Summary
Many proteins responsible for genome maintenance interact with one another via short sequence motifs. The best known of these are PIP motifs, which mediate interactions with the replication protein PCNA. Others include RIR motifs, which bind the translesion synthesis protein Rev1, and MIP motifs, which bind the mismatch repair protein Mlh1. Although these motifs have similar consensus sequences, they have traditionally been viewed as separate motifs, each with their own target protein. In this article, we review several recent studies that challenge this view. Taken together, they imply that these different motifs are not distinct entities. Instead, there is a single, broader class of motifs, which we call ‘PIP-like’ motifs, which have overlapping specificities and are capable of binding multiple target proteins. Given this, we must reassess the role of these motifs in forming the network of interacting proteins responsible for genome maintenance.
Keywords: DNA replication, DNA repair, DNA recombination, Rev1, translesion synthesis
Introduction
A large network of interacting proteins is responsible for genome maintenance. These proteins replicate DNA with high fidelity and repair damaged DNA through a variety of pathways (base excision repair, nucleotide excision repair, mismatch repair, etc.). Several proteins are hubs in this network and interact with many other factors participating in these processes. Proliferating cell nuclear antigen (PCNA), for example, is a global hub protein, which physically interacts with nearly one hundred proteins that are mostly involved in DNA replication and multiple DNA repair pathways [1–7]. Rev1, by contrast, is a local hub protein, which interacts with a dozen or so proteins that are primarily involved in the bypass of damage DNA during DNA replication [8–11]. Similarly, Mlh1 is a local hub protein, which interacts with approximately twenty proteins that are mainly involved in mismatch repair [12–15].
Many of the proteins that interact with these hubs do so via loosely conserved sequence motifs. The best known of these is the PCNA-interacting protein (PIP) motif, which is also known as a PIP box [1, 2, 16–18]. Others include the Rev1-interacting region (RIR) motif [9, 19, 20] and the Mlh1-interacting protein (MIP) motif [21–23]. These motifs all have similar consensus sequences that prominently feature two, adjacent aromatic residues (tyrosine or phenylalanine). Moreover, these motifs are usually located in intrinsically disordered regions of the proteins. Despite these obvious similarities, these different types of motifs have traditionally been considered to be distinct entities, each specific for binding a unique target hub protein. For example, PIP motifs are generally thought to be a narrow class of sequences whose sole function is to mediate interactions with PCNA. Similarly, RIR motifs and MIP motifs are generally thought to mediate interactions with only Rev1 and Mlh1, respectively.
Recently, a far more interesting picture has emerged from a number of studies by different research groups [24–26]. When taken in isolation these findings do not necessarily challenge the existing paradigm. When viewed together, however, they force us to re-think the nature of these motifs and the types of protein-protein interactions in which the proteins possessing them engage. In this article, we will review these recent studies and put forth this alternative picture. Specifically, we will argue that PIP motifs, RIR motifs, and MIP motifs do not represent distinct entities. Instead, there is a single, broad, loosely defined class of motifs (which we call ‘PIP-like’ motifs) that mediate interactions with multiple proteins involved in genome maintenance. This has important, practical implications regarding how the network of interacting proteins responsible for genome maintenance forms and functions.
PIP motifs bind PCNA
PCNA is an essential replication accessory protein that is perhaps most widely known for its role as the processivity factor for replicative DNA polymerases [3, 5–7]. It is a ring-shaped homotrimer that encircles and moves along double-stranded DNA [27]. In so doing, it acts as a sliding clamp that locks the replicative polymerases on their DNA substrates. PCNA, however, does far more than this. It also regulates the access of a wide range of enzymes involved in DNA replication, repair, and recombination to the DNA substrate and enhances their catalytic activities. For this reason, PCNA has been called the “maestro of the replication fork” [3].
