Abstract
Eukaryotic elongation factor 1 alpha (eEF1A) is an essential component of the translational apparatus. In the present study, eEF1A1b was isolated from the Nile tilapia. Real-time PCR and Western blot revealed that eEF1A1b was expressed highly in the testis from 90 dah (days after hatching) onwards. In situ hybridization and immunohistochemistry analyses showed that eEF1A1b was highly expressed in the spermatogonia of the testis. CRISPR/Cas9 mediated mutation of eEF1A1b resulted in spermatogenesis arrest and infertility in the F0 XY fish. Consistently, heterozygous mutation of eEF1A1b (eEF1A1b+/−) resulted in an absence of spermatocytes at 90 dah, very few spermatocytes, spermatids and spermatozoa at 180 dah, and decreased Cyp11b2 and serum 11-ketotestosterone level at both stages. Further examination of the fertilization capacity of the sperm indicated that the eEF1A1b+/− XY fish were infertile due to abnormal spermiogenesis. Transcriptomic analyses of the eEF1A1b+/− testis from 180 dah XY fish revealed that key elements involved in spermatogenesis, steroidogenesis and sperm motility were significantly down-regulated compared with the control XY. Transgenic overexpression of eEF1A1b rescued the spermatogenesis arrest phenotype of the eEF1A1b+/− testis. Taken together, our data suggested that eEF1A1b is crucial for spermatogenesis and male fertility in the Nile tilapia.
Eukaryotic translation elongation factor 1A (eEF1A) is one of the most abundant protein synthesis factors in eukaryotic cells. It is responsible for delivering aminoacylated tRNAs to the A site of the ribosome in a GTP-dependent reaction1. Eukaryotes possess a variable number of eEF1A genes with various expression patterns. Yeast (Saccharomyces cerevisiae) contains two eEF1A genes that are expressed almost equally in exponentially growing cells2. Fruit flies (Drosophila melanogaster) have two eEF1A genes, one of which is expressed only in certain life history stages3,4. In teleosts, only one eEF1A gene has been reported in zebrafish (Danio rerio), sea bream (Sparus aurata), Nile tilapia (Oreochromis niloticus) and medaka (Oryzias latipes)5,6,7,8. Recently, five eEF1A genes, referred to as eEF1A1-4 and 42Sp50, have been isolated and their relative expressions are different in ten tissues in flatfish (Solea senegalensis)9. In the African clawed frog (Xenopus laevis), three eEF1A genes are developmentally specific: the somatic form (eEF-1S) is present in embryos and most adult tissues, but is not detected in the germ cells; the oocyte form (eEF-1O), is present in germ cells and some adult tissues, but is not detected in the embryos; the 42Sp50 form, is detected only in oocytes10,11,12. In mammals, two eEF1A genes, named as eEF1A1 and eEF1A2, have been identified, eEF1A1, which is almost ubiquitously expressed, and eEF1A2, whose expression is restricted to some cell types in a few tissues13,14,15,16. Despite this, homozygous mutation of mammalian eEF1A2 is lethal during postnatal stages, possibly due to its critical function in translation elongation17,18,19.
Although eEF1A is reported to be ubiquitously expressed in all tissues examined, its role in gonads remains to be elucidated. Gametogenesis is a complex process, during which numerous proteins are synthesized in the testis and ovary. Mutation of the elongation factors for proteins synthesis, may result in infertility in vertebrates, as was demonstrated for eEF420. Besides its canonical function in translation elongation, eEF1A is also involved in other cellular processes, such as severing microtubule21, bundling F actin22, controlling cell apoptosis23 and regulating protein degradation24,25. To date, only one report showed that eEF1A genes might be involved in the gametogenesis via non-canonical function. Oral administration of the eEF1A1 inhibitor gamendazole resulted in infertility in male mice, probably via disruption of actin-filament bundles related to the Sertoli cell-spermatids ectoplasmic specialization junctions26. However, there have been no other reports on the role and mechanism of other eEFs, including eEF1A, in gametogenesis.
In addition to somatic cells, eEF1A is also expressed in germ cells. Proteins expressed in germ cells are directly related to gametogenesis. Disruption of genes expressed in germ cells often blocks gametogenesis27,28,29,30. Therefore, eEF1A might be directly involved in gametogenesis via protein synthesis in germ cells. Different eEF1A genes are reported to be expressed in the testis and ovary of Xenopus11,12. Our gonads transcriptomic data from gonads demonstrated that eEF1A1b and 42Sp50 are XY-enhanced and XX-specific genes in tilapia, respectively31. Taken together, we speculate that different elongation factors might be used for protein synthesis during spermatogenesis and oogenesis in lower vertebrates. Species with different eEF1A genes expressed in the testis and ovary would be a good model to study the diverse function of the eEF1A genes in spermatogenesis and oogenesis.
Nile Tilapia (Oreochromis niloticus), is a good model for the study of gene expression and function in fish due to the availability of monosex fish32, a short spawning cycle, many gonadal transcriptomes31 and published genome sequences33. In the present study, expression of eEF1A1b was found to been enriched in XY males during tilapia gonadal development. Ontogenetic studies were conducted to determine its expression profile by real-time PCR and Western blot. The cellular localization of eEF1A1b was documented by in situ hybridization (ISH) and immunohistochemistry (IHC). We also mutated eEF1A1b and analyzed the gonadal phenotype, gene expression, serum 11-ketotestosterone (11-KT) level and fertility of the eEF1A1b deficiency and eEF1A1b+/− fish.
Results
Phylogenetic and synteny analyses
As shown in the phylogenetic tree (supplemental Fig. S1), the phylogeny of eEF1A1 was split into two main clades. One clade included the eEF1A1 gene of tetrapods; and the other included the two distinct copies of eEF1A1 from teleosts that we named eEF1A1a and eEF1A1b.
Synteny analyses revealed the organization of the genomic region surrounding eEF1A1 genes in tetrapods and teleosts. eEF1A1a and its upstream gene slc17a5, its downstream gene ddx43 and kcnq5a showed conserved synteny both in teleosts and tetrapods. eEF1A1b and its upstream gene dtd1 showed conserved synteny only in teleosts, and its downstream gene kcnq5b and rims1 showed conserved synteny only in tetrapods. Except kcnq5, no other conserved synteny of gene loci surrounding eEF1A1a and eEF1A1b were found (supplemental Fig. S2).
Tissue distribution
Analysis by IHC showed that eEF1A1b was highly expressed in the brain, heart, liver, intestine, kidney and testis, weakly expressed in the ovary, but was undetectable in gill, spleen, head kidney and muscle (Supplemental Fig. S3).
Expression profile of eEF1A1b in developing gonads
Real-time PCR demonstrated that expression of eEF1A1b was low from 10 to 90 dah, increased sharply from 90 dah in testis and slightly decreased from 150 to 180 dah; while no significant changes were found during all stages in ovary (Fig. 1a).
Figure 1. Expression patterns of eEF1A1b in developing gonads.
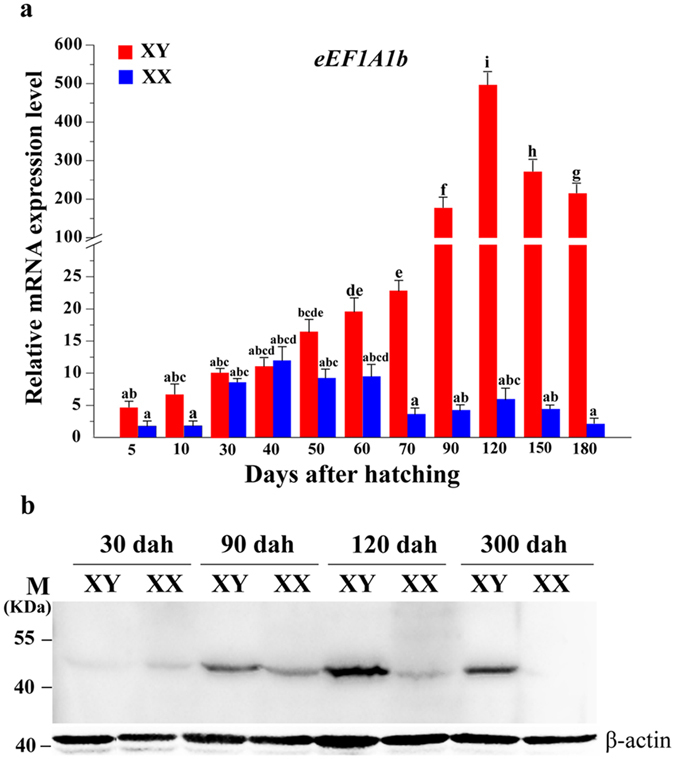
(a) Ontogenetic expression of eEF1A1b in tilapia gonads analyzed by real-time PCR. Data are expressed as mean ± SD of three different gonadal pools at each developmental stage. Different letters indicate statistical differences (P < 0.05) as determined by one-way ANOVA followed by post hoc test. (b) Expression of eEF1A1b in gonads of XX and XY fish at 30, 90, 120 and 300 dah by Western blot. Lanes 1, 3, 5 and 7, proteins extracted from testes; lanes 2, 4, 6 and 8, proteins extracted from ovaries. eEF1A1b was detected in both testis and ovary with significant higher expression in testis. 0.2 micrograms of protein were added per lane.
