Abstract
In GWAS, “generalization” is the replication of genotype-phenotype association in a population with different ancestry than the population in which it was first identified. Current practices for declaring generalizations rely on testing associations while controlling the Family Wise Error Rate (FWER) in the discovery study, then separately controlling error measures in the follow-up study. This approach does not guarantee control over the FWER or False Discovery Rate (FDR) of the generalization null hypotheses. It also fails to leverage the two-stage design to increase power for detecting generalized associations. We provide a formal statistical framework for quantifying the evidence of generalization that accounts for the (in)consistency between the directions of associations in the discovery and follow-up studies. We develop the directional generalization FWER (FWERg) and FDR (FDRg) controlling r-values, which are used to declare associations as generalized. This framework extends to generalization testing when applied to a published list of SNP-trait associations. Our methods control FWERg or FDRg under various SNP selection rules based on p-values in the discovery study. We find that it is often beneficial to use a more lenient p-value threshold than the genome-wide significance threshold. In a GWAS of Total Cholesterol (TC) in the Hispanic Community Health Study/Study of Latinos (HCHS/SOL), when testing all SNPs with p-values< 5 × 10−8 (15 genomic regions) for generalization in a large GWAS of whites, we generalized SNPs from 15 regions. But when testing all SNPs with p-values< 6.6 × 10−5 (89 regions), we generalized SNPs from 27 regions.
Keywords: Multiple testing, Shared genetics, One-sided p-values
Introduction
When presenting results from genome-wide association studies (GWAS), current standards require a “two-stage design” in which possible discoveries in the first stage are replicated in an independent study in a second stage (Cohen, 1999). ‘Generalization’ is the replication of a genotype-phenotype association in a population with different ancestry (or other characteristics) than the population in which it was first identified. As GWAS expand into populations of diverse ancestry, generalization testing is becoming more common. First, with non-white discovery populations, there tend to be fewer similar studies available, so only generalization and not replication is feasible. Second, when the discovery study population is admixed (e.g. Hispanics/Latinos), it is customary to seek generalization in some of its parental populations.
Although the current standard for GWAS mandates replication, error-controlling multiple testing adjustment procedures are often applied separately in the discovery and follow-up stages, without employing a replication- or generalization- based statistical framework. Bogomolov and Heller (2013) have shown that such approaches do not guarantee control over false generalization claims. Let the generalization null hypothesis state that a SNP is not associated with the trait in the discovery population, the follow-up population or both. This null is rejected if evidence of association exists for both populations. Define generalization testing as any multiple testing adjustment procedure that controls measures of generalization error such as the Family-Wise Error Rate (FWERg) or the False Discovery Rate (FDRg). In this paper, we propose methods to test the generalization null hypotheses in GWAS, by expanding and adapting recent statistical methods developed for replication.
Bogomolov and Heller (2013) considered replication testing using discovery and follow-up studies. They developed multiple testing procedures with protection against erroneous replicability claims by controlling the FWERg or the FDRg. A key result is that one must account for multiple testing in both the discovery and the follow-up studies to avoid a high number of erroneous replicability claims. Heller et al. (2014) suggested improvements to these procedures when used for GWAS, and developed r-values to quantify the evidence for replication while controlling FWERg or FDRg in GWAS. However, the r-values in Heller et al. (2014) do not account for the direction of the observed association. In this work we extend the r-values approach to incorporate the direction of observed associations. This acknowledges that we do not want to claim that an association generalizes if the direction of effect is different in the two populations. Our procedures achieve directional control by using one-sided p-values to compute directional r-values at the generalization testing stage, despite using two-sided tests in the discovery stage. This makes our procedures more powerful than the procedures of Heller et al. (2014) for discovering associations with the same direction in both studies. We perform extensive simulations to study fixed and data-adaptive rules for selecting SNPs based on their p-values in the discovery study, and compare multiple-testing adjustment procedures in combination with these selection rules.
Methods
Measures of false generalization
In multiple testing, there are two common measures of error: the FWER, and the FDR. In a single-stage GWAS, FWER is the probability of rejecting at least one null hypothesis corresponding to a SNP not associated with the trait. FDR is the expected proportion of falsely detected SNPs out of all those reported as associated with the trait.
