Abstract
Regulation of gene expression by cis-encoded riboswitches is a prevalent theme in bacteria. Of the hundreds of riboswitch families identified, the majority of them remain as orphans, without a clear ligand assignment. The ykkC orphan family was recently characterized as guanidine-sensing riboswitches. Herein we present a 2.3 Å crystal structure of the guanidine-bound ykkC riboswitch from Dickeya dadantii. The riboswitch folds into a boot-shaped structure, with a coaxially stacked P1/P2 stem forming the boot, and a 3′-P3 stem–loop forming the heel. Sophisticated base-pairing and cross-helix tertiary contacts give rise to the ligand-binding pocket between the boot and the heel. The guanidine is recognized in its positively charged guanidinium form, in its sp2 hybridization state, through a network of coplanar hydrogen bonds and by a cation–π stacking contact on top of a conserved guanosine residue. Disruption of these contacts resulted in severe guanidinium-binding defects. These results provide the structural basis for specific guanidine sensing by ykkC riboswitches and pave the way for a deeper understanding of guanidine detoxification—a previously unappreciated aspect of bacterial physiology.
Keywords: Dickeya dadantii, RNA structure, gene regulation, guanidine, orphan riboswitch
INTRODUCTION
Bacteria alter their gene expression program rapidly in response to nutrient level and environmental cues. Metabolite dependent cis-regulatory RNA structures called riboswitches are prominent regulators of bacterial gene expression. Riboswitches are typically embedded in the 5′-untranslated region (UTR) of a mRNA, where they modulate the transcription, translation, or occasionally, the integrity of the associated gene/operon (Serganov and Nudler 2013; Peselis and Serganov 2014; Price et al. 2014). In a typical scenario, the aptamer domain of the riboswitch adopts an alternative conformation upon ligand binding, which in turn exposes or sequesters the nearby expression platform of the riboswitch. The expression platform may contain a terminator stem–loop or a Shine–Dalgarno sequence, allowing the riboswitch to modulate transcription or translation, respectively (Mandal and Breaker 2004; Lu et al. 2008). Rare examples of eukaryotic riboswitches have also been reported, which were shown to regulate alternative splicing and mRNA degradation (Caron et al. 2012; Li and Breaker 2013). Although two dozen or so riboswitch families have been characterized, hundreds more remain as orphans, without a clear assignment of their cognate ligand (Barrick et al. 2004; Weinberg et al. 2010; Breaker 2011). This is usually due to a lack of knowledge about the function of riboswitch-associated genes or operons.
The ykkC motif is widely distributed across bacteria and is primarily associated with genes such as small multidrug resistance (SMR) efflux pumps and ATP-binding cassette (ABC) transporters (Barrick et al. 2004; Meyer et al. 2011). These transporters either have undefined function or appear to exert broad substrate specificity; hence, it is difficult to identify a common metabolite that could be regulating their expression (Jack et al. 2000; Paulsen 2003). Enzymes such as urea carboxylases, allophanate hydrolases, and numerous proteins with unknown function are also found to associate with ykkC, however, they cannot be generalized to a common metabolic pathway. Due to the combination of these factors, the ykkC riboswitch has resisted ligand identification. Sequence analysis shows that ykkC consists of two conserved stem–loop domains followed by a highly conserved 3′-region, which appears largely devoid of any secondary structure (Barrick et al. 2004). The 3′-region tends to overlap with a transcription terminator, suggesting that this riboswitch regulates the downstream operon at the level of transcription. Thus, it has been hypothesized that ykkC is involved in the response to intracellular toxins by controlling the expression of efflux pumps and other proteins involved in the detoxification process (Barrick et al. 2004).
Recently, the ligand of the ykkC riboswitch was identified as guanidine through in vivo screening of growth conditions that turn on the expression of a ykkC riboswitch-controlled reporter gene (Nelson et al. 2017). Guanidine is a known denaturant at high concentrations and a strong base that ionizes to its positively charged form (guanidinium) in the intracellular environment (Greenstein 1938). Although the guanidyl moiety is frequently found in larger metabolites such as arginine and agmatine, little is known about the physiological role of free guanidinium nor its homeostasis. The Nelson et al. (2017) study hypothesized that the ykkC riboswitch could respond to toxic levels of guanidinium present in bacteria by allowing the expression of efflux pumps and other detoxification enzymes. As the first step toward mechanistic characterization, here we provide the structural basis for guanidinium recognition by the ykkC riboswitch from Dickeya dadantii. Using isothermal titration calorimetry (ITC), we show that ykkC binds guanidinium with an apparent dissociation constant (Kdapp) of 39 micromolar (μM). The 2.3 Å crystal structure of this riboswitch reveals a complex binding pocket formed by the highly conserved 3′-region that accommodates a single guanidinium cation. The riboswitch exploits both the planar geometry and the positive charge of guanidinium for ligand discrimination. The involvement of the 3′-region in sensing guanidinium sequesters this region from participating in transcription terminator formation, thereby allowing transcription read-through of the associated genes. The structure and quantitative mutagenesis of the ykkC riboswitch set the foundation for an in-depth understanding of guanidine detoxification.