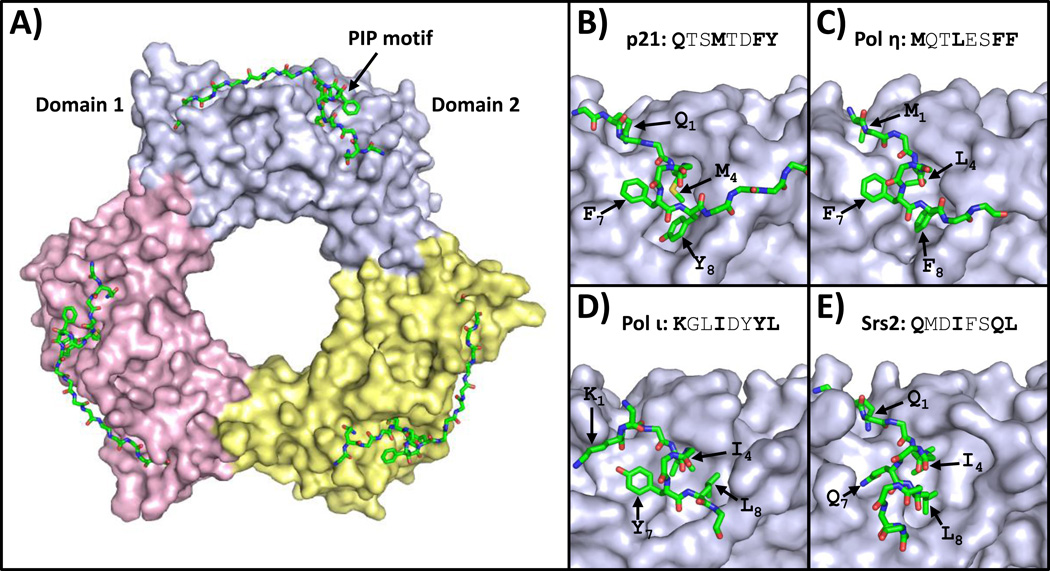
Many of the proteins that bind PCNA do so through partially conserved PIP motifs (Table 1) [1, 2, 16–18]. The consensus sequence for this eight-residue motif is a glutamine in position 1, a hydrophobic residue (usually leucine, isoleucine, or methionine) in position 4, and aromatic residues (phenylalanine or tyrosine) in positions 7 and 8. PIP motifs bind to the front face of the PCNA ring in a cleft between the two PCNA domains (Fig. 1A). Moreover, secondary contacts between PCNA and residues flanking the PIP motif often confer additional binding affinity.
Table 1.
PIP-like motifs.
| Protein | Organism | Residues | Sequence |
|---|---|---|---|
| PIP motifs | |||
| p21 | Human | 144–151 | QTSMTDFY |
| pol η | Human | 701–708 | MQTLESFF |
| pol ι | Human | 421–428 | KGLIDYYL |
| Srs2 | Yeast | 1149–1156 | QMDIFSQL |
| RIR motifs | |||
| pol η | Human | 531–536 | FFKQKS |
| pol ι | Human | 546–551 | FFSKKQ |
| Rad5 | Yeast | 12–17 | FFNDDL |
| MIP motifs | |||
| Exo1 | Yeast | 444–448 | RSKFF |
| Ntg2 | Yeast | 23–27 | RSKYF |
| F1 motifs | |||
| pol η | Human | 481–486 | ESFFQK |
Figure 1.
Structures of PCNA bound to PIP motifs. (A) An overview of the structure of human PCNA bound to peptides containing the PIP motif of p21 (PDB ID: 1AXC) is shown [28]. The three PCNA subunits are depicted in the surface representation (light blue, yellow, and pink). The PIP motif is depicted in the stick representation (with carbon, nitrogen, oxygen, and sulfur atoms colored green, blue, red, and yellow, respectively). (B) A close up view of human PCNA bound to the p21 PIP motif is shown [28]. The side chains of residues in positions 1, 4, 7, and 8 are indicated. (C) A close up view of human PCNA bound to the pol η PIP motif (PDB ID: 2ZVK) is shown [29]. (D) A close up view of human PCNA bound to the pol ι PIP motif (PDB ID: 2ZVM) is shown [29]. (E) A close up view of yeast PCNA bound to the Srs2 PIP motif (PDB ID: SV62) is shown [30].
The first X-ray crystal structure of PCNA bound to a consensus PIP motif was with the PIP motif from the cell-cycle checkpoint protein p21 (Fig. 1B) [28]. This structure clearly revealed how PCNA recognizes consensus PIP motifs. The consensus glutamine in position 1 binds in a small pocket on the front face of the PCNA ring called the “Q pocket”. Residues in positions 4 through 8 form a short 310 helix in which the consensus methionine in position 4, the consensus phenylalanine residue in position 7, and the consensus tyrosine in position 8 insert into a large hydrophobic pocket on the front of PCNA called the “three-forked plug” because of its resemblance to an electrical outlet. In addition, residues flanking the PIP motif on its C-terminal side form an extended anti-parallel β sheet with residues of the extended interdomain connector loop of PCNA.