Define the left-sided (right-sided) alternative as the scenario in which a given SNP allele is negatively (positively) associated with the trait in a given population. Let
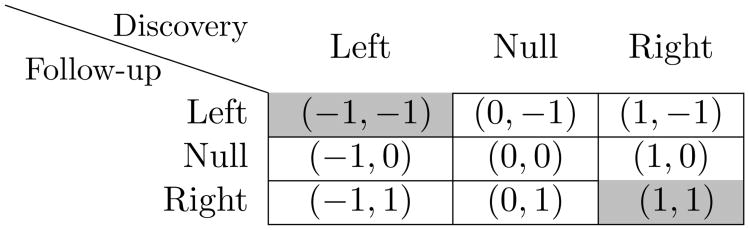
Let ℋj = {h = (h1j, h2j) : hij ∈ {−1, 0, 1}} be the set of 9 possible configurations of the vector Hj = (H1j, H2j) for two-sided alternatives for SNP j; these are depicted in Figure 1. The generalization null hypothesis for SNP j is true if Hj belongs to the set ℋ0 = {(−1, 1), (−1, 0), (1, −1), (1, 0), (0, 0), (0, −1), (0, 1)}. A SNP for which the generalization null is false has Hj ∈ ℋA = {(1, 1), (−1, −1)}. Thus, the generalization null hypothesis is rejected when a SNP is associated with the trait in both the discovery and the generalization populations, with the same directions of association.
Figure 1.
The set of possible configuration of the vector Hj = (H1j, H2j). The association of SNP j with the trait is defined as generalized association (marked as gray) when both alternatives are either left (negative direction of allele-trait association, Hj = (−1, −1)), or right (positive direction of allele-trait association, Hj = (1, 1)).
Suppose that R generalization claims are made by an analysis. Denote by and the indicators of whether a generalization null rejection (“generalization claim”) is made in the right or left direction, respectively, for SNP j. The number of true generalization claims is
and R – S is the number of false generalization claims. The directional generalization (and replication) FWER and FDR are given by:
Controlling for false generalizations
Definition: The directional FDRg/FWERg r-value for a SNP is the lowest FDR/FWER level at which we can say that the SNP association is generalized with the same direction of association in both the discovery and generalizing studies.
The directional p-values
Denote the left- and right-sided p-values for SNP association j in study i ∈ {1, 2} by , respectively. For continuous test statistics, . The p-values ( ) corresponding to variant j used in generalization analysis are defined as:
Thus, the one-sided p-values from both studies are guided by the estimated direction of association in the discovery study, so that if the evidence towards association is in the same direction in both studies, both , otherwise while .
Data and parameters required for FDRg/FWERg r-values computation
m, the number of SNPs examined in the discovery study.
ℛ1, the set of SNPs selected for follow-up based on discovery study results. Let R1 = |ℛ1| be their number.
The directional p-values for the followed-up SNPs { ( , ) : j ∈ ℛ1}.
l00 ∈ [0, 1), the user-specified lower bound on the fraction of SNP associations, out of the m SNPs examined in the discovery study, that are null in both studies. Default value for a GWAS is l00 = 0.8, following Heller et al. (2014).
c2 ∈ (0, 1), the emphasis given to the follow-up study (see Section Variations in Heller et al. (2014)), default value is c2 = 0.5.
Computation of the FDRg/FWERg r-values
- Defining functions , i ∈ ℛ1, x ∈ (0, 1):
- (a) Compute , the inverse weight function for the p-values from the discovery study.
- (b) For every SNP j ∈ ℛ1 compute the following e-values:
- (C) [FDRg] Let , where rank[ej(x)] is the rank of the e-value for a SNP j ∈ ℛ1 (with maximum rank for ties).
- (C) [FWERg] Let .
The FDRg (FWERg) r-value for SNP i ∈ ℛ1 is the solution to if a solution exists in (0, 1), and 1 otherwise. The solution is unique, see Lemma S1.1 in Heller et al. (2014).