RESULTS
D. dadantii ykkC riboswitch binds to guanidine with 39 µM affinity
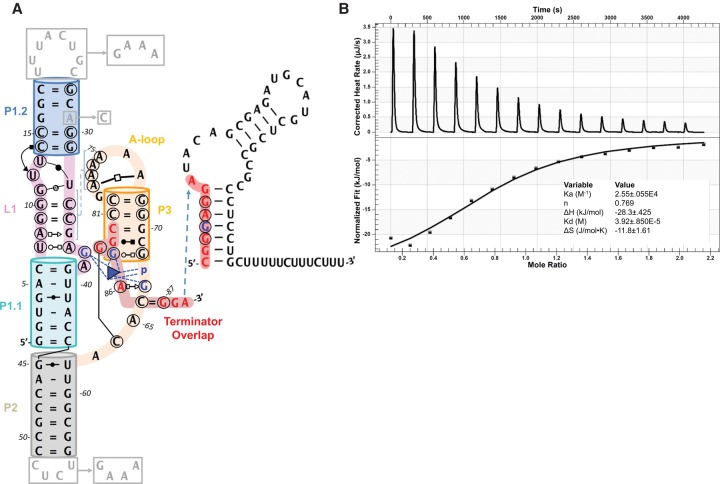
The D. dadantii ykkC (Dda_ykkC) riboswitch lacking the transcription terminator sequence was in vitro transcribed and purified for structural analysis. No significant mobility shift differences were observed in the native polyacrylamide (PAGE) analysis of Dda_ykkC in the presence or absence of 1 mM guanidine, suggesting that without the terminator sequence the riboswitch likely assumes the primed conformation, ready for ligand binding (data not shown). Strong heats were measured in an isothermal titration calorimetry (ITC) experiment when sixfold molar excess of guanidine hydrochloride (1.35 mM) was titrated into a calorimetric cell containing 0.22 mM prefolded Dda_ykkC riboswitch (Fig. 1B). The binding reaction is exothermic, with close to 1:1 ligand–RNA stoichiometry, and the fitting of the integrated injection heats yielded a Kdapp of 39 μM. Considering the large favorable binding enthalpy, ΔH value of −28.3 kJ/mol, and the smaller entropic component, TΔS of −11.8 J/mol * K, we concluded that the ligand–riboswitch interaction is enthalpically driven, presumably through the formation of favorable ligand–RNA contacts. Due to its high pKa value of 13.6, guanidine is expected to exist in its protonated isoform at neutral pH, as a positively charged guanidinium ion.
FIGURE 1.
Secondary structure model and ITC binding curve of Dda_ykkC. (A) 2D-representation of Dda_ykkC with Leontis–Westhof notation describing base interactions observed in the crystal structure. P1.1 (teal), L1 (violet), P1.2 (marine), P2 (gray), and P3 (orange) are shown. Circled residues are 97% conserved in subtype 1 ykkC riboswitches. Residues and phosphate contacting guanidinium (blue triangle) are highlighted in blue. Red residues participate in alternative terminator stem. Gray residues, boxes, and arrows show changes to WT sequence in the crystal construct. (B) ITC analysis of guanidinium binding by WT Dda_ykkC.
