Abstract
Medulloblastoma is the most frequent malignant pediatric brain tumor and is divided into at least four subgroups known as Wnt, SHH, Group 3 and Group 4. Here we characterized gene regulation mechanisms in the most aggressive subtype, Group 3 tumors, through genome-wide chromatin and expression profiling. Our results show that most active distal sites in these tumors are occupied by the transcription factor OTX2. Highly active OTX2 bound enhancers are often arranged as clusters of adjacent peaks and are also bound by the transcription factor NEUROD1. These sites are responsive to OTX2 and NEUROD1 knockdown and could also be generated de novo upon ectopic OTX2 expression in primary cells, showing that OTX2 cooperates with NEUROD1 and plays a major role in maintaining and possibly establishing regulatory elements as a pioneer factor. Among OTX2 target genes we identified the kinase NEK2, whose knockdown and pharmacological inhibition decreased cell viability. Our studies thus show that OTX2 controls the regulatory landscape of Group 3 medulloblastoma through cooperative activity at enhancer elements and contributes to the expression of critical target genes.
Keywords: Medulloblastoma, OTX2, epigenetic, enhancer, NEK2, NEUROD1
Introduction
Brain tumors are a leading cause of cancer related death in children and medulloblastoma is the most common malignant brain tumor type (1). Over the last decade, expression profiling of large cohorts of primary tumors led to the identification of at least four main molecular medulloblastoma variants (2-6), which differ in prognosis and demographic properties (7). DNA sequencing studies, focusing on both point mutations and copy number changes, have further reinforced these distinctions and show specific patterns of DNA alterations in each subtype (8-11). At a mechanistic level, the best understood subtypes are characterized by deregulation of either the Wnt or SHH pathways and are presumed to originate from specific precursor cell populations during development (12). The two remaining groups, known as Group 3 and Group 4, account for 60% of tumors but are poorly understood. Given the limited treatment options available for these tumors, further understanding of their biological basis could have major implications for therapy. This is particularly relevant for Group 3 tumors, which have the poorest outcomes.
Transcriptional regulation plays a central role in determining cell fate and identity in development and cancer. Master transcription factors have a major impact on transcriptional programs, as dramatically demonstrated by reprogramming experiments where differentiated cells are converted into pluripotent stem cells by the exogenous expression of a combination of transcription factors (13). Similar reprogramming strategies can also modify the tumor propagating potential of tumor cells (14-17). These transcriptional programs are controlled by promoters and by highly lineage-specific distal regulatory elements (18, 19), also called enhancers, that define the regulatory landscape of a given cell type. Rapidly advancing knowledge of the ‘histone code’ of post-translational modifications and the combined use of chromatin immunoprecipitation with next generation sequencing have greatly facilitated the large-scale identification of regulatory elements and have shown their importance in normal cells and cancer (18, 20-22). In the case of medulloblastoma, a recent study has shown that different subgroups have distinct sets of super-enhancers (23).
In this study we sought to gain insight into the mechanisms of gene regulation that drive Group 3 medulloblastoma. We find that the transcription factor OTX2, which has been implicated in cell proliferation and tumorigenicity in medulloblastoma (24-26), is present at most active enhancers in primary Group 3 tumors. In keeping with this finding, OTX2 is highly expressed in all Group 3 tumors and is in fact amplified in 8% of cases, making it the second most amplified gene after MYC (27). OTX2 is also expressed in the Wnt and Group 4 subtypes, suggesting that it may play an important role in the majority of medulloblastomas. While OTX2 binds a large number of active and inactive sites across the genome of Group 3 tumors, our data show a strong association between active enhancers and the presence of both OTX2 and the transcription factor NEUROD1. Downregulation of OTX2 or NEUROD1 results in reductions in markers of enhancer activity at these sites, demonstrating a significant contribution to chromatin states. Moreover, OTX2 is capable of operating as a pioneer factor at these active sites since introduction of OTX2 into primary mesenchymal stem cells results in de novo induction of enhancers. Finally, among highly responsive OTX2 target genes we identified the NEK2 kinase and found that Group 3 medulloblastoma cell lines are highly sensitive to either NEK2 knockdown or inhibition of its kinase activity. Thus, our studies show that OTX2 is a major activator of regulatory elements in Group 3 medulloblastoma and controls genes involved in cell growth and survival.
Results
A large set of enhancers is consistently active in primary Group 3 medulloblastomas
In order to define the regulatory landscape of Group 3 medulloblastoma, we mapped the genome-wide binding profiles of four histone modification marks from five fresh-frozen primary tumors and two cell lines using chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) and combined these epigenomic profiles with RNA expression levels measured by RNA-seq in the same samples. The histone modifications profiled were histone H3 lysine 4 trimethylation (H3K4me3, associated with active promoters); H3 lysine 4 monomethylation (H3K4me1, associated with enhancers); H3 lysine 27 trimethylation (H3K27me3, associated with Polycomb repression); and H3 lysine 27 acetylation (H3K27ac, associated with increased enhancer activity).
We first classified promoters into four groups based on active (H3K4me3) and repressive (H3K27me3) histone marks: active sites with H3K4me3 only, repressed sites with H3K27me3 only and sites that are either positive or negative for both marks (Figure S1A-B). As expected, gene expression in tumor tissues was associated with promoter chromatin states (Figure S1C).
Given the key role of enhancer elements in orchestrating transcriptional programs, we next identified a set of 9621 consistently active distal regulatory elements based on the presence of the H3K27ac activation mark (MACS q-value 0.01) in 4 out of 5 tumor samples and the absence of the H3K4me3 promoter mark (Figures 1A, Figures S1D-F and Table S1). The majority of these sites were either found in introns (53%) or intergenic regions (41%) (Figure S1G). Similar H3K27ac signals were also detected in more than 80% of these active enhancers in two Group 3 medulloblastoma cell lines (D341 and D283) (Figures 1A and S1H-I) in accordance with their previous classification based on expression profiling (28). Targets of active enhancers include genes known to be specifically expressed in Group 3 medulloblastoma such as NEUROG1 and TULP1 shown in Figure 1B. Overall the set of consistently active Group 3 enhancers was associated with higher average expression levels for their nearest genes compared to other transcripts (Figure 1C), indicating that these regulatory elements have significant effects on transcription in primary tumors and cell lines.
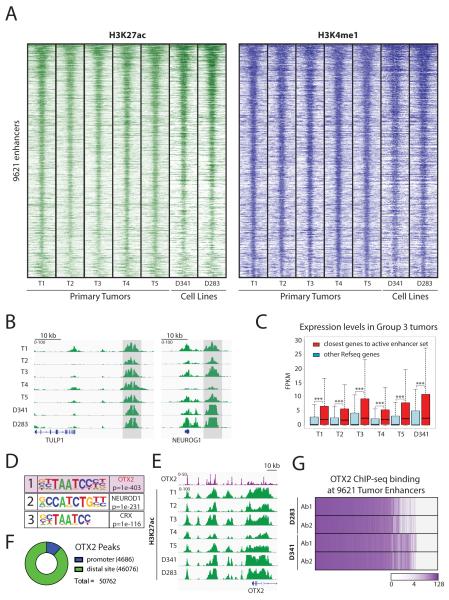
Figure 1. The majority of active enhancers shared by primary Group 3 medulloblastomas are bound by the transcription factor OTX2.
A. Identification of a set of 9621 shared active enhancers in primary Group 3 medulloblastoma tumors. Heatmaps depict H3K27ac (green) and H3K4me1 (blue) ChIP-seq signals in 5 frozen primary medulloblastoma tumors. Similar chromatin signals are found in two Group 3 cell lines (D341 and D283). Each row shows a 10 kb region centered on the active enhancer coordinates, ranked by average H3K27ac signals.