Establishing generalization with directional FDRg/FWERg control at level q
Denote the set of SNPs having directional FDRg/FWERg r-value at most q by ℛ2. If a SNP j ∈ ℛ2 has , it is declared as having a generalized left-sided alternative; otherwise, it is declared as having a generalized right-sided alternative.
Selection rules
In generalization analysis, SNP associations are first tested in the discovery study, and then a subset of these SNPs is selected for testing in the follow-up study, according to a selection rule. For instance, investigators often select SNPs with p-value< 5 × 10−8 in the discovery study. This is a Bonferroni correction for m = 106 tests, applied to control FWER in the single-stage design. We consider other selection rules for a generalization/replication-based study design.
- Selection rule 1, recommended by Heller et al. (2014) for FDRg control. Apply the FDR controlling BH procedure (Benjamini and Hochberg, 1995) on all p-values from the discovery study to obtain BH-adjusted p-values. Choose all SNPs with BH-adjusted p-value≤ t, where
Use q = 0.05 to control FDRg at the 0.05 level. The rationale here is that every SNP with BH-adjusted discovery p-value larger than t has no chance of generalizing. Heller et al. (2014) applied this selection rule in settings where either both discovery and replication used two-sided p-values, or both used one-sided p-values with pre-determined directions. We can also apply it to one-sided p-values used for generalization testing when the discovery study hypothesis tests were two-sided.(1) - Selection rule 2, recommended by Heller et al. (2014) for FWERg control. This rule selects all SNPs with discovery p-value< t′, where
As in selection rule 1, SNPs with p-value> t′ have no chance of generalizing using the FWERg controlling procedure and selecting them can only reduce power. Again, we can apply this selection rule on the one-sided p-values used for generalization testing.(2)
Selection rule 1 is data adaptive and depends on the distribution of signals in the discovery study. Selection rule 2 is fixed. In selection rule 2, if l00 = 0.8, q = 0.05, and m = 106, we get t′ = 1.14 × 10−7. When one-sided p-values are used for generalization testing, the original two-sided p-values passing this threshold are ≤ 2.28 × 10−7.
Linkage Disequilibrium (LD)
GWAS datasets may contain tens of millions of genotyped and imputed SNPs. Many of these SNPs are in linkage disequilibrium; that is, allelic variation within one SNP is correlated with allelic variation in another SNP. Often, when a discovery study association is detected, the region may contain tens of correlated SNPs with low p-values.
Since the patterns of LD vary between two different populations, and consequently different SNPs may best tag the underlying causal genetic variation, we recommend testing all SNPs satisfying the selection rule. This will increase the multiple testing burden for FWER-type control. However, this can be handled by calculating the effective number of independent SNPs based on the LD matrix of the SNPs in the generalization study. The increase in the number of tests is not a problem for FDR control, which is concerned with the fraction of discovered associations that are false positives.
Simulation studies: discovery and generalization GWAS
We assess our proposed methods in simulations. First, we simulated test statistics for two studies; second, we calculated p-values in the discovery study; third, we selected SNPs for generalization testing based on several selection rules; finally, we applied multiple testing adjustment procedures. We used selection rules 1 (for FDRg control) and 2 (for FWERg control), applied to both one- and two-sided p-values, and the selection rules that take all SNPs with p-values< 1 × 10−6, 1 × 10−7, and < 5 × 10−8 in the discovery study. The multiple testing adjustment procedures were FDRg r-values and BH on the follow-up study alone (for FDRg control), and FWERg r-values and Bonferroni on the follow-up study along (for FWERg control). We compared all methods with and without directional control.
In an additional simulation study provided in the supplementary material, we investigated GWAS of cohorts designed to mimic realistic data sets with differences in LD structure and MAFs between the discovery and the generalization cohorts in a smaller number of simulations. There, we also compared generalization testing of all SNPs satisfying the selection rule with a procedure that only tests the lead SNP from each detected region.
