Abstract
Recent advances have demonstrated the power of small molecules in promoting cellular reprogramming. Yet, the full potential of such chemicals in cell fate manipulation and the underlying mechanisms require further characterization. Through functional screening assays, we find that mouse embryonic fibroblast cells can be induced to trans-differentiate into a wide range of somatic lineages simultaneously by treatment with a combination of four chemicals. Genomic analysis of the process indicates activation of multi-lineage modules and relaxation of epigenetic silencing programs. In addition, we identify Sox2 as an important regulator within the induced network. Single cell analysis uncovers a novel priming state that enables transition from fibroblast cells to diverse somatic lineages. Finally, we demonstrate that modification of the culture system enables directional trans-differentiation towards myocytic, glial or adipocytic lineages. Our study describes a cell fate control system that may be harnessed for regenerative medicine.
Keywords: chemical combination, trans-differentiation, single cell analysis
Introduction
Cell fate decisions reflect complex processes in which numerous genes work in concert to encode hundreds of stable cell types. Recent advances have highlighted the remarkable ability to manipulate cellular states by forced expression of defined factors, starting from induced pluripotency1,2,3,4 to lineage trans-differentiation5,6,7,8,9,10,11. A molecular roadmap for reprogramming experiment has also been proposed12,13. These studies illustrate how combinatorial functions of master regulators may drive one cell type across epigenetic barriers to adopt an alternate fate.
Small molecules offer a complementary strategy for cell fate control. They are cell permeable, more tractable than transcription factors, and cost-effective. A few chemical compounds have been suggested to improve cellular reprogramming14,15,16,17,18,19. Remarkably, a combination of small molecules can replace all Yamanaka factors during the generation of induced pluripotent cells20,21. Enlightened by the idea, chemical cocktails that initiate neuronal trans-differentiation have been reported22,23,24,25. Similar approaches have been used to generate functional cardiac myocytes from both mouse and human fibroblasts26,27. Very recently, endodermal progenitors have also been obtained using the chemical reprogramming method28. Interestingly, many of these studies shared similar chemicals but described diverse outcomes. Yet, the mechanism behind this diversity is not known. The individual and combinatorial functions of small molecules should be further elucidated. The molecular pathways during chemical reprogramming process needs to be characterized at single cell level.
In this study, we aimed to systematically characterize the effects of a set of potent chemicals during cell fate conversions. Through phenotypic screening and targeting specific pathways, we found that mouse embryonic fibroblasts can be induced to trans-differentiate into diverse somatic cell types. Single cell profiling experiments suggest that expression stochasticity reflects chemically induced transcriptional programs rather than pre-existing cellular heterogeneity. We report a cell fate decision pathway through a stochastic priming state triggered by the combinatorial function of small molecules. We also show that modification of the culture system allows for efficient generation of lineage-specific cell types.
Results
Phenotypic screen identifies small molecule combinations that promote multi-lineage trans-differentiation
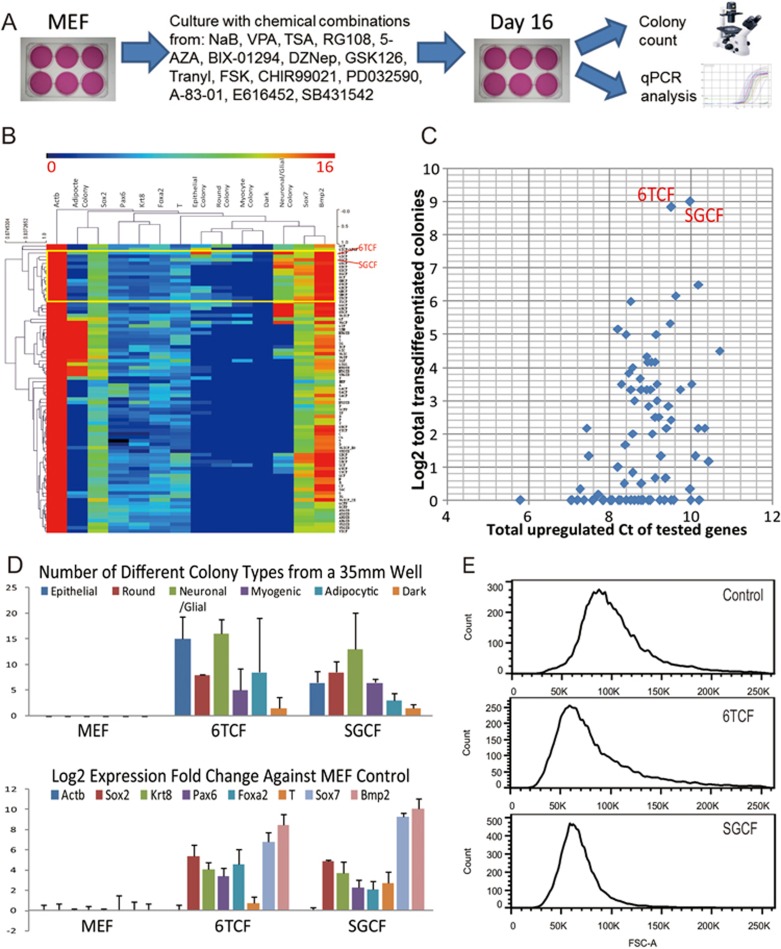
We started with a set of compounds reported to promote reprogramming (Supplementary information, Table S1). These chemicals include: HDAC inhibitors NaB (N), VPA (V) and TSA (A); DNMT inhibitors RG108 (R), 5-AZA (5); G9a inhibitor BIX-01294 (B); Ezh2 inhibitors DZNep (D), GSK126 (G); LSD1 inhibitor Tranylcypromine (T); AC activator FSK (F); GSK3 inhibitor CHIR99021 (C); MEK inhibitor PD032590 (P); ALK5 inhibitor A-83-01 (8), E616452 (6) and SB431542 (S). We tested the effects of these chemicals on mouse embryonic fibroblast (MEF) cells individually and in different combinations (Figure 1A). In designing different combinations, we sought to co-target different pathways and avoid potentially redundant chemicals (all tested combinations are listed in Supplementary information, Table S2). Cells were cultured on 6-well plates; media were changed every 4 days over a total culture time of 16 days. After the end of the treatment period, we performed careful morphological characterization in conjunction with qPCR analysis of 7 different lineage-specific markers for each sample. Assayed genes included the stem cell and neuronal marker Sox2, neuronal marker Pax6, epithelial marker Krt8, endoderm marker Sox7 and Foxa2, mesoderm marker T and Bmp2, as well as an endogenous control gene, Actb.
Figure 1.
Phenotypic screen identifies small molecule combinations that promote iMT process. (A) The scheme for functional screening of chemical combination with their effect on MEFs. (B) A heatmap summarizing combinatorial treatment of MEFs and the corresponding phenotypic data. In the heatmap each row corresponds to a chemical combination treatment; each column corresponds to a phenotypic measurement on Day 16 from a 35 mm well. (C) A scatter plot showing the correlation of colony forming data and qPCR data. Y-axis corresponds to the Log2 of total number of trans-differentiated colonies from a 35 mm plate after a specific treatment. X-axis corresponds to the total value of upregulated Ct from different qPCR measurement. (D) Bar charts showing the upregulation of lineage-specific markers or forming of lineage-specific colonies from MEFs upon 6TCF or SGCF treatment. Error bars correspond to standard deviation from three replicates. (E) FACS analysis of 6TCF and SGCF treated and control MEFs on Day6. Note significantly reduced cell size after treatment.
Among more than 100 different combinations, we identified several that elicited dramatic morphological changes in MEF culture. With 10 days of treatment, colonies with distinct morphologies emerged. We classified colony types as Epithelial, Round, Neuron-like (Branched), Myocytic (Big), Adipocytic, and Dark (Pigmented) colonies (Supplementary information, Table S2). These morphological changes were associated with significant upregulation of lineage-specific markers (Supplementary information, Tables S1 and S2). Most striking was the coexistence of colonies of distinct types within the same culture. Markers of different lineages were upregulated simultaneously upon the same treatment. In control MEF cultures, no significant morphological or gene expression changes were observed.