B. Two examples of the set of active enhancers shared by Group 3 medulloblastoma tumors. H3K27ac ChIP-seq signals are shown in green and consistently active regions are marked in light gray.
C. Genes associated with the Group 3 active enhancer set are expressed at higher levels in primary tumors and in the D341 cell line. Boxplot of RNA-seq FPKM expression values for genes closest to Group 3 active enhancers (red) compared to other loci (blue). *** Indicates p value < 1e-20.
D. Motif analysis of the active Group 3 enhancer set. OTX2 has the highest enrichment.
E. The OTX2 locus is highly active in primary tumors and cell lines and also contains several OTX2 peaks. H3K27ac ChIP-seq signals are shown in green. OTX2 ChIP-seq in D341 is shown in purple.
F. OTX2 is primarily localized at putative enhancer sites in Group 3 medulloblastoma cell lines. Pie chart showing OTX2 peaks annotated using the Refseq promoter database and H3K4me3 ChIP-seq data.
G. OTX2 is present at the majority of Group 3 active enhancers defined in primary tumors. The chart represents OTX2 ChIP-seq signals overlapping the genomic coordinates of the active Group 3 enhancer set. The color scale represents log2 ChIP-seq signals. Endogenous OTX2 was profiled in two Group 3 cell lines using two different antibodies.
OTX2 is present at most active enhancers in Group 3 medulloblastoma
To identify factors that control enhancer activity in Group 3 tumors we performed motif enrichment analysis of the underlying DNA sequences. The motif recognized by the transcription factor OTX2 was the most significantly enriched in our set of 9621 consistently active Group 3 medulloblastoma enhancers (Figure 1D). This was also the case in all active enhancers of each of the 5 primary tumors considered individually (Figure S2). Similar results were obtained for the two Group 3 cell lines D283 and D341 (Figure S2). In accordance with these findings, genome wide chromatin accessibility detected by Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) in the D283 cell line also showed OTX2 as the top motif for open chromatin locations (Figure S3A-D). The motifs recognized by the transcription factors CRX, NEUROD1 and NF1 were also enriched in these analyses albeit at considerably lower levels of significance when compared to OTX2 (Figures S2 and S3C). Thus, several methodologies show that OTX2 is a major feature of the Group 3 regulatory landscape.
OTX2 is a homeobox transcription factor essential for normal brain development that is induced during ESC differentiation, and plays a crucial function in enhancer activation in this setting (29-31). In medulloblastoma, OTX2 has been shown to be expressed in all non-SHH subtypes and the highest levels are observed in Group 3 tumors (25). In addition, amplifications have been demonstrated in some Group 3 cases (5, 24, 27, 32). More recently, the presence of a super-enhancer has also been reported in the OTX2 locus in medulloblastoma (23). Accordingly, we found that the OTX2 genomic region is covered by a large group of active enhancers in our tumor samples, as well as in the two cell lines D283 and D341 (Figure 1E). Several studies have shown that OTX2 plays an important role in cell proliferation and survival of medulloblastoma cells (24, 25) but the direct mechanisms responsible for these properties are not well defined.
To assess the role of OTX2 in shaping the Group 3 medulloblastoma epigenetic landscape, we first mapped endogenous OTX2 genome-wide binding sites by ChIP-seq. A total of 50762 highly reproducible OTX2 peaks were present in two Group 3 cell lines (D283 and D341) using two different antibodies (Figure S4A and Table S2). This is consistent with prior studies showing large numbers of binding sites for this transcription factor (33). The majority of OTX2 binding sites were found at putative enhancer regions (91%) while only a small fraction were present at promoters (9%) (Figures 1F and S4B-D). We next compared our set of consistently active Group 3 tumor enhancers to endogenous OTX2 binding sites and found a remarkable degree of overlap. Our data shows that a full 62% of active Group 3 enhancers contain OTX2 peaks in all our conditions (Figure 1G). Thus, as suggested by our in-silico motif analysis, direct profiling of OTX2 binding sites shows that OTX2 is present at the majority of consistently active enhancers in Group 3 medulloblastoma.
Given that OTX2 is most highly expressed in Group 3 tumors but is also expressed in WNT and Group 4 tumors we considered whether the latter subtypes share part of the active enhancer landscape observed in Group 3 tumors. Analysis of an independent dataset of medulloblastoma chromatin profiles (23) was highly consistent with our results and showed strong average H3K27ac signals in Group 3 tumors at OTX2 sites overlapping our set of active enhancers (85%) (Figure S4E). A significant but smaller fraction of these sites were also active in Wnt and Group 4 tumors (61% and 62% respectively). In contrast, only a minority of sites (30%) showed high H3K27ac levels in SHH tumors, which have low OTX2 levels (Figure S4E). Thus, OTX2 expression is associated with similarities in the enhancer landscape of non-SHH medulloblastoma tumors, suggesting that this transcription factor has a significant impact on regulatory activity in these medulloblastoma subtypes. Our comparison of H3K27ac signals also identified OTX2 sites that are specifically active in Group 3 tumors (Figure S4F). The majority of genes associated with these sites (69%) were expressed at higher levels in Group 3 tumors when compared to other medulloblastoma subtypes in an independent microarray expression dataset (10) (Figure S4G), suggesting that differential OTX2 activity is linked to differences in gene expression among medulloblastoma subtypes.
Active OTX2 bound enhancers are enriched for the transcription factor NEUROD1 and are often arranged in clusters
Despite its strong association with active enhancers, a large number of OTX2 binding sites were outside of the set of active Group 3 enhancers and contained low levels of the H3K27ac activation mark (Figures S4D and S5A). In order to test for differences between OTX2 peaks with either high or low H3K27ac, we defined two contrasting categories of OTX2 bound sites (Active and Inactive) based on consistently high H3K27ac (top third of the signal distribution in 4 out of 5 tumors and both cell lines, 7053 Active peaks) or the absence of significant H3K27ac signals (bottom third in 4 out of 5 tumors and both cell lines, 5751 Inactive peaks, Figure 2A). One example of each category is shown in Figure 2B. While H3K27ac levels varied over a thirty-fold range between these two categories, OTX2 signals were only modestly lower on Inactive sites (Figure S5B). H3K4me1 and open chromatin signals (measured by ATAC-seq) were lower but readily detectable in the Inactive set (Figure S5B-C-D) suggesting that OTX2 is generally associated with basal enhancer features. In contrast, RNA-seq data for cell lines and primary tumors showed strong differences in expression of neighboring genes between these two categories of OTX2 binding sites, pointing to a significant functional impact on gene regulation (Figure 2C). Thus, our data shows widespread binding of OTX2 on the genome that is linked to strikingly different effects on chromatin and gene regulation.
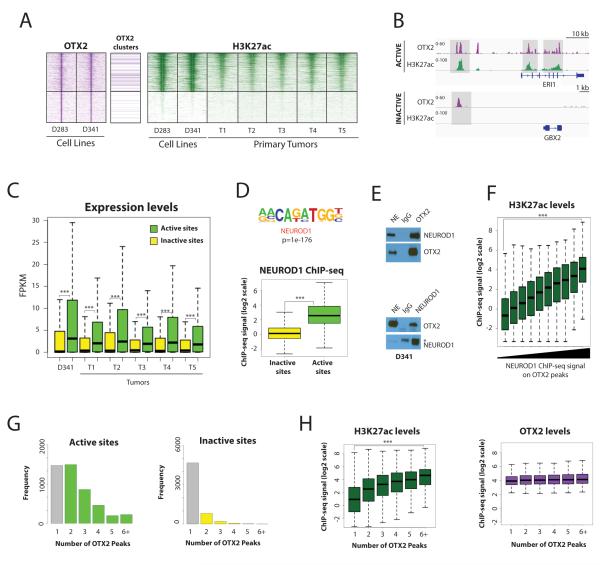
Figure 2. OTX2 is associated with higher levels of activity when paired with NEUROD1 and arranged in clusters.
