Abstract
Piwi proteins utilize small RNAs (piRNAs) to recognize target transcripts such as transposable elements (TE). However, extensive piRNA sequence diversity also suggests that Piwi/piRNA complexes interact with many transcripts beyond TEs. To determine Piwi target RNAs, we used ribonucleoprotein-immunoprecipitation (RIP) and cross-linking and immunoprecipitation (CLIP) to identify thousands of transcripts associated with the Piwi proteins XIWI and XILI (Piwi-protein-associated transcripts, PATs) from early stage oocytes of X. laevis and X. tropicalis. Most PATs associate with both XIWI and XILI and include transcripts of developmentally important proteins in oogenesis and embryogenesis. Only a minor fraction of PATs in both frog species displayed near perfect matches to piRNAs. Since predicting imperfect pairing between all piRNAs and target RNAs remains intractable, we instead determined that PAT read counts correlate well with the lengths and expression levels of transcripts, features that have also been observed for oocyte mRNAs associated with Drosophila Piwi proteins. We used an in vitro assay with exogenous RNA to confirm that XIWI associates with RNAs in a length- and concentration-dependent manner. In this assay, noncoding transcripts with many perfectly matched antisense piRNAs were unstable, whereas coding transcripts with matching piRNAs were stable, consistent with emerging evidence that Piwi proteins both promote the turnover of TEs and other RNAs, and may also regulate mRNA localization and translation. Our study suggests that Piwi proteins play multiple roles in germ cells and establishes a tractable vertebrate system to study the role of Piwi proteins in transcript regulation.
Keywords: Piwi, Xenopus, oocyte, XIWI, XILI
INTRODUCTION
Animal oocytes are generally large cells containing large quantities of maternally deposited mRNAs and proteins that are required to drive several rounds of cell division prior to the onset of zygotic transcription (Tadros and Lipshitz 2009). Among the most important maternally deposited ribonucleoproteins (RNPs) are germ cell enriched RNPs belonging to the RNA interference (RNAi) pathway, such as the Piwi pathway. Piwi proteins are conserved from basal animals to vertebrates (Grimson et al. 2008) and bind to short RNAs called Piwi-interacting RNAs (piRNAs), many of which have complementarity to transposable element (TE) sequences (Juliano et al. 2011; Siomi et al. 2011; Clark and Lau 2014). The base-pairing interactions between piRNAs and TE transcripts allows Piwi proteins to directly repress TEs, thus preserving the integrity of the germ cell genome (Han et al. 2015; Mohn et al. 2015; Senti et al. 2015; Wang et al. 2015; Yang et al. 2016).
However, a second, less understood function for Piwi proteins is to serve as a regulator of RNAs in the germ plasm (Harris and Macdonald 2001; Findley et al. 2003; Megosh et al. 2006), an asymmetrically localized compartment in animal oocytes composed of maternal RNAs and proteins that is later segregated into germ cell blastomeres (Strome and Updike 2015). Many important maternal RNAs in germ plasm are tightly regulated during oogenesis and embryogenesis to ensure proper development of the zygote. Although Piwi proteins and piRNAs are also enriched in animal germ plasms, it remains poorly understood if Piwi/piRNA complexes are involved in the regulation of these maternal RNAs, because many mutations in Piwi-pathway genes severely compromise early steps of gonadogenesis (Lin and Spradling 1997; Cox et al. 1998; Harris and Macdonald 2001; Findley et al. 2003), and thus hinder the study of later stage gametes.
Although many piRNAs contain TE sequences, an equally large if not greater number lack sequence homology with TEs because there are millions of unique piRNAs that are generated from many genomic loci that can be depleted of TE sequences (Girard et al. 2006; Lau et al. 2006; Li et al. 2013; Chirn et al. 2015). With this tremendous piRNA sequence diversity, the guiding mechanism of Piwi/piRNA complexes is unclear if imperfect base-pairing is considered (Post et al. 2014). Additionally, the intrinsic RNA-binding activities of Piwi proteins could further broaden the range of Piwi-protein associated transcripts (PATs) (Sytnikova et al. 2014). Therefore, understanding the regulatory mechanisms of animal Piwi proteins during gametogenesis requires a complete catalog of PATs across multiple animal species.
RNP-immunoprecipitation (RIP) and cross-linking immunoprecipitation (CLIP) are common methods that have provided tremendous insight into transcripts associated with various proteins and preferred binding sites of RNA-binding proteins (Ule et al. 2003; Keene et al. 2006; Licatalosi et al. 2008). A common goal of RNA target profiling experiments on Argonaute (AGO) and Piwi proteins (Chi et al. 2009; Hafner et al. 2010; Zisoulis et al. 2010; Leung et al. 2011; Vourekas et al. 2012, 2016; Sytnikova et al. 2014; Barckmann et al. 2015) has been to develop the set of sequence-recognition preferences between these RNP, the small RNAs bound within these RNPs, and their associated target mRNA transcripts. The AGO protein studies have reinforced mechanisms of miRNAs targeting 3′ untranslated regions (3′ UTRs) in mRNAs by the 5′ end “seed sequence” of the miRNAs, and revealed additional motifs in mRNAs that AGO proteins can bind independently of the miRNA (Chi et al. 2009, 2012; Hafner et al. 2010; Zisoulis et al. 2010; Leung et al. 2011; Baigude et al. 2012; Clark et al. 2014; Grosswendt et al. 2014). The immense diversity of piRNA sequences across animals continues to present a bioinformatics challenge in determining which piRNA-to-mRNA pairing is most meaningful (Vourekas et al. 2012, 2016; Post et al. 2014).
However, a bulk amount of piRNAs complementary to a nascent transcript is required to trigger Drosophila PIWI-dependent silencing (Post et al. 2014), which explains how TE transcripts are the most strongly regulated target and most abundant transcripts in PIWI CLIP-seq (Sytnikova et al. 2014). In addition, the majority of Drosophila piRNAs tend to be antisense to TE transcripts and therefore likely serve as targeting guides (Saito et al. 2006; Vagin et al. 2006; Brennecke et al. 2007; Ghildiyal et al. 2008; Malone et al. 2009). In spite of this, Piwi proteins in both Drosophila and mouse can associate broadly with protein-coding mRNAs as well as long-noncoding RNAs, including in a piRNA-independent fashion (Vourekas et al. 2012, 2016; Sytnikova et al. 2014; Barckmann et al. 2015). This broad association of mRNAs with Piwi proteins was most recently observed in Drosophila oocyte germ plasm (Barckmann et al. 2015; Vourekas et al. 2016), with a newly emerging hypothesis proposing that piRNA base-pairing requirements are so loose that transcript association with Piwi proteins may depend upon its length and its abundance (Vourekas et al. 2016).
We previously showed that several oocyte mRNAs associate with the X. tropicalis protein, XIWI (also known as PIWIL1 by the HUGO nomenclature) (Lau et al. 2009a). To comprehensively identify Xenopus oocyte PATs, we first characterized the expression and oocyte localization of the XIWI ortholog, XILI (also known as PIWIL2). We then utilized XIWI and XILI antibodies to perform RIP-seq and CLIP-seq in X. laevis and X. tropicalis oocytes, respectively, to generate extensive lists of Xenopus PATs and determine how many transcripts interact with both XIWI and XILI between these two frog species. Additionally, we characterized X. laevis oocyte piRNA populations and define intergenic piRNA cluster loci within the tetraploid genome of X. laevis. Finally, we demonstrate the capacity of an X. laevis oocyte extract to test for the stability of particular PATs with abundant amounts of mapping piRNAs as well as the capacity of exogenous RNA to interact with XIWI. This study expands our knowledge about the role of Piwi protein in Xenopus species, and provides a platform for future dissection of the Piwi-protein-dependent regulatory mechanisms for maternally deposited RNAs in Xenopus oocytes.
RESULTS
Characterization of X. laevis Piwi proteins, XIWI and XILI
Having previously characterized XIWI expression and localization in X. tropicalis (Lau et al. 2009a), we then turned our attention to its homolog, XILI, which is well expressed in X. laevis oocyte mRNA and protein profiles (Wilczynska et al. 2009; Wuhr et al. 2014). Using new gene models for X. laevis genes (James-Zorn et al. 2015), we examined the similarity of the X. laevis XIWI and XILI protein sequences with Piwi genes from other vertebrates and Drosophila. Phylogenetic analysis confirmed that Xenopus Piwi proteins are more evolutionarily similar to their mammalian homologs than to either fly or fish homologs (Fig. 1A). We raised antibodies for XILI as we had done previously for XIWI (Lau et al. 2009a), and found that both antibodies recognize a single predominant band in stage 1–3 oocyte extract (Fig. 1B). Additionally, we found no cross-reactivity between these antibodies in either IP or immunodepletion assays (Fig. 1C,D), indicating that these antibodies are highly specific. To determine the timing of XILI expression during oogenesis, we blotted for XILI in staged X. laevis oocytes (Dumont 1972) and observed that, similar to XIWI (Lau et al. 2009a), XILI is most highly expressed in early stage oocytes (Fig. 1E). Therefore, in all subsequent experiments, X. laevis oocyte extracts were made using only stage 1–3 oocytes.
FIGURE 1.
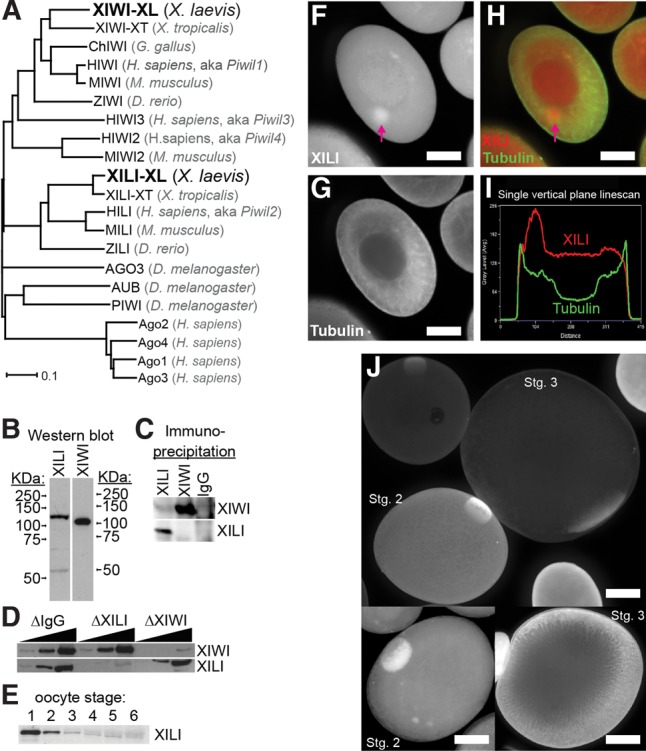
Characterization of XILI, homolog of XIWI, in X. laevis oocytes. (A) Phylogenetic tree of a selection of vertebrate Piwi proteins compared against the Drosophila Piwi proteins. The chicken homolog of PIWIL2/HILI was omitted because only a partial fragment of the protein sequence exists. Human Argonaute (Ago) proteins are used as an outgroup subclade to root the tree. (B) Western blot for XILI and XIWI in X. laevis stage 1–3 oocyte extract. (C) Western blot for XILI and XIWI in immuno-precipitates from XILI, XIWI, and preimmune rabbit IgG (mock precipitation). (D) Western blot for XILI and XIWI in immunodepleted X. laevis stage 1–3 oocyte extracts. (E) Western blot for XILI in a single X. laevis oocyte at the indicated stage of oogenesis. (F–I) Confocal imaging of XILI localization in a stage 1 X. laevis oocyte, with XILI and Tubulin patterns alone (F,G) and merged (H). Arrow points to the concentration of XILI in the Balbiani body. (I) Quantification of the XILI and Tubulin signals on a vertical plane that bisects the Balbiani body, supporting the XILI concentration in that organelle as well as in the germinal vesicle/nucleus of the oocyte. Scale bar represents 50 microns. (J) Confocal imaging of XILI localization in stage 2 and 3 oocytes.