Western blot analyses revealed that eEF1A1b was expressed in both testis and ovary, with significantly higher expression in the testis than in the ovary from 90 dah, peaking at approximately 120 dah, and decreased to a much lower level at 300 dah (Fig. 1b).
Cellular localization of eEF1A1b in gonads
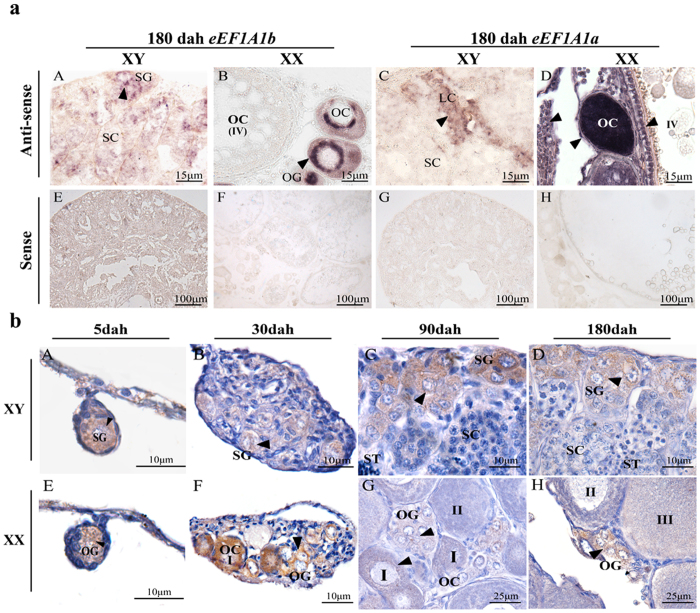
ISH analysis demonstrated that eEF1A1b was expressed in the spermatogonia of the testis, and in the oogonia and phase I oocytes of the ovary with eEF1A1b antisense probe (Fig. 2a, A and B). eEF1A1a, the paralogous gene of eEF1A1b, was expressed in the somatic cells of the testis, and in the oocytes and somatic cell of the ovary (Fig. 2a, C and D). In contrast, signal was detected in neither testis nor ovary with eEF1A1a and eEF1A1b sense probe (Fig. 2a, E- H).
Figure 2. Cellular localization of eEF1A1b in tilapia testis and ovary at different developmental stages by ISH and IHC.
(a) Cell type expressing eEF1A1a and eEF1A1b in tilapia gonads by ISH. eEF1A1b was detected in spermatogonia of the testis, while in the oogonia and phase I oocytes of the ovary at (A and B). eEF1A1a was detected in somatic cell of the testis, while in the oocytes and somatic cells of the ovary (C and D). (b) Cell type expressing eEF1A1b in tilapia gonads by IHC. Consistent with in situ hybridization results, eEF1A1b was detected in the spermatogonia of the testis from 5 to 180 dah (A–D), while in the oogonia of ovary at 5 dah, later in the oogonia and phase I oocytes of the ovary from 30 to 180 dah (E-H). SG, spermatogonia; SC, spermatocytes; ST, spermatids; OG, oogonia; OC, oocytes; I-IV, phase I to phase IV oocytes; Arrowheads indicate the positive signal.
Consistently, IHC analysis revealed that eEF1A1b was expressed exclusively in the spermatogonia of the testis from 5, 30, 90 and 180 dah (Fig. 2b, A-D), and was expressed in the oogonia of the ovary at 5 dah, later in the oogonia and phase I oocytes of the ovary at 30 to 180 dah (Fig. 2b, E-H).
Disruption of eEF1A1b by CRISPR/Cas9 and production of F1 generation
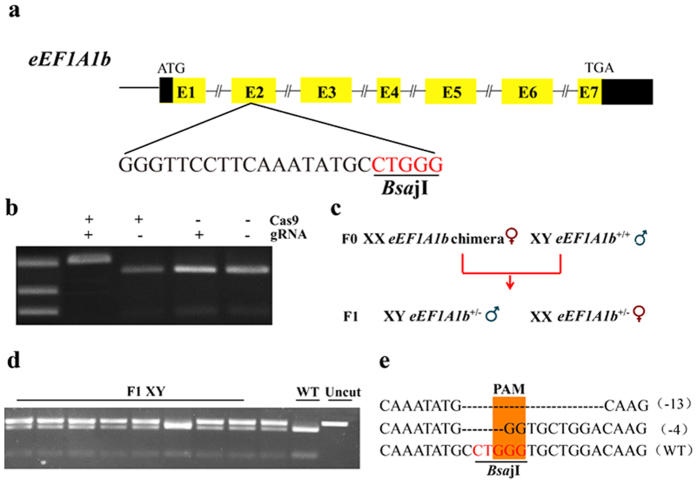
A guide RNA site containing BsajI adjacent to proto-spacer adjacent motif in the second exon of eEF1A1b was selected for mutation analysis (Fig. 3a). Genomic DNA from 20 pooled injected embryos and from fin of mutants was used as template for PCR amplification and mutation assays. Complete digestion produced fragments of 425 and 102 bp in the control groups whereas an intact DNA fragment was observed in embryos injected with both Cas9 mRNA and target guide RNA (Fig. 3b). The mutation rate in the pooled embryos was approximately 97%. In-frame and frame-shift deletions induced at the target site were confirmed by Sanger sequencing. We screened 40 individual microinjected fish by restriction enzyme digestion at 90 dah, and found 38 to be mutant for eEF1A1b. All mutant fish had mutation efficiency over 90% (Supplemental Tab. S3). Twenty mutant fish were further confirmed by Sanger sequencing. In total, we analyzed 200 sequences and identified 14 mutation types, including 11 types of deletion mutation and 3 types of insertion mutation. The majority (67.5%) were frame-shift mutations, while 32.5% were in-frame mutations (Supplemental Fig. S4). F1 offspring were obtained by outcross eEF1A1b chimeric XX with XY control fish, and two mutation types (4 and 13 bp deletion) were identified in F1 offspring by Sanger sequencing (Fig. 3c,d and e).
Figure 3. Design of CRISPR/Cas9 targeting eEF1A1b and establishment of the eEF1A1b mutation line.
(a,b) Disruption of tilapia eEF1A1b by CRISPR/Cas9. Gene structure of eEF1A1b showing the target site and the BsajI restriction site. 300 ng/μl of Cas9 mRNA and 150 ng/μl of gRNA were co-injected into one-cell stage embryos. At 72 hours after injection, 20 embryos were randomly selected and pooled to extract their genomic DNA for PCR amplification. The indels were confirmed by two assays, restriction enzyme digestion and Sanger sequencing. The Cas9 mRNA and gRNA were added as indicated. Clear undigested band was detected in embryos injected with both Cas9 mRNA and gRNA compared with the control. (c) Schematic diagram showing the breeding plan of F0 to F1 fish. (d) eEF1A1b+/− F1 generation detected by restriction enzyme digestion. (e) Sanger sequencing results from the uncleaved bands were listed. Deletions are marked by dashes, and the PAM is marked in light orange. Numbers to the right of the sequences indicate the loss or gain of bases for each allele, with the number of bases inserted (+) or deleted (−) indicated in parentheses. WT, wild type.
Effects of eEF1A1b deficiency on spermatogenesis and fertility in F0 fish
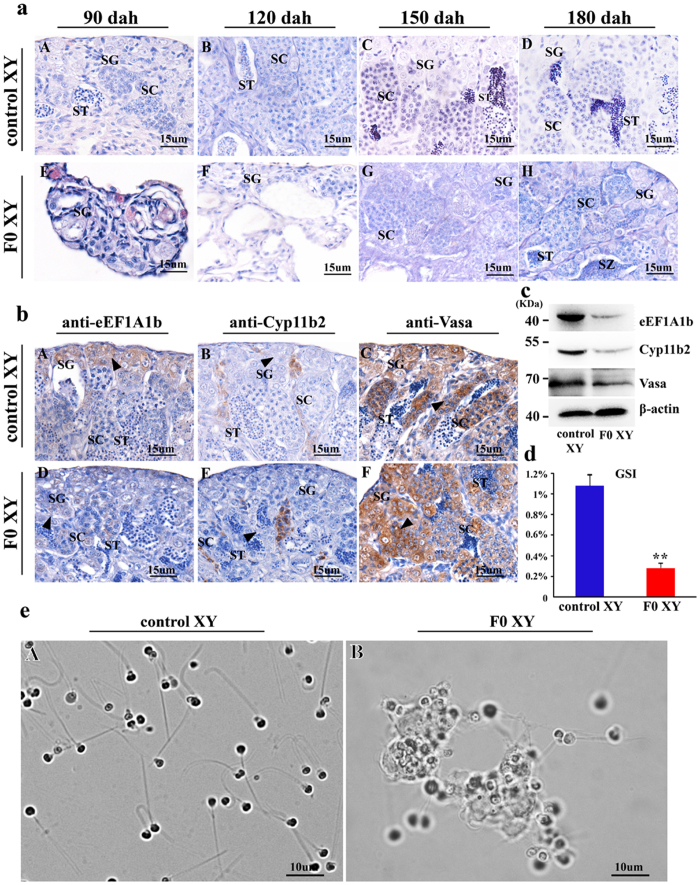
Morphologically, all kinds of germ cells including spermatogonia, spermatocytes, spermatids and spermatozoa were presented in the control testis at 90, 120, 150 and 180 dah (Fig. 4a, A-D). In contrast, in the eEF1A1b deficient testis only spermatogonia were observed at 90 and 120 dah, spermatocytes appeared at 150 dah and spermatids and spermatozoa appeared at 180 dah (Fig. 4a, E-H). Additionally, fewer spermatozoa were observed in the efferent duct of the eEF1A1b deficient testis at 180 dah compared with the control.
