Abstract
The Z-disc is a protein-rich structure critically important for the development and integrity of myofibrils, which are the contractile organelles of cross-striated muscle cells. We here used mouse C2C12 myoblast, which were differentiated into myotubes, followed by electrical pulse stimulation (EPS) to generate contracting myotubes comprising mature Z-discs. Using a quantitative proteomics approach, we found significant changes in the relative abundance of 387 proteins in myoblasts versus differentiated myotubes, reflecting the drastic phenotypic conversion of these cells during myogenesis. Interestingly, EPS of differentiated myotubes to induce Z-disc assembly and maturation resulted in increased levels of proteins involved in ATP synthesis, presumably to fulfill the higher energy demand of contracting myotubes. Because an important role of the Z-disc for signal integration and transduction was recently suggested, its precise phosphorylation landscape further warranted in-depth analysis. We therefore established, by global phosphoproteomics of EPS-treated contracting myotubes, a comprehensive site-resolved protein phosphorylation map of the Z-disc and found that it is a phosphorylation hotspot in skeletal myocytes, underscoring its functions in signaling and disease-related processes. In an illustrative fashion, we analyzed the actin-binding multiadaptor protein filamin C (FLNc), which is essential for Z-disc assembly and maintenance, and found that PKCα phosphorylation at distinct serine residues in its hinge 2 region prevents its cleavage at an adjacent tyrosine residue by calpain 1. Fluorescence recovery after photobleaching experiments indicated that this phosphorylation modulates FLNc dynamics. Moreover, FLNc lacking the cleaved Ig-like domain 24 exhibited remarkably fast kinetics and exceedingly high mobility. Our data set provides research community resource for further identification of kinase-mediated changes in myofibrillar protein interactions, kinetics, and mobility that will greatly advance our understanding of Z-disc dynamics and signaling.
The highly regular organization of myofibrils, the contractile organelles of cross-striated muscle cells, gives rise to the typical banding pattern of skeletal and cardiac muscle fibers. Myofibrils are mainly composed of an almost crystalline array of thin and thick filaments based on actin and myosin, respectively. The repeating contractile units of myofibrils are the sarcomeres, which are flanked by Z-discs. The latter protein-rich structures provide an essential structural framework by tethering actin filaments at their barbed ends, cross-linking them by antiparallel dimers of α-actinin and linking them to the giant protein titin at its amino terminus. Z-discs not only define the lateral boundaries of adjacent sarcomeres, but also help to connect myofibrils to each other, e.g. via intermediate filaments. In addition, they are involved in linking the contractile apparatus to the sarcolemma and the extracellular matrix via large, membrane-associated protein complexes, the costameres. The function of the Z-disc is not only limited to force transmission, but it is also an important hub for signal transduction events. To fulfil its dual role, Z-discs have to be dynamic and at the same time have to encompass numerous structural proteins.
Over the last years, the number of proteins with functions in mechanosensing and signal transduction identified to localize at least temporarily to the Z-disc has steadily increased (reviewed in (1, 2, 3)). To date, over 100 gene products are linked to the term “Z-disc” in the human or mouse NCBI gene database (http://www.ncbi.nlm.nih.gov/gene/). However, its precise protein inventory and phosphorylation landscape have not been coherently analyzed. Numerous signaling proteins such as protein kinase C (PKC)1 (4) and the protein phosphatase calcineurin (5) were shown to dynamically localize to the Z-disc. Notably, kinase- and phosphatase-mediated phosphorylation and dephosphorylation events may likely control the dynamic shuttling of proteins in and out of the Z-disc as recently revealed for myopodin (6), a protein interacting with F-actin, α-actinin, and filamin C (FLNc) (7, 8). The large cytoskeletal protein FLNc, in turn, constitutes an important hub in the Z-disc interactome with manifold binding partners such as myotilin (9), nebulette (10), the Xin actin-binding repeat containing proteins Xin (11) and XIRP2 (12), and the calsarcins/myozenins/FATZ proteins (13, 14, 15).
Distinct from its two other ubiquitously expressed family members FLNa and FLNb, FLNc is mainly expressed in cross-striated muscles (16). In healthy muscle, it predominantly localizes at Z-discs, whereas a minor portion is found beneath the sarcolemma in association with the dystrophin-associated glycoprotein complex (17). During myofibril development, FLNc assists in Z-disc assembly by acting as a molecular scaffold (18). Mutations in its gene cause severe myopathies and cardiomyopathies (reviewed in (19)).
All filamin isoforms feature an aminoterminal actin-binding domain (ABD) and a rod of 24 immunoglobulin-like (Ig-like) domains. Flexibility is mainly provided by hinge regions between Ig-like domains 15 and 16 (hinge 1) and 23 and 24 (hinge 2). Depending on cell type and differentiation stage, alternative splicing may remove hinge 1 in FLNc and FLNb (20, 21). The carboxyterminal Ig-like domain 24 mediates homodimerization, resulting in filamin dimers capable of cross-linking actin filaments (22, 23, 24), whereas hinge 2 was suggested to fulfil a regulatory role in dimerization (22). FLNc features a unique insertion of 82 amino acids in Ig-like domain 20, which is sufficient for Z-disc targeting (18). This insert is also likely important for establishing diverse protein interaction and scaffolding functions for cytoplasmic signaling processes. Compatible with its role in intracellular signaling events, FLNc was proposed to shuttle between the Z-disc and the sarcolemma or other compartments, particularly in disease states (17, 18, 25, 26). Phosphorylation events and calpain-dependent cleavage of FLNc may likely be involved in regulating functions and localization of the protein (27, 28, 29), however precise information concerning phosphorylation and cleavage sites and the physiological effects thereof have remained obscure.
In this work, we differentiated mouse C2C12 myoblasts into multinucleated myotubes by serum reduction followed by electrical pulse stimulation (EPS) to generate contracting myotubes comprising fully assembled sarcomeres with mature Z-discs. By quantitative proteomics, we determined changes in the abundance of proteins in EPS-treated contracting myotubes versus myoblasts and differentiated myotubes. We found that contracting C2C12 myotubes exhibited increased levels of proteins needed for ATP synthesis, reflecting their higher energy consumption because of contractile activity. Through phosphoproteomics analysis of contracting C2C12 myotubes, we further identified the myofibrillar Z-disc as major site of protein phosphorylation. Based on these data, we established a detailed site-specific phosphorylation map of the Z-disc proteome, highlighting the large number of highly phosphorylated Z-disc and Z-disc-associated proteins including FLNc. We focused on a phosphosite cluster in the hinge 2 region of FLNc for a detailed analysis. Through in vitro kinase assays coupled to high resolution MS we precisely mapped serine residues serving as specific PKCα substrate sites. These data were further confirmed by a cellular approach. In a newly established top-down and a targeted MS-based approach, we determined a distinct tyrosine residue positioned directly carboxyterminal to these phosphorylation sites as the major calpain 1 cleavage site in FLNc. PKCα-mediated phosphorylation not only controls cleavage by calpain 1, but also modulates FLNc dynamics. Interestingly, an FLNc variant lacking Ig-like domain 24, thus mimicking calpain 1 cleaved FLNc, exhibited remarkably fast kinetics and exceedingly high mobility.
EXPERIMENTAL PROCEDURES
Cell Culture
C2C12 and C2 myoblasts were cultured in high glucose DMEM medium (Life Technologies, Darmstadt, Germany) supplemented with 15% FCS (PAA, GE Healthcare Life Sciences, Freiburg, Germany), 1% nonessential amino acids, 1% penicillin/streptomycin and 1% sodium pyruvate (all Life Technologies) in six-well plates (Techno Plastic Products AG, Trasadingen, Switzerland) to a confluency of ∼90%. Differentiation was induced by reduction of the FCS content to 2% (30) in the absence of sodium pyruvate. Differentiation medium was changed every 48 h until complete myotube formation was observed (day 5–6). After myotube development, sarcomere formation was improved by EPS (0.5 Hz, 4 ms, 10–12 V) with a C-Pace EP Culture Pacer (IonOptix, Milton, MA) for 16–24 h. SILAC labeling of C2C12 myoblasts was performed with high glucose SILAC-DMEM medium (GE Healthcare Life Sciences) supplemented with dialyzed 15% FCS (PAA), 1% nonessential amino acids,1% sodium pyruvate, 1% proline (all Life Technologies), 84 mg/l arginine and 146 mg/l lysine (Cambridge Isotope Laboratories Inc., Tewksbury, MA) for at least nine cell doublings. Light, medium, and heavy stable isotope labeling by amino acids in cell culture (SILAC) was performed with 13C6 l-arginine and 12C6 l-lysine, 12C6 l-arginine and D4 l-lysine, and 13C615N4 l-arginine and 13C615N2 l-lysine, respectively. Labeled myoblasts were seeded into six-well plates and differentiation was induced by reduction of the dialyzed FCS content to 2% in the absence of sodium pyruvate (30). Immortalized mouse skeletal myoblasts (IMMs) were cultivated and transfected as described before ((31); (32)). IMMs with passage numbers of up to 40 were used for experiments. Human embryonic kidney cells (HEK293) were cultured in DMEM supplemented with 10% FCS and 1% sodium pyruvate. Transient transfections of HEK293 and C2 cells were performed using polyethylenimine (PEI, 1 μg/μl Polysciences Europe GmbH) and Lipofectamine2000 (Invitrogen, Darmstadt, Germany), respectively. To this end, cells were grown to a confluence of 70% in 6 cm dishes and for each transfection, a total of 4.5 μg DNA was used. PEI was mixed in a 3:1 ratio with DNA in a 6-fold volume of Opti-MEM (Life Technologies) and incubated for 15 min. C2 cells were transfected in solution according to the manufacture's protocol. Transfected cells were grown for 24 h before cell lysis. All cell lines were regularly tested to be mycoplasma-negative.
Cell Lysis and Sample Preparation
For lysis of C2C12 myotubes, plates were placed on ice and cells were washed twice with ice-cold PBS (CM-PBS; 0.75 mm CaCl2, 0.75 mm MgCl2, 155 mm NaCl2, 2.7 mm KCl, 2 mm KH2PO4, 10 mm Na2HPO4, pH 7.4). 150 μl precooled lysis buffer (7 m urea, 2 m thiourea, 1 mm sodium orthovanadate, 10 mm β-glycerophosphate, 9.5 mm sodium fluoride, 10 mm sodium pyrophosphate) were added per well, cells were scraped from the dish and sonified 2× for 10 s on ice for complete cell lysis. Insoluble material was removed by centrifugation for 20 min at 21,000 × g and 4 °C and the protein concentration was estimated using the Bradford assay (BioRad, München, Germany). Reduction and alkylation of proteins from phosphoproteome replicate 2 and dimethyl- and SILAC-labeled samples was performed as described (33) with slight modifications. Each reaction was carried out for 30 min using a final concentration of 1 mm dithiothreitol (DTT) and 55 mm 2-chloroacetamide before alkylation was quenched with a final concentration of 5 mm DTT. A lysate volume equal to a total protein amount of 100 μg (for each sample used for stable isotope dimethyl labeling), 7 mg (replicate 1), 8.1 mg (replicate 2), and 2 mg (for each SILAC replicate) was diluted 1:4 with 50 mm ammonium bicarbonate solution and digested with sequencing grade trypsin (1:50) (Promega, Mannheim, Germany) for 3.5 h at 200 rpm and 42 °C. Peptides were desalted using an Oasis HLB cartridge (Waters Corporation, Milford, MA) according to the manufacturer's protocol. Eluates were aliquoted, lyophilized and stored at −80 °C.
Stable Isotope Dimethyl Labeling
Triple stable isotope dimethyl labeling was performed as previously described (34). To this end, 100 μg tryptic digests of myoblasts (day 0), myotubes (day 5), and EPS-stimulated myotubes at day 5 were dissolved in 100 mm TEAB buffer and incubated for 1 h with triple dimethyl labeling reagents (all Sigma Aldrich, St. Louis, MO). A label switch was performed for each replicate (n = 3) and the completeness of the peptide labeling reaction was confirmed using an aliquot of 10 μg before mixing. Stable isotope-labeled peptide samples were mixed in a 1:1:1 ratio, desalted as described above, and dried in vacuo.
High-pH Reversed-Phase Chromatography
For each dimethyl and SILAC replicate, 300 μg and 2 mg of total peptide were dissolved in 200 μl buffer A (10 mm ammonium hydroxide, pH 10.5), sonicated for 3 min, centrifuged at 20,000 × g for 4 min and filtrated into a sample vial using a 0.2 μm syringe filter (Phenomenex, Aschaffenburg, Germany). Peptides samples were separated by high-pH reversed phase (hpH-RP) chromatography using a Dionex Ultimate 3000 system equipped with a RP Gemini C18 column (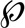 4.6 mm × 25 mm, 3 μ, 110 Å, Phenomenex) and operated at a flow rate of 200 μl/min and 750 μl/min, respectively. All separations were carried out using a binary solvent system (A: 10 mm ammonium hydroxide, pH 10.5; B: 10 mm ammonium hydroxide, pH 10.5, 90% acetonitrile) at a temperature of 40 °C. Peptide loading and binding was performed at 1% B for 5 min, followed by peptide elution by increasing buffer B to 20% in 35 min and further to 45% in 20 min. Ninety fractions were collected in a 96 well plate at 50 s intervals from minute 2 to 77. Every 30th fraction was pooled and acidified with TFA to a final concentration of 1%. From SILAC replicates, the collected fractions 19 and 20 were used for enrichment of phosphopeptides by TiO2 beads.
4.6 mm × 25 mm, 3 μ, 110 Å, Phenomenex) and operated at a flow rate of 200 μl/min and 750 μl/min, respectively. All separations were carried out using a binary solvent system (A: 10 mm ammonium hydroxide, pH 10.5; B: 10 mm ammonium hydroxide, pH 10.5, 90% acetonitrile) at a temperature of 40 °C. Peptide loading and binding was performed at 1% B for 5 min, followed by peptide elution by increasing buffer B to 20% in 35 min and further to 45% in 20 min. Ninety fractions were collected in a 96 well plate at 50 s intervals from minute 2 to 77. Every 30th fraction was pooled and acidified with TFA to a final concentration of 1%. From SILAC replicates, the collected fractions 19 and 20 were used for enrichment of phosphopeptides by TiO2 beads.
Strong Cation Exchange Chromatography
Tryptic digests were dissolved in 200 μl SCX buffer A (5 mm potassium dihydrogen phosphate, 20% acetonitrile (ACN, v/v), pH 2.8). The supernatant was loaded onto a Polysulfoethyl-A column (Ø 4.6 mm, 20 cm, 5 μm, 200 Å, PolyLC, Columbia, MD) equilibrated with SCX buffer A using a Dionex Ultimate 3000 UHPLC system. Peptides were separated at a flow rate of 700 μl/min applying a linear gradient of 0–30% SCX buffer B (5 mm potassium dihydrogen phosphate, 20% ACN (v/v), 500 mm KCl, pH 2.8) in 50 min starting 10 min after sample injection, followed by a gradient of 30–50% and 50–100% of solvent B in 10 min each. The column was washed with 100% B for 5 min and re-equilibrated for 20 min with 100% buffer A. 3 min fractions were collected during the gradient and 10 μl of each SCX fraction were dried in vacuo for LC/MS analysis. The remaining volume of each fraction was used for TiO2 enrichment. A total of 55 SCX fractions were collected for replicates 1 and 2 and further processed for phosphoproteomics analysis in a randomized procedure. Nonenriched and TiO2-enriched peptide samples were dried in vacuo and stored at −80 °C for further analysis.
Titanium Dioxide Enrichment
Phosphopeptides were enriched using titanium dioxide (TiO2) spherical beads as described before (35) with slight modifications. Briefly, 30 μl of TiO2 material (TiO2, 5 μm, GL Science Inc, Tokyo, Japan) resuspended in ACN 1:1 (v/v) were used for SCX fractions and hpH-RP fractions, whereas 15 μl were used for single protein digests. The TiO2 material was washed 2× with 200 μl washing buffer (80% ACN, 0.1% TFA) and 1× with 50 μl loading buffer (20% (v/v) acetic acid, 20 mg/ml 2,3-dihydroxybenzoic acid, 420 mm octasulfonic acid, 0.1% (v/v) heptafluorobutyric acid). SCX fractions or protein digests from in vitro kinase assays were mixed 1:1 (v/v) with 2× loading buffer, added to the prepared TiO2 material, vortexed and incubated for 20 min at 4 °C with slight agitation. Each sample was washed twice with 200 μl washing buffer. Subsequently, 50 μl elution buffer (50 mm ammonium dihydrogen phosphate, pH 10.5) were added to each sample followed by incubation for 10 min. Subsequently, 50 μl ACN were added, the suspension was loaded onto a gel loader tip (GE Healthcare) supplied with a 1 mm Teflon capillary and peptides were eluted by centrifugation at 5000 rpm. Eluates were placed on ice, acidified with 8 μl TFA and dried in vacuo.
