Abstract
There is ample evidence that crossing over is suppressed in heterochromatin associated with centromeres and nucleolus organizers (NORs). This characteristic has been attributed to all heterochromatin, but the generalization may not be justified. To investigate the relationship of crossing over to heterochromatin that is not associated with centromeres or NORs, we used a combination of fluorescence in situ hybridization of the maize 180-bp knob repeat to show the locations of knob heterochromatin and fluorescent immunolocalization of MLH1 protein and AFD1 protein to show the locations of MLH1 foci on maize synaptonemal complexes (SCs, pachytene chromosomes). MLH1 foci correspond to the location of recombination nodules (RNs) that mark sites of crossing over. We found that MLH1 foci occur at similar frequencies per unit length of SC in interstitial knobs and in the 1 µm segments of SC in euchromatin immediately to either side of interstitial knobs. These results indicate not only that crossing over occurs within knob heterochromatin, but also that crossing over is not suppressed in the context of SC length in maize knobs. However, because there is more DNA per unit length of SC in knobs compared to euchromatin, crossing over is suppressed (but not eliminated) in knobs in the context of DNA length compared to adjacent euchromatin.
Keywords: heterochromatin, crossing over, maize, knobs, synaptonemal complex
BASED on observations of the liverwort Pellia epiphylla, Emil Heitz first named and described “heterochromatin” as chromatin that remains condensed throughout the cell cycle and “euchromatin” as chromatin that is decondensed during interphase (Heitz 1928). Subsequently, heterochromatin has been observed in a wide variety of other eukaryotes, and many additional characteristics have been attributed to heterochromatin, including suppression of meiotic crossing over (Green 1966; Yunis and Yasmineh 1971; Comings 1972; John 1976; Ris and Korenberg 1979; Stack 1984; Szauter 1984; Loidl 1987; Sumner 2003; Grewal and Jia 2007; Lichten 2008).
Constitutive heterochromatin (subsequently referred to simply as heterochromatin) is most commonly found at telomeres and nucleolus organizers (NORs) and to either side of centromeres (i.e., pericentric heterochromatin), but heterochromatin can occur anywhere on chromosomes (Hsu and Arrighi 1971; Yunis and Yasmineh 1971; Dumas and Britton-Davidian 2002; Neves et al. 2005). A variety of evidence indicates that meiotic crossing over is suppressed, if not completely eliminated, in heterochromatin at NORs and in pericentric heterochromatin. This is based on observations of chiasmata (Mather 1933; Brown 1949; Natarajan and Gropp 1971; John 1976; Dumas and Britton-Davidian 2002; Lukaszewski et al. 2012), comparisons of molecular linkage maps with sequenced pseudomolecules and chromosome structure (Petes and Botstein 1977; Kota et al. 1993; Castiglioni et al. 1999; Islam-Faridi et al. 2002; Yu et al. 2003; Kim et al. 2005; van Os et al. 2006; Gore et al. 2009; Tomato Genome Consortium 2012), and observations of recombination nodules (RNs) and MLH1 foci on synaptonemal complexes (SCs, pachytene chromosomes) (Carpenter 1975; Sherman and Stack 1995; Froenicke et al. 2002; Anderson et al. 2003; Marcon and Moens 2003).
A variety of hypotheses have been proposed to explain how crossing over is suppressed in heterochromatin (John and Lewis 1965; Yunis and Yasmineh 1971; Carpenter 1975; Miklos and Nankivell 1976; Stack 1984; Civardi et al. 1994; Schnable et al. 1998; Bennetzen 2000; Fu et al. 2002; Blitzblau et al. 2007; Lichten 2008; Ellermeier et al. 2010; Vader et al. 2011). One hypothesis is that DNA double-strand break formation and repair is inhibited in heterochromatin, possibly due to DNA and histone modifications (such as methylation and deacetylation), the association of certain protein factors and small interfering RNAs (siRNAs) with DNA, or steric exclusion of crossover enzymes and RNs. Other hypotheses focus on differences in chromosome cores and cohesins, SC structure in heterochromatin compared to euchromatin, underrepresentation of heterochromatin in SC length, and interference with synapsis (SC formation) in heterochromatin. Also, assuming that crossing over usually takes place in or near genes, differences in DNA sequence may contribute, including a high concentration of tandem repeats and/or a lack of genes in heterochromatin. Recently, Vincenten et al. (2015) provided strong support for two of these hypotheses by showing that, in Saccharomyces cerevisiae, a kinetochore (centromere) protein complex called Ctf19 both inhibits nearby double-strand breaks needed for crossing over and promotes cohesion enrichment that also interferes with meiotic crossing over.
Does demonstrated suppression of crossing over in pericentric and NOR heterochromatin justify the assumption that crossing over is suppressed in all heterochromatin regardless of its chromosomal position? While there is no definitive evidence one way or the other, the assumption is supported by a report in Drosophila virilis that little if any crossing over occurred in a block of pericentric heterochromatin that was translocated into distal euchromatin (Baker 1958). However, it is possible that centromere-related factors (such as Ctf19) remained associated with this translocated block of pericentric heterochromatin, and that this accounts for the continued suppression of crossing over. Indeed, a cause and effect relationship between condensed chromatin and suppression of crossing over is clouded by two types of observations. First, crossover suppression need not be mediated by heterochromatin. For example, centromeres without heterochromatin can suppress nearby crossing over (the “centromere effect”), as can NORs (Beadle 1932; Mather 1939; Petes and Botstein 1977; Yamamoto and Miklos 1978; Lambie and Roeder 1986; Resnick 1987; Kota et al. 1993; Choo 1998; Blitzblau et al. 2007; Lichten 2008; Vincenten et al. 2015). Furthermore, most of the maize genome is comprised of retrotransposons where little if any crossing over occurs, even though many of the retrotransposons are present in distal euchromatin (Fu et al. 2002; Yao et al. 2002). Also, in some Allium species that lack pericentric heterochromatin, crossing over is preferentially localized near centromeres and suppressed in distal euchromatin (Levan 1933; Albini and Jones 1988; Stack and Roelofs 1996). Second, heterochromatin tends to form at locations in chromosomes where crossing over is infrequent (Charlesworth et al. 1994), so it is possible that pericentric and NOR heterochromatin result from suppressed crossing over rather than cause it (Topp and Dawe 2006).
Crossing over in heterochromatin has been reported occasionally, most often from observations of chiasmata and RNs rather than by linkage mapping, because there are few genetic markers in heterochromatin and molecular markers are difficult to map in regions of low recombination (Loidl 1987; Latos-Bielenska et al. 1990; Sudman and Greenbaum 1990; Sherman and Stack 1995; Anderson et al. 2003). In most cases, chiasmata and RNs that appeared to be in heterochromatin were either illustrated without comment or interpreted as crossover events that actually occurred in euchromatin and only appeared to be in heterochromatin. In the latter case, one of the three following explanations was offered.
Chiasma terminalization. Chiasmata observed in distal heterochromatin were proposed to have formed in more proximal euchromatin and then moved distally, i.e., terminalized, until they stopped in distal heterochromatin (John 1976; Jones 1978). Chiasma terminalization was an idea popularized by Darlington (1937) to explain the apparent terminal location of chiasmata in many organisms. While chiasma terminalization was once widely accepted, it never had strong experimental support, and subsequent experiments in a variety of species using differential labeling of sister chromatids with tritiated thymidine, differential staining of sister chromatids with bromodeoxyuridine, and heterozygous subterminal C-bands provide convincing evidence that chiasmata do not terminalize (Jones 1987; Loidl 1987).
A problem with resolution. Chiasmata observed in heterochromatin were suggested to have formed in tiny islands of euchromatin within heterochromatin or in euchromatin near heterochromatin, so that subsequent chromatin condensation obscured the euchromatin (Linnert 1955; Fontana and Vickery 1974; Klášterská et al. 1974; Jones 1978; Loidl 1979; de la Torre et al. 1986; Berger and Greilhuber 1991). This is difficult to prove or disprove, and small islands of euchromatin have been observed in some heterochromatin (Linnert 1955; Ramanna and Prakken 1967). Based on the assumption that crossing over does not occur in heterochromatin, this is a plausible explanation for observations of chiasmata that appear to be in heterochromatin, but it is not evidence that crossing over does not occur in heterochromatin.
Heterochromatin stickiness. Bivalent associations in heterochromatin at late prophase I were proposed to be due not to chiasmata, but to “stickiness” of heterochromatin (Drets and Stoll 1974; Fontana and Vickery 1974; Godin and Stack 1976; John and King 1980, 1985; Cermeno 1984; Carlson 1988). Chromatin stickiness has been observed in both mitosis and meiosis, between both homologous and nonhomologous chromosomes, and between all combinations of euchromatin and heterochromatin (Rhoades 1955; Yunis and Yasmineh 1971; Godin and Stack 1975, 1976). Indeed, in Drosophila oocytes, stickiness between pairs of achiasmatic homologs substitutes for chiasmata to encourage proper segregation (Giauque and Bickel 2016). However, distinguishing between chromatin stickiness and chiasmata is often difficult because, except for a few groups of organisms (especially grasshoppers), chiasmata are not well-defined structurally (John 1976; Crolla and Polani 1989; Stack 1991). Consequently, in late prophase I, euchromatic connections between corresponding sites on homologs are generally accepted as chiasmata, while heterochromatic connections between corresponding sites on homologs may be dismissed as stickiness (Klášterská et al. 1974). Although this interpretation could be correct, it is a double standard based on the assumption that crossovers and chiasmata form only in euchromatin.
