Abstract
N6-methyladenosine (m6A) is the most abundant internal modification in eukaryotic messenger RNAs (mRNAs), and plays important roles in cell differentiation and tissue development. It regulates multiple steps throughout the RNA life cycle including RNA processing, translation, and decay, via the recognition by selective binding proteins. In the cytoplasm, m6A binding protein YTHDF1 facilitates translation of m6A-modified mRNAs, and YTHDF2 accelerates the decay of m6A-modified transcripts. The biological function of YTHDF3, another cytoplasmic m6A binder of the YTH (YT521-B homology) domain family, remains unknown. Here, we report that YTHDF3 promotes protein synthesis in synergy with YTHDF1, and affects methylated mRNA decay mediated through YTHDF2. Cells deficient in all three YTHDF proteins experience the most dramatic accumulation of m6A-modified transcripts. These results indicate that together with YTHDF1 and YTHDF2, YTHDF3 plays critical roles to accelerate metabolism of m6A-modified mRNAs in the cytoplasm. All three YTHDF proteins may act in an integrated and cooperative manner to impact fundamental biological processes related to m6A RNA methylation.
Keywords: YTHDF3, N6-methyladenosine, translation, decay
Introduction
N6-methyladenosine (m6A), the most abundant internal modification in mRNA1, is emerging as a critical mRNA chemical mark that mediates post-transcriptional gene expression regulation. m6A is present in mammalian RNA at ∼0.1%-0.4% of all adenines, located within the consensus sequences G(m6A)C (70%) or A(m6A)C (30%)2,3. According to transcriptome-wide mapping results, transcripts from more than 7 000 human genes can be m6A-modified4,5.
m6A is dynamically deposited, removed, and recognized by m6A methyltransferases (“writers”), demethylases (“erasers”), and m6A-specific binding proteins (“readers”), respectively. The “writer” METTL3 is crucial in regulating stem cell pluripotency, cell differentiation, and circadian period6,7,8, whereas depletion of the “erasers” FTO and ALKBH5 has revealed their roles in energy homeostasis, adipocyte differentiation, and fertility in mice9,10,11.
Multiple cytoplasmic and nuclear m6A “reader” proteins have been identified4,12,13,14,15,16,17,18. Among them, the YTH domain family proteins directly bind and recognize m6A methylation on mRNA12,13,14,15,18. In the cytosol, YTHDF1 enhances translation of its targets by interacting with initiation factors and facilitating ribosome loading13; YTHDF2 promotes mRNA degradation by localizing m6A-modified mRNA to processing bodies in the cytoplasm12, and could change its own cellular localization in response to heat-shock stress18. The two proteins share a set of common target mRNAs, to which YTHDF1 binds earlier during the mRNA life cycle than YTHDF213. Together, YTHDF1 and YTHDF2 may contribute to generating a burst of protein synthesis, with increased translation from temporally controlled mRNAs13.
A third member of the YTH family, YTHDF3, shares over 65% protein sequence identity with both YTHDF1 and YTHDF2. Although YTHDF3 has been identified as a cytoplasmic m6A-binding protein4, its role in RNA metabolism is still unclear. Here we report the characterization of YTHDF3 as a partner of YTHDF1 and YTHDF2 to impact cytoplasmic metabolism of methylated mRNAs. We confirmed YTHDF3's affinity toward m6A-modified mRNA in cells, and identified its mRNA targets. It is found that YTHDF3 affects translation of its target mRNAs together with YTHDF1. Our results also indicate a spatiotemporal interplay among the three YTHDF proteins (YTHDFs) to cooperatively control translation and decay of their common mRNA targets in the cytosol.
Results
YTHDF3 binds m6A in cells and shares mRNA targets with YTHDF1 and YTHDF2
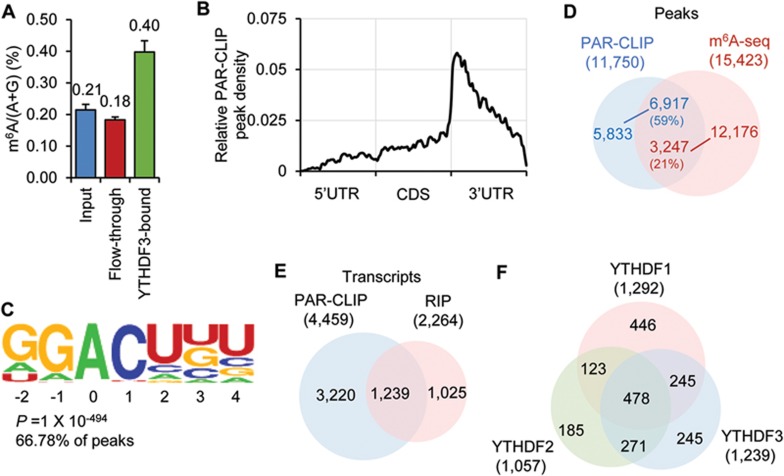
Previous studies have confirmed that YTHDF3 selectively binds m6A-modified mRNA in vitro12. To examine this selective binding activity inside cells, we performed RNA immunoprecipitation (RIP)-based experiments using cell lysates. We started by quantifying the m6A level of RNAs purified from the YTHDF3-RNA complex using liquid chromatography-tandem mass spectrometry (LC-MS/MS). Compared with input mRNA, we detected an over two-fold enrichment of m6A in the YTHDF3-bound portion (Figure 1A). Next, we mapped the binding sites of YTHDF3 in HeLa cells using photoactivatable ribonucleoside crosslinking and immunoprecipitation (PAR-CLIP). Three biological replicates of PAR-CLIP assays identified 11 750 common peaks as YTHDF3 binding sites and 4 459 mRNAs as target transcripts (Supplementary information, Figure S1A and Table S1). These binding sites cluster around the stop codon, coinciding with the distribution pattern of m6A along the length of mRNA (Figure 1B and Supplementary information, Figure S1B-S1C). Also, a GGACH motif, similar to the m6A consensus motif in the transcriptome, was identified in over 65% of the PAR-CLIP peaks (Figure 1C and Supplementary information, Figure S1D). Moreover, 59% of the PAR-CLIP peaks overlapped with m6A peaks profiled by using anti-m6A specific antibody in HeLa cells (Figure 1D). Therefore, we conclude that YTHDF3 recognizes the m6A modification on transcripts inside cells.
Figure 1.
YTHDF3 selectively binds m6A in cells. (A) LC-MS/MS quantification showing m6A is enriched in RNAs pulled down with YTHDF3 from the cell lysate. Error bars, mean ± sd, n = 2, technical replicates. (B) Distribution of common PAR-CLIP peaks identified in biological triplicates along the length of mRNA. (C) YTHDF3-binding motif identified by HOMER47 with common PAR-CLIP peaks identified in triplicates. P = 1× 10−494, the motif is identified in 67% of PAR-CLIP peaks. (D) Overlap of common YTHDF3 PAR-CLIP peaks and m6A-seq peaks from HeLa cells. PAR-CLIP peak numbers, blue; m6A peak numbers, red. (E) Overlap of target transcripts identified in PAR-CLIP triplicates and RIP replicates. (F) Overlap of target transcripts among YTHDF1-3.
To obtain high-confidence YTHDF3 target transcripts for further functional characterizations, we also sequenced the RNA that co-immunoprecipitated with YTHDF3 (RIP-seq). In two biological replicates of RIP-seq, 2 264 different transcripts were identified with over two-fold enrichment (Supplementary information, Table S1). About 1 239 of them overlap with transcripts identified by PAR-CLIP (Figure 1E and Supplementary information, Table S1), which were defined as high-confidence targets for YTHDF3 (CLIP + IP). Among these targets, genes related to the gene ontology (GO) term “regulation of transcription” are the most enriched (Supplementary information, Figure S1E and Table S1). YTHDF3 shares a large number of common targets with YTHDF1 and YTHDF2: 58% of YTHDF3 targets are also recognized by YTHDF1, and 60% by YTHDF2 (Figure 1F and Supplementary information, Table S1). These results suggest a potential coordinated action on common target transcripts among YTHDF proteins.