Simulating test statistics with null inflation
In each of 1,000 repetitions of the simulations, we sampled 106 independent test statistics for both the discovery and the follow-up studies. Of these SNPs, 100 were causal in the discovery study and 100 were causal in the follow-up study. 50 of the causal SNPs overlapped between the studies. We considered two common generalization scenarios. In the first setting the discovery study had relatively low power, and the follow-up study had high power. This may happen when discovery is performed in the Hispanics Community Health Study/Study of Latinos (HCHS/SOL) with follow-up in a large meta-analysis GWAS in individuals of European ancestry. In the second setting the discovery study had high power, and the follow-up study had low power. This happens when HCHS/SOL investigators study whether associations reported from large meta-analyses of whites generalize to Hispanics/Latinos. In both scenarios, we generated inflation (λgc = 1.21, Devlin and Roeder (1999)) in the test statistics of both the discovery and generalizing studies. Details of how test statistics were generated appear in the supplementary material, as well as details of additional simulation settings that vary the assumptions of the degree of overlap between the causal SNPs between the two populations, the number of causal SNPs, the powers of the two studies, and more.
The HCHS/SOL
The HCHS/SOL is a community based cohort study, following self-identified Hispanic/Latino individuals. Almost 13,000 study participants consented for genotyping. For references to the HCHS/SOL study and descriptions of genotyping, imputation and quality control see Conomos et al. (2016).
Identifying SNP-Total Cholesterol associations in the HCHS/SOL
We performed a GWAS of Total Cholesterol (TC) in the HCHS/SOL followed by generalization testing using publicly available GWAS results. Our GWAS was adjusted for sex, age, 5 principal components of ancestry, and study design variables (study center, sampling weights). Analysis was performed using a linear mixed effect model, with random effects corresponding to block groups, households, and kinship. As advocated by Kraft et al. (2009), the analysis plan mimicked the published analyses. Thus, we first regressed TC values on covariates, and then applied a rank-based inverse normal transformation on the residuals. The transformed residuals were the outcome variable in the GWAS.
We compared multiple generalization analyses of TC. We used results from the Global Lipids Genetics Consortium (GLGC) TC GWAS (Willer et al., 2013), which conducted a large meta-analysis of multiple cohorts of European ancestry comprising of over 180,000 individuals. First, we considered generalization geared towards establishing new associations, in which we perform a discovery GWAS in the HCHS/SOL, with generalization to whites. Second, we selected SNPs published by Teslovich et al. (2010) and Willer et al. (2013) and tested whether they generalize to Hispanics/Latinos.
We tested all SNPs satisfying the selection rule criterion, even if they were in LD with each other. However, we report generalization results both in terms of individual SNPs, and by genomic regions: after generalization testing, we identified the first region by taking the SNP with smallest discovery p-value (lead discovery SNP) to represent it. We then “removed” all SNPs in a region of 1Mbp around it, and continued to find other regions in a similar manner. A region with any SNP that generalized is declared a generalized region.
Results
Simulations
For each simulation setting, selection rule, and multiple testing adjustment method, Table 1 provides the power, defined as the average proportion of generalized SNPs, out of all generalizable SNPs in the simulation, and the estimated error control measure ( or ). We omitted the selection rule based on discovery two-sided p-value≤ 10−7, as this resulted in “intermediate” results in terms of both power and error control between selection rules of higher and lower p-value thresholds. Additional results are provided in the supplementary material.
Table 1.
Results from 1,000 simulations in two settings, of high discovery study power and low follow-up study power (“High disc”), and low discovery study power and high follow-up study power (“Low disc”). The left side of the table provides results for selection rules and multiple testing adjustment procedures aiming for FDRg control, and the right side provides analogous results when aiming for FWERg control. Power refers to the proportion of SNPs associated with the trait in both studies that were generalized. In adjustment methods, “one-s” and “two-s” refer to application of the method to either one- or two-sided p-values.