For these reasons, an investigation of the relationship between heterochromatin and crossover suppression should be carried out on homozygous dense masses of heterochromatin that lack interspersed islands of euchromatin, and which lie in euchromatin away from NORs and centromeres. Homozygous maize knobs meet these criteria because knobs are distinct masses of dense heterochromatin with no reported islands of euchromatin and knobs are located in distal, gene-rich euchromatin where crossing over is frequent (Buckler IV et al. 1999; Ghaffari et al. 2013). Knobs in maize are composed mainly of tandemly arranged blocks of a 180-bp repeat sequence interrupted by insertions of other repeated sequences including retrotransposons (Ananiev et al. 1998a).
In an earlier investigation of the distribution of RNs on maize SCs that were spread by hypotonic bursting, we found substantial numbers of RNs at the presumed sites of knobs and in euchromatin to either side of knobs (Figure 1) (Anderson et al. 2003). While we could not see knobs on spreads of maize SCs because the chromatin was dispersed, we knew where knobs were supposed to be based on the work of others (Chen et al. 2000). Because RNs occur at sites of crossing over (Carpenter 1975; Sherman and Stack 1995; Anderson et al. 2003, 2014; Marcon and Moens 2003), this result indicated that crossing over occurs along the SC in knob heterochromatin at rates similar to adjacent euchromatin. However, in deference to the assumption that crossing over is suppressed in heterochromatin, we suggested a model for the structure of homozygous knobs in which the knob on each homolog consists of a few long loops of condensed chromatin attached to a short segment of SC compared to the dimensions of the knob. The knob chromatin was proposed to mushroom over adjacent SC in euchromatin, so that most, if not all, RNs that appeared to be in knobs were really in euchromatin under an umbrella of knob heterochromatin (Figure 2).
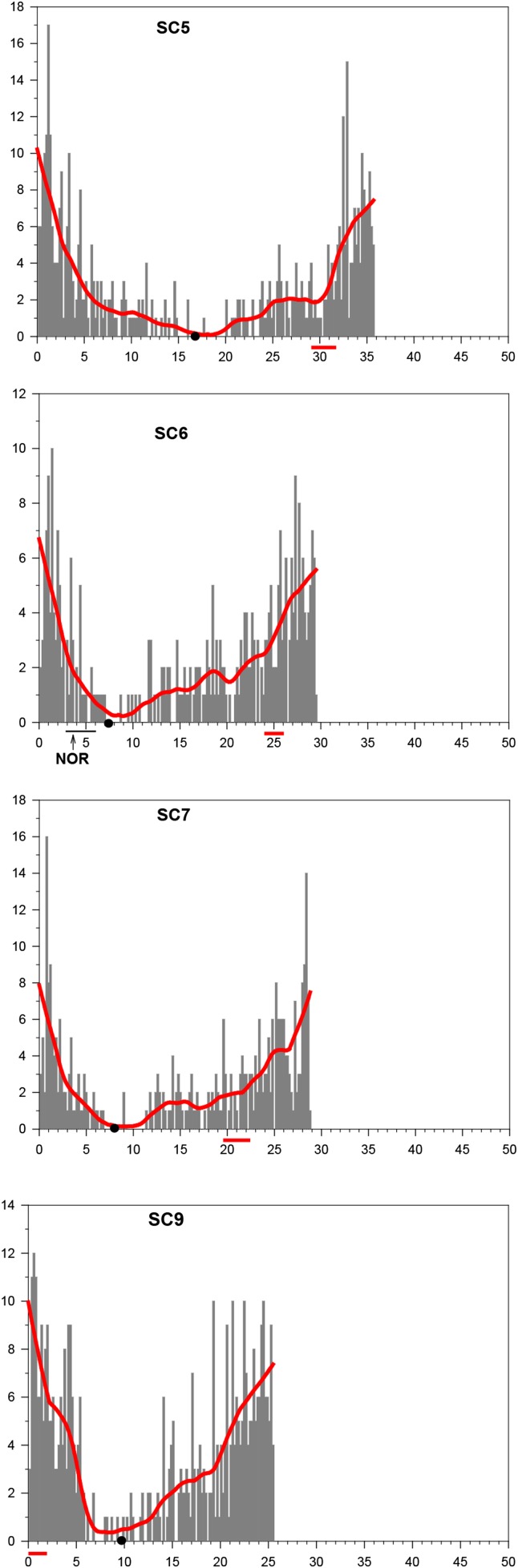
Figure 1.
Histograms showing the distribution of RNs along the length of KYS maize SCs 5, 6, 7, and 9. Each SC is represented on the x-axis in micrometers with the short arm to the left and the position of the kinetochore marked by a black dot. Beneath the x-axis, the locations of knobs are marked by horizontal, thick red lines, and the position of the NOR on SC 6 is marked by a horizontal, thin black line. The histogram bars on the y-axis represent the total number of RNs observed in each 0.2-μm segment of SC length for 203 SC 5’s, 176 SC 6’s, 178 SC 7’s, and 234 SC 9’s. A red smoothing line is superimposed over each histogram to show the general trend of RN distribution. These histograms are used with permission from GENETICS (Anderson et al. 2003). RN, recombination nodule; SC, synaptonemal complex.
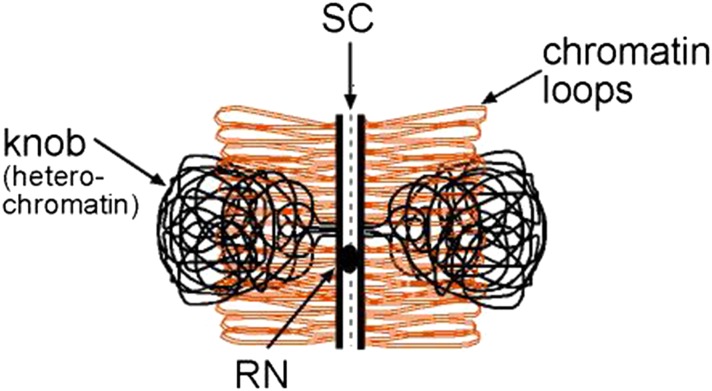
Figure 2.
Model to explain how crossing over could be suppressed in maize knob heterochromatin while RN frequencies on SC in knobs are similar to RN frequencies on SC in nearby euchromatin. In this frontal view of an SC, short, paired sister loops of euchromatin (orange) extend laterally from their regularly spaced attachment sites on lateral elements of the SC. Here, homozygous knob chromatin (black) consists of long, paired sister loops in each homolog that are collapsed to cover a length of SC well beyond their small attachment sites on the lateral elements. Thus, most of the SC covered by a knob is actually in euchromatin. A RN in the central region of the SC is shown located in euchromatin near the attachment sites of the knob loops. While this RN marks a crossover in euchromatin, it would look like the RN is in knob heterochromatin, thereby explaining our observations that RN frequencies “in knobs” are similar to RN frequencies in nearby euchromatin. This figure is used with permission from GENETICS (Anderson et al. 2003). RN, recombination nodule; SC, synaptonemal complex.
The research presented here tests our model by visualizing both knobs and crossover sites on the same spreads of SCs. This was accomplished by fluorescence in situ hybridization (FISH) of the 180-bp repeat found in maize knobs (Peacock et al. 1981; Ananiev et al. 1998a,b) and immunofluorescent labeling of maize SC lateral element protein AFD1 (Golubovskaya et al. 2006) and RN protein MLH1 to show crossover sites on SCs (Baker et al. 1996; Anderson et al. 1999, 2014). We found that long loops of knob heterochromatin attach to SCs along the entire length of knobs, not just at tiny sites, and that MLH1 foci occur on SCs within knob heterochromatin at frequencies similar to SCs in adjacent euchromatin.
Materials and Methods
Plants
KYS maize was grown in a greenhouse with controlled lighting and temperature.
Pachytene chromosome squashes
Anthers 2.0–2.2 mm in length were fixed for 1–18 hr at room temperature in freshly prepared 1:3 acetic ethanol. Immediately after clearing in 45% acetic acid, anthers were transferred to a drop of 45% acetic acid on a glass microscope slide, where they were bisected with a microscalpel. Primary microsporocytes in pachytene were squeezed out with steel dissecting needles, anther walls were removed, and the cells were gently squashed under a siliconized cover glass. The cover glass was removed by the dry ice method, and squashes were air-dried for 1 hr. For bright field microscopy, a drop of 2% aceto-orcein was placed on the squashes and covered with a cover glass. The slide was gently heated (without boiling) over an alcohol flame and then allowed to cool. The slide was inverted over 95% ethanol until the cover glass fell off. Before the ethanol dried, a cover glass was mounted using a drop of Euparal. For fluorescence microscopy, a cover glass was mounted using 15 µl of Vectashield (Vector Laboratories, Burlingame, CA) containing 5 µg/ml 4′,6-diamidino-2-phenylindole (DAPI).