YTHDF3 facilitates translation
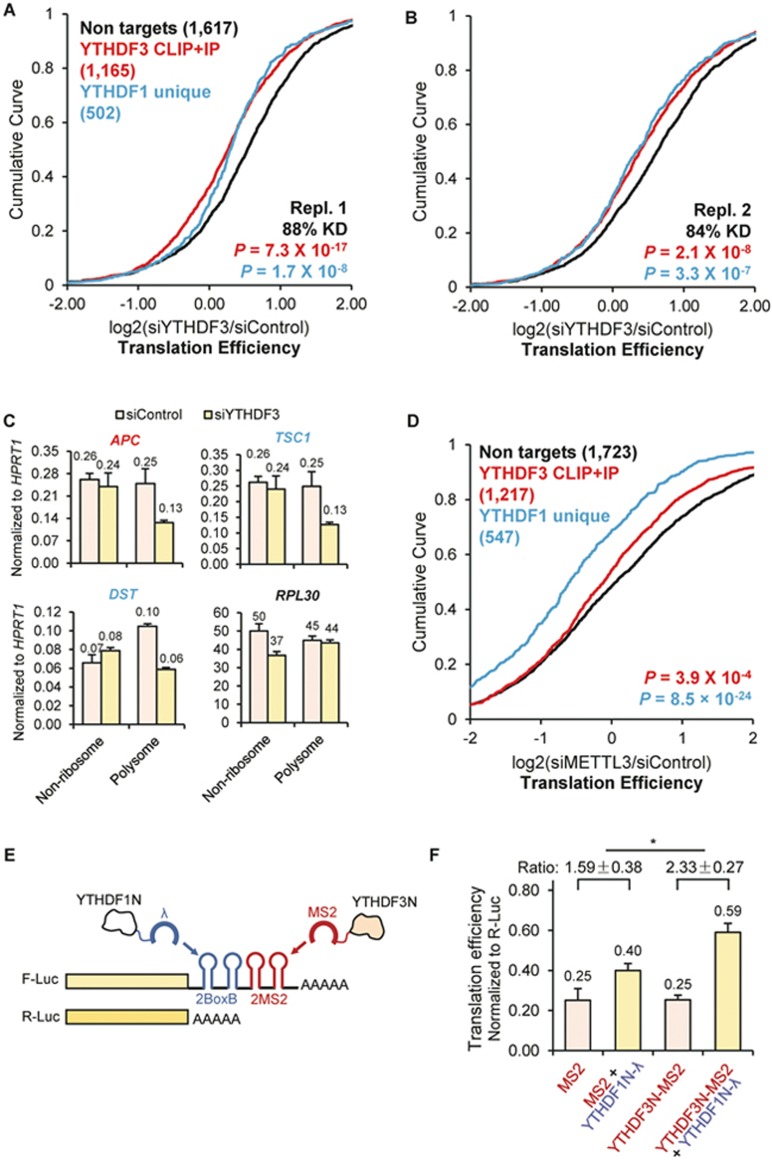
We next monitored mRNA targets of YTHDF3 upon depletion of YTHDF3 or an m6A methyltransferase component in HeLa cells, respectively. To evaluate whether YTHDF3 is involved in translation, we performed ribosome profiling in HeLa cells transfected with either siYTHDF3 or control siRNAs. To estimate translation efficiency, we investigated the ribosome density of each transcript by sequencing in parallel input mRNA and the ribosome footprint on mRNAs. Considering the established function of YTHDF1 in promoting translation13, we catalogued the evaluated transcripts into three groups: non-targets, YTHDF3 targets (CLIP + IP), and YTHDF1 targets that are not YTHDF3 targets (YTHDF1 unique). Compared with non-targets, a noticeable decrease of translation efficiency (TE) was observed in siYTHDF3 cells for both YTHDF3 targets (p < 2×10−8, Mann-Whitney U-test) and YTHDF1 unique targets (p < 3×10−7, Mann-Whitney U-test) (Figure 2A and 2B and Supplementary information, Figure S2A-S2D). This result suggests that YTHDF3 facilitates the translation of its RNA targets, and such effect has interplay with the role of YTHDF1.
Figure 2.
YTHDF3 promotes translation efficiency of its mRNA targets and facilitates the function of YTHDF1. (A-B, D) Cumulative distribution of log2-fold changes of translation efficiency (ratio between ribosome-bound fragments and input RNA) between siControl and siYTHDF3, biological replicate 1 (A), siYTHDF3, biological replicate 2 (B), and siMETTL3 (D). Three groups of genes were plotted: non-targets (neither targets of YTHDF1 nor targets of YTHDF3, black), YTHDF3 CLIP+IP (high-confidence YTHDF3 targets, red), and YTHDF1 unique (YTHDF1 targets that are not targets of YTHDF3, blue). Number of genes in each group was indicated in parentheses. P values were calculated from a two-sided Mann-Whitney test compared to non-targets. (C) Redistribution of representative targets in non-ribosome and polysome portions of mRNPs upon depletion of YTHDF3 measured by RT-qPCR. APC, a YTHDF3 target; TSC1 and DST, YTHDF1 unique targets; and RPL30, a non-target. Error bars, mean ± sd, n = 2, technical replicates. (E) Construct of the double tethering assay. A sequence of two BoxB followed by two MS2 stem loops was inserted at the 3′UTR of F-Luc (firefly luciferase) mRNA. The C-terminal YTH domains of YTHDF1 and YTHDF3 were respectively replaced with λ peptide (binding BoxB motif) and MS2 binding protein (binding MS2 motif). R-Luc (renilla luciferase) mRNA was used as an internal control for normalizing luciferase signals from different samples. (F) Translation efficiency of F-Luc normalized with R-Luc 4-hour post F-Luc induction, with the expression of effectors indicated at x-axis. The ratio between YTHDF1N-λ (yellow) and the corresponding control sample (grey) was calculated. Error bars, mean ± sd, n = 3. P = 0.05 (paired two-sided Student's t-test).
We then performed polysome profiling of YTHDF3-deficient cells to verify the observed trend. A different YTHDF3 siRNA sequence was used to eliminate any potential off-targeting effect of siRNAs. The three groups of mRNA-protein particles (mRNPs) were separated: non-ribosome (mRNPs without any ribosome), 40S-80S (mRNPs associated with ribosome but not being translated), and polysome (actively translating pool). Changes in the distribution of specific mRNAs among non-ribosome and polysome fractions were quantified via RT-qPCR. Noticeable decreases in the polysome fraction were observed for the selected YTHDF3 target (APC mRNA) and YTHDF1 unique targets (TSC1 mRNA and DST mRNA), but not for a non-target (RPL30 mRNA) (Figure 2C). This result validates the function of YTHDF3 in promoting translation of its mRNA targets and facilitating the function of YTHDF1.
After confirming the role of YTHDF3 in mRNA translation, we studied its dependence on m6A. On the basis of the published ribosome profiling data from HeLa cells depleted of the m6A methyltransferase complex component METTL319, we observed that after a global loss of m6A, YTHDF3 targets experienced a significant decrease in TE compared with non-targets (p < 4×10−4, Mann-Whitney U-test) (Figure 2D and Supplementary information, Figure S2I-S2J), supporting that YTHDF3 functions in an m6A-dependent manner.
We then evaluated the interplay between YTHDF1 and YTHDF3 in translation regulation of their common targets. A luciferase-based double tethering reporter assay was designed taking advantage of two orthogonal sets of an RNA motif and its binding protein12,20. A sequence composed of two BoxB stem loops followed by two MS2 stem loops was inserted to the 3′-UTR of the firefly luciferase (F-Luc) coding sequence. The C-terminal YTH domains of YTHDF1 and YTHDF3 were replaced with BoxB-binding protein λ peptide (YTHDF1N-λ) and MS2-binding protein (YTHDF3N-MS2), respectively. Thus, the assay mimics the binding of multiple m6A sites on one mRNA transcript by YTHDF1 and YTHDF3. Renilla luciferase (R-Luc) was used as an internal control to normalize the difference in transfection efficiency among samples (Figure 2E). The presence of YTHDF3N-MS2 magnified the translation-promoting effect of YTHDF1N-λ on F-Luc (Figure 2F, p = 0.05). The protein level of F-Luc was increased whereas its mRNA level remained unchanged (Supplementary information, Figure S2K). These results further support the role of YTHDF3 in cooperatively promoting translation with YTHDF1.