| High disc | Low disc | High disc | Low disc | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||
| adjustment | power |
|
power |
|
adjustment | power |
|
power |
|
||||
| Selection rule 1 (one-sided) | Selection rule 2 (one-sided) | ||||||||||||
|
| |||||||||||||
| BH (one-s) | 0.72 | 0.08 | 0.74 | 0.08 | Bonferroni (one-s) | 0.39 | 0.07 | 0.18 | 0.06 | ||||
| BH (two-s) | 0.65 | 0.08 | 0.74 | 0.08 | Bonferroni (two-s) | 0.34 | 0.08 | 0.16 | 0.06 | ||||
| FDRg r-values (one-s) | 0.54 | 0.01 | 0.48 | 0.00 | FWERg r-values (one-s) | 0.34 | 0.05 | 0.16 | 0.04 | ||||
| FDRg r-values (two-s) | 0.43 | 0.00 | 0.41 | 0.00 | FWERg r-values (two-s) | 0.27 | 0.04 | 0.12 | 0.03 | ||||
|
| |||||||||||||
| Selection rule 1 (two-sided) | Selection rule 2 (two-sided) | ||||||||||||
|
| |||||||||||||
| BH (one-s) | 0.77 | 0.05 | 0.58 | 0.05 | Bonferroni (one-s) | 0.37 | 0.07 | 0.16 | 0.06 | ||||
| BH (two-s) | 0.71 | 0.06 | 0.58 | 0.06 | Bonferroni (two-s) | 0.32 | 0.08 | 0.14 | 0.06 | ||||
| FDRg r-values (one-s) | 0.66 | 0.02 | 0.48 | 0.01 | FWERg r-values (one-s) | 0.32 | 0.05 | 0.14 | 0.04 | ||||
| FDRg r-values (two-s) | 0.56 | 0.01 | 0.42 | 0.01 | FWERg r-values (two-s) | 0.28 | 0.04 | 0.13 | 0.04 | ||||
|
| |||||||||||||
| 10−6 | 10−6 | ||||||||||||
|
| |||||||||||||
| BH (one-s) | 0.66 | 0.03 | 0.35 | 0.03 | Bonferroni (one-s) | 0.43 | 0.08 | 0.23 | 0.07 | ||||
| BH (two-s) | 0.63 | 0.04 | 0.35 | 0.03 | Bonferroni (two-s) | 0.38 | 0.09 | 0.21 | 0.08 | ||||
| FDRg r-values (one-s) | 0.63 | 0.02 | 0.35 | 0.02 | FWERg r-values (one-s) | 0.33 | 0.04 | 0.15 | 0.03 | ||||
| FDRg r-values (two-s) | 0.58 | 0.02 | 0.35 | 0.02 | FWERg r-values (two-s) | 0.26 | 0.04 | 0.11 | 0.02 | ||||
|
| |||||||||||||
| 5 × 10−8 | 5 × 10−8 | ||||||||||||
|
| |||||||||||||
| BH (one-s) | 0.48 | 0.03 | 0.18 | 0.02 | Bonferroni (one-s) | 0.33 | 0.07 | 0.14 | 0.06 | ||||
| BH (two-s) | 0.46 | 0.03 | 0.18 | 0.02 | Bonferroni (two-s) | 0.3 | 0.08 | 0.12 | 0.06 | ||||
| FDRg r-values (one-s) | 0.45 | 0.02 | 0.18 | 0.01 | FWERg r-values (one-s) | 0.3 | 0.05 | 0.12 | 0.03 | ||||
| FDRg r-values (two-s) | 0.42 | 0.02 | 0.18 | 0.01 | FWERg r-values (two-s) | 0.26 | 0.04 | 0.11 | 0.03 | ||||
In general, directional control had higher generalization power compared to using two-sided p-values. The difference was smaller when the selection rule was more stringent, i.e. most followed up SNPs were true associations. Higher discovery power resulted in higher generalization power, but also slightly higher error rates. Importantly, both FDRg and FWERg r-values always protected their target error measures.
FDRg control
Focusing on directional FDRg r-values, selection rule 1 applied to two-sided p-values was most powerful. Applying selection rule 1 to one-sided p-values resulted in a large proportion of null followed-up SNPs, and consequently, generalization testing using BH on the follow-up study alone did not control FDRg. BH on the follow-up study alone controlled FDRg in most (but not all) other settings. While BH on the follow-up study alone is less stringent than directional r-values, and therefore more “powerful”, the power differences between the two procedures are small when the selection rules are stringent and fewer null SNPs are selected for follow-up.