Figure 4. Effects of eEF1A1b deficiency on spermatogenesis and fertility in F0.
(a) Histological observations of testis from F0 and control XY fish at 90, 120, 150 and 180 dah. eEF1A1b deficiency resulted in spermatogenesis arrest. All kinds of germ cells including spermatogonia, spermatocytes, spermatids and spermatozoa were present at the control testis at 90, 120, 150 and 180 dah (A-D). In contrast, in the F0 testis only spermatogonia were observed at 90 and 120 dah, spermatocytes appeared at 150 dah, and spermatids and spermatozoa appeared at 180 dah (E-H). (b) Expression of eEF1A1b, Cyp11b2 and Vasa in the F0 and control XY fish by IHC. By IHC, eEF1A1b was observed in the control testis (A) while almost undetectable in F0 testis (D). Reduced Cyp11b2 expression was observed in Leydig cells in the F0 testis (B) compared with the control testis (E). Vasa positive signals were detected in both the control and F0 testis (C and F). Arrowheads indicate the positive signal. SG, spermatogonia; SC, spermatocytes; ST, spermatids; SZ, spermatozoa. (c) Expression of eEF1A1b, Cyp11b2 and Vasa in the F0 and control XY fish by Western blot. (d) Gonadal somatic index (GSI) of the F0 and control XY fish. (e) Morphology and motility of sperm from 180 dah F0 and control XY fish. The sperm from the control fish were with normal flagella, and displayed vigorous flagella activity and progressive movement (A); while the sperm from F0 fish were characterized by short or absent flagella. The motility of short-flagella sperms was weak, and the flagella-less sperms were stuck together, showing no motility (B). F0, eEF1A1b deficiency.
By IHC, eEF1A1b was detected in the spermatogonia, and Cyp11b2 was observed in the Leydig cells in the control testis at 180 dah (Fig. 4b, A and B). In contrast, reduced expression of eEF1A1b and Cyp11b2 was observed in the eEF1A1b deficient testis (Fig. 4b, D and E). However, Vasa was detected in the spermatogonia and spermatocytes in the eEF1A1b deficient and control testis (Fig. 4b, C and F).
Western blot analyses revealed that the expression of eEF1A1b, Cyp11b2 and Vasa was significantly reduced in the eEF1A1b deficient testis compared with those of the control (Fig. 4c). Additionally, the size of the eEF1A1b deficient testis was significantly smaller than that of the control testis as demonstrated by gonadal somatic index (GSI, gonad weight/body weight × 100%) (Fig. 4d).
Fertility test showed that the eEF1A1b deficient XY fish were infertile (supplemental Tab. S4). When examined under microscope, the sperm of the eEF1A1b deficient fish were abnormal. The sperm from the control fish were normal with a flagella length of 16.2 ± 2.4 μm, and displayed strong motility. In contrast, the sperm from eEF1A1b deficient fish were characterized by short or absent flagella. The short-flagella sperms with a flagella length of 7.55 ± 3.16 μm, displayed weak motility, and the flagella-less sperms were stuck together, showing no motility (Fig. 4e; supplemental Fig. S5).
Effects of eEF1A1b deficiency on ovarian development in F0 fish
In contrast to the significant phenotypes of eEF1A1b deficiency in the testis, the ovary of most mutant fish showed normal development at 120 and 180 dah. Histological analysis demonstrated that folliculogenesis was normal in the ovary of the eEF1A1b deficient XX fish (Supplemental Fig. S6).
Effects of eEF1A1b heterozygous mutation on spermatogenesis and fertility in F1 fish
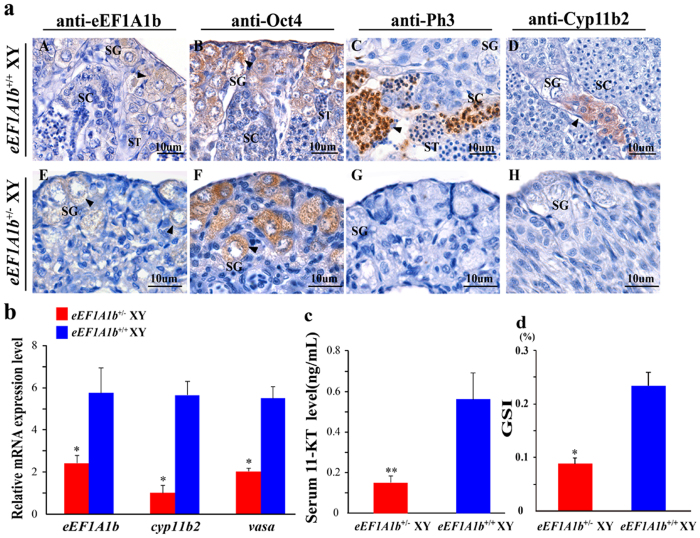
The heterozygous mutation of eEF1A1b (eEF1A1b+/−) XY fish with a 4 bp deletion were employed for phenotype analyses. Consistent with the phenotype of F0 generation, eEF1A1b+/− XY fish also displayed spermatogenesis arrest. Histologically, spermatogonia, spermatocytes and spermatids, were present at eEF1A1b+/+ testis, whereas only spermatogonia were observed in eEF1A1b+/− XY testis at 90 dah (Fig. 5a). By IHC, eEF1A1b and Oct 4 (spermatogonia maker), Phospho-histone h3 (Ph3, spermatocytes maker), Cyp11b2 were observed in the spermatogonia, spermatocytes and Leydig cells, respectively, of the eEF1A1b+/+ testis. In contrast, Oct 4 and reduced expression of eEF1A1b were observed in the spermatogonia, while no Ph3 and Cyp11b2 expression was detected in the eEF1A1b+/− testis. These results indicated that there were only spermatogonia, but no spermatocytes in the eEF1A1b+/− testis (Fig. 5a). In addition, the eEF1A1b, cyp11b2 and vasa mRNA levels were found to be significantly down-regulated in the eEF1A1b+/− testis by real-time PCR (Fig. 5b). Further, significantly lower serum 11-KT level was detected in the eEF1A1b+/− XY fish compared with that of the eEF1A1b+/+ XY fish at 90 dah (Fig. 5c). Furthermore, the size of the eEF1A1b+/− testis was significantly smaller than that of the eEF1A1b+/+ testis as reflected by the GSI.
Figure 5. Effects of the heterozygous mutation of eEF1A1b on spermatogenesis and fertility.
(a) IHC analyses of eEF1A1b, Oct 4 (spermatogonia maker), Ph3 (spermatocyte maker) and Cyp11b2 expression in the testis of eEF1A1b+/− and eEF1A1b+/+ XY fish at 90 dah. eEF1A1b and Oct 4, Ph3, Cyp11b2 were observed in the spermatogonia, spermatocytes and Leydig cells, respectively, of the eEF1A1b+/+ testis. In contrast, Oct 4 and reduced expression of eEF1A1b were observed in the spermatogonia, while no Ph3 and Cyp11b2 expression was detected in the eEF1A1b+/− testis. Arrowheads indicate the positive signal. SG, spermatogonia; SC, spermatocytes; ST, spermatids. (b) Expression of eEF1A1b, cyp11b2 and vasa in the eEF1A1b+/− and eEF1A1b+/+ XY fish at 90 dah by real-time PCR. (c) Serum 11-KT levels of the eEF1A1b+/− and eEF1A1b+/+ XY fish. (d) GSI of eEF1A1b+/− and eEF1A1b+/+ XY fish at 90 dah. Results were expressed as mean ± SD. Different letters indicate statistical differences at P < 0.05 as determined by one-way ANOVA followed by post hoc test.
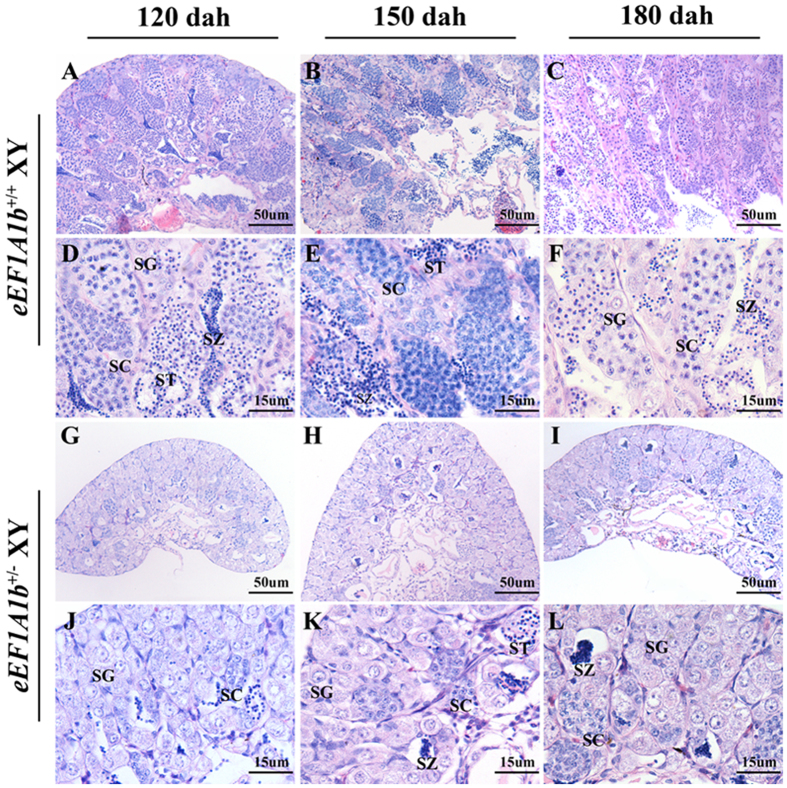
By histology, abundant spermatocytes, spermatids and spermatozoa were observed in the eEF1A1b+/+ testis at 120, 150 and 180 dah (Fig. 6, A-F). In contrast, in the testis of eEF1A1b+/− XY fish, just a few spermatocytes were observed at 120 dah, and a markedly reduced number of spermatocytes, spermatids and spermatozoa were observed at 150 and 180 dah (Fig. 6, G-L).