YTHDF proteins function cooperatively in the cytosol
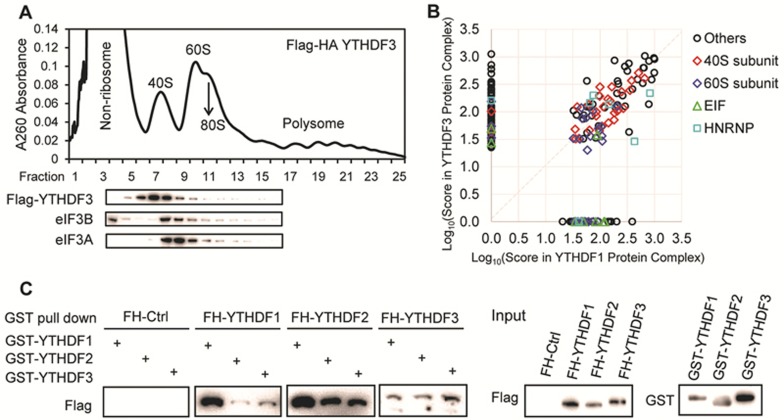
There are two mechanisms through which YTHDF3 could promote translation: it may directly recruit translation machineries such as initiation factors (eIFs), or it could interact with YTHDF1 and affect the function of YTHDF1. To investigate these possibilities, we studied protein partners of YTHDF3 using a HeLa stable cell line expressing YTHDF3 with a Flag-HA tandem epitope tag at the N-terminus. After polysome profiling of the stable cell line, western blotting of each fraction showed that YTHDF3 is distributed mainly in 40S mRNPs, suggesting YTHDF3 might regulate translation initiation (Figure 3A). However, neither eIF3A nor eIF3B was detected by western blotting in proteins specifically co-immunoprecipitated with YTHDF3 (Supplementary information, Figure S3A). Unlike YTHDF1, which interacts with eIF313, no direct interaction may exist between YTHDF3 and eIF3 components. In the reporter assay, YTHDF3N-MS2 itself did not promote translation (Figure 2F, first and third columns), further suggesting it might not recruit translation initiation factors directly.
Figure 3.
Protein partner analyses showing that YTHDF3 may interact with translation machineries through YTHDF1. (A) Polysome profiling of HeLa cells stably overexpressing Flag-HA tagged YTHDF3, and western blotting of Flag, eIF3A, and eIF3B in each fraction. (B) Comparison of components of protein complex co-immunoprecipitated with Flag-HA-YTHDF3 and Flag-HA-YTHDF1. Scores of each protein partner obtained from mass spectrometry were used in the x-y scatter plot. Subunits of 40S were labelled with red diamond; subunit of 60S, dark blue diamond; EIF components, green triangle; HNPNP proteins, light blue rectangle. See also Supplementary information, Table S2. (C) In vitro binding assay showing that YTHDFs co-immunoprecipitated with one another. Purified GST tagged YTHDF1-3 were incubated with cell lysates from Flag-HA (FH) tagged YTHDF1-3. FH tagged proteins were western blotted in the eluent after GST affinity pull down. FH-Ctrl, Flag-HA expression control cell line.
We next compared protein partners of YTHDF3 and YTHDF1. Components of the YTHDF3-containing protein complexs from tandem-affinity purification were subjected to protein mass spectrometry analysis (Supplementary information, Table S2). Protein partners reported for YTHDF1 using the same method were used for comparison13. YTHDF3 and YTHDF1 shared 86 out of 154 proteins identified to interact with YTHDF3, with 39 of the shared partners being 40S and 60S ribosomal proteins (Figure 3B and Supplementary information, Table S2). Together, these results suggest that YTHDF3 promotes translation through interacting with YTHDF1. In addition, immunoprecipitation and western blotting identified additional RNA-binding proteins as shared protein partners of YTHDF1 and YTHDF3 (Supplementary information, Figure S3B). They both interact with YBX1, which is known to impact mRNA splicing, localization, and translation21; and eIF4A3, a component of the exon junction complex that affects mRNA translation before being displaced by the first round of translating ribosomes22. These proteins may shuttle mRNAs to YTHDF3 and YTHDF1 for accelerated translation.
We examined the crosstalk among YTHDFs based on the observation that YTHDFs share hundreds of common targets and that YTHDF1 and YTHDF3 appear to have a synergistic effect on promoting translation. The direct binding between YTHDFs was tested with an in vitro binding assay using purified GST-tagged YTHDF1-3 and cell lysate containing Flag-HA-tagged YTHDF1-3. Flag blotting after GST pull down showed that YTHDFs directly interacted with each other (Figure 3C). In a similar co-immunoprecipitation assay, combinations of Flag-tagged YTHDFs and HA-tagged YTHDF3 were co-transfected into HeLa cells. Flag affinity pull down followed by HA blot validated that YTHDF3 interacts with YTHDF1 and YTHDF2 in an RNA-independent way, suggesting direct binding (Supplementary information, Figure S3C).
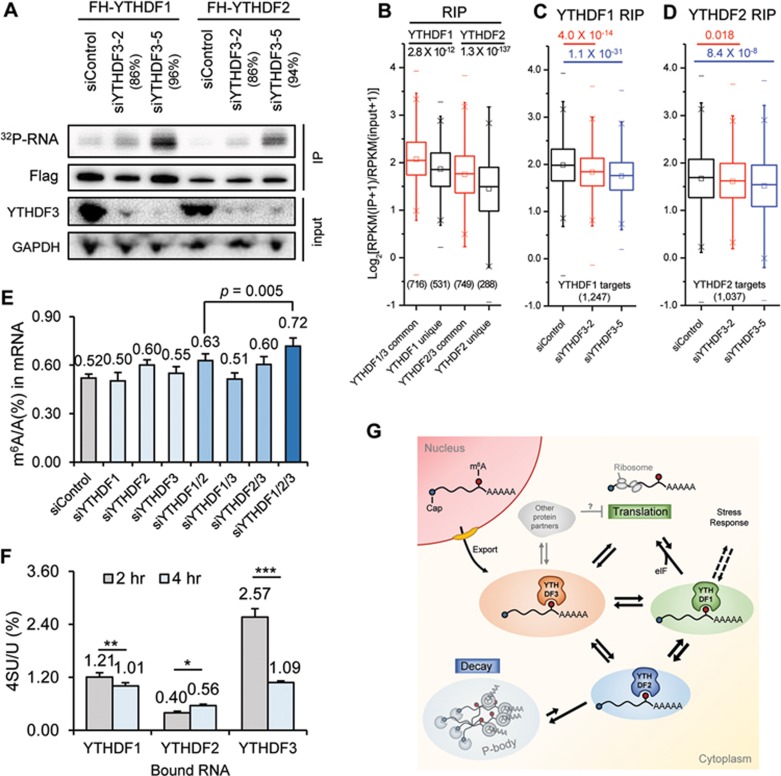
We then checked how depletion of YTHDF3 affects target binding activity of YTHDF1 and YTHDF2. PAR-CLIP was performed for YTHDF1 and YTHDF2 in cells depleted of YTHDF3, and RNAs pulled down were semi-quantified with 5′-32P labeling. Knockdown of YTHDF3 led to increased amount of RNA pulled down with YTHDF1 and YTHDF2, and this increased binding correlated with YTHDF3 knockdown efficiency (Figure 4A). To investigate how YTHDF3 affects RNA binding specificity of YTHDF1 and YTHDF2, we performed transcriptome-wide YTHDF1 and YTHDF2 RIP-seq in control cells and cells without YTHDF3. In control samples, YTHDF1/3 common targets showed greater enrichment than YTHDF1 unique targets, as was the case for YTHDF2 (Figure 4B). In the absence of YTHDF3, the RIP enrichment of YTHDF1 or YTHDF2 targets was reduced significantly (Figure 4C-4D). RIP followed by RT-qPCR of a number of specific transcripts confirmed that, when expression of YTHDF3 was reduced, YTHDF1 and YTHDF2 showed a decreased binding specificity, with reduced binding toward their target transcripts but increased binding toward non-target transcripts (Supplementary information, Figure S4A). The decreased binding toward targets was not caused by a change in protein levels of YTHDF1, as siYTHDF3 treatment increased the expression of YTHDF1 in HeLa cells (Supplementary information, Figure S4B). These results indicate that the binding specificity of YTHDF1 and YTHDF2 toward their target mRNAs could be affected by the presence of YTHDF3. In contrast, depletion of YTHDF1 or YTHDF2 led to a decreased amount of RNA bound by YTHDF3 (Supplementary information, Figure S4C). It is possible that YTHDF3 contributes more to the RNA binding specificity, whereas YTHDF1 and YTHDF2 contribute more to RNA binding affinity.
Figure 4.
