flag antibody and subjected to Western blot analysis with anti-HA antibody (upper panel). The same blot was reprobed with anti-Flag antibody (lower panel). Aliquots of total cell extracts from unprocessed cells (100 μg/lane) were also loaded on the gel (lanes 1 to 6).
To further characterize the interaction between p73 and PIAS-1, we overexpressed different HA-tagged p73 isoforms, including TAp73α, TAp73γ, and ΔNp73α (Fig. 2C and D) and Flag-tagged PIAS-1 in H1299 cells and performed coimmunoprecipitation experiments (Fig. 2C and D). Experiments were performed with immunoprecipitation with anti-Flag (PIAS-1) antibodies and staining with anti-HA antibodies (Fig. 2C) and in the reverse combination (Fig. 2D). As shown in Fig. 2C and D, full-length PIAS-1 interacted with all of the different p73 isoforms tested. These isoforms differ in their C and N termini but have the DBD and OD in common (Fig. 1A), suggesting that the binding site for PIAS-1 is contained in this region. Since the DBD is missing in the construct we used for the yeast two-hybrid experiments, our results demonstrate that the region of p73 required for the binding to PIAS-1 is the fragment from aa 345 to 450 that contains the OD (Fig. 1A). This is the only region common to all of the constructs used so far in our experiments. To confirm that this region is important, we performed coimmunoprecipitation experiments with the following additional deletion constructs: aa 1 to 319 (containing TA and DBD), aa 318 to 444 (containing the oligomerization domain alone), and aa 318 to 636 (containing the entire C terminus, including the oligomerization domain) (Fig. 2E). Our results show that PIAS-1 interacts only with the aa 318 to 636 construct. These data together with the data on the naturally occurring isomers β and γ show that the interaction requires a sequence between aa 318 and 389 that spans the oligomerization domain. The lack of interaction of the oligomerization domain alone (aa 318 to 444) is probably due to misfolding of this short sequence.
In order to map the region of PIAS-1 responsible for the binding to p73, we designed a construct expressing a region of PIAS-1 from aa 401 to 651, corresponding to the region of overlap of all of the clones that we obtained in the screening. As shown in Fig. 2F, PIAS-1(401-651) was still capable of interacting with p73 (lane 9). This interaction, however, appears to be weaker than the interaction of p73 with the full-length PIAS-1, suggesting that other parts of the molecule can contribute to the interaction. In particular, it has recently been shown that PIAS-1 interaction with p53 requires the N-terminal domain (33). We therefore generated a mutant of PIAS-1 lacking the first 300 aa [PIAS-1(300-651)] and performed coimmunoprecipitation experiments. Our results (Fig. 2G) show that PIAS-1(300-651) interacts with TAp73α, with an intensity comparable to that of the full-length PIAS-1, suggesting that the N-terminal region is not required for the interaction with p73. However, additional deletion mutants need to be tested to definitely exclude a role of the N terminus of PIAS-1 in the interaction.
Together, the above-mentioned data clearly demonstrate, for the first time, that p73 physically interacts with PIAS-1 and that the binding occurs between a C-terminal region of p73, likely containing the OD domain but not the SAM domain, and the region of PIAS-1 between aa 401 and 651.
PIAS-1 sumoylates p73.
Since it has been shown that PIAS-1 binds to p53 and promotes its sumoylation, probably functioning as a sumo E3 ligase (17, 38), we tested the possibility that p73 was also sumoylated by PIAS-1. Coexpression of TAp73α and SUMO-1 resulted in the sumoylation of TAp73α, as shown by the up-shifted band in Fig. 3B (lane 4). Overexpression of PIAS-1 under the same experimental conditions resulted in an increase in the sumoylation of TAp73α (Fig. 3A, lane 3). While TAp73α was clearly sumoylated, TAp73β and TAp73γ (which have completely different C termini but which also bind PIAS-1 [Fig. 2C and D]) were not sumoylated (Fig. 3A). ΔNp73α, sharing the same C terminus with TAp73α but lacking part of the N terminus, was still sumoylated (Fig. 3B), suggesting that the sumoylated residue resides in the carboxy-terminal part of p73α.
FIG. 3.
PIAS-1 sumoylates TAp73. (A) HA-TAp73α, HA-TAp73β, and HA-p73γ expression vectors were transiently transfected in H1299 cells together with Flag-SUMO-1 and full-length Flag-PIAS-1 or Flag-PIAS-1(401-651) expression vectors. Cells were subjected to Western blot analysis with anti-HA antibody (upper panel). The same blot was reprobed with anti-PIAS-1 antibody (second panel) and with an antiactin antibody (lower panel). The third panel shows a Western blot performed on proteins run on 15% polyacrylamide to detect the much shorter PIAS-1(401-651) mutant. Western blotting was performed with anti-PIAS-1 antibody as described above. Supershifted sumoylation bands (asterisks)are visible only when SUMO-1 is transfected into cells (lanes 3 and 4). The cotransfection of PIAS-1 produces a large increase of the sumoylated band (lane 3). TAp73β (lanes 5 to 8) and TAp73γ (lanes 9 to 12) are not sumoylated. The truncated form of PIAS-1, PIAS-1(401-651), is not capable of promoting p73 sumoylation. IB, immunoblot. (B) ΔNp73α is also sumoylated by PIAS-1. H1299 cells were transiently transfected with HA-TAp73α and HA-ΔNp73α expression vectors in combination with Flag-SUMO-1 and Flag-PIAS-1 expression vectors. Western blot analysis was performed as described for panel A. (C) H1299 cells were transiently transfected with HA-TAp73α mammalian expression vectors in combination with Flag-SUMO-1 and Flag-PIAS-1 expression vectors. Cell extracts were immunoprecipitated (IP) with anti-HA antibody. The immune complexes (lanes 5 to 8) were subjected to Western blot analysis with anti-SUMO antibody (FL-101; Santa Cruz) (upper panel). The same blot was reprobed with anti-HA antibody (lower panel). Aliquots of total cell extracts from unprocessed cells were also loaded on the gel (lanes 1 to 4).
To confirm that the higher-molecular-weight bands observed after coexpression of p73, PIAS-1, and SUMO-1 indeed represented sumoylated p73, we immunoprecipitated TAp73α and probed the blots with anti-SUMO antibodies (Fig. 3C and unpublished data). The higher-molecular-weight form observed was stained with anti-SUMO.
Sumoylation of TAp73α has previously been described to occur at lysine 627 (29). Our data are consistent with this report, since p73β and p73γ lack this residue and were not sumoylated by PIAS-1 (Fig. 3A). As expected, ΔNp73α, which differs only at the N terminus but which contains the C-terminal sumoylation site, was also sumoylated by PIAS-1 (Fig. 3B).
To confirm that PIAS-1 is capable of selectively sumoylating p73 only at lysine 627, we mutated this residue to arginine [HA-TAp73α(K627R)]. As shown in Fig. 4A, the TAp73α(K627R) mutant was not sumoylated even when both PIAS-1 and SUMO-1 were overexpressed. Figure 4B shows that the TAp73α(K627R) mutant was still capable of binding to PIAS-1 and therefore that the lack of sumoylation depends solely on the absence of lysine 627, confirming that this is the only p73 residue sumoylated by PIAS-1.