We normalized and averaged qPCR Ct and the colony counting data. We then performed hierarchical clustering of the aggregated data. The resulting heatmap revealed different categories of phenotypic changes (Figure 1B). Notably, a set of treatments, including 6TCF (E616452, Tranylcypromine, CHIR99021, FSK) and SGCF (SB431542, GSK126, CHIR99021, FSK) activated a wide range of lineage markers and promoted formation of colonies with diverse morphologies (Figure 1B). To identify the most potent combinations for multi-lineage cellular changes we plotted the total number of different colonies for each treatment against the total up-regulated Ct value for tested genes (Figure 1C). Two treatments, 6TCF and SGCF, emerged as most potent. Both conditions include bFGF and serum. As shown in Figure 1D, treatment of MEFs in standard MEF medium with bFGF, serum and 6TCF or SGCF generated all colony types and upregulated all tested lineage markers by culture day 16 (Figure 1D). In addition, FACS analysis revealed that by day 6 6TCF and SGCF treated MEFs exhibited reduced cell size (Figure 1E). These findings were highly reproducible. Due to its one-step but multi-directional feature, we term the process induced multi-lineage trans-differentiation (iMT).
A wide range of somatic cell types can be obtained through iMT
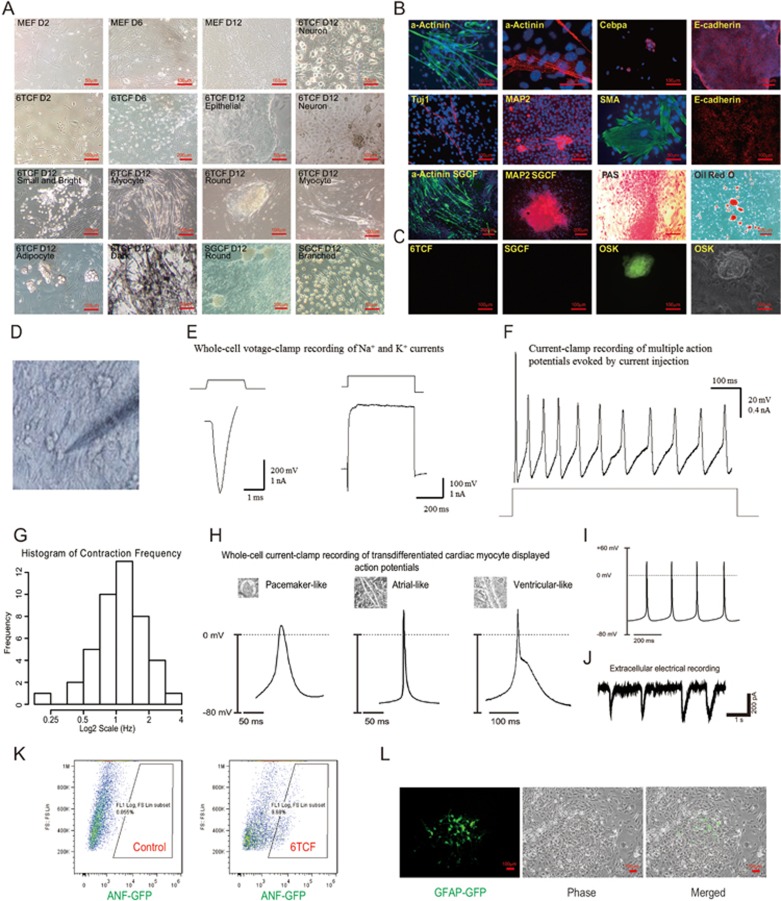
To verify iMT at a molecular level, we performed more detailed analyses. Upon 6TCF treatment of MEFs, some flat colonies started to emerge on day 2; epithelialized colonies appeared on day 6. On day 12, different cell and colony morphologies were easily recognized (Figure 2A). Strikingly, many myocytic cell colonies showed robust contraction, indicative of a cardiac phenotype (Supplementary information, Movie S1). We then stained 6TCF treated cultures with various antibodies to verify lineage-specific gene expression (Supplementary information, Table S3). As anticipated, we detected colonies characterized by very different gene signatures (Figure 2B, Supplementary information, Figure S1A and S1B). Beating cardiac myocyte colonies expressed high level of α-Actinin, with sarcomeric structures. Epithelial colonies expressed membrane localized E-cadherin. Round colonies and neuronal like cells are MAP2 positive. Some of the branched cells stained with neuronal markers Tuj1, Pax6 or glial marker GFAP. Smooth muscle actin (SMA) positive cells were found closely attached to the bottom of the culture well. Alpha-fetoprotein (AFP) antibody marked endodermal colonies, whereas Cebpa stained adipocytic colonies. PAS staining, which is used for identification of liver cells, labeled some large domed colonies. Oil red O staining confirmed the existence of adipocytes in the mixed culture.
Figure 2.
A wide ranges of somatic cell types can be obtained through iMT. (A) Pictures of various types of colonies generated via iMT from MEFs. (B) Molecular characterization of different iMT cell types by immunostaining, PAS staining or Oil red O staining on Day 16. DAPI staining is shown in blue. Staining of MEF controls are in Supplementary information, Figure S1D. (C) Oct4-GFP MEFs do not show any GFP signal after 6TCF or SGCF treatment on Day 16. (D) A picture showing the patch-clamp recording experiments. (E) Representative whole-cell voltage-clamp recording of Na+ and K+ currents from iMT neurons. (F) Representative current-clamp recording of multiple action potentials evoked by current injection from iMT neurons. (G) Distribution of contraction frequency for measured iMT cardiac myocytes. (H) Whole-cell electrical recording of iMT cardiac myocytes displaying action potentials representing different cardiac myocyte subtypes. (I) Representative action potentials of iMT cardiac myocytes. (J) Spontaneously contracting iMT cardiac myocytes have electrical activity measured by single cell extracellular electrodes. (K) FACS analysis of ANF-GFP MEF on Day 12 after 6TCF treatment suggests gene activation for embryonic cardiac trans-differentiation. (L) GFAP+ colonies generated by Day 20 of 6TCF treatment with GFAP-GFP MEFs.
Compared to 6TCF, SGCF treatment appeared to be more efficient in generating large areas of α-Actinin positive contracting colonies, MAP2 positive round colonies, as well as dark colonies (Figure 2B and Supplementary information, Figure S1B). The largest contracting colonies appeared in MEFs treated with only three chemicals SGF (Supplementary information, Figure S1B and Movie S2). Importantly, control MEFs failed to show significant signals for the tested markers (Supplementary information, Figure S1C). In order to determine whether iMT is applicable to cells other than MEFs, we derived tail tip fibroblasts (TTF) from 8-week mice. After treatment with 6TCF, we have also observed α-Actinin positive myocytes and Tuj1 positive neurons; although the efficiency was much lower (Supplementary information, Figure S1D).
In order to ascertain whether the 16 days of iMT process reflects a trans-differentiation process that bypasses the pluripotent state, we repeated experiments with Oct4-GFP MEFs. Although robust multi-lineage trans-differentiation was observed for 6TCF and SGCF treatments with Oct4-GFP MEFs by day 16, we did not detect an Oct4-GFP signal throughout the process. In contrast, GFP-positive colonies were readily derived upon overexpression of Oct4, Sox2 and Klf4 (Figure 2C). Moreover, continued culture of iMT colonies under ES cell medium (with LIF and 2i) failed to generate an Oct4-GFP signal or establish expandable clones. These findings suggest that Oct4 expression is not activated during the iMT process. Therefore, chemical induced multi-lineage trans-differentiation bypasses the pluripotent state.
To achieve functional characterization of the trans-differentiated cells, we performed electrophysiological analysis with selected cells in the day 16 iMT culture (Figure 2D). Whole-cell voltage-clamp recording experiments on candidate neuronal cells indicated that iMT neurons have appropriate Na+ currents and K+ currents (Figure 2E), as well as robust overshooting action potentials (Figure 2F and Supplementary information, Figure S1E). On the other hand, cardiac myocytes from iMT exhibited spontaneous contraction and Ca2+ waves with a frequency distributed around 1 Hz (Figure 2G, Supplementary information, Movie S3 and Figure S1F). Whole-cell electrical recording of representative iMT cardiac myocytes displayed action potentials that resemble reported cardiac subtypes29, including pacemaker-like, atrial-like and ventricular-like cardiac myocytes (Figure 2H and 2I). Single-cell extracellular recording revealed electrical activity of beating cardiac cells (Figure 2J).
