Abstract
Molecular epidemiological research suggests that mouse double minute 4 (MDM4) polymorphisms may be associated with cancer susceptibility, but results remain controversial. To derive a more precise evaluation, we performed a PRISMA compliant meta-analysis focused on five single nucleotide polymorphisms (rs11801299, rs1380576, rs10900598, rs1563828, and rs4245739) of MDM4. Overall, 23 studies involving 22,218 cases and 55,033 controls were analyzed. The results showed that rs4245739 was significantly associated with a decreased cancer risk in the allelic (C vs. A: odds ratio [OR] = 0.848, 95% confidence interval [CI] = 0.765–0.941, P = 0.002), heterozygous (AC vs. AA: OR = 0.831, 95% CI = 0.735–0.939, P = 0.003), and dominant (AC+CC vs. A: OR = 0.823, 95% CI = 0.727–0.932, P = 0.002) models. The association was more prominent in Asians. No significant association was found using any genetic model for the rs11801299, rs1380576, rs10900598, and rs1563828 SNPs. These results indicate that the rs4245739 polymorphism may contribute to a decreased cancer susceptibility and support the hypothesis that genetic variants in the MDM4 genes act as important modifiers of cancer risk.
Keywords: MDM4, polymorphism, cancer susceptibility, meta-analysis
INTRODUCTION
Mouse double minute 4 (MDM4, also known as MDMX) is a member of the MDM family that also includes MDM2 and its derivatives. MDM4 can bind directly to p53 and inhibit its transcriptional activation, as well as mediate various cellular pathways based on p53 [1]. Tumor suppressor protein p53 plays an important role in regulating cell growth, division, and apoptosis. Overactive MDM4 may reduce p53 tumor suppression function and contributes to tumor formation and progression [2]. MDM4 was found to be up-regulated in invasive breast carcinoma (by 14.2%), liver hepatocellular carcinoma (by 12.4%), retinoblastomas (by 65%), skin cutaneous melanomas (by 12%), and stage II–V melanomas (by > 65%) [3].
Molecular epidemiological research suggests that genetic variations in MDM4 gene may be associated with the cancer risk. Recently, a single nucleotide polymorphism (SNP) in the 3′ untranslated region of the MDM4 gene, rs4245739 A > C has been found to affect MDM4 mRNA stability and protein levels [4]. Genotype AA was recorded to be more frequent in patients with high-grade than low-grade ovarian carcinoma [5]. Furthermore, several studies indicated the rs4245739 C allele to be associated with a reduced risk for non-Hodgkin lymphoma [6], breast cancer [7], esophageal squamous cell carcinoma [8], and prostate cancer [9]. In 2009, Atwal and colleagues found that specific SNPs in MDM4 (rs10900594, rs2290853, rs2369244, and rs12039454) may affect p53 tumor-suppression activity [10]. Moreover, the presence of these SNPs in Ashkenazi Jewish and European cohorts has been associated with increased risks of early-onset breast and ovarian cancers. In short, several SNPs in the MDM4 have been associated with elevated or reduced cancer risk, but data are at variance. We therefore performed the PRISMA- compliant meta-analysis of the accumulated information and evaluated the associations of MDM4 polymorphisms with cancer susceptibility.
RESULTS
Characteristics of included studies
Figure 1 shows a flow chart of the studies selection procedure. From 567 initial studies, 497 were discarded at title or abstract level. Another 45 studies did not meet the prespecified inclusion criteria and were therefore excluded. Of the remaining 25 articles, 8 articles were also excluded due to some data being unavailable even after contacting the corresponding authors. Ultimately, 17 articles focusing on the association between MDM4 polymorphisms and cancer risk were identified. Only one of the studies involved rs116197192 and rs4252668, so we did not include these two SNPs in the subsequent meta-analysis [11]. The remaining 16 articles involved the following 5 SNPs: rs11801299 [12–14], rs10900598 [12–14], rs1380576 [12–15], rs1563828 [16–18], and rs4245739 [5–8, 19–23]. Five of the articles described multiple case–control studies of different types of cancer or different populations. Overall, 23 eligible case–control comparisons that involved 22,218 cancer patients and 55,030 controls were enrolled in this meta-analysis, with 3 studies considered to be of low quality (quality score < 10) [5, 11, 16] and 20 were of high quality (quality score ≥ 10) [6–8, 12–15, 17–23]. Within the distribution of genotypes in the control groups, all studies are consistent with Hardy-Weinberg equilibrium (HWE). Table 1 presents the characteristics of the individual studies.