Liquid Chromatography and High Resolution Mass Spectrometry
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) was performed using the UltiMate 3000 RSLCnano system (Dionex LC Packings/Thermo Fisher Scientific, Dreieich, Germany) coupled online to a Velos Orbitrap Elite, Q Exactive Plus or LTQ-Orbitrap XL instrument (all Thermo Fisher Scientific, Bremen, Germany). In general, UHPLC systems were equipped with two C18 μ-precolumns (Ø 0.3 mm × 5 mm; PepMap, Thermo Fisher Scientific) and an Acclaim® PepMap analytical column (ID: 75 μm × 250 mm, 2 μm, 100 Å, Dionex LC Packings/Thermo Fisher Scientific). High-resolution MS instruments were externally calibrated using standard compounds and equipped with a nanoelectrospray ion source and distal coated SilicaTips (FS360–20-10-D, New Objective, Woburn, MA) and MS/MS analyses were performed on multiply charged peptide ions.
For quantitative MS-based proteome analyses using a Q Exactive Plus, peptide mixtures from hpH-RP fractions were separated using a binary solvent system consisting of 0.1% formic acid (FA, solvent A, “A”) and 0.1% FA/86% ACN (solvent B, “B”). Samples were washed and pre-concentrated on a C18 μ-precolumn with 0.1% TFA for 5 min before switching the column in line with the analytical column. Replicates were analyzed applying a gradient of 78 min at a flow rate of 250 nl/min. Peptide samples were eluted with a gradient of 4–40% B in 50 min and 40–95% B in 5 min. After each gradient, the analytical column was washed with 95% B for 5 min and re-equilibrated for 5 min with 4% B. The instrument parameters were as follows: spray voltage 1.5 kV, capillary temperature 200 °C, tube lens voltage 100 V. Data dependent acquisition was performed using the software XCalibur 3.0.63. Mass spectra were acquired from m/z 370–1,700 with a resolution of 70,000 at m/z 200. Automatic gain control (AGC) was set to 3 × 106 ions and a maximum (max.) fill time of 60 ms. MS/MS analyses employing higher-energy collisional dissociation (HCD) in a TOP12 method was applied with the following parameters: AGC target, 1 × 105 ions; normalized collision energy (NCE), 28; dynamic exclusion time, 45 s; underfill ratio, 0.7%; max. fill time, 120 ms; isolation window, 3 m/z.
For large-scale phosphoproteome analysis, peptide mixtures from SCX fractions before and after TiO2 enrichment were separated using the solvent system described above. Before separation, samples were washed and pre-concentrated for 30 min on a C18 μ-precolumn with 0.1% TFA. Replicate 1 and 2 were analyzed with a 78 min-gradient at a flow rate of 300 nl/min and a 75-min gradient at 250 nl/min, respectively. For replicate 1, a gradient of 5–30% B in 60 min, 30–50% B in 20 min and 50–95% B in 5 min was used. After the gradient, the analytical column was washed with 95% B for 3 min and re-equilibrated for 5 min with 5% B. For replicate 2, the gradient used was as follows: 5–42% B in 65 min, 42–95% B in 5 min, and 95% B for 5 min and 5% B for 15 min for re-equilibration. MS analyses were performed on an LTQ-Orbitrap XL using CID for SCX fractions and multi-stage activation (MSA) with neutral loss (NL) masses of 32.7, 49 and 98 Da for TiO2-enriched samples. The instrument parameters were as follows: spray voltage, 1.5 kV; capillary voltage, 44 V; capillary temperature, 200 °C; tube lens voltage, 100 V; NCE, 35; activation q, 0.25; activation time, 30 ms. Data-dependent acquisition was performed using the software XCalibur 2.1.0 SP1.1160. Mass spectra were acquired from m/z 370–1,700 with a resolution of 60,000 at m/z 400. AGC was set to 2 × 105 (CID) and 5 × 105 ions (MSA) and a max. fill time of 500 ms. A TOP5 method was used with detection of fragment ions in the linear ion trap (LIT) using the following parameters: dynamic exclusion time: 45 s, AGC: 10,000 ions, max. fill time: 400 ms.
Samples from in vitro kinase assays were enriched for phosphopeptides by TiO2 and analyzed by LC-MS. RPLC separations were performed essentially as described above using 0.1% FA/86% ACN (solvent B) with 4% DMSO (Velos Orbitrap Elite) or without (Q Exactive) in a 1 h LC gradient. Velos Orbitrap Elite instrument parameters were as follows: spray voltage, 1.8 kV; capillary temperature, 200 °C; AGC, 1 × 106 ions; max. fill time, 200 ms. For MSA (NL, 32.7, 49 and 98 Da), a TOP10 method with a dynamic exclusion time of 45 s, NCE 35 and an AGC of 10,000 ions with a max. fill time of 200 ms was used. Mass spectra were acquired from m/z 370–1,700 with a resolution of 60,000 and 120,000 (at m/z 400) for MSA and electron transfer dissociation (ETD), respectively. For ETD, a TOP6 method with an exclusion time of 30 s and an AGC of 5,000 ions with a max. fill time of 100 ms and an activation time of 150 ms was employed. MS/MS analyses employing HCD were essentially performed as described above with the following modifications: spray voltage 1.8 kV, mass range from m/z 375–1700 and a max. fill time of 60 ms. For fragmentation, a TOP10 method was used with an exclusion time of 30 s, AGC of 1 × 106 ions, and a NCE of 30.
Enriched phosphopeptides from selected hpH-RP fractions were analyzed on a Velos Orbitrap Elite instrument. Samples were separated using a 2 h-gradient with a binary solvent system consisting of 0.1% FA and 0.1% FA/30% ACN/50% MeOH supplemented with 4% DMSO. The instrument was operated as described above using a TOP15 method (MSA; NL of 32.7, 49 and 98 Da) with a dynamic exclusion time of 45 s, an AGC of 1 × 105 ions and a max. fill time of 200 ms.
Targeted MS analyses were performed on a Q Exactive Plus using a binary solvent system consisting of solvent A and B as described above with a 5-min preconcentration step followed by separation applying a 45-min gradient at a flow rate of 250 nl/min. Peptide samples were eluted with a gradient of 4–40% B in 30 min and 40–95% B in 5 min. After each gradient, the analytical column was washed with 95% B for 5 min and re-equilibrated for 5 min with 4% B. The instrument parameters were as described above with a max. fill time of 120 ms. Targeted single ion monitoring (SIM) scans of the peptide SSIPKFSSDASKVVTR observed at m/z 854.96505 (+2), 570.31246 (+3), and 427.98616 (+4) were performed with an AGC of 5000 and an isolation window of 3 m/z. Targeted ions were selected for fragmentation using a data dependent MS/MS scan with the following parameters: AGC target 2 × 105 ions, NCE 28, dynamic exclusion time: 3 s, underfill ratio of 0.2%, max. fill time: 200 ms, isolation window 4 m/z.
For top-down analyses, a Velos Orbitrap Elite coupled online to an U3000 HPLC system equipped with a C18 μ-precolumn (Ø 0.3 mm × 5 mm; PepMap, Thermo Fisher Scientific) and an Acclaim PepMap300 analytical column (ID: 75 μm × 150 mm, 5 μm, 300 Å, Thermo Fisher Scientific) was used. Five microliters acidified sample were injected and separated using a binary solvent system consisting of 0.1% FA (solvent A) and 0.1% FA in 100% ACN (solvent B) with a gradient from 5% B to 90% B in 25 min at a flow rate of 313 nl/min. Subsequently, the column was washed for 2 min with 90% B and re-equilibrated with 5% B for 13 min. Data-dependent acquisition was performed using the software XCalibur 2.1.0 SP1.1160. Mass spectra were acquired from m/z 300–2000 with a resolution of 60,000 at m/z 400 and an AGC target value of 5 × 105 at a max. ion time of 200 ms. Multiply charged ions ≥ 3+ were selected for fragmentation. A TOP5 CID/ETD method was applied with the following parameters: dynamic exclusion time, 5 s; AGC, 1 × 106 ions; max. fill time, 500 ms; isolation width, m/z 4; CID activation time, 10 ms; ETD activation time, 100 ms with a fixed first mass at m/z 100.
Bioinformatics
For quantitative analysis of MS data from dimethyl labeling experiments, Andromeda integrated in MaxQuant 1.5.3.30 (36) was used to search peak lists against the UniProt ProteomeSet mouse database (release 2015_11, 57,276 protein entries). The precursor mass tolerance was set to 20 ppm for the first search and to 4.5 ppm for the main search. Trypsin was set as proteolytic enzyme allowing up to two missed cleavages. Cysteine carbamidomethylation and DimethylLys0, DimethylNter0, DimethlyLys4, DimethylNter4, DimethylLys8 and DimethylNter8 were set as fixed modification and oxidation of methionine as variable modification. Protein identification was based on at least one unique peptide with a minimum length of seven amino acids and a false discovery rate (FDR) of 1% was applied on both peptide and protein lists. The options “re-quantify” and “match between runs” were enabled. Proteins quantified in at least two biological replicates per experiment were considered for further bioinformatic and statistical data analyses. For processing of MaxQuant result files, a k-means hierarchical cluster analysis was performed using an in-house developed R script.
For global phosphoproteomics data, Andromeda integrated in MaxQuant 1.3.0.5 (36) was used to search peak lists against the UniProt ProteomeSet mouse database (release 2015_11, 57,276 protein entries). The precursor mass tolerance was set to 20 ppm for the first search and to 4.5 ppm for the main search. For MSA/CID data, the fragment mass tolerance was set to 0.5 Da. Trypsin was set as proteolytic enzyme allowing up to two missed cleavages. Oxidation of methionine and phosphorylation of serine, threonine, and tyrosine were set as variable modifications and cysteine carbamidomethylation as fixed modification in replicate 2. A FDR of 1% was applied on both peptide (on modified peptides separately) and protein lists. Numbers of unique phosphopeptides were counted based on the MaxQuant peptide ID in the Phospho(STY) sites table. Phosphosites scored with a MaxQuant localization probability of ≥ 0.75 were deemed “localized,” whereas sites with a localization probability of < 0.75 were counted as putative sites given that the amino acid sequence in combination with the number of phosphate groups was not identified with localized sites elsewhere in the data set.
For analysis of MS data from in vitro kinase assays, raw files were processed using Andromeda embedded in MaxQuant 1.4.1.2 and searched against the sequences of mouse and human FLNc d23–24 and their respective phosphosite mutants generated in this work using the IPI human decoy database (version 3.76,178,826 entries) as background for correct FDR calculation. Precursor and fragment mass tolerances were set to 10 ppm and 0.5 Da, respectively. Search parameters were as follows: proteolytic enzyme: trypsin, max. number of missed cleavages: 2, and variable modifications: methionine oxidation and phosphorylation of serine, threonine and tyrosine.
Result files from SILAC data were processed using MaxQuant 1.5.2.8 and searched against the UniProt ProteomeSet mouse database (release 2016_01, 58,239 protein entries). The parameters were set to the same value as for the analysis of data obtained from in vitro kinase assays with the addition of carbamidomethylation as fixed modification and multiplicity was set to three with Arg0, Lys0, Arg6, Lys4, Arg10, and Lys8. MaxQuant msms.txt files, all raw files and FLNc isoform sequences were imported into Skyline 2.6.0 (37). MS1 intensities were calculated as described by (38) using the MS1 filtering tutorial provided by the software developers. Skyline peptide settings were as follows: tryptic peptides with 1 missed cleavage, a time window of 3 min, min. and max. peptide length 8 and 30 amino acid, respectively, exclusion of cysteine-containing peptides, phosphorylation of serine, threonine, and tyrosine and oxidation of methionine as variable modifications, and max. number of variable modifications and neutral losses 3 and 1, respectively. Orbitrap default parameters were used for transition settings. Extracted ion chromatograms of the imported peptides were manually inspected for correct peak picking and peak integration was adjusted by hand, if necessary. Total MS1 areas for all peptides with an error less or equal to 3 ppm were exported into a pivot table and processed using Excel2010 and Origin 9.1. The mean and the standard error of the mean were calculated within the three biological replicates first and afterward within technical replicates. Intensities of all phosphopeptides were summed separately for human and mouse FLNc isoforms and phosphopeptides were normalized by the respective calculated summed intensity.
For top-down experiments, raw data were converted into 32-bit mzXML files using the MSConvert tool embedded in ProteoWizard 3.0.6965 (39) with default parameters. mzXML files were further processed with MSDeconv (40) into an msalign file with standard parameters and used for peptide search with TopPIC (41). Search parameters were as follows: fragmentation mode read from the input file, no cysteine protection group, an error tolerance of 10 ppm, an e-value of 0.01 as cut-off and a maximal PTM mass of 1000. Searches of the FLNc isoforms were performed against FASTA files containing the sequences of the corresponding proteins.
For Gene Ontology (GO) enrichment analysis, the Cytoscape 3.2.1 plugin BiNGO (42) was used. Enrichment was analyzed for the three main GO domains “cellular component” (CC), “molecular function” (MF), and “biological process” (BP). Benjamini-Hochberg FDR correction at a significance level of 0.05 was used for p value correction after the hypergeometric statistic test. Overrepresented categories after p value correction in comparison to the Mus musculus background dataset were evaluated for major differences between the two protein groups. Calpain cleavage sites were predicted using the program GPS-CCD with a cut-off score of 0.654 (43).
Mass Spectrometric Data Deposition
All raw data and original MaxQuant result files have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (http://www.ebi.ac.uk/pride/archive/login) (44) with the data set identifiers PXD004960 (quantitative myogenesis experiment), PXD003211 (large-scale phosphoproteomics analysis), PXD003185 (in vitro kinase assay) PXD003216 (top-down experiments), PXD004151 (SILAC-mFLNc S625) and PXD005097 (SRM FLNc analysis). Processed data of kinase assays and SILAC data analyzed with Skyline as well as their results are available on PanoramaWeb interface (45) https://panoramaweb.org/labkey/invitro_FLNc.url, https://panoramaweb.org/labkey/invivo_mFLNc.url and https://panoramaweb.org/labkey/SRM_hFLNc_calpain_GluC.url.
Design of cDNA Constructs and Site-Directed Mutagenesis
Site-directed mutagenesis was performed using the QuikChange® Lightning Site- Directed Mutagenesis Kit (Agilent Technologies, Waldbronn, Germany) as described in the manual. The AA (S2623/S2624 to alanine) and DD (S2623/S2624 to aspartate) mutants of hFLNc d23–24 in pET23a/EEF were obtained by PCR using the primer pairs CCTCAAGCCGGGGCGCCGCCTACAGCTCCATCC with GGATGGAGCTGTAGGCGGCGCCCCGGCTTGAGG, and CCTCCTCAAGCCGGGGCGACGACTACAGCTCCATCCCCA with TGGGGATGGAGCTGTAGTCGTCGCCCCGGCTTGAGGAGG, respectively, using the nonmutant variant as template.
Site-directed mutagenesis of mFLNc d23–24 and hFLNc d22–24 was performed by PCR in a two-step procedure. First, the primers GCGGTGCCGGCTACAGTTCC and GGAACTGTAGCCGGCACCGC were used for mFLNc23–24. Second, the primer pair GCGGTGCCGCCTACAGTTCC and GGAACTGTAGGCGGCACCGC or GCGGTGCCGACTACAGTTCC and GGAACTGTAGTCGGCACCGC was used to obtain the S2625A or S2625D point mutant of mFLNc d23–24. To generate the hFLNc d22–24 S2623/2624D mutant, the primers CGGGGCGACAGCTACAGCTC and GAGCTGTAGCTGTCGCCCCG were used and then the primer pair CGGGGCGACGACTACAGCTC and GAGCTGTAGTCGTCGCCCCG. For the hFLNc d22–24 S2623/2624A point mutant, GGGGCGCCAGCTACAGCTC and GAGCTGTAGCTGGCGCCCC were used followed by GGGCGCCGCCTACAGCTC and GAGCTGTAGGCGGCGCCC. To obtain full-length mutant FLNc variants in pEGFP vectors for FRAP, the wildtype (WT) Ig-like d23–24 were replaced by the respective mutant variant taking advantage of a unique AgeI restriction site in the cDNA encoding d23 and the SalI site in the multiple cloning site of the vector. Similarly, the cDNA encoding d23 and d24 was replaced by cDNA lacking the sequences encoding aa 2628–2725 (i.e. part of hinge 2 and complete Ig-like d24) to obtain an FLNc variant (ΔIg-like d24) similar to the calpain 1-digested full-length FLNc protein, but still retaining the calpain 1 cleavage site.