To harness the power of reporter cell lines for validating trans-differentiation process, we generated two reporter MEFs that mark cardiac and glial lineage-specific expression. For cardiac lineage, we chose the ANF-GFP system that labels both mouse embryonic heart (Supplementary information, Figure S1G and S1H) and repairing adult heart tissues (unpublished data)30. FACS analyses indicated that 6TCF treatment converts ∼9.7% of MEFs into ANF-GFP positive cells by day 12, suggesting robust activation of cardiac program (Figure 2K). For glial lineage, we chose the well-established GFAP-GFP system to mark astrocyte cells. After 20 days of 6TCF treatment, GFAP-GFP positive colonies are readily distinguishable (Figure 2L). The shape of the cells from GFAP-GFP positive colonies strongly resembled that of wild-type astrocytes. In addition, no detectable GFP signal or distinguishable colony type could be seen in the control GFAP-GFP MEF culture, even after 9 passages (Supplementary information, Figure S1I). We conclude that iMT process enables trans-differentiation of MEFs towards different functional somatic cell types.
A defined molecular roadmap for iMT
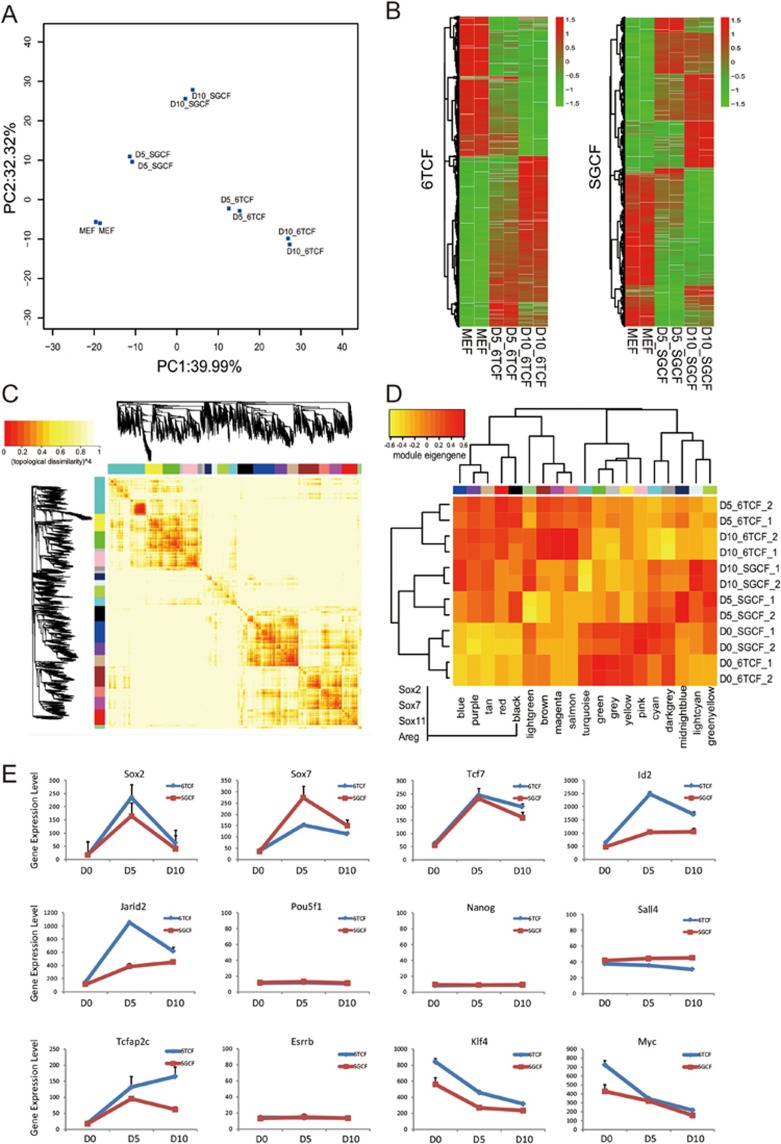
To investigate the molecular roadmap of iMT, we performed time course microarray analysis for both 6TCF and SGCF treatments on D0 (MEF), D5 and D10 (Table S4). PCA projection with differentially expressed genes at different time points highlighted cellular transitions during two treatments (Figure 3A). Gene expression heatmaps revealed global gene expression changes occurring during the iMT process (Figure 3B). Protein interaction network analysis using the STING database and pathway analysis using the KEGG database suggested several important signaling pathways were perturbed upon both treatments, including the WNT, TGF-β and MAPK pathways (Supplementary information, Figure S2A, S2B and Table S5). These pathways are directly associated with chemical functions of the cocktails. Both 6 (from 6TCF) and S (from SGCF) inhibit TGF-β signaling; while C inhibits GSK3 to activate WNT signaling. For both treatments, a large module composed of Cytochrome proteins was activated in common (Supplementary information, Figure S2B), indicating stimulated metabolic pathways. Importantly, the function and pathway enrichment analyses for the differentially expressed genes after 10 days of 6TCF treatment suggest regulation of a variety of developmental processes, such as blood vessel, heart, skeletal and neural development (Supplementary information, Table S5 and Figure S2C). The result fits well with our hypothesis of the iMT process.
Figure 3.
Gene expression analysis reveals the network dynamics during iMT. (A) A PCA projection plot showing microarray analysis of time course 6TCF and SGCF treatments. (B) A clustering heatmap showing differentially expressed genes during the time course experiment of 6TCF and SGCF treatments. (C) A gene expression correlation heatmap from the microarray data reveals multiple modules associated with iMT process. (D) A heatmap showing gene expression pattern of defined modules. (E) Gene expression dynamic for key regulators during the iMT process. Note that there is no significant upregulation of Pou5f1, Nanog, Esrrb or Sall4 during the iMT process.
In order to dissect different transcriptional modules that might hint at mechanisms of multi-lineage program activation, we carried out gene co-expression network analysis for our microarray data sets. Weighted correlation network analysis (R package WGCNA) was applied to differentially expressed genes in the time course experiments. 18 co-expression modules were found, among which 6 modules are linked to both 6TCF treated and SGCF treated samples. 4 modules are 6TCF specific and 5 modules SGCF specific. 7 modules are related to MEF-control samples (Figure 3C, Supplementary information, Table S5). Among the significantly activated modules after both 6TCF treatment and SGCF treatment, we found the black module to be the most relevant to lineage trans-differentiaton (Figure 3D). Interestingly, GSEA analysis showed that black module contains both neuronal and cardiac developmental genes including Sox2, Sox11, Sox7, Chd7, Tnnt1, Actc1 and Myl1. A Cytospace protein interaction network of this group of genes reveals that neuronal-specific genes and cardiac-specific genes are actually inter-connected (Supplementary information, Figure S2D). Areg, which is one of the most significantly activated genes from microarray data (Supplementary information, Tables S4 and S5), are at the center of this connection.
Among differentially expressed genes we identified several key regulators that are downstream of the perturbed pathways during both 6TCF and SGCF treatments (Figure 3E). Sox2, which is a central stem cell marker for many stem cell types, is strongly activated during the iMT process. Sox7, which is more endoderm specific, is also activated on Day 5. Other upregulated genes include trophoblast stem cell marker Tcfap2c, the WNT downstream transcription factor Tcf7, the TGF-β target Id2, and the PRC2 component Jarid2. The expression levels of most of these genes peak at day 5, and are still higher than control MEFs on day 10. Consistent with immunostaining and qPCR data, the expression of pluripotent markers Pou5f1, Nanog and Esrrb is below background and unchanged during iMT. In addition, two Yamanaka factors, Klf4 and Myc, are downregulated in the time course.