To determine the localization of XILI, we performed whole-mount immunofluorescence and confocal microscopy on stage 1–3 oocytes (Fig. 1F–J). Similar to XIWI (Lau et al. 2009a), XILI was present throughout the cytoplasm but was concentrated in the Balbiani body, which is a concentration of mitochondria, endoplasmic reticulum (ER), and mRNAs that will contribute to germ cell formation (Kloc et al. 2014). However, in contrast to XIWI that is predominantly cytoplasmic (Lau et al. 2009a), XILI was also observed in the nucleus (Fig. 1I). Our microscopy data provide three-dimensional resolution of XILI that differs with the biochemical fractionation studies of XIWI and XILI (Wilczynska et al. 2009). Nevertheless, our current and previous data on XILI and XIWI localization patterns (Lau et al. 2009a) are reminiscent of mouse MIWI2 being both cytoplasmic and nuclear, versus MIWI that is predominantly cytoplasmic (Aravin et al. 2008, 2009; Yang et al. 2016); and Drosophila PIWI's localization in ovarian cells, which is mainly nuclear but can also shuttle between the cytoplasm and the nucleus (Cox et al. 2000; Harris and Macdonald 2001; Brennecke et al. 2007; Gunawardane et al. 2007; Zamparini et al. 2011; Mani et al. 2014).
The majority of X. laevis piRNAs map to nonrepetitive, intergenic loci
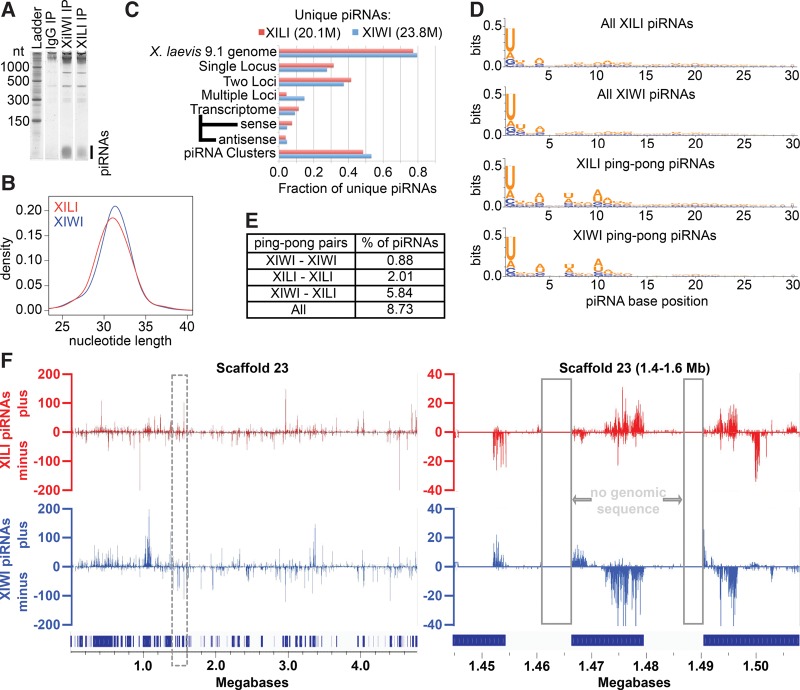
To further characterize X. laevis XIWI and XILI, we sequenced piRNAs bound by XIWI and XILI, which should guide these proteins to associate with target RNAs. Sequencing statistics of these and all other libraries are detailed in Supplemental Table S1. Both XIWI and XILI IPs from X. laevis oocyte extracts revealed stably bound piRNAs as well as large (>300 nt) RNAs associated with these Piwi/piRNA complexes (Fig. 2A). Neither of these coprecipitating RNA populations was present in mock IPs performed with total IgG from preimmune rabbit serum. Deep sequencing of the coprecipitating small RNAs (which we will refer to as the piRNA-seq libraries) produced over 20 million unique reads for both XIWI and XILI bound piRNAs. The length distributions were unimodal over a principal range of 29–34 nt, with XIWI piRNAs being slightly larger than XILI piRNAs on average (Fig. 2B), and also distinctly longer than X. tropicalis XIWI-associated piRNAs (27–31 nt) (Lau et al. 2009a). This result confirms the extensive variations in average piRNA lengths between different Piwi proteins within a species and between different animal species (Grivna et al. 2006a; Brennecke et al. 2007; Aravin et al. 2008; Gan et al. 2011; Williams et al. 2015). Notably, X. laevis piRNAs have the longest length for the mode peak at ∼32 nt, compared to other animal piRNAs (zebrafish: 27 nt, chicken: 25 nt, X. tropicalis: 29 nt, human: 30 nt) (Chirn et al. 2015).
FIGURE 2.
Distinctions between X. laevis oocyte XIWI and XILI piRNAs. (A) Coprecipitating RNAs from XIWI, XILI, and mock (preimmune rabbit IgG) IPs were separated on a 15% urea polyacrylamide gel and stained with SYBR Gold. (B) Histogram of XIWI and XILI piRNA strand lengths from the piRNA-seq library. (C) piRNAs mapped to X. laevis genome build 9.1 and counted against JGI v.1.8 transcripts (transcriptome) and piRNA cluster transcript models. (D) Base frequencies of all XIWI and XILI piRNAs as well as piRNAs with ping-pong signatures. (E) Percent of genomic mapping piRNAs with ping-pong signatures. (F) XIWI and XILI piRNAs mapping to Scaffold 23 in the X. laevis genome build 9.1, with a zoomed region from the dashed line box shown on the right panel. The piRNA cluster transcript models are denoted by dark blue rectangles above scaffold coordinates on the x-axis.
Nearly 80% of the piRNAs from XIWI and XILI IPs mapped to the X. laevis genome build 9.1 (Session et al. 2016), which is consistent with similar 90% and 82% proportions of mouse MIWI and MILI mapping piRNAs, respectively (Lau et al. 2009b; Chirn et al. 2015). This result suggests that the X. laevis genome tetraploidy does not significantly hamper our genomic analysis of X. laevis piRNAs. There are similar proportions of piRNAs in mammals and frogs that are un-mappable, possibly because they arise from repetitive sequences in heterochromatin that are refractory to genome assembly. However, the tetraploidy of nine pairs of homologous chromosomes (Uno et al. 2013; Session et al. 2016) might distort how frequently X. laevis piRNAs would map. Therefore, we searched for piRNAs that mapped to three or more loci, and found that only a minority of piRNAs map to multiple loci (Fig. 2C), suggesting that the vast majority of piRNAs in stage 1–3 oocytes do not bear the sequences of the most highly repetitive of sequences such as tRNA fragments that can also give rise to bonafide piRNAs (Lau et al. 2006; Houwing et al. 2007; Senti et al. 2015). Since nearly ∼70% of X. laevis piRNAs map to a single locus or two loci (accounting for polymorphisms between the tetraploid alleles), this suggests X. laevis oocyte piRNAs are similar to mouse pachytene piRNAs that also map uniquely to single sites in major intergenic piRNA cluster loci (Girard et al. 2006; Lau et al. 2006; Gan et al. 2011; Li et al. 2013; Han and Zamore 2014; Chirn et al. 2015).
In contrast to X. tropicalis and other animal genomes, there is still only an incomplete list of TE consensus sequences for the X. laevis genome build 9.1 (Session et al. 2016). This hampered our determination of the total proportion of X. laevis piRNAs directed at TEs, which could be similar to the minor fraction (<25%) of X. tropicalis piRNAs that derive from or are specifically targeting TEs (Armisen et al. 2009; Lau et al. 2009a). Nevertheless, the existence of multiply mapping X. laevis piRNAs suggest the existence of TE-derived piRNAs, with XIWI piRNAs more frequently mapping to multiple loci (18.4% of genome-mapping piRNAs) than XILI piRNAs (5.5% of genome-mapping piRNAs).
In insects and vertebrates, many TE-directed piRNAs are generated through a biogenesis mechanism called “ping-pong” (Aravin et al. 2007b, 2008; Brennecke et al. 2007; Gunawardane et al. 2007), which describes two opposing Piwi proteins engaging on complementary piRNA precursor transcripts and generating ping-pong piRNA pairs by slicing activity. Although the major bulk of X. laevis XIWI and XILI piRNAs display the most common bias for uracil at the 5′ end, the typical hallmark of primary piRNAs (Aravin et al. 2006; Girard et al. 2006; Brennecke et al. 2007; Gunawardane et al. 2007), a smaller but significant subset of piRNA pairs display ping-pong signature: a bias for adenosine at the tenth base position that is characteristic of secondary piRNAs (Fig. 2D). Nearly 9% of X. laevis oocyte piRNAs exhibited ping-pong signatures, two-thirds of which are between XIWI–XILI pairs, with ∼23% of these being between XILI–XILI pairs, and ∼10% between XIWI–XIWI pairs (Fig. 2E). Thus, ping-pong piRNAs are more readily detected in X. laevis oocytes compared to a smaller fraction in X. tropicalis oocytes (Lau et al. 2009a). In addition, the most numerous sets of pairs between XIWI and XILI are reminiscent of mouse MIWI2–MIILI complexes forming the most frequent ping-pong piRNA pairs in the testes (Aravin et al. 2008).
X. laevis piRNA cluster configurations can be large and display overlapping piRNAs from both genomic strands
The bulk of piRNAs derive from large, intergenic piRNA cluster loci that generate long-noncoding RNA (lncRNA) transcripts as one of the first precursors to piRNAs (Aravin et al. 2006; Girard et al. 2006; Lau et al. 2006; Li et al. 2013). To determine X. laevis piRNA clusters, we used the StringTie program (Pertea et al. 2015) to assemble the piRNA reads into transcripts based on the X. laevis genome build 9.1. Our StringTie assembly predicted 20,971 transcripts, of which nearly two-thirds of piRNAs that map to the genome also map to these conceptual piRNA clusters (Fig. 2C). These clusters, 97.4% of which are intergenic, range in size from 0.5–433 kb. However, due to the incomplete assembly of the X. laevis genome, we cannot yet determine the exact size and number of piRNA clusters. For example, piRNAs map along the entire length of a massive 4.7 Mb Scaffold 23 (Fig. 2F, left panel), but many large gaps in the genomic sequence renders the piRNA profiles into 76 separate piRNA clusters. Closer inspection of one of these clusters reveals peaks of piRNA coverage separated by genomic gaps and piRNAs mapping right up to the ends these gaps (Fig. 2F, right panel), suggesting that the inflated number of piRNA clusters is due to incomplete genome assembly.