Figure 1. Selection of studies of association between MDM4 genetic variants and cancer susceptibility.
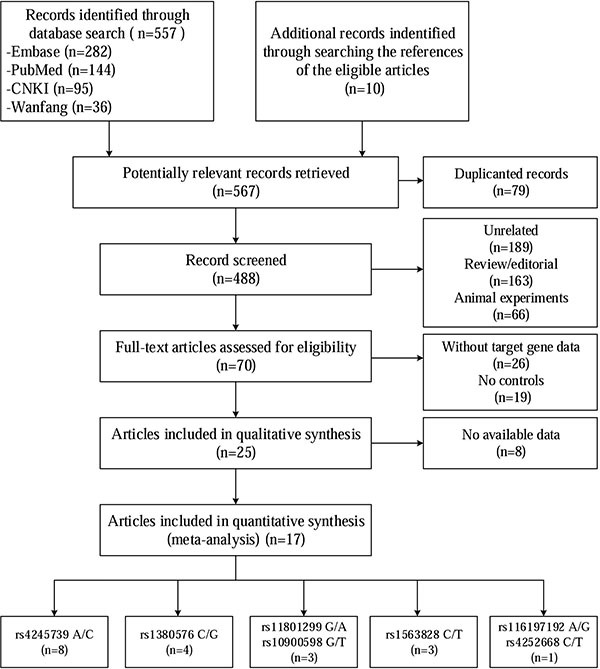
Table 1. Characteristics of studies in the meta-analysis.
| Author | Year | Ethnicity | Genotyping method | Cancer type | Case/control | Control source | HWE | Quality score (0–15) | Polymorphism site |
|---|---|---|---|---|---|---|---|---|---|
| Wynendaele [14] | 2010 | Caucasian | PCR-RFLP | OC | 154/154 | HB | P = 0.982 | 7 | rs4245739 |
| Yu [7] | 2011 | Caucasian | TaqMan | SCCHN | 1075/1079 | HB | P = 0.084 for rs11801299, P = 0.712 for rs1380576, P = 0.398 for rs10900598 | 11 | rs11801299, rs1380576, rs10900598 |
| Oliveira [6] | 2012 | Caucasian, Mullato, Black | PCR/RFLP | RB | 104/104 | PB | P = 0.683 for rs116197192, P = 0.802 for rs4252668 | 8 | rs116197192, rs4252668 |
| Song [11] | 2012 | Asian | MassArray | BC | 124/101 | HB | P = 0.862 | 7 | rs1563828 |
| Wang [8] | 2012 | Caucasian | TaqMan | Oral cancer | 320/321 | HB | Agreement with HWE* | 11 | rs11801299, rs1380576, rs10900598 |
| Yu [9] | 2012 | Caucasian | TaqMan | SCCHN | 380/335 | HB | P = 0.303 for rs11801299, P = 0.502 for rs1380576, P = 0.669 for rs10900598 | 10 | rs11801299, rs1380576, rs10900598 |
| Zhang [12] | 2012 | Asian | RT-PCR | NPC | 210/200 | PB | P = 0.944 | 10 | rs1563828 |
| Garcia [22] | 2013 | Caucasian | Illumina array | BC | 6512/41451 | Mixed | P = 0.183 | 11 | rs4245739 |
| Liu [15] | 2013 | Asian | PCR-RFLP | BC | 800/800 (Jinan); 300/600 (Huaian) | PB | P = 0.505 for Jinan, P = 0.483 for Huaian | 13 | rs4245739 |
| Zhou [16] | 2013 | Asian | PCR-RFLP | ESCC | 540/550 (Jinan); 588/600 (Huaian) | PB | P = 0.740 for Jinan, P = 0.379 for Huaian | 13 | rs4245739 |
| Fan [17] | 2014 | Asian | PCR-RFLP | NHL | 200/400 | PB | P = 0.487 | 12 | rs4245739 |
| Thunell [13] | 2014 | Caucasian | TaqMan | HCM | 50/799 | PB | P = 0.725 | 11 | rs1563828 |
| Feng [18] | 2014 | Asian | PCR-RFLP | GN | 419/494 | HB | P = 0.561 | 10 | rs4245739 |
| Gansmo [19] | 2015 | Caucasian | LightSNiP assay | BC (n = 1,717); LC (n = 1,331); CC (n = 1,531); PC (n = 2,500) | 7079/3747 | PB | P = 0.566 | 13 | rs4245739 |
| Gao [20] | 2015 | Asian | PCR-RFLP | LC | 320/640 (Jinan); 200/400 (Huaian) | PB | P = 0.399 for Jinan, P = 0248 for Huaian | 10 | rs4245739 |