FIG. 4.
p73 is sumoylated only at lysine 627. (A) H1299 cells were transiently transfected with either HA-TAp73α or its mutant HA-TAp73α(K627R) in combination with Flag-SUMO-1 and Flag-PIAS-1 expression vectors. Cell lysates were subjected to Western blot analysis performed with anti-HA antibody (upper panel). The same blot was reprobed with anti-PIAS-1 antibody (middle panel) and with an antiactin antibody (lower panel). The p73 mutant HA-TAp73α(K627R) cannot be sumoylated regardless of the coexpression of SUMO-1 and/or PIAS-1 (lanes 6 to 8). This mutant p73 is still capable of binding PIAS-1. Asterisks indicate sumoylated p73. IB, immunoblot. (B) HA-TAp73α and HA-TAp73α(K627R) expression vectors were transfected in H1299 cells together with the Flag-PIAS-1 expression vector. Cell extracts were immunoprecipitated (IP) with anti-Flag antibody and subjected to Western blot analysis performed with anti-HA antibody (upper panel). The same blot was reprobed with anti-HA antibody (lower panel). Aliquots of total cell extracts from unprocessed cells were also loaded on the gels (lanes 1 to 6). (C) The PIAS-1 mutant C350S is no longer capable of sumoylating p73. H1299 cells were transfected with HA-p73α in combination with Flag-SUMO-1 and wild-type Flag-PIAS-1 or mutant Flag-PIAS-1(C350S). Cells were subjected to Western-blot analysis performed with anti-HA antibody (upper panel). The same blot was reprobed with anti-PIAS-1 antibody (middle panel) and with an antiactin antibody (lower panel). (D) The PIAS-1 mutant (C350S) is still capable of binding p73. HA-p73α expression vectors were transiently transfected in H1299 cells, together with mutant Flag-PIAS-1(C350S). Cell extracts were immunoprecipitated with anti-Flag antibody and were subjected to immunoblot analysis with anti-HA antibody (upper panel). The same blot was reprobed with anti-Flag antibody (lower panel). Aliquots of total cell extract from unprocessed cells were also loaded on the gel (lanes 1 to 4).
PIAS-1 contains a central RING finger-like motif, called the SP-RING domain (17), containing two conserved cysteine residues, the latter of which is essential for ligase activity. Removal or mutation of the RING finger domain results in the loss of the ability of PIAS-1 to sumoylate p73. As shown in Fig. 3A (lanes 14 and 15), PIAS-1(401-651) lacking the RING finger domain, although still capable of binding p73 (Fig. 2F), has lost its ability to sumoylate p73. Similarly a PIAS-1 RING finger point mutant (C350S) retains its ability to bind p73 (Fig. 4D) but has completely lost its ability to sumoylate p73 (Fig. 4C). The faint high-molecular-weight band corresponding to sumoylated p73 observed in the presence of PIAS-1(C350S) is due to the activity of endogenous PIAS-1 and is visible in other blots when SUMO-1 but not PIAS-1 is overexpressed (Fig. 3B, lane 4, and 4A, lane 5).
PIAS-1 stabilizes the p73 protein.
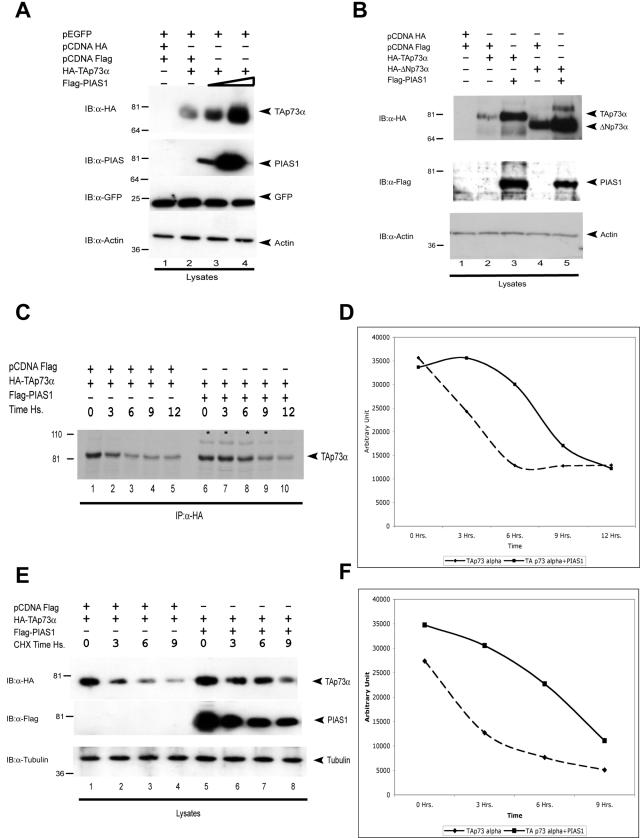
We noticed that in all of the experiments presented above, overexpression of PIAS-1 always resulted in increased levels of p73, suggesting that PIAS-1 affected TAp73α stability. In order to confirm that PIAS-1 stabilizes TAp73α, we overexpressed TAp73α with increasing amounts of PIAS-1. As shown in Fig. 5A, overexpression of increasing amounts of PIAS-1 resulted in increasing levels of TAp73α. As expected, like TAp73α, ΔNp73α, which is also capable of binding PIAS-1, was stabilized by overexpression of PIAS-1 (Fig. 5B). The half-life of TAp73α, measured by pulse-chase with 35S-labeled Met and Cys (Fig. 5C and D) and cycloheximide blockade (Fig. 5E and F) experiments, showed an increase from approximately 4 to 9 h in the presence of PIAS-1.
FIG. 5.
PIAS-1 stabilizes the p73 protein. (A) H1299 cells were transfected with HA-TAp73α alone or with increasing amounts of Flag-PIAS-1, and all samples were also cotransfected with equal amounts of a GFP-expressing plasmid. At 24 h after transfection, cell extracts were lysed andthe levels of p73 in whole-cell extracts were determined by immunoblotting (IB) with anti-HA antibody (upper panel). The same blot was reprobed with anti-Flag antibody (second panel) and with an antiactin antibody (lower panel). To demonstrate that there is no effect of PIAS-1 on unrelated proteins or on the promoter driving expression of p73, the blot was reprobed with anti-GFP antibody. (B) ΔNp73 is also stabilized by PIAS-1 coexpression. H1299 cells were transfected with either HA-TAp73α or HA-ΔNp73α together with PIAS-1. At 24 h after transfection, cell extracts were lysed and analyzed as described for panel A. (C) 35S pulse-chase. H1299 cells were transfected with the indicated plasmids. At 48 h posttransfection cells were labeled with l-35S in vitro cell labeling mix. Cells were collected at the indicated time points. Immunoprecipitation (IP) was performed with anti-HA (Y-11) polyclonal antibody (Santa Cruz), and then samples were run on a polyacrylamide gel and proteins were detected by autoradiography. (D) The relative amount of p73 protein was evaluated by densitometry. (E) Evaluation of TAp73 protein half-life. Cycloheximide was added to H1299 cells at 24 h after transfection with the indicated plasmids. p73 or tubulin protein levels were determined by collecting cells at the indicated time points and performing immunoblotting as described above. (F) The relative amount of p73 protein was evaluated by densitometry and normalized to tubulin.