Architecture and important tertiary features of the guanidinium-bound D. dadantii ykkC riboswitch
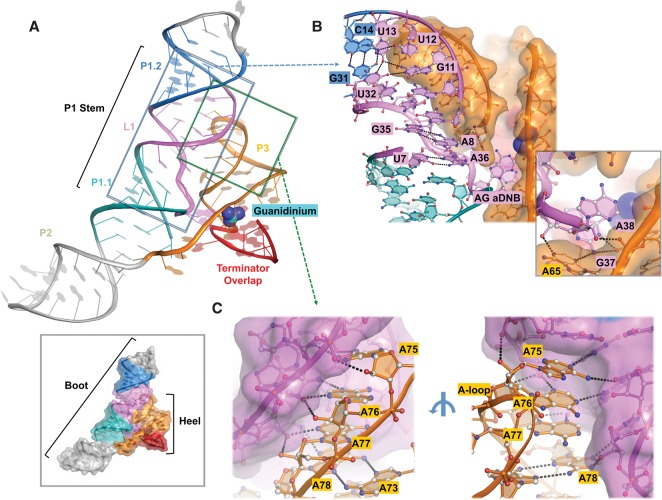
To understand how the ykkC riboswitch achieves specific recognition of its ligand, we determined a 2.3 Å crystal structure of Dda_ykkC bound to the guanidinium ion. Similar to many riboswitches (Batey et al. 2004; Smith et al. 2009; Price et al. 2015), the Dda_ykkC riboswitch could be roughly divided into two sets of RNA helices (P1/P2 and P3), juxtaposed and woven together by a set of conserved cross-helix contacts (Fig. 2A). In the context of the overall structure, P1/P2 forms a boot-like shape with P3 acting as the heel (Fig. 2A, inset). At the interface of the boot and heel, tertiary interactions between P1/P2 and P3 participate in the formation of the guanidinium-binding pocket. On one side of the interface, P1 and P2 coaxially stack into a curved pseudo-continuous helix. Secondary structure predictions of this region divided P1 into two helical portions (P1.1 and P1.2) separated by a large internal loop (L1) (Fig. 1A). As the structure reveals, this loop is continuously stacked, with the exception of an A38G37 asymmetric dinucleotide-bulge (aDNB) at the bottom of L1. Multiple base-triples and non-Watson-Crick (WC) base pairs are involved in maintaining the base stacking in L1, which explains the much elevated sequence conservation in this region (Fig. 2B). To accommodate the strand distortion caused by the aDNB, a C–G base pair (C6–G39) below the aDNB is highly conserved, presumably to impart stability to P1.1. Rising from the aDNB is a U7•A36 (WC–Hoogsteen [H]) pair, followed by an A8•G35 (H–sugar [S]) pair (Fig. 2B). These nonstandard pairings compensate for the backbone twist at the AG aDNB. Continuing up, two standard WC pairs (G9–C34 and G10–C33) are observed, after which the sugar–phosphate backbone distorts again with the formation of two base triples: G11•U32•U13 and G31–C14•U12 (Fig. 2B). As a result, the base of U13 is exposed and mediates a cross-helix stacking from P3 underneath. The AG aDNB allows the second cross-helix tertiary contact with P3; G37 forms two hydrogen bonds with A65 and a single bond to G67, while A38 makes a ribose–phosphate contact to C66, thereby weaving these domains of the riboswitch together (Fig. 2B, inset). Proximal to the aDNB, two magnesium ions coordinated by the phosphates of G37, C80, and C81 also act to connect L1 and P3 (Fig. 3A, inset; Supplemental Fig. S2). The P2 stem stacks underneath P1 and further contributes to the P1–P3 docking with its covalent tether that maintains P3 in close range of L1 (Fig. 2A).
FIGURE 2.
Overall structure of Dda_ykkC and detailed cross-helix interactions. (A) Cartoon representation of guanidinium (cyan) bound Dda_ykkC. Surface representation showing boot-like shape (bottom inset). (B) Detailed non-WC contacts in L1 region including AG aDNB and cross-helix contacts (inset). (C) Minor groove interaction between P3 A-loop and L1. Ribose zipper contacts are shown from the front (left) and base-specific contacts from the rear (right).
FIGURE 3.
Guanidinium recognition in the binding pocket. (A) Front (right) and rear (left) views of Dda_ykkC P3 stem. Base pairs are detailed with Leontis–Westhof notation in the middle with arrows indicating the location of the pair in front and rear views. Inset shows magnesium ions coordinated by the P1 and P3 backbones. (B) Side view of the binding pocket showing cation–π stacking interaction between guanidinium (cyan) and G67 (orange). Inset shows omit-map electron density of the binding pocket at 1.5 σ generated by simulated annealing refinement. Black dashes represent hydrogen bonds. Gray dashes represent weak electrostatic interactions. (C) Top view of the binding pocket showing hydrogen-bonding network. Mutants designed for binding affinity experiments are connected by lines to their corresponding residue labels. Interatomic distances are given next to dashes.