Pachytene SC spreads
Details for SC spreading can be found in Stack and Anderson (2009). Briefly, 40–80 living anthers 2.0–2.2 mm in length were gently removed from florets to avoid damage, i.e., bruising and cytomixis, and placed in a depression slide containing 0.2 ml of a digestion medium that was 0.56 mM monobasic potassium phosphate, 0.8 mM CaCl2, 0.1 mM piperazine-N,N′-bis(ethane sulfonic acid) (acid PIPES), 0.2% potassium dextran sulfate (MW ∼10,000 Da), 1% polyvinylpyrrolidone, and 0.7 M mannitol, with the pH adjusted to 4.1 using 0.1 N HCl and/or 0.1 N KOH. The depression slide was incubated at 18–20° in a closed Y-Petri plate with 5–10 ml of water in the bottom. After 5 min, 3 mg of desalted, lyophilized cytohelicase (Sigma [Sigma Chemical], St. Louis, MO) and 3 mg of desalted, lyophilized pectinase (Sigma) were added. The anthers were then bisected transversely to their long axes with a microscalpel. After 45 min of incubation, the contents of several anthers were squeezed out with steel dissecting needles. Living protoplasts in ≤ 0.5 µl of digestion medium were drawn (aspirated) into a siliconized micropipette and gently blown out into 10 µl of bursting medium [aqueous 0.05%, octylphenoxy polyethoxyethanol (IGEPAL, Sigma) that was 0.01% potassium dextran sulfate and 0.04% paraformaldehyde, pH 8.5] suspended at the tip of a plastic pipette. This mixture was touched-off onto the middle of a wiped, glow-discharged glass slide where the suspension spread over the surface. A further 10 μl of bursting medium were added. After 10–20 sec, the slide was taken to a hood, and the slide’s surface was immediately given 30 passes of nebulized aqueous 4% paraformaldehyde (pH 8.5). Then the slide was air-dried in the hood for 1 hr. Slides were passed through three washes consisting first of 10 sec in 140 ml of an aqueous 0.4% solution of Photo-Flo 200 with a drop of aqueous 0.05 M sodium borate added, followed by two 10 sec washes in 140 ml of distilled water. Slides were air-dried for 1 hr before storage at −80° in sealed plastic slide boxes.
Isolation of KYS maize genomic DNA
Young shoot tissue (100 mg) was diced into ∼0.5 cm fragments, placed in a 1.7 ml microtube containing 500 µl of freshly prepared extraction buffer (200 mM Tris-HCl, pH 7.5; 25 mM ethylenediaminetetraacetic acid [EDTA], pH 8.0; 250 mM NaCl; 0.5% SDS; and 5 mM dithiothreitol), and stored at −80°. Subsequently, the suspension was heated to 65° for 10 min and then gently macerated for 15–20 sec in the microtube using a disposable pestle. The suspension was centrifuged at maximum speed (16,000 rpm) in a microcentrifuge at room temperature for 10 min. The supernatant was mixed with an equal volume of isopropanol to precipitate DNA. The suspension was again centrifuged at maximum speed at room temperature. The supernatant was removed, and the pellet was cleaned with 70% ethanol and dried. The dried pellet was suspended in 100 µl of sterile deionized water and treated with 100 µg/ml RNase A for 5 min at room temperature before the DNA was precipitated with 70% ethanol at −20° for 10 min. After centrifugation, the pellet was resuspended in 200 µl of deionized water. The DNA was quantified and adjusted to a final concentration of 100 ng/µl.
Preparing the FISH probe for knobs
The 180-bp knob repeat was amplified and labeled according the manufacturer’s instructions using the Roche PCR DIG Labeling Mix, maize genomic DNA, and appropriate primers (F: 5ʹ-CAACGCCCATTTTTATCGAA-3ʹ, R: 5ʹ-CGACCAGAGGATCGTACACC-3ʹ) to yield a 164-bp digoxygenin-labeled PCR product. The probe was purified according to the manufacturer’s instructions using a Marligen PCR cleanup kit. SC spreads on glass microscope slides were treated in one of two ways (see below).
Silver staining:
Air-dried SC spreads on slides were fixed for 10 min in aqueous 4% paraformaldehyde pH 8.5 and then passed through three washes, consisting first of 10 sec in 140 ml of an aqueous 0.4% solution of Photo-Flo 200 to which one drop of aqueous 0.05 M sodium borate was added, followed by two 10 sec washes in 140 ml of distilled water. Slides were air-dried overnight at room temperature. Then a ∼25 × 50 mm rectangle of Tetko nylon screen was placed over the SC spreads, and two drops of aqueous 33% silver nitrate was added. The slide was placed in a Y-Petri dish so it was resting above 5 ml of water in the bottom of the dish. The petri dish was closed and floated in a closed water bath at 40° until the nylon screen turned light brown (∼25 min). The nylon screen was gently washed off with distilled water, and the slide was allowed to air-dry.
Immunolabeling:
Air-dried SC spreads on slides were immunolabeled using the procedure described by Lohmiller et al. (2008) with affinity-purified rat antibodies to maize (Zea mays) AFD1 protein diluted 1:50 and affinity-purified rabbit antibodies to tomato (Solanum lycopersicum) MLH1 protein diluted 1:50, followed by goat anti-rat 549 and goat anti-rabbit 488 (both from Jackson ImmunoResearch Laboratories), respectively, each at 1:500 dilution. Primary antibody incubations were done overnight at 4°, and secondary antibody incubations were done for 2 hr at 37°. The slides were stained with aqueous DAPI (10 µg/ml), washed with Tris-buffered saline (TBS) containing 0.05% Triton X, and cover glasses were mounted with Vectashield antifade mounting medium (Vector Laboratories). Slides were imaged before FISH, as described below.
FISH
FISH was performed as described (Zhong et al. 1996; Chang et al. 2007) on pachytene chromosome squashes, on SC spreads that were silver-stained and previously imaged, and on SC spreads that were immunostained with AFD1 and MLH1 and previously imaged. The cover glass and Vectashield mountant was removed from immunostained slides in deionized water, and then the slides were air-dried. For FISH, slides were incubated for 1 min in 45% v/v acetic acid, fixed with 1:3 acetic ethanol for 1 min, washed briefly in deionized water, and then digested with 100 µg/ml aqueous RNase A for 5 min at room temperature followed by pepsin (5 µg/ml in 0.01 M HCl) for 8 min at 37°. After briefly rinsing the slides in deionized water followed by 2 × SSC, the slides were fixed for 10 min in 1% w/v paraformaldehyde at pH 8.5, washed three times for 3 min each time in 2 × SCC, briefly rinsed in deionized water, dehydrated in 100% ethanol for 3 min at room temperature, and air-dried. Then, 20 µl of hybridization mixture [aqueous 2 × SSC containing 100 ng of labeled probe, 50% (v/v) formamide, 10% (w/v) sodium dextran sulfate, and 0.25% (w/v) sodium dodecyl sulfate] was placed on the slide, and a cover glass was added. Slides were heated for 2.5 min on an aluminum block at 80° to denature the DNA, and then slides were incubated at 37° in a moist chamber for at least 12 hr to permit hybridization. Slides were washed three times in aqueous 2 × SSC that was 50% v/v formamide at 42° for 80% stringency (Schwarzacher and Heslop-Harrison 2000). Slides were labeled with sheep anti-digoxygenin (Roche) followed by donkey anti-sheep conjugated to tetra-methyl rhodamine isothiocyanate (TRITC) (Jackson ImmunoResearch Laboratories) diluted 1:125 and 1:100, respectively, in 5% aqueous Roche blocking reagent. Antibody incubations were performed at 37° in 1-hr increments with three 3-min washes in an aqueous solution that was 100 mM Tris, 150 mM NaCl, and 0.05% v/v Tween-20, pH 7.5, after each incubation step. Donkey serum (5%) was added to the blocking buffer during the second secondary antibody incubation. After immunolabeling, slides were dehydrated through an ethanol series and air-dried. Cover glasses were mounted with Vectashield (Vector Laboratories) containing DAPI at 5 µg/ml.
Imaging
Images of partial and complete SC sets were captured using IP Lab software (version 4) using a 100 × 1.4 numerical aperture (NA) apochromatic objective and a cooled Hamamatsu monochrome 1344X1044 pixel camera attached to a Leica DM 5500B microscope with a Prior motorized stage. The microscope was equipped for bright field, phase contrast, and fluorescence microscopy, with zero pixel shift filter cubes for DAPI, FITC, and TRITC.
Measurements
The stage coordinates of individual nuclei were recorded so that the same nucleus could be imaged sequentially after air-drying, silver staining, fluorescence immunolabeling, and/or FISH, as appropriate. Images of the same nuclei were overlaid using CS2 Adobe Photoshop, but the layers were kept separate for later analysis. For each interstitial knob, the MLH1 layer alone was also examined to visualize and mark even dim MLH1 loci that are often difficult, if not impossible, to see in merged images. Bitmap images of the merged layers of the same nuclei were used to make measurements of SC lengths and mark the positions of kinetochores, knobs, and MLH1 foci using MicroMeasure (http://rydberg.biology.colostate.edu/MicroMeasure).
Data availability
Complete imaging data sets and reagents are available upon request.