Altered epigenetic status for key regulators during iMT
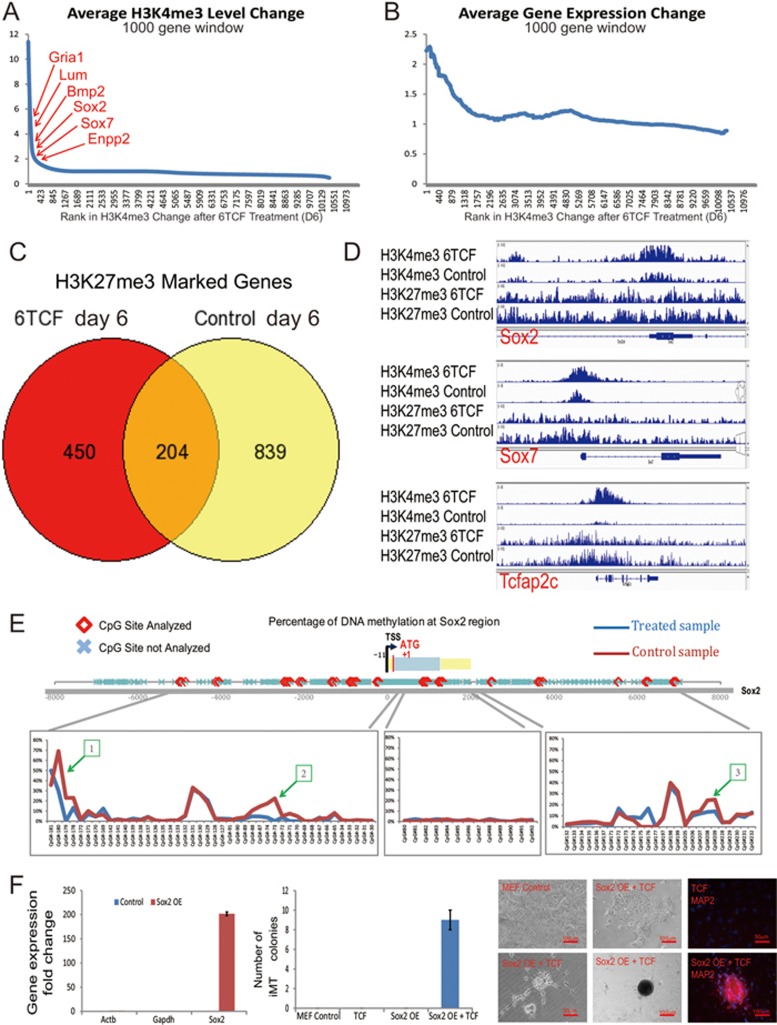
One of the key regulatory pathways perturbed by both chemical cocktails is the histone modification pathway. T from the combination 6TCF inhibits LSD1 and increases H3K4me3, while G from the combination SGCF inhibits Ezh2 and decreases H3K27me3. The action of both T and G would be predicted to relax chromatin structure, inhibit epigenetic silencing, and lead to gene expression activation. To identify global epigenetic changes during iMT, we performed ChIP-seq analysis for both H3K4me3 and H3K27me3 in 6TCF-treated and control MEFs (Supplementary information, Tables S6 and S7). We observe significant changes in H3K4me3 and H3K27me3 patterns upon 6 days treatment of 6TCF, as compared to control cells. Gene Ontology (GO) analysis of differentially marked genes for either H3K4me3 or H3K27me3 suggests regulation of development-related pathways (Supplementary information, Figure S3A). Among the dramatically upregulated genes, many are ranked top for increase in H3K4me3 level after 6 days of 6TCF treatment (Figure 4A). We observe a genome scale association of specific gene expression activation and regional increase in H3K4me3 level (Figure 4B). Notably, 6TCF treatment on day 6 significantly reduced the number of H3K27me3 genes (Figure 4C). Changes in H3K4me3 or H3K27me3 are key molecular events to alter MEF regulatory network during iMT (Supplementary information, Figure S3B and S3C). Figure 4D highlights the genomic regions of key regulators, such as Sox2, Sox7 and Tcfap2c, where we observe increased H3K4me3 and decreased H3K27me3, correlating well with their gene expression activation upon chemical induction.
Figure 4.
Epigenetic analyses reveal altered chromatin state of key regulators during iMT. (A) ChIP-seq analysis suggests increase of H3K4me3 level in multiple lineage markers. Genes are ordered by change in H3K4me3 enrichment after 6 days of 6TCF treatment; average H3K4me3 level changes of 1 000 gene window are then shown across the ordered genome. (B) ChIP-seq analysis suggests change in H3K4me3 level correlates with change in gene expression level upon 6TCF treatment. Genes are ordered by change in H3K4me3 enrichment after 6 days of 6TCF treatment; average gene expression changes of 1 000 gene window are then shown across the ordered genome. (C) ChIP-seq analysis suggests dramatic reduction of global H3K27me3 level upon 6TCF treatment on day 6 when compared with control MEFs. (D) IGV snapshots showing increased H3K4me3 and reduced H3K27me3 at Sox2, Sox7 and Tcfap2c regions during iMT process. (E) High-throughput DNA methylation screen identifies three differential methylation regions (−4 561 to −4 452, −502 to −494, 6 674 to 6 686 from transcriptional start site) at Sox2 loci between control and 6TCF-treated MEFs on Day 6. (F) Sox2 overexpression in MEF by retrovirus infection can partially replace the function of E616452 during iMT process.
Most of the trans-differentiated cells from iMT are present as colonies, suggesting that MEFs are first transformed into progenitor types before terminal cell fate decisions. We propose that activation of the stem cell marker Sox2 may be an important event for obtaining different progenitor identity. We searched for clues of differential DNA methylation that might account for Sox2 upregulation during iMT. We performed high throughput DNA methylation pyrosequencing across the Sox2 gene, and found three regions in the Sox2 gene with reduced DNA methylation levels in 6TCF treated MEFs compared to controls on day 6 (Figure 4E). In the same way, we have also discovered differentially methylated regions in the Sox7 locus (Supplementary information, Figure S3D). Further analysis of LINE-1 element suggests a small reduction of global DNA methylation (Supplementary information, Figure S3E).
The chemical E616452 (6) is also known as Repsox. 6, S or 8 (all target TGF-β signaling) turn out to be most essential for any trans-differentiation event throughout our screen (Supplementary information, Table S2). To functionally validate the importance of Sox2 activation during iMT, we overexpressed Sox2 in MEFs using retrovirus infection (Figure 4F). We first excluded 6 from the cocktail 6TCF, we found that TCF treatment alone did not produce trans-differentiation clones with control MEFs. However, Sox2 overexpression can partially rescue the phenotype, although with low efficiency (Figure 4F). Our results suggest that Sox2 activation is important within chemically induced genetic and epigenetic cascades.
Single cell analysis reveals an iMT priming cellular state
Genomic analyses of populations are informative, but are often complicated by cell heterogeneity. Characterization at the single cell level provides a means to probe cellular transitions and resolve stochastic processes31,32. We selected a set of 96 genes, including markers of different lineages, as well as top upregulated genes from the microarray time course experiment. We assayed single cells from control MEFs, control J1 embryonic stem cells (ESCs), day 6 MEFs treated with 6TCF, day 14 MEFs treated with 6TCF, as well as manually picked colonies with different morphologies (Supplementary information, Table S8).
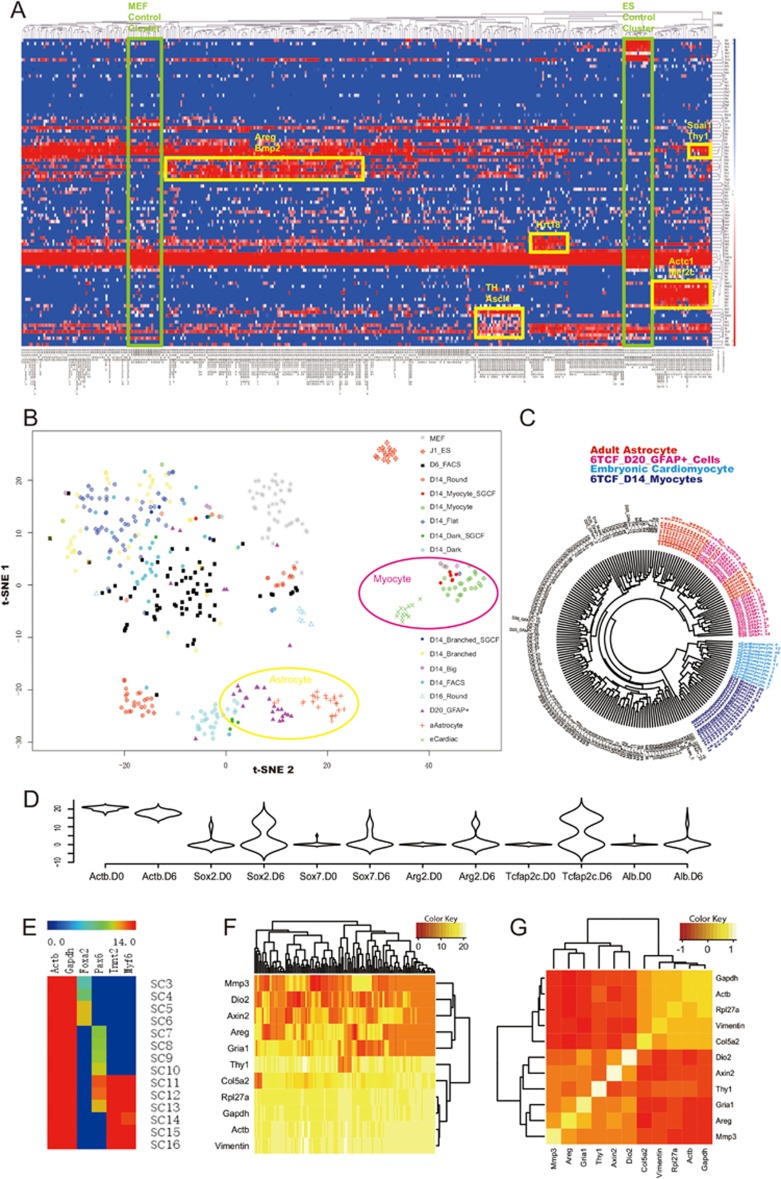
Hierarchical clustering of single cell level gene expression signatures revealed different groups of cell types expressing specific lineage modules (Figure 5A). In the t-SNE projection plot, we observe clearly separated cell clusters, including control MEFs and ESCs, iMT-derived myocytes and astrocytes, indicative of the robustness of the assay (Figure 5B). Interestingly, upon 6 days of 6TCF treatment, the relatively homogenous (when compared to treated cells) MEF single cells (labeled as grey dots in Figure 5B) become highly heterogeneous (labeled as black dots in Figure 5B), suggesting that stochasticity of iMT results from the chemical treatment rather than heterogeneity of the initial MEF culture. On day 6 the cellular network appears primed to a rather stochastic state. It is not until day 14 that cell fates of different lineages are fully resolved. In order to compare iMT cells with primary cells from tissues, we further profiled single cells from mouse embryonic heart and adult astrocytes as controls. Both the t-SNE projection and the circular dendrogram suggest that single cells from trans-differentiated GFAP+ colonies strongly resemble adult astrocytes, whereas single cells from trans-differentiated contracting myocytic colonies strongly resemble embryonic cardiac myocytes (Figure 5B and 5C). The single cell assay serves to confirm the trans-differentiated nature of the cells with more markers and higher resolution (Supplementary information, Table S8). It also helps to evaluate increased gene expression variability after chemical stimulation (Figure 5D), and the diversity of trans-differentiated single cells expressing lineage markers from endoderm, mesoderm or ectoderm (Figure 5E). Gene expression correlation analysis revealed several interesting markers that distinguish iMT-primed cells from fully trans-differentiated cells, including Areg, Mmp3, Dio2, Axin2 and Gria1 (Figure 5F, 5G, Supplementary information, Figure S4A and S4B). These genes are activated in a coordinated manner at the single cell level.