Nevertheless, XIWI and XILI bound piRNAs displayed an interesting mapping pattern to opposing genomic strands in Scaffold 23, between 1.48 Mb and 1.49 Mb (Fig. 2F, right panel). In contrast to dual-strand intergenic piRNA clusters in Drosophila, zebrafish, and chicken where many piRNAs overlap across genomic loci (Brennecke et al. 2007; Houwing et al. 2007, 2008; Chirn et al. 2015), mammalian and X. tropicalis intergenic piRNA clusters mainly generate piRNAs from a single genomic strand. Additionally, in dual-strand clusters stimulated for transcription by Myb factors (Li et al. 2013), there are two divergent lncRNA strands transcribed in opposite directions from a single central promoter region, generating piRNAs that do not overlap in the genome. Interestingly, X. laevis piRNA cluster configurations do not conform to outward transcription from a central promoter region such as mammalian dual-strand piRNA clusters (Fig. 2F, right panel), and this opposing mapping of X. laevis XIWI and XILI piRNAs is similar to the mapping patterns of piRNAs in zebrafish ZIWI and ZILI (Houwing et al. 2007, 2008) and for chicken piRNAs (Chirn et al. 2015). Thus, X. laevis piRNA clusters appear more similar to chicken and zebrafish clusters than to mammalian clusters, which also suggests a different mode of evolution on piRNA loci compared to the vertebrate Piwi protein phylogeny (Fig. 1A).
XIWI and XILI associate with a common pool of developmentally important transcripts
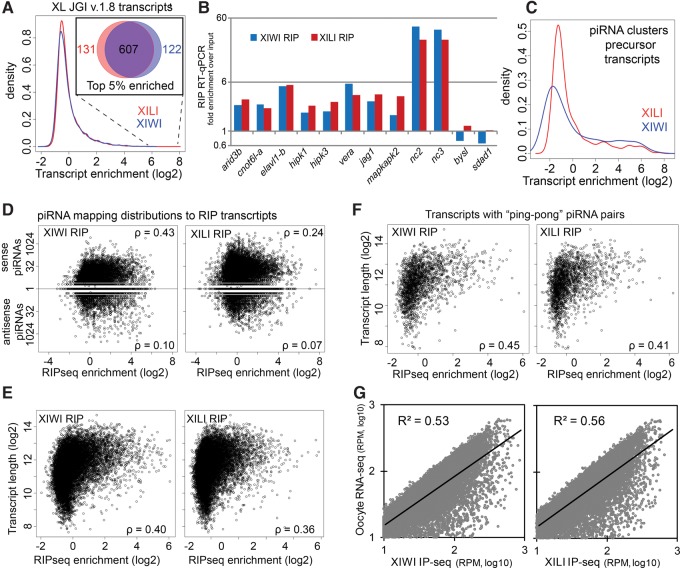
To discover the composition of additional RNAs associated with Xenopus Piwi proteins, we first identified the long (>300 nt) RNA transcripts bound by XIWI and XILI in X. laevis oocytes by deep sequencing (Fig. 2A). In each of the RIP-seq libraries, the majority of transcripts were only enriched in one of the IPs, and enrichment ratios exhibited a wide range from twofold to more than eightfold in log2 values (Fig. 3A). However, when comparing only the top 5% of enriched transcripts, this set indicated significant overlap in common transcripts associated with XIWI and XILI (Fig. 3A, inset). We confirmed the enrichment of several of these PATs by conducting reverse transcription and quantitative PCR on RNAs from two separate biological replicates of XIWI and XILI IPs (Fig. 3B). Lists of these gene transcripts associated with X. laevis XIWI and XILI are detailed in Supplemental Table S2. Gene ontology (GO) analysis revealed an enrichment of developmentally important functional categories including: protein kinases, transcriptional and translational regulation, signaling pathways, oogenesis, and apoptosis (Supplemental Table S3). Because the X. laevis genome build 9.1 still lacks a comprehensive list of TE consensus sequences, we examined piRNA cluster precursor transcripts instead as an expected positive control, and found a significant number (∼4%) of piRNA cluster transcripts in the XIWI and XILI RIP-seq libraries (Fig. 3D). However, the remaining majority of piRNA cluster precursor transcripts were absent from total oocyte extract and RIP-seq libraries, suggesting that either piRNA cluster transcripts are inherently unstable or occur during developmental stages even earlier than stage 1–3 oocytes.
FIGURE 3.
Features of X. laevis oocyte Piwi-protein associated transcripts (PATs). (A) Histogram of JGI v.1.8 transcript enrichments (defined as: IP fpkm/total RNA fpkm) from RIP-seq libraries. (Inset) Venn diagram of the top 5% of enriched transcripts from XIWI and XILI RIP-seq libraries. (B) Validation of transcripts that have high RIP-seq enrichment by RT-qPCR analysis of transcripts from XIWI and XILI IPs; bysl and sdad1 are negative controls. Bars represent an average of two biological replicates. (C) Histogram of piRNA cluster transcript enrichments (defined as: IP fpkm/total RNA fpkm) from RIP-seq libraries. (D–G) XIWI and XILI RIP-seq libraries counted against JGI v.1.8 transcripts. Spearman's rank correlation denoted by rho (ρ). RIP-seq enrichments plotted against: (D) the number of perfectly mapping piRNAs; (E) transcript length; (F) transcript length for only transcripts with mapping ping-pong piRNA pairs; (G) and transcript expression level.
To explore which particular feature might define the degree a target transcript associates with X. laevis XIWI and XILI, we examined the correlation between RIP-seq enrichment to the amount of base-paring piRNAs, transcript lengths, and transcript abundance. Consistent with other animal species, only a small fraction of piRNAs from either XIWI or XILI mapped to gene transcripts and among those that do, no bias was observed between sense and antisense orientations (Fig. 2C). When we compared the RIP-seq enrichment of a transcript with the number of piRNAs that map to it with nearly perfect complementarity (≤2 mismatches), we found a modest Spearman rank correlation (Fig. 3D). For both XIWI and XILI, the correlation was greater for sense mapping piRNAs than antisense. This difference may be partly due to detecting many fewer transcripts with antisense mapping piRNAs compared to transcripts with sense mapping piRNAs in the RNA-seq libraries (defined as having at least 1 full measured count by CuffDiff) (Supplemental Table S2). For example, only 21.4% and 25.1% of transcripts with antisense XIWI and XILI piRNAs, respectively, were present compared to 55.7% and 49.8% of transcripts with sense mapping XIWI and XILI piRNAs, respectively. This result suggested that transcripts with antisense mapping piRNAs are less stable in early stage oocytes and may contribute to genic piRNA biogenesis.
When we compared our RIP-seq enrichments to transcript length, we saw a Spearman rank correlation on par with the rank correlations to sense mapping piRNAs (Fig. 3E). When considering only transcripts with ping-pong piRNA pairs mapping to them, the correlations increased only slightly (Fig. 3F), suggesting that transcript length alone may be a major contributing factor for association. Finally, for the major bulk of the oocyte transcriptome, we observed a significant correlation between the RPM value of a transcript in the XIWI and XILI RIP-seq with its abundance in total RNA-seq (Fig. 3G). Together, these data support the conclusion that the features defining mRNA association with Piwi proteins in the Drosophila oocyte (Barckmann et al. 2015; Vourekas et al. 2016), such as transcript length and abundance, could also be conserved in the Xenopus oocyte.
X. tropicalis XIWI and XILI also associate with a wide range of transcripts in oocytes, including developmentally important gene mRNAs
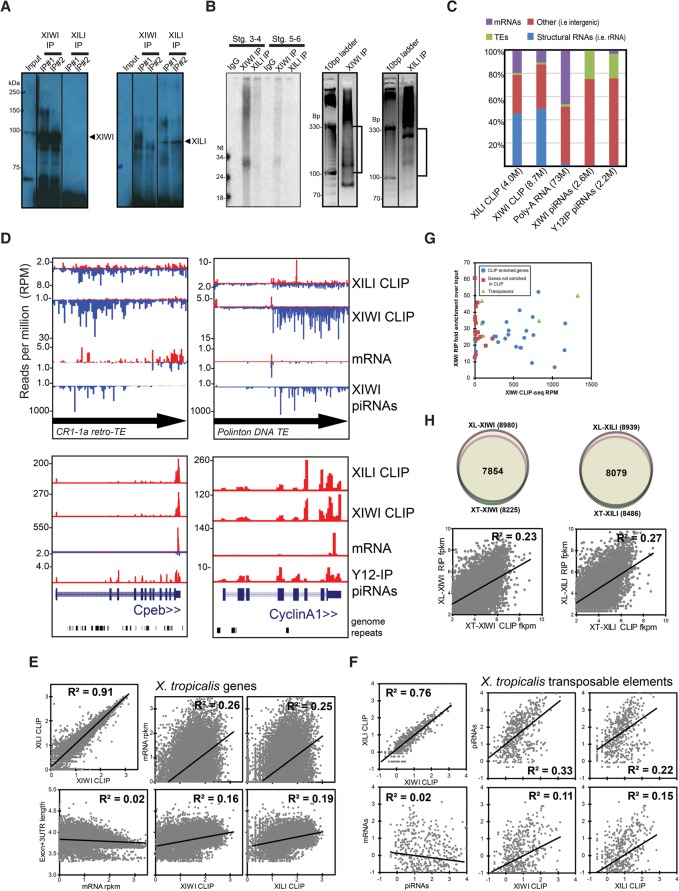
To compare PATs between X. laevis and X. tropicalis, we deeply sequenced transcripts from IPs of XIWI and XILI in X. tropicalis oocytes. Consistent with the X. laevis oocyte IPs, there was a much stronger Western blot signal and better efficiency in the XIWI IP in X. tropicalis compared to XILI (Fig. 4A). However, we also consistently observed poor radioactive labeling of piRNAs from the X. tropicalis XILI IP (Fig. 4B, left panel). For this reason, we sequenced small RNAs from an IP with the Y12 antibody that binds symmetrically dimethylated arginines on Piwi proteins (Kirino et al. 2009, 2010; Vagin et al. 2009), to provide an additional measure of XILI piRNAs. Nevertheless, sufficient amounts of other RNA transcripts besides piRNAs could be IP'ed by both antibodies.
FIGURE 4.
Features of X. tropicalis oocyte PATs and comparison to X. laevis oocyte PATs. (A) Western blots of IPs from stage 3–4 X. tropicalis oocytes. (B) Left panel shows radiolabeled RNAs from different staged X. tropicalis oocytes irradiated with UV and then subjected to IPs of XIWI and XILI resolved on a denaturing polyacrylamide gel. Right panel shows an ethidium bromide-stained agarose gel of the RNA fragments associating with XIWI and XILI IPs from extracts of UV-irradiated and RNase T1-treated X. tropicalis stage 1–4 oocytes; and brackets indicate the region excised for RNA sequencing. (C) The proportions of genomic functional annotations of the XIWI and XILI CLIP-seq reads, poly(A) RNAs, and piRNAs from X. tropicalis stage1–4 oocytes. (D) Genome browser tracks of reads from CLIP-seq, mRNA-seq, and piRNA-seq for two top represented TEs and two developmentally important gene transcripts. (E,F) Scatterplots and Pearson correlation (R2) values comparing between the various CLIP-seq, mRNA-seq, and piRNA-seq libraries for gene transcripts (E) and TEs (F). (G) Scatterplot comparing the amount of enrichment for candidate genes and TEs tested in RIP experiments versus their RPM counts of the CLIP-seq data sets. (H) Venn diagrams and scatterplots showing strong commonality and positive correlation in enrichment levels for XIWI- and XILI-associated transcripts compared between X. laevis and X. tropicalis oocytes. CLIP counts and piRNAs are log10 of reads per million, and fpkm is fragments per kilobase per million.