YTHDF proteins form an interconnected network in the cytosol. (A) Total RNAs bound by YTHDF1 and YTHDF2 quantified with PAR-CLIP followed by 5′-32P labelling in the control HeLa cells and cells depleted of YTHDF3 using two different siYTHDF3 oligos. Knockdown efficiency of the two siYTHDF3 oligoes was indicated, respectively. Samples loaded in the radioactivity gel were normalized with immunostaining of the Flag-tagged protein. (B-D) Genome-wide analysis of target affinity of YTHDF1 and YTHDF2 with or without YTHDF3. Box plot of RIP enrichment of different groups of YTHDF targets in siControl samples (B), and that of YTHDF1 targets (C) or of YTHDF2 targets (D) in siControl and siYTHDF3 samples. Box, 25%-75%; “−”, max and min; “×”, 1% and 99%; “□”, median. P values were calculated from a two-sided Mann-Whitney test. (E) LC-MS/MS quantification of m6A levels of HeLa cells treated with siControl, siYTHDF1, siYTHDF2, siYTHDF3, and combinations of those oligoes. Error bars, mean ± sd, n = 4 (two biological replicates × two technical replicates). (F) LC-MS/MS quantification of 4SU (4-thio-uridine) level in mRNAs pulled down with YTHDF1-3 2-hour and 4-hour post a 1-hour 4SU labeling of nascent RNAs. Error bars, mean ± sd, n = 3∼4. P values were calculated using paired two-sided Student's t-test. *P < 0.05; **P < 0.005; ***P < 0.0005; (G) A proposed model for an integrated partition network for m6A-modified transcripts mediated by YTHDFs in the cytosol. While YTHDF1 functions in translation regulation and YTHDF2 dominates in accelerating mRNA decay, YTHDF3 could serve as a hub for fine-tuning the RNA accessibility of YTHDF1-2. These three mRNA pools controlled by YTHDF1-3 could be interchangeable and highly dynamic, resulting in an interconnected and dynamic mRNA modulation through m6A. YTHDF3 might also interact with other protein partners (grey) to negatively impact translation.
We observed a 40% increase of the m6A level in mRNAs when all of the YTHDFs were depleted (Figure 4E); the additional knockdown of YTHDF3 further increases the m6A accumulation comparing with the double knockdown of YTHDF1 and YTHDF2. These results indicate that all three YTHDFs contribute collectively to accelerating the metabolism of m6A-modified mRNAs in the cytosol. By metabolically labeling nascent RNA with nucleoside-analogues, the occupancy of labeled RNA by YTHDFs could be quantified over time. It is observed that both YTHDF1 and YTHDF3 bound mRNA targets prior to YTHDF2 (Figure 4F), with YTHDF3 binding more nascent RNAs than YTHDF1 (Supplementary information, Figure S5A-S5C). In addition, we quantified m6A level changes of YTHDF-bound RNAs over time after transcription inhibition (Supplementary information, Figure S5D). The m6A level decreased in YTHDF3-bound RNAs, whereas it increased in YTHDF1-bound ones 2 h after transcription inhibition (Supplementary information, Figure S5E), which suggests a flow of m6A-modified transcripts from YTHDF3 to YTHDF1. Therefore, the m6A-modified transcripts might engage YTHDF3 and then YTHDF1 for translation promotion before being decayed through the YTHDF2-mediated pathway in the cytoplasm. Although both YTHDF1 and YTHDF3 facilitate translation of their target mRNAs, they also collectively affect the partitioning of methylated transcripts to YTHDF2 for accelerated decay.
Discussion
Our current understanding of m6A-mediated cytoplasmic mRNA regulation mainly came from functional and structural studies of YTHDF1 and YTHDF212,13,18,23,24,25. However, the role of YTHDF3, another cytoplasmic reader of the m6A modification, remains unclear. In this work, we present a systematic characterization of YTHDF3 in HeLa cells. Our studies on YTHDF3 binding sites and target mRNAs support the specific binding of m6A on cellular mRNAs by YTHDF3 (Figure 1B-1D), and revealed hundreds of common targets shared by YTHDFs (Figure 1F). YTHDF3 facilitated YTHDF1's role in promoting translation in the tethering assay (Figure 2E-2F and Supplementary information, Figure S2K). Knockdown of YTHDF3 resulted in: (i) reduced TE of mRNA targets of both YTHDF3 and YTHDF1 (Figure 2A-2B); (ii) decreased RNA-binding specificity of both YTHDF1 and YTHDF2 (Figure 4A-4B and Supplementary information, Figure S4A); and (iii) further increase of m6A levels in cells with depletion of both YTHDF1 and YTHDF2 (Figure 4E). Nascent RNA labeling and time-course assays revealed that YTHDF3 functions in the early time point of RNA life cycle comparing with YTHDF1 or YTHDF2 in the cytosol (Figure 4F and Supplementary information, Figure S5).
These results led us to propose an interconnected and dynamic model for the regulatory functions of YTHDFs in the cytosol (Figure 4G): after a target m6A-modified RNA is exported from the nucleus to the cytoplasm, it might be first recognized by YTHDF3 or a YTHDF3-YTHDF1 complex, which facilitates YTHDF1 binding for enhanced protein translation; the mRNA could be partitioned among all three YTHDF proteins and eventually bound by YTHDF2 for accelerated decay. The expression of YTHDF3 could act as a “buffering agent” for target access to YTHDF1 and YTHDF2. Fluctuation of YTHDF3 expression could affect the RNA-binding activity of the other two, impacting the TE and mRNA stability. YTHDF3 may thus add robustness to the network. Its presence and potential post-translational modifications could provide additional layers of regulation of methylated target mRNAs.
There are cases in which YTHDF3 is distinctly regulated among YTHDFs. For instance, it has been described that YTHDF3 is upregulated compared with YTHDF1 in acute myeloid leukemia clinical samples26,27, and coherently regulated with insulin receptors in aging processes28. These models could be probed in the future to further understand functions of YTHDF3 in different biological processes or systems.
Multiple nascent RNA labeling and time-course assays have suggested that YTHDF3 might interact with target mRNAs prior to YTHDF1 and YTHDF2 (Figure 4F and Supplementary information, Figure S5). This temporal order suggests the potential involvement of YTHDF3 in RNA transport and delivery, which is supported by the observation that knockdown of YTHDF3 leads to decreased TE (Figure 2A-2B), and increased cellular m6A level (Figure 4E) indicating decreased decay of m6A-modified RNAs. YTHDF3 may function as a hub for partitioning its common targets with YTHDF1 and YTHDF2, modulating the turnover rate of m6A-modified transcripts.
mRNA transport is crucial in polarized and long-lived cells like neurons. All YTHDFs have been identified in RNA granules assembled in vitro29, and the presence of YTHDF2 was reported in neuronal RNA granules isolated from rat brain lysate30. Of interest, YTHDF3 mRNA is found to be highly expressed in brain tissues compared with YTHDF1 and YTHDF231. The potential functions of YTHDF3 in RNA transport and delivery in neuronal or related systems merit further studies. mRNA delivery and localization are also important in processes such as cell differentiation and animal development32. YTHDF3 has been reported to be upregulated by p63 in skin development and differentiation33, and its transcript is m6A-methylated in ZFP217-mediated pluripotency and reprogramming regulation34. These biological contexts could be informative in investigating m6A-mediated spatial regulation of mRNAs.
It should be noted that, whereas a majority of YTHDF3 targets show decreased translation upon YTHDF3 depletion, there are still a group of transcripts behaving differently (Figure 2A-2B); 319 transcripts showed higher TE than the median of non-targets (TE > median) when YTHDF3 expression was disturbed in both biological replicates, whereas 614 transcripts showed lower TE (TE < median). The transcripts in the “TE > median” group tend to code proteins with different functions compared with those with reduced translation (Supplementary information, Figure S2E). They have lower base-level translation activity as revealed in the knockdown control sample, and are stronger YTHDF3 targets in terms of RIP enrichment (Supplementary information, Figure S2F-S2G). In fact, the top 40% YTHDF3 RIP targets were shown to be less actively translated in general compared with the low 20% ones (Supplementary information, Figure S2H). Besides promoting translation with YTHDF1 and facilitating mRNA decay through YTHDF2, it is possible that YTHDF3 is also involved in storage of its tightly bound RNA targets, or works as a chaperon protein to modulate the regulatory effect of other m6A-binding protein partners in specific biological contexts (Figure 4G).