Figure 5.
Single cell analysis uncovers an iMT priming state before stochastic cell fate decision. (A) A hierarchical clustering heatmap showing single cell gene expression analysis data of the iMT experiment. Non-treated MEFs and undifferentiated ESCs are also analyzed as controls. (B) t-SNE plot showing the iMT priming and fate decision pathway at single cell resolution. (C) Single cell profiles of iMT astrocytes and cardiac myocytes strongly resemble that of primary cells from respective lineages. (D) A violin plot showing expression level distribution of selected genes in MEF single cells before and after 6 days of 6TCF treatment. (E) A gene expression heatmap showing representative single cells from 6TCF-treated MEF on Day 14. Different cells express endoderm (Foxa2), ectoderm (Pax6) or mesoderm (Myf6) lineage markers. (F) A clustering heatmap showing the unique signature of the iMT priming cells. (G) A correlation heatmap highlighting the iMT priming module. See also Supplementary information, Figure S4.
Modification of the iMT culture system enables directional trans-differentiation towards myocytic, glial or adipocytic progenitors
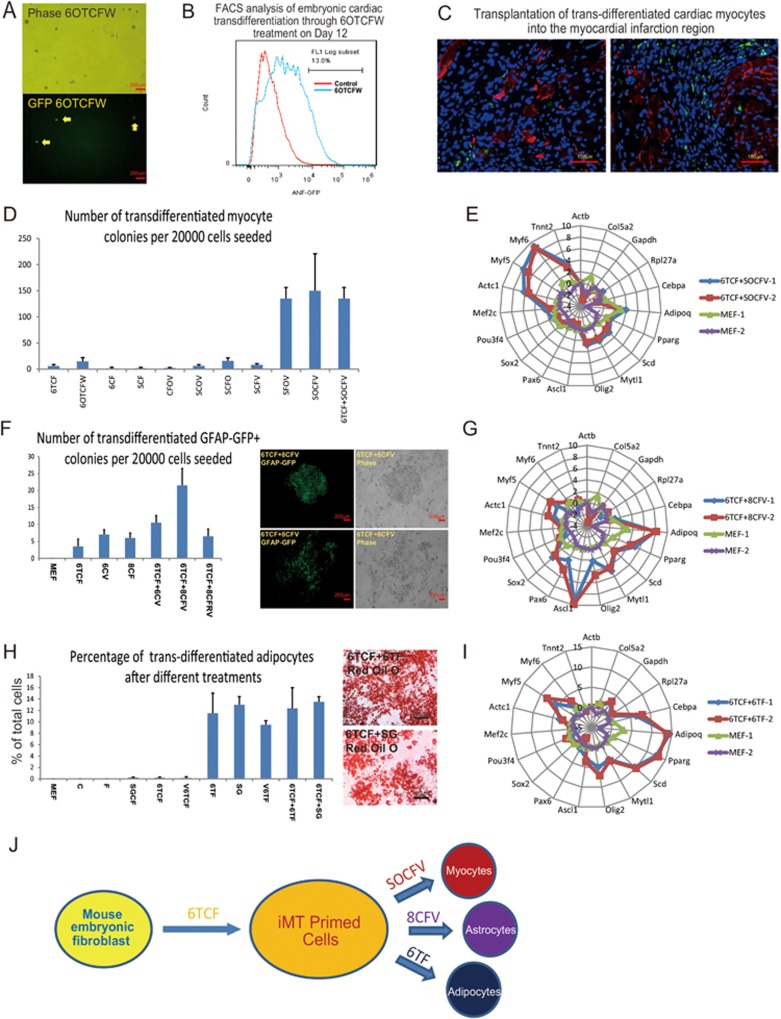
The iMT process produces a stochastic state with a flexible genetic network. We hypothesize that by modifying chemical combinations and stepwise culture conditions in iMT to favor a particular lineage, one should be able to achieve efficient directional trans-differentiation towards a single progenitor type. To examine this possibility, we first focused on cardiac trans-differentiation. We treated ANF-GFP MEFs with 6TCF together with two chemicals that are known to promote cardiac lineage differentiation, namely Dorsomorphin (O) and IWR-1 (W) (Supplementary information, Table S1)33,34. We also supplemented cultures with agents that support cardiac cell growth, including sodium pyruvate, holo-transferrin, vitamin C and Ciprofloxacin. Under this cardiac-promoting culture system, the embryonic cardiac marker ANF-GFP signal can be visualized as early as day 6 after cocktail induction (Figure 6A). FACS analyses indicated that 6OTCFW treatment converts ∼13% of MEFs to ANF-GFP cardiac progenitors by day 12 (Figure 6B). Either 6OTCFW treatment or 6TCF (4 days) followed by 6OTCFW treatment efficiently activates endodermal and cardiac programs (Supplementary information, Figure S5A). We transplanted day 8 treated cells into a mouse myocardial infarction (MI) model (see Materials and Methods). We observed successful integration of chemically induced cardiac progenitor cells in the repair region (Figure 6C and Supplementary information, Figure S5B). Continued optimizations suggest that replacing 6 with S and replacing W with V improve general cell viability. The use of combination SOCFV or 6TCF followed by SOCFV further increases the number of trans-differentiated myocytic colonies as well as contracting regions by more than 10-fold (Figure 6D). Gene expression profiling of cell cultures after stepwise treatment (6TCF 4 days + SOCFV 16 days) suggests specific gene expression activation of cardiac lineage markers such as Tnnt2, Actc1, Myf6 and Myf5 (Figure 6E).
Figure 6.