Recombinant Protein Expression and Purification
Expression of His6-tagged mouse and human FLNc fragments in E. coli BL21 (DE3) CodonPlus cells (Stratagene, Santa Clara, CA, USA) and their purification using Ni2+- NTA agarose (Qiagen, Venlo, the Netherlands) was performed essentially as described (7). Briefly, 50–100 ml of E. coli culture was allowed to express the recombinant proteins by addition of isopropyl β-d-1-thiogalactopyranoside (IPTG). Cells were centrifuged and the resulting pellet was resuspended in 4 ml lysis buffer (50 mm NaHPO4, 300 mm NaCl, 10 mm imidazole, 1 mg/ml lysozyme, pH 8.0) for 30 min on ice followed by ultrasonication for complete cell lysis. Cell debris was removed by centrifugation for 30 min at 4500 rpm and 4 °C and the supernatant was added to 500 μl Ni2+-NTA agarose prepared according to the manufacturer's protocol. The slurry was incubated for 2 h at 4 °C before the supernatant was removed following a centrifugation step. Subsequently, Ni2+-NTA agarose-coupled His6-tagged proteins were washed twice with precooled washing buffer (50 mm NaHPO4, 300 mm NaCl, 20 mm imidazole, pH 8.0) on a pre-equilibrated gravity-flow column (Mobicol, Boca Scientific, Boca Raton, FL). Proteins were eluted by adding elution buffer (50 mm NaHPO4, 300 mm NaCl, 250 mm imidazole, pH 8.0) and stored at 4 °C.
In Vitro Kinase Assays
Recombinantly expressed and purified mouse and human FLNc d23–24 WT and phosphosite mutants were dialyzed overnight at 4 °C in dialysis buffer (1 mm DTT, 100 mm KCl, 20 mm HEPES pH 7.4, 10 mm MgCl2). For MS-coupled in vitro kinase assays, a sample volume corresponding to 100 μg protein was added to a volume of 200 μl H2O and mixed with 5× PKC buffer (25 mm DTT, 100 mm KCl, 20 mm HEPES pH 7.4, 10 mm MgCl2, 0.5 mm CaCl2, 2.5 mm ATP, sodium fluoride, sodium pyrophosphate, sodium orthovanadate, β-glycerophosphate, 0.15% CHAPS, 1× PKC lipid activator (Merck Millipore, Darmstadt, Germany)). The assay was started by adding 200 ng PKCα (Sigma-Aldrich) and the reaction was performed under shaking at 200 rpm for 20 min at 30 °C for each FLNc construct. For further analysis, samples of three independent replicates were diluted 1:4 (v/v) with 50 mm ammonium bicarbonate and subjected to in-solution digestion using sequencing grade trypsin (1:50) (Promega) for 3.5 h at 200 rpm and 42 °C. Single protein digests were acidified with TFA (final concentration 1% (v/v)), subjected to TiO2 enrichment, and the resulting phosphopeptide-enriched fractions analyzed by LC-MS/MS using alternative fragmentation methods.
Radioactive in vitro kinase assays were performed with 1–2 μg protein in 1× PKC buffer. Samples were incubated with 5–10 μCi (γ-33P) ATP and 1 mm ATP for 20 min at 30 °C. In vitro phosphorylation was started by adding 10 ng or 20 ng PKCα. Subsequently, 4× Laemmli buffer was added to the samples followed by SDS-PAGE and autoradiography.
SILAC Analysis
SILAC-labeled C2C12 myotubes were starved overnight and subjected to EPS for 4 h. Subsequently, myotubes were subjected to treatment with phorbol-12-myristat-13-acetat (PMA, 200 μm, Sigma Aldrich) for 15 min, Gö6976 (10 μm, Merck) for 1 h, or DMSO for 1 h (control). A label switch was performed for each of the three biological replicates analyzed in this work. Protein concentrations were measured using the Bradford assay (BioRad, München, Germany) and control, PMA and Gö6976 treated samples were mixed in a 1:1:1 ratio. Protein digestion was performed as described above, peptides were separated using hpH-RP chromatography, and fractions 19–20 were subjected to TiO2-based phosphopeptide enrichment followed by LC-MS/MS analysis.
Calpainolysis Assays
In vitro calpainolysis assays were performed essentially as described (46). In brief, recombinantly expressed mouse and human FLNc d23–24 WT and phosphosite mutants with a carboxyterminal EEF- and His6-tag were coupled to Ni2+-NTA agarose beads and washed 2× with 1 ml calpain reaction buffer (20 mm HEPES, pH 7.4, 50 mm KCl, 2 mm MgCl2, 5 mm CaCl2, 1 mm DTT). Subsequently, the bound protein fragments were incubated for 30 min with 5 μg calpain 1 (Merck) in 200 μl reaction buffer at 37 °C and 200 rpm on a Thriller Thermoshaker (Peqlab, Erlangen, Germany). Pre-equilibrated gravity-flow columns were used to separate the beads from the supernatant. For top down analysis of FLNc proteolysis products, supernatants were acidified to a final concentration of 0.1% TFA. The remaining agarose beads were incubated with 150 μl Laemmli sample buffer for 5 min at 95 °C and samples were stored for Western blot analysis.
For further calpainolysis assays, HEK293 cells transiently expressing hFLNc d22–24 WT and the AA and DD phosphorylation site mutants were used or C2 cells expressing hFLNc d22–24 WT. Prior to lysis, cells were treated with PMA (200 nm for 15 min) or Gö6976 (10 μm for 1 h) as indicated. Cell lysis was performed 24 h after transfection on ice with precooled lysis buffer (10 mm Tris HCl, pH 7.4, 100 mm NaCl, 0.1% Triton X-100, 2 mm EGTA, sodium fluoride, sodium pyrophosphate, sodium orthovanadate, β-glycerophosphate). Calpain inhibitor IV (Merck) was directly added to the lysis buffer at a final concentration of 50 μm. Subsequent to lysis, cells were incubated in an ultrasonic bath for 20 min at 8 °C and centrifuged for 5 min at 1500 × g. Supernatants were adjusted to the same protein concentration and calpainolysis was started by addition of 5 mm CaCl2 and 0.85 μg calpain 1 (Merck) per 100 μl reaction volume. Samples were incubated for 20 min at 37 °C and 800 rpm and the reaction was stopped with the according volume of 5× Laemmli buffer. Cleavage products from three independent assays were analyzed by SDS-PAGE and immunoblotting. The percentage of cleaved hFLNc d23–24 WT and hFLNc d23–24 mutants was calculated based on quantified immunoblot signals of uncleaved hFLNc and the respective ∼13 kDa cleavage products.
For targeted MS analysis of the cleavage product, calpainolysis assays of mock-, Gö6976- and PMA-treated cells were performed as described above. Following incubation with recombinant calpain 1, the amino-terminal FLNc cleavage product was enriched by Myc-dynabeads (Thermo Fisher) according to the manufacture's protocol. Subsequently, on-bead digestion was performed with the protease GluC (Promega, 1:50) for 4 h at 42 °C. Supernatants were desalted, dried in vacuo and stored at −80 °C
Fluorescence Recovery After Photobleaching and Data Analysis
Fluorescence recovery after photobleaching (FRAP) experiments were performed with a Cell Observer Spinning Disk microscope (Carl Zeiss, Jena) equipped with an external 473 nm diode laser (Rapp OptoElectronic, Hamburg) allowing bleaching of precisely defined regions of interest (ROI) using a Plan-Apochromat 63×/1.4 oil objective. Cells were kept at 37 °C under 5% CO2. Image processing was carried out using Zen 2012 software (Carl Zeiss, Jena, Germany). For FRAP analyses, 15–30 experiments were performed. For each cell, 1–3 ROIs were chosen for bleaching with each region limited to a single Z-disc of neighboring myofibrils. Photobleaching was performed with 100% intensity of the 473-nm laser with a pulse time of 1 ms with 8 iterations. Images were taken before and immediately after bleaching. The fluorescence recovery was monitored for 400 s with an interval time of 0.1–5 s. FRAP experiments were performed with a Cell Observer S.D. (Carl Zeiss, Jena) equipped with an external 473 nm laser coupled via a scanner (UGA-40, Rapp OptoElectronic, Hamburg) allowing bleaching of precisely defined regions of interest (ROI), using a Plan-Apochromat 63×/1.4 oil objective. Cells were permanently kept at 37 °C and 5% CO2. Image processing was carried out using Zen2012 software (Carl Zeiss, Jena). For FRAP analyses 15–30 experiments were performed. For each cell, 1–3 ROIs were chosen for bleaching with areas of bleaching limited to one sarcomere. Photobleaching was done with 100% intensity of a 473-nm laser for a pulse time of 1 ms with 8 iterations. Images were taken before and immediately after bleaching. The fluorescence recovery was monitored for 400 s interval time of 0.1 to 2 s. The ImageJ package Fiji (47) was used to determine fluorescence intensity of bleached and unbleached areas at each time point. Normalized FRAP curves were generated from raw data as previously described (48). FRAP data are displayed as mean of 10–15 individual experiments. To generate corrected FRAP curves, the intensity in the bleached ROI (IROI(t)) and in the whole cell excluding the bleached area (Iwhole(t)) at each time point were initially subtracted by the corresponding background intensity (Ibg(t)) and the fraction formed.
Subsequently, normalization to the prebleach intensity was performed according to the following equation:
where Inorm(t) is the normalized and corrected intensity, Ipre corthe corrected intensity before photobleaching and I0 cor the corrected first data point after photobleaching. Normalized fluorescence intensity versus time was plotted using Prism 4.0 (GraphPad Software). All results are expressed as mean ± S.D. Before treatment cells were starved for 2 h in serum-free medium. For photobleaching upon inhibition of PKCα activity, 10 μm Gö6976 inhibitor was added to the cell culture medium 2 h before starting the FRAP experiments and cells were kept in the medium throughout the analysis.
Antibodies
Mouse monoclonal antibody T12 labels a titin epitope close to the Z-disc (49). Rabbit polyclonal antisera recognizing the carboxyterminal 16 amino acids of FLNc (50) and FLNc d22–24 were custom-made by BioGenes, Berlin, Germany. The latter antiserum was extensively cross-absorbed against recombinantly expressed FLNa and FLNb d22–24 to ascertain specificity for FLNc. Anti-GAPDH (#2118) and anticalpain 1 (#2556) were purchased from Cell Signaling Technology (Leiden, the Netherlands), anti-Myc-tag (66004–1-Ig) from Proteintech (Manchester, UK). Horseradish peroxidase (HRP)-conjugated anti-rabbit immunoglobulins and AlexaFluor596-conjugated rabbit anti-mouse immunoglobulins were purchased from Sigma Aldrich and Life Technologies (Darmstadt, Germany), respectively.
Immunofluorescence Staining
For characterization of different cell stages, C2C12 myoblasts were seeded on collagen-coated cover slips (Thermo Fischer Scientific) and cultivated for different time periods. Immunolocalization studies were performed 2 d after seeding of cells, 5–6 d of myocyte differentiation and additional 16–24 h of electrical stimulation of myotubes. Cell fixation was carried out for 10 min in methanol/acetone (1:1, v/v) at −20 °C. Cover slips were washed with CM-PBS, blocked with 10% normal goat serum and 1% bovine serum albumin for 20 min and stained with T12 antibody (1:20) for 45 min. After a washing step, samples were incubated with AlexaFluor596-conjugated anti-mouse immunoglobulins (1:250) for 45 min. DAPI staining (1:1000) was performed for 2 min before cells were mounted with ProLongGold (Life Technologies). Optical analysis of cell differentiation was performed using a Nikon Eclipse TS 100 fluorescence microscope equipped with NIS Elements Basic Research 4.00.03 software.
Miscellaneous
Information about the experimental design and statistical rationale for different analyses performed in this work are provided within the respective subsections in Material and Methods. Number of sample size, replicates, controls, and statistical tests were chosen according to published data with comparable methodology and generally accepted standards in the field. SDS-PAGE and Western blotting were performed by standard protocols; signals were detected using HRP-coupled secondary antibodies and an enhanced chemiluminescence system (Thermo Fisher Scientific) with a ChemoCam (Intas, Göttingen, Germany) equipped with a full-frame 3.2 megapixel Kodak KAF-3200ME camera. No image processing, other than cropping, scaling and contrast adjustment was applied. Quantification of Western blot signals was performed with Quantity One 4.6.9 (Bio-Rad, Hercules, CA). For statistical analysis, two sample t-tests were performed using OriginPro 9.1 (OriginLab, Northampton, MA). All quantitative Western blot data are presented as means ± S.E. of the mean (S.E.) or standard deviation (S.D.). To minimize the effects of subjective bias, Western blot data were generated and analyzed by two different experimentators. The sample size was estimated on previous experiments and statistical tests were chosen based on published data with comparable methodology.
RESULTS
Quantitative Proteomic Analysis of Contracting C2C12 Myotubes
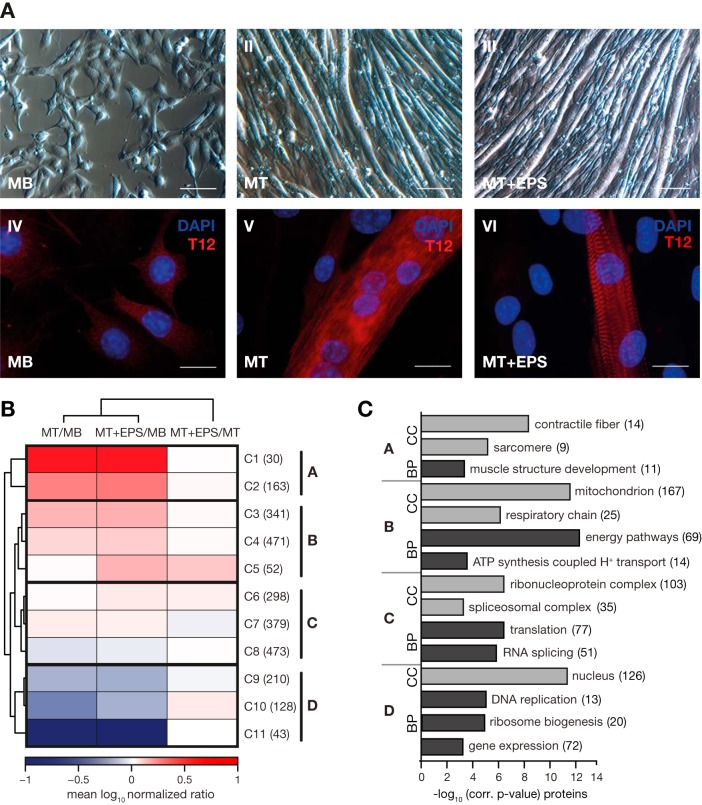
The coordinated assembly of sarcomeres, the basic contractile units of myofibrils, is an essential step of in vivo muscle differentiation. In mouse C2C12 myocytes, in vitro formation of contractile myofibrils comprising mature Z-discs can be induced by EPS of differentiated myotubes (51, 52). To follow the processes involved in the formation of contracting C2C12 myotubes using EPS, we here designed a global quantitative proteomic study comprising triple stable isotope dimethyl labeling, high-pH reversed-phase chromatography and LC-MS analyses (supplemental Fig. S1A). To this end, C2C12 myoblasts were grown to 90% confluence and differentiated for 5–6 days by serum reduction (Fig. 1A, I, II, IV and V). Subsequently, differentiated myotubes were subjected to EPS resulting in the formation of functional sarcomeres with mature Z-discs as highlighted by controlled myofibril contraction (Fig. 1A, III and VI, Movie 1). Our proteomic analysis of C2C12 myoblasts, myotubes and EPS-treated, contracting myotubes resulted in the quantification of 3520 proteins of which 2588 were quantified in at least two out of the three biological repicates of each cell state (supplemental Fig. S1B and supplemental Table S1). Through k-means cluster analysis, we identified 11 clusters which were further grouped into four main clusters (Fig. 1B and supplemental Fig. S1C–S1F, supplemental Table S1). We performed gene ontology (GO) enrichment analysis to reveal features overrepresented in the individual cluster (Fig. 1C, supplemental Table S2 and S3). As expected, proteins characterized by the GO terms “contractile fiber,” “sarcomer,” and “muscle structure development” were strongly up-regulated during differentitation of C2C12 myoblasts to myotubes, whereas levels of sarcomeric proteins were not altered by EPS (Fig. 1B, 1C1 and 1C2; Fig. 1C, main cluster A). The abundance of mitochondrial proteins including respiratory chain components significantly increased during myoblast differentiation (Fig. 1B and 1C, main cluster B). Interestingly, EPS-treated contracting myotubes showed increased protein levels of ATP synthase, which most likely reflects their higher energy demand in comparison with both myoblasts and “nonexercising” myotubes (Fig. 1B, C5). From 2588 quantified proteins, 1150 proteins (44%) (Fig. 1B, main cluster C) showed no change in abundance following the induction of myoblast differentiation. In the respective main cluster C, we found an overrepresenting of proteins with a role in gobal protein translation or mRNA splicing (Fig. 1B and 1C). As a consequence of the switch from cell proliferation to differentiation, the abundance of proteins with functions in DNA replication, ribosome biogenesis and gene expression was considerably decreased in both differentiated myotubes and contracting myotubes in comparison to myoblasts (Fig. 1B and 1C, main cluster D). For EPS-treated contracting myotubes, we observed a slight increase (+1.6-fold) in the abundance of proteins involved in transcription and translation, likely in consequence of the high metabolic and contractile activity of “exercising” skeletal myotubes (Fig. 1B, C10).