Recently X-ray crystal structures of PCNA bound to several non-consensus PIP motifs have been determined [29, 30]. These structures show how PCNA accommodates PIP motifs that diverge from the consensus sequence. For example, the non-classical DNA polymerase eta (pol η) possesses a PIP motif that lacks a glutamine in position 1 [29]. The non-consensus methionine at this position is still accommodated in the Q pocket on the front of PCNA. Likewise, positions 4 through 8 still retain the typical 310 helix and the consensus residues in positions 4, 7, and 8 still insert into the three-forked plug (Fig. 1C). By contrast, the non-classical polymerase iota (pol ι) possesses a PIP motif that lacks a glutamine in position 1 and an aromatic residue in position 8 [29]. The non-consensus arginine in position 1 does not bind in the Q pocket. Moreover, the PIP motif does not adopt a 310 helix but instead forms a β-bend-like structure that allows the consensus isoleucine and tyrosine residues at positions 4 and 7 and the non-consensus leucine in position 8 to insert into the three-forked plug (Fig. 1D).
The most extreme example of a non-consensus PIP motif whose structure has been determined is that of the anti-recombinogenic helicase Srs2 [30]. It lacks the two consensus aromatic residues in positions 7 and 8. The consensus glutamine in position 1 binds in the Q pocket. However, instead of forming a 310 helix, the residues spanning position 4 through 8 form a α-helix. This allows the consensus isoleucine at position 4 and the non-consensus glutamine and leucine residues at positions 7 and 8 to insert into the three-forked plug (Fig. 1E). Taken together, these structural studies show that PCNA can bind a wide range of PIP motifs that diverge significantly from the consensus sequence. This implies that identifying PIP motifs from sequence analysis alone is highly problematic.
RIR motifs bind Rev1
Rev1 is a protein that is best known for its role in translesion synthesis, a pathway for bypassing damaged DNA during DNA replication [8, 11, 31–36]. It has two distinct functions in translesion synthesis. First, it is a non-classical DNA polymerase that catalyzes the incorporation of nucleotides opposite abasic sites and several types of damaged guanines [8, 37]. Second, it is a scaffold that recruits other non-classical DNA polymerases, such as the aforementioned pol η and pol ι, to sites of DNA damage thereby allowing them to catalyze nucleotide incorporation opposite the damage [8, 9, 11, 38]. Recent evidence points to further roles of Rev1 in template switching [26], which is another pathway for bypassing DNA damage, and several DNA repair pathways, including base excision repair and interstrand crosslink repair [39].
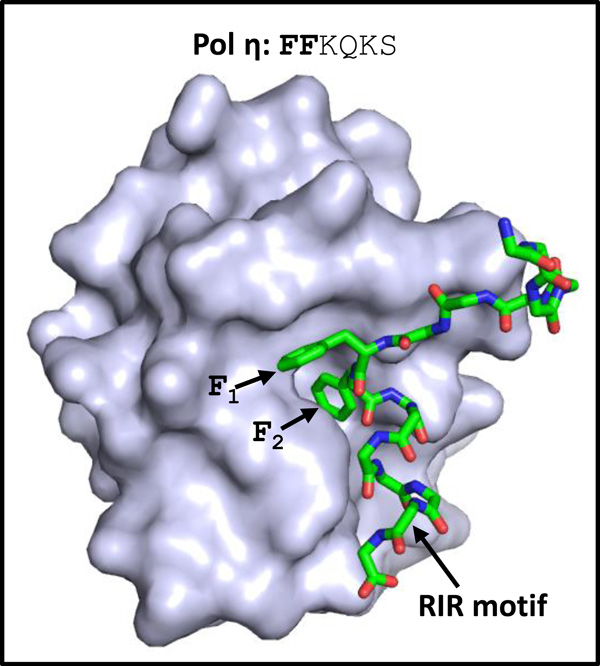
Many proteins that interact with Rev1 – including mammalian non-classical polymerases, the mammalian base excision repair protein XRCC1, the yeast template switching protein Rad5, and mammalian classical DNA polymerase delta (pol δ) – do so through partially conserved RIR motifs, which have a consensus sequence resembling that of PIP motifs (Table 1) [9, 10, 19, 26, 39–41]. The consensus sequence for this six-residue motif is phenylalanine residues in positions 1 and 2 and any amino acid residue except for proline in the remaining four positions. These RIR motifs bind to a four-helix bundle at the extreme C-terminus of Rev1 called the Rev1 C-terminal domain (CTD).
