Analysis of mouse and human B lineage acute lymphoblastic leukemia identifies evolutionarily conserved targets of the tumor-suppressive transcription factor IKAROS, implicating CTNND1 in leukemia maintenance.
Abstract
Genetic alterations disrupting the transcription factor IKZF1 (encoding IKAROS) are associated with poor outcome in B lineage acute lymphoblastic leukemia (B-ALL) and occur in >70% of the high-risk BCR-ABL1+ (Ph+) and Ph-like disease subtypes. To examine IKAROS function in this context, we have developed novel mouse models allowing reversible RNAi-based control of Ikaros expression in established B-ALL in vivo. Notably, leukemias driven by combined BCR-ABL1 expression and Ikaros suppression rapidly regress when endogenous Ikaros is restored, causing sustained disease remission or ablation. Comparison of transcriptional profiles accompanying dynamic Ikaros perturbation in murine B-ALL in vivo with two independent human B-ALL cohorts identified nine evolutionarily conserved IKAROS-repressed genes. Notably, high expression of six of these genes is associated with inferior event–free survival in both patient cohorts. Among them are EMP1, which was recently implicated in B-ALL proliferation and prednisolone resistance, and the novel target CTNND1, encoding P120-catenin. We demonstrate that elevated Ctnnd1 expression contributes to maintenance of murine B-ALL cells with compromised Ikaros function. These results suggest that IKZF1 alterations in B-ALL leads to induction of multiple genes associated with proliferation and treatment resistance, identifying potential new therapeutic targets for high-risk disease.
Introduction
Acute lymphoblastic leukemia (ALL) is the most frequent childhood cancer. Whereas ∼90% of pediatric ALL patients can be cured with current therapies, the prognosis for adult patients and children with relapsed disease is poor (Hunger and Mullighan, 2015). Over the last decade, our understanding of the genetic basis of B lineage ALL (B-ALL) pathogenesis and treatment response has been revolutionized by high-resolution genomic and transcriptional profiling of large patient cohorts (Roberts and Mullighan, 2015). These studies reveal that a majority of patients with B-ALL harbor somatic mutations or focal deletions that disable genes encoding lymphoid transcription factors, including EBF1, PAX5, and IKZF1 (Kuiper et al., 2007; Mullighan et al., 2007; Roberts and Mullighan, 2015).
Approximately 5% of pediatric B-ALL patients have disease harboring the t(9;22) translocation, forming what is commonly termed the Philadelphia (Ph) chromosome and resulting in expression of the oncogenic BCR-ABL1 fusion kinase. A novel disease subtype was recently defined based on a gene expression profile similar to Ph+ disease, termed Ph-like or BCR-ABL1-like B-ALL (Den Boer et al., 2009; Mullighan et al., 2009). The Ph+ and Ph-like disease subtypes have relatively high relapse rates and poor prognosis, and together comprise ∼15% of pediatric and ∼50% of adult B-ALL cases (Roberts et al., 2012, 2014; van der Veer et al., 2013). Somatic genetic alterations in IKZF1, which encodes the zinc finger transcription factor IKAROS, occur in 50–70% of Ph+ and Ph-like B-ALL cases (Mullighan et al., 2008a, 2009; Den Boer et al., 2009; Roberts et al., 2012, 2014; van der Veer et al., 2013, 2014; Churchman et al., 2015). These lesions commonly involve monoallelic loss of IKZF1 or deletion of internal exons, causing expression of dominant-negative protein isoforms, but also include deleterious point mutations and rare bi-allelic deletions. Although loss of normal IKAROS function clearly cooperates with BCR-ABL1 or other kinase-activating lesions during Ph+ and Ph-like B-ALL pathogenesis, the specific mechanistic basis for this genetic interaction remains unknown.
IKZF1 is unique among B-ALL transcription factor tumor suppressors in that its mutation or deletion is associated with adverse prognosis across multiple patient cohorts (Iacobucci et al., 2009; Martinelli et al., 2009; Mullighan et al., 2009; Kuiper et al., 2010; van der Veer et al., 2013). Furthermore, genomic studies of paired diagnosis and relapsed B-ALL samples demonstrate high rates of acquired IKZF1 alterations or positive selection of IKZF1-deficient subclones at relapse (Mullighan et al., 2008b; Yang et al., 2008; Kuiper et al., 2010; Dupuis et al., 2013; Krentz et al., 2013), and IKZF1 lesions also occur during progression from chronic phase CML to therapy-resistant lymphoid blast crisis disease (Mullighan et al., 2008a). IKZF1 deletions are also associated with high levels of residual disease after remission-induction therapy (Mullighan et al., 2009; Waanders et al., 2011; van der Veer et al., 2013). These studies suggest a role for IKZF1 loss in therapy resistance that may contribute to the poor treatment responses of Ph+ and Ph-like disease. Consistent with this idea, IKZF1 lesions independently predict high rates of relapse and decreased survival within Ph+ or Ph-like B-ALL cohorts after standard therapy (Martinelli et al., 2009; Mullighan et al., 2009; van der Veer et al., 2013; Roberts et al., 2014), and its loss is also associated with clinical resistance to the BCR-ABL1 inhibitor imatinib (van der Veer et al., 2014). Together, these studies point to a central role for IKZF1 deletions in B-ALL progression and treatment resistance.
Mouse models have identified critical Ikaros functions in lymphocyte development. Ikaros is required for normal function and lineage commitment of hematopoietic stem and progenitor cells (Nichogiannopoulou et al., 1999; Yoshida et al., 2006). Germline Ikaros-deficient mice have no B cells and defective T and myeloid lineage development (Wang et al., 1996), and conditional Ikaros inactivation in B lineage progenitors arrests their differentiation (Heizmann et al., 2013; Joshi et al., 2014; Schwickert et al., 2014). Hypomorphic Ikaros alleles promote development of aggressive T-ALL (Winandy et al., 1995; Dumortier et al., 2006) and also accelerate disease in models of Ph+ B-ALL (Virely et al., 2010; Schjerven et al., 2013; Churchman et al., 2015), verifying a conserved tumor suppressor function. Several recent studies have identified Ikaros-regulated genes through expression profiling of Ikaros-deficient B cell progenitors and B-ALL cells (Ferreirós-Vidal et al., 2013; Heizmann et al., 2013; Schjerven et al., 2013; Joshi et al., 2014; Schwickert et al., 2014; Churchman et al., 2015), linking their differentiation arrest to perturbed IL-7 and pre–BCR signaling (Heizmann et al., 2013; Schwickert et al., 2014). Ikaros-deficient B cell progenitors and B-ALL cells also show increased integrin signaling and adhesion to stromal cells (Joshi et al., 2014; Schwickert et al., 2014; Churchman et al., 2015). Although targeting these adhesion phenotypes attenuates proliferation of Ikaros-deficient cells (Joshi et al., 2014; Churchman et al., 2015), their relative role in B-ALL pathogenesis and treatment response is unclear.
Understanding the molecular basis of how IKZF1 deletions drive B-ALL development and therapy resistance can inform the development of improved treatments. Using a novel transgenic mouse model of Ikaros-low, BCR-ABL1+ B-ALL, we demonstrate that Ikaros suppression not only accelerates leukemia development but is subsequently required for its maintenance. Through integrated transcriptional profiling of multiple murine B-ALL models and human B-ALL patient cohorts, we identify several high confidence, conserved IKAROS-repressed genes. These include EMP1, encoding a known mediator of B-ALL treatment resistance, and CTNND1, for which we demonstrate a novel role in B-ALL maintenance.
Results
Ikaros knockdown accelerates onset of BCR-ABL1+ B-ALL
To generate a mouse model of high-risk, BCR-ABL1+, IKZF1 mutant human B-ALL, we adopted a well-characterized BCR-ABL1P190 transgenic mouse strain that succumbs to B-ALL after 3–6 mo (Heisterkamp et al., 1990). To allow reversible control of endogenous Ikaros expression in established B-ALL in vivo, we first generated a novel transgenic mouse strain where a tetracycline (tet)-regulated (TRE) promoter targeted to the type I collagen locus controls coexpression of GFP, along with an effective microRNA-based short hairpin RNA (shRNA) targeting murine Ikaros. We crossed these TRE-GFP-shIkaros mice to Vav-tTA transgenic mice, which express the tTA (tet-off) transactivator across the hematopoietic system (Kim et al., 2007; Takiguchi et al., 2013). Bi-transgenic animals succumbed to a GFP+ T lineage leukemia similar to our previous findings (Witkowski et al., 2015), verifying effective Ikaros knockdown in vivo (unpublished data).
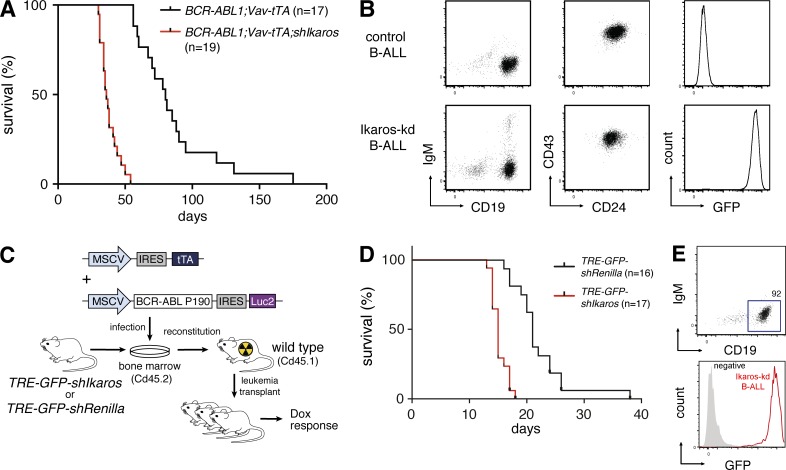
Using a mating strategy that produces mice of uniform strain background (Fig. S1, A and B), we generated a cohort of BCR-ABL1P190;Vav-tTA;TRE-GFP-shIkaros triple transgenic mice and BCR-ABL1P190;Vav-tTA bi-transgenic littermate controls. Triple transgenic mice succumbed to B-ALL of similar immunophenotype to BCR-ABL1P190 control disease, but with approximately half the latency (Fig. 1, A and B; median survival 36 d versus 80 d; P < 0.0001, log-rank test). Leukemias arising in triple transgenic mice (hereafter termed Ikaros-kd B-ALL) were uniformly GFP+, verifying that Ikaros knockdown contributes to accelerated leukemogenesis (Fig. 1 B).
Figure 1.
Reversible Ikaros knockdown promotes BCR-ABL1+ B cell leukemogenesis. (A) Kaplan-Meier survival analysis of BCR-ABL1P190;Vav-tTA;TRE-GFP-shIkaros mice (n = 19) and BCR-ABL1P190;Vav-tTA control mice (n = 17). Median survival 36 d versus 80 d, respectively; P < 0.0001, Log-rank (Mantel-Cox) test. (B) Flow cytometry of CD19/IgM and CD24/CD43 immunophenotype of splenocytes isolated from representative primary leukemic mice, and GFP expression of CD19+IgM– cells. (C) Schematic of adoptive transfer approach to generate BCR/ABL1+ B-ALL accelerated by reversible Ikaros knockdown. TRE-GFP-shIkaros or TRE-GFP-shRenilla transgenic bone marrow cells are co-transduced with MSCV vectors expressing BCR/ABL1 and tTA, and transplanted into lethally irradiated recipient mice. (D) Kaplan-Meier survival analysis of mice transplanted with TRE-GFP-shIkaros (n = 17) or control TRE-GFP-shRenilla (n = 16) cells infected as in C. Median survival 17 d versus 22 d, respectively; P < 0.0001, log-rank test. (E) Flow cytometry of CD19, IgM, and GFP expression of splenocytes isolated from representative leukemic primary recipient mice transplanted with infected TRE-GFP-shIkaros transgenic bone marrow cells.
In parallel, using previously established protocols (Li et al., 1999), we generated an additional B-ALL model through retroviral expression of BCR-ABL1P190 in bone marrow cells that were then used to reconstitute lethally irradiated recipient mice. To allow reversible Ikaros knockdown, we infected bone marrow cells from TREtight-GFP-shIkaros.4056 or control TREtight-GFP-shRenilla.713 transgenic mice (Witkowski et al., 2015) with retroviral vectors stably expressing BCR-ABL1P190 and tTA (Fig. 1 C). Recipients of co-transduced TREtight-GFP-shIkaros cells had significantly accelerated onset of a CD19+IgM– B-ALL compared with TREtight-GFP-shRenilla controls (median survival 17 d and 22 d, respectively; P < 0.0001), and again this Ikaros-kd disease was GFP+ (Fig. 1, D and E). Hence, transgenic shRNA-mediated knockdown of Ikaros cooperates with transgenic or retroviral expression of the BCR-ABL1P190 fusion oncogene to recapitulate the genetic interaction between IKAROS hypomorphism and BCR-ABL1 previously observed in human B-ALL and in germline Ikaros mutant mouse models (Mullighan et al., 2008a; Virely et al., 2010; Schjerven et al., 2013).