Since the X. tropicalis genome has a better annotation of genes and TEs compared to the X. laevis genome, and the smaller size of X. tropicalis oocytes facilitated UV crosslinking efficiency, we performed CLIP-seq to identify the binding sites of Piwi proteins in protein-coding RNAs. Although RIP is an efficient and simplistic technique (Keene et al. 2006), CLIP has additional capacity to reveal preferred RNP interaction regions within a transcript, such as microRNA (miRNA) binding sites for AGO proteins in the 3′ UTR of targeted mRNAs (Chi et al. 2009; Hafner et al. 2010). CLIP traps RNPs and associated RNAs with UV-light and then fragments the crosslinked sample with mild RNase treatment before the IP (Ule et al. 2003; Licatalosi et al. 2008). This increased stringency and added steps may also alter the proportion of transcripts identified between RIP and CLIP. Nevertheless, we deployed a similar CLIP-seq protocol previously applied to Drosophila PIWI (Sytnikova et al. 2014) on X. tropicalis XIWI and XILI from stage 1–3 oocytes, and sequenced the ∼100 to 300-nt-long RNA fragments as well as poly(A) tail selected RNA-seq libraries. These RNA reads were mapped to the X. tropicalis genome build Xentro3, the Repbase transposon consensus sequences, and Refseq transcriptome gene models (Hellsten et al. 2010).
Reads mapping to mRNAs represented ∼20% and ∼10% of the transcripts associated with XILI and XIWI, respectively (Fig. 4C). As expected, the CLIP-seq of XIWI and XILI contained significant amounts of ribosomal RNAs, which may be stabilized by cross-linking and is consistent with mouse MIWI associating with ribosomes (Grivna et al. 2006a,b; Unhavaithaya et al. 2009; Vourekas et al. 2012, 2016; Barckmann et al. 2015). TE transcripts were only a small fraction of X. tropicalis PATs, which may reflect their on-going depletion from the transcriptome by TE-directed piRNAs during oogenesis. Finally, many RNA reads remain uncharacterized as “intergenic” because of a lack of genome annotation.
The CLIP-seq libraries of XIWI and XILI afforded us a view of the distribution of CLIP reads across the length of TEs and mRNA transcripts. The two mostly highly represented TEs and two notable genes are shown in Figure 4D, while tables of the read counts are listed in Supplemental Table S4 (TEs) and Supplemental Table S5 (genes). The antisense bias of XIWI CLIP reads for TEs mirrored the bias of TE-directed piRNAs, whereas XILI CLIP reads were a balance of both sense and antisense TE reads, spread out across the TE consensus sequence. Although Class-II DNA transposons are notable for being highly represented in the X. tropicalis genome (Hellsten et al. 2010), the majority of piRNA and CLIP read distributions for DNA transposons were indistinguishable from the Class-I retrotransposons.
Gene transcripts were abundantly marked by both XIWI and XILI CLIP reads, with apparent peaks of reads at the genes’ 3′ UTRs. However, this concentration of reads is not an actual bias when normalized against the extensive lengths of 3′ UTRs of transcripts expressed in vertebrate oocytes (Smibert et al. 2012; Ulitsky et al. 2012; Lianoglou et al. 2013; Eichhorn et al. 2016; Lim et al. 2016). However, there is precedence for Piwi proteins preferring to bind gene 3′ UTRs as a potential mechanism of regulation and in piRNA biogenesis (Robine et al. 2009; Saito et al. 2009), but in Xenopus the 3′ UTR genic piRNAs are only observed in Y12 IPs while being poorly represented in XIWI IPs (Fig. 4C). Many of the XILI- and XIWI-associated transcripts have previously described roles in Xenopus oocyte and embryo developmental regulation, such as cpeb in modulating the translation regulation of maternal transcripts (Kim and Richter 2007; Radford et al. 2008), and cycA in regulating the cycle of embryonic cell divisions (Howe et al. 1995; Groisman et al. 2000; Mendez et al. 2002; Eckerdt et al. 2011).
Features of X. tropicalis XIWI- and XILI-associated transcripts
When we compared PAT counts in XIWI and XILI CLIP with their mRNA abundance and transcript length, we found a consistent positive correlation between the XIWI and XILI CLIP read gene counts and transcript abundance and length (Fig. 4E). This is consistent with the correlation trends observed for X. laevis XIWI and XILI RIP-seq data sets (Fig. 3E,G). The gene CLIP read counts between XIWI and XILI were also strongly correlated (Fig. 4E), possibly reflecting either similar target binding capacities or XILI's coprecipitation with XIWI in the UV-crosslinked X. tropicalis oocyte extract (Fig. 4A). In contrast to the lack of XIWI/XILI interactions in X. laevis IPs, we believe the UV light in CLIP-seq locked some proportion of X. tropicalis XIWI and XILI complexes, perhaps during the formation of “piRNA ping-pong pairs,” which is consistent with other groups observing other RNPs being cross-linked by UV light when engaged on the same transcript (Castello et al. 2016). Although few X. tropicalis piRNAs could be meaningfully matched to genes (i.e., beyond two mismatches), sufficiently abundant piRNAs could be matched to the TEs to show that TE CLIP reads positively correlated with piRNA amounts (Fig. 4F).
Since XIWI appeared to be much more abundant than XILI in the X. tropicalis oocyte extracts, we focused on testing RIP and RT-qPCR analyses on a subset of genes and TE transcripts most abundantly represented in the XIWI CLIP-seq lists (Supplemental Tables S3, S4). We consistently detected transcript enrichment from the XIWI RIP-RT-qPCR of genes and TE transcripts (Fig. 4G). However, several genes’ transcripts that were poorly represented in the XIWI CLIP were enriched in the XIWI RIP analysis, despite absolutely no detection of these transcripts in the RT-qPCR analysis of IgG negative control IPs. Currently, we conclude that some XIWI associated transcripts enriched in the RIP RT-qPCR experiment may have been depleted during the UV-light irradiation and RNase treatment of the extract during the CLIP procedure.
Finally, we compared the X. tropicalis CLIP-seq gene list with the X. laevis RIP-seq list, which we accomplished by cross-matching X. laevis gene models first to a human gene model to bridge to the X. tropicalis gene model (Supplemental Table S6). Given the differences in the RIP-seq and CLIP-seq protocols, we chose a compromise metric of FKPM (fragments per kilobase per million) to compare the two data sets, and required the genes display at least four FPKM counts. Interestingly, the vast majority of the same genes were represented in both species and Piwi protein IPs (Fig. 4H), and a positive correlation existed for these genes in terms of their FPKM counts between the two species Piwi protein IPs. These data suggest that there may be congruency in the Piwi pathway's biological influence on the oocyte transcriptomes in both of these frog species.
Exogenous transcript stability and XIWI association tests in X. laevis oocyte extract
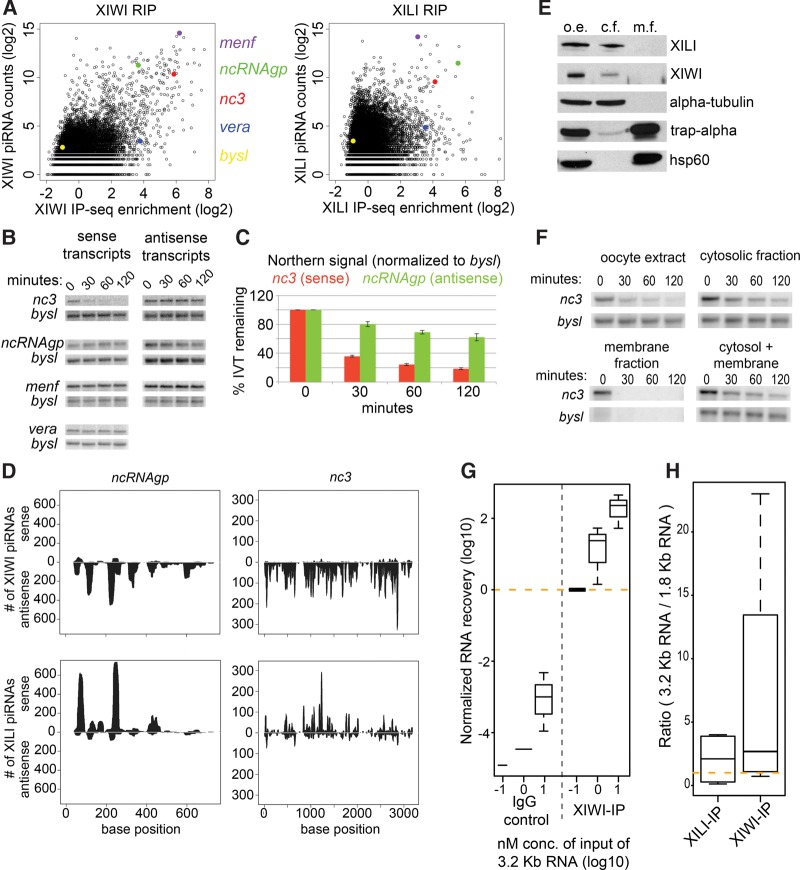
To gain insight into the potential fates of RNA transcripts associated with X. laevis Piwi/piRNA complexes, we first examined the stability of a handful of transcripts in stage 1–3 oocyte extract. We selected bysl as a negative control since it was neither enriched in the RIP-seq nor piRNA-seq libraries (Figs. 3B, 5A). For candidate targets, we selected transcripts enriched in our RIP-seq libraries, such as coding RNAs menf and vera, as well as the noncoding RNAs named ncRNAgp (Unigene ID: Xl.29596) and nc3 (Unigene ID: Xl.85152), which the latter was previously discovered to be expressed during tadpole tail regeneration (Fukazawa et al. 2009). To determine their stability in oocyte extract we developed a degradation assay which involved (i) in vitro transcribing and 5′ capping these transcripts, (ii) adding these transcripts to oocyte extract, (iii) extracting the total RNA at specific time points, and (iv) measuring the abundance of the transcripts via Northern blot. When we performed this assay on nc3, we observed degradation in a strand-specific manner: The sense strand of nc3 was degraded in oocyte extract over time while the antisense strand remained stable (Fig. 5B). We observed a similar but converse pattern for ncRNAgp—the sense strand was stable while the anti-sense strand was degraded over time in oocyte extract (Fig. 5B).
FIGURE 5.