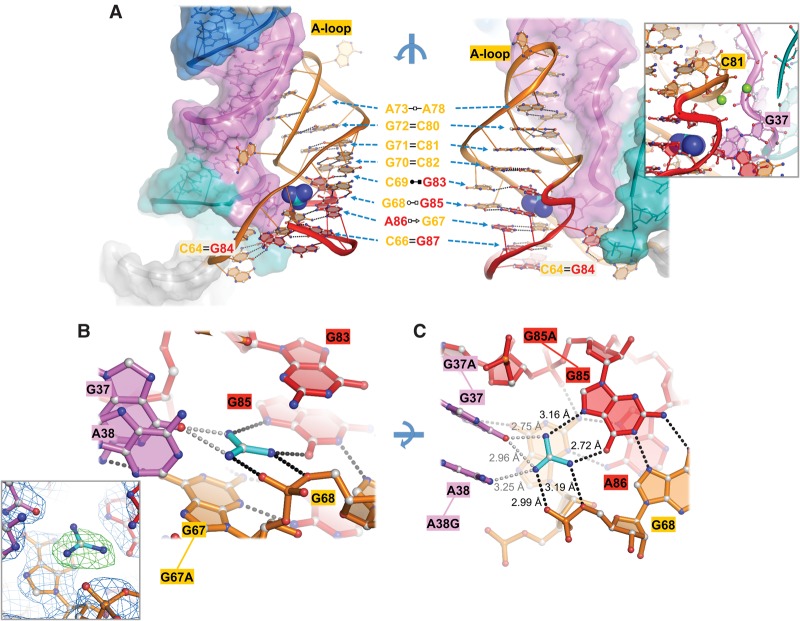
The 3′-portion of the riboswitch folds into the P3 helix, replete with backbone distortions and non-WC base pairs (Fig. 3A). The sequence in this region is highly conserved, but secondary structure could not be correctly predicted. Our structure reveals the presence of a 9-bp stem, capped by a highly conserved all-adenosine loop (A-loop) (Figs. 2C, 3A). Only three layers in P3 are continuous WC pairing: G70–C82, G71–C81, and G72–C80, which likely nucleate the formation of P3 (Fig. 3A). Moving upward to the A-loop region, A73•A78 form a H–H pair, A74 lacks specific contacts but mediates the strand reversal, and the next three adenosines continuously stack on top of A78 (Figs. 2C, 3A; Supplemental Fig. S2). In the crystal structure, four of the six consecutive adenosines in the A-loop mediate an important cross-helix tertiary contact to the minor groove of L1, which nicely explains their absolute conservation in the ykkC family. This starts from the tilted stacking of A75 underneath U13 of the L1 base triple, and continues with a set of tilted, base-specific minor groove contacts (A75•G11, A75•C33, A76•G10, and A78•G9), two type II A-minor interactions (A76•G11 and A77•G10) (Fig. 2C, left; Supplemental Fig. S1), and a continuous chain of ribose zipper contacts (A75•U12, A76•G11, and A77•G10) (Fig. 2C, right; Supplemental Fig. S1). Moving downward from the central WC pairs, the bottom half of the stem is less conventional. First, a weak single bond C69•G83 pair stacks over a H•WC pair (G68•G85) while G84 flips out from in between to form a long-range WC pair with C64 (Fig. 3A). Continuing down, the stem culminates in a S•H pair (G67–A86) followed by another long-range WC pair (C66–G87) (Fig. 3A). This nonstandard geometry sets the stage for two residues (G67 and G85) and a phosphate to create part of the binding pocket at the base of the “heel,” where the AG aDNB from L1 docks into P3. The floor of the pocket is sealed by the conserved G67, which is hydrogen bonded by A86 at the sugar edge, the phosphate of G85 at the WC edge, and G33 from the Hoogsteen edge (Fig. 3B,C). Meanwhile, the 5′-phosphate of G68, the Hoogsteen edge of G85, and the AG aDNB form the walls of the binding pocket (Fig. 3C). The C64–G84 pair causes an S-shaped twist in the backbone that further encloses the pocket, sealing one side of the interface between P3 and the AG aDNB (Fig. 3A, right). The conservation pattern in P3 is nicely explained by the structure. The highly conserved nucleotides in P3 are typically involved in tertiary contacts or non-WC pairing, whereas the less conserved or variable positions in P3 correspond to floppy flip-outs (i.e., G79 and A74) or weak pairing (i.e., C69•G83).