Ikaros knockdown is critical for BCR-ABL1+ B-ALL maintenance
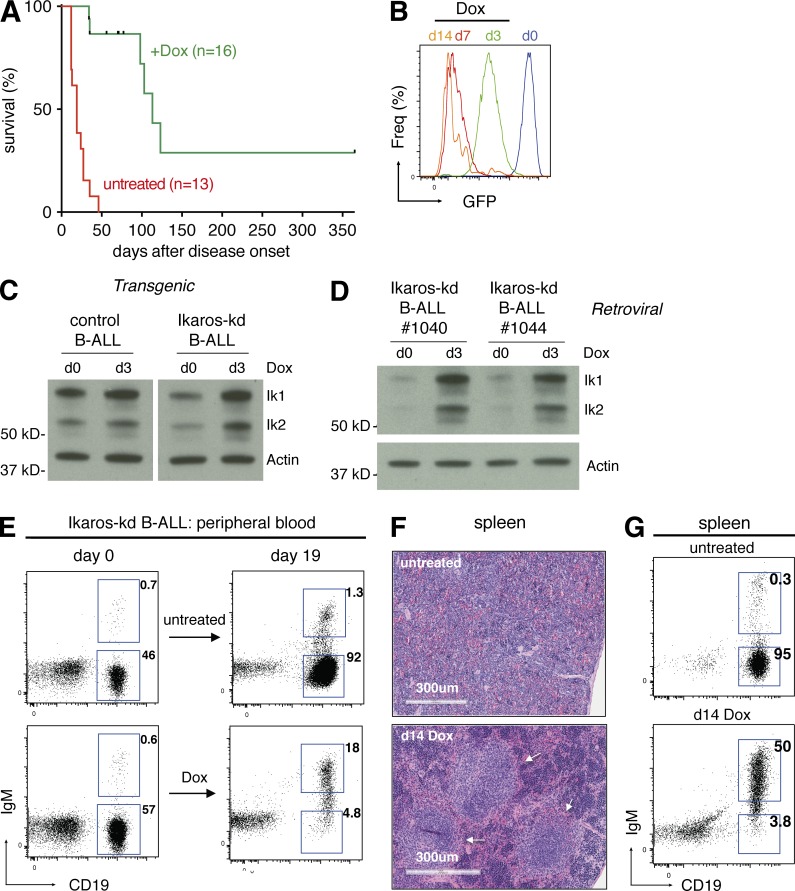
To address whether ongoing Ikaros suppression is required for B-ALL maintenance in vivo, we transplanted leukemia cells from several independent primary BCR-ABL1P190;Vav-tTA control or BCR-ABL1P190;Vav-tTA;TRE-GFP-shIkaros Ikaros-kd tumors (expressing the surface marker CD45.2) into cohorts of Rag1−/− CD45.1+ recipient mice. All untreated transplant recipient mice rapidly developed disease and become moribund within 2–4 wk (Fig. 2 A). Upon disease onset (donor-derived CD45.2+, CD19+IgM– cells exceeding ∼5% of peripheral leukocytes), subsets of recipients were given food supplemented with the tetracycline analogue doxycycline (Dox). In mice bearing Ikaros-kd B-ALL, 3 d of Dox treatment uniformly diminished GFP expression and increased Ikaros protein expression in leukemia cells in vivo, consistent with shutdown of tTA activity and loss of Ikaros shRNA (Fig. 2, B–D). Acute Dox treatment did not affect BCR-ABL1 mRNA expression or phosphorylation of the canonical BCR-ABL1 substrate STAT5 in B-ALL cells in vivo (Fig. S1, C–F). Although Dox treatment of mice bearing control B-ALL had no effect (Fig. S1, G–I), sustained Dox treatment of leukemic Ikaros-kd recipient mice resulted in significantly enhanced survival (median survival 19 d for untreated versus 113 d for Dox-treated mice; P < 0.0001; Fig. 2 A). After 2–3 wk of Dox treatment, B-ALL cells were almost completely cleared from the peripheral blood and spleen, and the remaining CD45.2+ cells had acquired IgM expression consistent with B lineage differentiation (Fig. 2, E–G). Of note, 2 Ikaros-kd B-ALL recipient mice remained disease-free 1 yr after Dox treatment commenced, suggesting that Ikaros restoration in established B-ALL can be curative in this leukemia transplant model (Fig. 2 A). Approximately half (6/16) of Dox-treated mice eventually relapsed with a GFP–CD45.2+CD19+IgM– B-ALL similar to the original disease (Fig. S2 A; described further below). Together these data demonstrate a critical role for Ikaros suppression in B-ALL maintenance despite the ongoing expression of oncogenic BCR-ABL1, providing rationale for restoring the Ikaros-regulated gene expression program as a potential therapeutic strategy in this context.
Figure 2.
Ikaros suppression is critical for maintenance of Ikaros-kd B-ALL. (A) Kaplan-Meier survival analysis of Ikaros-kd B-ALL transplant recipient mice (six independent primary BCR-ABL1;Vav-tTA;TRE-GFP-shIkaros leukemias, two to three recipients per condition) either untreated or Dox-treated upon disease establishment. Median survival 19 d for untreated versus 113 d for Dox treated; P < 0.0001, log-rank test. Censored events represent deaths unrelated to leukemia, including fighting and infection. (B) GFP expression of splenocytes from representative Ikaros-kd B-ALL–recipient mice that were untreated (d0) or Dox treated as indicated upon leukemia onset. (C) Western blot of Ikaros expression in control or Ikaros-kd B-ALL cells isolated from representative leukemic mice that were untreated or Dox treated as indicated. The Ikaros protein isoforms Ik1 and Ik2 are indicated. Actin is a loading control. (D) Western blot of Ikaros protein expression in Ikaros-kd B-ALL cells isolated from representative leukemic recipient mice that were untreated or Dox treated as indicated. (E) Flow cytometry of CD19 and IgM in CD45.2+ (donor-derived) peripheral blood cells from representative untreated (upper panels) or Dox-treated (lower panels) leukemic Ikaros-kd B-ALL transplant recipient mice. (F) Spleen histology of representative Ikaros-low B-ALL transplant recipient mice that were untreated or Dox treated for 14 d at disease onset, with arrows indicating follicular structures. (G) Flow cytometry of CD19 and IgM in CD45.2+ splenocytes from representative Ikaros-kd B-ALL transplant recipient mice with Dox treatments indicated.
Ikaros-regulated genes in murine BCR-ABL1+ B-ALL
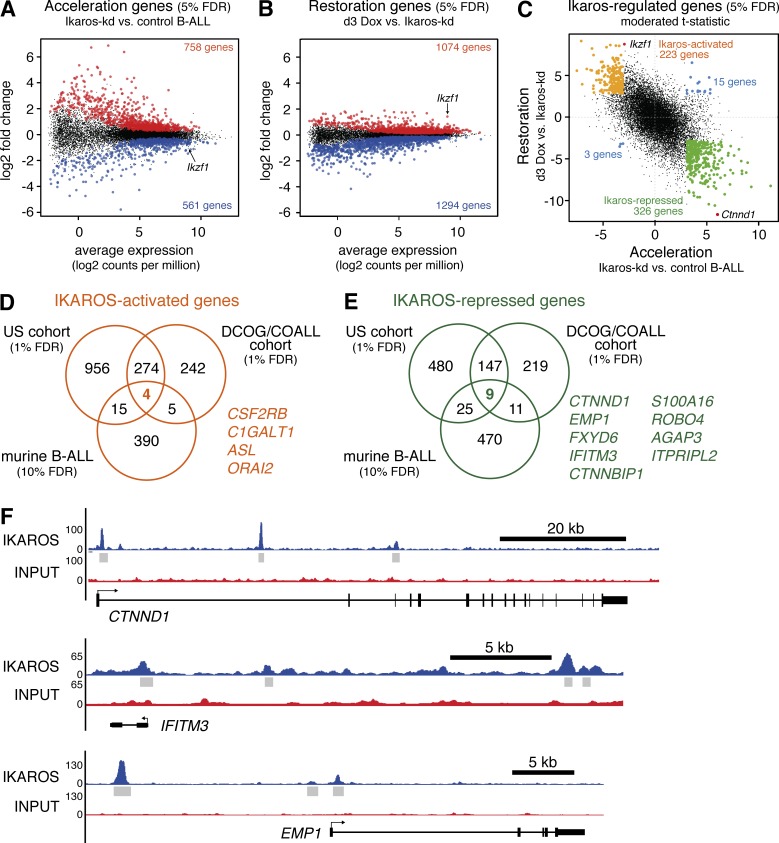
Having demonstrated that Ikaros suppression accelerates leukemogenesis and its restoration triggers dramatic disease regression, we used RNA sequencing (RNA-seq) to define the Ikaros-regulated transcriptome in these contexts. To identify transcriptional changes associated with accelerated onset of BCR-ABL1+ B-ALL caused by Ikaros knockdown, we compared expression profiles of B-ALL cells isolated by FACS from the spleens of recipient mice bearing independent primary Ikaros-kd leukemias (four BCR-ABL1P190;Vav-tTA;TRE-GFP-shIkaros leukemias and three TRE-GFP-shIkaros/retroviral BCR-ABL1P190 leukemias) to independent primary control leukemias (three BCR-ABL1P190;Vav-tTA bi-transgenic leukemias and one TRE-GFP-shRenilla/retroviral BCR-ABL1P190 leukemia). This identified 758 significantly up-regulated genes and 561 significantly down-regulated genes in Ikaros-kd B-ALL relative to control B-ALL at 5% false discovery rate (FDR). These were called Acceleration genes (Fig. 3 A and Table S1).
Figure 3.
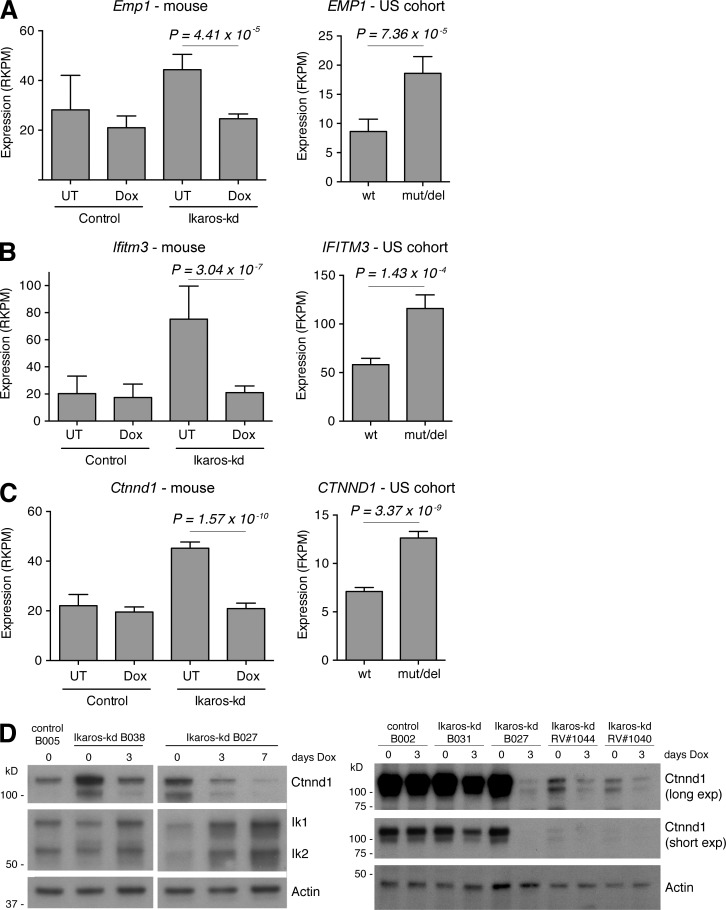
Identification of conserved IKAROS-regulated genes in BCR-ABL1+ B-ALL. (A) MA plot of average RNA-seq expression differences in B-ALL cells isolated from mice transplanted with primary Ikaros-kd (n = 7) versus control (n = 4) leukemias, highlighting genes with increased (red) or decreased (blue) expression at 5% FDR (Acceleration genes). (B) MA plot as in A for B-ALL cells isolated from mice bearing Ikaros-kd B-ALL (n = 7 independent primary leukemias), comparing 3 d Dox-treated mice to untreated mice. Genes with increased (red) or decreased (blue) expression at 5% FDR (Restoration genes) are highlighted. (C) Scatterplot of moderated t statistics of expressed genes in the Acceleration and Restoration experiments, highlighting genes that are differentially expressed (5% FDR) in both. Ikaros-activated genes are highlighted in orange, Ikaros-repressed genes in green, discordant genes in blue, and Ikaros/Ikzf1 and Ctnnd1 in red. (D and E) Venn diagrams of IKAROS-activated genes (D) and IKAROS-repressed genes (E) in the US and DCOG/COALL patient cohorts (both 1% FDR) intersected with Ikaros-activated and Ikaros-repressed genes identified in murine B-ALL (10% FDR). Conserved IKAROS-regulated genes are listed. (F) IKAROS ChIP-seq in human B-ALL cells showing significant binding peaks (gray bars) within and adjacent to the conserved IKAROS-repressed genes CTNND1, EMP1, and IFITM3.
To identify dynamic transcriptional changes accompanying acute Ikaros restoration in established B-ALL in vivo, we compared the expression profile of flow-sorted CD19+IgM– B-ALL cells harvested from the spleens of several mice bearing independent primary Ikaros-kd leukemias (four BCR-ABL1P190;Vav-tTA;TRE-GFP-shIkaros leukemias and three TRE-GFP-shIkaros/retroviral BCR-ABL1P190 leukemias) that were either untreated (GFPhi cells) or after 3-d in vivo Dox treatment (GFPmid cells, immunophenotypically indistinguishable from untreated). Endogenous Ikaros restoration in B-ALL in vivo led to induction of 1,074 genes and repression of 1,294 genes at 5% FDR, termed Restoration genes (Fig. 3 B and Table S2).