X. laevis PATs with high amounts of targeting piRNAs are degraded rapidly in oocyte extracts. (A) XIWI and XILI RIP-seq and piRNAseq libraries counted against UniGene transcripts. Specific transcripts denoted by color. (B) Degradation assays in X. laevis stage 1–3 oocyte extract. (C) Quantitated band intensities for degradation assay Northern blots. Sense nc3 transcript and antisense ncRNAgp transcript replicates where N = 4 and N = 5, respectively. Error bars denote the standard error of the mean (SEM). (D) XIWI and XILI piRNAs with perfect complementarity to ncRNAgp and nc3. (E) Western blot for molecular markers in X. laevis stage 1–3 oocyte extract (o.e.), cytosolic fractions (c.f.), and membrane fractions (m.f.). Trap-α marks endoplasmic reticulum membranes; hsp60 marks mitochondria; α-tubulin marks cytosol. (F) Degradation assay with nc3 sense strand in X. laevis stage 1–3 oocyte extract and fractionated cytosolic and membrane fractions. (G) Exogenous RNA association assay shows that a 3.2-kb-long RNA (encoding EMTB-3XGFP) is enriched in IPs with XIWI in a concentration-dependent and XIWI-dependent manner, since there is minimal RNA recovery in the IgG negative control. Box plots of experiments from three separate frog oocyte extracts. (H) The 3.2-kb-long RNA is more enriched in XIWI IPs compared to the 1.8-kb-long RNA (encoding EMTB-1XGFP), with much less distinction in XILI IPs. Box plots of experiments from four separate frog oocyte extracts.
Although the degradation rates for these transcripts differed, they were both highly reproducible (biological replicates for nc3 and ncRNAgp were N = 4 and N = 5, respectively) (Fig. 5C). Upon examination of the piRNAs that map to nc3 and ncRNAgp, we found that for both transcripts, XIWI-bound piRNAs mapped almost exclusively in the antisense orientation while XILI-bound piRNA mapped almost exclusively in the sense orientation (Fig. 5D). This led us to hypothesize that XIWI was promoting the degradation of the sense nc3 transcript while XILI may be responsible for degrading the antisense ncRNAgp transcript. We attempted to test this hypothesis by performing these assays in oocyte extracts immunodepleted of either XIWI or XILI. Unexpectedly, extracts immunodepleted of either XIWI or XILI (but not immunodepletion of other RNPs nor mock depletions) altered the extract composition such that all exogenously added RNAs and hundreds of endogenous RNAs were now unstable (data not shown). This observation complicates the determination of whether the X. laevis Piwi proteins directly caused nc3 or ncRNAgp degradation despite the piRNA-specific degradation patterns, or whether a general RNA-stabilization effect was removed when XIWI and XILI were depleted. RNA stabilization was conceivable because unlike the noncoding RNAs, the coding RNAs menf and vera showed no degradation in the extract (Fig. 5B), despite abundant piRNAs mapping to menf, whereas vera (also called igf2bp3) is a well-known germ plasm localized transcript with few piRNAs mapped (Elisha et al. 1995; Epstein et al. 1997; Yaniv et al. 2003; Kalous et al. 2014).
Another possibility for the observed degradation of the nc3 and ncRNAgp noncoding RNAs is that they may be portions of piRNA cluster precursor transcripts that are processed into piRNAs. We did not observe the appearance of piRNA-sized bands in our oocyte extract assays because the primary piRNA-processing pathway is not fully recapitulated in this extract. To test whether nc3 and ncRNAgp degradation might represent one intermediary step within primary piRNA processing, we took advantage of the ability to biochemically fractionate oocyte extract into a membrane fraction and a membrane-free cytosolic fraction (Fig. 5E). The endonuclease Zucchini is bound to the outer mitochondrial membrane (Haase et al. 2010; Saito et al. 2010; Huang et al. 2011; Watanabe et al. 2011; Ipsaro et al. 2012; Nishimasu et al. 2012; Voigt et al. 2012) and cleaves piRNA primary transcripts to generate piRNA 5′ ends (Huang et al. 2011; Watanabe et al. 2011; Han et al. 2015; Mohn et al. 2015). The ER marker trap-α and mitochondrial marker hsp60 were only in the membrane fraction, whereas both XIWI and XILI were only in the cytosolic fraction along with α-tubulin (Fig. 5E). The membrane fraction appeared to be hyperactive in degrading all RNAs, while the re-addition of cytosol to this membrane fraction restored the same rate of nc3 degradation and stability of bysl RNAs as the cytosol alone and the unfractionated oocyte extract (Fig. 5F). This result also suggests that nc3 degradation in the cytosolic fraction is unlikely to be caused by membrane-bound factors like Zucchini.
Finally, we used the oocyte extract to test the hypothesis suggested by our IP-seq data and from (Vourekas et al. 2016), that longer and more abundant RNAs may preferentially interact with Piwi proteins independently of piRNA-sequence abundance and direct targeting. We first transcribed a 1.8 kb and a 3.2 kb capped RNA, both encoding the human Ensconsin microtubule-binding domain fused to either one copy or three copies of the GFP sequence, respectively (Faire et al. 1999; von Dassow et al. 2009). When the 3.2 kb RNA was incubated in the oocyte extract, its recovery in XIWI IPs across a range of nonsaturating concentrations supported the conclusion that association scaled with the exogenous transcripts concentration in the extract (Fig. 5G). This RNA recovery was dependent on XIWI association since the signal was orders of magnitude greater than the IgG negative control. At 1 nM concentration, we also observed consistently greater enrichment in XIWI IPs for the longer 3.2 kb RNA over the 1.8 kb RNA (Fig. 5H), but no differential in RNA association was observed in XILI IPs. Since these two RNAs are highly similar except for the two extra copies of GFP sequences, the data support the conclusion that XIWI, being much more abundantly expressed in oocytes compared to XILI, has a greater capacity to associate with longer transcripts. Taken together, these two experiments confirm the prediction that Piwi proteins have an intrinsic nonspecific affinity for all RNAs and show length and concentration-dependent association with many different types of transcripts.
DISCUSSION
In this study, we characterized the localization of XILI in X. laevis oocytes, the configuration of X. laevis piRNA clusters, and identified extensive lists of Piwi-protein-associated transcripts (PATs) from the oocytes of X. laevis and X. tropicalis. From these lists, we conclude that: (i) XIWI and XILI interact with hundreds of mRNAs in both frogs; (ii) PATs are reasonably well conserved between frogs; (iii) each PAT's read counts correlate well with expression and transcript length. Finally, we describe target RNA degradation in an oocyte extract system, which will be a good first step toward understanding transcript regulation in the future. Our work establishes a foundation for future studies where we will leverage the technical advantages of each species to explore how piRNAs and the Piwi pathway ensure proper gametogenesis and embryogenesis.
Although we and others had first characterized X. tropicalis piRNAs due to the earlier release and better annotation of the X. tropicalis genome sequence (Armisen et al. 2009; Lau et al. 2009a; Hellsten et al. 2010), the draft X. laevis genome sequence build 9.1 (Session et al. 2016) and the JGI v1.8 transcript models were sufficient for us to conduct an analysis of piRNA clusters and RIP-seq enriched transcripts (James-Zorn et al. 2015). We acknowledge the genomics limitations with X. laevis, including the lack of a comprehensive TE consensus sequence list, and many sequence gaps within and between the genomic scaffolds (Session et al. 2016). Nevertheless, we could still map most of the X. laevis piRNAs to the genome and transcriptome (Fig. 2C), and discovered several notable new features. First, the ∼32-nt-long mode (peak in the distribution) of X. laevis piRNA lengths is the longest among vertebrates (Chirn et al. 2015). Second, in contrast to low ping-pong piRNA signatures and few piRNAs overlapping on genomic strands in X. tropicalis piRNA clusters (Lau et al. 2009a; Chirn et al. 2015), X. laevis piRNAs display a greater proportion of ping-pong piRNAs, and there is prominent plus-strand/minus-strand bias between XIWI/XILI bound piRNAs arising from the overlapping piRNAs in clusters such as on Scaffold 23 (Fig. 2F). With ∼40 million years of evolution separating X. laevis and X. tropicalis (Hellsten et al. 2010), our data suggest X. laevis piRNAs and piRNA clusters are more analogous to piRNA clusters in chicken and zebrafish, whereas most of the X. tropicalis piRNAs and clusters are analogous to mammalian intergenic piRNA clusters. Third, our data reveal that similar to other vertebrates like X. tropicalis and humans, the majority of X. laevis piRNAs also arise from nonrepetitive, intergenic cluster loci (Fig. 2C).
Our study identified thousands of XIWI- and XILI-associated mRNAs and noncoding RNAs that fall into two classes: the first class are unstable transcripts, possibly because of many perfectly matching, antisense piRNAs, whereas a second class are transcripts left unaffected in oocytes, even with significant numbers of matching piRNAs. The first class is exemplified by noncoding RNAs nc3 and ncRNAgp, which display significant numbers of matching piRNAs, and are degraded in a strand-specific manner in the X. laevis oocyte extract. The second class is exemplified by the protein coding RNAs vera and menf, which have many perfectly mapping piRNAs, yet both the sense and antisense mRNAs were stable in the oocyte extract. Future experiments will optimize the oocyte extract to resolve these differences in RNA stability, and will further dissect how Xenopus Piwi proteins may have a dual role in both RNA degradation and stabilization.
TE silencing is a major role for Piwi proteins and piRNAs that is deeply conserved from Drosophila to mammals (Saito et al. 2006; Vagin et al. 2006; Aravin et al. 2007a; Brennecke et al. 2007; Le Thomas et al. 2013; Post et al. 2014). Moreover, the Drosophila Piwi pathway can direct CCR4-mediated decay of maternal mRNAs during early embryogenesis (Rouget et al. 2010). However, the Piwi pathway also influences proper RNA localization in the Drosophila oocyte (Cook et al. 2004; Megosh et al. 2006; Klattenhoff et al. 2007; Becalska et al. 2011; Barckmann et al. 2015; Vourekas et al. 2016), as well as the mouse MIWI and MILI proteins in stabilizing mRNAs essential for spermiogenesis (Deng and Lin 2002; Grivna et al. 2006b; Unhavaithaya et al. 2009; Vourekas et al. 2012). An emerging hypothesis for this mRNA interaction mechanism is that the greater the abundance and length of a maternal transcript, the more likely that transcript will associate with the extensive diversity of piRNA sequences within Piwi protein complexes (Vourekas et al. 2016). Consistent with this hypothesis, we detected a positive correlation between a Xenopus PAT's frequency in RIP-seq libraries with higher expression levels and longer lengths (Figs. 3E,G, 4E,F), and confirmed these predictions experimentally using exogenously added RNAs in oocyte extract (Fig. 5G,H).
This study and our previous data show enrichment of XILI in the Balbiani body and XIWI in the vegetal cortex (Lau et al. 2009a). The Balbiani body later remodels into the germ plasm at the vegetal cortex, and both of these structures are concentrated with maternally deposited RNPs and RNAs destined to be segregated into future primordial germ cells (Kloc and Etkin 1995; King et al. 2005). These localized RNAs have been profiled by microarray (Claussen and Pieler 2010; Cuykendall and Houston 2010) and more recently by deep sequencing for both X. laevis and X. tropicalis (Claussen et al. 2015). Of the ∼60 X. laevis transcripts with experimentally verified vegetal localization (Claussen et al. 2015), only ∼40% were also enriched in either the XIWI or XILI RIP-seq libraries. This observation is reassuring but could also be due to both XIWI and XILI being expressed at high levels throughout the oocyte in addition to being enriched in vegetal cortex and Balbiani body. A new procedure to manually dissect Balbiani bodies may allow for future higher-resolution studies of XIWI and XILI PATs (Boke et al. 2016). Why XILI and XIWI become enriched at these locations also remains to be determined, but the event may also be to ensure that a set of maternal piRNAs is properly distributed in the embryo.