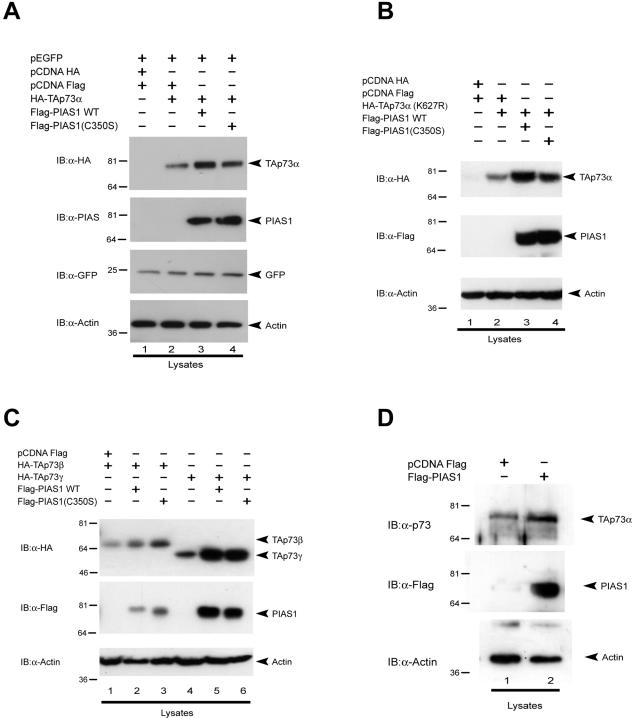
To test whether p73 stabilization requires its sumoylation, we performed cotransfection experiments similar to those described above with PIAS-1 and p73 mutants that affect sumoylation. As shown in Fig. 6A, the PIAS-1(C350S) mutant also stabilized TAp73α levels. Similarly, as shown in Fig. 6B, the TAp73α(K627R) mutant, which cannot be sumoylated, was stabilized by coexpression of PIAS-1. In addition, other naturally occurring isoforms (β and γ) were also stabilized by wild-type and mutant PIAS-1 (Fig. 6C). In order to demonstrate that this was not a nonspecific effect of PIAS-1 on any unrelated protein or on the promoter driving the expression of p73, we cotransfected the cells with a plasmid expressing GFP under the control of the same promoter and showed that overexpression of PIAS-1 did not lead to its stabilization (Fig. 5A and 6A). This also demonstrates that the transfection efficiency is comparable in all samples. In addition, we showed that overexpression of PIAS-1 resulted in stabilization of endogenous TAp73α (Fig. 6D), while reduction of endogenous PIAS-1 levels by siRNA resulted in decreased stability of overexpressed TAp73α (unpublished data). Finally, we showed that the effect pf PIAS-1 on p73 staining was independent of sumoylation, since a dotted nuclear p73 staining was also obtained when the TAp73α(K627R) mutant was coexpressed with PIAS-1 or when wild-type p73 was coexpressed with the PIAS-1(C350S) mutant (unpublished data).
FIG. 6.
Sumoylation is not required for p73 stabilization. (A) The PIAS-1 (C350S) mutant also stabilizes p73 protein. H1299 cells were transfected with HA-TAp73α together with either wild-type (WT) Flag-PIAS-1 or Flag-PIAS-1(C350S). At 24 h after transfection, cell extracts were lysed and the levels of p73 in whole-cell extracts were determined by immunoblotting (IB) with anti-HA antibody (upper panel). The same blot was reprobed with anti-Flag antibody (middle panel) and with an antiactin antibody to show expression of PIAS-1 and equal loading. (B) The p73 mutant (K627R), which cannot be sumoylated, is also stabilized by the interaction with PIAS-1. H1299 cells were transfected with the HA-TAp73α(K627R) mutant alone or together with wild-type or mutated PIAS-1. At 24 h after transfection, cell extracts were lysed and the levels of p73 in whole-cell extracts were determined by immunoblotting with anti-HA antibody (upper panel). The same blot was reprobed with anti-Flag antibody (middle panel) and with an antiactin antibody (lower panel). (C) Similarly, p73 isoforms that cannot be sumoylated (β and γ) are stabilized by the interaction with PIAS-1. H1299 cells were transfected with HA-TAp73β or HA-TAp73γ alone or together with wild-type PIAS-1. At 24 h after transfection, cell extracts were lysed and the levels of p73 in whole-cell extracts were determined by immunoblotting with anti-HA antibody (upper panel). The same blot was reprobed with anti-Flag antibody (middle panel) and with an antiactin antibody (lower panel). (D) PIAS-1 stabilizes endogenous p73. Hek293 cells were transfected with Flag-PIAS-1. At 24 h after transfection, cell extracts were lysed and the levels of p73 in whole-cell extracts were determined by immunoblotting with rabbit polyclonal anti-p73 antibody (upper panel). The same blot was reprobed with anti-Flag antibody (middle panel) and with an antiactin antibody (lower panel).
Together, these results demonstrate that the binding of PIAS-1 to p73 is sufficient to stabilize p73 protein levels and that this effect does not require p73 sumoylation.
Sumoylation results in p73 functional inactivation.