In summary, our results suggest a coordinated functional interaction of the three YTHDF proteins inside cells. YTHDFs share hundreds of common targets, and interact with the common targets with an apparent temporal order. YTHDF3 affects translation and decay of methylated mRNAs through cooperation with YTHDF1 and YTHDF2. YTHDF3 could noticeably affect functions performed by the other two YTHDFs. Therefore, besides the reversible, dynamic, and non-stoichiometric nature of the m6A modification itself, a divergent expression pattern of YTHDF proteins in different cell types or developmental stages could provide temporal-spatial control of protein production from m6A-modified transcripts. The integrated function of YTHDFs is further exemplified by recent studies on how the m6A modification on HIV RNA affects viral gene expression. Overexpression of any of the YTHDFs resulted in a similar effect of promotion of protein synthesis from HIV RNAs, and inhibition of HIV-1 virus infection by decreasing HIV-1 reverse transcription35,36.
Materials and Methods
Antibodies
The antibodies used in this study for western blotting were listed below in the format of name (catalogue; supplier; dilution fold): Rabbit anti-YTHDF1 (17479-1-AP; Proteintech; 2 000). Rabbit anti-YTHDF2 (24744-1-AP; Proteintech; 2 000). Mouse anti-YTHDF3 (sc-377119; Santa Cruz Biotech; 500). Goat anti-eIF3B (sc-16377; Santa Cruz Biotech; 2 000). Rabbit anti-eIF4A3 (17504-1-AP; Proteintech; 2 000). Rabbit anti-eIF3A (3411; Cell Signaling Technology; 2 000). Rabbit anti-YBX1 (ab76149; Abcam; 2 000). Mouse anti-Flag HRP conjugate (A8592; Sigma; 5 000). Mouse anti-HA HRP (sc-7392 HRP; Santa Cruz Biotech; 2 000). Donkey anti-Rabbit IgG-HRP (sc-2313; Santa Cruz Biotech; 2 000). Donkey anti-Goat IgG-HRP (sc-2033; Santa Cruz Biotech; 2 000). Goat anti-GAPDH HRP (A00192-100; GenScript; 5 000). Mouse anti-GST HRP (sc-138 HRP; Santa Cruz Biotech; 500).
Plasmid constructions
Flag-tagged YTHDF3 from commercial cDNA clones (Open Biosystems) was cloned into vector pcDNA 3.0 (BamHI, XhoI; forward primer (with coding sequence for Flag-tag), 5′-CGTACGGATCCGATTACAAGGACGACGATGACAAGATGTCAGCCACTAGCG-3′ reverse primer, 5′-CGTAGCTCGAGTCATTGTTTGTTTCTATTTCTCTCCCTAC-3′). Plasmids with high purity for mammalian cell transfection were prepared with a Maxiprep kit (Qiagen).
Double tethering reporter pmirGlo-Ptight-2BoxB-2MS2: 2BoxB-2MS2 sequence was designed as follows: 5′-CTCGACTAAGTCCAACTACTAAACTGGGCCCTGAAGAAGGGCCCATATAGGGCCCTGAAGAAGGGCCCTAGCAAGTTCAAATAAGGCTAGTCCGTTATCAACTTGGCCAACATGAGGATCACCCATGTCTGCAGGTCGACTCTAGAAAACATGAGGATCACCCATGTCTGCAGTATTCCCGGGTTCATTAGATCCTAA-3′37,38, synthesized, and PCR amplified and cloned to 3′-UTR of F-Luc in pmirGlo-Ptight vector reported12 (NheI, SbfI; forward primer, 5′-ATACGCTAGCCTCGACTAAGTCCAACTACTAAACTGGG-3′ reverse primer, 5′-GTATCCTGCAGGTTAGGATCTAATGAACCCGGGAATACTG-3′).
Tether effector: we first constructed pcDNA-Flag-MS2 vector by inserting Flag-MS2 protein coding sequence into pcDNA 3.0. MS2 coding sequence was amplified from pZS*12-MS2-GFP vector39, by two rounds of PCR (BamHI, XbaI; forward primer 1 (with EcoRV and XhoI sites), 5′-GGAGGTTCGGGGATATCGGGCTCGAGCATGGCTTCTAACTTTACTCAGTTCGTTCTC-3′ reverse primer 1, 5′-CATTCTAGACTAGTAGATGCCGGAGTTTGCTGC-3′ forward primer 2 (with coding sequence for Flag tag), 5′-CATGGATCCATGGATTACAAGGACGACGATGACAAGGGAGGTTCGGGGATATCGGG-3′ reverse primer 2, 5′-CATTCTAGACTAGTAGATGCCGGAGTTT-3′). The resulting plasmid was subjected to a second round of cloning by inserting N-terminal of YTHDF3, resulting in plasmid pcDNA-YTHDF3N-MS2 (EcoRV, XhoI; forward primer, 5′-CATGATATCGATGTCAGCCACTAGCGTGGATC-3′ reverse primer, 5′-CATCTCGAGCCTACACTAGAAGGTGAAGCACTGACAG-3′). The pcDNA-Flag-YTHDF1N-λ construct was reported previously13. Finally, inserted Flag-MS2, Flag-YTHDF3N-MS2, and Flag-YTHDF1N-λ were cloned to a modified pPB-CAG vector with MfeI and AgeI sites between BglII and XhoI sites for optimized expression in HeLa cells (MfeI, AgeI; forward primer, 5′-ATCGCAATTGATGGATTACAAGGACGACGATGACAAG-3′ Flag-MS2 and Flag-YTHDF3N-MS2: reserve primer, 5′-CATGACCGGTCTAGTAGATGCCGGAGTTTGCTG-3′ Flag-YTHDF1N-λ: reverse primer, 5′-CATGACCGGTTCAGTTTGCAGCTTTCCATTGAGC-3′).
Flag-tagged YTHDF1-3 and HA-tagged YTHDF1-3 were cloned by inserting CDS of YTHDF1-3 into modified pPB-CAG vectors where there is a Flag-tag or an HA-tag inserted upstream of the restriction enzyme site BglII. The vector was digested with BglII and XhoI, and the CDS of YTHDF1-3 were cloned with BamHI and XhoI (YTHDF1: forward primer: 5′-CGATGGATCCATGTCGGCCACCAGCGTG-3′ reverse primer: 5′-CGATCTCGAGTCATTGTTTGTTTCGACTCTGCCGTTCCTT-3′ YTHDF2: forward primer: 5′-GACTGGATCCATGTCGGCCAGCAG-3′ reverse primer: 5′-ATCGCTCGAGTTATTTCCCACGACCTTGAC-3′ YTHDF3: forward primer: 5′-CGATGGATCCATGTCAGCCACTAGCGTG-3′ reverse primer: 5′-CGATCTCGAGTTATTGTTTGTTTCTATTTCTCTCCCTACG-3′).
Flag-HA tandem-tagged YTHDF1-3 were cloned by inserting their CDS into a modified pPB-CAG vector where there is a Flag-HA-tandem tag inserted upstream of the restriction enzyme site BglII. The vector was digested with BglII and MfeI, and the CDS of YTHDF1-3 were cloned with BglII and MfeI (YTHDF1: forward primer: 5′-CGTACAGATCTATGTCGGCCACCAGCG-3′ reverse primer: 5′-CCATACTCGAGTCATTGTTTGTTTCGACTCTGCC-3′ YTHDF2: forward primer: 5′-CGTACAGATCTATGTCGGCCAGCAGCC-3′ reverse primer: 5′-CGATGCTCGAGTTATTTCCCACGACCTTGACG-3′ YTHDF3: forward primer: 5′-CGTACAGATCTATGTCAGCCACTAGCGTG-3′ reverse primer: 5′-CGTAGCTCGAGTTATTGTTTGTTTCTATTTCTCTCCCTAC-3′).
Mammalian cell culture, siRNA knockdown, and plasmid transfection
Human HeLa cell line used in this study was purchased from ATCC (CCL-2) and grown in DMEM (Gibco, 11995) media supplemented with 10% FBS and 1% 100 × Pen/Strep (Gibco). HeLa Tet-off cell line was purchased from Clontech and grown in DMEM (Gibco) media supplemented with 10% FBS (Tet system approved, Clontech), 1% 100 × Pen/Strep (Gibco) and 200 μg/ml G418 (Sigma).