Our study in Xenopus provides a critical vertebrate that bridges the spectrum from Drosophila to mammals, and our extensive list of Xenopus PATs indicate that the identity of these transcripts is conserved between X. laevis and X. tropicalis (Fig. 4H). The length and abundance trends of Xenopus PATs mirrors the trends of Drosophila AUB PATs (Barckmann et al. 2015; Vourekas et al. 2016), whereas the overlap of PATs in XIWI and XILI complexes contrasts to the more distinct set of PATs for mouse MIWI and MILI (Vourekas et al. 2012). These results can be reconciled by the fact that while XIWI and XILI are expressed simultaneously throughout oogenesis (Fig. 1E; Lau et al. 2009a), the temporal expression of MIWI and MILI is much more distinct in spermatogenesis (Vourekas et al. 2012). Another conserved theme is that many PATs are known to be developmentally important transcripts (Fig. 3C; Supplemental Tables S2, S4), such as cycB and dnc in Drosophila (Barckmann et al. 2015; Vourekas et al. 2016), and nc3, vera, cycA1, and Cpeb in Xenopus (Elisha et al. 1995; Howe et al. 1995; Epstein et al. 1997; Groisman et al. 2000; Mendez et al. 2002; Yaniv et al. 2003; Kim and Richter 2007; Radford et al. 2008; Fukazawa et al. 2009; Eckerdt et al. 2011; Kalous et al. 2014). In addition to its critical role in gametogenesis, the Piwi pathway may also influence embryonic development by ensuring these maternal transcripts are properly sequestered and then transmitted to future primordial germ cells. This role has not been examined yet because Piwi pathway mutations frequently disrupt gametogenesis and prevent fertilization. However, future experiments to delete a major piRNA cluster in Xenopus may change a subset of piRNAs that would reveal which transcripts in gametes become mis-regulated while still allowing some gametogenesis to occur.
MATERIALS AND METHODS
Antibodies preparation
The Xenopus Xili gene model was identified in the Ensembl v52 database for X. tropicalis (model ENSXETG00000010533). Amino acids 1–300 were amplified by PCR from Xenopus oocyte cDNA, and cloned into pET30a (pMB192). Protein was expressed at 22°C and purified using Ni–NTA resin (QIAGEN). Recombinant XIWI was used to generate antibodies in a rabbit (Covance), as described in Lau et al. (2009a). Laboratory antibodies were affinity purified before use. Other antibodies used in this study were α-tubulin (SIGMA T9026), trap-α (Covance, custom rabbit), and hsp60 (Abcam AB3080).
X. laevis oocyte extract preparation
Ovaries were removed from anesthetized and euthanized adult female X. laevis frog and cut into small (∼4 mm3) pieces and incubated in 0.1 M sodium phosphate pH 7.4 with 3 mg/mL collagenase at room temperature until follicle cells have dissociated from the oocytes, which typically takes 30–60 min. De-folliculated oocytes were washed in modified Barth's saline (MBS) three times and then filtered through a 450 µm screen to select the stage 1–3 oocytes (which passes through the screen) from the stage 4–6 oocytes (which remain behind the screen). Stage 1–3 oocytes were then washed into XB buffer (10 mM HEPES, pH 7.7, 50 mM sucrose, 0.1 mM CaCl2, 100 mM KCl, 0.1 mM MgCl2) three times and transferred into a Dounce homogenizer. Excess buffer was removed by aspiration. Protease inhibitors (leupeptin, pepstatin, and chymotrypsin) and Cytochalasin D was added to a final concentration of 10 μg/mL. Oocytes were homogenized with two passes of the pestle and then centrifuged in a 1.5 mL tube at 20,000g for 5 min at 4°C. The top fatty layer was removed by aspiration and the middle layer (oocyte extract) was transferred into individual tubes of 25–100 µL aliquots, flash frozen in liquid N2, and stored at −80°C.
Stage 1–3 oocyte extract was separated into cytosolic and light membrane fractions as described previously for X. laevis egg extract (Powers et al. 2001). In brief, stage 1–3 oocyte extract was centrifuged (250,000g for 70 min at 4°C). A clear cytosolic fraction was collected and spun again (250,000g for 25 min at 4°C) while the light membrane fraction was isolated, diluted, and spun over a sucrose gradient (26,000g for 20 min at 4°C). Light membranes were collected, and along with the cytosolic fraction they were aliquoted, flash-frozen in liquid nitrogen, and stored at −80°C.
Immunoprecipitation, immunodepletion, and immunostaining for microscopy
One hundred micrograms of anti-XIWI or anti-XILI antibody was prebound to 400 µL of Protein A Dynabeads according to manufacturer's instructions. Antibody/bead complex was then washed three times by resuspending with 1 mL of PBS, magnetically separating antibody/bead complexes and aspirating supernatant. Antibody/bead complex was then incubated with 100 µL of stage 1–3 oocyte extract for 1 h on ice with gentle agitation every 15 min to prevent beads from settling. Immunoprecipitation beads were then washed as before with 1 mL of PBS four times. The remaining extract after the IP was generally >90% depleted of XIWI or XILI, as judged by Western blotting. X. laevis stage 1–3 oocytes were fixed and processed for whole-mount immunofluorescence as previously described (Lau et al. 2009a). Oocytes were imaged using a spinning disc unit attached to an Olympus BX-61 microscope, using a 10× objective.
X. laevis XIWI and XILI RIP-seq and piRNA-seq
XIWI and XILI were immunoprecipitated from stage 1–3 oocyte extract. Coprecipitating RNAs were purified with TRIzol and built into libraries using an Illumina Tru-seq kit without any further selection. Total RNA was purified from stage 1–3 oocyte extract with TRIzol and poly(A) RNA was purified with oligo(dT) beads and built into libraries using the Illumina Tru-seq kit. Coprecipitating piRNAs were separated on a 15% polyacrylamide TBE-Urea gel. Small piRNA sized bands were excised from gel and built into libraries based on the Ribosome Profiling library method starting at step 18 (Ingolia et al. 2012).
X. tropicalis XIWI and XILI CLIP-seq
X. tropicalis females were anesthetized before euthanization, and ovaries were freshly dissected, teased apart, and then digested for ∼1 h in 3 mg/mL collagenase at room temperature. Stage 1–4 oocytes were recovered by consecutive filtration through 600 and 400 µm meshes. Oocytes were crosslinked by UV irradiation at 240 nm (2× 0.9 J/cm2 with mild agitation between the pulses) and lysed by dounce homogenizer (50 strokes) followed by sonication (two rounds for 30 sec; 2 sec pulse/2 sec pause cycle, Qsonica) in Q-buffer (20 mM HEPES-KOH, pH 7.9; 10% glycerol; 0.1 M KOAc; 0.2 mM EDTA; 1.5 mM MgCl2; 0.5 mM DTT; 1× Roche Complete EDTA-free Protease Inhibitor Cocktail; 0.5% NP40; 5× volume in relation to the dry pellet). The yolk was removed by spinning the lysate three times for 12,000g. The remaining steps in the CLIP-seq procedure were essentially conducted as described in Sytnikova et al. (2014) using XIWI and XILI antibodies. Specific modifications here include treating the clarified extract with RNase T1 (Fermentas) 1 U/mL final dilution for 10 min at 22°C, then cooling down for 5 min on ice. Protein A/G magnetic beads (Pierce) were coated with anti-XIWI and anti-XILI antibodies at 60 µg per 60 µL of beads for 2 h at 23°C, and washed with 1× PBS. Lysate from oocytes was added to the beads, and IP was conducted upon mild rotation for 1.5 h at 4°C.
For RIP-RT-qPCR, 25 μL Protein A/G Magnetic Beads were coupled to 20 μg antibody (anti Xiwi, anti IgG control) to IP XIWI from 100 µL of stage 1–4 oocyte extracts. Input total RNA and RNA from IPs were extracted with TRIzol, and RT and qPCR with SYBR Green was performed per standard procedures. Primer pairs for qPCR are available upon request.
Bioinformatics analyses of X. laevis RIP-seq and piRNA-seq libraries, and X. tropicalis CLIP-seq libraries
All the deep sequencing data sets analyzed in this paper are deposited at the NCBI Gene Expression Omnibus under the series accession: GSE63228.
A phylogenetic tree was generated by aligning full-length protein sequences with MUSCLE (Edgar 2004) and then computing branch distances using ClustalW2-Phylogeny (Larkin et al. 2007; Goujon et al. 2010). The RIP-seq genomic alignments used Bowtie2 (Langmead and Salzberg 2012) with the “--very-sensitive” preset. The piRNA-seq alignments used Bowtie1 (Langmead et al. 2009) -v2 after using custom Perl scripts to remove linker sequences Bowtie1 settings were -v2 -m4 --best, to allow up to four genomic alignments. To calculate enrichment values, we used CuffDiff (Trapnell et al. 2012, 2013) to count and generate fpkm values transcript enrichment (IPseq fpkm/Total RNAseq fpkm). Only reads above noise were counted (noise -> IPseq counts + Total RNAseq counts <100), because CuffDiff counts the number of reads against a gff file, and then weights multiple reads to a single gene so that the total equals 1. The strange decimals that are not quite 0.25 0.33, 0.5, 0.66, or 0.75 are due to the way CuffDiff weights the libraries and makes the calculations.
To identify ping-pong piRNA pairs we extracted unique 5′ mapping positions for all piRNAs and used these positions to identify pairs of piRNAs that fit the ping-pong signature (10-nt overlap on opposite strands). We then used the nucleotide sequences for identified ping-pong reads to calculate base frequencies at each position. Base frequencies were used as input to generate WebLogo 3 graphics (Crooks et al. 2004).
To detect XIWI and XILI interactions with piRNA cluster transcripts, we used CuffNorm to count our RIP-seq library reads against the piRNA clusters we generated with StringTie (Pertea et al. 2015). For X. laevis piRNA cluster loci determinations, we use StringTie parameters of -m50 -g50 merge10 kb 500 pb min, 50 piRNAs/kb min.
Gene ontology analysis used DAVID (Huang da et al. 2009a, 2009b) on a list of X. laevis JGI v.1.8 gene models queried with BLASTx against human UniProt protein homologs, then filtering for an E-value of 1 × 10−20 or lower. To assign orthologs using X. laevis and X. tropicalis protein coding sequences, we used the BLASTx results from Xenopus gene models to the human UniProt protein homologs and then linked the common human orthologs to bridge across other animal species. We then confirmed this output using a different program call Inparanoid, which can also directly link gene orthologs (Sonnhammer and Ostlund 2015).
To analyze the X. tropicalis CLIP-seq and RNA-seq libraries, we processed the files similarly as in Sytnikova et al. (2014). However, the major difference in this CLIP-seq analysis is that only straight read counts as RPMs were measured without a noise filtering step as previously described for HITS-CLIP (Chi et al. 2009), because the range of transcripts were so extensive, and there was no antigen to block the XIWI and XILI antibodies. In addition, we reasoned that the only way to directly compare the values of the RIP-seq data from X. laevis to the CLIP-seq data from X. tropicalis was to use as similar a read counting method as can be adapted between the two species.