FWERg control
The most powerful selection rule was always selection rule 2 applied to one-sided p-values. Applying Bonferroni correction to the follow-up study alone never controlled FWERg, and control was worse with two-sided p-values compared to one-sided p-values.
The HCHS/SOL Total Cholesterol GWAS
HCHS/SOL as the primary discovery study in a two-stage design
In Table 2, for each combination of selection rule and multiple testing adjustment method, we report the number of SNPs followed-up that are available in both the HCHS/SOL and the GLGC TC GWAS, the number of regions they correspond to, the number of generalized SNPs and generalized regions, and the number of regions with none of the SNPs having p-value< 5 × 10−8 in Willer et al. (2013)'s GWAS.
Table 2.
Generalization testing results from a set of analyses based on a HCHS/SOL GWAS as the discovery study, and GLGC GWAS as the follow-up study. For each selection rule we report the number of SNPs selected for follow-up testing, and the number of loci containing these SNPs. For combinations of selection rules and multiple testing adjustment method we report the number of generalized loci, and the number of generalized loci that did not contain any SNP with p-value< 5 × 10−8 in the GLGC GWAS.
| Selection rule | selected SNPs | loci | adjustment | gen SNPs | gen loci | # loci not sig Willer |
|---|---|---|---|---|---|---|
| Selection rule 1 - one-sided | 1,662 | 89 | FDRg r-values | 1,352 | 27 | 5 |
| Selection rule 1 - two-sided | 1,208 | 51 | FDRg r-values | 1,076 | 21 | 1 |
| 10−6 | 742 | 18 | FDRg r-values | 706 | 17 | 0 |
| 10−6 | 742 | 18 | FWERg r-values | 583 | 17 | 0 |
| Selection rule 2 - one-sided | 627 | 17 | FWERg r-values | 583 | 17 | 0 |
| Selection rule 2 - two-sided | 574 | 16 | FWERg r-values | 538 | 16 | 0 |
| 5 × 10−8 | 546 | 15 | FWERg r-values | 514 | 15 | 0 |
When the selection rule chose all SNPs with p-value< 5 × 10−8, the followed-up SNPs corresponded to 15 regions, all of which generalized under FWERg (and FDRg) control. For FWERg control, the highest number of generalized regions was obtained using both selection rule 2 (on one-sided p-values) and by choosing SNPs with p-value< 10−6. Indeed, SNPs with HCHS/SOL two-sided p-value> 2.28 × 10−7 (selection rule 2) cannot be generalized under FWERg control, so the higher threshold 10−6 could not increase power.
In the FDRg-controlling analysis applied on SNPs satisfying selection rule 1 on two-sided p-values, 21 regions generalized. These included a single generalized region that would not be reported in either the HCHS/SOL or the GLGC GWAS alone. The lead SNP, rs870992 on chromosome 5, had r-value= 0.008, HCHS/SOL p-value= 2 × 10−5, and GLGC p-value= 5.2 × 10−5. This SNP was formerly associated with concentration of liver enzymes in plasma in a GWAS (Chambers et al., 2011). In the FDRg-controlling analysis applied to SNPs satisfying selection rule 1 with one-sided p-values, there were 22 generalized regions with strong evidence of association in the GLGC GWAS (SNPs with p-values< 5 × 10−8), and 5 generalized regions that would not have been detected either in the HCHS/SOL or the GLGC GWAS alone. One of them was the region that includes rs870992. Another SNP, rs2072781 in chromosome 6, had r-value= 0.009 (HCHS/SOL p-value= 2.1 × 10−5, GLGC p-value= 1 × 10−4). This SNP is in the MYLIP gene, formerly associated with high TC in Mexicans (Weissglas-Volkov et al., 2011). Three additional regions had relatively higher p-values in the GLGC GWAS (0.007-0.05) and r-values in the range 0.01-0.05. The five regions are reported in the supplementary material.