One of the first structures of the Rev1 CTD bound to a RIR motif involved the RIR motif from human pol η (Fig. 2) [20]. This structure showed how the Rev1 CTD recognizes RIR motifs. The phenylalanine residues in positions 1 and 2 of the RIR motif insert in a hydrophobic pocket on the Rev1 CTD. The specificity of this interaction appears to come primarily from these two phenylalanine residues. This suggests that many motifs with two adjacent phenylalanine residues (or even tyrosine residues), including some PIP motifs and MIP motifs, may be capable of binding the Rev1 CTD.
Figure 2.
Structure of Rev1 bound to a RIR motif. The structure of the CTD of human Rev1 bound to a peptide containing the RIR motif of pol η (PDB ID: 2LSK) is shown [20]. The human Rev1 CTD is depicted in the surface representation (light blue). The peptide containing the RIR motif of human pol η is depicted in the stick representation (with carbon, nitrogen, and oxygen atoms colored green, blue, and red, respectively). The side chains of residues in positions 1 and 2 are indicated.
MIP motifs bind Mlh1
Mlh1 is a protein best known for its role in mismatch repair, a pathway for correcting errors made by DNA polymerases during DNA replication [12–15]. It also participates in crossing over during meiosis [42] and may play a role in base excision repair [21]. In mismatch repair in yeast, Mlh1 forms a heterodimer with Pms1 called MutLα. This dimer is believed to act as a scaffold that links the proteins that recognize the mismatches in the DNA with the proteins that carry out the subsequent steps of mismatch repair.
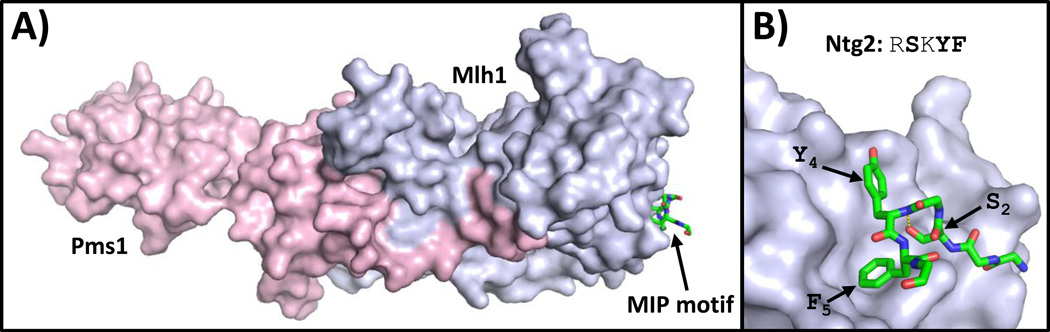
Mlh1 interacts with the exonuclease Exo1 and the DNA glycosylase Ntg2, both of which contain conserved MIP motifs (Table 1) [21–23]. The consensus sequence for this five-residue motif is an arginine or lysine in position 1, a serine in position 2, a lysine in position 3, an aromatic residue (phenylalanine or tyrosine) is position 4, and a phenylalanine is position 5. MIP motifs interact with the C-terminal region of Mlh1. In addition, secondary contacts between Mlh1 and residues flanking the MIP motif increase binding affinity. These are analogous to the secondary contacts observed between PIP motifs and PCNA.
X-ray crystal structures of Mlh1 bound to MIP motifs from Exo1 and Ntg2 show that the MIP peptides adopt an ST turn, a secondary structural element in which the side chain of the serine in position 2 forms a hydrogen bond with the main chain amine group of the residue in position 4 (Fig. 3) [23]. Residues in the MIP motif important for Mlh1 binding include the serine in position 2 and the adjacent aromatic residues in positions 4 and 5. These latter residues bind in a hydrophobic pocket similar to the mode of binding observed with PCNA and PIP motifs and with Rev1 and RIR motifs. Again, this suggests that other motifs with two adjacent aromatic residues, including some PIP motifs and RIR motifs, may be capable of binding to Mlh1.
Figure 3.