Modification of the iMT chemical combinations enables lineage-specific trans-differentiation. (A) Detection of ANF-GFP signal on day 6 upon 6OTCFW treatment. (B) FACS analysis on Day 12 suggests high efficiency of embryonic cardiac trans-differentiation from MEFs by 6OTCFW treatment. (C) Transplantation of 6OTCFW-treated MEFs into the myocardial infarction regions reveals integration of the trans-differentiated cardiac myocytes with the cardiac tissues. Some transplanted cells (green fluorescence by ANF-GFP) also express cardiac-specific marker Troponin (red fluorescence by immunostaining). (D) Modified iMT culture condition induces efficient cardiac myocytic trans-differentiation from MEFs. (E) Gene expression analysis reveals cardiac-specific gene activation after stepwise chemical treatment (6TCF 4 Days + SOCFV 16 Days). Expression value is in Log2 scale. MEFs were partially removed by 30 min of settling in tissue culture plate before analysis. (F) Modified iMT culture condition induces glial program from MEFs. (G) Gene expression analysis reveals neuronal or glial gene activation after stepwise chemical treatment (6TCF 6 Days + 8CFV 16 Days). Expression value is in Log2 scale. (H) Modified iMT culture condition induces efficient adipocytic trans-differentiation from MEFs. (I) Gene expression analysis reveals adipocytic-specific gene activation after stepwise chemical treatment (6TCF 6 Days + 6TF 12 Days). Expression value is in Log2 scale. MEFs were partially removed by 30 min of settling in tissue culture plate before analysis. (J) A diagram showing the roadmap for induced multi-lineage trans-differentiation by chemical combinations.
We then used GFAP-GFP MEFs to optimize neural/glial trans-differentiation. During our screens, we found that chemical 8 was effective in promoting neural/glial programs in MEFs (Supplementary information, Figure S5C). We show that 8CF induced high levels of expression of the neural stem cell marker gene Ascl1. We then found that adding V is beneficial for turning on GFP signal from the GFAP-GFP MEFs. On the other hand, adding R is beneficial for generating MAP2 positive round neural progenitor colonies (Supplementary information, Figure S5D). Stepwise culture condition with 6TCF iMT priming (6 Days) followed by 8CFV treatment (16 Days) turns out to be most effective in generating GFAP-GFP positive colonies (Figure 6F). Gene expression profiling of pooled cell cultures after stepwise treatment (6TCF 6 days + 8CFV 16 days) suggests gene expression activation of neural and glial lineage markers such as Ascl1, Olig2, Pax6 and Mytl1 (Figure 6G).
Moreover, we found that exclusion of C significantly increased efficiency of adipocytic trans-differentiation. Highly uniform adipocytes were obtained upon withdrawal of C for 12 days after initial 6TCF treatment. More than 10% of cells were adipocytes after treatment with 6TF, SG, 6TCF + 6TF or 6TCF + SG (Figure 6H, Supplementary information, Figure S5E, S5F and S5G, while colonies of other cell types became extremely rare (Supplementary information, Table S2). Gene expression profiling of cell cultures after stepwise treatment (6TCF 6 days + 6TF 12 days) suggest specific gene expression activation of adipocytic lineage markers such as Adipoq, Pparg, Scd and Cebpa (Figure 6I). These examples suggest that iMT can be modified for directional lineage trans-differentiation through different chemical combinations.
Discussion
Here we describe a chemically induced multi-lineage pathway in which MEFs are first stimulated into an iMT primed state, and then adopt specific lineage choices (Figure 6J). Because there are many directions to exit the iMT primed state, this cell fate decision process is multi-lineage in nature. Importantly both our genomic profiling and single cell analyses suggest that major gene expression activation and cellular heterogeneity arise upon the chemical treatments. Both lineage gene expression activation and colony level trans-differentiation are very dramatic when compared to untreated MEF cell culture. The reporter line experiment also provides evidence that lineage specific expressions are acquired after chemical treatment.
The iMT protocol resembles the first stage of the previously described three-step chemical induced pluripotent stem cell (CiPSC) protocol20,21. We believe that the use of serum in the iMT protocol improves somatic cell viability and trans-differentiation. It is possible that the iMT priming state is an important transitional state that lies upstream of chemical induced pluripotency. Our model is also supported by other published works on chemical induced trans-differentiation24,25. The iMT priming state is a synthetic, transient and unstable cellular state in which many unrelated lineage markers are coexpressed (Supplementary information, Table S5). In order to stabilize the genetic network, the cells have different options. With proper stimulation and stepwise culture conditions, primed cells may be guided to specific lineages (Figure 6J).
Remarkably, the chemical combinations that induce reprogramming or trans-differentiation processes target very similar pathways, including the WNT, TGF-β, cAMP, as well epigenetic pathways for global gene activation correlating with increased H3K4me3 or decreased H3K27me3. Microarray analysis also highlights activation of MAPK, which probably lies downstream of bFGF function. The synergistic effects of these pathways are essential to promote cellular transitions. The crosstalk among these pathways would be of great interest for studying the mechanisms of induced cell fate alteration.
Interestingly, we have used four chemicals (6, 8, S, O) to target the TGF-β pathway in our combinations, but the outcome turns out to be dramatically different. The use of 6 favors stochastic fate conversion; the use of 8 favors activation of neuronal and glial markers; whereas O works together with S to support cardiac trans-differentiation. Such difference might result from their preferential inhibitory effect over ALK family members. 6 specifically targets ALK5; 8 efficiently targets ALK5, 7; S targets ALK4, 5, 7; O targets ALK2, 3, 6 as well as AMPK. The balance of these ALK molecules may play critical roles during chemically induced cell fate decision process. Thus, it opens up the possibility of cellular conversion towards diverse cell types.
Finally, the chemical iMT process offers a novel approach for generating various progenitor cell types for regenerative medicine. It is cost-efficient and fast. By modifying chemical combinations and culture media, directional trans-differentiation can be achieved. Testing of lineage-specific culture media, as well as varying chemical concentrations will eventually enable generation of desired cell types with high efficiency and purity. The iMT process holds promise for generating cell lineages that may not be readily obtainable using traditional methods.
Materials and Methods
Cell culture
MEFs were isolated from E14.5 B6/C57, Oct4-GFP, ANF-GFP or GFAP-GFP mouse embryos. Head, vertebral column, dorsal root ganglia and all internal organs were removed and discarded and the remaining embryonic tissue was manually cut into pieces and incubated in 0.25% trypsin for 5 min. Cells from embryos were plated onto a 15cm tissue culture dish in MEF medium containing DMEM (Life Technologies) and 10% fetal bovine serum (FBS, Gemini). Mouse adult fibroblasts were isolated from tail tip samples. Tail tips were minced into pieces, placed on culture dishes, and incubated in MEF medium for 7days. MEFs and TTF cells were seeded at a density of 20 000 cells per 35 mm dish. The next day, MEF medium was replaced with the iMT media containing DMEM (Life Technologies) supplemented with 15% FBS (Gbico), 2 mM GlutaMax supplement (Life Technologies), 1% nonessential amino acids (Life Technologies), 0.1 mM β-mercaptoethanol (Sigma) and 40 ng/ml basic fibroblast growth factor (bFGF, Stemgent), as well as chemical combinations (Supplementary information, Table S2). Media were changed every 4 days. For directional trans-differentiation of MEFs into cardiac myocytes, iMT medium was further supplemented with 3 mM sodium pyruvate, 4 μg/ml Holo-transferrin (Sigma), 20 μg/ml Vitamin C (Sigma), 5 μg/ml Ciprofloxacin, 1 μΜ Dorsomorphoin (Sigma) and 2 μM IWR-1 (Sigma) to make iCM medium. Medium were changed every 4 days.
Immunofluorescence
Immunostaining was performed according to standard protocols. Cells were fixed with 4% paraformaldehyde for 30 min at room temperature. After washing with PBS, the cells were treated with PBS containing 0.2% Triton X-100 for 20 min. Then the cells were incubated in PBS supplemented with 3% bovine serum albumin (Sigma) and 0.02% Tween-20 for one hour. Cells were incubated in primary antibodies overnight at 4 °C, then washed five times with PBS and incubated for 1 h at room temperature with anti-rabbit or anti-mouse secondary antibodies Alexa Fluor-488 or Alexa Fluor-594 (1:1 000, Invitrogen).
PAS staining and oil red O staining
For Periodic Acid-Schiff (PAS) staining, cells were stained by PAS Kit (Sigma) following the manufacturer's instructions. For Oil Red O staining, cells were fixed with 4% paraformaldehyde, washed twice in ddH2O and once in 60% isopropanol, and then stained with 0.5% oil red O in propylene glycerol solution for 20 min at room temperature. Cells were washed four times with ddH2O before imaging.