We hypothesized that the global gene expression profile associated with Ikaros knockdown in steady-state B-ALL may be partially reversed upon acute Ikaros restoration. Indeed, a genuine association (genas) analysis, which estimates the underlying correlation between expression profiles correcting for technical features, confirmed a strong inverse correlation between global Acceleration and Restoration gene expression changes (correlation = –0.67; P < 10−16; Fig. 3 C). Comparing both experiments (5% FDR) enabled classification of Ikaros-regulated genes into two groups: (1) 223 genes with reduced expression in Ikaros-kd B-ALL relative to control B-ALL that are also up-regulated upon Ikaros restoration, designated Ikaros-activated genes; and (2) 326 genes with elevated expression in Ikaros-kd B-ALL relative to control B-ALL that are also repressed upon Ikaros restoration, designated Ikaros-repressed genes (Fig. 3 C and Table S3). Of the 567 total genes with significant differential expression in both experiments, only 18 (3%) showed discordant expression (Fig. 3 C), suggesting this strategy can effectively identify Ikaros-regulated genes in BCR-ABL1+ B-ALL in vivo. Roast gene set analysis comparing Ikaros-regulated genes to transcriptional changes occurring during normal murine B cell development in the bone marrow (Hoffmann et al., 2002) indicated that although Ikaros-kd leukemias are immunophenotypically similar to control leukemias driven by BCR-ABL1 alone, transcriptionally they appear arrested at an earlier progenitor stage, and this differentiation block is partially released upon acute Ikaros restoration (Fig. S1 J). Consistent with previous studies implicating Ikaros in cellular adhesion (Joshi et al., 2014; Schwickert et al., 2014; Churchman et al., 2015), gene ontology and KEGG pathway analysis indicated that Ikaros-repressed genes were significantly enriched for genes involved in cell–cell junctions/focal adhesions (Tables S4 and S5).
Identifying conserved transcriptional targets of Ikaros in human and mouse B-ALL
Recent RNA-seq of large human B-ALL sample cohorts has uncovered the transcriptional landscape of different B-ALL subtypes (Roberts and Mullighan, 2015). To identify transcriptomic changes associated with IKZF1 disruption in B-ALL, we examined a recently described cohort of 289 patients, including 252 patients with known IKZF1 mutation status (US cohort: St Jude Total studies, COG P9906, AALL0232, and adult ALL cohorts; Roberts et al., 2014). This cohort comprises multiple disease subtypes, but contains a high proportion (53%) of Ph+ and Ph-like cases with known poor outcome that are also enriched for IKZF1 alterations (Fig. S3 A; Roberts et al., 2014). Analysis of RNA-seq data from this cohort identified 661 up-regulated genes and 1,249 down-regulated genes (1% FDR) in 103 IKZF1 mutated/deleted (IKZF1mut/del) samples when compared with 144 IKZF1 wild-type (IKZF1wt) samples, designated IKAROS-repressed and IKAROS-activated genes, respectively (Fig. 3, D and E; and Table S6). We similarly analyzed microarray expression data from an independent cohort of 573 patients (DCOG/COALL cohort: Dutch ALL-8, -9, and -10, and German COALL-97 and -03–treated patients), including 139 cases with IKZF1 deletions and 371 cases without IKZF1 deletions (Den Boer et al., 2009; van der Veer et al., 2013). This cohort comprises major B-ALL subtypes at representative proportions, including 20% Ph+/BCR-ABL1-like cases (Fig. S4 A). Our analysis identified 386 IKAROS-repressed and 525 IKAROS-activated genes at 1% FDR (Fig. 3, D and E; and Table S7). Combined analysis identified 156 IKAROS-repressed genes and 278 IKAROS-activated genes common to the US and DCOG/COALL B-ALL cohorts (Fig. 3, D and E; and Table S8).
To identify conserved Ikaros target genes in B-ALL across species, we compared human common IKAROS-repressed and IKAROS-activated genes (1% FDR) with murine Ikaros-repressed and -activated genes (relaxing the threshold to 10% FDR to increase cross-species overlap). This stringent approach yielded nine conserved IKAROS-repressed genes and four conserved IKAROS-activated genes (Fig. 3, D and E).
Dynamic, conserved IKAROS-regulated genes are enriched for IKAROS binding
To investigate whether our high-confidence IKAROS-regulated genes were directly bound by IKAROS, we analyzed IKAROS chromatin immunoprecipitation-sequencing (ChIP-seq) data from human B-ALL cells (see Schjerven et al. in this issue). Notably, 212 of the 326 Ikaros-repressed genes (67%) and 147 of the 223 Ikaros-activated genes (66%) identified using our murine B-ALL models (Fig. 3 C) were IKAROS bound, a significant enrichment compared with 56% binding for other expressed genes (P = 0.0009 and P = 0.002, respectively, Fisher’s exact test; Table S3). Interestingly, all nine conserved IKAROS-repressed genes were bound by IKAROS (enrichment P = 0.006 relative to other expressed genes, Fisher’s exact test), indicating potential direct repression by IKAROS in B-ALL (Fig. 3 F).
High expression of the IKAROS-repressed genes CTNND1, EMP1, and IFITM3 in B-ALL is associated with decreased patient event–free survival
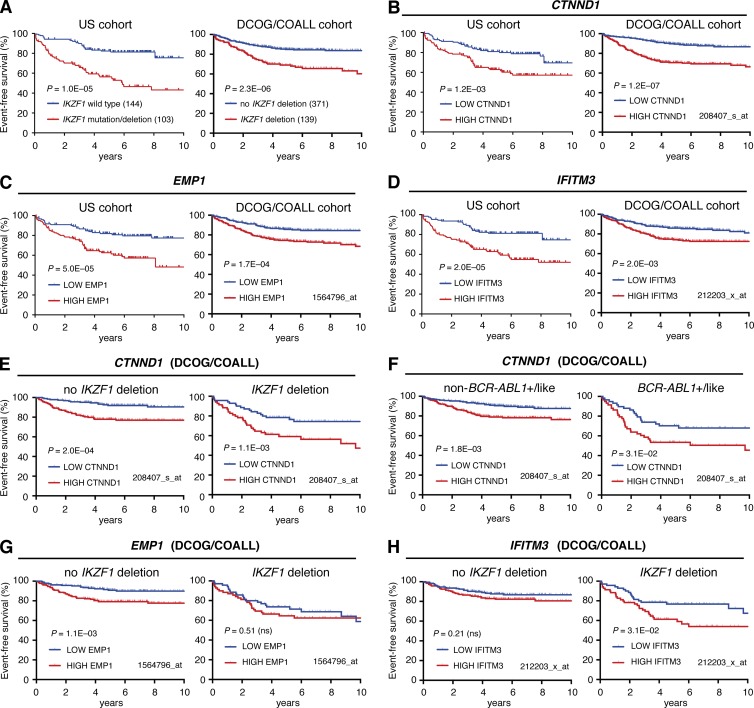
Consistent with previous studies (van der Veer et al., 2013; Roberts et al., 2014), IKZF1 mutation/deletion is associated with poor event–free survival (EFS) in the US patient cohort (hazard ratio [HR] = 3.21; 95% confidence interval [CI] = 2.23–5.83; log rank P = 1.0 × 10−5), and the DCOG/COALL patient cohort (HR = 2.47; 95% CI = 1.67–3.63; P = 2.3 × 10−6) we used to identify IKAROS-regulated genes (Fig. 4 A). We reasoned that altered expression of conserved IKAROS-activated and IKAROS-repressed genes may also have prognostic value in B-ALL. To address this, we examined EFS in patient cohorts after dividing them based on IKAROS-regulated gene expression into low (less than median) and high (greater than median) groups (Figs. S3, B and C; and S4, B and C). Low expression of the conserved IKAROS-activated genes CSF2RB, C1GALT1, and ORAI2 predicted poor EFS in the DCOG/COALL cohort; however, only ORAI2 was also predictive in the US cohort (Fig. S3, B and C; Fig. S4, B and C; and Table 1). High expression of six of the nine conserved IKAROS-repressed genes was associated with inferior EFS in both patient cohorts, with particularly strong effects observed for CTNND1, EMP1, and IFITM3 (Fig. 4, B–D; Fig. S3 C; Fig. S4 C; and Tables 1 and 2). Remarkably, stratifying DCOG/COALL patients by median CTNND1 expression was a better predictor of EFS than IKZF1 deletion status, with high expression conferring almost threefold higher risk (Fig. 4, A and B; and Table 2). Furthermore, high CTNND1 expression predicted poor outcome even within IKZF1 wild-type or IKZF1-deleted subgroups of that cohort, with 5-yr EFS <60% for patients with combined IKZF1 deletion and high CTNND1 expression (Fig. 4 E and Table 2). High CTNND1 expression was also associated with inferior EFS within BCR-ABL1+/BCR-ABL1-like or non-BCR-ABL1+/BCR-ABL1-like subgroups (Fig. 4 F). High EMP1 expression was also associated with decreased EFS in IKZF1 wild-type samples in both patient cohorts, whereas the predictive value of high IFITM3 expression appeared to depend on an association with IKZF1 deletion (Fig. 4, G and H; and Table 2).
Figure 4.
High CTNND1, EMP1, or IFITM3 expression predicts poor event-free survival. (A) Event-free survival analysis of US cohort (left) and DCOG/COALL cohort (right) B-ALL patients separated by IKZF1 mutation status as indicated. Log-rank p-values are shown. (B–D) EFS analysis as described for A, but separating cohorts based on median expression of (B) CTNND1, (C) EMP1, and (D) IFITM3. Expression of each gene across samples (determined by RNA-seq for US cohort and microarray for DCOG/COALL cohort with probeset indicated) are shown in Figs. S3 and S4. (E and F) EFS for DCOG/COALL patients grouped by IKZF1 deletion status (E) or BCR-ABL1+/like subgroup status (F), and then further separated based on median expression of CTNND1. (G and H) EFS for DCOG/COALL patients grouped by IKZF1 deletion status, and then further separated based on median expression of EMP1 (G) or IFITM3 (H).
Table 1. Conserved IKAROS-repressed and IKAROS-activated genes.
| Gene | Murine B-ALL | US cohort | DCOG/COALL cohort | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Differential expression (Acceleration) | Differential expression (Restoration) | Differential expression (combined Acceleration/ Restoration analysis) | Differential expression (IKZF1mut/del vs. IKZF1wt) | Event-Free Survivala (high vs. low for repressed, low vs. high for activated) | Differential expression (IKZF1del vs. IKZF1not del) | Event-Free Survivala (high vs. low for repressed, low vs. high for activated) | |||||||||
| Rank | P-value | Rank | P-value | Rankb | P-value | Rankc | P-value | poor outcome expression | P-value | Probesetd | Rank | P-value | poor outcome expression | P-value | |
| IKAROS-repressed | |||||||||||||||
| CTNND1 | 44 | 3.6 × 10−6 | 1 | 3.5 × 10−11 | 2 | 8.3 × 10−11 | 17 | 3.4 × 10−9 | high | 1.2 × 10−3 | 208407_s_at | 1 | 1.1 × 10−13 | high | 2.9 × 10−8 |
| EMP1 | 131 | 4.5 × 10−5 | 308 | 5.2 × 10−5 | 119 | 8.9 × 10−6 | 295 | 7.4 × 10−5 | high | 5.0 × 10−5 | 1564796_at | 129 | 4.5 × 10−6 | high | 1.2 × 10−4 |
| FXYD6 | 398 | 8.4 × 10−4 | 686 | 8.2 × 10−4 | 295 | 2.7 × 10−4 | 501 | 4.8 × 10−4 | high | 1.0 × 10−6 | 217897_at | 216 | 3.9 × 10−5 | high | 3.5 × 10−2 |
| IFITM3 | 628 | 2.8 × 10−3 | 85 | 6.9 × 10−7 | 77 | 2.0 × 10−6 | 368 | 1.4 × 10−4 | high | 2.0 × 10−5 | 212203_x_at | 96 | 1.2 × 10−6 | high | 1.7 × 10−3 |
| CTNNBIP1 | 253 | 2.5 × 10−4 | 1,154 | 5.7E-03 | 324 | 3.2 × 10−4 | 247 | 3.8 × 10−5 | high | 2.9 × 10−1 | 203081_at | 113 | 3.2 × 10−6 | high | 6.2 × 10−2 |
| S100A16 | 595 | 2.3 × 10−3 | 1,276 | 8.6 × 10−3 | 435 | 2.0 × 10−3 | 8 | 4.1E-10 | high | 3.4 × 10−2 | 227998_at | 95 | 1.1 × 10−6 | high | 2.0 × 10−1 |
| ROBO4 | 115 | 3.1 × 10−5 | 39 | 6.2 × 10−8 | 33 | 6.3 × 10−8 | 25 | 1.0 × 10−8 | high | 1.5 × 10−1 | 226028_at | 14 | 1.1 × 10−9 | high | 4.7 × 10−3 |
| AGAP3 | 820 | 6.1 × 10−5 | 1,132 | 5.4 × 10−3 | 472 | 2.9 × 10−3 | 24 | 1.0× 10−8 | high | 1.7 × 10−3 | 225789_at | 143 | 6.0 × 10−6 | high | 7.4 × 10−3 |
| ITPRIPL2 | 14 | 1.5 × 10−7 | 100 | 1.4 × 10−6 | 27 | 3.2 × 10−8 | 191 | 1.6 × 10−5 | high | 4.6 × 10−2 | 227514_at | 193 | 2.4 × 10−5 | high | 3.5 × 10−5 |
| IKAROS-activated | |||||||||||||||
| CSF2RB | 580 | 5.7 × 10−3 | 1,469 | 2.3 × 10−2 | 401 | 6.8 × 10−3 | 677 | 1.9 × 10−4 | low | 3.0 × 10−1 | 205159_at | 74 | 7.6 × 10−7 | low | 2.8 × 10−5 |
| C1GALT1 | 691 | 8.4E-03 | 53 | 2.5 × 10−6 | 64 | 7.9 × 10−6 | 903 | 4.5 × 10−4 | low | 9.0 × 10−1 | 226105_at | 92 | 1.6 × 10−6 | low | 6.7 × 10−3 |
| ASL | 543 | 4.7 × 10−3 | 238 | 1.0 × 10−4 | 157 | 1.5 × 10−4 | 666 | 1.8 × 10−4 | low | 6.4 × 10−1 | 204608_at | 81 | 9.5 × 10−7 | low | 1.8 × 10−1 |
| ORAI2 | 577 | 5.6 × 10−3 | 288 | 1.6 × 10−4 | 175 | 2.2 × 10−4 | 21 | 1.1 × 10−8 | low | 2.4 × 10−2 | 231406_at | 26 | 6.8 × 10−8 | low | 3.5 × 10−2 |
| IKAROS | |||||||||||||||
| IKZF1 | 653 (down) | 7.5 × 10−3 | 3 (up) | 1.4 × 10−8 | 4 | 6.6 × 10−8 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
N/A, not applicable.