Figure 6. Histological observations of testis from eEF1A1b+/− XY fish at 120, 150 and 180 dah.
Abundant spermatocytes, spermatids and spermatozoa were observed in the testis of eEF1A1b+/+ XY fish at 120, 150 and 180 dah (A-F), while just very few spermatocytes were observed at 120 dah, and a markedly reduced number of spermatocytes, spermatids and spermatozoa were observed at 150 and 180 dah in the testis of eEF1A1b+/− XY fish (G-L). D-F and J-L, Higher magnification of the A-C and G-I, respectively. SG, spermatogonia; SC, spermatocytes; ST, spermatids; SZ, spermatozoa.
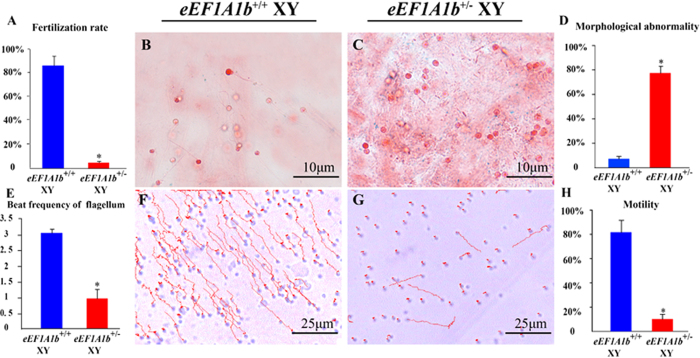
Fertility test showed that similar to the chimeric XY fish, eEF1A1b+/− XY fish were infertile (Fig. 7A). Morphological and biochemical analyses of sperm were performed to analyze the reason for male infertility in eEF1A1b+/− XY fish. The sperm of eEF1A1b+/+ XY fish were characterized by normal flagella, while the sperm of eEF1A1b+/− XY fish were flagella-less. Therefore, the ratio of sperm with morphological abnormalities to the total sperm was significantly higher in eEF1A1b+/− XY fish than that in the eEF1A1b+/+ XY fish (Fig. 7, B-D). Consistently, the activity of sperm flagella was significantly lower in eEF1A1b+/− XY fish than that in eEF1A1b+/+ XY fish (Fig. 7E). Consequently, the sperm forward motility was lost in the eEF1A1b+/− XY fish (Fig. 7, F-H).
Figure 7. Sperm analyses of eEF1A1b+/− XY fish at 180 dah.
The fertilization rate of eEF1A1b+/− XY fish was significantly lower than that of eEF1A1b+/+ XY fish (A). Sperm of eEF1A1b+/+ XY fish were with normal flagella, while most sperm of eEF1A1b+/− XY fish were flagella-less, and the ratio of sperm with morphological abnormalities to the total sperm was higher in eEF1A1b+/− XY fish than in eEF1A1b+/+ XY fish (B-D). The beat frequency of sperm flagella was higher in eEF1A1b+/+ XY fish than in eEF1A1b+/− XY fish (E). And the ratio of forward motility sperm was higher in eEF1A1b+/+ XY fish than in eEF1A1b+/− XY fish (F-H). The red line indicates the trajectories of sperm. Results were expressed as mean ± SD. Different letters indicate statistical differences at P < 0.05 as determined by one-way ANOVA followed by post hoc test.
Gonadal transcriptome analysis of eEF1A1b+/− XY fish
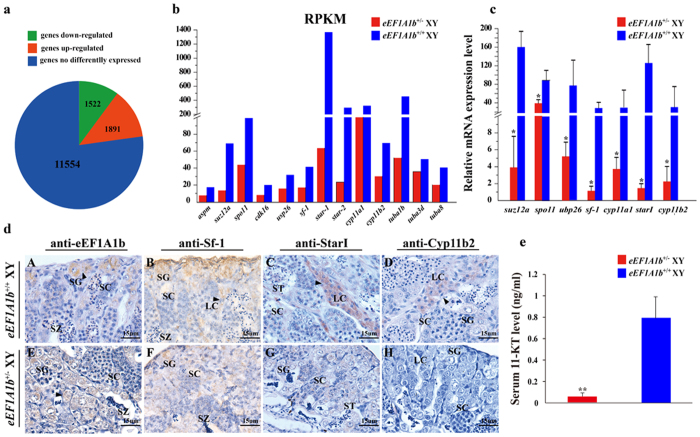
Transcriptome sequencing of the testes from the eEF1A1b+/− XY fish at 180 dah yielded a total of 40,039,839 reads. The eEF1A1b+/− XY transcriptome data were analyzed using the transcriptome data from testis of 180 dah XY fish obtained earlier by our group31 as a control. The total gene counts of the eEF1A1b+/− XY fish (22,284) were slightly more than those of the eEF1A1b+/+ XY fish (21,365). Compared with the control testis, 1,522 genes were down regulated, 1,891 genes were up regulated, 11,554 genes were not differentially expressed and 7,317 genes were at background expression level in the eEF1A1b+/− XY testis (Fig. 8a).
Figure 8. Transcriptomic analyses of gene expression profiles in eEF1A1b+/− testis at 180 dah.
(a) Testis expressed genes were divided into three parts: 1522 down-regulated genes, 1891 up-regulated genes, and 11554 non-differentially expressed genes. (b) Validation of genes with disrupted expression files from transcriptome data by real-time PCR. All examined genes displayed similar expression profiles to those from the transcriptome data. (c) Validation of genes with disrupted expression files from transcriptome data by IHC. Arrowheads indicate the positive signal. SG, spermatogonia; SC, spermatocytes; ST, spermatids; SZ, spermatozoa; LC, Leydig cells. (d) Serum 11-KT levels of the eEF1A1b+/− and eEF1A1b+/+ XY fish.
Further analysis revealed that of the 1522 down-regulated genes, 416 genes were involved in spermatogenesis process (e.g. aspm, cdk16, suz12a, usp26 and spo11), 86 genes were involved in steroidogenesis (e.g. sf-1, cyp11a1, star and cyp11b2) and 19 genes were involved in sperm motility (e.g. tuba1b, tuba3d and tuba8) (Fig. 8b). Real-time PCR analyses showed that mRNA levels of suz12a, spo11, usp26, sf-1, cyp11a1, star1 and cyp11b2 were significantly down-regulated in the eEF1A1b+/− testis compared with the control (Fig. 8c).
By IHC, eEF1A1b were observed in the spermatogonia, and Sf-1, StarI and Cyp11b2 were observed in the Leydig cells of the eEF1A1b+/+ testis (Fig. 8d, A-D). In contrast, reduced eEF1A1b and Sf-1 expression and very sparse StarI and Cyp11b2 were detected in the eEF1A1b+/− testis (Fig. 8d, E-H). Consequently, the serum 11-KT level of the eEF1A1b+/− XY fish was significantly lower than that of eEF1A1b+/+ XY fish (Fig. 8e).
Transgene mediated rescue of spermatogenesis in eEF1A1b +/− XY fish
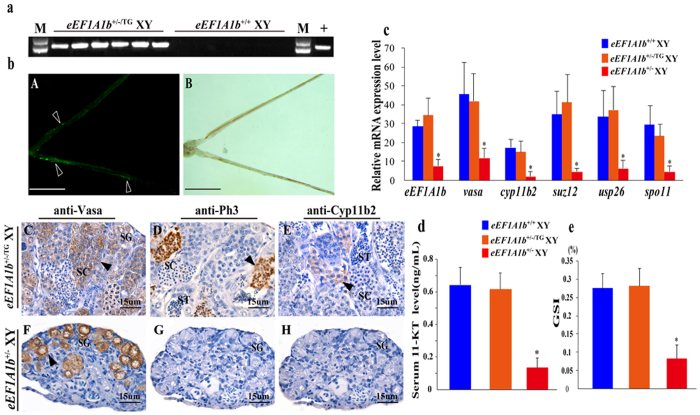
To rescue the spermatogenesis arrest phenotype of eEF1A1b+/− XY fish, transgenic overexpression of eEF1A1b was conducted in the eEF1A1b+/− XY fish using a CMV promoter-derived expression vector. A specific band of 720 bp was amplified from the genomic DNA of eEF1A1b transgenic (eEF1A1b+/−/TG) fish (Fig. 9a). GFP signals were detected in the eEF1A1b+/−/TG testis when examined at 90 dah (Fig. 9b, A and B).
Figure 9. Transgene mediated rescue of spermatogenesis in eEF1A1b+/− XY fish.