| Wu [10] | 2015 | Asian | TaqMan | GN | 642/720 | PB | P = 0.46 | 13 | rs1380576 |
| Gansmo [21] | 2016 | Caucasian | LightSNiP assay | EC (n = 1404); OC (n = 1385) | 2789/1870 | PB | P = 0.106 | 13 | rs4245739 |
This study only offered dominant-model data for rs11801299, rs1380576, and rs10900598, thus the p value of HWE were not calculated.
BC: breast cancer, CC: colon cancer, EC: endometrial cancer, ESCC: esophageal squamous cell carcinoma, GN: gastric neoplasms, HB: hospital based, HCM: hereditary cutaneous melanoma, LC: lung cancer, NHL: non-Hodgkin lymphoma, NPC: nasopharyngeal carcinoma, OC: ovarian carcinomas, PB: population based, PC: prostate cancer, RB: retinoblastoma, RT-PCR: reverse transcription-PCR, SCCHN: squamous cell carcinoma of the head and neck, SCLC: small cell lung cancer
Quantitative analysis
Table 2 presents the main results of this meta-analysis. The 16 studies of rs4245739 found that this SNP was significantly associated with a decreased cancer risk in the allelic (C vs. A: odds ratio [OR] = 0.848, 95% confidence interval [CI] = 0.765–0.941, P = 0.002), heterozygous (AC vs. AA: OR = 0.831, 95% CI = 0.735– 0.939, P = 0.003), and dominant (AC + CC vs. A: OR = 0.823, 95% CI = 0.727–0.932, P = 0.002) models [5–8, 19–23]. However, the relationship remains controversial in the other genetic models (Figure 2).
Table 2. The result of meta-analysis for various genotype models.
| SNP | Covariates | Variables | No. of studies | Sample size (case/control) | Allele | Homozygous | Heterozygous | Dominant | Recessive | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | Ph/I2 | OR (95%CI) | Ph/I2 | OR (95%CI) | Ph/I2 | OR (95%CI) | Ph/I2 | OR (95%CI) | Ph/I2 | |||||
| rs4245739 A/C | All | 16 | 19950/49914 | 0.848 (0.765-0.941) | <0.001/87.2% | 1.002 (0.866-1.159) | 0.008/52.3% | 0.831 (0.735-0.939) | <0.001/85.0% | 0.823 (0.727-0.932) | <0.001/86.8% | 0.981 (0.394-2.444) | <0.001/99.1% | |
| Ethnicity | Asian | 8 | 3416/4483 | 0.561 (0.439-0.718) | 0.001/71.1% | 0.782 (0.536-1.141) | 0.910/0.0% | 0.547 (0.428-0.698) | 0.007/64.0% | 0.544 (0.428-0.692) | 0.007/64.1% | 0.806 (0.562-1.156) | 0.918/0.0% | |
| Caucasian | 8 | 16534/45431 | 1.022 (0.948-1.101) | <0.001/78.6% | 1.028 (0.875-1.208) | 0.001/71.7% | 1.037 (0.960-1.119) | 0.060/64.3% | 1.034 (0.949-1.126) | <0.001/74.3% | 1.304(0.394-4.316) | <0.001/99.6% | ||