Log rank P-value is shown. Genes where EFS is not significantly different (P > 0.05) are italicized.
Genes are ranked by F statistic, with F P-value shown.
Genes are ranked within their expression category (e.g. IKAROS-repressed genes are ranked by FDR after excluding genes with negative logFC values from IKZFmut/del vs wt analysis).
Microarray probeset with the smallest P-value was selected, excluding probesets lacking a gene symbol.
Table 2. EFS association with conserved IKAROS-repressed and IKAROS-activated gene expression in patient cohorts separated by IKZF1 deletion/mutation status.
| Gene | US cohort | DCOG/COALL cohort | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total cohort (n = 289) | IKZF1 wild-type (n = 144) | IKZF1 mutated/deleted (n = 103) | Total cohort (n = 573) | no IKZF1 deletion (n = 371) | IKZF1 deletion (n = 139) | |||||||||||||
| Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value | |
| Ikaros-activated | ||||||||||||||||||
| CTNND1 | 1.98 | 1.26–3.15 | 1.2 × 10−3 | 1.67 | 0.80–3.44 | 1.7 × 10−1 | 1.34 | 0.75–2.42 | 3.2 × 10−1 | 2.92 | 1.93–3.96 | 2.9 × 10−8 | 2.85 | 1.60–4.52 | 2.0 × 10−4 | 2.13 | 1.19–3.77 | 1.1 × 10−2 |
| EMP1 | 2.48 | 1.54–3.85 | 5.0 × 10−5 | 2.42 | 1.19–5.23 | 1.7 × 10−2 | 1.96 | 1.11–3.61 | 2.2 × 10−2 | 2.06 | 1.41–2.89 | 1.2 × 10−4 | 2.50 | 1.42–4.01 | 1.0 × 10−3 | 1.21 | 0.68–2.14 | 5.1 × 10−1 |
| FXYD6 | 2.63 | 1.86–5.10 | 1.0 × 10−6 | 1.50 | 0.63–3.79 | 3.4 × 10−1 | 2.26 | 1.28–4.16 | 6.1 × 10−3 | 1.47 | 1.03–2.10 | 3.5 × 10−2 | 1.33 | 0.79–2.24 | 2.8 × 10−1 | 1.50 | 0.84–2.65 | 1.7 × 10−1 |
| IFITM3 | 2.54 | 1.60–4.01 | 2.0 × 10−5 | 1.39 | 0.68–2.90 | 3.7 × 10−1 | 2.06 | 1.15–3.72 | 1.6 × 10−2 | 1.79 | 1.24–2.54 | 1.7 × 10−3 | 1.39 | 0.83–2.33 | 2.2 × 10−1 | 1.89 | 1.06–3.34 | 3.1 × 10−2 |
| CTNNBIP1 | 1.32 | 0.84–2.08 | 2.9 × 10−1 | 1.32 | 0.64–2.75 | 4.5 × 101 | 1.09 | 0.61–1.95 | 7.8 × 10−1 | 1.41 | 0.98–2.01 | 6.2 × 10−2 | 1.05 | 0.62–1.76 | 8.6 × 10−1 | 1.11 | 0.62–1.96 | 7.3 × 10−1 |
| S100A16 | 1.70 | 1.08–2.70 | 3.4 × 10−2 | 1.02 | 0.49–2.10 | 9.7 × 10−1 | 0.87 | 0.48–1.56 | 6.3 × 10−1 | 1.26 | 0.88–1.81 | 2.0 × 10−1 | 2.06 | 1.20–3.39 | 8.0 × 10−3 | 0.92 | 0.52–1.64 | 7.8 × 10−1 |
| ROBO4 | 1.37 | 0.87–2.16 | 1.5 × 10−1 | 1.17 | 0.57–2.43 | 6.7 × 10−1 | 1.17 | 0.65–2.10 | 5.9 × 10−1 | 1.69 | 1.17–2.40 | 4.7 × 10−3 | 1.16 | 0.69–1.96 | 5.7 × 10−1 | 2.07 | 1.16–3.64 | 1.4 × 10−2 |
| AGAP3 | 1.93 | 1.24–3.11 | 1.7 × 10−3 | 1.91 | 0.92–3.95 | 8.5 × 10−2 | 1.48 | 0.84–2.72 | 1.8 × 10−1 | 1.64 | 1.14–2.33 | 7.4 × 10−3 | 1.36 | 0.81–2.28 | 2.5 × 10−1 | 1.54 | 0.87–2.74 | 1.4 × 10−1 |
| ITPRIPL2 | 1.56 | 0.99–2.47 | 4.6 × 10−2 | 1.87 | 0.90–3.88 | 9.4 × 10−2 | 0.90 | 0.50–1.62 | 7.3 × 10−1 | 2.17 | 1.49–3.05 | 3.5 × 10−5 | 1.93 | 1.13–3.19 | 1.6 × 10−2 | 2.01 | 1.13–3.58 | 1.8 × 10−2 |
| Ikaros-repressed | ||||||||||||||||||
| CSF2RB | 1.27 | 0.81–2.01 | 3.0 × 10−1 | 1.27 | 0.61–2.63 | 5.2 × 10−1 | 0.60 | 0.34–1.08 | 9.1 × 10−2 | 2.20 | 1.50–3.08 | 2.8 × 10−5 | 1.59 | 0.94–2.65 | 8.6 × 10−2 | 1.54 | 0.87–2.74 | 1.4 × 10−1 |
| C1GALT1 | 0.98 | 0.62–1.54 | 9.0× 10−1 | 0.81 | 0.39–1.68 | 5.7 × 10−1 | 0.90 | 0.50–1.61 | 7.2 × 10−1 | 1.66 | 1.15–2.35 | 6.7 × 10−3 | 1.56 | 0.92–2.61 | 9.8 × 10−2 | 1.47 | 0.83–2.60 | 1.9 × 10−1 |
| ASL | 0.90 | 0.57–1.42 | 6.4 × 10−1 | 1.03 | 0.50-2.13 | 9.4 × 10−1 | 1.11 | 0.57–1.82 | 9.6 × 10−1 | 1.28 | 0.89–1.82 | 1.8 × 10−1 | 1.67 | 0.99–2.80 | 5.6 × 10−1 | 0.85 | 0.48–1.50 | 5.7 × 10−1 |
| ORAI2 | 1.69 | 1.08–2.68 | 2.4 × 10−2 | 2.11 | 1.01-4.33 | 4.8 × | 0.89 | 0.50–1.59 | 6.9 × 10−1 | 1.47 | 1.03–2.10 | 3.5 × 10−2 | 1.08 | 0.64–1.81 | 7.8 × 10−1 | 1.56 | 0.88–2.76 | 1.3 × 10−1 |
Log rank P-values are shown. Genes where EFS is not significantly different (P > 0.05) are italicized.
Comparable deregulation of IKAROS-repressed genes in mouse and human B-ALL
The correlation between elevated CTNND1, EMP1, or IFITM3 expression with poor disease outcome in two independent patient cohorts is notable given that they are among the most highly expressed of the conserved IKAROS-regulated genes by RNA-seq in the US sample cohort and our mouse B-ALL model (Fig. 5, A–C; Fig. S3, B and C; and Fig. S5, A and B). Expression of each gene is elevated approximately twofold in IKAROS/Ikaros-disrupted B-ALL in both species, and is rapidly normalized upon acute Ikaros restoration in Ikaros-kd leukemia (Fig. 5, A–C). Together, these expression and outcome data implicate EMP1, IFITM3, and CTNND1, as IKAROS-repressed genes with potential functional importance in B-ALL pathogenesis and therapy response. Intriguingly, high EMP1 expression has recently been associated with poor outcome in B-ALL by promoting resistance to glucocorticoids (Ariës et al., 2014). Hence our identification of EMP1 as a novel and conserved IKAROS-repressed gene suggests a potential mechanism whereby IKZF1 disruption promotes B-ALL treatment resistance.
Figure 5.
Regulation of IKAROS-repressed genes in mouse and human B-ALL. (A–C) RNA-seq expression (RPKM) of (A) Emp1/EMP1, (B) Ifitm3/IFITM3, and (C) Ctnnd1/CTNND1 in control and Ikaros-kd murine B-ALL with Dox treatments indicated (left), and the in US B-ALL cohort stratified by IKZF1 mutation status (right). P-values from moderated Student’s t test. (D) Western blots of Ctnnd1 and Ikaros protein expression in multiple independent primary control and Ikaros-kd leukemias. Ikaros-kd samples B038, B027, and B031, and control samples B002 and B005 are transgenic leukemias (Fig. 1 and 2 and S1), whereas RV#1044 and #1040 were generated using the retroviral approach (Fig. S2). Actin is a loading control.
Also noteworthy among IKAROS-repressed genes was CTNND1, which encodes the cadherin-associated protein P120-catenin. When ranked by significance, CTNND1 was the top IKAROS-repressed gene in the DCOG/COALL patient cohort and among the top 20 such genes in the US cohort (Table 1). Ctnnd1 was the most significantly repressed gene in the murine B-ALL transcriptome upon Ikaros restoration in vivo, and ranked second among mouse Ikaros-repressed genes in our combined analysis (Table 1; Tables S2 and S3; and Fig. 5 C). Consistent with this RNA-seq analysis, we verified that Ikaros restoration triggered rapid loss of Ctnnd1 protein in multiple primary Ikaros-kd leukemias, with further Ctnnd1 down-regulation upon sustained Dox treatment (Fig. 5 D). Alternative pre-mRNA splicing produces multiple CTNND1/P120-catenin protein isoforms with cell type–specific expression and distinct subcellular localizations/functions (Schackmann et al., 2013). Examining RNA-seq data in mouse and human BCR-ABL+ B-ALL (driven by Ikaros knockdown and IKZF1 alteration, respectively) revealed that Ctnnd1/CTNND1 transcripts predominantly include exon A but exclude exons B and C (Fig. S5, C–E). These mRNA species encode protein isoforms harboring nuclear localization signals and lacking nuclear export signals, suggesting a potential nuclear role for CTNND1 in B-ALL (see Discussion).
Because Ctnnd1 mRNA consistently fell in Ikaros-kd leukemia upon Ikaros restoration, its expression rebounded to original high levels in multiple independent primary leukemias harvested from mice that relapsed after long-term Dox administration (Fig. S2, B–D). To examine this further, we performed RNA-seq of leukemia cells isolated from a series of mice bearing representative Ikaros-kd B-ALL B027 that were either untreated or acutely Dox treated, or after disease relapse after 103 d on Dox. Notably, elevated expression of Ctnnd1 and Emp1 at relapse contributed to an overall gene expression signature resembling the original untreated leukemia (Fig. S2, B–D), raising the possibility that renewed expression of these genes may contribute to the relapse phenotype.