Fig. 1.
Quantitative proteomic analysis of C2C12 myogenesis. A, Formation of contracting sarcomeres in C2C12 skeletal muscle cells studied by light (I–III) and fluorescence (IV–VI) microscopy. Cell fusion was demonstrated by DAPI staining of nuclei (blue) and sarcomere formation by staining using a T12 antibody (red) directed against the Z-disc-associated end of titin (IV–VI). Mononuclear myoblast (MB) at 50% confluence (I, IV) were grown to 90% confluence before cell fusion was induced by serum reduction. Following 5–6 days of differentiation, cells were fused to polynuclear myotubes (MT) with a diameter of 20–50 μm and a length of up to 1–2 mm (II, V). To generate contracting myotubes, cells were electrically stimulated for 16–24 h (MT+ESP). Sarcomere formation was confirmed by observing the typical striated pattern in fluorescence microscopy (VI); no morphological changes were observed in light microscopy (III). Scale bar: I-III, 100 μm; IV-VI, 10 μm. B, Hierarchical cluster analysis of 2588 proteins quantified at each state of C2C12 differentiation (MB, MT, MT+EPS) in at least two out of three biological replicates. Mean protein ratios were log10 transformed. K-means cluster analysis revealed 11 clusters which were further grouped to four main clusters: cluster A (C1 and C2), B (C3-C5), C (C6-C8), and D (C9-C11). C, GO-term enrichment analysis of proteins grouped in the main cluster A–D. p values after Benjamini-Hochberg false discovery rate (FDR < 0.05) correction were plotted against their corresponding GO-terms from the domains “cellular component” (CC) and “biological process” (BP). Numbers of identified proteins for each GO term are shown in brackets.
Study of the Phosphoproteome of Contracting Myotubes
With the aim to further obtain insight into sarcomeric signaling processes, we characterized the protein phosphorylation landscape of contracting C2C12 skeletal myotubes. To this end, EPS-treated contracting myotubes were subjected to phosphoproteomic analysis comprising SCX-based fractionation of tryptic digests, phosphopeptide enrichment using TiO2 beads and high resolution LC-MS (supplemental Fig. S2A). In addition, a small aliquot of each SCX fraction was directly analyzed to support peptide and protein identifications. From the analysis of two biological replicates, we identified a total of 6808 proteins comprising 2941 (43%) phosphoproteins (Fig. 2A, supplemental Table S4). The overlap of identified proteins and phosphoproteins between the two replicates was 83 and 63%, respectively. The total number of phosphosites was 11,369 with 8175 (72%) allocated to a distinct amino acid with a probability ≥ 0.75 (supplemental Table S5). The large majority (1914, 65%) of phosphoproteins comprised more than a single phosphosite with 529 proteins modified by more than five distinct phosphate groups (Fig. 2B).
Fig. 2.
The Z-disc is the main site of protein phosphorylation in sarcomeres. A, Numbers of (phospho-)proteins and phosphosites (p-sites) identified in contracting C2C12 myotubes. Localized p-sites were identified with a MaxQuant localization probability score ≥ 0.75. B, Distribution of p-sites according to the number of sites per protein. C, D, Distribution of p-sites identified in sarcomeric proteins of the I- or A-band (C). 532 p-sites (91%) were identified in proteins of the I-band. Of these, 412 p-sites (77%) were identified in true Z-disc and Z-disc-associated proteins, which is an essential structural part of the I-band (D). E, Classification of p-sites localized in 44 Z-disc-associated proteins. 264 p-sites (83%) were identified with high confidence with a MaxQuant localization probability ≥ 0.90 (red). Further 53 p-sites were localized with a probability ≥ 0.75 and < 0.90 (gray). F, G, GO-term enrichment analysis of the C2C12 myotube proteome (F) and phosphoproteome (G) data set established in this work. p values after Benjamini-Hochberg false discovery rate (FDR < 0.05) correction were plotted against their corresponding GO-terms from the three main domains “cellular component” (CC), “molecular function” (MF) and “biological process” (BP). Numbers of identified proteins for each term are shown in brackets.
The Z-disc is a Hotspot of Protein Phosphorylation in Sarcomeres
We assessed which and to what extent sarcomeric proteins contribute to the phosphoprylation landscape of contracting skeletal myotubes. Remarkably, 532 (91%) of all the 586 phosphosites in sarcomeric proteins were identified in 66 proteins of the I-band (Fig. 2C, supplemental Table S5). Within the I-band, 412 (77%) of all phosphosites were found in 31 genuine Z-disc proteins and 21 proteins peripherally associated with this structure (Fig. 2D), underscoring the suggested outstanding signaling capacity of Z-disc proteins in striated muscle cells (53, 2). The majority of localized phosphosites (264, 83%) in proteins associated with the myofibrillar Z-disc were assigned with high confidence (probability ≥ 0.9) and further 53 sites (17%) were scored with probabilities between 0.75 and 0.9 (Fig. 2E). Of these, 89 sites were not reported in the PhosphositePlus database (version 07.01.2016, supplemental Table S5).
We employed GO enrichment analysis to reveal functional features overrepresented in the myofibrillar proteome (Fig. 2F, supplemental Table S6) and phosphoproteome (Fig. 2G, supplemental Table S6). In both datasets, the terms “Z-disc,” “nucleus,” “gene expression,” “kinase activity,” and “cytoskeletal protein binding” were significantly enriched, whereas “mitochondrion” was only overrepresented in the proteome data. Interestingly, PKC-binding and processes related to phosphorylation and cytoskeletal organization were specifically enriched in the phosphoproteome (Fig. 2G). In summary, our data revealed the Z-disc as the major myofibrillar site of protein phosphorylation, supporting the notion that it acts as an important signaling platform within the sarcomere (53, 2).
A Detailed View On the Z-disc Phosphorylation Landscape
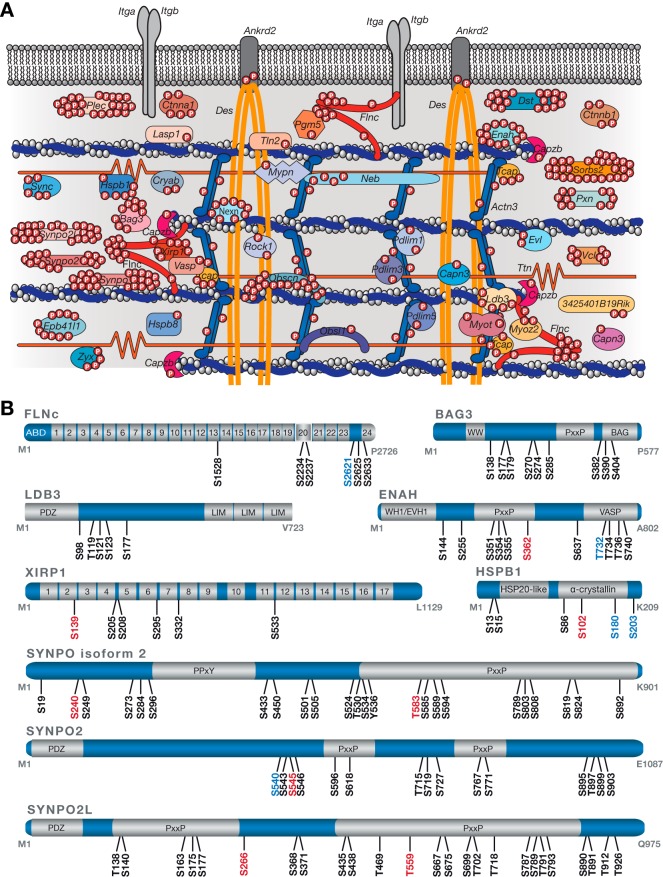
In the framework of this study, we defined a list of 42 true Z-disc proteins as well as 21 proteins directly associated with the Z-disc (associated and peripheral) and further 42 proteins connected to the Z-disc (membrane-associated, intercalated disc, intermediate filament) based on literature and expert knowledge (supplemental Table S7). The latter group comprises peripheral Z-disc proteins, including those connecting myofibrils with the sarcolemma, e.g. via costameric proteins like the dystrophin-associated protein complex. With the exception of a few Z-disc proteins only expressed in distinct muscle fibers such as nebulette and Fbxl22 in cardiac muscle (54, 55) or only located at the Z-disc under distinct conditions such as during embryonic development or during myofibrillar repair or remodeling processes (56), our data cover the complete Z-disc proteome known to date. This protein compendium includes a total of 31 specific (29 genes, 69%) and 21 associated (15 genes, 71%) Z-disc components with confident phosphosite localizations (Fig. 3A, supplemental Table S6). Remarkably, numerous Z-disc and -associated proteins such as FLNc, BCL2-associated athanogene 3 (BAG3), LIM domain binding 3 (LDB3), enabled homolog (ENAH, also referred to as MENA or ENA), Xin actin-binding repeat containing 1 (XIRP1), heat shock protein beta-1 (HSPB1), and the three members of the podin protein family synaptopodin (SYNPO), myopodin (SYNPO2) and tritopodin (CHAP; synaptopodin 2-like, SYNPO2L) were phosphorylated at multiple amino acid residues with up to a total number of 25 distinct phosphosites in SYNPO2L (Fig. 3B, supplemental Table S6). Interestingly, phosphosites often occurred in clusters, as observed in the podin protein family and FLNc.
Fig. 3.
A site-specific protein phosphorylation map of the myofibrillar Z-disc proteome from mouse skeletal muscle cells. A, Schematic overview of the Z-disc phosphoproteome. 44 genes (encoding for 51 proteins) were found to be modified by a single or multiple phosphate groups accounting for 70% of the known Z-disc proteome. Phosphosites localized in the Z-disc-associated regions of the two structural proteins titin (12 p-sites) and desmin (14 p-sites) are not depicted. B, Schematic illustration of highly phosphorylated Z-disc proteins. Localized phosphosites (mouse) are color-coded based on information provided by the PhosphoSitePlus database (version 07.01.2016) with black for known sites, blue for phosphosites known by similarity to orthologues sites in human proteins, and red for newly identified sites. Information about protein domains was retrieved from the UniProt and InterPro database. Immunoglobulin-like domains in FLNc and XIRP1 are numbered. ABD, actin-binding domain; BAG, Bcl2-associated athanogene; PDZ, post synaptic density protein, Drosophila disc large tumor suppressor, and zonula occludens-1 protein; PxxP, proline-rich; PPxY, proline-proline-x-tyrosine; VASP, vasodilator-stimulated phosphoprotein; WH1, WASP-homology/EVH1, Ena/VASP homology 1.
The Carboxyterminal Part of Filamin C is Multiply Phosphorylated In Myotubes
In this study, we identified six distinct phosphosites in mouse FLNc, two of which (pS2234, pS2237) were located in the isoform-specific insertion within the FLNc Ig-like domain 20 and three (pS2621, pS2625, pS2633) were clustered in its carboxyterminus (Fig. 3B, supplemental Table S6). Phosphorylation of FLNc at S2621 is reported for the first time in this work. Previous work suggested FLNc to be a potential substrate of PKCα in its carboxyterminal region (28). This prompted us to further analyze phosphorylation of FLNc at S2621 and S2625, both located in the hinge 2, as well as S2633 in the amino terminus of Ig-like domain 24 (Fig. 3B). We confirmed the correct assignment of these phosphosites located in a highly serine-rich stretch of amino acids (2618SSSSRGASYSSIPKFSSDASK2638) by manual interpretation of fragmentation spectra (supplemental Fig. S2B–S2D). For pS2621 and pS2625, we also identified the doubly phosphorylated peptide (supplemental Fig. S2E), indicating that FLNc can be concurrently phosphorylated at these serine residues in vivo. Kinase prediction analysis employing the algorithms NetPhosK and NetworKIN indicated that all three phosphorylated residues are putative substrate sites of PKC/PKCα (Fig. 2F). However, the exact PKC target site(s) in the carboxyterminal part of FLNc remained ambiguous.
Mouse FLNc S2625 and Human FLNc S2623/S2624 are Specific Substrate Sites of PKCα in vitro
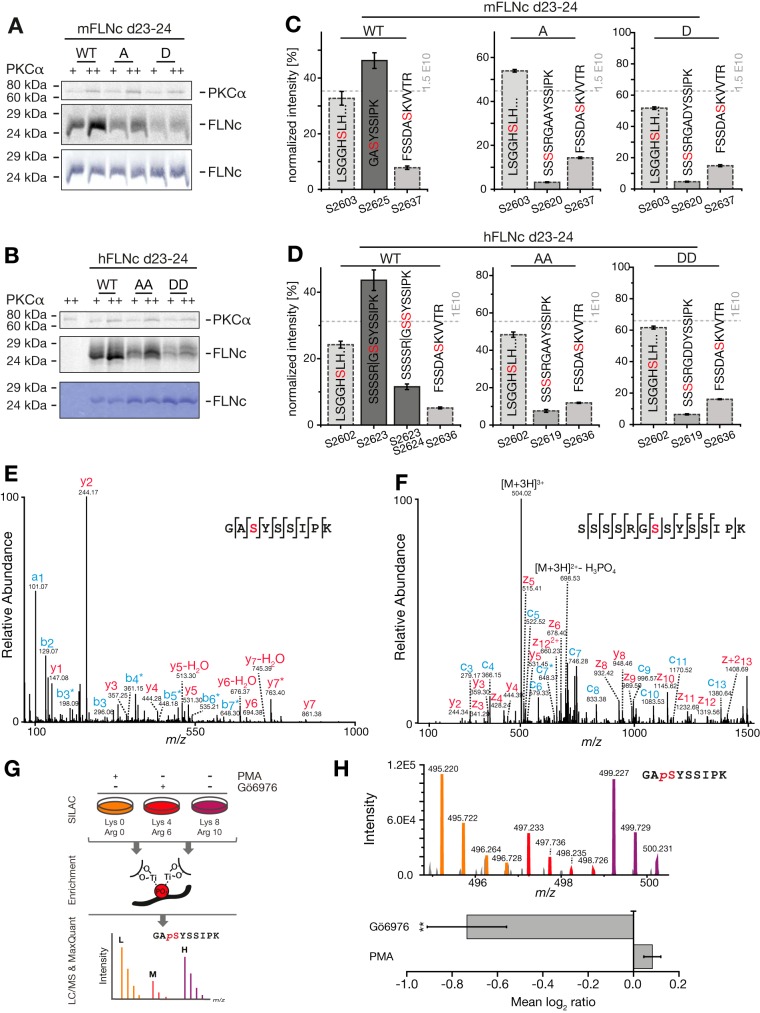
Our combined MS-based phosphoproteomics and kinase prediction data revealed for mouse FLNc (mFLNc) three in vivo sites potentially phosphorylated by PKC/PKCα (pS2621, pS2625 and pS2633). In human FLNc (hFLNc), the orthologous residues were consistently identified as in vivo phosphosites as well (57). Based on the presence of a PKCα consensus site motif (58), we hypothesized that PKCα likely phosphorylates mFLNC at S2625 and hFLNc at S2623 and/or S2624 (supplemental Fig. S3A). To test our hypothesis, we performed in vitro kinase assays using recombinantly expressed wildtype (WT) and phosphosite mutants of Ig-like domains 23–24 (d23–24) of mouse and human FLNc, each with a His6 and EEF tag at their carboxyterminal end (supplemental Fig. S3B). In mFLNc, we mutated S2625 to aspartate (D) or alanine (A) to mimic its constitutively phosphorylated or nonphosphorylated state, whereas in hFLNc the orthologous residues S2623 and S2624 were mutated. Incubation with PKCα in the presence of (γ-33P)-ATP showed that both WT constructs were phosphorylated by PKCα in a concentration-dependent manner, whereas signal intensities for the respective site mutants were considerably decreased, but not abolished (Fig. 4A and 4B). This indicated that PKCα is capable of phosphorylating additional sites in this in vitro assay. Phosphorylation of mFLNc d23–24 and hFLNc d23–24 WT and their respective phosphosite mutant forms was not observed following their incubation with (γ-33P)-ATP in the absence of PKCα (supplemental Fig. S4A).