In all analyses, there was a generalized region in which the HCHS/SOL lead SNP did not generalize (p-value= 0.92 in Willer et al. (2013)) but a different SNP in the same region did generalize (p-value= 1.4 × 10−46 in Willer et al. (2013)). This supports a strategy that analyzes all SNPs satisfying the selection rule, rather than an LD-pruned set.
Generalizing previously reported TC-SNP associations
There are 74 SNPs previously reported as associated with TC with p-values≤ 5 × 10−8 and are available for generalization testing in the HCHS/SOL data set. 51 SNPs were reported by Teslovich et al. (2010) and replicated by Willer et al. (2013). For these SNPs, we performed generalization analysis by treating the meta-analysis of the results of Teslovich et al. (2010) and Willer et al. (2013) as the discovery study. 33 of the these SNPs generalized to the HCHS/SOL. We performed a second generalization analysis on 23 SNPs reported only in Willer et al. (2013). None of these SNPs generalized.
In the supplementary material, we provide an additional analysis in which we pursue generalization for all SNPs with p-value< 10−6 in the GLGC GWAS, without any SNP pruning. This analysis generalized 9 more regions than the analysis that tested only the published lead SNPs.
Discussion
In this work, we propose to leverage two-stage design to increase generalization power in GWAS. We introduce procedures for calculating directional FDRg and FWERg r-values, computed based on one-sided p-values. We prove that r-values control their directional error measures when there is no genomic inflation, and show via simulations that errors are controlled in the presence of inflation. These procedures are, by construction, more powerful than those based on two-sided p-values when the direction of association is consistent between discovery and follow-up populations. We studied SNP selection rules that are geared towards generalization-based designs. Our simulation studies found that by choosing SNPs for generalization testing based on p-values less conservative than the genome-wide significance threshold, e.g. selection rules 1 and 2 for FDRg control and FWERg control, respectively, we are able to generalize more SNPs while controlling the desired error rate. Finally, we demonstrated our procedure on a GWAS of Total Cholesterol in the HCHS/SOL.
The proposed procedures require directional consistency between estimated associations for declaring generalization. Biologically, it makes sense that causal SNPs have the same effect direction in different populations. However, if a tag SNP has different direction of LD with the causal SNP in the discovery and follow-up populations, the estimated directions of association of the trait with this tag SNP will likely differ between the two populations and we could not declare generalization. Nevertheless, the gain in power and protection against false generalizations by requiring directional consistency outweighs such a rare possibility.
An approach that was promoted in the past to increase power in a two-stage design was to perform a joint analysis of the two studies via meta-analysis (Skol et al., 2006). However, this approach does not test the generalization null hypothesis, and an association may appear significant even if it exists only in one population. In contrast, our approach is focused on generalization testing, which allows for stronger conclusions about the underlying similarity in genetic associations between populations.
We provide practical recommendations based on our results. First, in terms of selection rules, we recommend selection rule 2 for FWERg control at the α level, which selects SNPs with two-sided discovery p-value< 2.28 × 10−7 for α = 0.05. For FDRg control at the α = 0.05 level, we recommend selecting SNPs with discovery p-value≤ 10−6, or based on selection rule 1 if it is more conservative. That is because selecting SNPs with p-value larger than selection rule 1 can only reduce power under FDRg control. Second, we recommend follow-up on all SNPs satisfying the selection rule. Limiting follow-up to lead SNPs from the discovery study may reduce generalization power due to different LD patterns between the discovery and follow-up populations. Finally, while FDRg control allows for more false positive generalizations compared to FWERg control, it also allows for more generalizations. While the GWAS culture favors caution and prioritizes FWER control, FDRg control may be more appropriate in generalization testing. In this setting an investigator may be willing to tolerate a small fraction of false positives among the generalizations, as the overall number of reported false associations may already be dramatically reduced, compared to reported associations from a discovery GWAS alone.