Inducible knockdown of IKAROS-repressed genes in BCR-ABL1+, Ikaros-kd B-ALL
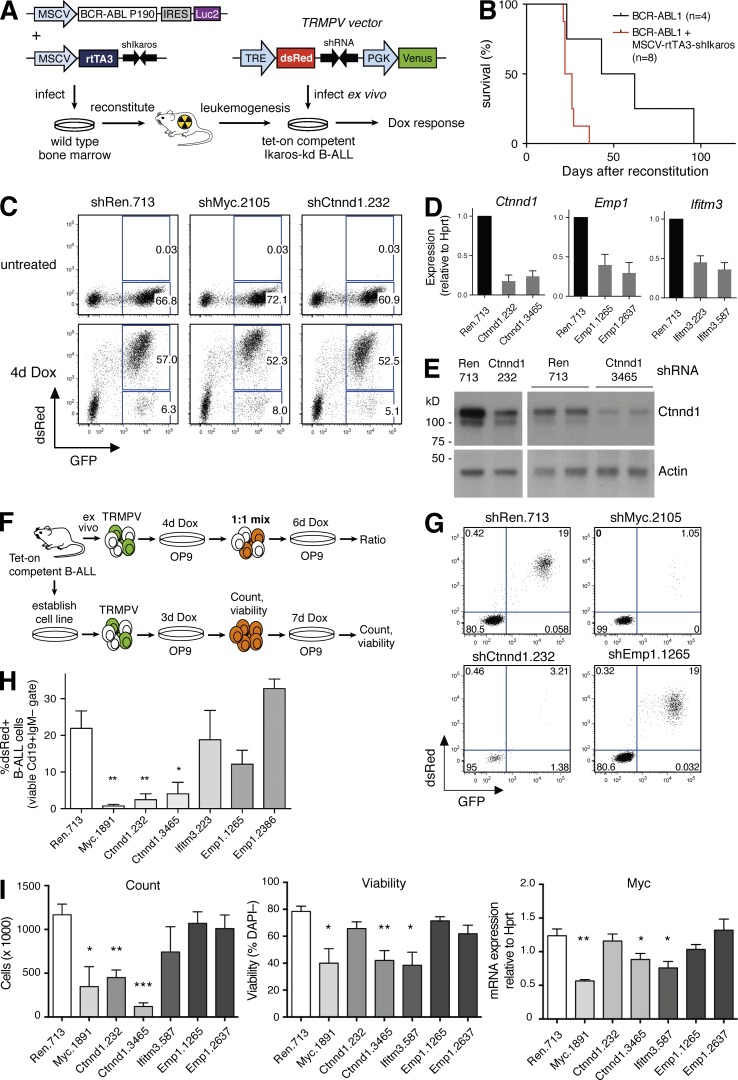
In BCR-ABL1–driven, Ikaros-kd B-ALL, we hypothesized that elevated expression of genes such as Ctnnd1, Emp1, and Ifitm3 may contribute to accelerated leukemogenesis, and the rapid repression of these genes upon acute Ikaros restoration may contribute to disease regression. To examine the dependency of Ikaros-kd B-ALL cells on Ctnnd1, Emp1, or Ifitm3, we used a novel strategy to generate primary leukemias in which endogenous genes can be dynamically inhibited using tet-regulated RNAi. First, we co-infected wild-type bone marrow cells with a retroviral BCR-ABL1P190 expression vector along with a novel vector encoding the tet-on transactivator rtTA3 linked to a stable shRNA targeting mouse Ikaros (MSCV-rtTA3-shIkaros; Fig. 6 A). MSCV-rtTA3-shIkaros accelerated BCR-ABL1-driven B lineage leukemogenesis when compared with bone marrow infected with the BCR-ABL1 vector alone (Fig. 6 B, median survival 24 d versus 53 d; P < 0.05, log-rank test), yielding Ikaros-kd leukemias that also express rtTA3 (tet-on competent B-ALL).
Figure 6.
Ctnnd1 knockdown compromises growth of Ikaros-kd, BCR-ABL1+ B-ALL in vitro. (A) Strategy for generating tet-on competent, Ikaros-kd, BCR-ABL1+ B-ALL, allowing inducible target gene knockdown. (B) Kaplan-Meier survival analysis of mice reconstituted with infected bone marrow cells as shown in A. Recipients of cells co-transduced with MSCV-BCR-ABL1-IRES-Luc2 and MSCV-rtTA3-shIkaros (n = 8) had median survival of 24 d, versus 53 d for MSCV-BCR-ABL1-IRES-Luc2 alone (n = 4; P = 0.022, log-rank test). (C) Representative dsRed and GFP flow cytometry of tet-on competent Ikaros-kd BCR-ABL1+ B-ALL derived from B cells infected with TRMPV vectors as shown in A. Cells were cultured on OP9 stroma and Dox treated as indicated, with dsRed marking shRNA expressing cells. (D) RT-qPCR expression of Ctnnd1, Emp1, and Ifitm3 relative to Hprt in ex vivo cultured tet-on competent B-ALL cells infected with TRMPV vectors expressing the indicated shRNAs. Cells were Dox treated for 4 d and GFP+dsRed+ cells were isolated by FACS. Mean ± SEM for three independent experiments. (E) Western blots of Ctnnd1 expression in splenic tumor cells harvested from two independent tet-on competent primary B-ALL transplant recipients infected with TRMPV vectors expressing shRen.713 or two different Ctnnd1 shRNAs as indicated. Before organ harvest, mice were Dox treated for 4 d and GFP+dsRed+ cells were isolated by FACS. Actin is a loading control. (F) Schematic of the culture strategy for (G–I) below. (G) Representative dsRed and GFP flow cytometry of primary tet-on competent Ikaros-kd BCR-ABL1+ B-ALL cells (viable CD19+IgM– cells) after ex vivo infection with TRMPV vectors, 4 d Dox treatment, and plating at a 1:1 ratio of uninfected (CD19+GFP–mCherry–) and infected (CD19+GFP+mCherry+) B-ALL cells as shown in F. (H) Proportion of dsRed+ B-ALL cells within the viable CD19+IgM– gate as described in G. Mean ± SEM for four experiments in two independent primary tet-on–competent leukemias. (I) Analysis of an established cell line derived from continuous passage of a primary tet-on–competent Ikaros-kd BCR-ABL1+ B-ALL, infected with indicated TRMPV-shRNAs as shown in F. After 3 d of Dox treatment on OP9 stroma, 50,000 sorted GFP+dsRed+ B-ALL cells were replated in Dox on OP9 stroma for a further 7 d before analysis. Plots show overall cell counts (left), the proportion of viable DAPI– cells (middle), and RT-qPCR analysis of Myc expression (right). Mean ± SEM, n = 5 independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001, relative to Ren.713, unpaired Student’s t test.
To inducibly inhibit murine Ctnnd1, Emp1, and Ifitm3 we cloned effective miR30-based shRNAs into the retroviral TRMPV vector (Zuber et al., 2011), which stably expresses the GFP derivative Venus and allows tet-inducible coexpression of a shRNA linked to the fluorescent marker dsRed (Fig. 6 A). We infected primary tet-on competent B-ALL cells ex vivo with TRMPV vectors targeting Renilla luciferase (negative control), the essential proliferative driver Myc (positive control), and two independent shRNAs targeting each IKAROS-repressed gene. Dox treatment induced dsRed fluorescence in GFP+ B-ALL cells in vitro and in vivo, and we verified depletion of target gene mRNA and protein in these cells (Fig. 6, C–E).
Ctnnd1 promotes primary Ikaros-kd B-ALL growth and viability ex vivo
To assess the role of conserved IKAROS-repressed genes in Ikaros-kd B-ALL maintenance, we plated freshly harvested BCR-ABL1-rtTA3-shIkaros B-ALL cells on OP9 stromal cells, which support primary lymphoblast proliferation (Nakano et al., 1994). After immediate ex vivo infection of two independent primary leukemias with TRMPV vectors and growth in Dox medium for 4 d, infected (CD19+GFP+mCherry+) cells were mixed at a 1:1 ratio with uninfected (CD19+GFP–mCherry–) cells and plated in Dox for an additional 6 d (Fig. 6 F). In this ex vivo competition assay, B-ALL cells expressing the positive control Myc shRNA were almost completely depleted from the culture (Fig. 6, G and H). Cells expressing either of the two effective shRNAs targeting Ctnnd1 were also significantly depleted, whereas inhibition of Emp1 or Ifitm3 had no effect in this culture system (Fig. 6, G and H).
We then established a BCR-ABL1-rtTA3-shIkaros B-ALL cell line through continuous culture (∼3 wk) on OP9 cells. Infection with TRMPV vectors and 3 d of Dox treatment induced dsRed/shRNA, at which point cells expressing the Ctnnd1.3465 shRNA showed a minor decrease in viability (Fig. S5, F and G). 50,000 viable GFPhidsRedhi B-ALL cells were then sorted and replated on stromal support for an additional 7 d in the presence of Dox to maintain shRNA expression and target gene knockdown (Figs. 6 F and S5, H and I). As anticipated, Myc knockdown decreased B-ALL proliferation and viability relative to negative control cells expressing an shRNA-targeting Renilla luciferase (Fig. 6 I). Notably, both Ctnnd1 shRNAs also inhibited B-ALL cell expansion (associated with loss of viability for the Ctnnd1.3465 shRNA), but with no significant change in cell cycle (Figs. 6 I and S5, H and I). Previous studies have implicated Ctnnd1 in the regulation of Wnt pathway target genes, including the proliferative drivers Myc and Ccnd1/Cyclin D1 (Daniel and Reynolds, 1999; van Roy and McCrea, 2005; Schackmann et al., 2013). Although we observed a modest decrease in Myc expression in Ctnnd1.3465 shRNA cells, Ccnd1 expression was not affected by Ctnnd1 knockdown (Fig. 6 I and S5 J). Although Ifitm3 and Emp1 knockdown did not reduce overall cell numbers in these assays, Ifitm3 suppression modestly decreased B-ALL viability associated with Myc and Ccnd1 down-regulation (Figs. 6 I and S5 J). Together, these experiments demonstrate that Ctnnd1 promotes growth and viability of Ikaros-kd BCR-ABL1+ B-ALL cells ex vivo.
Discussion
Disruption of genes encoding hematopoietic transcription factors occurs in the majority of B-ALL cases; however, identifying transcription factor target genes relevant to tumor suppression remains a major challenge. In this study, we have taken a unique approach to identifying high-confidence IKAROS-regulated genes in B-ALL based on an integrated analysis of gene expression profiles accompanying IKAROS loss in murine and human disease. This approach includes customary comparisons of IKAROS-deficient and IKAROS-proficient samples in multiple primary murine B-ALL models and two independent human B-ALL cohorts, but importantly also incorporates gene expression profiling of murine B-ALL upon dynamic restoration of endogenous Ikaros in vivo. Including this transcriptional filter mutes the influence of co-occurring oncogenic lesions, cell type differences, or adaptive gene expression changes associated with steady-state Ikaros loss, and enriches for genes particularly sensitive to Ikaros status in established disease.
In the novel mouse models of BCR-ABL1+ B-ALL described here, RNAi-based Ikaros knockdown significantly accelerates leukemogenesis, consistent with previous studies using Ikaros germline mutant mice (Virely et al., 2010; Schjerven et al., 2013; Churchman et al., 2015). By exploiting the reversibility of RNAi, we further demonstrate that Ikaros-low B-ALL undergoes sustained and in some cases indefinite regression when Ikaros function is re-engaged. The sensitivity of established leukemia to Ikaros restoration provides proof-of-principle for therapeutic approaches that engage components of the Ikaros-regulated transcriptome. In addition, the fact that B-ALL can be eradicated by Ikaros restoration despite ongoing BCR-ABL1 activity implicates Ikaros in pathways that cooperate with BCR-ABL1 during disease pathogenesis in addition to providing BCR-ABL1–independent functions in established disease. BCR-ABL1–independent functions of IKAROS also appear likely in pediatric BCR-ABL1+ B-ALL, where IKZF1 mutation/deletion is associated with poor clinical response to standard therapies and the BCR-ABL1 inhibitor imatinib (van der Veer et al., 2014).
The B-ALL mouse models described here and elsewhere (Virely et al., 2010; Schjerven et al., 2013; Churchman et al., 2015) functionally recapitulate the high rates of IKZF1 alteration observed in human BCR-ABL1+ B-ALL, suggesting that the transcriptomic changes underlying this genetic interaction may be evolutionarily conserved. We approached this possibility by applying stringent statistical thresholds to identify ∼400 IKAROS-regulated genes common to two independent human B-ALL sample cohorts comprising multiple disease subtypes, then intersecting them with genes that behave concordantly in our mouse B-ALL models. This unbiased, cross-species analysis yields 13 conserved IKAROS-regulated genes in B-ALL. Of particular interest are the nine conserved IKAROS-repressed genes, which are all directly bound by IKAROS and may represent disease drivers and therapeutic targets in IKZF1-deficient disease. High expression of 6 of these genes (CTNND1, EMP1, FXYD6, IFITM3, AGAP3, and ITPRIPL2) also individually predict inferior event-free survival in both patient cohorts, suggesting a potential role in B-ALL therapy resistance.
Recent studies in mice show that Ikaros mutant B cell progenitors and B-ALL cells rely on cell–cell adhesion for survival (Joshi et al., 2014; Churchman et al., 2015). Disrupting adhesion may be a rational approach to treating Ikaros mutant B-ALL (Joshi et al., 2014; Churchman et al., 2015), however the adhesive mechanisms of Ikaros mutant cells remain poorly understood. Notably, of the 9 conserved IKAROS-repressed genes identified in our study, IFITM3, EMP1, and CTNND1 all encode transmembrane or membrane-associated proteins with established roles in cell–cell adhesion. IFITM3 encodes interferon inducible transmembrane protein 3, required for germ cell migration (Tanaka et al., 2005), which plays an essential role in cellular resistance to viral infection (Everitt et al., 2012) and promotes colon cancer pathogenesis and metastasis (Andreu et al., 2006; Li et al., 2011). Of note, IFITM3 was recently implicated in B-ALL pathogenesis for the first time, emerging as a central mediator of B-ALL proliferation and survival by promoting CD19-mediated PI3K–AKT signaling (Lee et al., 2015). EMP1 encodes epithelial membrane protein 1, which in epithelial cells resides in tight junctions (Durgan et al., 2015). EMP1 was also recently implicated in B-ALL, identified among genes with high expression in B-ALL samples that are resistant to therapeutic glucocorticoids (Ariës et al., 2014). In that study, elevated EMP1 confers glucocorticoid resistance and enhanced adhesion of ALL cell lines in vitro, and is associated with inferior event–free survival of B-ALL patients (Ariës et al., 2014). Hence, our identification of EMP1 and IFITM3 as conserved IKAROS-repressed genes suggests novel mechanisms whereby disruption of IKZF1 promotes B-ALL development and treatment resistance. EMP1 is also unique among conserved IKAROS-regulated genes in that its high expression predicts poor EFS within the IKZF1 wild-type subset of both patient cohorts, suggesting an additional, IKAROS-independent influence on disease outcome.