(a) Confirmation of eEF1A1b transgene insertion by genomic PCR. Lane 1 and 14, DNA marker; Lane 2-7, XY fish carrying eEF1A1b-transgene; Lane 8-13, XY control fish; Lane 15, empty plasmid as template. (b) A, the GFP signals in the testis of eEF1A1b+/−/TG XY fish in a dark field; Arrowheads indicate the positive signal. B, bright field; C-H, IHC analyses of Vasa, PH3 and Cyp11b2 expression in the testis of eEF1A1b+/−/TG and eEF1A1b+/− XY fish; SG, spermatogonia; SC, spermatocytes; ST, spermatids. (c) Expression of eEF1A1b, cyp11b2, vasa, suz12, usp26 and spo11 in the eEF1A1b+/−/TG, eEF1A1b+/− and eEF1A1b+/+ XY fish by real-time PCR. (d) Serum 11-KT levels of the eEF1A1b+/−/TG, eEF1A1b+/− and eEF1A1b+/+ XY fish. (e) GSI of eEF1A1b+/−/TG, eEF1A1b+/− and eEF1A1b+/+ XY fish. Results were expressed as mean ± SD. Different letters indicate statistical differences at P < 0.05 as determined by one-way ANOVA followed by post hoc test.
By IHC, Vasa and Ph3 were observed in the spermatogonia/spermatocytes and spermatocytes, and Cyp11b2 were detected in the Leydig cells of the eEF1A1b+/−/TG testis (Fig. 9b, C-E). In contrast, reduced Vasa, and no Ph3 and Cyp11b2 were observed in the eEF1A1b+/− testis (Fig. 9b, F-H).
By real-time PCR, the expression of eEF1A1b, cyp11b2, vasa, suz12 and usp26 in the eEF1A1b+/−/TG testis showed no significant differences compared with the eEF1A1b+/+ testis, but significantly higher than that of the eEF1A1b+/− testis (Fig. 9c). Consistently, the serum 11-KT level and GSI of the eEF1A1b+/− XY fish showed no significant differences compared with the eEF1A1b+/+ testis, but significantly higher than those of the eEF1A1b+/− testis (Fig. 9d and e)
Discussion
Evolutionary implication of eEF1A1b
In the present study, eEF1A1a and eEF1A1b genes were successfully identified from the Nile tilapia, zebrafish, medaka and fugu. Phylogenetic analyses of eEF1A1s from vertebrates demonstrated that eEF1A1a and eEF1A1b arose from the teleost genome duplication34. eEF1A1a has retained the ancestral functions, while eEF1A1b has evolved novel function/expression patterns. Moreover, conserved synteny of eEF1A1a with kcnq5a and eEF1A1b with kcnq5b was observed in teleosts. Taken together, these results suggested that eEF1A1a and eEF1A1b were derived from the teleost-specific genome duplication.
The expression pattern and functional relevance of eEF1A1b in tilapia
Spatial-temporal gene expression patterns are important aspects of gene function analysis. Our previous data showed that eEF1A1a displayed no gender difference in expression, while eEF1A1b was a male-biased gene in tilapia gonad31. In the present study, both real-time PCR and Western blot analyses demonstrated that eEF1A1b showed sexually dimorphic expression from 70 dah, and was highly expressed in the testis from 90 dah onwards, correlated with the initiation of spermatogenesis and the appearance of spermatocytes. The expression of eEF1A1b reached its highest levels with the increase of the spermatogonia at 120 dah. Further, eEF1A1b was expressed exclusively in spermatogonia, while eEF1A1a was found to be mostly expressed in somatic cells of the testis. In the ovary, the expression of eEF1A1b was low and constant. eEF1A1 is a housekeeping gene required for survival of all eukaryotic cells35,36, and therefore, at least one eEF1A gene must be expressed in the germ cells and somatic cells of the gonad in vertebrates. In mammals, eEF1A1 is a sex unbiased gene, and is expressed in the germ cell and somatic cell of the gonad13,14,15,16. In Xenopus, eEF-1S and eEF-1O are expressed in somatic cells and germ cells respectively, and 42sp50 is detected only in oocytes10,11. In tilapia, eEF1A1a was expressed in the somatic cells of the testis and ovary, while eEF1A1b and 42Sp50 were expressed in the spermatogonia and oogonia/oocytes, respectively. Taken together, different elongation factor isoforms are used for male and female germ cells, as well as germ cells and somatic cells in lower vertebrates, while only one (eEF1A1) is used in both male and female germ cells and somatic cells of gonad in mammals.
In the present study, eEF1A1b deficiency resulted in spermatogenesis arrest and consequent infertility in F0 XY fish. This was reflected by defective of meiosis, reduced GSI and abnormal spermiogenesis. Heterozygous mutants of eEF1A1b displayed a phenotype similar to that of the F0 XY fish. The eEF1A1b+/− testis showed no and very few spermatocytes at 90 and 120 dah, respectively. Spermatocytes, spermatids and spermatozoa were remarkably reduced at 150 and 180 dah. Further, most sperm from the eEF1A1b+/− XY fish were flagella-less, and therefore, incapable of fertilization. Anatomical examination of eEF1A1b+/− XY fish showed mostly normal organs and tissues, including the heart and brain, the organ with high eEF1A1b abundance (supplemental Fig. S7). Thus, our results indicated the impairment of testis development, and failure of fertilization in mutant fishes were probably not the result of defects in these tissues, but a specific effect of the eEF1A1b deficiency in testis. Both the eEF1A1b deficient and eEF1A1b+/− XY fish were infertile, clearly indicating that eEF1A1b is critical for spermatogenesis and fertility in male tilapia.
Of all eEF1A isoforms, only eEF1A1b was expressed in the spermatogonia in tilapia. Consequently, mutation of eEF1A1b caused spermatogenesis arrest and infertility. It is well documented that eEF1A is indispensable for protein synthesis. Recently, a report showed that genetic ablation of eEF4, which is a key quality-control factor in translation, causes testis-specific dysfunction in oxidative phosphorylation, leading to male infertility in mice20. Besides elongation factors, disruption of other genes expressed in the spermatogonia also blocks spermatogenesis27,29,37. Consistently, in the present study, the mRNA level of suz12a, aspm, and usp26, which were expressed in spermatogonia and critical for spermatogenesis28,37,38,39, were significantly down regulated in the eEF1A1b+/− testis. It is reasonable to believe that they were also down regulated in protein levels although we did not check the protein levels of these genes due to lack of specific antibodies. On the other hand, eEF1A1 might also be involved in spermatogenesis via other cellular processes26. However, the non-canonical function of eEF1A1b in spermatogenesis of tilapia remains to be elucidated.
In addition, suppression of steroidogenesis was observed in eEF1A1b mutant XY fish, as reflected by the decrease of Leydig cells, down-regulation of the expression of Sf-1 and the steroidogenic enzyme StarI, StarII, Cyp11a1, Cyp11b2 and 11β-HSD and a decrease in serum 11-KT level. It is well known that 11-KT is essential for spermatogenesis in fish40, and therefore, spermatogenesis arrest and infertility in eEF1A1b deficient and eEF1A1b+/− XY fish might be the resulted of low levels of 11-KT. Nevertheless, it is an indirect effect of the eEF1A1b mutation as this gene is not expressed in the Leydig cells of tilapia. These results suggested that spermatogonia-derived signals might affect the Leydig cells directly or indirectly. It is widely accepted that, during the development of testis, male germ cells are influenced by signals from the surrounding somatic cells, but not vice versa41. But this conventional perspective was challenged recently. For example, mutation dominant-white spotting locus, which is expressed in spermatogonia of testis42, resulted in absence of germ cells, decreased Leydig cell population and disordered expression profiles of Leydig cells key markers (cyp11a1, hsd3β and insl3) in mice43. Moreover, overexpression of figla resulted in up-regulation of 11-KT in male tilapia44. Taken together, we speculate that the Leydig cells in the testis might be influenced by signals from the male germ cells. However, the mechanisms involved remain to be elucidated.
The phenotype of eEF1A1b+/− testis was rescued by eEF1A1b overexpression as reflected by the normal testis size and appearance of all types of male germ cells, including spermatocytes and spermatids, which were absent in the eEF1A1b+/− testis. Moreover, eEF1A1b transgene also rescued the expression of those genes involved in the spermatogenesis process and steroidogenesis altered in the eEF1A1b+/− testis. These results validated that the spermatogenesis arrest phenotype is exclusively caused by eEF1A1b haploinsufficiency rather than off-target effects.
Previous reports showed that asthenozoospermia is a common cause of human male infertility, diagnosed by shortened sperm flagella, reduced sperm concentration and motility45 similar to the infertile phenotype of eEF1A1b mutant XY fish. The exact cause of asthenozoospermia is unclear so far, and mutation of eEF1A1 might be lethal in mammals. Mutation of eEF1A1b in tilapia could be used as a model for investigating the possible involvement of eEF1A1 in asthenozoospermia.
In summary, phylogenetic and syntenic analyses revealed that eEF1A1b is a duplicated copy of eEF1A1. In the testis it is exclusively expressed in the spermatogonia. Loss of function and transgenic overexpression studies indicated that the spermatogenesis arrest and infertility phenotypes observed in the eEF1A1b+/− XY fish was indeed caused by eEF1A1b haploinsufficiency. Taken together, our results suggested that eEF1A1b is essential for spermatogenesis and male fertility in the Nile tilapia. In addition, mutation of eEF1A1b in tilapia may provide an excellent model for studying human asthenozoospermia.