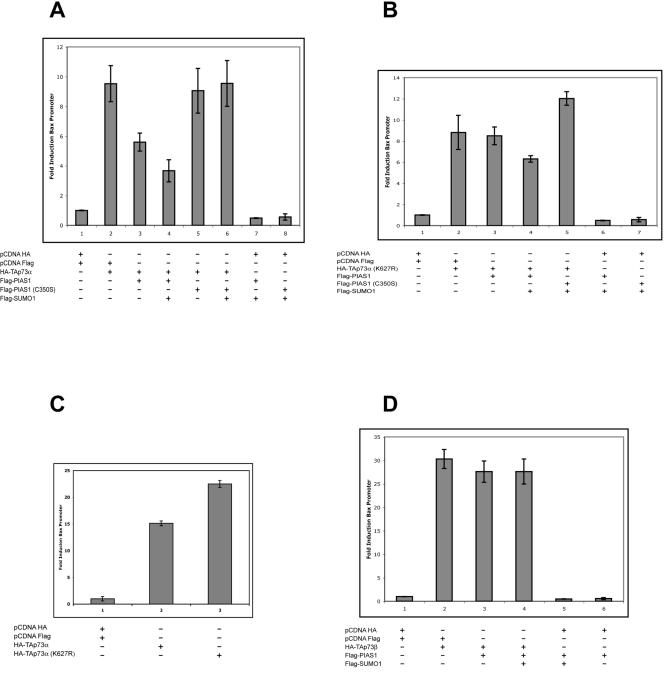
We next tested whether sumoylation of p73 results in a modification of its transcriptional activity. To this end, we cotransfected the Bax promoter cloned upstream of the luciferase gene (Bax-luc) together with different combinations of p73 isoforms, PIAS-1 and SUMO-1, and some of their mutants. As shown in Fig. 7A, cotransfection of PIAS-1 or PIAS-1 and SUMO-1 with TAp73α resulted in a decrease in the luciferase activity. Consistently, cotransfection with the PIAS-1(C350S) mutant with or without SUMO-1 had no effect on TAp73α transcriptional activity. As expected, the transcription mediated by the TAp73α(K627R) mutant, which cannot be sumoylated, was not affected by coexpression of PIAS-1 with or without SUMO-1 (Fig. 7B). The slight reduction of transcriptional activity seen with cotransfection of PIAS-1 and SUMO-1 is probably because of the reduction of the basal activity of the promoter (compare bars 1 and 6) due to an effect on endogenous p73 levels or other factors regulating the Bax promoter. Figure 7C shows that TAp73α(K627R) has a slightly but consistently higher transcriptional activity than wild-type TAp73α, consistent with the ability of endogenous PIAS-1 to negatively regulate the activity of p73. Similarly, PIAS-1 expression has no effect on TAp73β transcriptional activity (Fig. 7D). This may also explain why the beta isoform is often described as more active than the alpha isoform (5, 16, 28). Similar results were obtained with other p53-responsive promoters, namely, p21 and MDM2 (see below and data not shown).
FIG. 7.
PIAS-1-mediated sumoylation of p73α inhibits its transcriptional activity. H1299 cells were seeded in 96-well plates, transfected at 50 to 60% confluence with different combinations of expression plasmids together with a Bax-luciferase reporter plasmid, and harvested after 24 h. (A) p73α increases Bax reporter activity (bar 2), and this is reduced by cotransfection with PIAS-1 (bar 3) and, more markedly, by additional cotransfection with SUMO-1 (bar 4). The PIAS-1 mutant C350S, which can bind to p73 but cannot mediate its sumoylation, does not reduce p73α transcriptional activity (bars 5 and 6). (B) The p73α mutant K627R, which cannot be sumoylated, increases Bax reporter activity (bar 2), but this is unaffected by PIAS-1 (bar 3) and is only slightly reduced by cotransfection with PIAS-1 and SUMO-1 (bar 4). The PIAS-1 mutant C350S, which cannot mediate sumoylation, is also inactive on K627R (bar 5). (C) TAp73α(K627R) is constitutively more active than the wild type in activating p73-responsive promoters. (D) p73β increases Bax reporter activity (bar 2), which is unaffected by PIAS-1 alone (bar 3) or PIAS-1 plus SUMO-1 (bar 4). Error bars indicate standard deviations.
Together these results demonstrate that the final effect of p73 sumoylation is a reduction of its transcriptional activity.
Sumoylated p73 is associated with the nuclear matrix.
We next investigated whether sumoylation of p73 results in a change of its subcellular localization. As shown in Fig. 1B, p73 is localized to the nucleus with or without overexpression of PIAS-1. Since it has recently been shown that p73 can be associated with the nuclear matrix (2) and since sumoylation has been associated with protein compartmentalization (reviewed in reference 39), we investigated whether sumoylation of p73 results in changes to its distribution within the nuclei. To this end, H1299 cells were transfected with HA-p73α expression vectors alone or with Flag-SUMO-1 and Flag-PIAS-1 expression vectors. After 24 h cells were lysed by using a high-salt nuclear matrix extraction procedure to separate various nuclear fractions (2). Our results show that p73 was detected in nucleocytoplasm, high-salt wash, and nuclear matrix fractions. Interestingly, the sumoylated form of p73 was detected only in the nuclear matrix fraction (data not shown).
PIAS-1 expression determines cell cycle reentry.
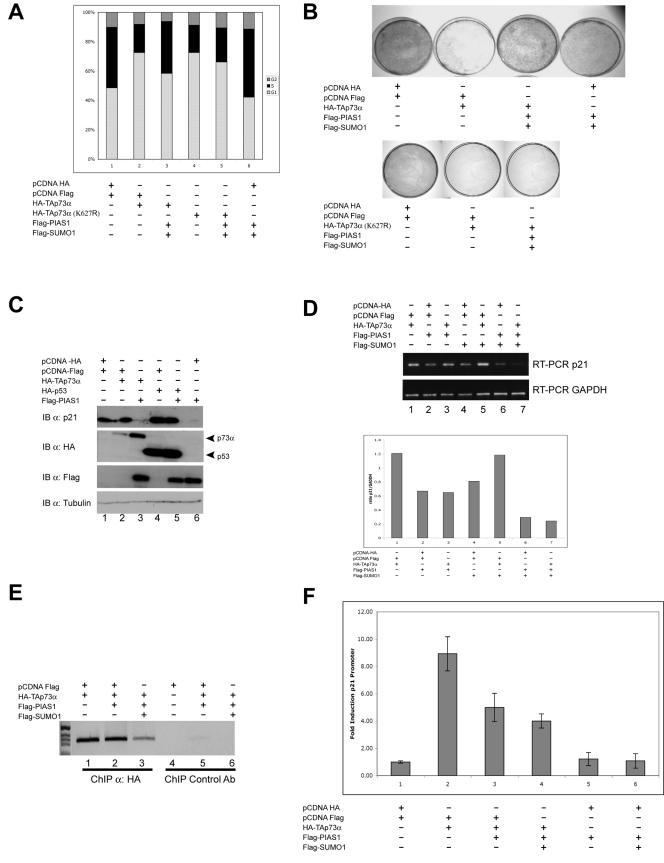
We next addressed whether binding and sumoylation of p73 by PIAS-1 and the inhibition of p73 transcriptional activity by luciferase assay have functional significance. Since p73 blocks cells in the G1 phase of the cell cycle, we investigated whether PIAS-1 expression could overcome this block. As shown in Fig. 8A, coexpression of PIAS-1 and SUMO-1 with p73 produced a reduction of the p73-dependent G1 block (compare bars 2 and 3). In addition, transfection of PIAS-1 and SUMO-1 alone resulted in a reduction of the number of cells in G1 and an increase of cells in S compared to the case for cells transfected with an empty vector (compare bar 6 with bar 1). This is possibly due to an effect of PIAS-1 on endogenous p73. The colony suppression assay shown in Fig. 8B (upper panel) confirms the results of the cell cycle analysis and demonstrates that PIAS-1 transfected cells are actively cycling and are not blocked in a different phase of the cell cycle. Consistently, overexpression of the TAp73α(K627P) mutant resulted in cell cycle arrest and a reduction in the number of colonies, and as expected, PIAS-1 expression had no effect on the activity of this mutant (Fig. 8B, lower panel).
FIG. 8.