Molecular mechanism of guanidinium sensing by ykkC
The high resolution of this crystal structure greatly aided our ability to unambiguously assign the guanidinium ligand. At the center of the binding pocket described above, we observed a flat, triangular-shaped electron density, indicative of a sp2-hybridized planar guanidinium ion (Fig. 3B, inset; Supplemental Fig. S2). The shape of the density and the ligand–RNA interaction distances ruled out the possibility of fitting a water or metal ion in the pocket. This guanidinium ion forms a cation–π stack with G67 at the floor of the pocket, and participates in a network of coplanar hydrogen bond contacts to the residues constituting the walls of the pocket (Fig. 3B). Although a cation–π stack could take place between a metal ion and a base, the interaction is expected to be stronger in the case of a guanidinium ion, due to its delocalized sp2 hybridization state (Zarić 2003; Blanco et al. 2013). Coplanar with guanidinium, the riboswitch accepts a total of four hydrogen bonds from the ligand in the form of two bidentate interactions: one with the Hoogsteen edge of G85, the other with the bridging and nonbridging phosphoryl oxygens of G68 (Fig. 3C). At ∼120° apart, the G37/A38 aDNB approaches guanidinium from a tilted angle. Judging by the orientation, the partially negative O6 of G37 is involved in an electrostatic contact with the two amines (Fig. 3C). Notably, N1 of A38 is also in position for an electrostatic interaction. However, considering its sub-optimal distance (3.3 Å), weaker electronegativity, and less sequence conservation, we hypothesize that A38 may not contribute substantially to guanidinium binding. Overall, the directionality of the hydrogen-bonding network allows the riboswitch to distinguish between guanidinium and similar shaped metabolites such as urea. Moreover, the size of the binding pocket provides little extra room for larger molecules, explaining why this riboswitch only responds to free guanidinium, but not guanidyl-containing metabolites (Nelson et al. 2017).
Structure-guided mutagenesis of D. dadantii ykkC evaluated by ITC analysis
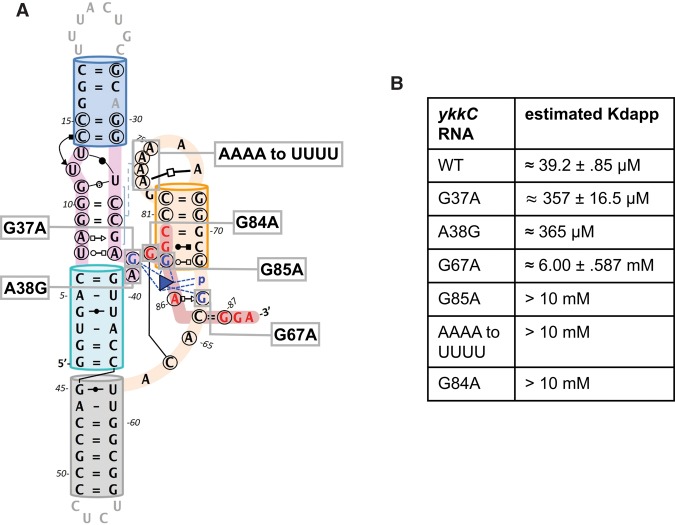
Structure-guided mutagenesis was carried out to evaluate the importance of the observed tertiary contacts and ligand–RNA interactions (Fig. 4A). Functional perturbations were read out from guanidine-binding affinity changes measured using ITC, as described in Figure 1. Given that all mutations significantly impacted binding affinity, Kdapps of mutants are calculated from low c-value curves and should be considered estimates. Two mutants were designed to target the AG aDNB. G37A is expected to disrupt both a cross-helix contact to G67 and H-bond contacts to guanidinium directly, whereas A38G is expected to introduce a steric clashing to G67. Both mutations reduced the guanidine-binding affinity by nearly 10-fold (Fig. 4B). G67 mediates a network of H-bonds to form the floor of the binding pocket, and forms the cation–π interaction to guanidinium. Hence, it is not surprising to find that the G67A mutation drastically reduced guanidinium binding by ∼200-fold (Fig. 4B). Mutations designed to disrupt a bidentate hydrogen bond to guanidinium (G85A), the L1–P3 minor groove interaction (AAAA to UUUU), and P3 folding (G84A) all produced flat binding curves despite titrating a great excess of guanidinium (Supplemental Fig. S3). The best fitting curves assign a Kd value of 10 mM to each of these mutants, though it should be noted that this is the maximum value the software can assign and the true binding constant is likely even worse. This large disruption in binding is expected for G85A and AAAA to UUUU as they perturb either guanidinium contacts or crucial tertiary contacts. The detrimental effect of G84A emphasizes the importance of this long-range G–C pair to the stability of P3. Overall, the mutagenesis results indicate that residues observed in our structure participating in guanidinium sensing and/or proper folding are indeed necessary for guanidinium binding.
FIGURE 4.
Structure-guided mutagenesis evaluated by ITC analysis. (A) 2D-representation of Dda_ykkC showing the positions with which each mutant corresponds. (B) Table displaying estimates of WT and mutant binding affinities for guanidinium.