Electrophysiology
After 16 days of iMT treatment, selected cells were recorded in extracellular buffer of the following composition (in mM): 145 NaCl, 3 KCl, 3 CaCl2, 2 MgCl2, 10 HEPES, and 8 glucose, pH 7.3. The patch pipette solution contained (in mM): 136.5 K-gluconate, 17.5 KCl, 10 HEPES, 0.2 EGTA, 9 NaCl, 4 MgATP and 0.3 Na2ATP, pH was adjusted to 7.3 with KOH. Glass patch pipettes were directly attached on single cells for extracellular recording. Intracellular action potentials were recorded in current-clamp whole-cell configurations. The iMT cardiac myocytes were held at −70 mV with a stimulation of 3 nA for 5 ms to elicit an action potential. The iMT neurons were held at −60 mV, and step currents from +0.2 nA to 1.8 nA were injected to elicit action potentials. Na+ currents and composite K+ currents were recorded in the voltage-clamp configuration by delivering voltage steps ranging from −70 mV to +100 mV in cells held at −70 mV. Delayed rectifier K1 currents were activated by 0.5 s voltage steps from −40mV to +20 mV after a 0.5-s-long step to −40 mV. Recordings were performed using an Axon Multiclamp 700 B and Clampex 10 software. Data were digitized at 10 kHz and analyzed with Clampfit 10 software.
Microarray analysis
Total RNA was extracted with the Rneasy Mini Kit (Qiagen). cDNA was synthesized and then amplified through in vitro transcription. Labeled cRNA was fragmented and then hybridized to GeneChip Mouse Genome 430A 2.0 Arrays (Affymetrix). Washing, detection, and scanning were performed as described by manufacturer protocols (Affymetrix). Signal intensities of genes for each genechip were computed using the Affymetrix Microarray Suite 5.0. The raw data were firstly summarized, and then processed using the standard RMA algorithm with quantile normalization (Supplementary information, Table S4).
For data analysis, the signal intensities for all probes of each gene were averaged and Log2 transformed. The transformed data were subjected to one-factor analysis of variance using Limma (Linear Models for Microarray Data) package in R. For differentially expressed genes, the false positives were controlled by a cut-off value of false discovery rate (FDR) ≤ 0.001. PCA projection analysis (Figure 3A) and Clustering heatmap (Figure 3B) were generated using R software (http://www.r-project.org/) with differentially expressed genes between each two groups from days 0, 5 and 10 samples for 6TCF or SGCR treatment (FDR ≤ 0.01). PCA were performed using prcomp function; Clustering heatmap were made using pheatmap package. The function and pathway enrichment analyses of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) in Supplementary information, Table S5 were conducted with DAVID Microarray software (http://david.abcc.ncifcrf.gov/home.jsp) using cut-off value of P-value ≤ 0.05.
For network construction in Supplementary information, Figure S2A and S2B, differentially expressed genes between each two group from days 0, 5 and 10 samples during 6TCF or SGCF treatment (FDR ≤ 0.01) were firstly projected onto the KEGG pathway database, then the dataset of genetic network were constructed according to gene-gene interaction in KEGG Pathway. Finally, the genetic networks were visually constructed by Cytoscape 3.2 using the above gene-gene interaction dataset.
For gene co-expression network analysis, R package WGCNA was applied to the most variable microarray probes across samples (based on ranking of cv2 = variance/means^2). A signed gene co-expression network was built by measuring topological dissimilarity among all gene pairs and 18 co-expression modules were found, which relatively linked to MEF-control, 6TCF- and SGCF-treated samples. Module eigengenes to samples were then calculated to show correlation between sample and module. GO enrichment and KEGG pathway analyses were further conducted with DAVID Bioinformatics Resources 6.8. Genes of interest were extracted from the whole network and inputted into Cytospace for visualization.
DNA methylation analysis
A multiplex bisulfite PCR was designed by PyroMark Assay Design Software (Qiagen) to interrogate majority of the CpGs in Sox2 gene. Sequences in repeat regions were excluded from the design. 500 ng genomic DNA was bisulfite treated using the EZ DNA Methylation Kit (Zymo Research). 24-plex PCR was performed using TaKaRa EpiTaq HS PCR (Takara Bio). The PCR were performed using this protocol: 95 °C 15 min; 18 × (95 °C 30 s; 64 to 55 °C 30 s, −0.5 °C; 72 °C 30 s); 27 × (95 °C 30 s; 54 °C 30 s; 72 °C 30 s); 72 °C 5 min; 4 °C. The PCR products were QCed on Bioanalyzer (Agilent), purified using QIAquick PCR Purification Kit (Qiagen). Libraries were prepared using the KAPA Library Preparation Kit (Roche) for Ion Torrent. Libraries were quantified, pooled and sequenced using Ion PGM™. The percentage of methylation of each CpG site being sequenced was calculated by the number of methylated reads dividing by the number of total reads.
ChIP-seq analysis
Cells were fixed in 1% formaldehyde for 10 min, quenched with glycine and washed 2 times with PBS. Cells were then resuspended in lysis buffer and sonicated to shear the chromatin to an average length of 600 bp. Supernatants were precleared with Protein-A/G Dynabeads (Invitrogen) and 10% input was collected. Immunoprecipitations were performed with antibodies to H3K4me3 and H3K27me3 (Millipore). DNA-protein complexes were pulled down using Protein-A/G Dynabeads (Invitrogen) washed. DNA was recovered by overnight incubation at 65°C to reverse crosslinks and purified using QIAquick PCR purification columns (QIAGEN). Libraries were prepared from ChIP DNA experiments with the TruSeq® DNA LT/HT Sample Prep Kit (Illumina) following manufacturer's instructions. The libraries were then sequenced with single end for 50 bp in HiSeq 2500 (Illumina) at RiboBio. Raw reads were filtered with standard quality control measures and reads that passed this prefiltering step were aligned with the Bowtie software to the mouse genome (Mm9 build). Peak calling was performed with MACS. Homer software was used to annotate ChIP-seq peaks, and to find differential peaks between treated and control samples.
Single cell qPCR
Individual primer sets (total of 96) were pooled to a final concentration of 0.1 μM for each primer. Individual cells were sorted or manually picked into 96 well PCR plates loaded with 5 μL RT-PCR master mix (2.5 μL 2× Reaction Mix, Vazyme Single Cell Sequence Specific Amplification Kit; 0.5 μL primer pool; 0.1 μL RT/Taq Enzyme, Vazyme Single Cell Sequence Specific Amplification Kit; 1.9 μL nuclease free water) in each well. Sorted plates were immediately frozen on dry ice. After brief centrifugation at 4 °C, the plates were immediately placed on PCR machine. Cell lyses and sequence-specific reverse transcription were performed at 50 °C for 60 min. Then reverse transcriptase inactivation and Taq polymerase activation were achieved by heating to 95 °C for 3 min. Subsequently, in the same tube, cDNA went through 20 cycles of sequence-specific amplification by denaturing at 95 °C for 15 s, annealing and elongation at 60 °C for 15 min. After pre-amplification, PCR plates were stored at −80 °C to avoid evaporation. Pre-amplified products were diluted 5-fold prior to analysis. Amplified single cell samples were analyzed with Universal PCR Master Mix (Applied Biosystems), EvaGreen Binding Dye (Biotium) and individual qPCR primers using 96.96 Dynamic Arrays on a BioMark System (Fluidigm). Ct values (Supplementary information, Table S6) were calculated using the BioMark Real-Time PCR Analysis software (Fluidigm). A background Ct of 28 is used to generate Log2 scale gene expression levels for each gene. Hierarchical clustering analysis was performed using MultiExperiment Viewer (MeV, http://www.tm4.org/mev.html) with the average linkage method and Pearson correlation distance. The t-SNE analysis was performed using R software. Briefly, PCA analysis was done with “prcomp” package, the first 45 PCs that account for 90% of variation was selected to calculate Euclidean distance among cells. Visualization of the distance was performed with “Rtsne” package. Gene to gene correlation analysis is processed with “UsingR” package in R.
Transplantation of iMT cardiac myocytes into myocardial infarction (MI) mouse model
B6/C57 mice (12 weeks old) were anesthetized. Chests were opened between the left 4th and 5th intercostals space and the heart was exposed. MI was induced by permanent ligation of the left anterior descending coronary artery proximal to the first branch. 10 000 6OTCFW treated (on day 8) ANF-GFP MEFs or 10 000 untreated ANF-GFP MEFs were injected near the ligation region. One week after cell transplantation, heart tissues were collected, dehydrated in 30% sucrose solution, embedded in Tissue-Tek OCT compound and frozen on dry ice. Frozen tissue slices of 6.0 μm thick were then prepared and tissue slices were then fixed with 4% formaldehyde solution for ten minutes, permeabilized with 0.02% Triton X-100 PBS and blocked with PBS containing 5% goat serum. The slides were stained with primary antibody against Troponin, followed by specific secondary antibody, and then imaged with Leica fluorescence microscope.