Fig. 4.
S2625 and S2623/S2624 are the specific substrates sites of PKCα in the hinge 2 region of mFLNC and hFLNc, respectively. A, B, Radioactive in vitro kinase assays. Recombinant wildtype and phosphosite mutants of mouse (A) and human (B) FLNc Ig-like domains 23–24 (d23–24) were treated with PKCα in the presence of (γ-33P]ATP and analyzed by SDS-PAGE followed by autoradiography or Coomassie staining. S2625 of mFLNC d23–24 was replaced by A or D; S2623/S2624 of hFLNC d23–24 by AA or DD. As a control, PKCα was incubated in (γ-33P)ATP-containing kinase buffer without hFLNc d23–24 (B). WT, wildtype; A, alanine; D, aspartate; +, 10 ng PKCα; ++, 20 ng PKCα. C, D, MS-based in vitro kinase assays. Reactions were performed as described in (A, B) using unlabeled ATP and PKCα. MS data from three independent kinase experiments for mFLNc d23–24 WT, A and D sites mutants (C) and hFLNc WT, AA, and DD site mutants (D) were quantified. Intensities of phosphopeptides distinctive for a specific phosphorylation site (red) were added up per experiment and represented as normalized mean ± S.E. E, F, Fragmentation spectra of mono-phosphorylated peptides of mouse (E) and human (F) FLNc d23–24 WT forms. PKCα-dependent phosphorylation of mFLNc-S2625 and hFLNc-S2623 was determined by higher-collisional dissociation and electron transfer dissociation, respectively. Fragment ions exhibiting a neutral loss of phosphoric acid (H3PO4; 97.9768 u) are marked with an asterisk (*); loss of water (H2O) as indicated. Phosphorylated residues are depicted in red; b/c- and y/z-ion series in red and blue, respectively. G, Experimental workflow to study mouse FLNc S2625 phosphorylation in C2C12 myotubes by SILAC and quantitative MS. Three biological replicates were performed including label-swaps. H, Changes in the phosphorylation of mouse FLNc S2625 in C2C12 myotubes treated with PMA or Gö6976 in comparison to control. Top: SILAC triplet of the mouse FLNc phosphopeptide 2623GApSYSSIPK2631. Bottom: Relative quantification of the mouse FLNc phosphopeptide 2623GApSYSSIPK2631 using Skyline. Shown is the mean log2 ratio of the area under the curve ± S.E. **p ≤ 0.006, n = 3.
To obtain residue-resolved information about PKCα-mediated phosphorylation of FLNc, we performed in vitro kinase assays in conjunction with quantitative MS. Experiments were performed in triplicates using equal protein amounts (supplemental Fig. S4B) followed by phosphopeptide analysis using complementary fragmentation methods and MS1-based quantification using Skyline (37) (supplemental Table S8). This comprehensive analysis unequivocally established PKCα-mediated phosphorylation of mFLNc at S2625 (Fig. 4C, left panel and Fig. 4E). In addition, we found that PKCα also phosphorylated S2603 and S2637 in vitro, although to a lesser extent (Fig. 4C, left panel). The latter two phosphosites have not been identified in vivo, suggesting that they occurred nonspecifically under in vitro conditions. Interestingly, phosphorylation was partially “swapped” to S2620 when S2625 was mutated to alanine or aspartate in mFLNc (Fig. 4C, middle and right panel). For hFLNc d23–24 WT and the respective AA and DD mutants, we observed largely identical PKCα-dependent phosphorylation patterns (compare Fig. 4D and 4C). Here, S2623 served as main substrate site of PKCα, but phosphorylation of both S2623 and S2624 was observed to a minor extent as well (Fig. 4D, left panel and Fig. 4F). In summary, we revealed S2625 and S2623/S2624 localized in the hinge 2 region of mouse and human FLNc, respectively, as specific substrate sites of PKCα in vitro.
FLNc S2625 is an In Vivo Substrate of PKCα in Mouse Skeletal Muscle Cells
To demonstrate that mFLNc S2625 is a substrate of PKC in vivo, we determined the relative abundance of the FLNc phosphopeptide 2623GApSYSSIPK2631 in C2C12 myotubes treated with PMA, an activator for the conventional PKC isoforms α,β,γ, or Gö6976, an inhibitor for PKCα and PKCβ1 (59) in comparison to DMSO-treated cells (control). To this end, we performed SILAC labeling of C2C12 cells and specifically analyzed the mFLNc phosphopeptide 2623GApSYSSIPK2631 by quantitative MS following TiO2 enrichment (Fig. 4G). The experiment was performed in three biological replicates including a label swap to determine significant changes in S2625 phosphorylation. As a result, we found phosphorylation of mFLNc S2625 to be significantly decreased upon treatment of C2C12 myotubes with Gö6976 (Fig. 4H, supplemental Fig. S4C). Because PKCα is the conventional PKC isoform predominantly expressed (97%) in mouse skeletal muscle cells (60), these data strongly indicate that FLNc S2625 is phosphorylated by PKCα in vivo. However, we cannot exclude that PKCβ1 may further contribute to mFLNc S2625 phosphorylation in vivo. Our data also indicate that PKCα is active in contracting myotubes as we only observed a slight increase in FLNc S2625 phosphorylation in PMA-treated cells (Fig. 4H, supplemental Fig. S4C). In summary, SILAC-MS analysis shows that FLNc is a target of PKCα at S2625 in mouse skeletal muscle cells.
Tyrosine Carboxyterminally to the PKCα Substrate Site Is the Main Calpain 1 Cleavage Site
The carboxyterminal part of FLNc was reported to be a substrate of the proteases calpain 3 and calpain 1 (also referred to as μ-calpain) active in skeletal muscle fibers (61, 62, 29). In silico data indicated that calpain 1 preferentially cleaves hFLNc carboxyterminally to Y2625 in addition to S2626 as suggested previously (29) and, correspondingly, mFLNc at Y2626 and S2627 (supplemental Table S9). Interestingly, mutation of the neighboring PKCα substrate sites S2623/S2624 (hFLNc) and S2625 (mFLNc) to alanine or aspartate decreased the cleavage probability at several sites. For the phosphomimetic mutants, this effect was even more pronounced and also led to a clear decrease in the number of predicted calpain 1 cleavage sites.
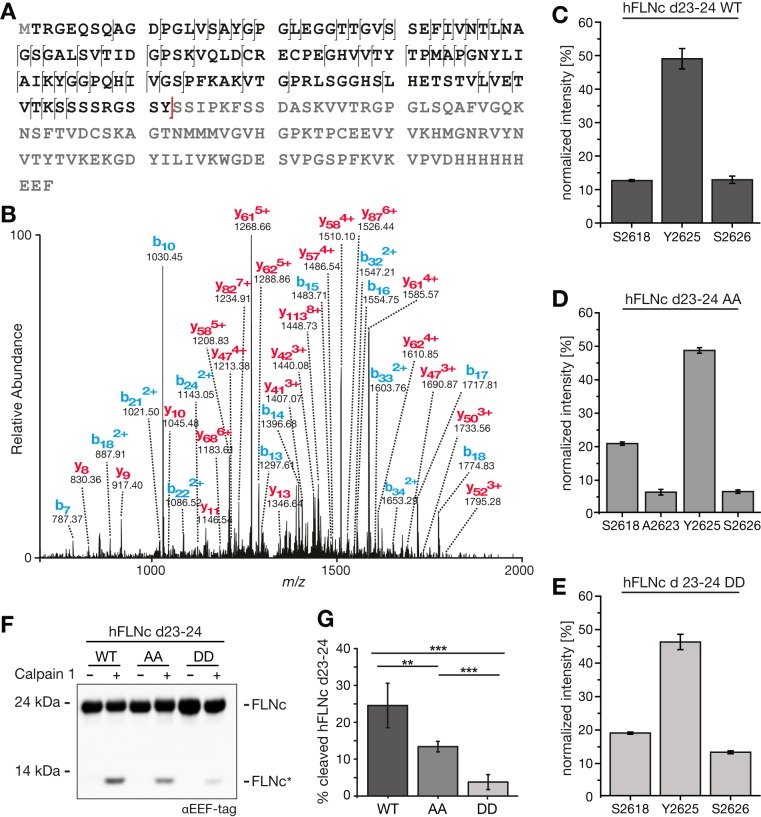
To unequivocally identify which site is the main cleavage site in FLNc, we established a novel site-resolving MS-based in vitro calpainolysis assay. To this end, carboxyterminally His6- and EEF-tagged d23–24 constructs of human and mouse FLNc WT and the respective phosphosite mutants (supplemental Fig. S3B) were recombinantly expressed, immobilized on Ni2+-NTA beads and incubated with recombinant calpain 1. Resulting aminoterminal cleavage products were characterized as intact protein fragments by high resolution LC-MS following a top-down approach. For hFLNc d23–24 WT, we determined Y2625 as the main calpain 1 cleavage site, whereas cleavage at S2618 and S2626 was only minor (Fig. 5A–5C, supplemental Fig. S5A, supplemental Table S10). The AA and DD site mutants showed virtually identical cleavage patterns with an additional cleavage site only occurring in the nonphosphorylatable AA variant (Fig. 5D and 5E, supplemental Fig. S5B and S5C, supplemental Table S10). Consistent with these findings, top-down MS analysis of calpain 1-dependent aminoterminal cleavage products of mFLNc revealed Y2626 as main cleavage site supplemental Fig. S6A–S6G, supplemental Table S10). For the S2625D mutant, we observed S2627 as an additional prominent cleavage site. However, this altered specificity is most likely the consequence of a considerably reduced efficiency of calpain 1 for cleaving this variant as observed by immunoblot analysis using an antibody against the His6-tag to detect both intact (∼24 kDa) and cleaved forms (∼13 kDa) supplemental Fig. S6H). Thus, a lower cleavage specificity may be indicative of an effective site-specific protection against proteolysis.
Fig. 5.
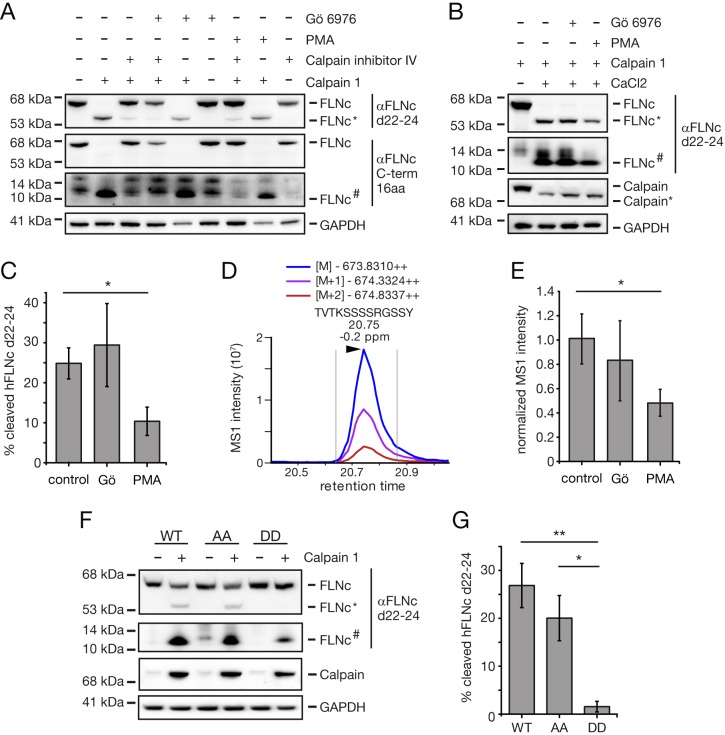
Y2625 is the major calpain 1-dependent cleavage site in the hinge 2 region of hFLNc. A, B, Top-down MS analysis. Recombinant hFLNc d23–24 fused carboxyterminally to a His6- and EEF-tag was treated with recombinant human calpain 1 and aminoterminal cleavage products were subjected to LC-MS/MS using low-energy collision-induced dissociation (CID) as fragmentation method. A, Amino acid sequence of hFLNc d23–24-His6-EEF. B, Top down CID spectrum of the main cleavage product observed at m/z 13,355.66; sequence and fragmentation pattern are depicted in black in (A) with the calpain 1-dependent cleavage site in red. C–E, Determination of the main calpain 1 cleavage site. In vitro calpain 1 assays and top-down analyses were performed as in (A, B). Data from three independent experiments for hFLNc d23–24 WT (C) and S2623/S2624 phosphosite mutants (AA mutant, (D); DD mutant, (E)) were quantified and represented as normalized mean ± S.E. Calpain 1-dependent cleavage sites are denoted. F, Samples were analyzed by immunoblotting. FLNc*, carboxyterminal cleavage product. G, Quantification of data shown in (F) for n = 3 experiments. Error bars represent the S.D., a two-tailed Student's t test was used. **p ≤ 0.0034, ***p ≤ 0.0003.
In both human and mouse FLNc, the main calpain 1 cleavage site (hFLNc-Y2625 and mFLNc-Y2626) was found in direct vicinity of the PKCα substrate site(s) identified in this work. Because the introduction of negative charges at these sites might affect the susceptibility of the respective substrate to calpain 1 cleavage, we studied the efficiency of calpain 1-dependent cleavage of all hFLNC variants by SDS-PAGE and immunoblotting (Fig. 5F, supplemental Fig. S3B). We quantified immunoblot signals from three independent in vitro calpainolysis assays to calculate the percentage of cleaved hFLNc (Fig. 5G). Approximately 25% of hFLNc d23–24 WT was cleaved by calpain 1, whereas the phosphomimetic DD mutant was almost completely protected from proteolysis, and the AA mutant appeared only slightly less susceptible to degradation. These data suggest that exchange of two hydrophilic by two negatively charged/phosphorylated residues causes a conformational change around the cleavage site, rendering the substrate less prone to calpain 1 binding. Indeed, the rate of calpain cleavage directly depends on polypeptide conformation rather than the linear sequence of amino acids surrounding a cleavage site (63).
Taken together, we showed that Y2625 and Y2626 are the major calpain 1 cleavage sites in human and mouse FLNc d23–24, respectively, whereas the introduction of negative charges at the aminoterminal serine residues (i.e. S2623/S2624 or S2625) significantly reduces its susceptibility to cleavage.