We examined generalization testing of associations from European ancestry populations to Hispanics/Latinos, and vice versa. Hispanics/Latinos are admixed and have large proportion of European ancestry; therefore we expect a large overlap in genetic architecture between the two populations. However, we expect our conclusions to hold also when studying generalizations between other populations. We performed additional simulations studies with varying degrees of overlap between causal SNPs and distributions of test statistics, corresponding to many plausible generalization scenarios. The conclusions remained the same.
While our methodology focuses on generalization of variants, in the data analysis we also reported results by regions, where we reported a region as generalized if at least one of its associated SNPs generalized. However, we did not offer a measure of region-generalization evidence. Assigning a r-value for this null hypothesis is a topic of future work.
Software
An R package to perform generalization analysis can be installed using the R commands
library(devtools)
install_github(“tamartsi/generalize”, subdir = “generalize”)
and the manual can be viewed in https://github.com/tamartsi/generalize/blob/Package_update/generalize-manual.pdf. Also, a web applet that computes r-values based on one-sided p-values from the discovery and follow-up study, and does not require any software installation, is available in http://www.math.tau.ac.il/∼ruheller/App.html
Supplementary Material
Acknowledgments
The authors thank the staff and participants of HCHS/SOL for their important contributions. This work was supported in part by NHLBI HHSN268201300005C. The Hispanic Community Health Study/Study of Latinos was carried out as a collaborative study supported by contracts from the National Heart, Lung, and Blood Institute (NHLBI) to the University of North Carolina (N01-HC65233), University of Miami (N01-HC65234), Albert Einstein College of Medicine (N01-HC65235), Northwestern University (N01-HC65236), and San Diego State University (N01-HC65237). The following Institutes/Centers/Offices contribute to the HCHS/SOL through a transfer of funds to the NHLBI: National Institute on Minority Health and Health Disparities, National Institute on Deafness and Other Communication Disorders, National Institute of Dental and Craniofacial Research, National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Neurological Disorders and Stroke, NIH Institution-Office of Dietary Supplements. The research of YB has received funding from the European Research Council under the European Community's Seventh Framework Programme (FP7/2007-2013) / ERC grant agreement n° [294519] (PSARPS).
References
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995:289–300. [Google Scholar]
- Bogomolov M, Heller R. Discovering findings that replicate from a primary study of high dimension to a follow-up study. Journal of the American Statistical Association. 2013;108:1480–1492. [Google Scholar]
- Chambers JC, Zhang W, Sehmi J, Li X, Wass MN, Van der Harst P, Holm H, Sanna S, Kavousi M, Baumeister SE, et al. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet. 2011;43:1131–1138. doi: 10.1038/ng.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen B. Freely associating. Nat Genet. 1999;22:1–2. doi: 10.1038/8702. [DOI] [PubMed] [Google Scholar]
- Conomos M, Laurie C, Stilp A, Gogarten S, McHugh C, Nelson S, Sofer T, Fernandez-Rhodes L, Justice A, Graff M, et al. Genetic Diversity and Association Studies in US Hispanic/Latino Populations: Applications in the Hispanic Community Health Study/Study of Latinos. The American Journal of Human Genetics. 2016;98:165–184. doi: 10.1016/j.ajhg.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Heller R, Bogomolov M, Benjamini Y. Deciding whether follow-up studies have replicated findings in a preliminary large-scale omics study. Proceedings of the National Academy of Sciences. 2014;111:16262–16267. doi: 10.1073/pnas.1314814111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft P, Zeggini E, Ioannidis JP. Replication in genome-wide association studies. Statistical Science: A review journal of the Institute of Mathematical Statistics. 2009;24:561. doi: 10.1214/09-STS290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nature genetics. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissglas-Volkov D, Calkin AC, Tusie-Luna T, Sinsheimer JS, Zelcer N, Riba L, Tino AMV, Ordoñez-Sánchez ML, Cruz-Bautista I, Aguilar-Salinas CA, et al. The N342S MYLIP polymorphism is associated with high total cholesterol and increased LDL receptor degradation in humans. The Journal of clinical investigation. 2011;121:3062–3071. doi: 10.1172/JCI45504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, et al. Discovery and refinement of loci associated with lipid levels. Nature Genetics. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.