PIAS-1 expression determines cell cycle reentry. (A) Cell cycle profile of Saos-2 cells transfected with the indicated combination of plasmids, collected 48 h after transfection, stained with PI, and analyzed by flow cytometry. Cells were cotransfected with a GFP-spectrin (18) construct at a ratio of 1:20 ratio (GFP/other plasmids), and only the GFP-positive cells were analyzed. Expression of PIAS-1 with or without p73 results in a reduction of the number of cells in G1 and an increase in the number of cells in S phase. Expression of TAp73α(K627R) also results in a G1 arrest. Coexpression of PIAS-1 does not rescue the block induced by the p73 mutant. (B) Colony suppression assay of H-1299 cells transfected with TAp73α either alone or in combination with PIAS-1 and SUMO-1 together with pBabe-Puro. Overexpression of p73 results in the expected reduction in the number of colonies; coexpression of PIAS-1 results in an increased number of colonies, while coexpression of both PIAS-1 and SUMO-1 produces an even larger increase (upper panel). Expression of TAp73α(K627R) also results in a reduction in the number of colonies; coexpression of PIAS-1 does not rescue the cells (lower panel). (C) Western blot showing p21 expression in cells transfected with the indicated combinations of plasmids and collected 24 h after transfection. Overexpression of p73 increases p21 protein levels, while coexpression of PIAS-1 results in the complete disappearance of p21 protein. PIAS-1 also produces the disappearance of basal p21 levels in cells that do not overexpress p73 (lane 6). PIAS-1 overexpression has no effect on p21 levels when p53 is also overexpressed (compare lanes 4 and 5). IB, immunoblot. (D) RT-PCR of p21 mRNAs of cells transfected with the indicated combinations of plasmids. Overexpression of TAp73α increases p21 mRNA levels, while coexpression of PIAS-1 with or without SUMO-1 produces a marked reduction in p21 mRNA abundance. PIAS-1 also causes disappearance of basal p21 levels in cells that do not overexpress p73. The PIAS-1/GAPDH ratio from the densitometric analysis of the PCR bands is shown in the lower panel. (E) ChIP with anti-HA antibodies in cells overexpressing HA-TAp73α alone or in combination with PIAS-1 or PIAS-1 and SUMO-1, showing reduced binding of TAp73α to the p21 promoter in the presence of PIAS-1. The effect is even more striking when SUMO-1 is also coexpressed. Immunoprecipitation with an unrelated antibody (Ab) (anti-keratin 5) was performed to show the specificity of the reaction (lanes 4 to 6). (F) Luciferase assay of cells transfected with a p21 reporter plasmid (p21-Luc) and the indicated combinations of plasmids. p73 activates the p21 promoter, coexpression of PIAS-1 reduces p73 transcriptional activity, and the effect is more pronounced in the presence of overexpressed SUMO-1. Error bars indicate standard deviations.
This effect of PIAS-1 is due to a down-regulation of p21 protein expression, as shown by Western blot analysis (Fig. 8C) of cells transfected with both TAp73 and PIAS-1. Expression of PIAS-1 alone also lowered basal p21 protein levels, consistent with the observation that PIAS-1-transfected cells have a reduced number of cells blocked in G1 (Fig. 8A, bar 6), and may again reflect an effect of PIAS-1 on endogenous TAp73. As expected, overexpression of p53 also increased p21 protein levels (Fig. 8C, lane 4), although this increase was unaffected by coexpression of PIAS-1 (Fig. 8C, lane 5). PIAS-1 with and without SUMO-1 reduced p21 mRNA abundance as shown by the reduction of p21 mRNA levels measured by RT-PCR in cells transfected with PIAS-1 alone or in combination with TAp73 (Fig. 8D). The corresponding densitometric ratios of PIAS-1 and GAPDH are shown in Fig. 8D. In agreement with the RT-PCR data, PIAS-1 also reduced TAp73 binding to the endogenous p21 promoter, as demonstrated by the ChIP shown in Fig. 8E, and inhibited up-regulation of the p21 promoter by TAp73 (Fig. 8F).
In conclusion, these data demonstrate that PIAS-1 expression influences cell cycle reentry by down-regulating basal and p73-induced p21 expression, although a direct effect of PIAS-1 on p21 expression cannot be completely excluded.
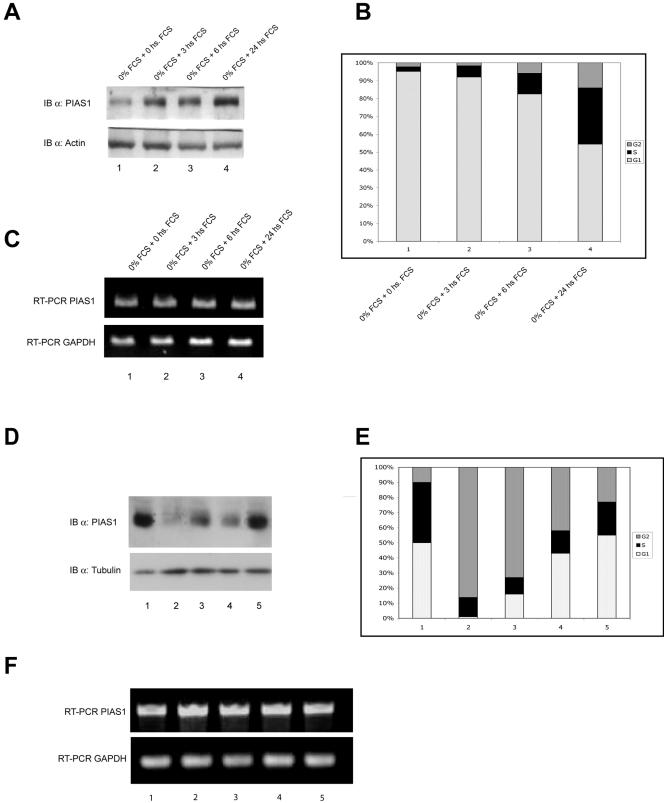
PIAS-1 expression is regulated during the cell cycle.
To further investigate the role of PIAS-1 in cell cycle regulation, we have studied changes in expression of endogenous PIAS-1 during the cell cycle. As shown in Fig. 9A, growth-arrested Saos-2 cells showed low levels of PIAS-1 protein compared with cycling cells grown in 10% serum. Serum-starved cells were almost totally blocked in G1 (>95%), as shown by flow cytometry analysis (Fig. 9B). Readdition of serum resulted in cells reentering the cell cycle, paralleled by an increase of PIAS-1 levels (Fig. 9A) and a decrease in p21 (unpublished data). Similarly, when cells were blocked in G2 by nocodazole treatment, no PIAS-1 protein could be detected (Fig. 9D, lane 2, and E, bar 2). Again, an increase of the S phase (Fig. 9E, bars 3, 4, and 5) parallels an increase of PIAS-1 expression (Fig. 9D, lanes 3, 4, and 5).
FIG. 9.