Along with EMP1 and IFITM3, CTNND1 is notable among conserved IKAROS-repressed genes for several reasons. Ctnnd1 is the most significantly down-regulated gene upon Ikaros restoration in our murine B-ALL model, displaying rapid and sustained mRNA and protein repression. In the DCOG/COALL patient cohort, CTNND1 is the top-ranked up-regulated gene in IKZF1-deleted versus IKZF1 wild-type disease, and its elevated expression is a more powerful predictor of poor EFS than IKZF1 deletion. Our identification of CTNND1 as a novel and direct IKAROS-repressed gene provides a mechanistic basis for previous studies listing CTNND1 among genes highly expressed in IKZF1-deleted versus IKZF1 wild-type, Ph-negative B-ALL (Vitanza et al., 2014), and among genes highly expressed in Ph+ B-ALL (a subtype enriched for IKZF1 alterations; Mullighan et al., 2008a) versus other B-ALL subtypes (Juric et al., 2007). Importantly, we find that high CTNND1 expression is associated with inferior EFS, even within molecular subgroups of the DCOG/COALL cohort (including those defined by IKZF1 status or combined BCR-ABL1/BCR-ABL1-like status), suggesting that high CTNND1 expression influences B-ALL outcome regardless of whether it occurs as a result of IKZF1 deletion.
Using a novel tet-on competent murine B-ALL model driven by combined BCR-ABL1 expression and Ikaros knockdown, we have demonstrated a novel requirement for Ctnnd1 in leukemia maintenance. In epithelial cells, the Ctnnd1 gene product P120-catenin regulates cell–cell adhesion by direct stabilization of E-cadherin (Ireton et al., 2002; Davis et al., 2003), but in many cell types, it also activates β-catenin (Ctnnb1) by directly interfering with the transcriptional repressor Kaiso in the nucleus (Daniel and Reynolds, 1999; van Roy and McCrea, 2005; Schackmann et al., 2013). We have shown for the first time that mouse and human B-ALL cells predominantly express Ctnnd1/CTNND1 mRNA splice forms predicted to encode nuclear localized P120-catenin. Although it is tempting to speculate that P120-catenin could contribute β-catenin activation, which has been implicated in acute leukemia self-renewal and relapse (Wang et al., 2010; Dandekar et al., 2014; Giambra et al., 2015), we did not observe consistent down-regulation of the β-catenin target genes Myc and Ccnd1 upon Ctnnd1 knockdown in our B-ALL model system. Hence the mechanism whereby P120-catenin promotes B-ALL maintenance remains to be determined.
In summary, we have taken advantage of novel mouse models and recent major advances in the molecular annotation of large B-ALL patient cohorts to perform the first cross-species comparison of IKAROS-regulated genes in B-ALL. Among gene products with conserved, abnormally high expression in IKZF1-mutated/deleted B-ALL are EMP1 and IFITM3, both recently implicated in B-ALL pathogenesis and therapy resistance, and CTNND1, for which we demonstrate a novel role in B-ALL maintenance. Elevated CTNND1, EMP1, or IFITM3 expression in B-ALL is associated with increased risk of relapse in 2 independent patient cohorts. These findings suggest novel mechanisms whereby IKZF1 mutations confer poor prognosis in B-ALL and identify potential new therapeutic targets in high risk disease.
Materials and methods
Transgenic mice
TREtight-GFP-shIkaros.4056 and TREtight-GFP-shRenilla.713 mice were previously described (Witkowski et al., 2015). TRE-GFP-shIkaros.2709 transgenic mice harbor a different shRNA (Ikaros.2709 not Ikaros.4056) and a different tet-regulatable promoter (TRE not TREtight) that affords improved shRNA expression in B cell progenitors, and were generated using previously described protocols (Premsrirut et al., 2011). BCR-ABL1P190 and Vav-tTA transgenic mice were described previously (Heisterkamp et al., 1990; Kim et al., 2007). TRE-GFP-shIkaros.4056 transgenic mice (Witkowski et al., 2015) and TRE-GFP-shIkaros.2709 transgenic mice were PCR genotyped using a common forward primer that anneals to the endogenous Col1a1 locus (5′-AATCATCCCAGGTGCACAGCATTGCGG-3′) and a reverse primer that either anneals to the Col1a1 locus (5′-CTTTGAGGGCTCATGAACCTCCCAGG-3′) to amplify a 238-bp wild-type allele product, or anneals to the transgene SAdpA cassette (5′-ATCAAGGAAACCCTGGACTACTGCG-3′) to amplify a 300-bp transgene product. PCR conditions were 95°C for 5 min, 35 × (95°C for 40 s, 62°C for 45 s, and 72°C for 60 s), and 72°C for 60 s. The BCR-ABL1P190 transgene was detected using PCR primers 5′-AGAGATCAAACACCCTAACCT-3′ and 5′-CCAAAGCCATACTCCAAATGC-3′, using PCR conditions 95°C 6’, 35 × (95°C for 30 s, 55°C for 40 s, and 72°C for 40 s), 72°C 60”. The Vav-tTA transgene was detected as described previously (Liu et al., 2014). Mice were on an inbred C57BL6/J background apart from Vav-tTA mice, which were on an FVB/N background. Doxycycline (Dox; Sigma-Aldrich) was administered in the diet at 600 mg/kg food (Specialty Feeds). All mouse experiments were approved by the Walter and Eliza Hall Institute Animal Ethics Committee.
Retroviral infection
We adapted an established protocol (Li et al., 1999) to preferentially drive B-ALL rather than CML upon retroviral expression of BCR-ABL1P190. Bone marrow harvested from both femurs of 4-wk-old CD45.2+ mice was resuspended in ice-cold PBS, centrifuged at 1,500 rpm for 5 min at 4°C, and red blood cells were briefly lysed using red cell lysis buffer. Cells were resuspended in IMDM supplemented with 15% FCS, 10 ng/ml mIL-7 (PeproTech), 100 ng/ml mSCF (PeproTech), 50 ng/ml Flt3L and TPO (in-house), and 2 mM l-glutamine (Life Technologies), and incubated at 37°C, 10% CO2 overnight. Cells were then reresuspended in fresh media at 2 × 106 cells/ml. Retronectin-coated, non–tissue culture treated plates were coated with 1 ml of MSCV-BCR-ABL1-IRES-Luc2 and MSCV-IRES-tTA retrovirus. Viral supernatant was aspirated, and 1 ml of bone marrow cell suspension was plated per well followed by incubation cells at 37°C, 10% CO2 overnight. Infected cells were then injected intravenously into lethally irradiated CD45.2+ recipient mice. Similar methods were used to generate tet-on competent B-ALL; however, bone marrow was co-infected with MSCV-BCR-ABL1-IRES-Luc2 and MSCV-rtTA3-shIkaros.4056 before transplant.
Leukemia cell culture and Western blotting
OP9 stromal feeder cells were cultured in IMDM supplemented with 10% FBS, 100 µg/ml streptomycin, 100 U/ml penicillin, 100 µM l-glutamine, and 50 µM β-mercaptoethanol and grown at 37°C in 10% CO2. B-ALL tumor cells were cultured on an OP9 layer in media conditioned with 1ng/µl IL-7 (PeproTech). For tet-on–competent B-ALL transduction, 5 × 106 freshly harvested secondary leukemic spleen cells were spin-infected at 1,200 rpm at 22°C onto TRMPV-shRNA retrovirus-coated plates for 2 h. After centrifugation, cells were incubated in medium for 6 h at 37°C in 10% CO2 and plated onto OP9 stroma layer supplemented with Doxycycline and IL-7 (PeproTech). Sorted cells were cultured on OP9 in the presence of Doxycycline and analyzed by flow cytometry using BD Fortessa. Cell lysates were Western blotted with anti-Ikaros antibody E-20 (sc-9861; Santa Cruz Biotechnology, Inc.), anti–P120-catenin antibody 98/pp120 (BD), and anti-actin antibody I-19 (sc-1616; Santa Cruz Biotechnology, Inc.).
Leukemia transplantation and flow cytometry analysis
Splenocytes (predominantly CD19+IgM– leukemia cells) from primary leukemic mice were injected into the tail vein of immunocompromised CD45.1+Rag1−/− recipient mice (106 cells/mouse), which generally developed overt signs of leukemia (e.g., anemia and splenomegaly) after ∼2–3 wk. Single-cell suspensions were prepared from bone marrow, spleen, and peripheral blood of secondary leukemic mice. After red blood cell lysis, cells were stained with APC-conjugated anti-IgM (eBioscience), PE-Cy7–conjugated anti-CD19 (BD), Alexa Fluor700–conjugated anti-CD45.2 (WEHI), biotin-conjugated CD43 (BD), PE-conjugated CD24 (BD), and PerCP-Cy5.5–conjugated Streptavidin. Stained cells were analyzed on a fluorescence-activated cell sorter (Fortessa; BD). To assess viability, cells were stained with Fluoro-Gold (Sigma-Aldrich). For OP9 co-culture, cells were harvested and removed from the stromal layer before antibody staining. Stat5 activation (pStat5) analysis was performed as previously described (Tremblay et al., 2016) on single cell spleen or bone marrow suspensions stained with antibodies against mouse CD45.1 (A20) and CD45.2 (104; BD). Cells were fixed in 4% paraformaldehyde for 10 min at 4°C in the dark, washed twice with 2% FCS in PBS, and permeabilized in pre-chilled Phosflow Perm Buffer III (BD) for 30 min at 4°C. Permeabilized cells were washed in 2% FCS in PBS and stained overnight at 4°C using rabbit anti–mouse phospho-Stat5 (Tyr694) antibody (#9351; Cell Signaling Technology). Stained cells were washed twice and incubated in 2% FCS in PBS with donkey Alexa Fluor 546–conjugated anti–rabbit secondary antibody (A10040; Molecular Probes) for 1 h on ice. Cells were washed twice in ice-cold PBS, and flow analysis was performed using a LSRII cytometer.
Gene expression analysis
RNA was extracted from FACS-sorted CD45+CD19+IgM– tumor cells using an RNeasy kit (QIAGEN, CA). For RT-qPCR analysis, 4.5 µg of total RNA was converted to cDNA, 1 µl of which was used in triplicate 10 µl SYBR Green (Roche) PCR reactions using a LightCycler 480 Instrument (Roche). Primer sequences (5′-3′): Hprt-FWD 5′-CAGTACAGCCCCAAAATGGT-3′; Hprt-REV 5′-CAAGGGCATATCCAACAACA-3′; Ikzf1-FWD 5′-CAATGTCGCCAAACGTAAGA-3′; Ikzf1-REV 5′-GTTGATGGCATTGTTGATGG-3′; Ctnnd1-FWD 5′-CAGGACAGATTGTGGAAACCTA-3′; Ctnnd1-REV 5′-GCTGTACTGTCCGAGTTGTCAT-3′; Emp1-FWD 5′-GTTGGTGCTACTGGCTGGTC-3′; Emp1-REV 5′-TACCACCAGTGCAGTTCTTCC-3′; Ifitm3-FWD 5′-ATGTGGTCTGGTCCCTGTTC-3′; Ifitm3-REV 5′-CTTAGCAGTGGAGGCGTAGG-3′; Myc-FWD 5′-ACAGGACTCCCCAGGCTCCG-3′; Myc-REV 5′-CGTGGCTGTCTGCGGGGTTT-3′; Ccnd1-FWD 5′-TCTGTGAGGAGCAGAAGTGC-3′; Ccnd1-REV 5′-CTTAGAGGCCACGAACATGC-3′. Ct values were calculated using LightCycler 480 software and relative mRNA expression levels determined using Standard Curve Method.
For RNA-seq analysis, RNA-seq libraries were prepared according to Illumina TruSeq RNA preparation protocols. Single-index RNA adapters were ligated to RNA samples run on the Illumina HiSeq 2000 sequencing platform, whereas dual-indexed sequencing adapters were ligated to samples for the Illumina Next-Seq 500 platform. 26 RNA samples were sequenced in three different batches. Eight samples from tumors B891, B1030, B1040, and B1044 (retroviral system) produced between 2–12 million single-end 100-bp reads on Illumina HiSeq. 6 samples from tumors B011, B031, and B035 (transgenic system) produced 14–17 million paired-end 100 bp reads on Illumina HiSeq. 12 samples from tumor B005, B014, B027, B031, B038, and B041 (transgenic system) produced 5–7 million paired-end 76-bp reads on Illumina NextSeq with counts from four technical replicates summed for each sample. Samples from tumor B1030, B011, B005, and B041 contain two controls, one at day 0 and the other at day 3 Dox treatment. For the remaining tumors, each has an Ikaros knockdown sample and an Ikaros restoration sample after Dox treatment.
Bioinformatic analysis
RNA sequence reads were mapped to the mouse genome (mm10) using the subread aligner (Liao et al., 2013) implemented in the Rsubread software package. Read counts for each Entrez Gene were obtained using featureCounts (Liao et al., 2014) and its inbuilt mm10 annotation. Sequence reads and counts are available from Gene Expression Omnibus under accession no. GSE75976.