Results
Hybridization of the 180-bp knob repeat
Using traditional pachytene squashes, pericentric heterochromatin and knobs in distal euchromatin are visible by both phase contrast microscopy and by fluorescence microscopy after staining DNA with DAPI (Figure 3, A and B). FISH using the 180-bp knob repeat demonstrated that the probe hybridized selectively to all five of the knobs observed in KYS primary microsporocytes (Figure 3C). Three of the knobs are located interstitially in the long arms of chromosomes 5, 6, and 7, and two of the knobs are located terminally on the short arms of chromosomes 1 and 9 (McClintock et al. 1981; Kato et al. 2004).
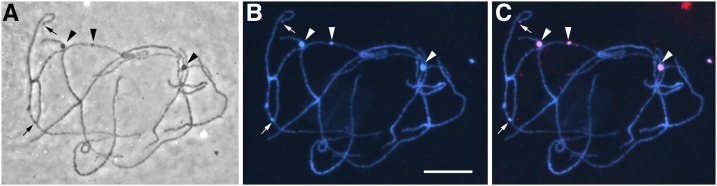
Figure 3.
KYS maize pachytene chromosome squash. (A) By phase microscopy, large interstitial knobs (arrowheads) on the unstained chromosomes are visible along with smaller terminal knobs (small arrows). Pericentric heterochromatin is visible as darker segments of bivalents that are often about one-third of the length of each bivalent, e.g., observe the bivalent at the lower center. (B) The same squash after DAPI staining. Knobs are prominent bright blue enlargements along bivalents indicating a high concentration of DNA in knobs. Pericentric heterochromatin is also brighter than the distal euchromatin indicating higher concentrations of DNA in pericentric heterochromatin (but less than in knobs). (C) The same squash after FISH with the 180-bp knob repeat (red). Large interstitial knobs (arrowheads) and the smaller terminal knobs (small arrows) are specifically labeled with the 180-bp repeat probe. Bar, 5 µm.
We first tested whether maize knobs are attached to segments of SC that are short compared to the length of their corresponding knobs, as proposed in our model (Figure 2). For this, maize SC spreads were stained with silver to reveal SCs as long, dark threads with distinct kinetochores (Figure 4A). This permitted each of the 10 maize SCs to be identified on the basis of its relative length and arm ratio (Anderson et al. 2003). Next, DAPI staining showed a diffuse blue sheath around each SC as a result of the hypotonic dispersion of chromatin loops (Figure 4B). Finally, FISH using the 180-bp knob repeat showed numerous loops of knob chromatin (red) extending far from their distinct attachment sites on the SCs. At least some of these knob loops are a great deal longer than the blue-stained chromatin loops extending from the nonknob parts of the SCs (Figure 4, C and D). The interstitial knobs on chromosomes 5, 6, and 7 consisted of multiple loops of chromatin attached along well-defined segments of SC that averaged 1.7, 0.8, and 1.7 µm in length, respectively (Table 1). Measurements of interstitial knobs on traditional squashed pachytene chromosomes stained with DAPI or aceto-orcein were similar in length (0.5–2.0 µm). Thus, the length of SC occupied by these interstitial knobs in SC spreads is similar to the length of the same knob observed in a squash. We could not make similar comparisons for the terminal knobs on SCs 1 and 9 because the loops appeared to extend from the ends of these SCs (Figure 4, C and D).
Figure 4.
Maize pachytene SC spread. (A) SCs and kinetochores stained with silver. Each SC has been identified on the basis of relative length and arm ratio, and numbered at its kinetochore. Kinetochores on SCs 1 and 7 are stuck together as are the kinetochores on SCs 2 and 5. SC 8 is partially asynapsed. (B) The same SC spread visualized with DAPI fluorescence. Due to the hypotonic spreading procedure, the chromatin loops around the SCs are dispersed laterally as a fuzzy blue coat along the length of each SC. (C) The FISH signal (red) of the 180-bp repeat for this SC spread. (D) Merged images of silver-stained SCs (inverted to appear white), DAPI-stained chromatin (blue), and the 180-bp knob repeat (red). The chromatin of the interstitial knobs (on the long arms of SCs 5, 6, and 7) occupies well-defined SC segments, and at least some chromatin loops from the knobs (red) extend much farther from their SC attachment sites than chromatin loops in distal euchromatin and pericentric heterochromatin. For the terminal knobs (on the short arms of SCs 1 and 9) the loops also extend far from their attachment sites, but the loops appear to extend from the tips of the SCs without defining distinct terminal SC segments. Bar, 5 µm. SC, synaptonemal complex.
Table 1. Position and length of interstitial heterochromatic knobs on the long arms of hypotonically spread SCs 5, 6, and 7.
| SC ID | Number Observed SC sets | Average Length (µm) | Average Knob Position on the Long Arm as Measured from the Centromere | Average Length (µm) of Knob on SC (SD) | ||||
|---|---|---|---|---|---|---|---|---|
| SC | Long Arm | Proximal Edge | Distal Edge | |||||
| % | µm | % | µm | |||||
| 5 | 6 | 37.0 | 19.2 | 62.9 | 12.0 | 71.7 | 13.7 | 1.7 (0.2) |
| 6 | 6 | 30.8 | 22.2 | 72.8 | 16.1 | 76.2 | 16.9 | 0.8 (0.3) |
| 7 | 6 | 30.8 | 22.5 | 63.5 | 14.3 | 71.0 | 16.0 | 1.7 (0.3) |
SC, synaptonemal complex; ID, identifier.
Localization of MLH1 foci on SCs in relation to knobs localized by FISH
Next, we examined spread maize SCs that had been labeled with antibodies to maize AFD1 protein and crossover sites on these SCs that had been labeled with antibodies to tomato MLH1 protein (Golubovskaya et al. 2006; Lhuissier et al. 2007). Most SCs have one or two MLH1 foci, and we found an average of 13.9 MLH1 foci per SC set (n = 18) (Figure 5). Individual MLH1 foci varied in brightness, possibly depending on the amount of the MLH1 protein present in individual RNs (Anderson et al. 2014). Using the 180-bp knob repeat for FISH on these previously immunolabeled SC spreads allowed us to compare the location of knobs and MLH1 fluorescent foci simultaneously on the same SCs (Figure 5). Unfortunately, the immunolocalization procedure for AFD1 and MLH1 that preceded FISH blurred the edges of knobs compared with their appearance without the immunolocalization procedure (compare Figure 4, C and D to Figure 5). Even so, we observed MLH1 foci near and apparently within interstitial knobs on the long arms of SCs 5, 6, and 7. We observed a total of 255 SCs with interstitial knobs (Table 2) and MLH1 foci. Of the total 367 MLH1 foci observed on these SCs, 25 MLH1 foci were near a knob (defined as within 1 µm to either side of a knob) and 16 MLH1 foci were within a knob. Using data on average SC lengths plus the average SC length occupied by the knobs on these chromosomes (Table 1), we calculated frequencies of MLH1 foci/μm for whole SCs, in knobs, and near knobs (1 µm in euchromatin to either side of the knob = 2 µm SC length for each SC) (Table 2). We found similar frequencies of MLH1 foci/μm for each SC (0.042–0.046). For MLH1 foci in knobs, frequencies ranged from 0.026 to 0.056 µm−1, and for MLH1 foci near knobs frequencies ranged from 0.039 to 0.056 µm−1 (Table 2). While the frequencies of MLH1 foci within and near knobs for SCs 6 and 7 are close (0.052 vs. 0.056 µm−1 and 0.056 vs. 0.053 µm−1, respectively), the frequencies of RNs in and near the knob on SC 5 are lower (0.026 and 0.039 µm−1, respectively.
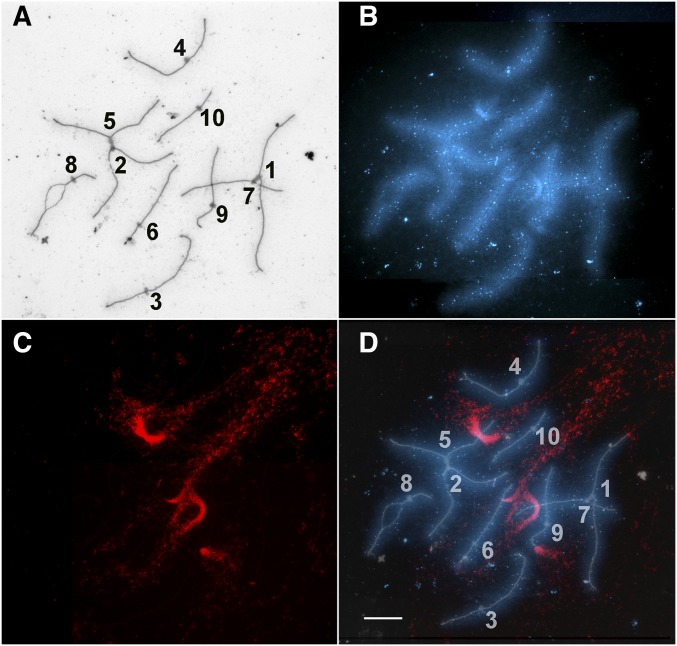
Figure 5.