A. OTX2 bound distal sites show a wide range of H3K27ac levels. Heatmaps depicting OTX2 ChIP-seq (purple) in cell lines and H3K27ac ChIP-seq (green) in cell lines and tumors. Sites where OTX2 is arranged as clusters of at least three peaks are marked. Rows are ordered by the average H3K27ac levels in cell lines and tumors and show a 10 kb region centered on each OTX2 binding site. Two categories of sites are shown: consistently Active sites (top; n=7053) and consistently Inactive sites (bottom; n=5751).
B. One example of each category of OTX2 binding sites in D341 cell line: Active site (top) Inactive site (bottom). OTX2 ChIP-seq signals are shown in purple and H3K27ac ChIP-seq signals are shown in green.
C. OTX2 enhancer activity is linked to higher gene expression levels in medulloblastoma primary tumors and D341 medulloblastoma cells. FPKM levels of the closest genes in each category of OTX2 peak: Active (green) and Inactive (yellow). *** Indicates p value < 1e-20.
D. NEUROD1 is enriched at active OTX2 sites. Top: Motif analysis comparing Active vs Inactive OTX2 binding sites shows that the NEUROD1 motif is the most highly enriched in Active sites. Bottom: Boxplot of endogenous NEUROD1 ChIP-seq signal intensities shows higher levels of NEUROD1 on Active (green) OTX2 sites compared to Inactive sites (yellow) in D341 cells. *** Indicates p value < 1e-20.
E. OTX2 and NEUROD1 are part of the same endogenous protein complexes in medulloblastoma cells. Co-immunoprecipitation experiments in D341 nuclear extracts (NE) show an interaction between OTX2 and NEUROD1. NEUROD1 signals are detected after OTX2 immunoprecipitation (top) and reciprocal experiments show OTX2 signals after NEUROD1 immmunoprecipitation (bottom). * indicates signals for IgG Heavy chains used for immunoprecipitation.
F. NEUROD1 signals on OTX2 binding sites are associated with higher levels of enhancer activity. Boxplot showing H3K27ac ChIP-seq signal levels on OTX2 peaks categorized by NEUROD1 ChIP-seq signal levels in D341 cells. *** Indicates p value < 1e-20.
G. OTX2 binds as clusters of peaks on Active enhancers. Histograms show the number of OTX2 peaks identified in a 5 kb window around each OTX2 peak in Active (Left) and Inactive distal sites (Right).
H. OTX2 peak clusters have higher levels of enhancer activity. Boxplots show H3K27ac (green) and OTX2 (purple) ChIP-seq levels on OTX2 binding sites according to the number of neighboring OTX2 peaks. *** Indicates p value < 1e-20.
We then searched for factors that may contribute to different H3K27ac levels at OTX2 binding sites. Upon scanning the underlying sequences of active and inactive OTX2 peaks, we found similar frequencies of OTX2 motif occurrences (> 95% peaks in each category contained at least one motif occurrence). In contrast, motif enrichment analysis comparing active and inactive OTX2 binding sites showed a strong enrichment for co-occurrence of the binding motif for NEUROD1, a bHLH transcription factor implicated in neural development (Figure 2D, top panel and S5E), and which has been shown to be expressed in medulloblastoma (34). This cooperation was confirmed by direct profiling of endogenous NEUROD1 by ChIP-seq (Figure 2D, bottom panel) and reciprocal co-immunoprecipation experiments for the endogenous proteins (Figure 2E). Indeed, NEUROD1 signals measured at OTX2 peaks were strongly associated with the levels of H3K27ac, suggesting that these transcription factors cooperate to activate enhancers in medulloblastoma (Figure 2F). Similarly, H3K27ac ChIP-seq signals were significantly higher at sites with both NEUROD1 and OTX2 compared to NEUROD1 alone (Figure S5F).
Finally, we also noticed that OTX2 was often organized as clusters of three or more peaks on active enhancers while it was mostly found as isolated peaks on inactive distal sites (Figure 2G and 2A, 37% of active sites have OTX2 clusters vs 5% of inactive sites). Indeed, H3K27 acetylation increased with the number of OTX2 bound sites within 5kb of each other (Figure 2H), suggesting that the genomic organization of OTX2 binding sites may also contribute to activation levels of bound enhancers.
Together, our results show that, while OTX2 has a widespread presence in the genome of Group 3 medulloblastomas, only a fraction of OTX2 binding sites are associated with enhancer and neighboring gene activity. Strong marks of enhancer activity, measured by H3K27ac levels, are consistently present in a subset of OTX2 binding sites, which are distinguished by the coordinated presence of NEUROD1 and a clustered arrangement of OTX2 signals.
Both OTX2 and NEUROD1 contribute to enhancer activation levels
The wide range of H3K27ac levels associated with OTX2 binding sites prompted us to assess the direct contribution of OTX2 to this chromatin mark. In accordance with previous studies, shRNA or CRISPR-Cas9 mediated depletion of OTX2 results in striking decreases in cell growth and viability of medulloblastoma cells (Figure S6A-B). We thus focused our analysis on the earliest time point after infection and selection (5 days), to highlight the most responsive OTX2 binding sites and their associated target genes.
The results of two independent shRNA knockdown experiments were highly consistent, and showed that decreased OTX2 levels were associated with strong reductions in H3K27ac in D341 cells (Figure 3A, correlation coefficient 0.76). While OTX2 has been previously associated with transcriptional repression in some systems (26, 35, 36) we did not observe significant increases in H3K27ac at direct binding sites to suggest a direct repressive effect on this chromatin mark (Figure 3A). Active OTX2 sites showed marked decreases in H3K27ac upon OTX2 loss, while sites initially devoid of H3K27ac remained mostly unaffected in D341 and D283 cells (Figure 3B-C and S6C-D). Similar chromatin results were obtained after CRISPR-Cas9 mediated OTX2 knockout in D341 cells (Figure S6E-H, correlation coefficient 0.69). OTX2 thus operates primarily as an activator of regulatory elements and makes major contributions to H3K27ac levels at its binding sites.
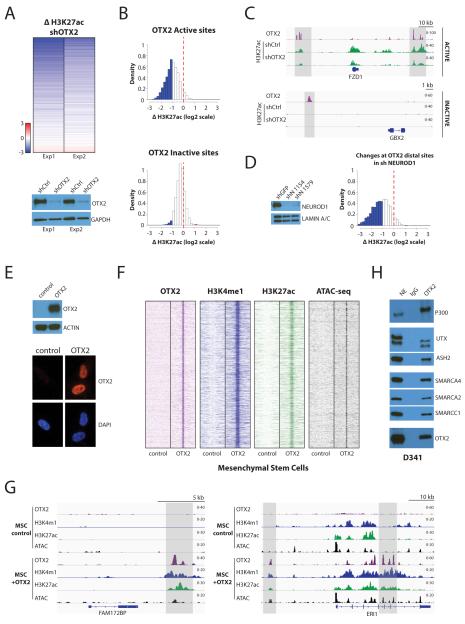
Figure 3. OTX2 maintains enhancer activity in Group 3 medulloblastoma and can operate as a pioneer factor in primary cells.
A. OTX2 is required to maintain marks of enhancer activity in D341 medulloblastoma cells. Top: A heatmap shows changes in H3K27ac ChIP-seq at distal sites with decreased OTX2 binding following knockdown with lentiviral shRNA (19513 sites are shown). Bottom: Immunoblotting for OTX2 in the two D341 knockdown experiments shows decreases in OTX2 protein levels.