PKCα-mediated Phosphorylation of S2623/S2624 Protects Human FLNc from Limited Proteolysis by Calpain 1
To address the question whether PKCα-mediated phosphorylation of hFLNc-S2623/S2624 is the determining factor for protection against calpain 1 cleavage under cellular conditions, we transiently expressed aminoterminally tagged hFLNc d22–24 in HEK293 or C2 mouse skeletal muscle cells supplemental Fig. S3C) and treated the cells either with PMA to stimulate classical PKC activity, or with Gö6976 to inhibit PKCα. Subsequently, cell lysates were treated with recombinant calpain 1 in the absence or presence of calpain inhibitor IV and intact hFLNc d22–24 and cleavage products thereof were detected by immunoblotting using antibodies directed against either d22–24 or the last carboxyterminal 16 amino acids of hFLNC. As expected, we observed efficient proteolysis of hFLNc d22–24 in both HEK293 and C2 cells following treatment of lysates with calpain 1 and this effect was abolished with calpain inhibitor IV (Fig. 6A, lanes 1–3; supplemental Fig. S7, lanes 1–3). Pretreatment of lysates with the PKCα inhibitor Gö6976 slightly increased proteolysis, whereas incubation with the PKC stimulant PMA reduced the levels of cleavage products (Fig. 6A, lane 5 and 8; supplemental Fig. S7, lane 5 and 8). Consistently, cleavage of hFLNc d22–24 by activated calpain 1 was more than 2-fold reduced following activation of PKC by PMA (Fig. 6B and 6C). Inhibition of PKCα by Gö6976 resulted in no significant increase in the amount of cleavage products. The latter finding can be explained by assuming that only a rather small fraction of overexpressed hFLNc d22–24 is phosphorylated by endogenous PKCα in HEK293 cells, underscoring the strong dependence of FLNc's susceptibility to calpain 1 on its phosphorylation status. We further confirmed these findings as well as the calpain 1-specific cleavage site Y2625 in hFLNc by a targeted MS-based approach. To this end, we identified and quantitatively monitored by targeted SIM the doubly protonated peptide 2613TVTKSSSSRGSSY2625 generated by calpain 1 cleavage of hFLNc, followed by proteolytic digestion of the N-terminal hFLNc cleavage product with GluC (Fig. 6D). Consistent with our quantitative Western blot data shown in 6C, quantification by targeted SIM revealed a significant decrease in calpain 1 cleavage of hFLNc following treatment of cells with PMA (Fig. 6E). Moreover, comparative quantitative analysis of calpain 1-dependent cleavage products of hFLNc d22–24 WT and S2623/S2624 site mutants (DD, AA; supplemental Fig. S3C) expressed in HEK293 cells demonstrated that the phosphomimetic DD variant was almost completely protected from proteolysis (Fig. 6F and 6G). In line with the effect observed for the recombinant hFLNc d23–24 AA variant (Fig. 5F and 5G), mutation of S2623/S2624 to alanine resulted in a lower percentage of cleaved hFLNc d22–24 in HEK293 cells (Fig. 6F and 6G).
Fig. 6.
PKC protects hFLNc from calpain 1-dependent proteolysis through phosphorylation of S2623/S2624. A, HEK293 cells transiently expressing hFLNc d22–24 were treated with Gö6976 or PMA as indicated. Subsequently, cell lysates supplemented with 5 mm CaCl2 were incubated with human calpain 1 and the calpain inhibitor IV as indicated. Samples were analyzed by immunoblotting. FLNc*, aminoterminal cleavage product; FLNc#, carboxyterminal cleavage product. B, HEK293 cells expressing hFLNc d22–24 were treated with Gö6976 or PMA as indicated. Calpain 1 was added to cell lysates and activated with CaCl2 or not as indicated. Samples were analyzed by immunoblotting. FLNc*, aminoterminal cleavage product; FLNc#, carboxyterminal cleavage product. C, Quantification of data shown in (B). Percentage of cleaved hFLNc d22–24 was calculated for n = 3 independent experiments. Shown is the mean ± S.E. *p = 0.0442. D, Extracted ion chromatogram of the peptide 2613TVTKSSSSRGSSY2625 derived from calpain-cleaved hFLNc d22–24.For targeted MS analysis using SIM scans, the N-terminal hFLNc d22–24 cleavage product was further digested using GluC. E, Relative quantification of the calpain 1 cleavage-specific hFLNc d22–24 peptide shown in (D). Quantification is based on normalized MS1 signal intensities observed in calpain 1 cleavage experiments using untreated (control), Gö6976- or PMA-treated HEK293 cells expressing hFLNc d22–24. Shown are the mean ± S.E. (n = 4). *p = 0.0345. F, HEK293 cells were transfected with constructs for hFLNc d22–24 WT and respective S2623/S2624 phosphosite mutants (AA, DD mutant). Calpain 1 was added to cell lysates (containing 5 mm CaCl2) as indicated and samples were analyzed by immunoblotting. FLNc*, aminoterminal cleavage product; FLNc#, carboxyterminal cleavage product. G, Quantification of data shown in (F). Percentage of cleaved hFLNc d22–24 was calculated for n = 3 independent experiments. Shown is the mean ± S.E. *p = 0.0245, **p ≤ 0.0078.
PKC Modulates the Dynamic Behavior of Human FLNc Through Phosphorylation of S2623/S2624 in Skeletal Myotubes
FLNc fulfils important roles in the assembly, repair and maintenance of myofibrils (17, 64, 32), functions that suggest that FLNc must be highly dynamic and mobile under distinct physiological conditions. We therefore continued to investigate whether PKC-mediated phosphorylation and calpain 1-dependent proteolysis of hFLNc have a physiological effect on its dynamic behavior. To study the mobility and dynamics of hFLNc in living cells, we employed fluorescence recovery after photobleaching (FRAP) using immortalized mouse myoblasts expressing hFLNc WT fused to EGFP or mutant forms in which S2623/S2624 were changed to alanine (AA) or aspartate (DD) or the Ig-like domain 24 (amino acids 2628–2725) was deleted supplemental Fig. S3D). Following photobleaching of defined regions of myotubes expressing EGFP-fusion proteins in a cross-striated pattern (Fig. 7A), the recovery profiles of hFLNC WT (in cells treated with or without the PKC inhibitor Gö6976) and mutant forms were recorded. hFLNc-EGFP recovery profiles were biphasic with a lateral diffusion-dependent initial, fast recovery as determined by diffusion testing supplemental Fig. S8). The subsequent slower phase of the recovery profile depends on the exchange process of bound protein with the soluble fraction and, thus, allows for the calculation of protein halftimes as shown in Fig. 7B. Mean halftimes were significantly increased for the AA mutant (54 s) and, to an even higher extent, following inhibition of PKC activity (93 s) in myocytes expressing hFLNc WT. In stark contrast, the DD and ΔIg-like d24 mutants of hFLNc exhibited significantly shorter mean halftimes (11 and 10 s) compared with the WT form (25 s). Remarkably, we did not observe changes in the mobile fraction of hFLNc when cells were treated with Gö6976 to inhibit PKC or double site mutants were expressed (Fig. 7C). The only exception was the ΔIg-like d24 mutant of hFLNc showing an increase in the mobile fraction (92%) compared with FLNc WT (78%), which points to a lower affinity of this truncated FLNc form to Z-disc structures. In summary, our data suggest that PKC precisely controls the dynamics of hFLNc in skeletal myotubes through phosphorylation of S2623/S2624. Removal of Ig-like domain 24 mimicking cleavage by calpain 1 led to a considerably shorter halftime along with an increase in mobility, possibly because of the inability of the truncated hFLNc form to dimerize.
Fig. 7.
Dynamics and mobility of hFLNc is controlled by phosphorylation and cleavage of its Ig-like domain 24. A, Immortalized mouse myoblasts (IMMs) were transfected with constructs for EGFP-tagged full-length hFLNc WT, S2623/2624 phosphosite mutants (AA, DD), or hFLNc lacking Ig-like domain 24. IMMs expressing full-length hFLNc WT were treated with PKC inhibitor Gö6976 for 1 h or not before fluorescence recovery after photobleaching (FRAP) analysis. Shown is a representative FRAP experiment with IMMs after 5 days of differentiation before bleaching (prebleach), immediately (postbleach), 5 and 20 s after bleaching, and after full recovery (recovery). Boxes indicate the region bleached. Scale bar: 5 μm. B, Quantification of data from FRAP studies described in (A). Statistical data are depicted in box and whisker plots. Each calculated median halftime is shown as a line surrounded by a box, which represents the interquartile range comprising the median ± 25% of the data. Whiskers extend at most two standard deviations from the median. Data from n ≥ 10 independent experiments are shown. **p ≤ 0.0011, ***p ≤ 0.0003. C, Percentage of mobile fractions calculated from FRAP studies described in (A). Shown is the mean ± S.D. n ≥ 10, *p = 0.0233.
DISCUSSION
The formation of mature Z-discs during sarcomere assembly is an important step in myogenesis which requires and is induced by contractile activity (65, 66). In this work, we differentiated C2C12 myoblasts to myotubes and further subjected these multinucleated cells to EPS to generate contracting myotubes with mature Z-discs. Although previous studies focused on proteomic changes during myoblast differentiation (30, 53, 54), we here performed for the first time a quantitative proteomic analysis of EPS-treated contracting C2C12 myotubes in comparison to proliferating myoblasts and differentiated “nonexercising” myotubes (Fig. 1A–1B, supplemental Table S1). Consistent with earlier work (30, 67, 68), proteins associated with sarcomere assembly and mitochondrial functions were significantly increased in abundance as a consequence of skeletal muscle cell differentiation induced by serum reduction. Interestingly, EPS of differentiated myotubes to induce contraction had no effect on the abundance of sarcomeric proteins, indicating that all components needed for Z-disc maturation and full assembly of the contractile apparatus are already expressed in “nonexercising” myotubes (Fig. 1B and 1C, cluster A). EPS-treated contracting myotubes exhibited a higher abundance of proteins involved in ATP synthesis in addition to a slight increase in a set of proteins associated with transcriptional and translational processes (Fig. 1B, C5 and C10, supplemental Table S2), highlighting the ability of EPS to alter function and activity of cultured skeletal myotubes. By global phosphoproteomic analysis, we further present a detailed site-resolved protein phosphorylation map of EPS-treated contracting C2C12 myotubes with 8175 localized sites in 2941 proteins (Fig. 2A, supplemental Table S4 and S5). Evaluation of our large phosphoproteome dataset revealed the Z-disc as major protein phosphorylation hot spot in myofibrils, whereas only 9% of the identified sarcomeric phosphosites were located in A-band proteins (Fig. 2C, 2D, and 2G). The vast majority of genuine Z-disc and Z-disc-associated proteins (44, 70%) were modified by a single or even multiple phosphate groups, establishing phosphorylation of Z-disc proteins as a central theme for intracellular signal transduction processes in myofibrils (Fig. 3A). Remarkably, we found clustered multisite phosphorylations in numerous Z-disc-associated proteins, including FLNc, BAG3, ENAH, LDB3, and the three podin protein family members SYNPO, SYNPO2, and SYNPO2L (Fig. 3B). These findings hint at distinct regulatory regions that may control their protein interactions, dynamics, localization and/or other functions. Our data strengthen the emerging view of the Z-disc as a way station and molecular scaffold for protein kinases and phosphatases (69, 2, 53).
GO enrichment analysis revealed PKC signaling as a significant feature in the phosphoproteome of skeletal myotubes (Fig. 2G, supplemental Table S6). Several proteins were reported to anchor PKC at the Z-disc following activation, highlighting the importance of PKC signaling in the regulation of highly dynamic Z-disc protein assemblies essential for skeletal and cardiac muscle function and growth (53, 70, 71). For example, the actin-capping protein CapZ anchors PKCβ to myofilaments in cardiomyocytes (72). Under hypertrophic conditions, PKC signaling decreases the binding affinity of CapZ to actin filaments, thereby destabilizing the actin filaments and allowing sarcomere remodeling (73). Further PKC anchoring proteins at the Z-disc are LDB3, also called cypher or ZASP (74, 75, 76), and enigma homologue protein ENH (PDLIM5) (77), which together with calsarcin-1 (MYOZ2) form a complex with an important role in linking Z-discs to the extracellular matrix via the multi-adaptor protein FLNc (78). All these proteins were characterized as phosphoproteins in striated muscle cells in this work (Fig. 3A, supplemental Table S4). A detailed analysis of FLNc revealed phosphorylation at six distinct serine residues (Fig. 3B, supplemental Table S5). Two of these phosphosites are located in the unique 82 amino acid insert within Ig-like domain 20, which facilitates targeting of FLNc to the Z-disc (9). Earlier work identified FLNc as a bona fide substrate of PKB/Akt at S2233 in rabbit skeletal muscle (79). Here we consistently identified phosphorylation of murine FLNc at S2234 (corresponding to S2233 in human FLNc) (79) as well as concurrent phosphorylation of S2234 and S2237 in mouse myotubes. Interestingly, binding of aciculin (also called PGM5; a dystrophin-binding protein involved in myofibril development, maintenance and repair) to FLNc requires the insertion in Ig-like domain 20 (32). Likewise, only FLNc containing this insertion interacts with Xin (XIRP1) and XIRP2 (12). Other proteins reported to specifically bind this FLNc region include myopodin (7) and myotilin (9). With the exception of XIRP2, we identified all these FLNc interactors also as strongly phosphorylated proteins (Fig. 3A and 3B). In this context, it is interesting to note that FLNc, aciculin, and Xin are proteins whose phosphorylation increased within a few minutes after inducing pressure overload in mouse hearts (80), suggesting their close interplay in signaling pathways underlying the adaptation of striated muscle to mechanically induced stress. Thus, phosphorylation of FLNc at S2234/S2237 may provide a mechanism for controlling such dynamic and competing protein interactions depending on the physiological condition.
In addition, we identified a cluster of three phosphosites in the carboxyterminus of FLNc in hinge 2 and at the beginning of repeat 24 (Fig. 3B and supplemental Fig. S2). Similar to Ig-like domain 20, this region of FLNc is involved in multiple protein interactions, including dimerization (domain 24; (22, 23)), binding to calsarcin-2 (domain 23; (15)) and δ- and γ-sarcoglycans (domain 24; (17)). The interaction with both sarcoglycans is abolished by calpain 3-dependent cleavage of FLNc at its hinge 2 region (61). Moreover, phosphorylated FLNc was found in consequence of a phospholipid-dependent interaction of PKCα via its carboxyterminal end (28). However, PKCα-dependent phosphorylation sites in FLNc and their physiological relevance have remained uncharacterized so far.
Our in vitro kinase assays revealed S2625 and S2623/S2624, located in the carboxyterminus of mouse and human FLNc, respectively, as specific PKCα sites (Fig. 4, supplemental Table S8). In contrast, phosphorylation events of mFLNC at S2621 and S2633 were not mediated by PKCα, implying involvement of additional kinases. Selective PKCα signaling to the carboxyterminal Ig-like domain 24 of FLNa and FLNc, but not FLNb was proposed previously (28). Here we unequivocally show that PKCα targets the hinge 2 region of FLNc in vitro. This serine-rich sequence stretch (2621SRGASYSSIPKFSS2633)is uniquely present in FLNc, implying that the PKCα site in FLNa differs. Previous work reported three phosphosites (p2523, pS2526, and pS2599) in hinge 2 and Ig-like domain 24 of FLNa in nonmuscle mouse tissues (81). We here identified four FLNa phosphosites (pS968, pS1084, pS1459, pS2327), all not residing within Ig-like domains 23 to 24 supplemental Table S5), indicating that only the carboxyterminus of FLNc is specifically targeted by PKCα in skeletal muscle cells under the conditions tested.
What may the functional impact of PKCα phosphorylation of FLNc's hinge 2 region be? The carboxyterminus of FLNc was identified as substrate of calpain 1 (29), and FLNc and calpain 1 colocalize at the edges of the Z-disc ((18); (29)). FLNa phosphorylated by PKA was more resistant to calpain 1 cleavage, thereby inhibiting cytoskeletal reorganization (82). A similar PKCα-dependent mechanism was proposed for protecting FLNc from calpain 1 cleavage (29). Interestingly, active PKCα itself is further cleaved to a constitutively active form by calpain, which might increase FLNc phosphorylation and its resistance to calpain cleavage (83, 63). Chicken gizzard filamin is rapidly degraded to 240- and 10-kDa fragments by calpain, thereby severing its cross-linking ability, however the exact cleavage site remained unidentified (84). The detection of calpain cleavage sites is generally hampered by the lack of known consensus sequences, rendering predictions unreliable (85). To overcome this issue, we combined here for the first time in vitro proteolysis assays with top-down MS to accurately determine calpain 1 cleavage sites in FLNc. Our in vitro data show that the main cleavage site is a tyrosine residue located directly carboxyterminally to the PKCα substrate site(s) in the hinge 2 region of FLNc (Fig. 5A–5C, supplemental Table S10). This cleavage specificity is retained for the respective phosphosite mutants (Fig. 5D and 5E, supplemental Table S10), but its efficiency was significantly decreased in the phosphomimetic form (Fig. 5F and 5G, supplemental Table S10). Our experimental data generally support earlier findings suggesting that calpain preferentially cleaves in disordered segments between structured domains between a tyrosine (P1) and a carboxyterminal serine (P1') with proline in position P2'- P4' or P3 (86).