Materials and Methods
Animals
Nile tilapia, Oreochromis niloticus, was kept in recirculating aerated freshwater tanks at 26 °C under a natural photoperiod. All-XX progenies were obtained by crossing a pseudomale (XX male, producing sperm after hormonal sex reversal) with a normal female (XX). All-XY progenies were obtained by crossing a supermale (YY) with a normal female. Animal experiments were conducted in accordance with the regulations of the Guide for Care and Use of Laboratory Animals and were approved by the Committee of Laboratory Animal Experimentation at Southwest University.
Phylogenetic and synteny analysis of eEF1A1b
A phylogenetic tree of eEF1A1s was constructed using tilapia 42Sp50 as the outgroup. Sequences were obtained from the NCBI (http://blast.ncbi.nlm.nih.gov/) and Ensembl (http://www.ensembl.org/index.html) databases. The accession numbers of the sequences used are listed in the Supplemental Tab.S1. Full-length amino acid sequences were aligned using MEGA5.046. The credibility of the branching was tested using bootstrap resampling with 1000 pseudo-replicates.
For syntenic analysis, the location of eEF1A1 (including eEF1A1a and eEF1A1b) and their adjacent genes were determined for human, rat, chicken, green anole, clawed frog, Nile tilapia, medaka, fugu and zebrafish using the Ensembl genome browser (http://www.ensembl.org/index.html).
Tissue distribution of eEF1A1b by IHC
The various tissues obtained from XX and XY tilapia at 180 dah tilapia were fixed in Bouin’s solution for 12 hours at room temperature, dehydrated and embedded in paraffin. All tissue blocks were sectioned at 5 μm and used for IHC analysis, which was performed as described previously27. The antibody against eEF1A1b was diluted 1:1000 for use.
Production and characterization of eEF1A1b antibody
The peptide antigen (SGWNGDNMLEPSPNMT) of tilapia eEF1A1b was prepared and injected into rabbits. eEF1A1b antiserum (a-DTc) was collected and affinity purified. To confirm the specificity of the polyclonal antibody, total protein was extracted from XX and XY gonads of 30, 90, 120 and 300 dah tilapia. Western blots were carried out using the purified antibody at dilution of 1:500.
Expression profile of eEF1A1b in tilapia gonad by real-time PCR and Western blot
For ontogenic expression analysis, three parallel gonadal samples were prepared from the fish at 5, 10, 30, 40, 50, 60, 70, 90, 120, 150 and 180 dah. For sampling points from 5 to 60 dah, fish were dissected and viscera were removed under a stereoscopic microscope. RNAlater reagent (Ambion, TX) was poured on the coelomic epithelium to stabilize the RNA in the gonads, and then the gonads were removed using fine forceps. Gonads were pooled in a tube with RNAlater reagent and stored at −80 °C before RNA extraction.
Total RNAs (2.0 μg) were extracted and reverse transcribed into cDNA using PrimeScript RT Master Mix Perfect Real Time Kit according to the manufacturer’s instructions (Takara, Japan). Real-time PCR was carried out on an ABI-7500 real-time PCR machine according to the protocol of SYBR® Premix Ex Taq II. The relative abundance of eEF1A1b mRNA transcripts was evaluated using the formula: R = 2−∆∆Ct 47. The geometric mean of the copy numbers of the three reference genes (β-actin, gapdh and eEF1A1a) was used to normalize the expression of eEF1A1b. Primer sequences used for real-time PCR are listed in Supplemental Tab. S2. Data were expressed as the mean ± S.D. Statistical analysis was performed using GraphPad Prism4 software (GraphPad Software, USA). Significant differences between groups were tested by one-way ANOVA with post-hoc test. P < 0.05 was considered to be statistically significant.
Total proteins extracted from testes and ovaries of 30, 90, 120 and 300 dah tilapia were separated using 12% SDS-PAGE under reducing condition. Western blot was carried out as reported previously48 using the purified antibodies at dilution of 1:500.
Cellular location of eEF1A1b in gonads analysis by ISH and IHC
ISH was performed using tilapia gonads collected 180 dah from monosex (XX or XY) fish. Gonads were dissected and fixed in 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4, 4% PFA) at 4 °C overnight. After fixation, gonads were embedded in paraffin. Cross-sections of 5 μm were cut with a sliding microtome and adhered to polylysine-treated slides with diethylpyrocarbonate-treated water. Slides were dried at 180 °C for 4 hours to remove the RNase contamination and stored at 4 °C until use. Probes for both sense and antisense digoxigenin-labeled RNA strands were transcribed in vitro from a linearized pGEM-Teasy-eEF1A1b cDNA clone using the RNA labeling kit (Roche, Germany). ISH was performed as described previously49.
For IHC analysis, the gonads of 5, 30, 90 and 180 dah monosex (XX and XY) fish were dissected, fixed in Bouin’s solution for 12 hours at room temperature, dehydrated and embedded in paraffin. All tissue blocks were sectioned at 5 μm for IHC analysis, which was performed as described previously27. The antibody against eEF1A1b was diluted 1:1000 for use.
Disruption of eEF1A1b by CRISPR/Cas9
CRISPR/Cas9 was performed to knockout eEF1A1b in tilapia27. The gRNA target site was selected from eEF1A1b sequences corresponding to GGN18NGG on the sense or antisense strand of DNA (http://zifit.partners.org/ZiFiT/). Candidate target sequences were compared with the entire tilapia genome using the Basic Local Alignment Search Tool to avoid cleavage of off-target sites. One-cell stage embryos were divided into two batches, one for microinjection and the other for control. The gRNA and Cas9 mRNA were co-injected into one-cell stage embryos with the optimal concentration of 150 ng/μl and 300 ng/μl, respectively. Twenty injected embryos were collected 72 hours after injection. Genomic DNA was extracted from pooled injected embryos and control embryos, and used to access the mutations. DNA fragments spanning the eEF1A1b target site was amplified using gene specific primers (Supplemental Tab. 2). The indels were confirmed by restriction enzyme digestion with BsajI and Sanger sequencing. In addition, the percentage of uncleaved band was measured by quantifying the band intensity using Quantity One Software (Bio-Rad, USA)50. The indel (insertion and deletion) frequency was calculated by dividing uncleaved band intensity to the total band intensity from single digestion experiment.
The remaining micro-injected fish were reared until sampling for phenotypic assays. To screen the mutant fish, a piece of tail fin was clipped from each individual, and genomic DNA was extracted as described above. The target genomic locus was amplified using the primers eEF1A1b-cas-F/R. Mutations were assessed by restriction enzyme digestion and Sanger sequencing. The genetic sex of the mutants was determined using a sex-linked DNA marker (marker5) reported previous by our group32. The eEF1A1b-deficient fish were used for gonad histology, gene expression (real-time PCR, IHC, and Western blot), serum androgen (11-KT) analyses and fertility test at 90 and 180 dah.
eEF1A1b deficiency on spermatogenesis and fertility
Gonads of eEF1A1b deficient fish were sampled at 90, 120, 150 and 180 dah. Samples were fixed in Bouin’s solution for 24 hours at room temperature, dehydrated and embedded in paraffin. Tissue blocks were sectioned at 5 μm and stained with hematoxylin and eosin (H.E) or used for IHC analysis. Vasa (germ cell marker) antibody was donated by Prof. Nagahama, the National Institute of Basic Biology, Okazaki, Japan. The specificity of the antibody has been analyzed previously51. eEF1A1b and Cyp11b2 (11β-hydroxylase, the key enzyme for androgen 11-KT synthesis) antibody were prepared by our laboratory. The specificity of Cyp11b2 antibody was checked previously52. Antibodies against eEF1A1b, Vasa and Cyp11b2 were diluted 1:1000, 1:1000 and 1:500 for use, respectively. IHC analyses were performed as described above. Photographs were taken under Olympus BX51 light microscope.
Western blots were performed to confirm the expression change of eEF1A1b, Cyp11b2 and Vasa in the eEF1A1b deficient and the control testis at 180 dah.
Serum 11-KT level was measured using the EIA Assay kits (Cayman Chemical Co, USA) following the manufacturers’ instructions. Blood samples were collected from the caudal vein of the eEF1A1b deficient and control XY fish at 90 dah and kept at 4 °C overnight. Serum was collected after centrifugation and stored at −20 °C until use.
Sperm collected from eEF1A1b deficient and the control XY fish at 180 dah were diluted 1:1000 with phosphate buffered saline. Examination of morphology and motility of sperm was performed under Olympus BX51 light microscope.
To test fertility, 1200 mature eggs from an XX mother fish were divided into six groups (each with 200 eggs), and artificial insemination was performed using sperm from eEF1A1b deficient and control XY fish (n = 3). 12 hours later, the number of gastrula-stage embryos was scored under a light microscope to calculate fertilization rate. Hatching rates were calculated at 1 dah (5 days post fertilization). The concentration of sperm from eEF1A1b deficient and control XY fish was calculated using cell count data from flow cytometry (n = 3).
Production of F1
The eEF1A1b heterozygous F1 offspring were obtained by mating the F0 female founder with a wild-type male. F1 fish were screened by restriction enzyme digestion of PCR amplifications, as described above. Mutate sequences were confirmed by Sanger sequencing.