B. Depletion of OTX2 has a strong effect on chromatin states of Active OTX2 sites compared to Inactive sites. Histograms show changes in H3K27ac at sites with decreased OTX2 after infection of D341 medulloblastoma cells with shRNA lentiviruses. Top: H3K27ac decreases in Active sites after OTX2 depletion (n=2649). Bottom: Inactive sites are mostly unaffected by OTX2 depletion (n=2240). The red line indicates no variation; blue bars correspond to more than two-fold decrease and red bars to more than two-fold increase.
C. Examples of ChIP-seq tracks of H3K27ac on Active and Inactive OTX2 distal sites following OTX2 knockdown in D341 medulloblastoma cells. OTX2 is shown in purple and H3K27ac in green. Regions of interest are shown in light gray.
D. Depletion of NEUROD1 affects chromatin states at OTX2 bound enhancers. Left: NEUROD1 immunoblotting in D341 knockdown experiments. Right: Histogram plot showing H3K27ac ChIP-seq changes at OTX2 distal sites with decreased NEUROD1 after infection of D341 medulloblastoma cells with lentiviral shRNA (3377 sites are shown). The red line indicates no variation; blue bars correspond to more than two-fold decrease and red bars to more than two-fold increase.
E. Expression of ectopic OTX2 in primary mesenchymal stem cells (MSCs). Top: OTX2 protein is detected in MSCs after lentiviral infection. Bottom: Immunofluorescence demonstrates the absence of OTX2 signals in control cells and the presence of nuclear signals after introduction of ectopic OTX2 in MSCs (72 hours post-infection).
F. OTX2 creates medulloblastoma enhancers de novo in MSCs. Heatmaps depict OTX2 (purple), H3K4me1 (blue) and H3K27ac (green) ChIP-seq signals as well as ATAC-seq (black) on 12286 de novo enhancers 72 hours after OTX2 expression in MSCs. Each row shows a 10 kb region centered on OTX2 binding sites in medulloblastoma.
G. Examples of OTX2 induced de novo enhancer creation in MSCs. OTX2 (purple), H3K4me1 (blue) and H3K27ac (green) ChIP-seq as well as ATAC-seq (black). Regions of interest are shown in light gray.
H. OTX2 interacts with chromatin modifying complexes associated with enhancer chromatin states. Co-immunoprecipitation experiments were performed in D341 nuclear extracts and show interactions with components involved in chromatin opening (SMARCA4, SMARCA2, SMARCC1), and deposition of the enhancer marks H3K4me1 (UTX, ASH2) and H3K27ac (p300).
Given our finding that active OTX2 binding sites are enriched for NEUROD1 we also tested whether NEUROD1 can contribute to H3K27ac levels at these locations. As in the case of OTX2, knockdown of NEUROD1 strongly decreased the viability of medulloblastoma cells suggesting that NEUROD1-mediated gene regulation is critical for tumor survival (Figure S6I). Similarly to OTX2, we focused our analysis on early timepoints after NEUROD1 knockdown, and noted marked decreases of H3K27ac levels at many active OTX2 binding sites (Figure 3D and S6J-K). Thus, NEUROD1 makes significant contributions to H3K27ac levels at a subset of OTX2 sites, consistent with cooperation between these transcription factors in the activation of enhancers. Taken together, data from our knockdown experiments shows that OTX2 makes major contributions to enhancer activation and cooperates with NEUROD1 to maintain enhancer function.
OTX2 can operate as a pioneer factor to activate enhancer sites de novo
To further test the direct effects of OTX2 on chromatin we ectopically expressed this transcription factor using lentiviral vectors in mesenchymal stem cells (MSCs), a non-neural cell type which has been shown to have a permissive chromatin environment in various reprogramming studies. In contrast to other primary stem cell models such as embryonic stem cells and neural stem cells, MSCs do not express either OTX2 or NEUROD1 (Figure 3E and Figure S7A) and thus allowed us to test the effects of OTX2 in isolation. After expression, OTX2 binding was detected at a majority of sites (65%) identified in medulloblastoma (Figure S7B). To highlight OTX2-dependent events we focused our attention on approximately 19000 genomic sites bound by OTX2 and devoid of detectable H3K4me1 basal enhancer marks in control MSC conditions. These sites were also characterized by the absence of signals for open chromatin by ATAC-seq (Figure 3F). Remarkably, the introduction of OTX2 was followed by profound chromatin changes at 66% of these sites including chromatin opening and the widespread de novo induction of enhancer activation marks resembling those observed in medulloblastoma (Figure 3F-G). In keeping with these results, co-immunoprecipitation experiments showed that OTX2 can interact with members of several complexes known to be involved in chromatin remodeling and enhancer activation including the acetyltransferase EP300 (associated with H3K27ac deposition at enhancers), as well as members of the H3K4 methyltransferase MLL complexes (UTX and ASH2, associated with H3K4me1) and the BAF chromatin remodeling complex (SMARCA4, SMARCA2 and SMARCC1, associated with chromatin opening) (Figures 3H and S7C). Thus, OTX2 is capable of interacting with a variety of chromatin remodeling complexes that may account for its ability to activate medulloblastoma enhancers de novo.
Using a similar approach, we also tested the contribution of each transcription factor at enhancer sites co-occupied by NEUROD1 and OTX2 in medulloblastoma. Our results show that OTX2 was able to bind 59% of these sites while NEUROD1 could only bind 20% after introduction into MSCs (Figure S7D), indicating a difference in the ability of the these transcription factors to access these locations. Among sites devoid of detectable H3K4me1 basal enhancer marks in control conditions, OTX2 could activate almost four fold more sites than NEUROD1 (n=1788 vs n=479) and a subset could be activated by either OTX2 or NEUROD1 (n=477) (Figure S7E). These results show that NEUROD1 can make contributions to enhancer activity by itself but is significantly less powerful than OTX2 at inducing chromatin remodeling at enhancers defined in medulloblastoma.
The mitotic kinase NEK2 is a critical OTX2 target gene and is associated with cell survival in medulloblastoma
Since it is well established that OTX2 regulates cell growth in medulloblastoma cells we next sought to identify specific target genes that may contribute to this property by analyzing coordinated changes in chromatin and gene expression. To achieve this we performed RNA-seq following acute OTX2 knockdown in D341 medulloblastoma cells, and integrated H3K27ac changes at OTX2 binding sites with gene expression changes at their nearest genes. As expected, changes in H3K27ac at the set of active OTX2 distal sites were associated with decreases in expression levels of neighboring genes (Figure 4A).
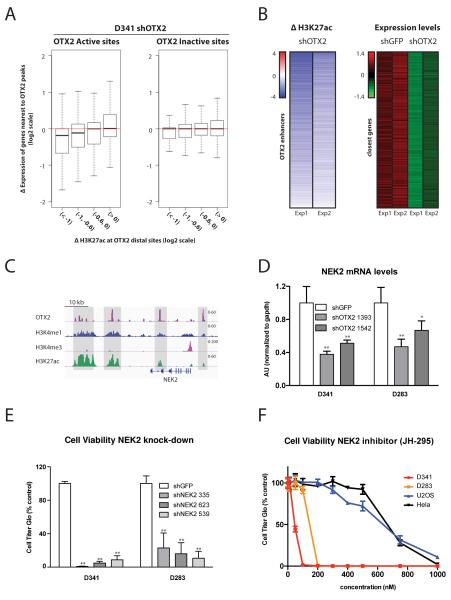
Figure 4. OTX2 dependent enhancer activity is associated with changes in the expression of target genes, including the mitotic kinase NEK2.
A. Genes close to OTX2 distal sites are sensitive to decreased OTX2 expression. Boxplot comparing changes in H3K27ac at sites with decreases in OTX2 binding after shRNA depletion with changes in the expression of closest genes in D341 cells. H3K27ac decreases are associated with reductions in expression of target genes. The effect is most evident in OTX2 Active sites (p-value= 5.65e-12 for Wilcoxon rank test between the first and fourth intervals).