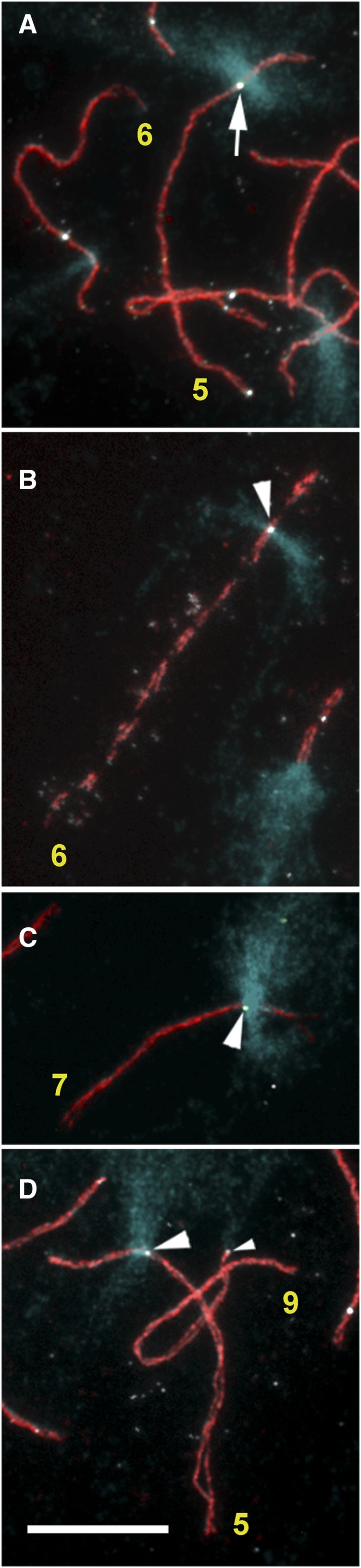
Portions of maize SC spreads labeled with antibodies to MLH1 (white foci) and AFD1 (red, to show the SC axes) and hybridized to the 180-bp knob repeat (blue). MLH1 foci are observed near (arrow) and in (arrowheads) knobs. (A) Portion of a pachytene SC spread showing an MLH1 focus near the edge of the knob on SC 5. Other MLH1 foci in this image are not in or near knobs. (B) Portion of an SC spread in early diplotene showing an MLH1 focus in the interstitial knob of the long arm of SC 6. (C) Portion of a pachytene SC spread showing an MLH1 focus in the interstitial knob on the long arm of SC 7. (D) Portion of a pachytene SC spread showing an MLH1 focus in the interstitial knob of SC 5 and a small MLH1 focus at the tip of the short arm of SC 9 (small arrowhead) where the terminal knob is located. Bar, 5 µm. SC, synaptonemal complexes.
Table 2. Number of MLH1 foci on SCs in or near interstitial knob heterochromatin for SCs 5, 6, and 7.
| SC ID | Total Number of SCs Observed | Number MLH1 Foci | Number of MLH1 Foci per Micrometer of SC Length | ||||
|---|---|---|---|---|---|---|---|
| SC | Near Knoba | In Knob | SC | Near Knoba | In Knob | ||
| 5 | 89 | 139 | 7 | 4 | 0.042 | 0.039 | 0.026 |
| 6 | 72 | 96 | 8 | 3 | 0.043 | 0.056 | 0.052 |
| 7 | 94 | 132 | 10 | 9 | 0.046 | 0.053 | 0.056 |
| Total | 255 | 367 | 25 | 16 | |||
SC, synaptonemal complex; ID, identifier.
Near is defined as within 1 µm of SC to either side of the knob or a total of 2 µm of SC length.
In addition, we observed 92 MLH1 foci on 52 SC 1s and found only one MLH1 focus at the site of the small terminal knob, and we observed 94 MLH1 foci on 73 SC 9s and found three MLH1 foci at the site of SC 9’s larger terminal knob (Figure 5D). Because we were unable to measure the length of SC in these terminal knobs, we were also unable to determine frequencies of MLH1 foci/μm of SC for these knobs.
Discussion
Our earlier observations of RNs on KYS maize SCs indicated that crossing over occurs in knob heterochromatin at frequencies similar to the surrounding euchromatin (Figure 1) (Anderson et al. 2003). However, this work had the potential problem that knobs were not observed directly on the SC spreads used for RN mapping, so the exact location and size of knobs had to be inferred from other data. However, even if the projected knob locations on SCs were not precisely correct, it was clear that there were no 0.5–2.0 µm segments of SCs near knob sites that lacked RNs, as would be expected if knob heterochromatin blocked meiotic crossing over. Therefore, we offered a model for knob structure that would make our observations compatible with the assumption that crossing over is suppressed in heterochromatin. In this model, a knob consists of a few long loops of condensed chromatin, which have a tiny site of insertion on the SC compared to the size of the knob (Figure 2).
In the research described here, we used FISH of the maize 180-bp knob repeat to determine the location of knobs on SC spreads and the length of SC into which loops of knob heterochromatin were inserted. In agreement with our model, DAPI staining and FISH showed that many (possibly all) of the hypotonically dispersed loops of knob chromatin are much longer than typical hypotonically dispersed nonknob chromatin loops (Figure 4). However, in disagreement with our model, chromatin loops of the interstitial knobs on SCs 5, 6 and 7 did not have tiny insertion sites but were inserted all along well-defined SC segments that were similar in length to the corresponding knobs measured on squashed pachytene chromosomes (Results and Table 1). The observation that knob chromatin occupies a significant segment of SC length is also consistent with earlier reports that knobs increase the length of pachytene chromosomes by the length of the knobs, and with reports that knobs buckle out in heterozygotes, presumably due to the different lengths of SC lateral elements (Longley 1937; Dempsey 1994). Thus, our earlier report (Anderson et al. 2003) that RNs occur at presumed heterochromatic knob sites in maize together with our current observation that loops of knob chromatin are inserted all along the length of interstitial knobs strongly suggests that meiotic crossing over occurs regularly within these knobs.
We also used immunofluorescent localization of AFD1 protein and MLH1 protein, and FISH localization of the 180-bp knob repeat, to directly observe the locations of MLH1 foci and knobs on the same SCs. All of the interstitial knobs on SCs 5, 6, and 7 occur within distal euchromatin. The frequency of MLH1 foci/μm was essentially the same in and to either side of the knobs on SCs 6 and 7 (0.052–0.056 MLH1 foci/μm of SC), indicating that these knobs have no effect on the rate of crossing along the SC (Table 2). For SC 5, the frequency of MLH1 foci/μm of SC is lower in the knob than to either side of the knob (0.026 vs. 0.039 MLH1 foci/μm of SC, respectively), and both of these values are lower than the frequencies for the knobs on SCs 6 and 7. However, because of the small sample sizes, it is unclear whether these values represent real differences between the knobs, but in support of a difference, there is a dip in RN frequency at the knob site on SC 5 in Figure 1. From the data, we conclude that crossing over occurs in all of these knobs, and the knobs either have no or only a modest effect on the frequency of crossing over along SC nearby or within their boundaries.
Crossovers visualized by MLH1 foci represent type I crossovers, i.e., those that show interference (Mercier et al. 2015). In maize, mathematical modeling indicated that about 80% of the crossovers as assessed by RNs are type I crossovers with the remaining RNs (not labeled by MLH1) being type II crossovers, i.e., those that do not show interference (Falque et al. 2009; Mercier et al. 2015). Based on these results and an average of about 20 RNs per nucleus (Anderson et al. 2003), one would expect an average of about (0.8 × 20) 16 MLH1 foci per nucleus in maize. Instead, we observed an average of about 14 MLH1 foci per nucleus. While it is not clear why fewer MLH1 foci were observed than expected, possible explanations include transient detection of MLH1 foci as observed for mice (Anderson et al. 1999), a lower efficiency of immunolabeling (perhaps caused by using antibodies raised against tomato MLH1 protein), and/or an error in modeling. In any case, the lower than expected number of MLH1 foci could indicate that we may be underestimating the total number of type I crossovers. In addition, type II crossovers that may comprise 20% of all crossovers are not detected by MLH1 antibodies, so the total amount of crossing over in and near knobs may be ∼20% higher than we observed based on MLH1 foci (Table 2). Together, these results indicate that the reported frequencies of MLH1 foci in and around knobs are probably minimal estimates of crossing over, and that the actual amount of crossing over in these locations may be substantially higher. Furthermore, in tomato, type I and type II crossovers have different distributions, with type II crossovers being observed more often than expected in the short arms of acrocentric chromosomes and in pericentric heterochromatin compared to type I crossovers (Anderson et al. 2014). If the same pattern applies to maize, it could explain why only a few MLH1 foci were observed at the ends of the terminal knob-carrying short arms of SC 1 and SC 9, with most of the many RNs observed at those positions presumably representing type II crossovers (Figure 1) (Anderson et al. 2003).