DISCUSSION
Riboswitches are shown to be remarkably versatile biosensors with the ability to discriminate between different small molecules with similar chemical properties. Here we present high-resolution structure analysis of the ykkC riboswitch to detail its guanidinium-sensing mechanisms. The molecular recognition strategy includes the following: (i) A network of coplanar hydrogen bonds and an electrostatic interaction to specify the sp2-hybridized guanidinium form; closely related molecules such as urea do not satisfy the charge and hydrogen-bonding pattern. (ii) A cation–π contact favoring a flat rather than round cation; metal ions are disfavored. (iii) A form-fitting pocket that repels the binding of larger molecules containing guanidyl groups. Guanidine is a strong nucleic acid and protein denaturant at high concentrations. Interestingly, molecular simulation revealed that it interacts with nucleobases preferentially in two fashions: coplanar edge contacts through bidentate H-bond formation and cation–π stacking onto purine bases (Blanco et al. 2013). Both schemes are exploited by ykkC to achieve specific guanidinium recognition (Fig. 3B,C). The positioning of a guanosine residue underneath the ligand differs from protein–RNA interactions where arginine guanidyl groups tend to form the more favorable bidentate interaction instead of cation–π stacking (Luscombe et al. 2001; Morozova et al. 2006). The conservation of a guanosine at this position in ykkC is likely due to a combination of its contributions to guanidinium sensing and its centrality in the hydrogen-bonding network of the P3 domain.
The structure of ykkC also sheds light into its possible conformational dynamics. The extensive helix distortion and nonstandard base-pairing in P3 seem to imply less thermostability and greater conformation flexibility. A likely cotranscriptional folding scenario may be that the riboswitch assumes the coaxially stacked P1/P2 structure first, with the 3′-region subsequently nucleating from the 3-bp G–C helix. Key events leading to the stable structure include the A-loop docking into P1, the AG dinucleotide docking into P3, and the formation of several long-range G–C pairs to solidify the P3 structure. It appears that most ykkC family riboswitches are transcriptional regulators; sequence at the end of the 3′-region often overlaps with a competing transcription terminator. In the case of Dda_ykkC, the last eight residues of P3 are predicted to form alternative base pairs in a terminator stem. The partial sequestration of these residues seen in our structure (Fig. 3A) suggests that the presence of guanidinium imparts additional stability to P3, promoting transcription read-through of the downstream operon by perturbing the formation of the terminator. Interestingly, ykkC continues structural and mechanistic themes observed in other riboswitches. Similar to ykkC, the Mn2+-sensing riboswitch ybpY–ykoY and the guanine riboswitch adopt folds resembling adjacent helical domains with a ligand-binding pocket formed in between by cross-helix contacts (Batey et al. 2004; Price et al. 2015). In both cases, the regulatory mechanism is reliant upon the ligand stabilizing local structure in one of the helical domains, as we have proposed above for ykkC.
The new functional characterizations of ykkC from this structural study open the door to unknown territories of bacterial physiology. In D. dadantii, the ykkC element precedes an operon containing the three components of an ABC transporter, two putative urea carboxylase-related proteins, and a putative urea carboxylase. Even with the correct ligand assigned, it is still difficult to understand how guanidinium connects these seemingly disparate cellular functions. The most obvious hypothesis is that the transporters downstream from ykkC are guanidinium pumps, and the rest of the enzymes are involved in the chemical transformation of guanidine. Indeed, an SMR protein controlled by this riboswitch has been shown to selectively bind guanidine, while a ykkC-associated urea carboxylase has been characterized as a guanidine carboxylase (Nelson et al. 2017). Thus, ykkC senses the intracellular concentration of guanidinium and turns on the expression of these enzymes and transporters to modify and/or remove guanidinium before it reaches toxic levels. Overall, ykkC riboswitches appear to control processes involved in the removal or modification of guanidinium, hinting at a hitherto unappreciated area of bacterial physiology involving free guanidine.
Members of the ykkC family of riboswitches exhibit notable variations in sequence conservation. The Nelson et al. (2017) study describes two subtypes of ykkC that each regulates a unique set of genes: “subtype 1” includes Dda_ykkC and is associated with the efflux pumps and carboxylases described above, “subtype 2” is less common and controls genes involved in purine metabolism. Subtype 2 riboswitches display different sequence preferences at positions observed in our structure to be intimately involved in guanidinium sensing. For example, at the positions analogous to G67 and A38, pyrimidines are highly conserved (Nelson et al. 2017). Likewise, the critical G84 and G85 positions are not strictly conserved as guanosines, but tend to be purines. These differences suggest that the subtype 2 ykkC riboswitches adopt a different binding pocket conformation. Additionally, mini-ykkC and ykkC-III elements have been described that associate with similar genes as subtype 1 ykkC, but unlike subtype 2, share no sequence conservation, and are predicted to form completely different structures (Weinberg et al. 2010). The structural and biochemical analysis of these distinct ykkC family elements would certainly reveal new aspects of bacterial physiology related to guanidinium sensing.