B. Identification of a set of highly OTX2 responsive genes through integration of epigenomic and expression profiles. Heatmaps show significant chromatin changes and significant decreases in expression in genes regulated by OTX2 bound distal elements (FDR q-value=0.2). Left: H3K27ac changes at distal sites in two experiments (n=1347 OTX2 enhancers). Right: Z scores of gene expression levels in matching genes for two control (shGFP) and two OTX2 shRNA depletion experiments (n=510 genes).
C. Chromatin states at the NEK2 locus. ChIP-seq tracks showing OTX2 (purple), H3K4me1 (blue), H3K4me3 (pink) and H3K27ac (green) in D341 cells.
D. NEK2 expression is sensitive to OTX2 levels. RT-qPCR measuring NEK2 mRNA levels in D341 and D283 cell lines five days post-infection with lentiviral shRNAs targeting OTX2. ** Indicates p value < 0.01 and * Indicates p value < 0.05.
E. NEK2 knockdown affects medulloblastoma cell growth. Cell viability assays 14 days post-infection in D341 and D283 cell lines with three specific shRNA targeting NEK2. ** Indicates p value < 0.01.
F. Medulloblastoma cell lines are sensitive to NEK2 inhibition. Cell viability assays five days post-treatment with the indicated concentrations of NEK2 inhibitor JH-295 in the medulloblastoma cell lines D341 and D283 and the non-medulloblastoma cell lines U2OS and Hela.
We next selected loci with significant changes in chromatin and significant decreases in gene expression (Figure 4B and Table S3). Gene Set Enrichment Analyses (GSEA) showed enrichment in genes involved in controlling cell growth such as POLA2, RPA3, WEE1 and ECT2 (Figure S8A). We further refined this list by focusing on the set of genes that was highly expressed in Group 3 tumors compared to normal cerebellum in a previously published microarray dataset (4)(Figure S8B and Table S4). Among these genes we identified WEE1 and NEK2 (Figure S8B-C), two kinases that represent potential therapeutic targets. WEE1 inhibitors have been recently shown to significantly reduce the survival of medulloblastoma cells (37), supporting the notion that OTX2 target genes may play important roles in cell survival in this disease.
To further support this concept we set out to test the role of NEK2 in the survival of medulloblastoma cells. NEK2, which has been implicated in the regulation of centrosome separation and microtubule organization (38), is highly expressed in various cancers but has not been studied in medulloblastoma. The NEK2 locus is associated with several OTX2 bound active enhancers (Figure 4C), which showed a rapid decrease in H3K27 acetylation levels after knockdown (Figure S8D). NEK2 downregulation observed in RNA-seq experiments was confirmed by RT-qPCR in both D341 and D283 cell lines upon OTX2 knock-down (Figure 4D) and in D341 upon OTX2 knockout (Figure S8E), showing that the expression of this gene is highly responsive to OTX2 levels. We then tested the effects of NEK2 depletion using three different shRNAs and observed strong decreases in medulloblastoma cell viability upon knockdown (Figure 4E and S8F). In contrast, only a minority of 216 cancer cell lines in a recent shRNA screen (39) were sensitive to depletion of NEK2 in experiments that included one of the shRNAs used in our studies (Figure S8G). Finally, as a proof of concept, we tested the sensitivity of medulloblastoma cell lines to the recently described small-molecule NEK2 inhibitor JH-295 (40) and found that D341 and D283 cells were significantly more sensitive than unrelated control cells to NEK2 inhibition (Figure 4F). Our experiments thus show that OTX2 target genes can have significant effects on the growth of medulloblastoma cells and may represent an attractive set of candidate therapeutic targets for the treatment of Group 3 medulloblastoma and possibly other non-SHH medulloblastoma subtypes.
Discussion
Our genome-wide maps of chromatin states in Group 3 medulloblastoma show that OTX2 is a major feature of the regulatory landscape of these tumors. In keeping with OTX2 binding data from prior studies, our profiles show a large number of OTX2 binding sites across the genome (33). The combination of these profiles with our data on enhancer activation marks and knockdown studies further shows that OTX2 is found at 62% of consistently active enhancers in primary Group 3 tumors and that it makes significant contributions to enhancer activation states. Together with the fact that OTX2 is required for the survival of medulloblastoma cells, our studies point to a critical functional role for OTX2 in the regulatory landscape of Group 3 medulloblastoma.
While OTX2 expression is highest in Group 3 tumors, significant levels are also observed in the Wnt and Group 4 subtypes. In accordance with this expression pattern, our data show that a considerable fraction of OTX2 bound enhancers identified in Group 3 tumors are also active in Wnt and Group 4 tumors and thus suggest that OTX2 may also play an important role in the regulation of gene expression programs in these other medulloblastoma subtypes. In addition to these shared sites we also identified approximately 1000 enhancers that are highly active specifically in Group 3 tumors, indicating that the effects of OTX2 are dependent on differences in expression levels among subtypes or other modifying factors.
Remarkably, ectopic expression in primary cells also shows that OTX2 is capable of operating as a pioneer factor to induce active enhancers resembling those observed in Group 3 tumors. The fact that many of these sites start with a closed chromatin conformation implies that the introduction of OTX2 can initiate a set of coordinated chromatin remodeling activities, including chromatin opening and the deposition of marks of enhancer activity. In support of this, immunoprecipitation experiments show interactions between OTX2 and components of several complexes involved in enhancer function such as BAF, MLL and p300. The pioneer function of OTX2 also raises the possibility that a large fraction of the regulatory landscape of medulloblastoma cells could be the result of the pathogenic overexpression of OTX2. Given that the cell of origin of Group 3 tumors has not been defined, the widespread presence of OTX2 in the genome could be either a reflection of high expression in a cell of origin or a pathogenic event that alters the normal expression patterns of this transcription factor. Amplification of OTX2, which has been observed in a fraction of Group 3 medulloblastomas, may represent such an alteration but other mechanisms may also be involved since high OTX2 expression levels are observed in all non-SHH tumors (5, 24, 27, 32). Importantly, our data also suggests that OTX2 may maintain its own expression by a feed-forward mechanism in tumors since we identified multiple OTX2 bound active enhancers near the OTX2 locus.
The high degree of heterogeneity for activation marks revealed at OTX2 binding sites by our studies further highlights the importance of coordinated chromatin profiling in testing the direct impact of transcriptional regulators. In this case, the finding that the signal intensity of OTX2 peaks is not a good predictor for the level of chromatin activation points to key contributions by additional factors. In addition, as opposed to other transcription factors that may have both activation and repressive properties, our measurement of chromatin changes after OTX2 knockdown did not show a pattern compatible with direct repression at inactive sites. OTX2 thus appears to be a strong contributor to enhancer activation only at a subset of its binding sites.
Our data shows that the presence of the transcription factor NEUROD1 and a clustered arrangement of OTX2 signals are both linked to increased activation levels at OTX2 bound distal regulatory elements. The identification of NEUROD1 as a collaborating transcription factor is of particular significance because, together with our knockdown studies showing decreased cell survival, it points to NEUROD1 as a key transcriptional mediator in Group 3 medulloblastoma. NEUROD1 expression in medulloblastoma has been documented (34) but its function in this context was unknown. Like OTX2, NEUROD1 is expressed in cerebellar precursors and affects the development of this organ (41-43) but a cooperative relationship between these transcription factors has not been established. Both transcription factors have been shown to induce and activate enhancers in separate studies in embryonic stem cells (30, 31, 44) and, in the case of OTX2, a collaborative relationship with Oct4 in these cells was proposed based on increased H3K27ac signals (30, 31). This is analogous to our finding of cooperation with NEUROD1 and suggests that OTX2 function may be modulated by specific interactions in different settings. Given that our immunoprecipitation experiments show that endogenous OTX2 and NEUROD1 are present in the same complexes in medulloblastoma, screening for agents that alter this interaction may provide a new direction for the development of future therapies.