Calpains were suggested to play a role in integrin signaling (87) by cleaving specifically β-integrin family members at a conserved NPXY/NXXY motif in the carboxyterminal half of the cytoplasmic domains involved in talin and filamin binding (88). Involvement of calpains in signaling has been emphasized by in vitro studies revealing kinases (e.g. focal adhesion kinase, FAK), phosphatases (e.g. protein tyrosine phosphatase 1B, PTP1B) and cytoskeletal proteins (e.g. paxillin, vinculin) as substrates (63). Nonetheless, virtually no information on how calpain cleavage modulates distinct signaling events in vivo is available to date. In this work, we showed that stimulation of PKCα activity by PMA significantly attenuates the susceptibility of FLNc to cleavage by calpain 1 (Fig. 6A–6E, supplemental Fig. S7). The effect was even more pronounced for a FLNc variant in which the serine residues identified as substrate sites of PKCα (S2623/2624) were mutated to aspartic acid, thus mimicking human FLNc in its constitutively phosphorylated form (Fig. 6F and 6G). We conclude that phosphorylation of these serines is the mechanism by which FLNc is protected from limited proteolysis by calpain 1 in its hinge 2 region.
Finally, we used FRAP experiments to study the effect of phosphorylation and calpain 1 cleavage on FLNc dynamics in living skeletal muscle cells. Most importantly, an FLNc variant mimicking calpain 1-cleaved FLNc showed a highly significant reduction of the mean halftime and an increase of the mobile fraction (Fig. 7B and 7C). This indicates that FLNc without its dimerization domain, but including all its known Z-disc targeting domains, is still targeted to Z-discs. The lack of Ig-like domain 24 apparently reduces the stability of the interactions with its Z-disc ligands, rendering the protein less stable (89).
Our FRAP data also revealed a highly significant decrease in the mean halftime for the phosphomimetic (S2623D/S2624D) variant of FLNc. Consistently, the effect was reciprocal for the double site mutant mimicking FLNc in its dephosphorylated form (S2623A/S2624A), whereas inhibition of PKCα even further increased the mean halftime (Fig. 7B). The latter observation may indicate that α phosphorylates FLNc at an additional site, potentially beyond Ig-like domains 23–24, which further affects its dynamics. A strong candidate is S2237, located in the insertion of Ig-like domain 20, which is involved in targeting FLNc to Z- discs (9). These data, in accordance with the observation that binding of phosphorylated filamin to isolated myofibrils is significantly lower (90) convincingly show that phosphorylation of FLNc is important for increasing its mobility, a prerequisite for initiating signaling processes.
In summary, our extensive analysis of the phosphoproteome of the sarcomeric Z-disc identified this structure as a myofibrillar phosphorylation hotspot. Our data confirm recent suggestions that the Z-disc is highly involved in signaling processes. Our finding that calpain 1 cleavage of FLNc is regulated by phosphorylation of one (mouse) or two (human) serine residues directly flanking the cleavage site is a paradigmatic example of a phosphorylation-dependent mechanism identified by a follow-up study based on these data. We expect that the vast amount of phosphorylation sites uncovered in this work will lead to identification of further events regulated by protein phosphorylation.
Supplementary Material
Acknowledgments
We thank Andreas Schummer, Caroline Gerbeth and Kurt Lobenwein for technical assistance and Zacharias Orfanos and Chris Meisinger for discussions. Deposition of the data to the ProteomeXchange Consortium was supported by the PRIDE Team, EBI, and publication to the Panorama public server was supported by the Skyline/Panorama Team.
Footnotes
Author contributions: L.R. performed phosphoproteomics, mass spectrometric and biochemical analyses with the support of H.W., B.K., A.F., and A.S. FRAP measurements were performed by Y.L. and A.R. All authors analyzed data, designed and interpreted experiments. B.W. and D.F. supervised the study. B.W. conceived the project and wrote the manuscript with the input of other authors.
* This research was supported by the Deutsche Forschungsgemeinschaft (FOR 1352 to B.W. and D.O.F., and FOR 1228 to D.O.F.) and the Excellence Initiative of the German Federal & State Governments (EXC 294 BIOSS to BW).
 This article contains supplemental material.
This article contains supplemental material.
The authors declare that they have no conflict of interest.
1 The abbreviations used are:
- PKC
- protein kinase C
- ABD
- actin-binding domain
- AGC
- automatic gain control
- BAG
- Bcl2-associated athanogene
- BP
- biological process
- CC
- cellular component
- D
- domain
- ENAH
- enabled homolog
- EPS
- electrical pulse stimulation
- ETD
- electron transfer dissociation
- FA
- formic acid
- FDR
- false discovery rate
- FLNc
- filamin C
- FRAP
- fluorescence recovery after photobleaching
- GO
- gene ontology
- HCD
- higher-energy collisional dissociation
- HEK293
- human embryonic kidney 293
- hpH-RP
- high pH reversed phase chromatography
- HRP
- horseradish peroxidase
- HSPB1
- heat shock protein beta-1
- Ig
- immunoglobulin
- IMMs
- immortalized mouse skeletal myoblasts
- IPTG
- isopropyl β-d-1-thiogalactopyranoside
- LDB3
- LIM domain binding 3
- LIT
- linear ion trap
- MF
- molecular function
- MSA
- multi-stage activation
- NL
- neutral loss
- PDZ
- post synaptic density protein, Drosophila disc large tumor suppressor, and; zonula occludens-1 protein
- PEI
- polyethylenimine
- PMA
- phorbol-12-myristat-13-acetat
- PPxY
- proline-proline-x-tyrosine
- PxxP
- proline-rich
- ROI
- regions of interest
- SCX
- strong cation exchange
- SD
- standard deviation
- SEM
- standard error of the mean
- SIM
- single ion monitoring
- SYNPO
- synaptopodin
- SYNPO2
- myopodin
- SYNPO2L
- tritopodin (CHAP; synaptopodin 2-like)
- TiO2
- titanium dioxide
- VASP
- vasodilator-stimulated phosphoprotein
- WH1
- WASP-homology/EVH1, Ena/VASP homology 1
- WT
- wildtype
- XIRP1
- Xin actin-binding repeat containing protein 1.
REFERENCES
- 1. Luther P. K. (2009) The vertebrate muscle Z-disc: sarcomere anchor for structure and signalling. J. Muscle Res. Cell Motil. 30, 171–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Frank D., and Frey N. (2011) Cardiac Z-disc Signaling Network. J. Biol. Chem. 286, 9897–9904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Knöll R., Buyandelger B., and Lab M. (2011) The sarcomeric Z-disc and Z-discopathies. J. Biomed. Biotechnol. 2011, 569–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gu X., and Bishop S. P. (1994) Increased protein kinase C and isozyme redistribution in pressure-overload cardiac hypertrophy in the rat. Circ. Res. 75, 926–931 [DOI] [PubMed] [Google Scholar]
- 5. Heineke J., Ruetten H., Willenbockel C., Gross S. C., Naguib M., Schaefer A., Kempf T., Hilfiker-Kleiner D., Caroni P., Kraft T., Kaiser R. A., Molkentin J. D., Drexler H., and Wollert K. C. (2005) Attenuation of cardiac remodeling after myocardial infarction by muscle LIM protein-calcineurin signaling at the sarcomeric Z-disc. Proc. Natl. Acad. Sci. U.S.A. 102, 1655–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Faul C., Dhume A., Schecter A. D., and Mundel P. (2007) Protein kinase A, Ca2+/calmodulin-dependent kinase II, and calcineurin regulate the intracellular trafficking of myopodin between the Z-disc and the nucleus of cardiac myocytes. Mol. Cell. Biol. 27, 8215–8227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Linnemann A., van der Ven P. F., Vakeel P., Albinus B., Simonis D., Bendas G., Schenk J. A., Micheel B., Kley R. A., and Fürst D. O. (2010) The sarcomeric Z-disc component myopodin is a multiadapter protein that interacts with filamin and alpha-actinin. Eur. J. Cell Biol. 89, 681–692 [DOI] [PubMed] [Google Scholar]
- 8. Linnemann A., Vakeel P., Bezerra E., Orfanos Z., Djinović-Carugo K., van der Ven P. F. M., Kirfel G., and Fürst D. O. (2013) Myopodin is an F-actin bundling protein with multiple independent actin-binding regions. J. Muscle Res. Cell Motil. 34, 61–69 [DOI] [PubMed] [Google Scholar]
- 9. van der Ven P. F., Wiesner S., Salmikangas P., Auerbach D., Himmel M., Kempa S., Hayess K., Pacholsky D., Taivainen A., Schröder R., Carpén O., and Fürst D. O. (2000) Indications for a novel muscular dystrophy pathway. gamma-filamin, the muscle-specific filamin isoform, interacts with myotilin. J. Cell Biol. 151, 235–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eulitz S., Sauer F., Pelissier M. C., Boisguerin P., Molt S., Schuld J., Orfanos Z., Kley R. A., Volkmer R., Wilmanns M., Kirfel G., van der Ven P. F., and Fürst D. O. (2013) Identification of Xin-repeat proteins as novel ligands of the SH3 domains of nebulin and nebulette and analysis of their interaction during myofibril formation and remodeling. Mol. Biol. Cell 24, 3215–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van der Ven P. F., Ehler E., Vakeel P., Eulitz S., Schenk J. A., Milting H., Micheel B., and Fürst D. O. (2006) Unusual splicing events result in distinct Xin isoforms that associate differentially with filamin c and Mena/VASP. Exp. Cell Res. 312, 2154–2167 [DOI] [PubMed] [Google Scholar]
- 12. Kley R. A., Maerkens A., Leber Y., Theis V., Schreiner A., van der Ven P. F., Uszkoreit J., Stephan C., Eulitz S., Euler N., Kirschner J., Müller K., Meyer H. E., Tegenthoff M., Fürst D. O., Vorgerd M., Müller T., and Marcus K. (2013) A combined laser microdissection and mass spectrometry approach reveals new disease relevant proteins accumulating in aggregates of filaminopathy patients. Mol. Cell. Proteomics 12, 215–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Faulkner G., Pallavicini A., Comelli A., Salamon M., Bortoletto G., Ievolella C., Trevisan S., Kojic S., Dalla Vecchia F., Laveder P., Valle G., and Lanfranchi G. (2000) FATZ, a filamin-, actinin-, and telethonin-binding protein of the Z-disc of skeletal muscle. J. Biol. Chem. 275, 41234–41242 [DOI] [PubMed] [Google Scholar]
- 14. Frey N., and Olson E. N. (2002) Calsarcin-3, a novel skeletal muscle-specific member of the calsarcin family, interacts with multiple Z-disc proteins. J. Biol. Chem. 277, 13998–14004 [DOI] [PubMed] [Google Scholar]
- 15. Takada F., Vander Woude D. L., Tong H. Q., Thompson T. G., Watkins S. C., Kunkel L. M, and Beggs A. H. (2001) Myozenin: An alpha -actinin- and gamma -filamin-binding protein of skeletal muscle Z lines. Proc. Natl. Acad. Sci. U.S.A. 98, 1595–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maestrini E., Patrosso C., Mancini M., Rivella S., Rocchi M., Repetto M., Villa A., Frattini A., Zoppè M., and Vezzoni P. (1993) Mapping of two genes encoding isoforms of the actin binding protein ABP-280, a dystrophin like protein, to Xq28 and to chromosome 7. Hum. Mol. Genet. 2, 761–766 [DOI] [PubMed] [Google Scholar]
- 17. Thompson T. G., Chan Y. M., Hack A. A., Brosius M., Rajala M., Lidov H. G., McNally E. M., Watkins S., and Kunkel L. M. (2000) Filamin 2 (FLN2): A muscle-specific sarcoglycan interacting protein. J. Cell Biol. 148, 115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van der Ven P. F., Obermann W. M., Lemke B., Gautel M., Weber K., and Fürst D. O. (2000) Characterization of muscle filamin isoforms suggests a possible role of gamma-filamin/ABP-L in sarcomeric Z-disc formation. Cell Motil. Cytoskelet. 45, 149–162 [DOI] [PubMed] [Google Scholar]
- 19. Fürst D. O., Goldfarb L. G., Kley R. A., Vorgerd M., Olivé M., and van der Ven P. F. (2013) Filamin C-related myopathies: pathology and mechanisms. Acta Neuropathol. 125, 33–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xie Z., Xu W., Davie E. W., and Chung D. W. (1998) Molecular cloning of human ABPL, an actin-binding protein homologue. Biochem. Biophys. Res. Commun. 251, 914–919 [DOI] [PubMed] [Google Scholar]
- 21. van der Flier A., Kuikman I., Kramer D., Geerts D., Kreft M., Takafuta T., Shapiro S. S., and Sonnenberg A. (2002) Different splice variants of filamin-B affect myogenesis, subcellular distribution, and determine binding to integrin [beta] subunits. J. Cell Biol. 156, 361–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Himmel M., Van Der Ven P. F., Stöcklein W., and Fürst D. O. (2003) The limits of promiscuity: isoform-specific dimerization of filamins. Biochemistry 42, 430–439 [DOI] [PubMed] [Google Scholar]
- 23. Pudas R., Kiema T.-R., Butler P. J. G., Stewart M., and Ylänne J. (2005) Structural basis for vertebrate filamin dimerization. Structure 13, 111–119 [DOI] [PubMed] [Google Scholar]
- 24. Sjekloća L., Pudas R., Sjöblom B., Konarev P., Carugo O., Rybin V., Kiema T. R., Svergun D., Ylänne J., and Djinović Carugo K. (2007) Crystal structure of human filamin C domain 23 and small angle scattering model for filamin C 23–24 dimer. J. Mol. Biol. 368, 1011–1023 [DOI] [PubMed] [Google Scholar]
- 25. Bönnemann C. G., Thompson T. G., van der Ven P. F., Goebel H. H., Warlo I., Vollmers B., Reimann J., Herms J., Gautel M., Takada F., Beggs A. H., Fürst D. O., Kunkel L. M., Hanefeld F., and Schröder R. (2003) Filamin C accumulation is a strong but nonspecific immunohistochemical marker of core formation in muscle. J. Neurol. Sci. 206, 71–78 [DOI] [PubMed] [Google Scholar]
- 26. Nilsson M. I., Nissar A. A., Al-Sajee D., Tarnopolsky M. A., Parise G., Lach B., Fürst D. O., van der Ven P. F., Kley R. A., and Hawke T. J. (2013) Xin is a marker of skeletal muscle damage severity in myopathies. Am. J. Pathol. 183, 1703–1709 [DOI] [PubMed] [Google Scholar]
- 27. Kawamoto S., and Hidaka H. (1984) 1-(5-Isoquinolinesulfonyl)-2-methylpiperazine (H-7) is a selective inhibitor of protein kinase C in rabbit platelets. Biochem. Biophys. Res. Commun. 125, 258–264 [DOI] [PubMed] [Google Scholar]
- 28. Tigges U., Koch B., Wissing J., Jockusch B. M., and Ziegler W. H. (2003) The F-actin Cross-linking and Focal Adhesion Protein Filamin A Is a Ligand and in Vivo Substrate for Protein Kinase C. J. Biol. Chem. 278, 23561–23569 [DOI] [PubMed] [Google Scholar]
- 29. Raynaud F., Jond-Necand C., Marcilhac A., Fürst D., and Benyamin Y. (2006) Calpain 1-gamma filamin interaction in muscle cells: a possible in situ regulation by PKC-alpha. Int. J. Biochem. Cell Biol. 38, 404–413 [DOI] [PubMed] [Google Scholar]
- 30. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., and Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 31. Winter L., Staszewska I., Mihailovska E., Fischer I., Goldmann W. H., Schröder R., and Wiche G. (2014) Chemical chaperone ameliorates pathological protein aggregation in plectin-deficient muscle. J. Clin. Investig. 124, 1144–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Molt S., Buhrdel J. B., Yakovlev S., Schein P., Orfanos Z., Kirfel G., Winter L., Wiche G., van der Ven P. F., Rottbauer W., Just S., Belkin A. M., and Furst D. O. (2014) Aciculin interacts with filamin C and Xin and is essential for myofibril assembly, remodeling and maintenance. J. Cell Sci. 127, 3578–3592 [DOI] [PubMed] [Google Scholar]