AllStars negative control siRNA from Qiagen (SI03650318) was used as control siRNA in knockdown experiments. YTHDF3 siRNAs were ordered from Qiagen. (siYTHDF3-2: Hs_YTHDF3_2 with the target sequence 5′-ATGGATTAAATCAGTATCTAA-3′ siYTHDF3-5: Hs_YTHDF3-5 with the target sequence 5′-TAAGTCAAAGAAGACGTATTA-3′). YTHDF1, YTHDF2, and METTL3 siRNA were reported previously12,13,19. Transfection was achieved by using Lipofectamine RNAiMAX (Invitrogen) for siRNAs, Lipofectamine 2000 (Invitrogen) for transfection of one or two plasmids, and Lipofectamine LTX Plus (Invitrogen) for transfection of multiple plasmids in tethering assay following the manufacturer's protocols.
Stable overexpression in cell lines: stable expression in HeLa cells with double-tagged YTHDF1, YTHDF2, and YTHDF3 individually (N terminal Flag and HA in tandem) was created by puromycin selection (2 μg/ml) with the modified pPB-CAG vector. The control cell line with expression of only tandem Flag and HA peptides was created similarly.
LC-MS/MS
About 50-100 ng of mRNA were digested by nuclease P1 (1 U, Wako) in 25 μl of buffer containing 20 mM of NH4OAc (pH = 5.3) at 42 °C for 2 h, followed by the addition of NH4HCO3 (1 M, 3 μl, freshly made) and alkaline phosphatase (1 U, sigma). After an additional incubation at 37 °C for 2 h, the sample was diluted to 50 μl and filtered (0.22 μm pore size, 4 mm diameter, Millipore), and 5 μl of the solution was subjected to LC-MS/MS. Nucleosides were separated by reverse phase ultra-performance liquid chromatography on a C18 column with on-line mass spectrometry detection using an Agilent 6410 QQQ triple-quadrupole LC mass spectrometer in positive electrospray ionization mode. The nucleosides were quantified by using retention time and the nucleoside to base ion mass transitions of 284-152 (G), 282.1-150.1 (m6A), 268-136 (A), 245-113.1 (U), 261.3-129 (4SU, 4-thio-uridine), and 244-112 (C). Quantification was performed in comparison with the standard curve obtained from pure nucleoside standards running with the same batch of samples. The m6A level was calculated as the ratio of m6A to A or the ratio of m6A to (A+G) based on the calibrated concentrations40. For 4SU quantification, 0.25 mM DTT was supplemented in the digestion buffer and each digestion step lasted for only 1 h in order to avoid oxidation of 4SU. The 4SU level was calculated as the ratio of 4SU to U based on the calibrated concentrations.
RIP-LC-MS/MS
The RIP procedure was reported previously12. Sixty million HeLa cells stably overexpressing Flag-HA-tagged YTHDF3 were subjected to RIP procedure. Input, flow-through, and YTHDF3-bound RNA were purified with Trizol reagent. mRNAs of the three portions were further purified by depleting rRNA with RiboMinus Eukaryote Kit v2 (Ambion) followed by depleting tRNA with RNA Clean and Concentrator-5 (Zymo Research, 200 nt cutoff protocol). About 50 ng purified mRNA of each sample were subjected to LC-MS/MS quantification of m6A levels as described above12,40.
Tethering assay
The tethering assay procedure was adapted from the previous report12,13. About 50 ng reporter plasmid (pmirGlo-Ptight-2BoxB-2MS2) and 250 ng of each effecter plasmid (Flag-MS2, Flag-YTHDF3N-MS2, Flag-YTHDF1N-λ, or the combination indicated) were used to transfect HeLa Tet-Off Advanced Cell Line (613356, Clontech) in each well of six-well plate at 60%-80% confluency under doxycycline (DOX, 100 ng/ml) inhibition. Six hours later, transfection mixture was replaced with fresh media containing DOX (100 ng/ml). Eighteen hours later, each well was trypsin-digested, extensively washed with PBS by suspending and spinning down for three times, and re-seeded into 96-well plate (1:30) and 12-well plate (1:3) in media without DOX to induce transcription of F-luc. Four hours after F-luc induction, cells in the 96-well plate were analyzed by Dual-Glo Luciferase Assay Systems (E2920, Promega). F-luc activity was normalized by R-luc to evaluate protein production from the reporter. At the same time, samples in the 12-well plate were processed to extract total RNA (DNase I digested) by TRIzol® reagent (Invitrogen), followed by RT-qPCR quantification. The amount of F-luc mRNA was also normalized by that of R-luc mRNA. TE of F-Luc mRNA was calculated as the ratio of normalized F-Luc activity (protein level) to normalized F-Luc mRNA level.
RNA isolation
mRNA isolation for LC-MS/MS: total RNA was isolated from wild-type or transiently transfected cells with TRIzol reagent (Invitrogen). mRNA was extracted using Dynabeads® mRNA DIRECT kit (Ambion) followed by further removal of remaining rRNA using RiboMinus Eukaryote Kit v2 (Ambion). mRNA concentration was measured by Qubit® RNA HS Assay Kit with Qubit® 2.0 fluorometer.
RNA isolation for RT-qPCR: we followed the instruction of Direct-zol™ RNA Miniprep kit (Zymo Research) or RNeasy Miniprep kit (Qiagen) both with an in-column DNase I digestion step. For RT-qPCR following polysome profiling, RNA was extracted according to the manufacturers manual of TRizol® reagent (added 1:1), and contaminate DNA was removed using RNA clean and concentrator-25 (Zymo Research) with additional in-column DNase I digestion.
Ethanol precipitation: to the RNA solution being purified or concentrated, 1/10 volume of 3 M NaOAc, pH 5.5, 1 μl GlycoBlue™ coprecipitant (15 mg/ml, Ambion), and 2.7 volume of 100% ethanol were added. The mixture was stored at −20 °C overnight, and then centrifuged at 15 000 × g for 15 min. After removing the supernatant, the pellet was washed twice with 1 ml 75% ethanol, and dissolved in the appropriate amount of RNase-free water as indicated.
PAR-CLIP
PAR-CLIP for sequencing: we followed the previously reported protocol12 except starting with 150-200 million Flag-HA-tagged YTHDF3 stable expressioned HeLa cells instead of transient overexpression cells. YTHDF3-RNA complex was SDS-PAGE purified with a size-selection range from 60 to 95kDa, and the RNA fragments were extracted via ethanol precipitation after protease K digestion of the gel slices. The purified RNA pellet was dissolved in 12 μl RNase-free water, of which 6 μl was subjected to small RNA library preparation with NEBNext® Multiplex Small RNA Library Prep Set for Illumina® (E7300S, NEB).
PAR-CLIP for quantification of protein-bound RNAs: 20 million Flag-HA-tagged YTHDF1 or YTHDF2 stable expression HeLa cells treated with either siControl or siYTHDF3 were subjected to the same PAR-CLIP procedure while using γ-32P-ATP in T4 PNK 5′ end-repairing. After stringent washing following radioactive-labeling, the samples were subjected to SDS-PAGE and the gel was exposed to a blanked phosphorimager screen overnight. The screen was then imaged with the Molecular Imager FX™ (Bio-Rad). Same protocol was used for 20 million Flag-HA-tagged YTHDF3 stable expression HeLa cells treated with siControl, siYTHDF1, or siYTHDF2.
RIP
RIP-seq: the procedure was reported previously12. Input mRNAs were prepared by either Poly(A) selection (replicate 1, FastTrack MAG Micro mRNA isolation kit, Invitrogen) or rRNA removal (replicate 2, RiboMinus Eukaryote Kit v2, Ambion). Input mRNA and IP with 150-200 ng RNA of each sample were used to generate the library using TruSeq stranded mRNA sample preparation kit (Illumina). For RIP-seq of YTHDF1 and YTHDF2 with or without YTHDF3 depletion, 60 million HeLa cells were subjected to Flag-IP. Input RNAs and IP RNAs were subjected to rRNA removal in parallel before library preparation.
RIP-RT-qPCR: 20 million HeLa cells stably overexpressing Flag-HA YTHDF1 or Flag-HA YTHDF2 were collected 48 h after the treatment of siControl or siYTHDF3. The RIP procedure was the same as that in RIP-seq till before eluting. About 0.5 ml Trizol® reagent was added to anti-Flag M2 magnetic beads (Sigma) to extract YTHDF1- or YTHDF2-bound (IP) RNAs. Amount of non-target or target transcripts in both the input and IP RNAs were analyzed with RT-qPCR, and IP enrichment ratio of a transcript was calculated as the ratio of its amount in IP to that in the input yielded from same amount of cells.