Taken together, our studies demonstrate that the direct characterization of regulatory networks in cancer has great potential to reveal critical aspects of oncogenic programs. By focusing on consistent activation patterns at distal enhancers our results show that the activity of OTX2 and its interaction with NEUROD1 are key regulatory events in Group 3 medulloblastoma and possibly in all non-SHH tumors. Given the importance of these transcription factors we also expect that their direct targets would be enriched for genes with critical functions. Accordingly, cell growth pathways were enriched among the most responsive OTX2 target genes and testing of the mitotic kinase NEK2 as a top candidate showed a strong dependency of medulloblastoma cells on the activity of this enzyme. While more extensive testing will be necessary to validate the use of NEK2 inhibitors in medulloblastoma, our analysis of this kinase further supports the concept that genome-wide characterization of chromatin states in cancer can uncover cellular dependencies on transcription factors and their target genes that may be exploited for the development of future cancer therapies.
Methods
Primary tumors and primary mesenchymal stem cells
Primary medulloblastoma specimens and primary mesenchymal stem cells were collected with approval from the Institutional Review Boards of Massachusetts General Hospital, Children’s Hospital Boston and Centre Hospitalier Universitaire Vaudois (CHUV, University of Lausanne). Samples were de-identified prior to our analysis. Group 3 subgroup classification was validated through gene expression.
Cell culture conditions and drugs
Cell lines were obtained directly from ATCC and not further authenticated. Media were obtained from Life Technologies. Human mesenchymal stem cells were cultured in Iscove’s modified Dulbecco’s medium (IMDM) containing 10% fetal calf serum (FCS) and 10 ng/ml platelet-derived growth factor BB (PeproTech), as described previously (45). The medulloblastoma cell lines D341 and D283 were grown in suspension in Ultra-Low attachment cell culture flasks (Corning) in iMEM and EMEM medium respectively. Hela and U2OS were grown in DMEM. All media were supplemented with 10% FCS and cells were cultivated at 37C with 5% CO2. Cells were maintained and split every 3-4 days according to ATCC recommendations. The NEK2 inhibitor JH295 was purchased from Millipore, reconstituted in DMSO and single use aliquots were conserved at −20C.
Lentiviral Generation
Lentiviruses were produced in 293T LentiX cells (Clontech) by co-transfecting the vector of interest with the packaging vectors GAG/POL and VSVg using LT1 reagent (Mirus Bio) according to manufacturer instructions. Viral supernatants were collected 72h after transfection and concentrated using LentiX concentrator (Clontech). Virus containing pellets were resuspended in PBS and added dropwise on cells in presence of media supplemented with 6 μg/mL polybrene. Selection of lentivirally-infected cells was achieved with puromycin used at 0.75-2 μg/ml (hpMSC and Medulloblastoma cell lines respectively). Overexpression or knockdown efficiency was determined by Western blot analysis and RT-qPCR.
Real-Time Quantitative RT-PCR and Western Blot Analysis
Total RNA was isolated from cells using NucleoSpin RNA (Clontech). cDNA was obtained starting from 500 ng of template and using a high-capacity RNA to-cDNA kit (Applied Biosystems) and random hexamers. Real-time PCR amplification was performed using fast SYBR Green Master Mix (Life Technologies) and specific PCR primers in a Lightcycler 480 instrument (Roche). Oligonucleotides used are provided as a supplementary file. Relative quantification of each target, normalized to an endogenous control (GAPDH), was performed using the comparative Ct method (Applied Biosystems). Error bars indicate SD of three technical replicates and represent at least two independent biological experiments. Statistical analyses were performed by Student’s t-test. Western blotting was performed using standard protocols. Primary antibodies used for Western blotting are listed in a supplementary table. Secondary antibodies were goat anti-rabbit and goat anti-mouse immunoglobulin G-horseradish peroxidase-conjugated (Bio-Rad, 1:10,000 dilution). Membranes were developed using Western Lightning Plus-ECL enhanced chemiluminescence substrate (PerkinElmer) and visualized using photographic film.
ChIP-seq
ChIP assays were carried out on D341, D283 and MSCs cultures of approximately 2–5 million cells per sample and per epitope, following the procedures described previously (46, 47). In brief, chromatin from formaldehyde-fixed cells was fragmented to a size range of 200–700 bases with a Branson 250 sonifier. Solubilized chromatin was immunoprecipitated with antibodies overnight at 4C. Antibody-chromatin complexes were pulled down with protein G-Dynabeads (Life Technologies), washed, and then eluted. After crosslink reversal, RNase A, and proteinase K treatment, immunoprecipitated DNA was extracted with AMP Pure beads (Beckman Coulter). ChIP DNA was quantified with Qubit. 2-5 ng ChIP DNA samples were used to prepare sequencing libraries, and ChIP DNA and input controls were sequenced with the Nextseq 500 Illumina genome analyzer. For primary tissue preparations, 10-30 mg of tumor was first cut on dry ice and chopped on ice with a razor blade. Samples were then resuspended in 1 mL cold PBS and fixed 15 minutes at room temperature by adding formaldehyde to a final concentration of 1%. Glycine was added for 5 minutes at room temperature. Samples were first washed in 1 mL cold PBS and resuspended in 1 mL cold PBS before manual homogenization with a syringe and processing as described above.
Processing of ChIP-seq data
ChIP-seq reads were aligned to the reference genome (hg19) using BWA (48). Aligned reads were extended to 200 bp to approximate fragment sizes and density maps were derived by counting the number of fragments overlapping each position with IGV tools (49). Density maps were normalized to 10M reads. ChIP-seq coverage was visualized with IGV (50). Peaks were identified with the MACS2 peak caller (51) using matched input controls and q-values of 10−2 for H3K27ac and OTX2, and 10−3 for NEUROD1. Peaks in blacklisted genomic regions identified by the ENCODE consortium (22) were excluded. Peak intersections were calculated using the Genomic Ranges Bioconductor package (52). Average signals for ChIP-seq data were calculated over genomic intervals using bwtools (53). Shared Group 3 medulloblastoma enhancers were defined by the overlap of H3K27ac peaks in at least 4 primary tumors. Regions within 1 kb of Refseq TSS locations (transcriptional start sites) and peaks with strong H3K4me3 signals typical of active promoters were subtracted from these intervals. OTX2 peaks were defined by the overlap of ChIP-seq signals obtained using two antibodies in two Group 3 medulloblastoma cell lines (D341 and D283). OTX2 peaks were classified as promoters if present within 1kb of Refseq TSSs or if they were associated with strong H3K4me3 signals. Promoter chromatin states for primary tumors were determined using H3K4me3 and H3K27me3 signals within 1kb of each TSS to define 4 categories of sites: Active (at least 4 tumors with H3K4me3 and no tumors with H3K27me3), Repressed (at least 4 tumors with H3K27me3 and no tumors with H3K4me3), Bivalent (at least 4 tumors with both H3K4me3 and H3K27me3 signals) or lacking all signals. Positive H3K4me3 loci were defined at a relative density of 2 based on the observed signal distribution. Loci were considered positive for H3K27me3 if signals were above 2 standard deviations from the mean ChIP input signal. OTX2 distal sites were classified as consistently Active or Inactive if they were in the top third (Active) or bottom third (Inactive) of the distribution of H3K27ac signals for at least three primary tumors and two cell lines. Differences in peak signals between tumor types or changes after depletion of OTX2 or NEUROD1 were compared using a t-test and corrected for multiple hypothesis testing to obtain FDR q-values. For knockdown and CRISPR experiments, sites with 1.5 fold decreases in OTX2 and 3 fold in NEUROD1 were considered for analysis. NEUROD1 peaks were called in two Group 3 medulloblastoma cell lines (D341 and D283). Regions cobound by OTX2 and NEUROD1 were defined by the overlap of OTX2 distal sites with NEUROD1 peaks in medulloblastoma. For experiments demonstrating ‘de novo’ enhancer induction in MSCs (mesenchymal stem cells), we selected OTX2 or NEUROD1 bound distal sites with H3K4me1 signals below 2 standard deviations from the mean ChIP input signal in control cells. To identify motif occurrences in peaks, we ran TFBSTools (54) (searchSeq function with min.score threshold set to 90%) in the Bioconductor package. Motif analysis and detailed peak annotation were performed with HOMER (55). For analysis of gene expression changes linked to chromatin states, peaks were assigned to the nearest transcriptional start sites (TSS) of hg19 Refseq genes (excluding miRNAs).