| Cancer type | BC | 3 | 9329/44721 | 0.766 (0.573-1.025) | <0.001/94.3% | 0.937 (0.545-1.608) | <0.001/83.3% | 0.776 (0.571-1.055) | <0.001/92.1% | 0.759 (0.547-1.054) | <0.001/93.7% | 0.945 (0.590-1.512) | 0.002/79.3% | |
| LC | 3 | 1128/1150 | 0.607 (0.308-1.194) | <0.001/90.4% | 1.075 (0.844-1.370) | 0.762/0.0% | 0.584 (0.288-1.185) | <0.001/90.0% | 0.585 (0.839-1.068) | <0.001/90.5% | 1.067 (0.843-1.352) | 0.814/0.0% | ||
| ESCC | 2 | 1851/4787 | 0.616 (0.478-0.793) | 0.438/0.0% | 1.197 (0.318-4.500) | 0.759/0.0% | 0.565 (0.431-0.741) | 0.484/0.0% | 0.580 (0.445-0.757) | 0.464/0.0% | 1.273 (0.339-4.785) | 0.763/0.0% | ||
| OC | 2 | 1539/2023 | 1.065 (0.958-1.183) | 0.767/0.0% | 1.009 (0.778-1.308) | 0.641/0.0% | 1.141 (0.993-1.312) | 0.917/0.0% | 1.119 (0.980-1.278) | 0.841/0.0% | 0.954 (0.741-1.229) | 0.653/0.0% | ||
| Other | 6 | 6103/8314 | 0.960 (0.878-1.049) | 0.080/52.1% | 0.964 (0.841-1.105) | 0.917/0.0% | 0.949 (0.835-1.079) | 0.044/59.1% | 0.947 (0.839-1.068) | 0.049/57.9% | 0.972 (0.851-1.110) | 0.912/0.0% | ||
| Source of controls | HB | 2 | 622/646 | 0.935 (0.790-1.106) | 0.648/0.0% | 0.803 (0.547-1.180) | 0.985/0.0% | 0.999 (0.791-1.262) | 0.598/0.0% | 0.960 (0.769-1.199) | 0.588/0.0% | 0.812 (0.563-1.170) | 0.895 /0.0% | |
| PB | 13 | 12816/18898 | 0.803 (0.714-0.903) | <0.001/83.0% | 0.969 (0.874-1.075) | 0.869/0.0% | 0.768 (0.664-0.890) | <0.001/83.4% | 0.768 (0.667-0.886) | <0.001/83.6% | 0.967 (0.874-1.069) | 0.906 /0.0% | ||
| Mixed | 1 | 6512/41451 | 1.159 (1.112-1.207) | – | 1.355 (1.229-1.494) | – | 1.148 (1.087-1.213) | – | 1.180 (1.120-1.243) | – | 1.277 (1.162-1.405) | – | ||
| rs11801299 G/A | All (Caucasian) | 2 | 1446/1400 | 1.715 (0.531-5.545) | <0.001/98.5% | 3.549 (0.302-41.765) | <0.001/97.2% | 1.583 (0.523-4.794) | <0.001/97.1% | 1.816 (0.471-7.003) | <0.001/98.2% | 2.817 (0.401-19.803) | <0.001/95.6% | |
| rs1380576 C/G | All | 3 | 2088/2120 | 1.018 (0.931-1.114) | 0.393/0.0% | 1.002 (0.831-1.208) | 0.558/0.0% | 1.094 (0.958-1.250) | 0.918/0.0% | 1.070 (0.944-1.212) | 0.771/0.0% | 0.943 (0.797-1.116) | 0.461/0.0% | |
| Ethnicity | Asian | 1 | 642/720 | 0.939 (0.807-1.091) | – | 0.895 (0.676-1.184) | – | 1.093 (0.846-1.411) | – | 1.008 (0.798-1.273) | – | 0.849 (0.671-1.075) | – | |