Reads were sorted according to the barcode sequence in their 5′ adaptor, and then adaptor sequences were trimmed by FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). Reads were mapped to the X. tropicalis genome Xentro3 build by using Bowtie1 (allowing maximum two mismatches) (Langmead et al. 2009). Structural RNAs were determined by cross-mapping to a custom database, and removed from subsequent analyses. TE reads mapping was performed against a list of X. tropicalis consensus TE sequences obtained from the Repbase database (Kapitonov and Jurka 2008). Within each data set of mapped reads, a signal merge count for unique reads was obtained (read frequency information was dismissed to avoid a jackpot effect). This basic processing pipeline is written in shell script (process.sh).
Next, the gene counts were determined by mapping the reads to the RefSeq gene models for the X. tropicalis genome Xentro3 build (Hellsten et al. 2010). WIG files with step size of 50 bp were generated and viewed by the UCSC Browser. Read counts for each gene were calculated by overlapping the RefSeq track of the UCSC Browser to the genome, and RNA gene counts were also broken down to intron and exon reads within a transcript. The transcript isoform with the highest count was selected as representative for a gene. The script for counting the RNA and CLIP reads for each gene interval is called ngs_genecentric.c.
Oocyte extract RNA degradation assay
Candidates PATs such as bysl, menf, vera, nc3, and ncRNAgp were cloned into pBluescript SK- with a 5′ EcoRI site and a 3′ PstI site. Sense transcripts were in vitro transcribed with T7 RNA polymerase while antisense transcripts were in vitro transcribed with T3 RNA polymerase (New England Biolabs). Transcripts were then capped with Vaccinia capping enzyme. Target transcripts (menf, nc3, ncRNAgp, or vera) along with the control transcript bysl were added to stage 1–3 oocyte extract to a final concentration of 1 nM each. At time points 0, 30, 60, and 120 min, 2 µL of oocyte extract was removed and the total RNA was processed with 500 µL of TRIzol. From each time point, 2 µg of RNA was electrophoresed on a 1% agarose, 6% formaldehyde gel in 1× MOPS buffer. RNAs were transferred onto nitrocellulose membrane and probed by Northern blot. Northern probes were generated with Stratagene Prime-It RmT random primer labeling kit with 32P α-dCTP. Probe sequences are available upon request.
XIWI and XILI binding assays with EMTB-GFP RNAs
The 1.8 kb EMTB-1XGFP and 3.2 kb EMTB-3XGFP mRNAs encode the ensconsin microtubule binding domain fused to 1× or 3× GFP sequences (Faire et al. 1999; von Dassow et al. 2009). Transcripts were made with the Sp6 Megascript kit (Ambion), capped with the Vaccinia virus capping system (NEB), and purified by lithium chloride precipitation. Capped RNAs were diluted to 250 nM and stored at −80°C. To compare the relative enrichment of RNAs in XIWI, XILI, and IgG negative control IPs, EMTB-1XGFP and EMTB-3XGFP RNAs were added to stage 1–3 oocyte extracts at 1 nM concentration, and incubated at room temperature for 30 min. This experiment was repeated from extracts prepared from four different frogs. To examine the concentration dependence of EMTB-3XGFP binding to XIWI, capped RNA was added to stage 1–3 oocyte extract at 10 nM, 1 nM, and 0.1 nM and incubated for 30 min at room temperature. This experiment was repeated in extracts prepared from three different frogs. IPs were performed as described above and washed four times with 1 mL of 1× PBS, and total RNA was purified from each IP and total extract prior to IP using the TRIzol and Directzol kit (Zymo Research). RNA was reverse transcribed using random hexamers and used as input for quantitative PCR. Primers for qPCR are AGTGCGAAGCGAAACAGCA and TGCTAATGACGGGATCAACA, which amplifies an equivalent amplicon from both EMTB-1XGFP and EMTB-3XGFP.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Ulandt Kim and the MGH sequencing core for all Illumina sequencing of X. laevis sequences, members of the Blower laboratory for critical comments on this work, Michael Rosbash's laboratory for assistance in deep-sequencing X. tropicalis libraries, and Dianne Schwarz for critical comments on the manuscript. This work was supported by the National Institutes of Health via Core Facilities grant P30 NS045713 to the Brandeis Biology Department; R00HD057298 and R01AG052465 to N.C.L.; and R01GM086434 and R01GM108548 to M.D.B. N.C.L. was a Searle Scholar, and M.D.B. was also supported by a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund.
Author contributions: J.A.T. and M.D.B. conducted the experiments and bioinformatics analyses in X. laevis, while Y.A.S., I.A., G.W.C., and N.C.L. conducted the experiments and bioinformatics analyses in X. tropicalis. J.A.T., M.D.B., and N.C.L. wrote the paper.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.058859.116.
REFERENCES
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J. 2007a. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318: 761–764. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. 2007b. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316: 744–747. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, van der Heijden GW, Castaneda J, Vagin VV, Hannon GJ, Bortvin A. 2009. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet 5: e1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armisen J, Gilchrist MJ, Wilczynska A, Standart N, Miska EA. 2009. Abundant and dynamically expressed miRNAs, piRNAs, and other small RNAs in the vertebrate Xenopus tropicalis. Genome Res 19: 1766–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baigude H, Ahsanullah, Li Z, Zhou Y, Rana TM. 2012. miR-TRAP: a benchtop chemical biology strategy to identify microRNA targets. Angew Chem Int Ed Engl 51: 5880–5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barckmann B, Pierson S, Dufourt J, Papin C, Armenise C, Port F, Grentzinger T, Chambeyron S, Baronian G, Desvignes JP, et al. 2015. Aubergine iCLIP reveals piRNA-dependent decay of mRNAs involved in germ cell development in the early embryo. Cell Rep 12: 1205–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becalska AN, Kim YR, Belletier NG, Lerit DA, Sinsimer KS, Gavis ER. 2011. Aubergine is a component of a nanos mRNA localization complex. Dev Biol 349: 46–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boke E, Ruer M, Wuhr M, Coughlin M, Lemaitre R, Gygi SP, Alberti S, Drechsel D, Hyman AA, Mitchison TJ. 2016. Amyloid-like self-assembly of a cellular compartment. Cell 166: 637–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103. [DOI] [PubMed] [Google Scholar]
- Castello A, Fischer B, Frese CK, Horos R, Alleaume AM, Foehr S, Curk T, Krijgsveld J, Hentze MW. 2016. Comprehensive identification of RNA-binding domains in human cells. Mol Cell 63: 696–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. 2009. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460: 479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Hannon GJ, Darnell RB. 2012. An alternative mode of microRNA target recognition. Nat Struct Mol Biol 19: 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirn GW, Rahman R, Sytnikova YA, Matts JA, Zeng M, Gerlach D, Yu M, Berger B, Naramura M, Kile BT, et al. 2015. Conserved piRNA expression from a distinct set of piRNA cluster loci in eutherian mammals. PLoS Genet 11: e1005652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JP, Lau NC. 2014. Piwi Proteins and piRNAs step onto the systems biology stage. Adv Exp Med Biol 825: 159–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark PM, Loher P, Quann K, Brody J, Londin ER, Rigoutsos I. 2014. Argonaute CLIP-seq reveals miRNA targetome diversity across tissue types. Sci Rep 4: 5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussen M, Pieler T. 2010. Identification of vegetal RNA-localization elements in Xenopus oocytes. Methods 51: 146–151. [DOI] [PubMed] [Google Scholar]
- Claussen M, Lingner T, Pommerenke C, Opitz L, Salinas G, Pieler T. 2015. Global analysis of asymmetric RNA enrichment in oocytes reveals low conservation between closely related Xenopus species. Mol Biol Cell 26: 3777–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook HA, Koppetsch BS, Wu J, Theurkauf WE. 2004. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell 116: 817–829. [DOI] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. 1998. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev 12: 3715–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Lin H. 2000. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development 127: 503–514. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuykendall TN, Houston DW. 2010. Identification of germ plasm-associated transcripts by microarray analysis of Xenopus vegetal cortex RNA. Dev Dyn 239: 1838–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lin H. 2002. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2: 819–830. [DOI] [PubMed] [Google Scholar]
- Dumont JN. 1972. Oogenesis in Xenopus laevis (Daudin). I. Stages of oocyte development in laboratory maintained animals. J Morphol 136: 153–179. [DOI] [PubMed] [Google Scholar]
- Eckerdt F, Yamamoto TM, Lewellyn AL, Maller JL. 2011. Identification of a polo-like kinase 4-dependent pathway for de novo centriole formation. Curr Biol 21: 428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhorn SW, Subtelny AO, Kronja I, Kwasnieski JC, Orr-Weaver TL, Bartel DP. 2016. mRNA poly(A)-tail changes specified by deadenylation broadly reshape translation in Drosophila oocytes and early embryos. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elisha Z, Havin L, Ringel I, Yisraeli JK. 1995. Vg1 RNA binding protein mediates the association of Vg1 RNA with microtubules in Xenopus oocytes. EMBO J 14: 5109–5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein M, Pillemer G, Yelin R, Yisraeli JK, Fainsod A. 1997. Patterning of the embryo along the anterior-posterior axis: the role of the caudal genes. Development 124: 3805–3814. [DOI] [PubMed] [Google Scholar]
- Faire K, Waterman-Storer CM, Gruber D, Masson D, Salmon ED, Bulinski JC. 1999. E-MAP-115 (ensconsin) associates dynamically with microtubules in vivo and is not a physiological modulator of microtubule dynamics. J Cell Sci 112 (Pt 23): 4243–4255. [DOI] [PubMed] [Google Scholar]
- Findley SD, Tamanaha M, Clegg NJ, Ruohola-Baker H. 2003. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development 130: 859–871. [DOI] [PubMed] [Google Scholar]
- Fukazawa T, Naora Y, Kunieda T, Kubo T. 2009. Suppression of the immune response potentiates tadpole tail regeneration during the refractory period. Development 136: 2323–2327. [DOI] [PubMed] [Google Scholar]
- Gan H, Lin X, Zhang Z, Zhang W, Liao S, Wang L, Han C. 2011. piRNA profiling during specific stages of mouse spermatogenesis. RNA 17: 1191–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, et al. 2008. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320: 1077–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. 2006. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442: 199–202. [DOI] [PubMed] [Google Scholar]
- Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. 2010. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 38: W695–W699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. 2008. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature 455: 1193–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin H. 2006a. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev 20: 1709–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna ST, Pyhtila B, Lin H. 2006b. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci 103: 13415–13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman I, Huang YS, Mendez R, Cao Q, Theurkauf W, Richter JD. 2000. CPEB, maskin, and cyclin B1 mRNA at the mitotic apparatus: implications for local translational control of cell division. Cell 103: 435–447. [DOI] [PubMed] [Google Scholar]
- Grosswendt S, Filipchyk A, Manzano M, Klironomos F, Schilling M, Herzog M, Gottwein E, Rajewsky N. 2014. Unambiguous identification of miRNA:target site interactions by different types of ligation reactions. Mol Cell 54: 1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590. [DOI] [PubMed] [Google Scholar]
- Haase AD, Fenoglio S, Muerdter F, Guzzardo PM, Czech B, Pappin DJ, Chen C, Gordon A, Hannon GJ. 2010. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev 24: 2499–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BW, Zamore PD. 2014. piRNAs. Curr Biol 24: R730–R733. [DOI] [PubMed] [Google Scholar]
- Han BW, Wang W, Li C, Weng Z, Zamore PD. 2015. Noncoding RNA. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 348: 817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AN, Macdonald PM. 2001. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development 128: 2823–2832. [DOI] [PubMed] [Google Scholar]
- Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, et al. 2010. The genome of the Western clawed frog Xenopus tropicalis. Science 328: 633–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. 2007. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell 129: 69–82. [DOI] [PubMed] [Google Scholar]
- Houwing S, Berezikov E, Ketting RF. 2008. Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J 27: 2702–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe JA, Howell M, Hunt T, Newport JW. 1995. Identification of a developmental timer regulating the stability of embryonic cyclin A and a new somatic A-type cyclin at gastrulation. Genes Dev 9: 1164–1176. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Huang H, Gao Q, Peng X, Choi SY, Sarma K, Ren H, Morris AJ, Frohman MA. 2011. piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev Cell 20: 376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. 2012. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc 7: 1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GJ. 2012. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature 491: 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James-Zorn C, Ponferrada VG, Burns KA, Fortriede JD, Lotay VS, Liu Y, Brad Karpinka J, Karimi K, Zorn AM, Vize PD. 2015. Xenbase: core features, data acquisition, and data processing. Genesis 53: 486–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano C, Wang J, Lin H. 2011. Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Annu Rev Genet 45: 447–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalous A, Stake JI, Yisraeli JK, Holt CE. 2014. RNA-binding protein Vg1RBP regulates terminal arbor formation but not long-range axon navigation in the developing visual system. Dev Neurobiol 74: 303–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. 2008. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat Rev Genet 9: 411–412; author reply 414. [DOI] [PubMed] [Google Scholar]
- Keene JD, Komisarow JM, Friedersdorf MB. 2006. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc 1: 302–307. [DOI] [PubMed] [Google Scholar]
- Kim JH, Richter JD. 2007. RINGO/cdk1 and CPEB mediate poly(A) tail stabilization and translational regulation by ePAB. Genes Dev 21: 2571–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King ML, Messitt TJ, Mowry KL. 2005. Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol Cell 97: 19–33. [DOI] [PubMed] [Google Scholar]
- Kirino Y, Kim N, de Planell-Saguer M, Khandros E, Chiorean S, Klein PS, Rigoutsos I, Jongens TA, Mourelatos Z. 2009. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol 11: 652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Vourekas A, Kim N, de Lima Alves F, Rappsilber J, Klein PS, Jongens TA, Mourelatos Z. 2010. Arginine methylation of vasa protein is conserved across phyla. J Biol Chem 285: 8148–8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, Theurkauf WE. 2007. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev Cell 12: 45–55. [DOI] [PubMed] [Google Scholar]
- Kloc M, Etkin LD. 1995. Two distinct pathways for the localization of RNAs at the vegetal cortex in Xenopus oocytes. Development 121: 287–297. [DOI] [PubMed] [Google Scholar]
- Kloc M, Jedrzejowska I, Tworzydlo W, Bilinski SM. 2014. Balbiani body, nuage and sponge bodies—term plasm pathway players. Arthropod Struct Dev 43: 341–348. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. 2006. Characterization of the piRNA complex from rat testes. Science 313: 363–367. [DOI] [PubMed] [Google Scholar]
- Lau NC, Ohsumi T, Borowsky M, Kingston RE, Blower MD. 2009a. Systematic and single cell analysis of Xenopus Piwi-interacting RNAs and Xiwi. EMBO J 28: 2945–2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Robine N, Martin R, Chung WJ, Niki Y, Berezikov E, Lai EC. 2009b. Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res 19: 1776–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Toth KF. 2013. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev 27: 390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Young AG, Bhutkar A, Zheng GX, Bosson AD, Nielsen CB, Sharp PA. 2011. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat Struct Mol Biol 18: 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XZ, Roy CK, Dong X, Bolcun-Filas E, Wang J, Han BW, Xu J, Moore MJ, Schimenti JC, Weng Z, et al. 2013. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol Cell 50: 67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lianoglou S, Garg V, Yang JL, Leslie CS, Mayr C. 2013. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev 27: 2380–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. 2008. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456: 464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Lee M, Son A, Chang H, Kim VN. 2016. mTAIL-seq reveals dynamic poly(A) tail regulation in oocyte-to-embryo development. Genes Dev 30: 1671–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Spradling AC. 1997. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development 124: 2463–2476. [DOI] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SR, Megosh H, Lin H. 2014. PIWI proteins are essential for early Drosophila embryogenesis. Dev Biol 385: 340–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megosh HB, Cox DN, Campbell C, Lin H. 2006. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr Biol 16: 1884–1894. [DOI] [PubMed] [Google Scholar]
- Mendez R, Barnard D, Richter JD. 2002. Differential mRNA translation and meiotic progression require Cdc2-mediated CPEB destruction. EMBO J 21: 1833–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Handler D, Brennecke J. 2015. Noncoding RNA. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science 348: 812–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani MK, Bonnefond L, Matsumoto N, Nishizawa T, Nakanaga K, Aoki J, et al. 2012. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature 491: 284–287. [DOI] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33: 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post C, Clark JP, Sytnikova YA, Chirn GW, Lau NC. 2014. The capacity of target silencing by Drosophila PIWI and piRNAs. RNA 20: 1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers M, Evans EK, Yang J, Kornbluth S. 2001. Preparation and use of interphase Xenopus egg extracts. Curr Protoc Cell Biol 9: 11.10.1–11.1.24. [DOI] [PubMed] [Google Scholar]
- Radford HE, Meijer HA, de Moor CH. 2008. Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim Biophys Acta 1779: 217–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robine N, Lau NC, Balla S, Jin Z, Okamura K, Kuramochi-Miyagawa S, Blower MD, Lai EC. 2009. A broadly conserved pathway generates 3′UTR-directed primary piRNAs. Curr Biol 19: 2066–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouget C, Papin C, Boureux A, Meunier AC, Franco B, Robine N, Lai EC, Pelisson A, Simonelig M. 2010. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 467: 1128–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. 2006. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev 20: 2214–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC. 2009. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461: 1296–1299. [DOI] [PubMed] [Google Scholar]
- Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, Siomi H, Siomi MC. 2010. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev 24: 2493–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senti KA, Jurczak D, Sachidanandam R, Brennecke J. 2015. piRNA-guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear piRNA repertoire. Genes Dev 29: 1747–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Session AM, Uno Y, Kwon T, Chapman JA, Toyoda A, Takahashi S, Fukui A, Hikosaka A, Suzuki A, Kondo M, et al. 2016. Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538: 336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. 2011. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol 12: 246–258. [DOI] [PubMed] [Google Scholar]
- Smibert P, Miura P, Westholm JO, Shenker S, May G, Duff MO, Zhang D, Eads BD, Carlson J, Brown JB, et al. 2012. Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Rep 1: 277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer EL, Ostlund G. 2015. InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res 43: D234–D239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S, Updike D. 2015. Specifying and protecting germ cell fate. Nat Rev Mol Cell Biol 16: 406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sytnikova YA, Rahman R, Chirn GW, Clark JP, Lau NC. 2014. Transposable element dynamics and PIWI regulation impacts lncRNA and gene expression diversity in Drosophila ovarian cell cultures. Genome Res 24: 1977–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. 2009. The maternal-to-zygotic transition: a play in two acts. Development 136: 3033–3042. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. 2003. CLIP identifies Nova-regulated RNA networks in the brain. Science 302: 1212–1215. [DOI] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Subtelny AO, Koppstein D, Bell GW, Sive H, Bartel DP. 2012. Extensive alternative polyadenylation during zebrafish development. Genome Res 22: 2054–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unhavaithaya Y, Hao Y, Beyret E, Yin H, Kuramochi-Miyagawa S, Nakano T, Lin H. 2009. MILI, a PIWI-interacting RNA-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. J Biol Chem 284: 6507–6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y, Nishida C, Takagi C, Ueno N, Matsuda Y. 2013. Homoeologous chromosomes of Xenopus laevis are highly conserved after whole-genome duplication. Heredity 111: 430–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324. [DOI] [PubMed] [Google Scholar]
- Vagin VV, Wohlschlegel J, Qu J, Jonsson Z, Huang X, Chuma S, Girard A, Sachidanandam R, Hannon GJ, Aravin AA. 2009. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev 23: 1749–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt F, Reuter M, Kasaruho A, Schulz EC, Pillai RS, Barabas O. 2012. Crystal structure of the primary piRNA biogenesis factor Zucchini reveals similarity to the bacterial PLD endonuclease Nuc. RNA 18: 2128–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dassow G, Verbrugghe KJ, Miller AL, Sider JR, Bement WM. 2009. Action at a distance during cytokinesis. J Cell Biol 187: 831–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vourekas A, Zheng Q, Alexiou P, Maragkakis M, Kirino Y, Gregory BD, Mourelatos Z. 2012. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat Struct Mol Biol 19: 773–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vourekas A, Alexiou P, Vrettos N, Maragkakis M, Mourelatos Z. 2016. Sequence-dependent but not sequence-specific piRNA adhesion traps mRNAs to the germ plasm. Nature 531: 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Han BW, Tipping C, Ge DT, Zhang Z, Weng Z, Zamore PD. 2015. Slicing and binding by Ago3 or Aub trigger Piwi-bound piRNA production by distinct mechanisms. Mol Cell 59: 819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Chuma S, Yamamoto Y, Kuramochi-Miyagawa S, Totoki Y, Toyoda A, Hoki Y, Fujiyama A, Shibata T, Sado T, et al. 2011. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev Cell 20: 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczynska A, Minshall N, Armisen J, Miska EA, Standart N. 2009. Two Piwi proteins, Xiwi and Xili, are expressed in the Xenopus female germline. RNA 15: 337–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams Z, Morozov P, Mihailovic A, Lin C, Puvvula PK, Juranek S, Rosenwaks Z, Tuschl T. 2015. Discovery and characterization of piRNAs in the human fetal ovary. Cell Rep 13: 854–863. [DOI] [PubMed] [Google Scholar]
- Wuhr M, Freeman RM Jr, Presler M, Horb ME, Peshkin L, Gygi SP, Kirschner MW. 2014. Deep proteomics of the Xenopus laevis egg using an mRNA-derived reference database. Curr Biol 24: 1467–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Chen KM, Pandey RR, Homolka D, Reuter M, Janeiro BK, Sachidanandam R, Fauvarque MO, McCarthy AA, Pillai RS. 2016. PIWI slicing and EXD1 drive biogenesis of nuclear piRNAs from cytosolic targets of the mouse piRNA pathway. Mol Cell 61: 138–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv K, Fainsod A, Kalcheim C, Yisraeli JK. 2003. The RNA-binding protein Vg1 RBP is required for cell migration during early neural development. Development 130: 5649–5661. [DOI] [PubMed] [Google Scholar]
- Zamparini AL, Davis MY, Malone CD, Vieira E, Zavadil J, Sachidanandam R, Hannon GJ, Lehmann R. 2011. Vreteno, a gonad-specific protein, is essential for germline development and primary piRNA biogenesis in Drosophila. Development 138: 4039–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zisoulis DG, Lovci MT, Wilbert ML, Hutt KR, Liang TY, Pasquinelli AE, Yeo GW. 2010. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat Struct Mol Biol 17: 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.