PIAS-1 expression is regulated during the cell cycle. (A) Western blot of Saos-2 cells grown in serum-free medium for 48 h (0 h) and then grown in medium containing 10% fetal calf serum (FCS) for 3, 6, and 24 h, showing that PIAS-1 is down-regulated in growth-arrested cells and is up-regulated in cells reentering the cell cycle. The same blot was reprobed with an antiactin antibody to show equal loading. IB, immunoblot. (B) Graph showing the distribution of cells in the different cell cycle phases evaluated by flow cytometry on an aliquot of the cells used for the Western blot in panel A. An increase of PIAS-1 expression parallels the increased number of cells in the S phase. (C) RT-PCR of RNA extracted from cells treated as described above, showing that there are no changes in PIAS-1 mRNA levels. (D) Hek293 cells untreated (lane 1) or blocked in G2 by treatmentwith 0.2 μg of nocodazole per ml for 16 h (lane 2) and then released by culture in nocodazole-free medium and analyzed after 2, 4, and 8 h (lanes 3, 4, and 5, respectively). The same blot was reprobed with an antitubulin antibody to show equal loading. (E) Graph showing the distribution of cells in the different cell cycle phases evaluated by flow cytometry on an aliquot of the cells used for the Western blot in panel B. Nocodazole-treated cells are completely blocked in G2 and show no expression of PIAS-1; again, an increase of PIAS-1 expression parallels the increased number of cells in the S phase. (F) RT-PCR of RNA extracted from cells treated as described above, showing that there are no changes in PIAS-1 mRNA levels.
To investigate whether the changes in PIAS-1 protein levels were secondary to changes in transcription, we performed RT-PCR (Fig. 9C to F) and real time RT-PCR (data not shown) on samples taken at the same time points. Our results show that under these experimental conditions there was no correlation between changes in PIAS-1 protein levels and PIAS-1 mRNA abundances, suggesting that changes in the protein levels depend on posttranscriptional regulation (Fig. 9C to F).
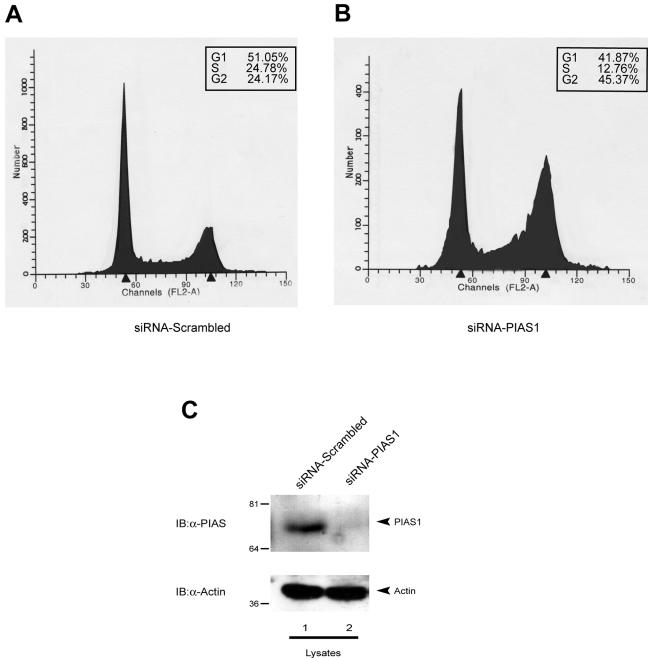
To further clarify the role of PIAS-1 in cell cycle regulation, we used an RNA interference approach. Transfection with specific anti-PIAS-1 oligonucleotides reduced the proportion of cells in S phase by roughly half, while increasing the proportion of cells in G2 from 24 to 45% (Fig. 10).
FIG. 10.
Reduction of PIAS-1 expression results in G2 arrest. Transfection of siRNA against PIAS-1 mRNA results in a G2 block. Saos-2 cells were electroporated either with a nonsense siRNA (A) or with a specific siRNA targeted against PIAS-1 mRNA (B). Cells were collected 48 h after transfection, fixed, stained with PI, and analyzed by flow cytometry. The cell cycle profile shows a reduction of cells in S and G1 and an increase of cells in G2/M. The box shows the percentage of cells in each phase of the cell cycle. (C) Western blot of a cell extract from an aliquot of the samples analyzed for the cell cycle, showing that transfection of siRNA down-regulates PIAS-1 expression. IB, immunoblot.
Together, these data show that PIAS-1 is expressed mainly in cells that are in the S phase of the cell cycle, while when cells are blocked either in G1 or in G2, PIAS-1 protein levels are completely down-regulated. Furthermore, the data also suggest that reduction in PIAS-1 expression leads to an arrest in G2. Although this might not fully reflect the situation in actively proliferating cells and should therefore be interpreted with caution, it nevertheless shows a clear dependence of PIAS-1 levels on cell cycle status and an effect of PIAS-1 down-regulation on cell cycle progression.
DISCUSSION
p73 shares high sequence homology with its more famous sibling, p53, but this corresponds to only a partial overlap in the function of the two proteins (1, 3, 5, 9, 16, 27). Very little is known about the regulation of p73 activity, although phosphorylation, acetylation, and ubiquitination have been shown to modulate p73 stability and function (reviewed in reference 28). Using a yeast two hybrid screening approach, we have identified PIAS-1 as an important regulator of p73 activity. PIAS family members have previously been shown to sumoylate a number of proteins. PIAS-1 inhibits DNA binding by activated STAT-1 and therefore inhibits its transcriptional activity (24). However, although STAT-1 is weakly sumoylated by PIAS-1 on Lys 703, this does not seem to be responsible for the inhibition of STAT-1 transactivation (36). Another family member, PIASy, sumoylates the Wnt-responsive transcription factor LEF1 and redirects it to the nuclear matrix (37), in a fashion similar to the effects of PIAS-1-mediated sumoylation of p73 on its subnuclear localization.
PIAS-1 has recently been shown to act as a SUMO ligase for p53 (17, 38). Here we show that PIAS-1 also binds and sumoylates p73. The binding of PIAS-1 alone to p73 is sufficient to stabilize the p73 protein, thus increasing its steady-state levels. Stabilization clearly does not require sumoylation of p73, since the sumoylation-defective TAp73α(K627R) mutant is still stabilized by PIAS-1 overexpression. Similarly, a PIAS-1 RING finger mutant that has lost the ability to sumoylate is still capable of stabilizing p73 protein levels. However, sumoylation of p73 by PIAS-1 results in its functional inactivation.
p73 sumoylation results in the loss of its ability to transactivate responsive promoters; consistently, TAp73α(K627R), which cannot be sumoylated, shows a higher basal activity. Similarly, the naturally occurring, C-terminally truncated β form has always been shown to be more active than the full-length α form (5, 16, 28). We suggest that this is because this isoform cannot be sumoylated.