Statistical analysis used the edgeR (Robinson et al., 2010) and limma (Ritchie et al., 2015) software packages. Genes were filtered as not expressed if they failed to achieve 0.5 read counts per million (cpm) in at least four samples. Genes from chromosome Y and the gene Xist were filtered out to avoid gender effects. Predicted genes, immunoglobulin genes, mitochondrial genes, and genes without official gene symbols were also filtered. Trimmed Mean of M-values (TMM) scale normalization (Robinson and Oshlack, 2010) was applied and read counts were transformed to log2-cpm with a prior count of 1 using edgeR’s cpm function. Linear models were used to test for expression differences between Ikaros restoration versus Ikaros knockdown and between Ikaros knockdown versus control at day 0 while adjusting for batch effects. An extra surrogate variable was estimated to adjust for unwanted nuisance technical effects by performing a singular value decomposition of the residuals, with emphasis on highly variable genes. Empirical array quality weights were estimated to allow for differences in quality between the RNA samples (Ritchie et al., 2006). Each tumor was treated as a random block, allowing for correlation between the two samples from the same tumor (Smyth et al., 2005). Differential expression were assessed using empirical Bayes moderated t-statistics (Smyth, 2004), allowing for an abundance trend in the standard errors and for robust estimation of the Bayesian hyperparameters. The underlying biological correlation between the Acceleration and Restoration expression profiles was estimated using limma’s genas function (Ritchie et al., 2015). This method corrects for the technical correlation that arises from sharing controlling samples between the two profiles.
To identify conserved IKAROS-repressed and -activated genes, human gene symbols from the US cohort were mapped to Entrez Gene IDs using NCBI annotation and probeset IDs from the DCOG/COALL cohort were mapped to Entrez Gene IDs using Affymetrix annotation. Murine gene lists were mapped to human Entrez Gene IDs using the Jackson Laboratory mouse-human ortholog table and the NCBI mouse-human homologue table. Gene ontology analysis used limma’s goana function. Tumors with relapse samples were analyzed separately for Fig. S2. Log2 reads per kilobase per million reads (log2-RPKM) were computed using edgeR’s rpkm function with prior count of 1. The heat map was created using the heatmap.2 function in the gplots package.
IKAROS ChIP in human B-ALL cells as detailed in Schjerven et al. (2016) identified 11,446 human IKAROS-bound genes corresponding to 11,525 mouse homologues. Overlap analysis identified IKAROS-regulated genes bound by IKAROS.
B-ALL patient cohort analysis
US cohort patients were from the St. Jude Children’s Research Hospital Total XV and Total XVI protocols, the COG P9906 high-risk B-ALL study, the COG AALL0232 high-risk ALL study, the ECOG E2993 trial, the MD Anderson Cancer Centre protocols, and the Alliance (Cancer and Leukemia Group B) protocols C19802 and C10102. Single-nucleotide polymorphism (SNP) 6.0 microarray analysis (Affymetrix) and RNA sequencing (Illumina Tru-seq) were performed as previously described (Roberts et al., 2014). Event-free survival was estimated using Kaplan-Meier, with Peto’s estimator of standard deviation and the log-rank test. An event was defined as a failure to achieve remission, a relapse after remission, or the development of a second malignancy. Analysis was performed using Prism software (GraphPad), R software, and SAS software version 9.1.2 (SAS Institute).
The DCOG/COALL cohort comprises children with newly diagnosed BCP-ALL enrolled in consecutive Dutch Childhood Oncology Group trials (DCOG ALL-8, ALL-9, and ALL-10) and German Cooperative ALL trials (COALL 06–97 and 07–03). This combined patient cohort was described previously (Den Boer et al., 2009; van der Veer et al., 2013). Event-free survival included relapse, secondary malignancy or death as an event and was estimated using the actuarial Kaplan-Meier method and data were compared using the log-rank test. EFS was estimated in R version 3.0.1 using the package survival 2.37–4.
Online supplemental material
Figs. S1 and S2 describe generation of primary and relapsed mouse B-ALL models. Figs. S3 and S4 detail human B-ALL cohort data. Fig. S5 documents Ikaros target gene expression in mouse B-ALL models and CTNND1 splicing in B-ALL. Tables S1–S3 list murine B-ALL Acceleration, Restoration, and Ikaros-regulated genes. Tables S4 and S5 include gene ontology and KEGG analysis. Tables S6 and S7 list human B-ALL IKAROS-regulated genes. Table S8 lists IKAROS-regulated genes common to both human B-ALL cohorts. Tables S1–S8 are available as Excel Files.
Acknowledgments
We thank K. Stoev, E. Lanera, C. Alvarado, E. Simankowicz, and WEHI Bioservices staff for mouse work; E. Viney and J. Sarkis at the Australian Phenomics Network Transgenic RNAi service; M. Everest and M. Tinning at the Australian Genome Research Facility; S. Wilcox for help with next generation sequencing; A. Hoogkamer for bioinformatics assistance; and M. Faux for P120-catenin advice. We thank J. Zuber and S. Lowe for Myc shRNAs, N. Heisterkamp for BCR-ABL1P190 mice, D. Largaespada for Vav-tTA mice, and H. Schjerven and M. Muschen for sharing unpublished ChIP data. We thank B. Kile and members of the Dickins laboratory for advice and discussions.
This work was supported by the National Health and Medical Research Council of Australia (NHMRC) project grants 1024599 and 1080183 (R.A. Dickins), Senior Research Fellowship (G.K. Smyth), and Early Career Fellowship (L. Cimmino), NHMRC IRIISS, and Victorian State Government OIS grants. This work was supported in part by American Lebanese Syrian Associated Charities and National Cancer Institute grants CA21765 and CA145707 of St. Jude Children’s Research Hospital; a Stand Up to Cancer Innovative Research Grant and St. Baldrick’s Foundation Scholar Award (C.G. Mullighan); and an American Society of Hematology Scholar Award (K.G. Roberts). This work was also supported by Dutch Cancer Society grant EMCR 2007-3718 (M.L. den Boer), the Pediatric Oncology Foundation Rotterdam (M.L. den Boer and J.M. Boer), and the European Union's Seventh Framework Program/European Network for Cancer Research in Children and Adolescents grant HEALTH-F2-2011-261474 (M.L. den Boer). The work was also funded by the Leukaemia Foundation of Australia (scholarships to M.T. Witkowski and G.J. Liu; fellowship to M.D. McKenzie), a Sylvia and Charles Viertel Charitable Foundation Fellowship (R.A. Dickins), and a Victorian Endowment for Science, Knowledge and Innovation Fellowship (R.A. Dickins).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- B-ALL
- B lineage acute lymphoblastic leukemia
- BCR-ABL1
- breakpoint cluster region-ABL proto-oncogene 1
- CTNND1
- catenin delta 1
- EFS
- event-free survival
- EMP1
- epithelial membrane protein 1
- IFITM3
- interferon induced transmembrane protein 3
- IKZF1
- IKAROS family zinc finger 1
- Ph+
- Philadelphia-chromosome positive
- Ph-like
- Philadelphia-chromosome-like
References
- Andreu P., Colnot S., Godard C., Laurent-Puig P., Lamarque D., Kahn A., Perret C., and Romagnolo B.. 2006. Identification of the IFITM family as a new molecular marker in human colorectal tumors. Cancer Res. 66:1949–1955. 10.1158/0008-5472.CAN-05-2731 [DOI] [PubMed] [Google Scholar]
- Ariës I.M., Jerchel I.S., van den Dungen R.E., van den Berk L.C., Boer J.M., Horstmann M.A., Escherich G., Pieters R., and den Boer M.L.. 2014. EMP1, a novel poor prognostic factor in pediatric leukemia regulates prednisolone resistance, cell proliferation, migration and adhesion. Leukemia. 28:1828–1837. 10.1038/leu.2014.80 [DOI] [PubMed] [Google Scholar]
- Churchman M.L., Low J., Qu C., Paietta E.M., Kasper L.H., Chang Y., Payne-Turner D., Althoff M.J., Song G., Chen S.C., et al. 2015. Efficacy of Retinoids in IKZF1-Mutated BCR-ABL1 Acute Lymphoblastic Leukemia. Cancer Cell. 28:343–356. 10.1016/j.ccell.2015.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandekar S., Romanos-Sirakis E., Pais F., Bhatla T., Jones C., Bourgeois W., Hunger S.P., Raetz E.A., Hermiston M.L., Dasgupta R., et al. 2014. Wnt inhibition leads to improved chemosensitivity in paediatric acute lymphoblastic leukaemia. Br. J. Haematol. 167:87–99. 10.1111/bjh.13011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J.M., and Reynolds A.B.. 1999. The catenin p120(ctn) interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol. Cell. Biol. 19:3614–3623. 10.1128/MCB.19.5.3614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M.A., Ireton R.C., and Reynolds A.B.. 2003. A core function for p120-catenin in cadherin turnover. J. Cell Biol. 163:525–534. 10.1083/jcb.200307111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Boer M.L., van Slegtenhorst M., De Menezes R.X., Cheok M.H., Buijs-Gladdines J.G.C.A.M., Peters S.T.C.J.M., Van Zutven L.J.C.M., Beverloo H.B., Van der Spek P.J., Escherich G., et al. 2009. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 10:125–134. 10.1016/S1470-2045(08)70339-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumortier A., Jeannet R., Kirstetter P., Kleinmann E., Sellars M., dos Santos N.R., Thibault C., Barths J., Ghysdael J., Punt J.A., et al. 2006. Notch activation is an early and critical event during T-Cell leukemogenesis in Ikaros-deficient mice. Mol. Cell. Biol. 26:209–220. 10.1128/MCB.26.1.209-220.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis A., Gaub M.P., Legrain M., Drenou B., Mauvieux L., Lutz P., Herbrecht R., Chan S., and Kastner P.. 2013. Biclonal and biallelic deletions occur in 20% of B-ALL cases with IKZF1 mutations. Leukemia. 27:503–507. 10.1038/leu.2012.204 [DOI] [PubMed] [Google Scholar]
- Durgan J., Tao G., Walters M.S., Florey O., Schmidt A., Arbelaez V., Rosen N., Crystal R.G., and Hall A.. 2015. SOS1 and Ras regulate epithelial tight junction formation in the human airway through EMP1. EMBO Rep. 16:87–96. 10.15252/embr.201439218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everitt A.R., Clare S., Pertel T., John S.P., Wash R.S., Smith S.E., Chin C.R., Feeley E.M., Sims J.S., Adams D.J., et al. GenISIS Investigators. MOSAIC Investigators . 2012. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 484:519–523. 10.1038/nature10921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreirós-Vidal I., Carroll T., Taylor B., Terry A., Liang Z., Bruno L., Dharmalingam G., Khadayate S., Cobb B.S., Smale S.T., et al. 2013. Genome-wide identification of Ikaros targets elucidates its contribution to mouse B-cell lineage specification and pre-B-cell differentiation. Blood. 121:1769–1782. 10.1182/blood-2012-08-450114 [DOI] [PubMed] [Google Scholar]
- Giambra V., Jenkins C.E., Lam S.H., Hoofd C., Belmonte M., Wang X., Gusscott S., Gracias D., and Weng A.P.. 2015. Leukemia stem cells in T-ALL require active Hif1α and Wnt signaling. Blood. 125:3917–3927. 10.1182/blood-2014-10-609370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisterkamp N., Jenster G., ten Hoeve J., Zovich D., Pattengale P.K., and Groffen J.. 1990. Acute leukaemia in bcr/abl transgenic mice. Nature. 344:251–253. 10.1038/344251a0 [DOI] [PubMed] [Google Scholar]
- Heizmann B., Kastner P., and Chan S.. 2013. Ikaros is absolutely required for pre-B cell differentiation by attenuating IL-7 signals. J. Exp. Med. 210:2823–2832. 10.1084/jem.20131735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann R., Seidl T., Neeb M., Rolink A., and Melchers F.. 2002. Changes in gene expression profiles in developing B cells of murine bone marrow. Genome Res. 12:98–111. 10.1101/gr.201501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunger S.P., and Mullighan C.G.. 2015. Acute Lymphoblastic Leukemia in Children. N. Engl. J. Med. 373:1541–1552. 10.1056/NEJMra1400972 [DOI] [PubMed] [Google Scholar]
- Iacobucci I., Storlazzi C.T., Cilloni D., Lonetti A., Ottaviani E., Soverini S., Astolfi A., Chiaretti S., Vitale A., Messa F., et al. 2009. Identification and molecular characterization of recurrent genomic deletions on 7p12 in the IKZF1 gene in a large cohort of BCR-ABL1-positive acute lymphoblastic leukemia patients: on behalf of Gruppo Italiano Malattie Ematologiche dell’Adulto Acute Leukemia Working Party (GIMEMA AL WP). Blood. 114:2159–2167. 10.1182/blood-2008-08-173963 [DOI] [PubMed] [Google Scholar]