Structure of Mlh1 bound to a MIP motif. (A) An overview of the structure of the CTD of yeast MutLα bound to a peptide containing the MIP motif of Ntg2 (PDB ID: 4FMN) is shown [23]. The Mlh1 and Pms1 subunits are depicted in the surface representation (light blue and pink, respectively). The peptide containing the MIP motif of Ntg2 is depicted in the stick representation (with carbon, nitrogen, and oxygen atoms colored green, blue, and red, respectively). The side chains of residues in positions 2, 4, and 5 and the hydrogen bond between the serine at position 2 and the backbone at position 4 are indicated.
Recent studies challenge the traditional paradigm
In this article, we have described the traditional paradigm regarding PIP-like motifs – that there are (so far) three distinct classes of motifs, each specific for a different target protein. PIP motifs mediate interactions with PCNA, RIR motifs mediate interactions with Rev1, and MIP motifs mediate interactions with Mlh1. These motifs have consensus sequences that involve two adjacent aromatic residues, and structural studies show that these aromatic residues facilitate the interactions with target proteins by binding within hydrophobic pockets.
Several recent studies have challenged this paradigm by showing that some of these motifs are specific for more than one target protein. The first such study identified a novel interaction between human non-classical polymerases pol η and a non-catalytic subunit of classical pol δ [24]. Pol δ is responsible for lagging strand replication and participates in several DNA repair pathways [43, 44]. In humans, it consists of four subunits: POLD1, POLD2, POLD3, and POLD4. The POLD1 subunit is the catalytic subunit, and the POLD2 subunit is a scaffold onto which the other subunits of pol δ are assembled [24].
This study showed that pol η binds the POLD2 subunit of pol δ [24]. Moreover, this interaction is mediated by two adjacent phenylalanine residues in a motif that the authors of this study called an F1 motif (Table 1). Their main conclusion is that pol η possesses an F1 motif, a new PIP-like motif that mediates interactions with pol δ. It is surprising, however, that the authors did not note that this F1 motif is one of the two previously identified RIR motifs of pol η [9]. To our knowledge, this is the first instance of any PIP-like motif – in this case an RIR motif – mediating interactions with multiple target proteins – in this case Rev1 and pol δ. This was the first hint that the different classes of PIP-like motifs may not be distinct entities.
A second study poses an even more serious challenge to the traditional paradigm [25]. As described above, mammalian non-classical polymerases such as pol η, pol ι, and pol κ all possess both PIP motifs, which are believed to mediate their interactions with PCNA, and RIR motifs, which are thought to mediate their interactions with Rev1 [9]. Yeast pol η, by contrast, possesses only a single PIP motif, which is believed to mediate its interaction with PCNA [45]. Yeast pol η was known to interact with Rev1 [46], but the structural basis of this interaction was not known, as yeast pol η was not thought to posses a RIR motif.
This study surprisingly showed that the PIP motif of yeast pol η mediates the interaction with both PCNA and Rev1 [25]. In fact, the PIP motif binds the Rev1 CTD with greater affinity than it binds PCNA. Moreover, it binds the Rev1 CTD in the same hydrophobic pocket as RIR motifs do. In other words, this PIP motif is in reality both a PIP motif and a RIR motif. This is the first example of a canonical PIP motif interacting with a target protein other than PCNA. This same study also showed that some of the PIP and RIR motifs in the mammalian non-classical polymerases also have overlapping binding specificities [25]. The RIR motif of pol κ, for example, binds both the Rev1 CTD and PCNA with similar affinities.
Even more recently, an X-ray crystal structure of the complex between the yeast Rev1 CTD and a newly discovered RIR motif from the template switching protein Rad5 was determined [26]. This particular RIR motif is nearly a consensus PIP motif (Table 1). Thus, it is likely that the RIR motif of Rad5 also mediates its interactions with PCNA, although this has yet to be experimentally tested. Taken together, these studies demonstrate that the lines between PIP motifs, RIR motifs, and F1 motifs are blurred and it is very likely that these classes of motifs do not represent distinct entities.
A new paradigm emerges
In the previous section, we reviewed some recent studies showing that several PIP-like motifs are specific for more than one target protein. For example, the RIR motif of human pol η interacts with both Rev1 and pol δ [24]. Similarly, the PIP motif of yeast pol η interacts with both PCNA and Rev1 [25]. These findings show that PIP motifs, RIR motifs, and F1 motifs have partially overlapping specificities and the lines separating these motifs have become blurred.