We have shown that the rate of crossing over per unit length of SC is not suppressed in knob heterochromatin compared to euchromatin. On the other hand, chromatin loops in knobs are much longer than chromatin loops in euchromatin, and assuming that the chromatin loops occur at the same frequency per micrometer of SC in euchromatin and heterochromatin (Zickler and Kleckner 1999), there must be more DNA per micrometer of SC in knobs compared to SC in euchromatin. If so, there must be proportionately less crossing over per unit length (base pairs) of DNA in knobs compared to nearby euchromatin. A similar effect of DNA loop length on crossover frequency was suggested earlier based on observations in other species (Stack 1984). To date, no accurate measurements have been made of the amount of DNA in maize knobs. The most similar comparison comes from the work of Peterson et al. (1996), who used Feulgen staining and microdensitometry of tomato pachytene chromosomes to estimate that there is at least fivefold more DNA per micrometer of SC in pericentric heterochromatin compared to distal euchromatin. If this estimate is applied to maize knobs, the crossover rate per base pair of knob DNA would be only one-fifth the crossover rate per base pair of DNA in nearby euchromatin. This ratio of crossovers is similar to that reported for euchromatin compared to knob-like heterochromatin in Arabidopsis thaliana (The Cold Spring Harbor Laboratory, Washington University Genome Sequencing Center, and PE Biosystems Arabidopsis Sequencing Consortium 2000; Fransz et al. 2000). Thus, it seems that whether or not crossing over is suppressed in knobs compared to euchromatin depends on the context: crossing over is suppressed in the context of DNA amount but not suppressed in the context of SC length.
Knob heterochromatin has been reported to influence recombination and preferential segregation, but no genes have been localized in maize knobs, and for that reason knobs do not appear on linkage maps (Rhoades 1978; Ananiev et al. 1998b; Ghaffari et al. 2013). Recently, Ghaffari et al. (2013) were able to map three knobs in gene-rich regions onto the reference genome for maize B73. Ghaffari et al. (2013) observed a reduction in crossing over in markers closely linked to a small knob in the homozygous condition, although without accurate estimates of the amount of DNA in the knob they were unable to quantify the reduction on a centiMorgan per megabase (cM/Mb) scale. For knobs in the heterozygous condition (knob/knobless), they found as much as a twofold reduction in euchromatic crossing over in cM/Mb near knob sites. However, given previous evidence that heterozygous knobs often cause local asynapsis, this reduction of crossing over is more likely due to interference with synapsis than to the effect of knob heterochromatin per se (Rhoades and Dempsey 1966; Rhoades 1978; Dempsey 1994; Fransz et al. 2000). So far, there is no molecular evidence for crossing over in knobs, but the variability in the sizes of knobs within maize populations (Kato 1976), “suggests recombination and unequal crossing over is frequent within knobs…” (Buckler IV et al. 1999). More generally, changes in the number of tandem repeats, such as those found in knobs, are often explained by unequal crossing over, although such changes would not necessarily have to occur during meiosis (Smith 1976; Yamamoto and Miklos 1978). Recent sequencing of the special centromeric chromatin in the B73 maize chromosome 10 indicates that it has been involved in many double-strand break-induced rearrangements (including inversions that have moved three genes into this centromere). However, because the rearrangements appear to be mediated by microhomology that typically involves ∼5 bp overlaps, it is unlikely that they are due to homologous meiotic recombination that involves matching sequences hundreds of base pairs long (Paques and Haber 1999; Wolfgruber et al. 2016). With further improvements in sequencing difficult genomic regions, it may become possible to determine whether similar types of rearrangements also occur in knob heterochromatin and whether they are mediated by meiotic homologous recombination.
Our observations of RNs at knob sites (Anderson et al. 2003) and of MLH1 foci in knobs (this work) do not necessarily mean that crossing over is occurring in the 180-bp tandem repeats that comprise the bulk of DNA in most maize knobs. Other sequences have been observed in knobs, including retrotransposons and 350-bp TR-1 element repeats that are related to the 180-bp repeats (Ananiev et al. 1998a; Ghaffari et al. 2013; Wolfgruber et al. 2016). In addition, maize genome assemblies are not good enough to rule out the presence of other interspersed low copy sequences scattered among 180-bp knob repeats. Thus, our results indicate only that crossing over is occurring within knob heterochromatin, but not which sequences are involved in crossing over or in attachment of knob loops to SC lateral elements.
Conclusions
Although there is no doubt that crossing over is suppressed in some heterochromatin, it has not been clear whether all heterochromatin suppresses crossing over. Considering the multiple factors involved in crossing over (Ellermeier et al. 2010; Vader et al. 2011; Vincenten et al. 2015) and that there are different types of heterochromatin based on behavior (constitutive vs. facultative), staining characteristics, degrees of condensation, chemical modifications, and various constituents and locations (Loidl 1987; Carlson 1988; Craig 2004; Roudier et al. 2011; van Steensel 2011), it should not be surprising that different heterochromatins could vary in the characteristic of meiotic crossing over. Based on our observations of RNs and MLH1 foci in maize knobs, it seems likely that crossing over occurs at a similar rate per unit length of SC within knobs and in euchromatin to either side of knobs. However, because there is more DNA per unit length of SC within a knob compared to SC in adjacent euchromatin, there is proportionately less crossing over per base pair within knobs compared to adjacent euchromatin (Gaut et al. 2007). Thus, crossing over in knob heterochromatin is not suppressed in the context of SC length, while crossing over is suppressed, but not eliminated, in the context of DNA length.
Acknowledgments
We thank Zac Cande for the antibodies to maize AFD1 protein and Song-Bin Chang for selecting the primers for PCR amplification of the 180-bp maize knob repeat sequence.
Footnotes
Communicating editor: J. A. Birchler
Literature Cited
- Albini S. M., Jones G. H., 1988. Synaptonemal complex spreading in Allium cepa and Allium fistulosum. II. Pachytene observations: the SC karyotype and the correspondence of late recombination nodules and chiasmata. Genome 30: 399–410. [Google Scholar]
- Ananiev E. V., Phillips R. L., Rines H. W., 1998a Complex structure of the knob DNA on maize chromosome 9: retrotransposon invasion into heterochromatin. Genetics 149: 2025–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananiev E. V., Phillips R. L., Rines H. W., 1998b A knob-associated tandem repeat in maize capable of forming fold-back DNA segments: are chromosome knobs megatransposons? Proc. Natl. Acad. Sci. USA 95: 10785–10790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L. K., Reeves A., Webb L. M., Ashley T., 1999. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 151: 1569–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L. K., Doyle G. G., Brigham B., Carter J., Hooker K. D., et al. , 2003. High resolution crossover maps for each bivalent of Zea mays using recombination nodules. Genetics 165: 849–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L. K., Lohmiller L. D., Tang X., Hammond D. B., Javernick L., et al. , 2014. Combined fluorescent and electron microscopic imaging unveils the specific properties of two classes of meiotic crossovers. Proc. Natl. Acad. Sci. USA 111: 13415–13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S. M., Plug A. W., Prolla T. A., Bronner C. E., Harris A. C., et al. , 1996. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat. Genet. 13: 336–342. [DOI] [PubMed] [Google Scholar]
- Baker W. K., 1958. Crossing over in heterochromatin. Am. Nat. 92: 59–60. [Google Scholar]
- Beadle G. W., 1932. A possible influence of the spindle fibre on crossing-over in Drosophila. Proc. Natl. Acad. Sci. USA 18: 160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., 2000. The many hues of plant heterochromatin. Genome Biol. 1: 107.1–107.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger R., Greilhuber J., 1991. C-band and chiasma distribution in Scilla siberica (Hyacinthaceae). Genome 34: 179–189. [Google Scholar]
- Blitzblau H. G., Bell G. W., Rodriguez J., Bell S. P., Hochwagen A., 2007. Mapping of meiotic single-stranded DNA reveals double-strand-break hotspots near centromeres and telomeres. Curr. Biol. 17: 2003–2012. [DOI] [PubMed] [Google Scholar]
- Brown S. W., 1949. The structure and meiotic behavior of the differentiated chromosomes of tomato. Genetics 34: 437–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler E. S., IV, Phelps-Durr T. L., Buckler C. S. K., Dawe R. K., Doebley J. F., et al. , 1999. Meiotic drive of chromosomal knobs reshaped the maize genome. Genetics 153: 415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson W. R., 1988. The cytogenetics of corn, pp. 259–331 in Corn and Corn Improvement, edited by Sprague G. F., Dudley J. W. Crop Science Society, Madison, WI. [Google Scholar]