| Caucasian | 2 | 1446/1400 | 1.065 (0.953-1.191) | 0.735/0.0% | 1.098 (0.853-1.413) | 0.850/0.0% | 1.095 (0.937-1.279) | 0.680/0.0% | 1.096 (0.945-1.270) | 0.681/0.0% | 1.052 (0.827-1.339) | 0.948/0.0% | ||
| rs10900598 G/T | All (Caucasian) | 2 | 1446/1400 | 0.530 (0.163-1.729) | <0.001/98.7% | 0.253 (0.019-3.363) | <0.001/98.2% | 0.559 (0.193-1.615) | <0.001/96.6% | 0.481 (0.126-1.832) | <0.001/98.1% | 0.341 (0.043-2.713) | <0.001/97.4% | |
| rs1563828 C/T | All | 3 | 384/1100 | 0.909 (0.738-1.120) | 0.750/0.0% | 0.768 (0.474-1.244) | 0.642/0.0% | 0.972 (0.728-1.297) | 0.867/0.0% | 0.929 (0.706-1.223) | 0.825/0.0% | 0.776 (0.490-1.229) | 0.657/0.0% | |
| Ethnicity | Asian | 2 | 334/301 | 0.948 (0.750-1.199) | 0.928/0.0% | 0.859 (0.510-1.445) | 0.921/0.0% | 0.999 (0.718-1.392) | 0.678/0.0% | 0.969 (0.708-1.325) | 0.764/0.0% | 0.855 (0.522-1.402) | 0.804/0.0% | |
| Caucasian | 1 | 50/799 | 0.776 (0.488-1.236) | – | 0.412 (0.096-1.770) | – | 0.890 (0.494-1.603) | – | 0.808 (0.456-1.434) | – | 0.434 (0.103-1.823) | – | ||
| Cancer type (Source of controls) | BC (HB) | 1 | 124/101 | 0.962(0.651-1.421) | – | 0.830(0.358-1.926) | – | 1.100(0.627-1.932) | – | 1.034(0.608-1.757) | – | 0.791(0.358-1.747) | – | |
| Other (PB) | 2 | 260/999 | 0.889(0.695-1.138) | 0.493/0.0% | 0.740(0.411-1.335) | 0.350/0.0% | 0.930(0.664-1.301) | 0.859/0.0% | 0.893(0.647-1.232) | 0.680/0.0% | 0.768(0.437-1.352) | 0.358/0.0% | ||
Figure 2.
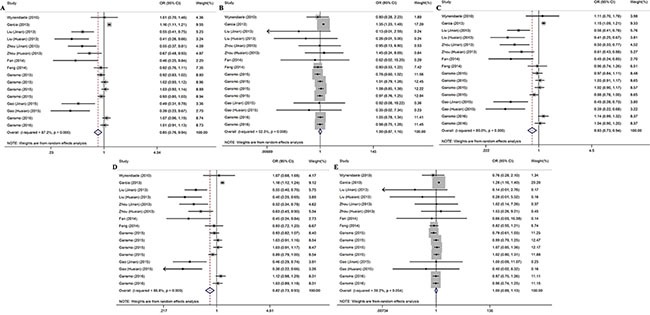
Forest plots for the association between the rs4245739 polymorphisms and cancer risk under five genetic models (A) allelic model; (B) homozygous model; (C) heterozygous model; (D) dominant model; (E) recessive model.
Seven studies examined the associations of the other four SNPs (rs11801299, rs10900598, rs1380576, and rs1563828) with the risk of cancer [12–18], but no significant associations were found (Table 2 and Supplementary Table S2).
Meta regression
The Q statistic and the point estimate (I2) indicated the presence of high heterogeneity between studies in the meta-analysis of rs4245739, rs11801299, and rs10900598 (i.e., P < 0.10 and/or I2 > 50%).
For the meta-analysis of rs4245739, which involved more than 10 studies, we performed a meta-regression to determine the potential source of heterogeneity. Table 3 indicates that the main sources of significant heterogeneity were ethnicity (P < 0.001) and genotyping methods (P < 0.010).