Ribosome profiling
We followed the procedure reported previously13. Ribosome profiling was conducted using TruSeq Ribo Profile (Mammalian) Library Prep Kit (Illumina, RPHMR12126). Oligo siYTHDF3-5 was used in two biological replicates. The sequencing data obtained from ribosome profiling were denoted as “ribosome-bound fragments” and those from input as “input RNA”. TE was defined as the ratio of “ribosome-bound fragments” and “input RNA”, which reflected the relative occupancy of 80S ribosome per mRNA species41.
Polysome profiling
Polysome profiling was performed to verify genome-wide analysis results from ribosome profiling. We followed the procedure reported previously13,42, with following modifications. (1) Before collection, cycloheximide (CHX) was added to the media at 100 μg/ml for 7 min. (2) The lysis buffer was formulated as 20 mM HEPES, pH 7.6, 100 mM KCl, 5 mM MgCl2, 100 μg/ml CHX, 1% Triton-X-100, freshly add 1:100 protease inhibitor (Roche), 40 U/ml SUPERasin (Ambion).
Oligo siYTHDF3-2 was used in this experiment to perturb YTHDF3 expression. siControl, siYTHDF3-2, Flag-HA-tagged YTHDF3 stable expression HeLa cells, and stable expression control HeLa cells were analyzed with polysome profiling.
Sequencing data analysis
General pre-processing of reads: all samples were sequenced by illumine Hiseq2000 or Hiseq4000 with single end 50 bp read length. For libraries that generated from small RNA (PAR-CLIP and ribosome profiling), the adapters were trimmed by using FASTX-Toolkit43. The deep sequencing data were mapped to Human genome version hg38 by Tophat version 2.044 without any gaps and allowed for at most two mismatches. RIP and ribosome profiling were analyzed by DESeq45 to generate RPKM (reads per kilobase, per million reads).
Data analysis for each experiment: (1) PAR-CLIP data were analyzed by PARalyzerv1.146 with default settings. Binding motif was analyzed by HOMER (v4.7)47; (2) for RIP, enrichment fold was calculated as log2(IP/input); (3) for ribosome profiling, only genes with RPKM > 1 in input RNA sample and RPKM > 0 in ribosome-bound fragments were used for analysis and the change fold was calculated as log2(siYTHDF3/siControl) or log2(siMETTL3/siControl);
Integrative data analysis and statistics: PAR-CLIP targets were defined as gene targets reproducible among three biological replicates (4 459). RIP targets (2 264) were genes with log2(IP/input) > 1 in both biological replicates. The overlap of PAR-CLIP and RIP targets was defined as high-confidence targets (1 239). For Figure 2 and Supplementary information, Figure S2, the three groups of transcripts are defined as the following: (1) YTHDF3 targets: high confidence targets of YTHDF3; (2) YTHDF1 unique: high-confidence targets of YTHDF1 reported previously as “CLIP+IP”, but not high-confidence target of YTHDF313; (3) Non-target: non-target for a protein is defined as (a) complementary set of PAR-CLIP targets; (b) RIP enrichment fold < 0; Non-target in the figure is defined as the intersection of the non-targets of YTHDF1 and that of YTHDF3. For the comparison of PAR-CLIP and m6A peaks, at least 1 bp overlap was applied as the criteria of overlap peaks. Nonparametric Mann-Whitney U-test (Wilcoxon rank-sum test, two sided, significance level = 0.05) was applied in ribosome profiling data analysis as previous reported48. GO term analyses were performed by DAVID49,50. Top terms were then selected for visualizations by the interactive graph function of REVIGO51.
Data accession: all the raw and processed data can be accessed under GSE86214 (https://www.ncbi.nlm.nih.gov/geo/). The analysis of Figure 1D, Figure 2D, and Supplementary information, Figure S2I-S2J utilized data from GSE46705. The analysis of Figure 1F, Figure 2A-2B, and Supplementary information, Figure S2A-S2D utilized data from GSE49339 and GSE63591.
RT-qPCR
All RNA templates used in RT-qPCR were digested with DNaseI during purification step to avoid false signal from genomic DNA. RT-qPCR primers were designed with Primer 352,53 to cover exon-exon junctions shared in all isoforms of the matured mRNAs. About 500 ng RNAs were first reverse-transcribed into cDNAs with PrimeScript™ RT reagent Kit (Takara), and then cDNAs were subjected to qPCR analysis with FastStart SYBR Green Master Mix (Roche) in machine LightCycler 96 (Roche). HPRT1 was chosen as an internal control as reported previously, and the primer sequences for SON, HPRT1, INTS5, F-luc, and R-luc have been reported12,13. Primers for other genes are listed as follows (mRNA: forward primer; Reserve primer):
YTHDF3: 5′-TGACAACAAACCGGTTACCA-3′ 5′-TGTTTCTATTTCTCTCCCTACGC-3′
APC: 5′-AGGCTGCATGAGAGCACTTGTG-3′ 5′-CACACTTCCAACTTCTCGCAACG-3′
DBF4: 5′-GGACATTAAGGATCTGGGAGG-3′ 5′-GGACTTGGTACAGGAGAAATTCG-3′
RPL30: 5′-ACAGCATGCGGAAAATACTAC-3′ 5′-AAAGGAAAATTTTGCAGGTTT-3′
NFRKB: 5′-ATCTTAGCCTTGTTCAGTGGG-3′ 5′-CGCTTGTACTGTGACTTGAAG-3′
DST: 5′-AGAGAAACACCAACATAGCTAGG-3′ 5′-GGCCTCAATAGTTAACCGGG-3′
MYC: 5′-TTCGGGTAGTGGAAAACCAG-3′ 5′-AGTAGAAATACGGCTGCACC-3′
EPM2AIP1: 5′-GCTGAGGTGATCTGGCG-3′ 5′-CGTCGACTTCCATCTTGCTTC-3′
BTG2: 5′-GCAGAGGCTTAAGGTCTTCAG-3′ 5′-CTTGTGGTTGATGCGAATGC-3′
IL17RB: 5′-AACTTCACAACCACTCCCC-3′ 5′-GCACCTTCACTATCCCCAG-3′
TSC1: 5′-CTGGACAGACTGATACAGCAGG-3′ 5′-TGCGGATCTCATCTGAAGGAGG-3′
Tandem affinity purification of YTHDF3 protein interactome and mass spectrometry identification
We followed the procedure reported previously13,54. The protein mass spectroscopy was performed by Institutes of Biomedical Sciences at Fudan University, Shanghai.
Protein co-immunoprecipitation and western blotting
Sixty million of Flag-HA-tagged YTHDF3 or Flag-HA peptide stable overexpression HeLa cells were collected for one immunoprecipitation. The cell pellet was re-suspended with 2-3 volumes of lysis buffer (10 mM HEPES pH 7.6, 150 mM KCl, 2 mM EDTA, 0.5% NP-40, 0.5 mM DTT, 1:100 protease inhibitor cocktail, 400 U/ml SUPERasein), and incubated on ice for 10 min. The lysate solution cleared by centrifuge at 15 000× g for 15 min at 4 °C. Although 50 μl of cell lysate was saved as input, the rest was incubated with the anti-Flag M2 magnetic beads (Sigma) in ice-cold NT2 buffer (50 mM HEPES, pH 7.6, 200 mM NaCl, 2 mM EDTA, 0.05% NP-40, 0.5 mM DTT, 200 U/ml SUPERasein) at 4 °C at 4h. The beads were then subjected to extensive wash with 6 × 1 ml portions of ice-cold NT2 buffer, and then separated into two portions. One portion was eluted with 3 × Flag peptide (0.5 mg/ml in NT2 buffer, Sigma) at room temperature for 1 h, whereas the other portion was subjected to RNase treatment with 1% RNase A (Thermo Fisher, 10 mg/ml) and 1% RNase T1 (Thermo Fisher, 1 000 U/ul) at room temperature for 30 min. After RNase treatment, the second portion was washed with ice-cold NT2 buffer for another six times and eluted under the same condition. About 1% input and half amount of the eluted IP samples were loaded for one western blotting.
As shown in Supplementary information, Figure S3C, 20 million of HeLa cells were transfection with equal amount of HA-YTHDF3 and one of Flag-YTHDFs plasmids. Flag-IP was performed as described above.