Sumoylation of p53 has been shown to result in either activation or repression of p53 activity (10, 17, 20, 35, 38), possibly as a result of the different promoters and the different cell lines used, suggesting that the role of sumoylation may vary depending on the cellular context. Our results with p73 consistently show that sumoylation reduces transcriptional activity on different promoters. In addition, transfection of the PIAS-1 mutant that has lost its ability to sumoylate has no effect on p73 activity. These data are apparently in contrast with a previous report showing that sumoylation of p73 has no effect on its transcriptional activity (29). However, both in our experiments and in those reported by Minty and coworkers (29), when only SUMO-1 is cotransfected with p73, at best a very modest effect on p73 transcriptional activity is observed. This is in accordance with our results showing that overexpression of SUMO-1 alone results in very low, often undetectable sumoylation of p73. The addition of PIAS-1 allows the reaction to occur, with or without the addition of SUMO-1, demonstrating that endogenous levels of SUMO-1 are probably sufficient to give a functional effect.
p73 can also be ubiquitinated, although the target residue is not yet known, and this, perhaps surprisingly, results in stabilization (as does non-sumoylation-dependent binding of PIAS-1) together with transcriptional enhancement (30). Sumoylation does not affect p73 protein levels, but sumoylated p73 is restricted to the nuclear matrix fraction. Sumoylation is not required for p73 localization to the nuclear matrix, as suggested also in previous reports (2), although all of the transcriptionally inactive sumoylated p73 is localized to the nuclear matrix.
We also show that the functional consequence of inactivating p73 by sumoylation is a reduction of cells in G1, suggesting that PIAS-1 may function as a checkpoint switch promoting cell cycle progression. Consistently, PIAS-1 is up-regulated in proliferating cells and down-regulated in growth-arrested cells in a cell cycle-dependent fashion. PIAS-1 starts to be expressed at the end of the G1 phase and peaks during S phase, disappearing again completely in G2. PIAS-1-mediated G1 exit involves a reduction in p21 mRNA abundance, promoter activity, and protein expression secondary to a reduction in p73 binding to the p21 promoter. Therefore, PIAS-1-mediated sumoylation of p73 reduces its binding to the p21 promoter with a reduction in p21 expression and release from G1. Since PIAS-1 has also been reported to sumoylate and inhibit p53, it is possible that PIAS-1 also exerts its effect on the cell cycle through p53 inactivation in p53 wild-type cells, although PIAS-1 did not affect the p53-mediated increase in p21 protein levels (Fig. 9C).
The RNA interference experiments support an important role for PIAS-1 in cell cycle regulation. A specific reduction of PIAS-1 expression results in a profound reduction in the proportion of cells in S phase, with an increase in cells blocked in G2. The observed small reduction of the numbers of cells in G1 may merely reflect the reduction in cells proceeding through M and a dominant effect of PIAS-1 on the G2 checkpoint. This would be consistent with the disappearance of endogenous PIAS-1 in cells blocked in G2. In conclusion, we show that up-regulation of PIAS-1 is an important event in cell cycle control resulting in cell cycle progression, whereas reduced expression results in G2 arrest. This is partly achieved through sumoylation of p73, although clearly sumoylation of other target proteins may also be involved. Among these are other members of the p53 family (17, 38), together with STAT (36, 42) and SMAD (32) proteins.
Acknowledgments
This work was supported by grants from Telethon (E1224), Ministero Sanita', and MIUR to V.D.L. and from AIRC, the EU (QLK-CT-2002-01956), Progetto Genomica Funzionale COMETA, FIRB-2001, MIUR-2002, MinSan, Telethon, and the Medical Research Council to G.M.
REFERENCES
- 1.Agami, R., G. Blandino, M. Oren, and Y. Shaul. 1999. Interaction of c-Abl and p73alpha and their collaboration to induce apoptosis. Nature 399:809-813. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Yehoyada, M., I. Ben-Dor, and Y. Shaul. 2003. c-Abl tyrosine kinase selectively regulates p73 nuclear matrix association. J. Biol. Chem. 278:34475-34482. [DOI] [PubMed] [Google Scholar]
- 3.Catani, M. V., A. Costanzo, I. Savini, M. Levrero, V. de Laurenzi, J. Y. Wang, G. Melino, and L. Avigliano. 2002. Ascorbate up-regulates MLH1 (Mut L homologue-1) and p73: implications for the cellular response to DNA damage. Biochem. J. 364:441-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darnell, J. E., Jr. 1997. STATs and gene regulation. Science 277:1630-1635. [DOI] [PubMed] [Google Scholar]
- 5.De Laurenzi, V., A. Costanzo, D. Barcaroli, A. Terrinoni, M. Falco, M. Annicchiarico-Petruzzelli, M. Levrero, and G. Melino. 1998. Two new p73 splice variants, gamma and delta, with different transcriptional activity. J. Exp Med. 188:1763-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Laurenzi, V. D., M. V. Catani, A. Terrinoni, M. Corazzari, G. Melino, A. Costanzo, M. Levrero, and R. A. Knight. 1999. Additional complexity in p73: induction by mitogens in lymphoid cells and identification of two new splicing variants epsilon and zeta. Cell Death Differ. 6:389-390. [DOI] [PubMed] [Google Scholar]
- 7.Duval, D., G. Duval, C. Kedinger, O. Poch, and H. Boeuf. 2003. The ‘PINIT’ motif, of a newly identified conserved domain of the PIAS protein family, is essential for nuclear retention of PIAS3L. FEBS Lett. 554:111-118. [DOI] [PubMed] [Google Scholar]
- 8.Gietz, D., A. St. Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong, J. G., A. Costanzo, H. Q. Yang, G. Melino, W. G. Kaelin, Jr., M. Levrero, and J. Y. Wang. 1999. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature 399:806-809. [DOI] [PubMed] [Google Scholar]
- 10.Gostissa, M., A. Hengstermann, V. Fogal, P. Sandy, S. E. Schwarz, M. Scheffner, and G. Del Sal. 1999. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 18:6462-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottifredi, V., G. Pelicci, E. Munarriz, R. Maione, P. G. Pelicci, and P. Amati. 1999. Polyomavirus large T antigen induces alterations in cytoplasmic signalling pathways involving Shc activation. J. Virol. 73:1427-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grob, T. J., M. F. Fey, and A. Tobler. 2002. The two faces of p73. Cell Death Differ. 9:229-230. [DOI] [PubMed] [Google Scholar]