The structural basis of the interactions between these PIP-like motifs and their target proteins likewise calls into question the rigid distinctions between these motifs. Structures of PCNA bound to PIP motifs show that sequences diverging from the PIP consensus sequence can still bind PCNA with high affinity [29, 30]. Structures of Rev1 bound to RIR motifs show that the specificity comes largely from the two aromatic residues of the RIR motif [20, 40]. Thus, one expects that many PIP-like sequences from any of these classes would bind both PCNA and Rev1.
Given the very recent studies showing overlapping specificities of different types of PIP-like motifs and given the way divergent sequences can bind multiple target proteins, a new paradigm is emerging. In this new view, PIP motifs, RIR motifs, MIP motifs, and F1 motifs do not represent distinct entities. Instead, there is a single, broader class of motif, which we suggest be called PIP-like motifs, that subsumes these previous classes. Some individual PIP-like motifs may be specific for only one target protein, while others are likely specific for multiple target proteins.
Several important points need to be made regarding this shift in paradigm. First, one should not rely on consensus sequences alone when ascribing a function to a particular PIP-like motif. Instead, experimental verification of target protein binding will be necessary for determining the specificity and function of any given motif. Second, a careful experimental study of the specificity determinants of PIP-like motifs needs to be undertaken to ascertain if there is a set of rules that can allow one to better predict target protein specificity. For example, does the glutamine in position 1 of consensus PIP motifs provide greater specificity for binding PCNA? Can non-consensus PIP motifs lacking aromatic residues in positions 7 or 8 bind other target proteins? Lastly, it is important to consider other potential target proteins of PIP-like motifs. It is very likely that other target proteins exist beyond PCNA, Rev1, Mlh1, and pol δ.
Conclusions and outlook
PCNA, Rev1, Mlh1, pol δ, and other proteins are hubs in the large network of interacting proteins responsible for genome maintenance. Many proteins interact with these hubs via short sequence motifs such as PIP motifs, RIR motifs, MIP motifs, and F1 motifs. The traditional paradigm is that these motifs are distinct entities, each specific for binding a single target protein. Recent studies have made it clear that the distinctions between them are no longer useful and ought to be abandoned. In fact, preserving the notion of these separate classes of motifs may be misleading and counter-productive.
We believe that abandoning them in favor of the more general class of PIP-like motifs will benefit the field in several important ways. First, it will open the door to new hypotheses regarding how various proteins fit together in this network. This includes considering other potential target proteins of PIP-like motifs beyond the hub proteins described above. Second, it will emphasize the need to experimentally examine the specificity and function of any individual PIP-like motif more fully.
What we are left with after rejecting the traditional paradigm is a much more interesting and nuanced picture of the network of interacting proteins necessary for genome maintenance. Hundreds of PIP-like motifs, each with specificity for one or more target hub proteins, hold this network together in more ways than previously considered. Much work remains as we reassess the role of these motifs in forming this network. Ultimately, these future studies will lead to a deeper understanding of the fundamental mechanisms by which genome stability is maintained.
Finally, it should be noted that the structural basis for these interactions are quite simple in nature. They are essentially aromatic residues inserting into hydrophobic pockets on target proteins. Given this, one wonders whether these types of interactions might also occur between proteins beyond those necessary for genome maintenance. It is possible, perhaps even likely, that PIP-like motifs play key roles in mediating protein-protein interactions and organizing networks of interacting proteins necessary to carry out a much wider range of biological functions.