- Ireton R.C., Davis M.A., van Hengel J., Mariner D.J., Barnes K., Thoreson M.A., Anastasiadis P.Z., Matrisian L., Bundy L.M., Sealy L., et al. 2002. A novel role for p120 catenin in E-cadherin function. J. Cell Biol. 159:465–476. 10.1083/jcb.200205115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi I., Yoshida T., Jena N., Qi X., Zhang J., Van Etten R.A., and Georgopoulos K.. 2014. Loss of Ikaros DNA-binding function confers integrin-dependent survival on pre-B cells and progression to acute lymphoblastic leukemia. Nat. Immunol. 15:294–304. 10.1038/ni.2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juric D., Lacayo N.J., Ramsey M.C., Racevskis J., Wiernik P.H., Rowe J.M., Goldstone A.H., O’Dwyer P.J., Paietta E., and Sikic B.I.. 2007. Differential gene expression patterns and interaction networks in BCR-ABL-positive and -negative adult acute lymphoblastic leukemias. J. Clin. Oncol. 25:1341–1349. 10.1200/JCO.2006.09.3534 [DOI] [PubMed] [Google Scholar]
- Kim W.I., Wiesner S.M., and Largaespada D.A.. 2007. Vav promoter-tTA conditional transgene expression system for hematopoietic cells drives high level expression in developing B and T cells. Exp. Hematol. 35:1231–1239. 10.1016/j.exphem.2007.04.012 [DOI] [PubMed] [Google Scholar]
- Krentz S., Hof J., Mendioroz A., Vaggopoulou R., Dörge P., Lottaz C., Engelmann J.C., Groeneveld T.W., Körner G., Seeger K., et al. 2013. Prognostic value of genetic alterations in children with first bone marrow relapse of childhood B-cell precursor acute lymphoblastic leukemia. Leukemia. 27:295–304. 10.1038/leu.2012.155 [DOI] [PubMed] [Google Scholar]
- Kuiper R.P., Schoenmakers E.F., van Reijmersdal S.V., Hehir-Kwa J.Y., van Kessel A.G., van Leeuwen F.N., and Hoogerbrugge P.M.. 2007. High-resolution genomic profiling of childhood ALL reveals novel recurrent genetic lesions affecting pathways involved in lymphocyte differentiation and cell cycle progression. Leukemia. 21:1258–1266. 10.1038/sj.leu.2404691 [DOI] [PubMed] [Google Scholar]
- Kuiper R.P., Waanders E., van der Velden V.H., van Reijmersdal S.V., Venkatachalam R., Scheijen B., Sonneveld E., van Dongen J.J., Veerman A.J.P., van Leeuwen F.N., et al. 2010. IKZF1 deletions predict relapse in uniformly treated pediatric precursor B-ALL. Leukemia. 24:1258–1264. 10.1038/leu.2010.87 [DOI] [PubMed] [Google Scholar]
- Lee J.-W., Geng H., Chen Z., Park E., Klemm L., Bailey C.C., and Muschen M.. 2015. IFITM3 (CD225) links the B cell antigen CD19 to PI3K-AKT signaling in human ALL cells. Blood. 126:1325 10.5045/br.2015.50.3.126 [DOI] [Google Scholar]
- Li D., Peng Z., Tang H., Wei P., Kong X., Yan D., Huang F., Li Q., Le X., Li Q., and Xie K.. 2011. KLF4-mediated negative regulation of IFITM3 expression plays a critical role in colon cancer pathogenesis. Clin. Cancer Res. 17:3558–3568. 10.1158/1078-0432.CCR-10-2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Ilaria R.L. Jr., Million R.P., Daley G.Q., and Van Etten R.A.. 1999. The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J. Exp. Med. 189:1399–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., and Shi W.. 2013. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41:e108 10.1093/nar/gkt214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., and Shi W.. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30:923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Liu G.J., Cimmino L., Jude J.G., Hu Y., Witkowski M.T., McKenzie M.D., Kartal-Kaess M., Best S.A., Tuohey L., Liao Y., et al. 2014. Pax5 loss imposes a reversible differentiation block in B-progenitor acute lymphoblastic leukemia. Genes Dev. 28:1337–1350. 10.1101/gad.240416.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli G., Iacobucci I., Storlazzi C.T., Vignetti M., Paoloni F., Cilloni D., Soverini S., Vitale A., Chiaretti S., Cimino G., et al. 2009. IKZF1 (Ikaros) deletions in BCR-ABL1-positive acute lymphoblastic leukemia are associated with short disease-free survival and high rate of cumulative incidence of relapse: a GIMEMA AL WP report. J. Clin. Oncol. 27:5202–5207. 10.1200/JCO.2008.21.6408 [DOI] [PubMed] [Google Scholar]
- Mullighan C.G., Goorha S., Radtke I., Miller C.B., Coustan-Smith E., Dalton J.D., Girtman K., Mathew S., Ma J., Pounds S.B., et al. 2007. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 446:758–764. 10.1038/nature05690 [DOI] [PubMed] [Google Scholar]
- Mullighan C.G., Miller C.B., Radtke I., Phillips L.A., Dalton J., Ma J., White D., Hughes T.P., Le Beau M.M., Pui C.-H., et al. 2008a BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 453:110–114. 10.1038/nature06866 [DOI] [PubMed] [Google Scholar]
- Mullighan C.G., Phillips L.A., Su X., Ma J., Miller C.B., Shurtleff S.A., and Downing J.R.. 2008b Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 322:1377–1380. 10.1126/science.1164266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan C.G., Su X., Zhang J., Radtke I., Phillips L.A.A., Miller C.B., Ma J., Liu W., Cheng C., Schulman B.A., et al. Children’s Oncology Group . 2009. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N. Engl. J. Med. 360:470–480. 10.1056/NEJMoa0808253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T., Kodama H., and Honjo T.. 1994. Generation of lymphohematopoietic cells from embryonic stem cells in culture. Science. 265:1098–1101. 10.1126/science.8066449 [DOI] [PubMed] [Google Scholar]
- Nichogiannopoulou A., Trevisan M., Neben S., Friedrich C., and Georgopoulos K.. 1999. Defects in hemopoietic stem cell activity in Ikaros mutant mice. J. Exp. Med. 190:1201–1214. 10.1084/jem.190.9.1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premsrirut P.K., Dow L.E., Kim S.Y., Camiolo M., Malone C.D., Miething C., Scuoppo C., Zuber J., Dickins R.A., Kogan S.C., et al. 2011. A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell. 145:145–158. 10.1016/j.cell.2011.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Diyagama D., Neilson J., van Laar R., Dobrovic A., Holloway A., and Smyth G.K.. 2006. Empirical array quality weights in the analysis of microarray data. BMC Bioinformatics. 7:261 10.1186/1471-2105-7-261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., and Smyth G.K.. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts K.G., and Mullighan C.G.. 2015. Genomics in acute lymphoblastic leukaemia: insights and treatment implications. Nat. Rev. Clin. Oncol. 12:344–357. 10.1038/nrclinonc.2015.38 [DOI] [PubMed] [Google Scholar]
- Roberts K.G., Morin R.D., Zhang J., Hirst M., Zhao Y., Su X., Chen S.C., Payne-Turner D., Churchman M.L., Harvey R.C., et al. 2012. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 22:153–166. 10.1016/j.ccr.2012.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts K.G., Li Y., Payne-Turner D., Harvey R.C., Yang Y.L., Pei D., McCastlain K., Ding L., Lu C., Song G., et al. 2014. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N. Engl. J. Med. 371:1005–1015. 10.1056/NEJMoa1403088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., and Oshlack A.. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11:R25 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schackmann R.C., Tenhagen M., van de Ven R.A., and Derksen P.W.. 2013. p120-catenin in cancer - mechanisms, models and opportunities for intervention. J. Cell Sci. 126:3515–3525. 10.1242/jcs.134411 [DOI] [PubMed] [Google Scholar]
- Schjerven H., McLaughlin J., Arenzana T.L., Frietze S., Cheng D., Wadsworth S.E., Lawson G.W., Bensinger S.J., Farnham P.J., Witte O.N., and Smale S.T.. 2013. Selective regulation of lymphopoiesis and leukemogenesis by individual zinc fingers of Ikaros. Nat. Immunol. 14:1073–1083. 10.1038/ni.2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjerven H., Ayongaba E.F., Aghajanirefah A., McLaughlin J., Cheng D., and Geng H.. 2016. Genetic analysis of Ikaros target genes and tumor suppressor function in BCR-ABL1+ pre-B ALL. J. Exp. Med. 214:xxx–xxx. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwickert T.A., Tagoh H., Gültekin S., Dakic A., Axelsson E., Minnich M., Ebert A., Werner B., Roth M., Cimmino L., et al. 2014. Stage-specific control of early B cell development by the transcription factor Ikaros. Nat. Immunol. 15:283–293. 10.1038/ni.2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. 2004. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:e3. [DOI] [PubMed] [Google Scholar]
- Smyth G.K., Michaud J., and Scott H.S.. 2005. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 21:2067–2075. 10.1093/bioinformatics/bti270 [DOI] [PubMed] [Google Scholar]
- Takiguchi M., Dow L.E., Prier J.E., Carmichael C.L., Kile B.T., Turner S.J., Lowe S.W., Huang D.C., and Dickins R.A.. 2013. Variability of inducible expression across the hematopoietic system of tetracycline transactivator transgenic mice. PLoS One. 8:e54009 10.1371/journal.pone.0054009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S.S., Yamaguchi Y.L., Tsoi B., Lickert H., and Tam P.P.. 2005. IFITM/Mil/fragilis family proteins IFITM1 and IFITM3 play distinct roles in mouse primordial germ cell homing and repulsion. Dev. Cell. 9:745–756. 10.1016/j.devcel.2005.10.010 [DOI] [PubMed] [Google Scholar]
- Tremblay C.S., Brown F.C., Collett M., Saw J., Chiu S.K., Sonderegger S.E., Lucas S.E., Alserihi R., Chau N., Toribio M.L., et al. 2016. Loss-of-function mutations of Dynamin 2 promote T-ALL by enhancing IL-7 signalling. Leukemia. 30:1993–2001. 10.1038/leu.2016.100 [DOI] [PubMed] [Google Scholar]
- van der Veer A., Waanders E., Pieters R., Willemse M.E., Van Reijmersdal S.V., Russell L.J., Harrison C.J., Evans W.E., van der Velden V.H., Hoogerbrugge P.M., et al. 2013. Independent prognostic value of BCR-ABL1-like signature and IKZF1 deletion, but not high CRLF2 expression, in children with B-cell precursor ALL. Blood. 122:2622–2629. 10.1182/blood-2012-10-462358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veer A., Zaliova M., Mottadelli F., De Lorenzo P., Te Kronnie G., Harrison C.J., Cavé H., Trka J., Saha V., Schrappe M., et al. 2014. IKZF1 status as a prognostic feature in BCR-ABL1-positive childhood ALL. Blood. 123:1691–1698. 10.1182/blood-2013-06-509794 [DOI] [PubMed] [Google Scholar]
- van Roy F.M., and McCrea P.D.. 2005. A role for Kaiso-p120ctn complexes in cancer? Nat. Rev. Cancer. 5:956–964. 10.1038/nrc1752 [DOI] [PubMed] [Google Scholar]
- Virely C., Moulin S., Cobaleda C., Lasgi C., Alberdi A., Soulier J., Sigaux F., Chan S., Kastner P., and Ghysdael J.. 2010. Haploinsufficiency of the IKZF1 (IKAROS) tumor suppressor gene cooperates with BCR-ABL in a transgenic model of acute lymphoblastic leukemia. Leukemia. 24:1200–1204. 10.1038/leu.2010.63 [DOI] [PubMed] [Google Scholar]
- Vitanza N.A., Zaky W., Blum R., Meyer J.A., Wang J., Bhatla T., Morrison D.J., Raetz E.A., and Carroll W.L.. 2014. Ikaros deletions in BCR-ABL-negative childhood acute lymphoblastic leukemia are associated with a distinct gene expression signature but do not result in intrinsic chemoresistance. Pediatr. Blood Cancer. 61:1779–1785. 10.1002/pbc.25119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waanders E., van der Velden V.H., van der Schoot C.E., van Leeuwen F.N., van Reijmersdal S.V., de Haas V., Veerman A.J., van Kessel A.G., Hoogerbrugge P.M., Kuiper R.P., and van Dongen J.J.. 2011. Integrated use of minimal residual disease classification and IKZF1 alteration status accurately predicts 79% of relapses in pediatric acute lymphoblastic leukemia. Leukemia. 25:254–258. 10.1038/leu.2010.275 [DOI] [PubMed] [Google Scholar]
- Wang J.H., Nichogiannopoulou A., Wu L., Sun L., Sharpe A.H., Bigby M., and Georgopoulos K.. 1996. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 5:537–549. 10.1016/S1074-7613(00)80269-1 [DOI] [PubMed] [Google Scholar]
- Wang Y., Krivtsov A.V., Sinha A.U., North T.E., Goessling W., Feng Z., Zon L.I., and Armstrong S.A.. 2010. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 327:1650–1653. 10.1126/science.1186624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winandy S., Wu P., and Georgopoulos K.. 1995. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell. 83:289–299. 10.1016/0092-8674(95)90170-1 [DOI] [PubMed] [Google Scholar]
- Witkowski M.T., Cimmino L., Hu Y., Trimarchi T., Tagoh H., McKenzie M.D., Best S.A., Tuohey L., Willson T.A., Nutt S.L., et al. 2015. Activated Notch counteracts Ikaros tumor suppression in mouse and human T-cell acute lymphoblastic leukemia. Leukemia. 29:1301–1311. 10.1038/leu.2015.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.J., Bhojwani D., Yang W., Cai X., Stocco G., Crews K., Wang J., Morrison D., Devidas M., Hunger S.P., et al. 2008. Genome-wide copy number profiling reveals molecular evolution from diagnosis to relapse in childhood acute lymphoblastic leukemia. Blood. 112:4178–4183. 10.1182/blood-2008-06-165027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Ng S.Y., Zuniga-Pflucker J.C., and Georgopoulos K.. 2006. Early hematopoietic lineage restrictions directed by Ikaros. Nat. Immunol. 7:382–391. 10.1038/ni1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J., McJunkin K., Fellmann C., Dow L.E., Taylor M.J., Hannon G.J., and Lowe S.W.. 2011. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat. Biotechnol. 29:79–83. 10.1038/nbt.1720 [DOI] [PMC free article] [PubMed] [Google Scholar]