- 13.Grob, T. J., U. Novak, C. Maisse, D. Barcaroli, A. U. Luthi, F. Pirnia, B. Hugli, H. U. Graber, V. De Laurenzi, M. F. Fey, G. Melino, and A. Tobler. 2001. Human delta Np73 regulates a dominant negative feedback loop for TAp73 and p53. Cell Death Differ. 8:1213-1223. [DOI] [PubMed] [Google Scholar]
- 14.Ihle, J. N. 2001. The Stat family in cytokine signaling. Curr. Opin. Cell Biol. 13:211-217. [DOI] [PubMed] [Google Scholar]
- 15.Irwin, M. S., and W. G. Kaelin. 2001. p53 family update: p73 and p63 develop their own identities. Cell Growth Differ. 12:337-349. [PubMed] [Google Scholar]
- 16.Kaghad, M., H. Bonnet, A. Yang, L. Creancier, J. C. Biscan, A. Valent, A. Minty, P. Chalon, J. M. Lelias, X. Dumont, P. Ferrara, F. McKeon, and D. Caput. 1997. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 90:809-819. [DOI] [PubMed] [Google Scholar]
- 17.Kahyo, T., T. Nishida, and H. Yasuda. 2001. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell 8:713-718. [DOI] [PubMed] [Google Scholar]
- 18.Kalejta, R. F., T. Shenk, and A. J. Beavis. 1997. Use of a membrane-localized green fluorescent protein allows simultaneous identification of transfected cells and cell cycle analysis by flow cytometry. Cytometry 29:286-291. [DOI] [PubMed] [Google Scholar]
- 19.Kotaja, N., U. Karvonen, O. A. Janne, and J. J. Palvimo. 2002. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell. Biol. 22:5222-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwek, S. S., J. Derry, A. L. Tyner, Z. Shen, and A. V. Gudkov. 2001. Functional analysis and intracellular localization of p53 modified by SUMO-1. Oncogene 20:2587-2599. [DOI] [PubMed] [Google Scholar]
- 21.Levrero, M., V. De Laurenzi, A. Costanzo, J. Gong, G. Melino, and J. Y. Wang. 1999. Structure, function and regulation of p63 and p73. Cell Death Differ. 6:1146-1153. [DOI] [PubMed] [Google Scholar]
- 22.Levrero, M., V. De Laurenzi, A. Costanzo, J. Gong, J. Y. Wang, and G. Melino. 2000. The p53/p63/p73 family of transcription factors: overlapping and distinct functions. J. Cell Sci. 113:1661-1670. [DOI] [PubMed] [Google Scholar]
- 23.Liao, J., Y. Fu, and K. Shuai. 2000. Distinct roles of the NH2- and COOH-terminal domains of the protein inhibitor of activated signal transducer and activator of transcription (STAT) 1 (PIAS1) in cytokine-induced PIAS1-Stat1 interaction. Proc. Natl. Acad. Sci. USA 97:5267-5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, B., J. Liao, X. Rao, S. A. Kushner, C. D. Chung, D. D. Chang, and K. Shuai. 1998. Inhibition of Stat1-mediated gene activation by PIAS1. Proc. Natl. Acad. Sci. USA 95:10626-10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Long, J., I. Matsuura, D. He, G. Wang, K. Shuai, and F. Liu. 2003. Repression of Smad transcriptional activity by PIASy, an inhibitor of activated STAT. Proc. Natl. Acad. Sci. USA 100:9791-9796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Megidish, T., J. H. Xu, and C. W. Xu. 2002. Activation of p53 by protein inhibitor of activated Stat1 (PIAS1). J. Biol. Chem. 277:8255-8259. [DOI] [PubMed] [Google Scholar]
- 27.Melino, G., V. De Laurenzi, and K. H. Vousden. 2002. p73: friend or foe in tumorigenesis. Nat. Rev. Cancer 2:605-615. [DOI] [PubMed] [Google Scholar]
- 28.Melino, G., X. Lu, M. Gasco, T. Crook, and R. A. Knight. 2003. Functional regulation of p73 and p63: development and cancer. Trends Biochem. Sci. 28:663-670. [DOI] [PubMed] [Google Scholar]
- 29.Minty, A., X. Dumont, M. Kaghad, and D. Caput. 2000. Covalent modification of p73alpha by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J. Biol. Chem. 275:36316-36323. [DOI] [PubMed] [Google Scholar]
- 30.Miyazaki, K., T. Ozaki, C. Kato, T. Hanamoto, T. Fujita, S. Irino, K. Watanabe, T. Nakagawa, and A. Nakagawara. 2003. A novel HECT-type E3 ubiquitin ligase, NEDL2, stabilizes p73 and enhances its transcriptional activity. Biochem. Biophys. Res. Commun. 308:106-113. [DOI] [PubMed] [Google Scholar]
- 31.Nicoletti, I., G. Migliorati, M. C. Pagliacci, F. Grignani, and C. Riccardi. 1991. A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J. Immunol. Methods 139:271-279. [DOI] [PubMed] [Google Scholar]
- 32.Ohshima, T., and K. Shimotohno. 2003. Transforming growth factor-beta-mediated signaling via the p38 MAP kinase pathway activates Smad-dependent transcription through SUMO-1 modification of Smad4. J. Biol. Chem. 278:50833-50842. [DOI] [PubMed] [Google Scholar]
- 33.Okubo, S., F. Hara, Y. Tsuchida, S. Shimotakahara, S. Suzuki, H. Hatanaka, S. Yokoyama, H. Tanaka, H. Yasuda, and H. Shindo. 2004. NMR structure of the N-terminal domain of SUMO ligase PIAS1 and its interaction with tumor suppressor p53 and A/T-rich DNA oligomers. J. Biol. Chem. 279:31455-31461. [DOI] [PubMed] [Google Scholar]
- 34.O'Shea, J. J., M. Gadina, and R. D. Schreiber. 2002. Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell 109(Suppl.):S121-S131. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez, M. S., J. M. Desterro, S. Lain, C. A. Midgley, D. P. Lane, and R. T. Hay. 1999. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 18:6455-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rogers, R. S., C. M. Horvath, and M. J. Matunis. 2003. SUMO modification of STAT1 and its role in PIAS-mediated inhibition of gene activation. J. Biol. Chem. 278:30091-30097. [DOI] [PubMed] [Google Scholar]
- 37.Sachdev, S., L. Bruhn, H. Sieber, A. Pichler, F. Melchior, and R. Grosschedl. 2001. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 15:3088-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt, D., and S. Muller. 2002. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl. Acad. Sci. USA 99:2872-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seeler, J. S., and A. Dejean. 2003. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell. Biol. 4:690-699. [DOI] [PubMed] [Google Scholar]
- 40.Tirard, M., J. Jasbinsek, O. F. Almeida, and T. M. Michaelidis. 2004. The manifold actions of the protein inhibitor of activated STAT proteins on the transcriptional activity of mineralocorticoid and glucocorticoid receptors in neural cells. J. Mol. Endocrinol. 32:825-841. [DOI] [PubMed] [Google Scholar]
- 41.Ueda, Y., M. Hijikata, S. Takagi, T. Chiba, and K. Shimotohno. 1999. New p73 variants with altered C-terminal structures have varied transcriptional activities. Oncogene 18:4993-4998. [DOI] [PubMed] [Google Scholar]
- 42.Ungureanu, D., S. Vanhatupa, N. Kotaja, J. Yang, S. Aittomaki, O. A. Janne, J. J. Palvimo, and O. Silvennoinen. 2003. PIAS proteins promote SUMO-1 conjugation to STAT1. Blood 102:3311-3313. [DOI] [PubMed] [Google Scholar]
- 43.Yang, A., and F. McKeon. 2000. P63 and P73: P53 mimics, menaces and more. Nat. Rev. Mol. Cell. Biol. 1:199-207. [DOI] [PubMed] [Google Scholar]