RNA-Seq and expression analysis
Total RNA was isolated from cells using NucleoSpin RNA (Clontech). 1 μg of total RNA was treated with Ribogold zero to remove ribosomal RNA. Remaining RNA was used to construct Illumina sequencing libraries with random primers according to manufacturer instructions using the Truseq Stranded RNA LT Kit. Reads were aligned with the STAR aligner to hg19 and expression values were calculated using either Cufflinks (for FPKM values) or HTseq (for counts). Differential expression for knockdown experiments was calculated using DESeq2. Microarray expression data for figure S2 were obtained from GEO (GSE37418)(10) and for figure S8 (http://archive.broadinstitute.org/pubs/medulloblastoma/cho) (4).
ATAC-seq genome-wide DNA accessibility assay
ATAC-seq analysis was performed as recently described (56). Briefly, 5×104 cells for each condition were first incubated in hypotonic buffer then resuspended in lysis buffer, centrifugated and resuspended in Transposase reaction mix for additional 30 min at 37C, following manufacturer recommendations (Nextera DNA sample Prep Kit, Illumina). After DNA purification, adaptor sequences were added to the fragmented DNA by PCR. Purified PCR products were sequenced using the Illumina Nextseq device. Paired end reads were aligned to hg19 using BWA (48). Read start sites were adjusted to represent the center of the transposon binding event (+4 bp in the plus strand and −5 bp in the minus strand). Signal densities were calculated over a sliding 150 bp window at 20 bp steps and normalized to 10M reads in each experiment.
Nuclear Extract Preparation and co-immunoprecipitation experiments
D341 cells were first washed in PBS and swollen on ice in Buffer A (10 mM Tris-HCl pH 8, 10 mM KCL, 1.5 mM MgCl2 supplemented with 1 mM DTT, protease/phosphatase inhibitors (Pierce) and 1mM PMSF). Cells were then lysed on ice in Buffer A containing 0.15 % NP-40. Nuclei were sedimented by centrifugation and resuspended in IPH Buffer (50 mM Tris-HCl pH 8, 150 mM NaCl, 5 mM EDTA, 0.5 % NP-40 and 10% glycerol supplemented with protease/phosphatase inhibitors (Pierce) and 1mM PMSF) before sonication in a QSONICA 800 R instrument. Nuclear protein supernatant was then collected after centrifugation for 15 min at 14 000 rpm and 4C. Proteins were quantified using a Bradford assay (Pierce).
For immunoprecipitation experiments, 500 μg of nuclear lysate were diluted in IPH buffer to a final volume of 1 mL and incubated overnight at +4°C with 2 μg of the indicated antibodies in the presence of magnetic G-Dynabeads (Life Technologies) and 100 μg/mL Ethidium Bromide (SIGMA-ALDRICH). Beads were washed 5 times with IPH buffer and eluted by boiling in loading Laemmli buffer.
Plasmids and Antibodies
shRNAs were cloned in the pLKO.1 lentiviral backbone. The coding sequence of OTX2 mRNA (NM_172337) was PCR amplified from D341 cells and cloned into a pENTR-D-TOPO vector before Gateway reaction into an expression vector. gRNAs targeting OTX2 were cloned into the lentiviral CRISPR-Cas9 vector (Addgene #52961) following standard procedures. Sequence information and antibody references are provided in Table S5.
Immunofluorescence staining
Staining was performed using standard protocols. Briefly, cells were fixed in a 1X PBS solution containing 4% paraformaldehyde 15 minutes at room temperature (RT), washed with 1X PBS and stored at 4C. Cells were permeabilized 10 minutes at RT in 1X PBS containing 0.5 % Triton X-100 then blocked for 30 minutes at RT, stained with OTX2 antibody for 2 hours at RT and with Alexa-Fluor 546-conjugated secondary antibody (Life Technologies) for 1h at RT. Cells were washed with 1X PBS between each step of the protocol. Nuclei were stained with DAPI solution.
Cell viability assays
Medulloblastoma and non-medulloblastoma cell lines were seeded in triplicates and grown under log phase growth conditions in Ultra-Low attachment 24 well plate and cell culture treated 96 well plate respectively. After the indicated incubation times, cell viability was measured in triplicates using the CellTiter-Glo luminescent assay (Promega) as described by the manufacturer. Endpoint luminescence was measured on a SpectraMax M5 plate reader (Molecular Devices). The data displayed are representative of at least two biological experiments performed in duplicates or triplicates. Statistical analyses were performed by Student’s t-test.
Supplementary Material
Statement of significance.
The gene regulation mechanisms that drive medulloblastoma are not well understood. Using chromatin profiling, we find that the transcription factor OTX2 acts as a pioneer factor and, in cooperation with NEUROD1, controls the Group 3 medulloblastoma active enhancer landscape. OTX2 itself or its target genes, including the mitotic kinase NEK2, represent attractive targets for future therapies.
Acknowledgements
We would like to thank Ivan Stamenkovic, Shawn Gillespie, Leah Escalante, Mario Suva, David Ebb, Nancy Tarbell, Torunn Yock and Howard Weinstein for valuable advice and support.
Financial support:
This study was supported by research grants from A Kids’ Brain Tumor Cure Foundation, aka The PLGA Foundation (M.N.R). M.N.R. receives research support from the V Foundation for Cancer Research. N.R. is supported by the Swiss National Science Foundation Professorship grant (PP00P3-157468/1) and the MEDIC Foundation. S.P. and J.P.M were supported by a research grant from NCI (R01 CA109467).
Footnotes
Disclosure of potential conflict of interest:
M.N.R. receives research support from Affymetrix. No potential conflicts of interest were disclosed by the other authors.
G.B. and M.N.R designed the study and wrote the manuscript.
G.B., M.E.A., N.R., W.E.B., N.E.R., B.N., S.R. and A.V. performed the experiments.
M.N.R, M.J.A., S.I. and G.B. conducted bioinformatics analyses.
T.C.A, J.C.K, J.P.M., P.T. and S.L.P provided necessary reagents and conceptual advice.
Accession Numbers
The data accompanying this paper have been deposited into GEO under accession number GSE92585.