Acknowledgments
We thank Christine Kondratick, Kyle Powers, Melissa Gildenberg, Brittany Ripley, Marc Wold, Maria Spies, and Lynne Dieckman for discussions. The project described was supported by award number GM081433 and GM108027 from the National Institute of General Medicine to M.T.W. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Abbreviations
- BRCT
BRCA1 C-terminal
- CTD
C-terminal domain
- MIP
Mlh1-interacting protein
- PCNA
proliferating cell nuclear antigen
- PIP
PCNA-interacting protein
- pol δ
DNA polymerase delta
- pol η
DNA polymerase eta
- pol ι
DNA polymerase iota
- pol κ
DNA polymerase kappa
- RIR
Rev1-interacting region
References
- 1.Tsurimoto T. PCNA binding proteins. Front Biosci. 1999;4:D849–D858. doi: 10.2741/tsurimoto. [DOI] [PubMed] [Google Scholar]
- 2.Maga G, Hubscher U. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J Cell Sci. 2003;116:3051–3060. doi: 10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- 3.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Naryzhny SN. Proliferating cell nuclear antigen: a proteomics view. Cell Mol Life Sci. 2008;65:3789–3808. doi: 10.1007/s00018-008-8305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhuang Z, Ai Y. Processivity factor of DNA polymerase and its expanding role in normal and translesion DNA synthesis. Biochim Biophys Acta. 2010;1804:1081–1093. doi: 10.1016/j.bbapap.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dieckman LM, Freudenthal BD, Washington MT. PCNA structure and function: insights from structures of PCNA complexes and post-translationally modified PCNA. Subcell Biochem. 2012;62:281–299. doi: 10.1007/978-94-007-4572-8_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boehm EM, Gildenberg MS, Washington MT. The Many Roles of PCNA in Eukaryotic DNA Replication. Enzymes. 2016;39:231–254. doi: 10.1016/bs.enz.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawrence CW. Cellular roles of DNA polymerase zeta and Rev1 protein. DNA Repair. 2002;1:425–435. doi: 10.1016/s1568-7864(02)00038-1. [DOI] [PubMed] [Google Scholar]
- 9.Ohmori H, Hanafusa T, Ohashi E, Vaziri C. Separate roles of structured and unstructured regions of Y-family DNA polymerases. Adv Protein Chem Struct Biol. 2009;78:99–146. doi: 10.1016/S1876-1623(08)78004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wojtaszek J, Lee CJ, D'Souza S, Minesinger B, et al. Structural basis of Rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of Rev1, heterodimeric polymerase (Pol) zeta, and Pol kappa. J Biol Chem. 2012;287:33836–33846. doi: 10.1074/jbc.M112.394841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pryor JM, Dieckman LM, Boehm EM, Washington MT. Eukaryotic Y-Family Polymerases: A Biochemical and Structural Perspective. Nucleic Acids Mol Biol. 2014;30:85–108. [Google Scholar]
- 12.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 13.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 14.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 15.Kunz C, Saito Y, Schar P. Mismatched repair: variations on a theme. Cell Mol Life Sci. 2009;66:1021–1038. doi: 10.1007/s00018-009-8739-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jonsson ZO, Hindges R, Hubscher U. Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen. EMBO J. 1998;17:2412–2425. doi: 10.1093/emboj/17.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warbrick E. The puzzle of PCNA's many partners. Bioessays. 2000;22:997–1006. doi: 10.1002/1521-1878(200011)22:11<997::AID-BIES6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 18.Hingorani MM, O'Donnell M. Sliding clamps: a (tail)ored fit. Curr Biol. 2000;10:R25–R29. doi: 10.1016/s0960-9822(99)00252-3. [DOI] [PubMed] [Google Scholar]
- 19.Ohashi E, Hanafusa T, Kamei K, Song I, et al. Identification of a novel REV1-interacting motif necessary for DNA polymerase kappa function. Genes Cells. 2009;14:101–111. doi: 10.1111/j.1365-2443.2008.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pozhidaeva A, Pustovalova Y, D'Souza S, Bezsonova I, et al. NMR structure and dynamics of the C-terminal domain from human Rev1 and its complex with Rev1 interacting region of DNA polymerase eta. Biochemistry. 2012;51:5506–5520. doi: 10.1021/bi300566z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gellon L, Werner M, Boiteux S. Ntg2p, a Saccharomyces cerevisiae DNA N-glycosylase/apurinic or apyrimidinic lyase involved in base excision repair of oxidative DNA damage, interacts with the DNA mismatch repair protein Mlh1p. Identification of a Mlh1p binding motif. J Biol Chem. 2002;277:29963–29972. doi: 10.1074/jbc.M202963200. [DOI] [PubMed] [Google Scholar]