Table 3. The results of meta-regression for rs4245739.
| Covariates | Number of dummy variablesa | C vs. A | CC vs. AA | AC vs. AA | AC + CC vs. AA | CC vs. AC + AA |
|---|---|---|---|---|---|---|
| Publication year | – | 0.518 | 0.279 | 0.536 | 0.536 | 0.133 |
| Ethnicity | 2 | < 0.001 | 0.238 | < 0.001 | < 0.001 | 0.281 |
| Cancer type | 5 | 0.166 | 0.065 | 0.086 | 0.119 | 0.061 |
| Genotyping methods | 3 | 0.006 | 0.001 | 0.008 | 0.004 | 0.002 |
| Source of controls | 3 | 0.204 | – | 0.229 | 0.200 | 0.002 |
The Bonferroni correction was used according the number of dummy variables. The statistical significance level that should be used for each covariate separately is 0.050, 0.025, 0.010, 0.017, and 0.017 respectively.
Subgroup analysis
The subgroup analysis stratified by ethnicity indicated that rs4245739 decreased the risk of cancer in the allelic (C vs. A: OR = 0.561, 95% CI = 0.439–0.718, P < 0.001), heterozygous (AC vs. AA: OR = 0.547, 95% CI = 0.428–0.698, P < 0.001), and dominant (AC+CC vs. A: OR = 0.544, 95% CI = 0.428–0.692, P < 0.001) models in Asian but not Caucasian populations. Similarly, significant correlations with reduced cancer risk were also observed with three genetic models in population-based control groups (C vs. A: OR = 0.803, 95% CI = 0.714–0.903, P < 0.001; AC vs. AA: OR = 0.768, 95% CI = 0.664–0.890, P < 0.001; AC + CC vs. A: OR = 0.768, 95% CI = 0.667–0.886, P < 0.001). Moreover, reduced risks of esophageal squamous cell carcinoma was detected (Table 2).
Subgroup analyses were also performed for the other four SNPs, but no significant association was found (Table 2).
Sensitivity analysis
The results of the leave-one-out analysis of rs4245739 indicated that no individual study excessively influenced the pooled effect in any genetic models of the meta-analysis (Figure 3). Removing the three studies [5, 11, 16] considered to be of low quality from the meta-analysis of rs4245739 and rs1563828 did not change the results significantly (Supplementary Table S3). For rs11801299, rs1380576, and rs10900598, we added a study [13] that only produced dominant-model data, but no conspicuous change in the pooled ORs was detected (Supplementary Table S3).
Figure 3.
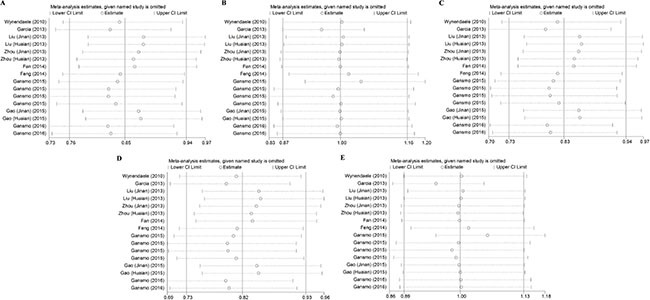
Leave-one-out analysis of association between the rs4245739 polymorphisms and cancer risk under five genetic models (A) allelic model; (B) homozygous model; (C) heterozygous model; (D) dominant model; (E) recessive model.
Publication bias
Figure 4 shows funnel plots for the meta-analysis of rs4245739. The funnel plots are asymmetrical and Egger's test indicated the presence of significant publication bias in five genetic models (Egger's test: P < 0.001 for allele, heterozygous and dominant models; P = 0.010 for homozygous model; P = 0.011 for recessive model). The results did not change after the correction using the trim and fill method. We also calculated the fail-safe numbers for the positive results, which were 80 for the allelic, heterozygous, and dominant genetic models. This suggests there would need to be 80 unpublished studies to render the findings of the meta-analysis nonsignificant.
Figure 4.