Accession number
Genomic data are accessible at GEO: GSE91374
Author Contributions
Experiments were designed by G Guo and X Han, carried out by X Han, H Yu, D Huang, Y Xu, A Saadatpour, X Li, L Wang, J Yu, L Pinello, S Lai, M Jiang, X Tian, F Zhang, Y Cen, Y Fujiwara and W Zhu. Data was analyzed by B Zhou, T Zhou, H Ouyang, J Wang and G Yuan. The paper was written by G Guo, SH Orkin and S Duan.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
We thank L Yan, Z Zhong, Y Wang, X Tian, M Nguyen, HE Benjamin, E Macro, Z Shao, M Jiang, L Sun for help on experiments. We thank EpigenDx for DNA methylation analysis and RiboBio for ChIP-seq analysis. We thank Y Zhou, J Ji, W Cai, J Chen, Z Li for insightful discussions on the project. This work was supported by funding from Fundamental Research Funds for the Central Universities (GG), 1000 Youth Talent Plan (GG) and Harvard Stem Cell Institute Pilot Grant (SHO). This work was supported by Zhejiang University Stem Cell Institute. Patent applications have been filed related to the chemical reprogramming method reported in this paper.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Material
Small molecule combinations activate lineage specific regulators in the mouse embryonic fibroblast (MEF) cells.
Phenotypic data from screened chemicals or chemical combinations.
List of antibodies used for molecular characterization of lineage specific cell types.
Microarray data for 6TCF and SGCF treatment time course.
Analysis of differentially expressed genes during iMT process.
MACS peak files from H3K4me3 or H3K27me3 ChIP-seq experiments for 6TCF treated or control MEF cells.
Annotation of peaks from H3K4me3 or H3K27me3 ChIP-seq experiments for 6TCF treated or control MEF cells.
Single cell qPCR data during iMT process as well as used qPCR primer.
Spontaneous contraction of trans-differentiated cardiac myocytes from 6TCF treated MEF.
Spontaneous contraction of trans-differentiated cardiac myocytes from SGF treated MEF.
Spontaneous Ca2+ waves observed in trans-differentiated cardiac myocytes from 6TCF treated MEF.
Immunocytochemistry identifies trans-differentiated cells from MEF and TTF cells.
Analysis with differentially expressed genes during iMT process reveals involvement of several key pathways.
iMT process is associated with changes in chromatin structure.
A unique gene expression module marks the iMT priming state before cell fate decision.
Modification of the iMT chemical combinations enables efficient lineage specific trans-differentiation.
References
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126:663–676. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 2007; 448:318–324. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007; 131:861–872. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007; 318:1917–1920. [DOI] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 2010; 463:1035–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo E, Rampalli S, Risueno RM, et al. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature 2010; 468:521–526. [DOI] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 2010; 142:375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazzo M, Dell'Anno MT, Dvoretskova E, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature 2011; 476:224–227. [DOI] [PubMed] [Google Scholar]
- Han DW, Tapia N, Hermann A, et al. Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell Stem Cell 2012; 10:465–472. [DOI] [PubMed] [Google Scholar]
- Pereira CF, Chang B, Qiu J, et al. Induction of a hemogenic program in mouse fibroblasts. Cell Stem Cell 2013; 13:205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, He Z, Ji S, et al. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature 2011; 475:386–389. [DOI] [PubMed] [Google Scholar]
- Polo JM, Anderssen E, Walsh RM, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 2012; 151:1617–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature 2008; 454:49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Ambasudhan R, Yuan X, et al. A chemical platform for improved induction of human iPSCs. Nat Methods 2009; 6:805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wei W, Zhu S, et al. Generation of rat and human induced pluripotent stem cells by combining genetic reprogramming and chemical inhibitors. Cell Stem Cell 2009; 4:16–19. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang Q, Yin X, et al. Generation of iPSCs from mouse fibroblasts with a single gene, Oct4, and small molecules. Cell Res 2011; 21:196–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Maehr R, Guo W, et al. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol 2008; 26:795–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B, Ng JH, Heng JC, Ng HH. Molecules that promote or enhance reprogramming of somatic cells to induced pluripotent stem cells. Cell Stem Cell 2009; 4:301–312. [DOI] [PubMed] [Google Scholar]
- Zhu S, Rezvani M, Harbell J, et al. Mouse liver repopulation with hepatocytes generated from human fibroblasts. Nature 2014; 508:93–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou P, Li Y, Zhang X, et al. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science 2013; 341:651–654. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Zhao T, Guan J, et al. A XEN-like state bridges somatic cells to pluripotency during chemical reprogramming. Cell 2015; 163:1678–1691. [DOI] [PubMed] [Google Scholar]
- Cheng L, Hu W, Qiu B, et al. Generation of neural progenitor cells by chemical cocktails and hypoxia. Cell Res 2014; 24:665–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Qiu B, Guan W, et al. Direct conversion of normal and Alzheimer's disease human fibroblasts into neuronal cells by small molecules. Cell Stem Cell 2015; 17:204–212. [DOI] [PubMed] [Google Scholar]
- Li X, Zuo X, Jing J, et al. Small-molecule-driven direct reprogramming of mouse fibroblasts into functional neurons. Cell Stem Cell 2015; 17:195–203. [DOI] [PubMed] [Google Scholar]
- Zhang M, Lin YH, Sun YJ, et al. Pharmacological reprogramming of fibroblasts into neural stem cells by signaling-directed transcriptional activation. Cell Stem Cell 2016; 18:653–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Huang C, Xu X, et al. Direct reprogramming of mouse fibroblasts into cardiomyocytes with chemical cocktails. Cell Res 2015; 25:1013–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao N, Huang Y, Zheng J, et al. Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science 2016; 352:1216–1220. [DOI] [PubMed] [Google Scholar]
- Wang Y, Qin J, Wang S, et al. Conversion of human gastric epithelial cells to multipotent endodermal progenitors using defined small molecules. Cell Stem Cell 2016; 19:449–461. [DOI] [PubMed] [Google Scholar]
- Hescheler J, Fleischmann BK, Lentini S, et al. Embryonic stem cells: a model to study structural and functional properties in cardiomyogenesis. Cardiovasc Res 1997; 36:149–162. [DOI] [PubMed] [Google Scholar]
- Zeller R, Bloch KD, Williams BS, Arceci RJ, Seidman CE. Localized expression of the atrial natriuretic factor gene during cardiac embryogenesis. Genes Dev 1987; 1:693–698. [DOI] [PubMed] [Google Scholar]
- Guo G, Huss M, Tong GQ, et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell 2010; 18:675–685. [DOI] [PubMed] [Google Scholar]
- Guo G, Luc S, Marco E, et al. Mapping cellular hierarchy by single-cell analysis of the cell surface repertoire. Cell Stem Cell 2013; 13:492–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J, Daleo MA, Murphy CK, et al. Dorsomorphin, a selective small molecule inhibitor of BMP signaling, promotes cardiomyogenesis in embryonic stem cells. PLoS One 2008; 3:e2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson J, Titmarsh D, Hidalgo A, Wolvetang E, Cooper-White J. Primitive cardiac cells from human embryonic stem cells. Stem Cells Dev 2012; 21:1513–1523. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Small molecule combinations activate lineage specific regulators in the mouse embryonic fibroblast (MEF) cells.
Phenotypic data from screened chemicals or chemical combinations.
List of antibodies used for molecular characterization of lineage specific cell types.
Microarray data for 6TCF and SGCF treatment time course.
Analysis of differentially expressed genes during iMT process.
MACS peak files from H3K4me3 or H3K27me3 ChIP-seq experiments for 6TCF treated or control MEF cells.
Annotation of peaks from H3K4me3 or H3K27me3 ChIP-seq experiments for 6TCF treated or control MEF cells.
Single cell qPCR data during iMT process as well as used qPCR primer.
Spontaneous contraction of trans-differentiated cardiac myocytes from 6TCF treated MEF.
Spontaneous contraction of trans-differentiated cardiac myocytes from SGF treated MEF.
Spontaneous Ca2+ waves observed in trans-differentiated cardiac myocytes from 6TCF treated MEF.
Immunocytochemistry identifies trans-differentiated cells from MEF and TTF cells.
Analysis with differentially expressed genes during iMT process reveals involvement of several key pathways.
iMT process is associated with changes in chromatin structure.
A unique gene expression module marks the iMT priming state before cell fate decision.
Modification of the iMT chemical combinations enables efficient lineage specific trans-differentiation.