- 22.Dherin C, Gueneau E, Francin M, Nunez M, et al. Characterization of a highly conserved binding site of Mlh1 required for exonuclease I-dependent mismatch repair. Mol Cell Biol. 2009;29:907–918. doi: 10.1128/MCB.00945-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gueneau E, Dherin C, Legrand P, Tellier-Lebegue C, et al. Structure of the MutLalpha C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site. Nat Struct Mol Biol. 2013;20:461–468. doi: 10.1038/nsmb.2511. [DOI] [PubMed] [Google Scholar]
- 24.Baldeck N, Janel-Bintz R, Wagner J, Tissier A, et al. FF483-484 motif of human Poleta mediates its interaction with the POLD2 subunit of Poldelta and contributes to DNA damage tolerance. Nucleic Acids Res. 2015;43:2116–2125. doi: 10.1093/nar/gkv076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boehm EM, Powers KT, Kondratick CM, Spies M, et al. The Proliferating Cell Nuclear Antigen (PCNA)-interacting Protein (PIP) Motif of DNA Polymerase eta Mediates Its Interaction with the C-terminal Domain of Rev1. J Biol Chem. 2016;291:8735–8744. doi: 10.1074/jbc.M115.697938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu X, Lin A, Zhou C, Blackwell SR, et al. Involvement of budding yeast Rad5 in translesion DNA synthesis through physical interaction with Rev1. Nucleic Acids Res. 2016;44:5231–5245. doi: 10.1093/nar/gkw183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krishna TS, Fenyo D, Kong XP, Gary S, et al. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 28.Gulbis JM, Kelman Z, Hurwitz J, O'Donnell M, et al. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 29.Hishiki A, Hashimoto H, Hanafusa T, Kamei, et al. Structural basis for novel interactions between human translesion synthesis polymerases and proliferating cell nuclear antigen. J Biol Chem. 2009;284:10552–10560. doi: 10.1074/jbc.M809745200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Armstrong AA, Mohideen F, Lima CD. Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2. Nature. 2012;483:59–63. doi: 10.1038/nature10883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prakash S, Prakash L. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 2002;16:1872–1883. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- 32.Lehmann AR. Replication of damaged DNA. Cell Cycle. 2003;2:300–302. [PubMed] [Google Scholar]
- 33.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 34.Lehmann AR. Replication of damaged DNA by translesion synthesis in human cells. FEBS Lett. 2005;579:873–876. doi: 10.1016/j.febslet.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 35.Washington MT, Carlson KD, Freudenthal BD, Pryor JM. Variations on a theme: eukaryotic Y-family DNA polymerases. Biochim Biophys Acta. 2010;1804:1113–1123. doi: 10.1016/j.bbapap.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol. 2012;13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 38.Nelson JR, Gibbs PE, Nowicka AM, Hinkle DC, et al. Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol Microbiol. 2000;37:549–554. doi: 10.1046/j.1365-2958.2000.01997.x. [DOI] [PubMed] [Google Scholar]
- 39.Gabel SA, DeRose EF, London RE. XRCC1 interaction with the REV1 C-terminal domain suggests a role in post replication repair. DNA Repair. 2013;12:1105–1113. doi: 10.1016/j.dnarep.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wojtaszek J, Liu J, D'Souza S, Wang S, et al. Multifaceted recognition of vertebrate Rev1 by translesion polymerases zeta and kappa. J Biol Chem. 2012;287:26400–26408. doi: 10.1074/jbc.M112.380998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pustovalova Y, Magalhaes MT, D'Souza S, Rizzo AA, et al. Interaction between the Rev1 C-Terminal Domain and the PolD3 Subunit of Polzeta Suggests a Mechanism of Polymerase Exchange upon Rev1/Polzeta-Dependent Translesion Synthesis. Biochemistry. 2016;55:2043–2053. doi: 10.1021/acs.biochem.5b01282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Argueso JL, Kijas AW, Sarin S, Heck J, et al. Systematic mutagenesis of the Saccharomyces cerevisiae MLH1 gene reveals distinct roles for Mlh1p in meiotic crossing over and in vegetative and meiotic mismatch repair. Mol Cell Biol. 2003;23:873–886. doi: 10.1128/MCB.23.3.873-886.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, et al. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nick McElhinny SA, Kissling GE, Kunkel TA. Differential correction of lagging-strand replication errors made by DNA polymerases {alpha} and {delta} Proc Natl Acad Sci U S A. 2010;107:21070–21075. doi: 10.1073/pnas.1013048107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haracska L, Kondratick CM, Unk I, Prakash S, et al. Interaction with PCNA is essential for yeast DNA polymerase eta function. Mol Cell. 2001;8:407–415. doi: 10.1016/s1097-2765(01)00319-7. [DOI] [PubMed] [Google Scholar]
- 46.Acharya N, Haracska L, Prakash S, Prakash L. Complex formation of yeast Rev1 with DNA polymerase eta. Mol Cell Biol. 2007;27:8401–8408. doi: 10.1128/MCB.01478-07. [DOI] [PMC free article] [PubMed] [Google Scholar]