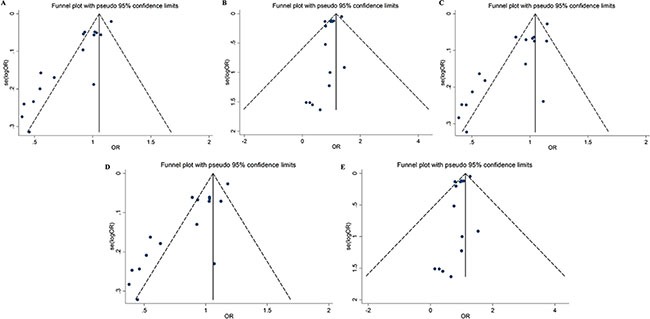
Funnel plots for the association between the rs4245739 polymorphisms and cancer risk under five genetic models (A) allelic model; (B) homozygous model; (C) heterozygous model; (D) dominant model; (E) recessive model.
Publication bias was not tested for the meta-analysis of the other SNPs due to the small number of included studies (i.e., less than five each).
DISCUSSION
How genetic factors influence the susceptibility of patients to cancer is receiving increasing attention [24, 25]. Prompted by the important role of MDM4 in the development of cancer, we have conducted the first comprehensive meta-analysis of the relationships between MDM4 polymorphisms and the risk of cancer. The results showed that the MDM4 rs4245739 polymorphism is associated with a significantly decreased risk of cancer. A subgroup analysis by ethnicity revealed that carriers of the C allele and mutated genotypes had a significantly lower cancer risk than Asian wild-type carriers, suggesting that the decreased cancer risk is ethno-specific. In addition, the other SNPs of MDM4 analyzed (i.e., rs11801299, rs1380576, rs10900598, and rs1563828) were not found to be associated with the risk of cancer.
The MDM4 gene is located on chromosome 1q32, which is an important regulator of the p53 pathway in vivo [2]. Elevated expression of MDM4 has been seen in both relatively rare (e.g., retinoblastoma and ocular melanoma) and more common (e.g., cutaneous melanoma and breast cancer) types of tumor [26]. There is also an increasing recognition that MDM4 is a promising and relatively safe therapeutic target for p53 reactivation therapy.
SNPs are the most common variations in genetic sequences and can alter the splicing of primary transcripts or gene expression, and may further affect the susceptibility, progression, and prognosis of diseases [27]. Functional SNPs have been identified in the MDM4 gene since 2009. Rs2279744 was reported under positive evolutionary selection to be associated with the risk of breast and ovarian cancers and with human fertility in Caucasian populations [10, 28], while rs1563828 was found to be associated with an earlier age at the diagnosis of estrogen-receptor-negative but not estrogen-receptor-positive breast cancers[29]. In the last decades, rs4245739 A > C was widely studied, which was found to create a functional target site for hsa-miR-191 and hsa-miR-887. Both miRs bind to the C-allele with higher affinity than to the A-allele, leading to miR-mediated decrease in MDM4 protein levels in cells carrying the C variant [4, 5]. Several case–control studies assessing this mutation in various cancer forms (esophageal squamous cell carcinoma, non-Hodgkin lymphoma, breast cancer, and prostate cancer), have found the C-allele to be associated with reduced risk, but have all been performed in Chinese populations [6–9]. Notably, there is a substantial difference in the distribution of this SNP between Europeans and Asians with a MAF of 0.26 and 0.05, respectively [30]. The somewhat variable results regarding MDM4 rs4245739 and cancer risk may also be explained by yet unknown functional SNP (s) that are in linkage disequilibrium with rs4245739 [20].
This is the first meta-analysis to evaluate whether SNPs in the MDM4 gene are associated with cancer risk. We screened more than 500 titles and abstracts, and found that the relevant literature is focused on 5 SNPs (rs11801299, rs1380576, rs10900598, rs1563828, and rs4245739), and especially rs4245739. The reported research has involved various types of cancer, including ovaries, lung, stomach and other cancers. When performing meta-analyses, it is strongly recommended to investigate the influence of potential heterogeneity factors in order to avoid drawing simplistic and potentially misleading conclusions [31]. We therefore performed meta-regression, sensitivity analysis, and subgroup analysis in the present study. The main finding of the meta-analysis is that rs4245739 is significantly associated with a decreased cancer risk in the allelic, heterozygous, and dominant models. Due to the presence of significant heterogeneity, we conducted meta-regression by publication year, ethnicity, cancer type, genotyping method, and source of controls; the results highlighted ethnicity and genotyping method as a major driver of heterogeneity. Accordingly, subgroup analyses were performed based on ethnicity, and significant associations were only found in Asian populations. We also performed a sensitivity analysis to evaluate the robustness of the results, which revealed that no single study substantially changed the corresponding pooled ORs and 95% CIs. Five indices of publication bias were examined: the funnel plots and Begg's and Egger's linear regression tests indicated significant publication bias; however, using the trim-and-fill analysis correction results did not change, and the fail-safe number was larger than the number of included studies, which suggests that the positive results were robust despite the existence of publication bias.