- Carpenter A. T. C., 1975. Electron microscopy of meiosis in Drosophila melanogaster females: II: the recombination nodule - a recombination-associated structure at pachytene? Proc. Natl. Acad. Sci. USA 72: 3186–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castiglioni P., Ajmone-Marsan P., van Wijk R., Motto M., 1999. AFLP markers in a molecular linkage map of maize: codominant scoring and linkage group distribution. Theor. Appl. Genet. 99: 425–431. [DOI] [PubMed] [Google Scholar]
- Cermeno M. C., 1984. Evidence of nonchiasmate bonds at metaphase I in inbred rye. Can. J. Genet. Cytol. 26: 409–414. [Google Scholar]
- Chang S. B., Anderson L. K., Sherman J. D., Royer S. M., Stack S. M., 2007. Predicting and testing physical locations of genetically mapped loci on tomato pachytene chromosome 1. Genetics 176: 2131–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Sniegowski P., Stephan W., 1994. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371: 215–220. [DOI] [PubMed] [Google Scholar]
- Chen C. C., Chen C. M., Hsu F. C., Wang C. J., Yang J. T., et al. , 2000. The pachytene chromosomes of maize as revealed by fluorescence in situ hybridization with repetitive DNA sequences. Theor. Appl. Genet. 101: 30–36. [Google Scholar]
- Choo K. H. A., 1998. Why is the centromere so cold? Genet. Res. 8: 81–82. [DOI] [PubMed] [Google Scholar]
- Civardi L., Xia Y., Edwards K., Schnable P. S., Nikolau B. J., 1994. The relationship between genetic and physical distances in the cloned al-sh2 interval of Zea mays L. genome. Proc. Natl. Acad. Sci. USA 91: 8268–8272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comings D. E., 1972. The structure and function of chromatin, pp. 237–431 in Advances in Human Genetics, edited by Harris H., Hirschhorn K. Plenum Press, New York. [DOI] [PubMed] [Google Scholar]
- Craig J. M., 2005. Heterochromatin–many flavours, common themes. Bioessays 27: 17–28. [DOI] [PubMed] [Google Scholar]
- Crolla J. A., Polani P. E., 1989. Meiosis in trisomic female mice with Robertsonian translocations II. Chromosome behavior at first and second meiotic prophases. Cytogenet. Cell Genet. 52: 118–123. [DOI] [PubMed] [Google Scholar]
- Darlington C. D., 1937. Recent Advances in Cytology. The Blakiston Company, Philadelphia. [Google Scholar]
- de la Torre J., Lopez-Fernandez C., Nichols R., Gosalvez J., 1986. Heterochromatin readjusting chiasma distribution in two species of the genus Arcyptera: the effect among individuals and populations. Heredity 56: 177–184. [Google Scholar]
- Dempsey E., 1994. Traditional analysis of maize pachytene chromosomes, pp. 432–441 in The Maize Handbook, edited by Freeling M., Walbot V. Springer-Verlag, New York. [Google Scholar]
- Drets M. E., Stoll M., 1974. C-banding and non-homologous associations in Gryllus argentinus. Chromosoma 48: 367–390. [DOI] [PubMed] [Google Scholar]
- Dumas D., Britton-Davidian J., 2002. Chromosomal rearrangements and evolution of recombination: comparison of chiasma distribution patterns in standard and Robertsonian populations of the house mouse. Genetics 162: 1355–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier C., Higuchi E. C., Phadnis N., Holm L., Geelhood J. L., et al. , 2010. RNAi and heterochromatin repress centromeric meiotic recombination. Proc. Natl. Acad. Sci. USA 107: 8701–8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falque M., Anderson L. K., Stack S. M., Gauthier F., Martin O., 2009. Two types of meiotic crossovers coexist in maize. Plant Cell 21: 3915–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana P. G., Vickery V. R., 1974. Heterochromatin content and chiasma distribution in the megameric chromosomes of Stethophyma gracile and S. lineatum (Orthoptera: Acrididae). Chromosoma 46: 375–395. [DOI] [PubMed] [Google Scholar]
- Fransz P. F., Armstrong S., de Jong J. H., Parnell L. D., van Drunen C., et al. , 2000. Integrated cytogenetic map of chromosome arm 4S of A. thaliana: structural organization of heterochromatic knob and centromere region. Cell 100: 367–376. [DOI] [PubMed] [Google Scholar]
- Froenicke L., Anderson L. K., Weinberg J., Ashley T., 2002. Male mouse recombination maps for each autosome identified by chromosome painting. Am. J. Hum. Genet. 71: 1353–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H., Zheng Z., Dooner H. K., 2002. Recombination rates between adjacent genic and retrotransposon regions in maize vary by 2 orders of magnitude. Proc. Natl. Acad. Sci. USA 99: 1082–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut B. S., Wright S. I., Rizzon C., Dvorak J., Anderson L. K., 2007. Recombination: an underappreciated factor in the evolution of plant genomes. Nat. Rev. Genet. 8: 77–84. [DOI] [PubMed] [Google Scholar]
- Ghaffari R., Cannon E. K. S., Kanizay L. B., Lawrence C. J., Dawe R. K., 2013. Maize chromosomal knobs are located in gene-dense areas and suppress local recombination. Chromosoma 122: 67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giauque C. C., Bickel S. E., 2016. Heterochromatin-associated proteins HP1a and Piwi collaborate to maintain the association of achiasmate homologs in Drosophila oocytes. Genetics 203: 173–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin D. E., Stack S. M., 1975. Heterochromatic connectives between the chromosomes of Secale cereale. Can. J. Genet. Cytol. 17: 269–273. [Google Scholar]
- Godin D. E., Stack S. M., 1976. Homologous and non-homologous chromosome associations by interchromosomal chromatin connectives in Ornithogalum virens. Chromosoma 57: 309–318. [Google Scholar]
- Golubovskaya I. N., Hamant O., Timofejeva L., Wang C. J. R., Braun D., et al. , 2006. Alleles of afd1 dissect Rec8 functions during meiotic prophase I. J. Cell Sci. 119: 3306– 3315. [DOI] [PubMed] [Google Scholar]
- Gore M. A., Chia J. M., Elshire R. J., Sun Q., Ersoz E. S., et al. , 2009. A first-generation haplotype map of maize. Science 326: 1115–1117. [DOI] [PubMed] [Google Scholar]
- Green M. M., 1966. The effects of autosomal euchromatin on crossing over in the X chromosome of Drosophila melanogaster. Am. Nat. 100: 159–162. [Google Scholar]
- Grewal S. I. S., Jia S., 2007. Heterochromatin revisited. Nat. Rev. Genet. 8: 35–46. [DOI] [PubMed] [Google Scholar]
- Heitz E., 1928. Das heterochromatin der Moose. I. Jahrb. Wiss. Botanik. 69: 762–818. [Google Scholar]
- Hsu T. C., Arrighi F. E., 1971. Distribution of constitutive heterochromatin in mammalian chromosomes. Chromosoma 34: 243–253. [DOI] [PubMed] [Google Scholar]
- Islam-Faridi M. N., Childs K. L., Klein P. E., Hodnett G., Menz M. A., et al. , 2002. A molecular cytogenetic map of sorghum chromosome 1: fluorescence in situ hybridization analysis with mapped bacterial artificial chromosomes. Genetics 161: 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B., 1976. Myths and mechanisms of meiosis. Chromosoma 54: 295–325. [DOI] [PubMed] [Google Scholar]
- John B., Lewis K. R., 1965. The meiotic system. Protoplasmatologia 6: 1–335. [Google Scholar]
- John B., King M., 1980. Heterochromatin variation in Cryptobothrus chrysophorus III. Synthetic hybrids. Chromosoma 78: 165–186. [Google Scholar]
- John B., King M., 1985. Pseudoterminalization, terminalization, and non-chiasmate modes of terminal association. Chromosoma 92: 89–99. [Google Scholar]
- Jones G. H., 1978. Giemsa C-banding of rye meiotic chromosomes and the nature of “terminal” chiasmata. Chromosoma 66: 45–57. [Google Scholar]
- Jones G. H., 1987. Chiasmata, pp. 213–244 in Meiosis, edited by Moens P. B. Academic Press, New York. [Google Scholar]
- Kato Y. T. A., 1976. Cytological studies of maize [Zea Mays L.] and Teosinte [Zea Mexicana Schrader Kuntze] in relation to their origin and evolution. Mass. Agr. Exper. Stat. Bul. 635: 1–185. [Google Scholar]
- Kato A., Lamb J. C., Birchler J. A., 2004. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. USA 101: 13554–13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. J., Islam-Faridi M. N., Klein P. E., Stelly D. M., Price H. J., et al. , 2005. Comprehensive molecular cytogenetic analysis of sorghum genome architecture: distribution of euchromatin, heterochromatin, genes and recombination in comparison to rice. Genetics 171: 1963–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klášterská I., Natarajan A. T., Ramel C., 1974. Heterochromatin distribution and chiasma localization in the grasshopper Bryodema tuberculata (Fabr.) (Acrididae). Chromosoma 44: 393–404. [DOI] [PubMed] [Google Scholar]
- Kota R. S., Gill K. S., Gill B. S., 1993. A cytogenetically based physical map of chromosome 1B in common wheat. Genome 36: 467–475. [DOI] [PubMed] [Google Scholar]
- Lambie E. J., Roeder G. S., 1986. Repression of meiotic crossing over by a centromere (CEN3) in Saccharomyces cerevisiae. Genetics 114: 769–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latos-Bielenska A., Trautmann T., Vogel W., 1990. Visualization of DNA in pachytene by monoclonal antibodies against BrdU reveals synaptonemal complex-like structures. Cytogenet. Cell Genet. 54: 24–28. [DOI] [PubMed] [Google Scholar]
- Levan A., 1933. Cytological studies in Allium. IV. Allium fistulosum. Sven. Bot. Tidskr. 27: 211–232. [Google Scholar]