While our manuscript was under review, an independent structural study on guanidinium sensing by a subtype 1 ykkC riboswitch from Sulfobacillus acidophilus was reported (Reiss et al. 2017). In general, the important tertiary interactions and guanidinium-sensing mechanisms detailed therein agree with what we observed in Dda_ykkC. Minor structural differences are noticed. These two studies therefore provide two reference points to distill the essential elements governing RNA-based guanidinium sensing.
MATERIALS AND METHODS
Constructs and plasmids
The sequences of the constructs used in the biochemical and crystallization experiments are documented in Supplemental Table S1.
RNA preparation
RNA constructs were cloned and produced as described previously (Ke and Doudna 2004; Grigg and Ke 2013). Sequences were cloned into the pUC19 plasmid and were preceded by a T7 RNA polymerase promoter and followed by the hepatitis δ virus ribozyme (HDV) to produce homogeneous ends. Guanosine residues were added at the beginning of each to increase expression. For crystal constructs, plasmid templates for transcription were prepared with QIAGEN MegaPrep kits and linearized by restriction digestion after the HDV sequence. For ITC analysis, transcription templates were prepared using PCR amplification. Ten milliliters of in vitro transcription reactions were performed as previously (Ke and Doudna 2004). RNA was gel purified by urea denaturing PAGE. The ykkC bands were excised from the gel, crushed, and eluted into water at 4°C overnight. RNA was buffer-exchanged to 10 mM HEPES, pH 7.0, 50 mM NaCl and refolded at 5 µM in 10 mL of the same buffer by heating at 65° C for 15 min, adding 10 mM MgCl2 and (when appropriate) 1 mM guanidine HCl. RNA was left at 65°C for an additional 5 min and then placed on ice. Cooled samples were concentrated to 0.2 mM and then used for crystallization.
Crystallization and data collection
RNA constructs were screened for crystallization by hanging drop vapor diffusion at 0.1 and 0.2 mM RNA, at 16°C with 1.0 mM Guan. HCl. Optimized conditions for the D. dadantii ykkC riboswitch were: 0.110 mM RNA in a mother liquor of 20% (+/−)-2-methyl-2,4-pentanediol (MPD), 40 mM Na cacodylate, pH 6.0, 80 mM NaCl, 12 mM spermine tetra-HCl, at 16°C, with 1:1 RNA:mother liquor drop ratio. For phasing, 20 mM iridium hexamine (IrHex) and 20% MPD were added to the crystals for 1.5 h prior to flash freezing in liquid N2.
Data were collected at the Advanced Photon Source (APS) 24 ID-C Northeastern Collaborative Access Team (NE-CAT), as indicated in Supplemental Table S2. Data sets were processed using HKL-2000 (Otwinowski and Minor 1997) or by XDS (Kabsch 2010) as part of NE-CAT's RAPD pipeline. The D. dadantii structure was phased by the single-wavelength anomalous dispersion (SAD) method from iridium using PHENIX AutoSol (Adams et al. 2010). The hkl2map interface for the SHELX suite was used (Pape and Schneider 2004; Sheldrick 2010). A model using the SAD data was built by alternating rounds of manual building in COOT (Emsley et al. 2010) followed by refinement in phenix.refine. The final model was built by molecular replacement of the SAD model into a higher resolution native data set using PHENIX Phaser-MR followed by additional refinement.
ITC analysis
ITC was conducted using a TA instruments Nano ITC with a low volume cell. The binding curve of the D. dadantii WT riboswitch for guanidinium was obtained at 25°C with a cell concentration of 0.224 mM RNA and syringe concentration of 1.35 mM guan. HCl. For mutants, cell concentrations between 0.100 and 0.200 mM RNA and syringe concentrations 6, 12, 30, and 60 times the cell concentration were used. Model fitting and data analysis were done using Nanoanalyze software. The first injection data point was excluded for all model fitting except a single WT run. For the severely impaired binding curves of the mutants, the n-value parameter was fixed at 1.0 for model fitting, following the method for analyzing low affinity binding (Turnbull and Daranas 2003).
DATA DEPOSITION
Atomic coordinates and structure factor amplitudes have been deposited with the PDB with accession code 5U3G.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant GM118174 to A.K. This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P41 GM103403). The Pilatus 6M detector on 24-ID-C beamline is funded by a NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract no. DE-AC02-06CH11357.