In vitro binding assay using purified protein
GST-tagged YTHDF1-3 were purified as described previously12. Sixty million of Flag-HA YTHDF1-3 stable expression cell lines and control cell lines were lysed in 1 ml IPP buffer (150 mM NaCl, 0.2% NP-40, 10 mM Tris, pH 7.5, 1:100 protease inhibitor cocktail, 0.5 mM DTT). The cell lysate was aliquoted into three portions, and concentrated GST-tagged YTHDF1-3 were diluted to the final concentration of 2.5 μM. The mixture was rotated at 4 °C for 2 h before 50 μl of pre-washed GST-affinity magnetic beads (Pierce) was added for another 2-h rotation. The GST-affinity beads were then washed with 400 μl IPP buffer for four times. Proteins bound by GST-affinity beads were eluted with 25 μl 2× Laemmli Sample Buffer (Bio-Rad) at 95 °C for 5 min and subjected to Flag blotting analysis.
Pulse-chase experiment by metabolic labeling of nascent mRNA
LC-MS/MS test with 4SU labeling, and colocalization and qPCR test with 5-ethinyluridine labeling were performed with the same procedure described previously13. Flag-HA-tagged YTHDF1-3 stable expression HeLa cells were used in these experiments.
Author Contributions
HS and XW contributed equally to this work. HS, XW, and CH designed the experiments. HS and XW performed the experiments and analyzed the data. BSZ assisted with performing experiments and provided valuable discussion. HS, XW, and ZL processed the high-throughput RNA sequencing and proteomics data. HM helped in stable line generation and protein mass spectrometry experiment. PJH and CL assisted with performing experiments. HS, XW, and CH wrote the manuscript. All the authors provided comments and suggestions to the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
We thank Professor T Pan (The University of Chicago) for kindly providing the polysome profiling instrument. We thank Dr J Tauler (The University of Chicago) for editing the manuscript. We also thank Institutes of Biomedical Sciences at Fudan University for the protein mass spectrometry experiments. The work was supported by the National Institutes of Health grants GM071440 and GM113194 to CH. CH is an investigator of the Howard Hughes Medical Institute (HHMI). HS is supported by the Gustavus F Swift Fellowship from the Chemistry Department at University of Chicago. HM is supported by the Postdoctoral International Exchange Program of the China Postdoctoral Council (CPC). The Mass Spectrometry Facility of the University of Chicago is funded by the National Science Foundation (CHE-1048528).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Analyses of YTHDF3 PAR-CLIP data.
Ribosome profiling and polysome profiling of HeLa cells depleted of YTHDF3 (A-J), and protein and mRNA levels of reporters in the double tethering assay (K).
YTHDF3 and YTHDF1 co-immunoprecipitate and share protein partners.
YTHDF3 regulates binding specificity and affinity of THDF1 and YTHDF2 towards target RNAs.
Temporal order of YTHDFs' interaction with m6A-modified transcripts.
Target transcripts of YTHDF3 and Comparison of YTHDF1-3 targets
YTHDF3 protein interactome and Comparison of YTHDF1 and YTHDF3 protein partners
References
- Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA 1974; 71:3971–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C, Gershowitz A, Moss B. 5′-Terminal and internal methylated nucleotide sequences in HeLa cell mRNA. Biochemistry 1976; 15:397–401. [DOI] [PubMed] [Google Scholar]
- Wei C, Moss B. Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry 1977; 16:1672–1676. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012; 485:201–206. [DOI] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012; 149:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Molinie B, Wang J, et al. M6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 2014; 15:707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, et al. M6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 2015; 347:1002–1006. [DOI] [PubMed] [Google Scholar]
- Fustin JM, Doi M, Yamaguchi Y, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 2013; 155:793–806. [DOI] [PubMed] [Google Scholar]
- Fischer J, Koch L, Emmerling C, et al. Inactivation of the Fto gene protects from obesity. Nature 2009; 458:894–898. [DOI] [PubMed] [Google Scholar]
- Zhao X, Yang Y, Sun BF, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res 2014; 24:1403–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 2013; 49:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014; 505:117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, et al. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 2015; 161:1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Wang X, Liu K, et al. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol 2014; 10:927–929. [DOI] [PubMed] [Google Scholar]
- Xiao W, Adhikari S, Dahal U, et al. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol Cell 2016; 61:507–519. [DOI] [PubMed] [Google Scholar]
- Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell 2015; 162:1299–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015; 518:560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian S-B. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 2015; 526:591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 2014; 10:93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4: NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev 2006; 20:1885–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliseeva IA, Kim ER, Guryanov SG, Ovchinnikov LP, Lyabin DN. Y-box-binding protein 1 and its functions. Biochemistry 2011; 76:1402–1433. [DOI] [PubMed] [Google Scholar]
- Le Hir H, Séraphin B. EJCs at the heart of translational control. Cell 2008; 133:213–216. [DOI] [PubMed] [Google Scholar]
- Li F, Zhao D, Wu J, Shi Y. Structure of the YTH domain of human YTHDF2 in complex with an m6A mononucleotide reveals an aromatic cage for m6A recognition. Cell Res 2014; 24:1490–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Roundtree IA, Wang P, et al. Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res 2014; 24:1493–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Liu K, Ahmed H, Loppnau P, Schapira M, Min J. Structural basis for the discriminative recognition of N6-methyladenosine RNA by the human YT521-B homology domain family of proteins. J Biol Chem 2015; 290:24902–24913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, He X, Zhong S. Cross-species microarray analysis with the OSCAR system suggests an INSR->Pax6->NQO1 neuro-protective pathway in aging and Alzheimer's disease. Nucleic Acids Res 2007; 35:105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 2012; 149:753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsche R, Karra D, Bennett KL, et al. Interactome of two diverse RNA granules links mRNA localization to translational repression in neurons. Cell Rep 2013; 5:1749–1762. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 2007; 445:168–176. [DOI] [PubMed] [Google Scholar]
- Medioni C, Mowry K, Besse F. Principles and roles of mRNA localization in animal development. Development 2012; 139:3263–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkaya B, Ortt K, Sinha S. Novel in vivo targets of ΔNp63 in keratinocytes identified by a modified chromatin immunoprecipitation approach. BMC Mol Biol 2007; 8:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilo F, Zhang F, Sancho A, et al. Coordination of m6A mRNA methylation and gene transcription by ZFP217 regulates pluripotency and reprogramming. Cell Stem Cell 2015; 17:689–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraja Tirumuru, Boxuan Simen Zhao, Wuxun Lu, Zhike Lu CH, Li W. N6-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. eLife 2016; 5:e15528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EM, Bogerd HP, Kornepati AVR, et al. Posttranscriptional m6A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe 2016; 19:675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell 1998; 2:437–445. [DOI] [PubMed] [Google Scholar]
- Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE. Y14 and hUpf3b form an NMD-activating complex. Mol Cell 2003; 11:939–949. [DOI] [PubMed] [Google Scholar]
- Guet CC, Bruneaux L, Min TL, et al. Minimally invasive determination of mRNA concentration in single living bacteria. Nucleic Acids Res 2008; 36:e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 2011; 7:885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 2009; 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandin V, Sikström K, Alain T, et al. Polysome fractionation and analysis of mammalian translatomes on a genome-wide scale. J Vis Exp 2014; 87:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, Wood T, Zhang Z, Miller W. Comparison of DNA sequences with protein sequences. Genomics 1997; 46:24–36. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009; 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010; 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran DL, Georgiev S, Mukherjee N, et al. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol 2011; 12:R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 2012; 336:233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Lempicki RA, Sherman BT. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009; 37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fran Supek, Matko Bošnjak, Nives Škunca TŠ. Revigo summarizes and visualizes long lists of gene ontology terms. PLoS One 2011; 6: e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics 2007; 23:1289–1291. [DOI] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, et al. Primer3-new capabilities and interfaces. Nucleic Acids Res 2012; 40: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Sawada J, Sui G, et al. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 2003; 422:735–738. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analyses of YTHDF3 PAR-CLIP data.
Ribosome profiling and polysome profiling of HeLa cells depleted of YTHDF3 (A-J), and protein and mRNA levels of reporters in the double tethering assay (K).
YTHDF3 and YTHDF1 co-immunoprecipitate and share protein partners.
YTHDF3 regulates binding specificity and affinity of THDF1 and YTHDF2 towards target RNAs.
Temporal order of YTHDFs' interaction with m6A-modified transcripts.
Target transcripts of YTHDF3 and Comparison of YTHDF1-3 targets
YTHDF3 protein interactome and Comparison of YTHDF1 and YTHDF3 protein partners