- Lhuissier F. G. P., Offenberg H. H., Wittich P. E., Vischer N. O. E., Heyting C., 2007. The mismatch repair protein MLH1 marks a subset of strongly interfering crossovers in tomato. Plant Cell 19: 862–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten M., 2008. Meiotic chromatin: the substrate for recombination initiation, pp. 165–193 in Recombination and Meiosis: Models, Means and Evolution, edited by Egel R., Lankenau D. H. Springer-Verlag, Berlin. [Google Scholar]
- Linnert G., 1955. Die struktur der pachytanchromosomen in euchromatin und heterochromatin und ihre auswirkung auf die chiasmabildung bei salvia-arten. Chromosoma 7: 90–128. [DOI] [PubMed] [Google Scholar]
- Lohmiller L. D., De Muyt A., Howard B., Offenberg H. H., Heyting C., et al. , 2008. Cytological analysis of MRE11 protein during early meiotic prophase I in Arabidopsis and tomato. Chromosoma 117: 277–288. [DOI] [PubMed] [Google Scholar]
- Loidl J., 1979. C-band proximity of chiasmata and absence of terminalisation in Allium flavum (Liliaceae). Chromosoma 73: 45–51. [Google Scholar]
- Loidl J., 1987. Heterochromatin and differential chromosome staining in meiosis. Biol. Zent. 106: 641–662. [Google Scholar]
- Longley A. E., 1937. Morphological characters of teosinte chromosomes. J. Agric. Res. 54: 835–862. [Google Scholar]
- Lukaszewski A. J., Kopecky D., Line G., 2012. Inversions of chromosome arms 4AL and 2BS in wheat invert the patterns of chiasma distribution. Chromosoma 121: 201–208. [DOI] [PubMed] [Google Scholar]
- Marcon E., Moens P., 2003. Mlh1p and Mlh3p localize to precociously induced chiasmata of okadaic acid treated mouse spermatocytes. Genetics 165: 2283–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather K., 1933. The relation between chiasmata and crossing over in diploid and triploid Drosophila melanogaster. J. Genet. 27: 343–360. [Google Scholar]
- Mather K., 1939. Crossing over and heterochromatin in the X chromosome of Drosophila melanogaster. Genetics 24: 413–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B., Kato T. A., Blumenschein A., 1981. Chromosome Constitution of Races of Maize. Colegio de Postgraduados, Chapingo, Mexico. [Google Scholar]
- Mercier R., Mézard C., Jenczewski E., Macaisne N., Grelon M., 2015. The molecular biology of meiosis in plants. Annu. Rev. Plant Biol. 66: 297–327. [DOI] [PubMed] [Google Scholar]
- Miklos G. L. G., Nankivell R. N., 1976. Telomeric satellite DNA functions in regulating recombination. Chromosoma 56: 143–167. [DOI] [PubMed] [Google Scholar]
- Natarajan A. T., Gropp A., 1971. The meiotic behaviour of autosomal heterochromatic segments in hedgehogs. Chromosoma 35: 143–152. [DOI] [PubMed] [Google Scholar]
- Neves N., Delgado M., Silva M., Caperta A., Morais-Cecílio L., et al. , 2005. Ribosomal DNA heterochromatin in plants. Cytogenet. Genome Res. 109: 104–111. [DOI] [PubMed] [Google Scholar]
- Paques F., Haber J. E., 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock W. J., Dennis E. S., Rhoades M. M., Pryor A. J., 1981. Highly repeated DNA squence limited to knob heterochromatin in maize. Proc. Natl. Acad. Sci. USA 78: 4490–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson D. G., Price H. J., Johnston J. S., Stack S. M., 1996. DNA content of heterochromatin and euchromatin in tomato (Lycopersicon esculentum) pachytene chromosomes. Genome 39: 77–82. [DOI] [PubMed] [Google Scholar]
- Petes T. D., Botstein D., 1977. Simple Mendelian inheritance of the reiterated ribosomal DNA of yeast. Proc. Natl. Acad. Sci. USA 74: 5091–5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanna M. S., Prakken R., 1967. Structure and homology between pachytene and somatic metaphase chromosomes of the tomato. Genetica 38: 115–133. [Google Scholar]
- Resnick M. A., 1987. Investigating the genetic control of biochemical events in meiotic recombination, pp. 157–210 in Meiosis, edited by Moens P. B. Academic Press, Orlando. [Google Scholar]
- Rhoades M. M., 1955. The cytogenetics of maize, pp. 123–219 in Corn and Corn Improvement, edited by Sprague G. F. Academic Press, New York. [Google Scholar]
- Rhoades M. M., 1978. Genetic effects of heterochromatin in maize, pp. 641–671 in Maize Breeding and Genetics, edited by Walden D. B. John Wiley and Sons, New York. [Google Scholar]
- Rhoades M. M., Dempsey E., 1966. The effect of abnormal chromosome 10 on preferential segregation and crossing over in maize. Genetics 53: 989–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ris H., Korenberg J., 1979. Chromosome structure and levels of chromosome organization. Cell Biol. 2: 267–361. [Google Scholar]
- Roudier F., Ahmed I., Berard C., Sarazin A., Mary-Huard T., et al. , 2011. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 30: 1928–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P. S., Hsia A. P., Nikolau B. J., 1998. Genetic recombination in plants. Curr. Opin. Plant Biol. 1: 123–129. [DOI] [PubMed] [Google Scholar]
- Schwarzacher T., Heslop-Harrison P., 2000. Practical In Situ Hybridization. BIOS Scientific Publishers Ltd., New York. [Google Scholar]
- Sherman J. D., Stack S. M., 1995. Two-dimensional spreads of synaptonemal complexes from solanaceous plants. VI. High-resolution recombination nodule map for tomato (Lycopersicon esculentum). Genetics 141: 683–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. P., 1976. Evolution of repeated DNA sequences by unequal crossover. Science 191: 528–535. [DOI] [PubMed] [Google Scholar]
- Stack S. M., 1984. Heterochromatin, the synaptonemal complex, and crossing over. J. Cell Sci. 71: 159–176. [DOI] [PubMed] [Google Scholar]
- Stack S. M., 1991. Staining plant cells with silver. II. Chromosome cores. Genome 34: 900–908. [Google Scholar]
- Stack S., Roelofs D., 1996. Localized chiasmata and meiotic nodules in the tetraploid onion Allium porrum. Genome 39: 770–783. [DOI] [PubMed] [Google Scholar]
- Stack S. M., Anderson L. K., 2009. Electron microscopic immunogold localization of recombination-related proteins in spreads of synaptonemal complexes from tomato microsporocytes, pp. 147–169 in Meiosis Volume 2 Cytological Methods, edited by Keeney S. Humana Press, Inc., Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- Sudman P. D., Greenbaum I. F., 1990. Unequal crossing over and heterochromatin exchange in the X-Y bivalents of the deer mouse, Peromyscus beatae. Chromosoma 99: 183–189. [DOI] [PubMed] [Google Scholar]
- Sumner A. T., 2003. Chromosomes: Organization and Function. Blackwell Science Ltd., Malden, MA. [Google Scholar]
- Szauter P., 1984. An analysis of regional constraints on exchange in Drosophila melanogaster using recombination-defective meiotic mutants. Genetics 106: 45–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cold Spring Harbor Laboratory, Washington University Genome Sequencing Center, and PE Biosystems Arabidopsis Sequencing Consortium, 2000. The complete sequence of a heterochromatic island from a higher eukaryote. Cell 100: 377–386. [PubMed] [Google Scholar]
- Tomato Genome Consortium , 2012. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp C. N., Dawe R. K., 2006. Reinterpreting pericentromeric heterochromatin. Curr. Opin. Plant Biol. 9: 647–653. [DOI] [PubMed] [Google Scholar]
- Vader G., Blitzblau H. G., Tame M. A., Falk J. E., Curtin L., et al. , 2011. Protection of repetitive DNA borders from self-induced meiotic instability. Nature 477: 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Os H., Andrzejewski S., Bakker E., Barrena I., Bryan G. J., et al. , 2006. Construction of a 10,000-marker ultradense genetic recombination map of potato: providing a framework for accelerated gene isolation and genomewide physical map. Genetics 173: 1075–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B., 2011. Chromatin: constructing the big picture. EMBO J. 30: 1885–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincenten N., Kuhl L.-M., Lam I., Oke A., Kerr A. R. W., et al. , 2015. The kinetochore prevents centromere-proximal crossover recombination during meiosis. Elife 4: e10850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgruber T. K., Nakashima M. M., Schneider K. L., Sharma A., Xie Z., et al. , 2016. High quality maize centromere 10 sequence reveals evidence of frequent recombination events. Front. Plant Sci. 7: 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Miklos G. L. G., 1978. Genetic studies on heterochromatin in Drosophila melanogaster and their implication for the functions of satellite DNA. Chromosoma 66: 71–98. [DOI] [PubMed] [Google Scholar]
- Yao H., Zhou Q., Li J., Smith H., Yandeau M., et al. , 2002. Molecular characterization of meiotic recombination across the 140-kb multigenic a1-sh2 interval of maize. Proc. Natl. Acad. Sci. USA 99: 6157–6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.-K., Tang S., Slabaugh M. B., Heesacker A., Cole G., et al. , 2003. Toward a saturated molecular genetic linkage map for cultivated sunflower. Crop Sci. 43: 367–387. [Google Scholar]
- Yunis J. J., Yasmineh W. G., 1971. Heterochromatin, satellite DNA, and cell function. Science 174: 1200–1209. [DOI] [PubMed] [Google Scholar]
- Zhong X., Fransz P. F., Wennekes-van Eden J., Zabel P., van Kammen A., et al. , 1996. High-resolution mapping on pachytene chromosomes and extended DNA fibres by fluorescence in-situ hybridization. Plant Mol. Biol. Report. 14: 232–242. [Google Scholar]
- Zickler D., Kleckner N., 1999. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 33: 603–754. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Complete imaging data sets and reagents are available upon request.