References
- 1.Ostrom QT, Gittleman H, Fulop J, Liu M, Blanda R, Kromer C, et al. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2008-2012. Neuro Oncol. 2015;17(Suppl 4):iv1–iv62. doi: 10.1093/neuonc/nov189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson MC, Fuller C, Hogg TL, Dalton J, Finkelstein D, Lau CC, et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24:1924–31. doi: 10.1200/JCO.2005.04.4974. [DOI] [PubMed] [Google Scholar]
- 3.Kool M, Koster J, Bunt J, Hasselt NE, Lakeman A, van Sluis P, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One. 2008;3:e3088. doi: 10.1371/journal.pone.0003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho YJ, Tsherniak A, Tamayo P, Santagata S, Ligon A, Greulich H, et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol. 2011;29:1424–30. doi: 10.1200/JCO.2010.28.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29:1408–14. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123:465–72. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kool M, Korshunov A, Remke M, Jones DT, Schlanstein M, Northcott PA, et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012;123:473–84. doi: 10.1007/s00401-012-0958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–10. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Northcott PA, Shih DJ, Peacock J, Garzia L, Morrissy AS, Zichner T, et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature. 2012;488:49–56. doi: 10.1038/nature11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–8. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones DT, Jager N, Kool M, Zichner T, Hutter B, Sultan M, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–5. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibson P, Tong Y, Robinson G, Thompson MC, Currle DS, Eden C, et al. Subtypes of medulloblastoma have distinct developmental origins. Nature. 2010;468:1095–9. doi: 10.1038/nature09587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 14.Gidekel S, Pizov G, Bergman Y, Pikarsky E. Oct-3/4 is a dose-dependent oncogenic fate determinant. Cancer Cell. 2003;4:361–70. doi: 10.1016/s1535-6108(03)00270-8. [DOI] [PubMed] [Google Scholar]
- 15.Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–77. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 16.Suva ML, Rheinbay E, Gillespie SM, Patel AP, Wakimoto H, Rabkin SD, et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157:580–94. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suva ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339:1567–70. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leung D, Jung I, Rajagopal N, Schmitt A, Selvaraj S, Lee AY, et al. Integrative analysis of haplotype-resolved epigenomes across human tissues. Nature. 2015;518:350–4. doi: 10.1038/nature14217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsankov AM, Gu H, Akopian V, Ziller MJ, Donaghey J, Amit I, et al. Transcription factor binding dynamics during human ES cell differentiation. Nature. 2015;518:344–9. doi: 10.1038/nature14233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–61. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Consortium EP An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin CY, Erkek S, Tong Y, Yin L, Federation AJ, Zapatka M, et al. Active medulloblastoma enhancers reveal subgroup-specific cellular origins. Nature. 2016;530:57–62. doi: 10.1038/nature16546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di C, Liao S, Adamson DC, Parrett TJ, Broderick DK, Shi Q, et al. Identification of OTX2 as a medulloblastoma oncogene whose product can be targeted by all-trans retinoic acid. Cancer Res. 2005;65:919–24. [PubMed] [Google Scholar]
- 25.Adamson DC, Shi Q, Wortham M, Northcott PA, Di C, Duncan CG, et al. OTX2 is critical for the maintenance and progression of Shh-independent medulloblastomas. Cancer Res. 2010;70:181–91. doi: 10.1158/0008-5472.CAN-09-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bunt J, Hasselt NE, Zwijnenburg DA, Hamdi M, Koster J, Versteeg R, et al. OTX2 directly activates cell cycle genes and inhibits differentiation in medulloblastoma cells. Int J Cancer. 2012;131:E21–32. doi: 10.1002/ijc.26474. [DOI] [PubMed] [Google Scholar]
- 27.Northcott PA, Jones DT, Kool M, Robinson GW, Gilbertson RJ, Cho YJ, et al. Medulloblastomics: the end of the beginning. Nat Rev Cancer. 2012;12:818–34. doi: 10.1038/nrc3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weeraratne SD, Amani V, Teider N, Pierre-Francois J, Winter D, Kye MJ, et al. Pleiotropic effects of miR-183~96~182 converge to regulate cell survival, proliferation and migration in medulloblastoma. Acta Neuropathol. 2012;123:539–52. doi: 10.1007/s00401-012-0969-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beby F, Lamonerie T. The homeobox gene Otx2 in development and disease. Exp Eye Res. 2013;111:9–16. doi: 10.1016/j.exer.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 30.Buecker C, Srinivasan R, Wu Z, Calo E, Acampora D, Faial T, et al. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell. 2014;14:838–53. doi: 10.1016/j.stem.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang SH, Kalkan T, Morissroe C, Marks H, Stunnenberg H, Smith A, et al. Otx2 and Oct4 drive early enhancer activation during embryonic stem cell transition from naive pluripotency. Cell Rep. 2014;7:1968–81. doi: 10.1016/j.celrep.2014.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boon K, Eberhart CG, Riggins GJ. Genomic amplification of orthodenticle homologue 2 in medulloblastomas. Cancer Res. 2005;65:703–7. [PubMed] [Google Scholar]
- 33.Hovestadt V, Jones DT, Picelli S, Wang W, Kool M, Northcott PA, et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature. 2014;510:537–41. doi: 10.1038/nature13268. [DOI] [PubMed] [Google Scholar]
- 34.Rostomily RC, Bermingham-McDonogh O, Berger MS, Tapscott SJ, Reh TA, Olson JM. Expression of neurogenic basic helix-loop-helix genes in primitive neuroectodermal tumors. Cancer Res. 1997;57:3526–31. [PubMed] [Google Scholar]
- 35.Bai RY, Staedtke V, Lidov HG, Eberhart CG, Riggins GJ. OTX2 represses myogenic and neuronal differentiation in medulloblastoma cells. Cancer Res. 2012;72:5988–6001. doi: 10.1158/0008-5472.CAN-12-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bunt J, Hasselt NA, Zwijnenburg DA, Koster J, Versteeg R, Kool M. OTX2 sustains a bivalent-like state of OTX2-bound promoters in medulloblastoma by maintaining their H3K27me3 levels. Acta Neuropathol. 2013;125:385–94. doi: 10.1007/s00401-012-1069-2. [DOI] [PubMed] [Google Scholar]
- 37.Harris PS, Venkataraman S, Alimova I, Birks DK, Balakrishnan I, Cristiano B, et al. Integrated genomic analysis identifies the mitotic checkpoint kinase WEE1 as a novel therapeutic target in medulloblastoma. Mol Cancer. 2014;13:72. doi: 10.1186/1476-4598-13-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia J, Franqui Machin R, Gu Z, Zhan F. Role of NEK2A in human cancer and its therapeutic potentials. Biomed Res Int. 2015;2015:862461. doi: 10.1155/2015/862461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cowley GS, Weir BA, Vazquez F, Tamayo P, Scott JA, Rusin S, et al. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci Data. 2014;1:140035. doi: 10.1038/sdata.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henise JC, Taunton J. Irreversible Nek2 kinase inhibitors with cellular activity. J Med Chem. 2011;54:4133–46. doi: 10.1021/jm200222m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frantz GD, Weimann JM, Levin ME, McConnell SK. Otx1 and Otx2 define layers and regions in developing cerebral cortex and cerebellum. J Neurosci. 1994;14:5725–40. doi: 10.1523/JNEUROSCI.14-10-05725.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyata T, Maeda T, Lee JE. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 1999;13:1647–52. doi: 10.1101/gad.13.13.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vernay B, Koch M, Vaccarino F, Briscoe J, Simeone A, Kageyama R, et al. Otx2 regulates subtype specification and neurogenesis in the midbrain. J Neurosci. 2005;25:4856–67. doi: 10.1523/JNEUROSCI.5158-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pataskar A, Jung J, Smialowski P, Noack F, Calegari F, Straub T, et al. NeuroD1 reprograms chromatin and transcription factor landscapes to induce the neuronal program. EMBO J. 2016;35:24–45. doi: 10.15252/embj.201591206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riggi N, Suva ML, De Vito C, Provero P, Stehle JC, Baumer K, et al. EWS-FLI-1 modulates miRNA145 and SOX2 expression to initiate mesenchymal stem cell reprogramming toward Ewing sarcoma cancer stem cells. Genes Dev. 2010;24:916–32. doi: 10.1101/gad.1899710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–92. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pohl A, Beato M. bwtool: a tool for bigWig files. Bioinformatics. 2014;30:1618–9. doi: 10.1093/bioinformatics/btu056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan G, Lenhard B. TFBSTools: an R/bioconductor package for transcription factor binding site analysis. Bioinformatics. 2016;32:1555–6. doi: 10.1093/bioinformatics/btw024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–8. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.