Author contributions: R.A.B. and A.K. designed the experiments. R.A.B. performed RNA crystallization, structure determination, and ITC. I.R.P. contributed extensively in RNA phasing, model building, and structure refinement. Each author contributed to manuscript preparation.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.060186.116.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr Sect D Biol Crystallogr 66: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I, et al. 2004. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci 101: 6421–6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey RT, Gilbert SD, Montange RK. 2004. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature 432: 411–415. [DOI] [PubMed] [Google Scholar]
- Blanco F, Kelly B, Sánchez-Sanz G, Trujillo C, Alkorta I, Elguero J, Rozas I. 2013. Non-covalent interactions: complexes of guanidinium with DNA and RNA nucleobases. J Phys Chem B 117: 11608–11616. [DOI] [PubMed] [Google Scholar]
- Breaker RR. 2011. Prospects for riboswitch discovery and analysis. Mol Cell 43: 867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron MP, Bastet L, Lussier A, Simoneau-Roy M, Massé E, Lafontaine DA. 2012. Dual-acting riboswitch control of translation initiation and mRNA decay. Proc Natl Acad Sci 109: E3444–E3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr Sect D Biol Crystallogr 66: 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenstein JP. 1938. Sulfhydryl groups in proteins. II. J Biol Chem 125: 501. [Google Scholar]
- Grigg JC, Ke A. 2013. Structural determinants for geometry and information decoding of tRNA by T box leader RNA. Structure 21: 2025–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack DL, Storms ML, Tchieu JH, Paulsen IT, Saier MH. 2000. A broad-specificity multidrug efflux pump requiring a pair of homologous SMR-type proteins. J Bacteriol 182: 2311–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66: 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke A, Doudna JA. 2004. Crystallization of RNA and RNA–protein complexes. Methods 34: 408–414. [DOI] [PubMed] [Google Scholar]
- Li S, Breaker RR. 2013. Eukaryotic TPP riboswitch regulation of alternative splicing involving long-distance base pairing. Nucleic Acids Res 41: 3022–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Smith AM, Fuchs RT, Ding F, Rajashankar K, Henkin TM, Ke A. 2008. Crystal structures of the SAM-III/SMK riboswitch reveal the SAM-dependent translation inhibition mechanism. Nat Struct Mol Biol 15: 1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscombe NM, Laskowski RA, Thornton JM. 2001. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res 29: 2860–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Breaker RR. 2004. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat Struct Mol Biol 11: 29–35. [DOI] [PubMed] [Google Scholar]
- Meyer MM, Hammond MC, Salinas Y, Roth A, Sudarsan N, Breaker RR. 2011. Challenges of ligand identification for riboswitch candidates. RNA Biol 8: 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova N, Myers J, Shamoo Y. 2006. Protein-RNA interactions: exploring binding patterns with a three-dimensional superposition analysis of high resolution structures. Bioinformatics 22: 2746–2752. [DOI] [PubMed] [Google Scholar]
- Nelson JW, Atilho RM, Sherlock ME, Stockbridge RB, Breaker RR. 2017. Metabolism of free guanidine in bacteria is regulated by a widespread riboswitch class. Mol Cell 65: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Pape T, Schneider TR. 2004. HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J Appl Crystallogr 37: 843–844. [Google Scholar]
- Paulsen IT. 2003. Multidrug efflux pumps and resistance: regulation and evolution. Curr Opin Microbiol 6: 446–451. [DOI] [PubMed] [Google Scholar]
- Peselis A, Serganov A. 2014. Themes and variations in riboswitch structure and function. Biochim Biophys Acta 1839: 908–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price IR, Grigg JC, Ke A. 2014. Common themes and differences in SAM recognition among SAM riboswitches. Biochim Biophys Acta 1839: 931–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price IR, Gaballa A, Helmann JD, Ke A. 2015. Mn2+-sensing mechanisms of yybP-ykoY orphan riboswitches. Mol Cell 57: 1110–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss CW, Xiong Y, Strobel SA. 2017. Structural basis for ligand binding to the guanidine-I riboswitch. Structure 25: 195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Nudler E. 2013. A decade of riboswitches. Cell 152: 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick GM. 2010. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr 66: 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, Lipchock SV, Ames TD, Wang J, Breaker RR, Strobel SA. 2009. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat Struct Mol Biol 16: 1218–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull WB, Daranas AH. 2003. On the value of c: can low affinity systems be studied by isothermal titration calorimetry? J Am Chem Soc 125: 14859–14866. [DOI] [PubMed] [Google Scholar]
- Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR. 2010. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol 11: R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarić SD. 2003. Metal ligand aromatic cation−π interactions. Eur J Inorg Chem 2003: 2197–2209. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.