- 33. Francavilla C., Hekmat O., Blagoev B., and Olsen J. V. (2014) SILAC-based temporal phosphoproteomics. Methods Mol. Biol. 1188, 125–148 [DOI] [PubMed] [Google Scholar]
- 34. Boersema P. J., Raijmakers R., Lemeer S., Mohammed S., and Heck A. J. (2009) Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protocols 4, 484–494 [DOI] [PubMed] [Google Scholar]
- 35. Wiese H., Kuhlmann K., Wiese S., Stoepel N. S., Pawlas M., Meyer H. E., Stephan C., Eisenacher M., Drepper F., and Warscheid B. (2014) Comparison of alternative MS/MS and bioinformatics approaches for confident phosphorylation site localization. J. Proteome Res. 13, 1128–1137 [DOI] [PubMed] [Google Scholar]
- 36. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 37. MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., and MacCoss M. J. (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schilling B., Rardin M. J., MacLean B. X., Zawadzka A. M., Frewen B. E., Cusack M. P., Sorensen D. J., Bereman M. S., Jing E., Wu C. C., Verdin E., Kahn C. R., Maccoss M. J., and Gibson B. W. (2012) Platform-independent and Label-free Quantitation of Proteomic Data Using MS1 Extracted Ion Chromatograms in Skyline: Application to Protein Acetylation and Phosphorylation. Mol. Cell. Proteomics 11, 202–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chambers M. C., Maclean B., Burke R., Amodei D., Ruderman D. L., Neumann S., Gatto L., Fischer B., Pratt B., Egertson J., Hoff K., Kessner D., Tasman N., Shulman N., Frewen B., Baker T. A., Brusniak M. Y., Paulse C., Creasy D., Flashner L., Kani K., Moulding C., Seymour S. L., Nuwaysir L. M., Lefebvre B., Kuhlmann F., Roark J., Rainer P., Detlev S., Hemenway T., Huhmer A., Langridge J., Connolly B., Chadick T., Holly K., Eckels J., Deutsch E. W., Moritz R. L., Katz J. E., Agus D. B., MacCoss M., Tabb D. L., and Mallick P. (2012) A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu X., Inbar Y., Dorrestein P. C., Wynne C., Edwards N., Souda P., Whitelegge J. P., Bafna V., and Pevzner P. A. (2010) Deconvolution and database search of complex tandem mass spectra of intact proteins: a combinatorial approach. Mol. Cell. Proteomics 9, 2772–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu X., Segar M. W., Li S. C., and Kim S. (2014) Spectral probabilities of top-down tandem mass spectra. BMC Genomics 15, S9–S9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maere S., Heymans K., and Kuiper M. (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 [DOI] [PubMed] [Google Scholar]
- 43. Liu Z., Cao J., Gao X., Ma Q., Ren J., and Xue Y. (2011) GPS-CCD: a novel computational program for the prediction of calpain cleavage sites. PLoS ONE 6, e19001–e19001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vizcaíno J. A., Côté R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Pérez-Riverol Y., Reisinger F., Ríos D., Wang R., and Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sharma V., Eckels J., Taylor G. K., Shulman N. J., Stergachis A. B., Joyner S. A., Yan P., Whiteaker J. R., Halusa G. N., Schilling B., Gibson B. W., Colangelo C. M., Paulovich A. G., Carr S. A., Jaffe J. D., MacCoss M. J., and MacLean B. (2014) Panorama: a targeted proteomics knowledge base. J. Proteome Res. 13, 4205–4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mandic A., Viktorsson K., Strandberg L., Heiden T., Hansson J., Linder S., and Shoshan M. C. (2002) Calpain-mediated Bid cleavage and calpain-independent Bak modulation: two separate pathways in cisplatin-induced apoptosis. Mol. Cell. Biol. 22, 3003–3013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., and Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Meth. 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Al Tanoury Z., Schaffner-Reckinger E., Halavatyi A., Hoffmann C., Moes M., Hadzic E., Catillon M., Yatskou M., and Friederich E. (2010) Quantitative kinetic study of the actin-bundling protein L-plastin and of its impact on actin turn-over. PLoS ONE 5, e9210–e9210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fürst D. O., Osborn M., Nave R., and Weber K. (1988) The organization of titin filaments in the half-sarcomere revealed by monoclonal antibodies in immunoelectron microscopy: a map of ten nonrepetitive epitopes starting at the Z line extends close to the M line. J. Cell Biol. 106, 1563–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chevessier F., Schuld J., Orfanos Z., Plank A.-C., Wolf L., Maerkens A., Unger A., Schlötzer-Schrehardt U., Kley R. A., von Hörsten S., Marcus K., Linke W. A., Vorgerd M., van der Ven P. F. M., Fürst D. O., and Schröder R. (2015) Myofibrillar instability exacerbated by acute exercise in filaminopathy. Hum. Mol. Genet., ddv421–ddv421 [DOI] [PubMed] [Google Scholar]
- 51. Langelaan M. L., Boonen K. J., Rosaria-Chak K. Y., van der Schaft D. W., Post M. J., and Baaijens F. (2011) Advanced maturation by electrical stimulation: Differences in response between C2C12 and primary muscle progenitor cells. J. Tissue Engineering Regenerative Med. 5, 529–539 [DOI] [PubMed] [Google Scholar]
- 52. Fujita H., Nedachi T., and Kanzaki M. (2007) Accelerated de novo sarcomere assembly by electric pulse stimulation in C2C12 myotubes. Exp. Cell Res. 313, 1853–1865 [DOI] [PubMed] [Google Scholar]
- 53. Pyle W. G., and Solaro R. J. (2004) At the crossroads of myocardial signaling: the role of Z-discs in intracellular signaling and cardiac function. Circ. Res. 94, 296–305 [DOI] [PubMed] [Google Scholar]
- 54. Moncman C. L., and Wang K. (1995) Nebulette: a 107 kD nebulin-like protein in cardiac muscle. Cell Motil. Cytoskelet. 32, 205–225 [DOI] [PubMed] [Google Scholar]
- 55. Spaich S., Will R. D., Just S., Spaich S., Kuhn C., Frank D., Berger I. M., Wiemann S., Korn B., Koegl M., Backs J., Katus H. A., Rottbauer W., and Frey N. (2012) F-box and leucine-rich repeat protein 22 is a cardiac-enriched F-box protein that regulates sarcomeric protein turnover and is essential for maintenance of contractile function in vivo. Circ. Res. 111, 1504–1516 [DOI] [PubMed] [Google Scholar]
- 56. Hawke T. J., Atkinson D. J., Kanatous S. B., Van der Ven P. F., Goetsch S. C., and Garry D. J. (2007) Xin, an actin binding protein, is expressed within muscle satellite cells and newly regenerated skeletal muscle fibers. Am. J. Physiol. Cell Physiol. 293, C1636-C1644 [DOI] [PubMed] [Google Scholar]
- 57. Zhou H., Di Palma S., Preisinger C., Peng M., Polat A. N., Heck A. J., and Mohammed S. (2013) Toward a comprehensive characterization of a human cancer cell phosphoproteome. J. Proteome Res. 12, 260–271 [DOI] [PubMed] [Google Scholar]
- 58. Nishikawa K., Toker A., Johannes F. J., Songyang Z., and Cantley L. C. (1997) Determination of the specific substrate sequence motifs of protein kinase C isozymes. J. Biol. Chem. 272, 952–960 [DOI] [PubMed] [Google Scholar]
- 59. Martiny-Baron G., Kazanietz M. G., Mischak H., Blumberg P. M., Kochs G., Hug H., Marmé D., and Schächtele C. (1993) Selective inhibition of protein kinase C isozymes by the indolocarbazole Gö 6976. J. Biol. Chem. 268, 9194–9197 [PubMed] [Google Scholar]
- 60. Jensen T. E., Maarbjerg S. J., Rose A. J., Leitges M., and Richter E. A. (2009) Knockout of the predominant conventional PKC isoform, PKCalpha, in mouse skeletal muscle does not affect contraction-stimulated glucose uptake. Am. J. Physiol. Endocrinol Metab 297, E340–E348 [DOI] [PubMed] [Google Scholar]
- 61. Guyon J. R., Kudryashova E., Potts A., Dalkilic I., Brosius M. A., Thompson T. G., Beckmann J. S., Kunkel L. M., and Spencer M. J. (2003) Calpain 3 cleaves filamin C and regulates its ability to interact with a- and b-sarcoglycans. Muscle Nerve 28, 472–483 [DOI] [PubMed] [Google Scholar]
- 62. Taveau M., Bourg N., Sillon G., Roudaut C., Bartoli M., and Richard I. (2003) Calpain 3 is activated through autolysis within the active site and lyses sarcomeric and sarcolemmal components. Mol. Cell. Biol. 23, 9127–9135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Goll D. E., Thompson V. F., Li H., Wei W., and Cong J. (2003) The calpain system. Physiol. Rev. 83, 731–801 [DOI] [PubMed] [Google Scholar]
- 64. Dalkilic I., Schienda J., Thompson T. G., and Kunkel L. M. (2006) Loss of FilaminC (FLNc) Results in severe defects in myogenesis and myotube structure. Mol. Cell. Biol. 26, 6522–6534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sanger J. W., Kang S., Siebrands C. C., Freeman N., Du A., Wang J., Stout A. L., and Sanger J. M. (2005) How to build a myofibril. J. Muscle Res. Cell Motil. 26, 343–354 [DOI] [PubMed] [Google Scholar]
- 66. Geach T. J., Hirst E. M., and Zimmerman L. B. (2015) Contractile activity is required for Z-disc sarcomere maturation in vivo. genesis 53, 299–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cui Z., Chen X., Lu B., Park S. K., Xu T., Xie Z., Xue P., Hou J., Hang H., Yates J. R. 3rd, and Yang F. (2009) Preliminary quantitative profile of differential protein expression between rat L6 myoblasts and myotubes by stable isotope labeling with amino acids in cell culture. Proteomics 9, 1274–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Le Bihan M. C., Barrio-Hernandez I., Mortensen T. P., Henningsen J., Jensen S. S., Bigot A., Blagoev B., Butler-Browne G., and Kratchmarova I. (2015) Cellular proteome dynamics during differentiation of human primary myoblasts. j. Proteome Res. 14, 3348–3361 [DOI] [PubMed] [Google Scholar]
- 69. Frank D., Kuhn C., Katus Ha., and Frey N. (2006) The sarcomeric Z-disc: a nodal point in signalling and disease. J. Mol. Med. 84, 446–468 [DOI] [PubMed] [Google Scholar]
- 70. Sanger J. M., and Sanger J. W. (2008) The dynamic Z bands of striated muscle cells. Sci. Signal. 1, pe37–pe37 [DOI] [PubMed] [Google Scholar]
- 71. Wang J., Shaner N., Mittal B., Zhou Q., Chen J., Sanger J. M., and Sanger J. W. (2005) Dynamics of Z-band based proteins in developing skeletal muscle cells. Cell Motil. Cytoskelet. 61, 34–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pyle W. G., La Rotta G., de Tombe P. P., Sumandea M. P., and Solaro R. J. (2006) Control of cardiac myofilament activation and PKC-betaII signaling through the actin capping protein, CapZ. J. Mol. Cell. Cardiol. 41, 537–543 [DOI] [PubMed] [Google Scholar]
- 73. Hartman T. J., Martin J. L., Solaro R. J., Samarel A. M., and Russell B. (2009) CapZ dynamics are altered by endothelin-1 and phenylephrine via PIP2- and PKC-dependent mechanisms. Am. J. Physiol. Cell Physiol. 296, C1034–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhou Q., Ruiz-Lozano P., Martone M. E., and Chen J. (1999) Cypher, a striated muscle-restricted PDZ and LIM domain-containing protein, binds to alpha-actinin-2 and protein kinase C. J. Biol. Chem. 274, 19807–19813 [DOI] [PubMed] [Google Scholar]
- 75. Arimura T., Hayashi T., Terada H., Lee S. Y., Zhou Q., Takahashi M., Ueda K., Nouchi T., Hohda S., Shibutani M., Hirose M., Chen J., Park J. E., Yasunami M., Hayashi H., and Kimura A. (2004) A Cypher/ZASP mutation associated with dilated cardiomyopathy alters the binding affinity to protein kinase C. J. Biol. Chem. 279, 6746–6752 [DOI] [PubMed] [Google Scholar]
- 76. Cheng H., Zheng M., Peter A. K., Kimura K., Li X., Ouyang K., Shen T., Cui L., Frank D., Dalton N. D., Gu Y., Frey N., Peterson K. L., Evans S. M., Knowlton K. U., Sheikh F., and Chen J. (2011) Selective deletion of long but not short Cypher isoforms leads to late-onset dilated cardiomyopathy. Hum. Mol. Genet. 20, 1751–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Nakagawa N., Hoshijima M., Oyasu M., Saito N., Tanizawa K., and Kuroda S. (2000) ENH, containing PDZ and LIM domains, heart/skeletal muscle-specific protein, associates with cytoskeletal proteins through the PDZ domain. Biochem. Biophys. Res. Commun. 272, 505–512 [DOI] [PubMed] [Google Scholar]
- 78. Cheng H., Kimura K., Peter A. K., Cui L., Ouyang K., Shen T., Liu Y., Gu Y., Dalton N. D., Evans S. M., Knowlton K. U., Peterson K. L., and Chen J. (2010) Loss of enigma homolog protein results in dilated cardiomyopathy. Circ. Res. 107, 348–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Murray J. T., Campbell D. G., Peggie M., Mora A., Alfonso M., and Cohen P. (2004) Identification of filamin C as a new physiological substrate of PKBalpha using KESTREL. Biochem. J. 384, 489–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Chang Y. W., Chang Y. T., Wang Q., Lin J. J., Chen Y. J., and Chen C. C. (2013) Quantitative phosphoproteomic study of pressure-overloaded mouse heart reveals dynamin-related protein 1 as a modulator of cardiac hypertrophy. Mol. Cell. Proteomics 12, 3094–3107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villén J., Haas W., Sowa M. E., and Gygi S. P. (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Chen M., and Stracher A. (1989) In situ phosphorylation of platelet actin-binding protein by cAMP-dependent protein kinase stabilizes it against proteolysis by calpain. J. Biol. Chem. 264, 14282–14289 [PubMed] [Google Scholar]
- 83. Kishimoto A., Mikawa K., Hashimoto K., Yasuda I., Tanaka S., Tominaga M., Kuroda T., and Nishizuka Y. (1989) Limited proteolysis of protein kinase C subspecies by calcium-dependent neutral protease (calpain). J. Biol. Chem. 264, 4088–4092 [PubMed] [Google Scholar]
- 84. Davies P. J., Wallach D., Willingham M. C., Pastan I., Yamaguchi M., and Robson R. M. (1978) Filamin-actin interaction. Dissociation of binding from gelation by Ca2+-activated proteolysis. J. Biol. Chem. 253, 4036–4042 [PubMed] [Google Scholar]
- 85. Franco S. J., and Huttenlocher A. (2005) Regulating cell migration: calpains make the cut. J. Cell Sci. 118, 3829–3838 [DOI] [PubMed] [Google Scholar]
- 86. Tompa P., Buzder-Lantos P., Tantos A., Farkas A., Szilágyi A., Bánóczi Z., Hudecz F., and Friedrich P. (2004) On the sequential determinants of calpain cleavage. J. Biol. Chem. 279, 20775–20785 [DOI] [PubMed] [Google Scholar]
- 87. Fox J. E. B., and Saido T. C. (1999) Calpain in signal transduction. In: Wang, K. K. W., and Yuen, P., eds. Calpain: Pharmacology and Toxicology of Calcium-Dependent Protease, pp. 103–126, Taylor & Francis; Philadelphia. [Google Scholar]
- 88. Pfaff M., Du X., and Ginsberg M. H. (1999) Calpain cleavage of integrin beta cytoplasmic domains. FEBS Lett. 460, 17–22 [DOI] [PubMed] [Google Scholar]
- 89. Gontier Y., Taivainen A., Fontao L., Sonnenberg A., van der Flier A., Carpen O., Faulkner G., and Borradori L. (2005) The Z-disc proteins myotilin and FATZ-1 interact with each other and are connected to the sarcolemma via muscle-specific filamins. J. Cell Sci. 118, 3739–3749 [DOI] [PubMed] [Google Scholar]
- 90. Chiang W., and Greaser M. L. (2000) Binding of filamin isoforms to myofibrils. J. Muscle Res. Cell Motil. 21, 321–333 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.