Heterozygous mutant of eEF1A1b on spermatogenesis and fertility
The gonads of heterozygous mutant of eEF1A1b (eEF1A1b+/−) XY fish were sampled at 90, 120, 150 and 180 dah for H.E and IHC analysis. eEF1A1b, Oct 4 (spermatogonia maker) and Cyp11b2 antibodies were prepared by our laboratory. Phospho-histone h3 (spermatocytes maker) antibody was obtained from the Cell signaling Technology (Beverly, MA, USA). Antibodies against eEF1A1b, Oct4, Histone 3 and Cyp11b2 were diluted 1:1000, 1:1000, 1:1000 and 1:500 for use, respectively. Sf-1 and StarI antibodies were prepared by our laboratory. The specificity of Sf-1 and StarI antibodies was checked previously48,53. Antibodies against Sf-1 and StarI were diluted 1:1000 and 1:200 for use, respectively. Serum 11-KT level of eEF1A1b+/− and eEF1A1b+/+ fish at 90 and 180 dah were measured using the EIA Assay kits as described above.
Sperm of eEF1A1b+/− and eEF1A1b+/+ XY fish was sampled 180 dah to assay sperm quality. Fertility tests were performed as described above. Sperm analysis, including sperm count, morphological abnormality, trajectory, flagellum beat frequency, motility and path velocity, were conducted using Sperm Quality Analyzer (ZKPACS-E) in The Ninth People’s Hospital of Chongqing.
Transcriptome analysis
The transcriptomes of 180 dah control testes was sequenced previously by our group54. Six testes from eEF1A1b+/− XY fish were used for transcriptome analyses. Total RNA was extracted from testes using the RNeasy Mini Kit (50) (Qiagen) according to the manufacturer’s instructions. The extracted RNA was further treated with deoxyribonuclease 1 (ribonuclease free) to eliminate genomic DNA contamination. The oligo (dT) bead-enriched mRNA was disrupted into short fragments (200–700 nt) using fragmentation buffer. These short fragments were used as templates for first- and second-strand cDNA synthesis using a DNA synthesis kit (Invitrogen). A QiaQuick PCR purification kit (Qiagen) was used to purify these fragments, and the elution buffer was used for end repair and addition of the poly (A) tail. Then, these short fragments were ligated to sequencing adapters. After agarose gel electrophoresis, fragments between 320 and 370 nt were cut from the gel for PCR amplification. cDNA libraries were constructed from the two samples and sequenced on an Illumina HiSeq2000 instrument. Clean reads from each library were aligned to the reference genome (Orenil1.0, http://www.ensembl.org/Oreochromis_niloticus/Info/Index) using Tophat with default parameters, and the reads per kb per million reads (RPKM) method was used to calculated gene expression level55. The assembled transcripts were merged with the reference annotation (Oreochromis niloticus. Orenil1.0.78.gtf, downloaded from Ensembl) using Cuffmerge, and differential expression analysis was performed using Cuffdiff. Transcriptomes of control-XY and eEF1A1b+/− XY were used to analyze the genes expression profiles. The threshold for the p-value was determined using false discovery rate (FDR) set at 0.0131. In this study, genes with RPKM < 1 in the transcriptome were considered to be background expression and were excluded from further analysis. Gonadal expressed genes were divided into non-differentially expressed genes between control and eEF1A1b+/− XY (NDGs), eEF1A1b+/− XY up-regulated genes (URGs), and eEF1A1b+/− XY down-regulated genes (DRGs). ‘FDR > 10−2’ and ‘−1 < log2 |(control-XY_RPKM/eEF1A1b+/− XY_RPKM)| < 1’ were used to identify NDGs, ‘FDR > 10−2’ and log2 (control-XY_RPKM/eEF1A1b+/− XY_RPKM) > 1 were used to identify DRGs, and ‘FDR > 10−2’ and (control-XY_RPKM/eEF1A1b+/− XY_RPKM) < −1 were used to identify (URGs). To study the biological pathways of the up/down-regulated genes involved, we mapped these differentially expressed genes to pathways in the Kyoto Encyclopedia of Genes and Genomes (KEGG) using KOBAS web server (http://kobas.cbi.pku.edu.cn/).
Transgene mediated rescue of spermatogenesis in eEF1A1b+/− XY fish
In vivo transgenic overexpression of the eEF1A1b in eEF1A1b+/− XY individuals was performed according to the methods of our previous study49. An expression plasmid for eEF1A1b was prepared as follows. Complementary DNA encoding the ORF of eEF1A1b was amplified by PCR with a primer set introducing the BamHI and EcoRI sites (Supplemental Tab. 2). The amplified fragment was digested by BamHI and EcoRI and ligated into the multiple cloning sites downstream of the cytomegalovirus (CMV) sequence of the pIRES-hrGFP-1a vector (Stratagene, La Jolla, CA). The prepared construct was microinjected into eEF1A1b+/− XY fertilized eggs at the one-cell stage. Genomic DNA was extracted from injected fish and used to screen transgenic fish at 90 dah. DNA fragments were amplified using gene specific primers (Supplemental Tab. 2). The testis of the injected fish was examined by monitoring the GFP signal, and later the testis was subjected to both histological and immunohistochemical analyses. Antibodies against Ph3, Vasa and Cyp11b2 were diluted as described above. Serum 11-KT level was measured using the EIA Assay kits as described above.
Additional Information
How to cite this article: Chen, J. et al. Heterozygous mutation of eEF1A1b resulted in spermatogenesis arrest and infertility in male tilapia, Oreochromis niloticus. Sci. Rep. 7, 43733; doi: 10.1038/srep43733 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The authors cordially thank Prof. Yoshitaka Nagahama from the National Institute of Basic Biology, Okazaki, Japan for providing the Vasa antibody. We are grateful to Prof. Kocher TD, Department of Biology, University of Maryland, USA for his critical reading of the manuscript. This work was supported by grants 91331119, 31630082 and 31602134 from the National Natural Science Foundation of China; grant 2011 AA100404 from the National High Technology Research and Development Program (863 program) of China; grant 20130182130003 from the Specialized Research Fund for the Doctoral Program of Higher Education of China; grants cstc2015jcyjB80001 and cstc2013kjrc-tdjs80003 from the Natural Science Foundation Project of Chongqing, Chongqing Science and Technology Commission; grant 2016T90830 and 2015M570765 from China Postdoctoral Science Foundation; grant Xm2015028 from Chongqing Postdoctoral Science Foundation; grant XDJK2016C157 from the Fundamental Research Funds for the Central Universities.
Footnotes
The authors declare no competing financial interests.
Author Contributions D.W. conceived and designed the experiments; J.C., D.J., D.T., Z.F. and Y.W. performed the experiments; D.W. and J.C. analyzed the data; D.J. and M.L. contributed reagents/materials/analysis tools. D.W. and J.C. wrote the manuscript. All authors read and approved the manuscript.
References
- Mateyak M. K. & Kinzy T. G. eEF1A: thinking outside the ribosome. Journal of Biological Chemistry 285, 21209–21213 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koji N., Masataka K., Shigekazu N. & Yoshito K. Structure of the two genes coding for polypeptide chain elongation factor lα (EF-lα) froni Saccharomyces cerevisiae. Gene 45, 265–273 (1986). [DOI] [PubMed] [Google Scholar]
- Hovemann B., Richter S., Walldorf U. & Cziepluch C. Two genes encode related cytoplasmic elongation factors 1α (EF-1α) in Drosophila melanogaster with continuous and stage specific expression. Nucleic Acids Research 16, 3175–3194 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danforth B. N. & Ji S. Elongation factor-1 alpha occurs as two copies in bees: implications for phylogenetic analysis of EF-1 alpha sequences in insects. Molecular biology and evolution 15, 225–235 (1998). [DOI] [PubMed] [Google Scholar]
- Gao D., Li Z., Murphy T. & Sauerbier W. Structure and transcription of the gene for translation elongation factor 1 subunit alpha of zebrafish (Danio rerio). Biochimica et Biophysica Acta (BBA)-Gene Structure and Expression 1350, 1–5 (1997). [DOI] [PubMed] [Google Scholar]
- Kinoshita M. et al. cDNA Cloning of Polypeptide Chain Elongation Factor 1. ALPHA. from Medaka Oryzias latipes. Fisheries science 65, 133–137 (1999). [Google Scholar]
- Nowell M. A. et al. Cloning and expression of an elongation factor-1α in sea bream (Sparus aurata) larvae and adult tissue. Marine Biotechnology 2, 173–179 (2000). [DOI] [PubMed] [Google Scholar]
- Mochida K. & Matsubara T. Molecular cloning of an elongation factor 1α and its mRNA localization in testis of the Nile tilapia Oreochromis niloticus. Fisheries science 68, 830–837 (2002). [Google Scholar]
- Infante C., Asensio E., Cañavate J. P. & Manchado M. Molecular characterization and expression analysis of five different elongation factor 1 alpha genes in the flatfish Senegalese sole (Solea senegalensis Kaup): differential gene expression and thyroid hormones dependence during metamorphosis. BMC molecular biology 9, 1 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdallah B., Hourdry J., Krieg P. A., Denis H. & Mazabraud A. Germ cell-specific expression of a gene encoding eukaryotic translation elongation factor 1 alpha (eEF-1 alpha) and generation of eEF-1 alpha retropseudogenes in Xenopus laevis. Proceedings of the National Academy of Sciences 88, 9277–9281 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps S. et al. Two forms of elongation factor 1 alpha (EF-1 alpha O and 42Sp50), present in oocytes, but absent in somatic cells of Xenopus laevis. The Journal of cell biology 114, 1109–1111 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeber J., Claußen M., Jahn O. & Pieler T. Interaction of 42Sp50 with the vegetal RNA localization machinery in Xenopus laevis oocytes. FEBS Journal 277, 4722–4731 (2010). [DOI] [PubMed] [Google Scholar]
- Lee S., Francoeur A.-M., Liu S. & Wang E. Tissue-specific expression in mammalian brain, heart, and muscle of S1, a member of the elongation factor-1 alpha gene family. Journal of Biological Chemistry 267, 24064–24068 (1992). [PubMed] [Google Scholar]
- Knudsen S. M., Frydenberg J., Clark B. F. & Leffers H. Tissue-dependent variation in the expression of elongation factor-1α isoforms: Isolation and characterisation of a cDNA encoding a novel variant of human elongation-factor 1α. European Journal of Biochemistry 215, 549–554 (1993). [DOI] [PubMed] [Google Scholar]
- Lee S., Wolfraim L. A. & Wang E. Differential expression of S1 and elongation factor-1 alpha during rat development. Journal of Biological Chemistry 268, 24453–24459 (1993). [PubMed] [Google Scholar]
- Lee S., LeBlanc A., Duttaroy A. & Wang E. Terminal differentiation-dependent alteration in the expression of translation elongation factor-1α and its sister gene, S1, in neurons. Experimental cell research 219, 589–597 (1995). [DOI] [PubMed] [Google Scholar]
- Chambers D. M., Peters J. & Abbott C. M. The lethal mutation of the mouse wasted (wst) is a deletion that abolishes expression of a tissue-specific isoform of translation elongation factor 1α, encoded by the Eef1a2 gene. Proceedings of the National Academy of Sciences 95, 4463–4468 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalyfa A. et al. Characterization of elongation factor-1A (eEF1A-1) and eEF1A-2/S1 protein expression in normal and wasted mice. Journal of Biological Chemistry 276, 22915–22922 (2001). [DOI] [PubMed] [Google Scholar]
- Newbery H. et al. Translation elongation factor eEF1A2 is essential for post-weaning survival in mice. Journal of Biological Chemistry 282, 28951–28959 (2007). [DOI] [PubMed] [Google Scholar]
- Gao Y. et al. Mammalian elongation factor 4 regulates mitochondrial translation essential for spermatogenesis. Nature structural & molecular biology 23, 441–449 (2016). [DOI] [PubMed] [Google Scholar]
- Shiina N., Gotoh Y., Kubomura N., Iwamatsu A. & Nishida E. Microtubule severing by elongation factor 1 alpha. Science 266, 282–285 (1994). [DOI] [PubMed] [Google Scholar]
- Yang F., Demma M., Warren V., Dharmawardhane S. & Condeelis J. Identification of an actin-binding protein from Dictyostelium as elongation factor 1a. Nature 347, 494–496 (1990). [DOI] [PubMed] [Google Scholar]
- Abbas W., Kumar A. & Herbein G. The eEF1A proteins: at the crossroads of oncogenesis, apoptosis, and viral infections. Frontiers in oncology 5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotokezaka Y. et al. Interaction of the eukaryotic elongation factor 1A with newly synthesized polypeptides. Journal of Biological Chemistry 277, 18545–18551 (2002). [DOI] [PubMed] [Google Scholar]
- Chuang S.-M. et al. Proteasome-mediated degradation of cotranslationally damaged proteins involves translation elongation factor 1A. Molecular and cellular biology 25, 403–413 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tash J. S. et al. Gamendazole, an orally active indazole carboxylic acid male contraceptive agent, targets HSP90AB1 (HSP90BETA) and EEF1A1 (eEF1A), and stimulates Il1a transcription in rat Sertoli cells. Biology of reproduction 78, 1139–1152 (2008). [DOI] [PubMed] [Google Scholar]
- Li M. et al. Efficient and heritable gene targeting in tilapia by CRISPR/Cas9. Genetics 197, 591–599 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wosnitzer M. S. et al. Ubiquitin specific protease 26 (USP26) expression analysis in human testicular and extragonadal tissues indicates diverse action of USP26 in cell differentiation and tumorigenesis. PloS one 9, e98638 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargelius A. et al. Dnd knockout ablates germ cells and demonstrates germ cell independent sex differentiation in Atlantic salmon. Scientific reports 6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. Prmt5 is required for germ cell survival during spermatogenesis in mice. Scientific reports 5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W. et al. Characterization of gonadal transcriptomes from Nile tilapia (Oreochromis niloticus) reveals differentially expressed genes. PloS one 8, e63604 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y.-L. et al. Screening and characterization of sex-linked DNA markers and marker-assisted selection in the Nile tilapia (Oreochromis niloticus). Aquaculture 433, 19–27 (2014). [Google Scholar]
- Brawand D. et al. The genomic substrate for adaptive radiation in African cichlid fish. Nature 513, 375–381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. S., Braasch I., Frickey T., Meyer A. & Van de Peer Y. Genome duplication, a trait shared by 22,000 species of ray-finned fish. Genome research 13, 382–390 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshi R. et al. Overexpression of translation elongation factor 1A affects the organization and function of the actin cytoskeleton in yeast. Genetics 157, 1425–1436 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S. R. & Kinzy T. G. Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nature structural & molecular biology 12, 772–778 (2005). [DOI] [PubMed] [Google Scholar]
- Mu W., Starmer J., Fedoriw A. M., Yee D. & Magnuson T. Repression of the soma-specific transcriptome by Polycomb-repressive complex 2 promotes male germ cell development. Genes & development 28, 2056–2069 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B. et al. Establishment of a proteomic profile associated with gonocyte and spermatogonial stem cell maturation and differentiation in neonatal mice. Proteomics 14, 274–285 (2014). [DOI] [PubMed] [Google Scholar]
- Pulvers J. N. et al. Mutations in mouse Aspm (abnormal spindle-like microcephaly associated) cause not only microcephaly but also major defects in the germline. Proceedings of the National Academy of Sciences 107, 16595–16600 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura C., Miura T., Yamashita M., Yamauchi K. & Nagahama Y. Hormonal induction of all stages of spermatogenesis in germ-somatic cell coculture from immature Japanese eel testis. Development, growth & differentiation 38, 257–262 (1996). [DOI] [PubMed] [Google Scholar]
- M Escott G., A da Rosa L. & da Silveira Loss E. Mechanisms of Hormonal Regulation of Sertoli Cell Development and Proliferation: A Key Process for Spermatogenesis. Current molecular pharmacology 7, 96–108 (2014). [DOI] [PubMed] [Google Scholar]
- Manova K., Nocka K., Besmer P. & Bachvarova R. F. Gonadal expression of c-kit encoded at the W locus of the mouse. Development 110, 1057–1069 (1990). [DOI] [PubMed] [Google Scholar]
- Rios-Rojas C., Spiller C., Bowles J. & Koopman P. Germ cells influence cord formation and leydig cell gene expression during mouse testis development. Developmental Dynamics (2015). [DOI] [PubMed] [Google Scholar]
- Qiu Y. et al. Figla Favors Ovarian Differentiation by Antagonizing Spermatogenesis in a Teleosts, Nile Tilapia (Oreochromis niloticus). PloS one 10, e0123900 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curi S. et al. Asthenozoospermia: analysis of a large population. Archives of andrology 49, 343–349 (2003). [DOI] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Yu X. et al. Characterization of two paralogous StAR genes in a teleost, Nile tilapia (Oreochromis niloticus). Molecular and cellular endocrinology 392, 152–162 (2014). [DOI] [PubMed] [Google Scholar]
- Wang D.-S. et al. Foxl2 up-regulates aromatase gene transcription in a female-specific manner by binding to the promoter as well as interacting with ad4 binding protein/steroidogenic factor 1. Molecular Endocrinology 21, 712–725 (2007). [DOI] [PubMed] [Google Scholar]
- Ostyn A., De Buyser M. L., Guillier F., Krys S. & Hennekinne J. A. Benefits of the combined use of immunological-and PCR-based methods for determination of staphylococcal enterotoxin food safety criteria in cheeses. Food Analytical Methods 5, 173–178 (2012). [Google Scholar]
- Kobayashi T., Kajiura-Kobayashi H. & Nagahama Y. Two isoforms of vasa homologs in a teleost fish: their differential expression during germ cell differentiation. Mechanisms of development 111, 167–171 (2002). [DOI] [PubMed] [Google Scholar]
- Zhang X. et al. Isolation of doublesex-and Mab-3-related transcription factor 6 and its involvement in spermatogenesis in tilapia. Biology of reproduction, biolreprod. 114.121418 (2014). [DOI] [PubMed] [Google Scholar]
- Xie Q.-P. et al. Haploinsufficiency of SF-1 Causes Female to Male Sex Reversal in Nile Tilapia, Oreochromis niloticus. Endocrinology, en. 2015–2049 (2016). [DOI] [PubMed] [Google Scholar]
- Sun L.-N. et al. Transdifferentiation of differentiated ovary into functional testis by long-term treatment of aromatase inhibitor in Nile tilapia. Endocrinology 155, 1476–1488 (2014). [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L. & Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods 5, 621–628 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.