Some limitations of this meta-analysis should be considered when interpreting its findings. Firstly, although we applied a highly sensitive search strategy to retrieve potentially eligible studies, we cannot rule out the possibility that some relevant studies were overlooked. Secondly, the number of eligible studies for the analyzed SNPs was small, which may have resulted in the statistical power being insufficient to detect weak but significant associations. Thirdly, most of the studies included in this meta-analysis involved Caucasian and Asian populations, and so further studies involving other ethnic populations are required. Finally, this study was a meta-analysis with a case–control design, and so the presence of confounding should be considered.
In summary, this meta-analysis has demonstrated that the MDM4 rs4542739 polymorphism was associated with decreased cancer risk, especially in Asian populations. However, due to the limitations listed above, the findings of this investigation should be interpreted with caution. Well-designed, multicenter, and large-cohort studies are needed to confirm our findings in the future.
MATERIALS AND METHODS
This meta-analysis was performed according to the guidelines described in the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-analyses) statement [32] (PRISMA Checklist see Supplementary Table S1).
Search strategy and study selection
To identify all published studies related to the relationships between MDM4 polymorphisms and cancer risk, we searched the following databases up to June 23, 2016: PubMed, Embase, China National Knowledge Infrastructure (CNKI, http://www.cnki.net/), and Wanfang Data (WD, https://www.wanfangdata.net/). The following MeSH (Medical Subject Heading) terms and/or text words were used in PubMed: “MDM4 protein, human,” “polymorphism, single nucleotide,” “genotype,” “mutation,” “alleles,” “genetic variation,” “neoplasms,” and “carcinoma.” The following Emtree terms were also used in Embase: “protein mdmx,” “genetic polymorphism,” “single nucleotide polymorphism,” “genotype phenotype correlation,” “mutation,” “allele,” “genetic variability,” “neoplasm,” and “carcinoma” (The full search strings see Supplementary Methods). We searched the CNKI and WD databases using the Chinese characters corresponding to these keywords. The reference lists in articles retained for review were also examined manually to further identify potentially relevant reports.
All of the studies included in the current analysis needed to meet the following criteria: (i) involved an assessment of the relationship between MDM4 polymorphism and cancer risk, (ii) had a case–control design, and (iii) provided sufficient information to estimate OR and 95% CI values.
Data extraction
The following information about each study was extracted: first author's name, publication year, race, genotyping methods, cancer type, numbers of cases and controls, source of controls, and P value for HWE in controls. Publications were classified as involving different studies if they contained subjects with different cancer types or populations. All SNPs included in the subsequent meta-analysis were represented using dbSNP identifiers (i.e., rs numbers). For papers that did not report genotype or allele distributions, we sought the genotype information by directly e-mailing the first or corresponding author.
Data extraction was performed independently by two of the authors (Y.J.Z. and Z.J.D.), with any disagreements resolved by consensus.
Quality score assessment
Two independent investigators (Y.J.Z. and Z.J.D.) assessed the quality of eligible studies using quality scoring criteria modified from those used in previous meta-analyses (Supplementary Table S4) [33, 34]. These modified criteria were based on traditional quality scoring protocols used for observational studies involving genetic epidemiology, and the scores ranged from 0 points (worst quality) to 15 points (best quality) [35]. The studies were then dichotomized into those of low quality (score < 10) and high quality (score ≥ 10).
Statistical analysis
The associations between MDM4 polymorphisms and the risk of cancer were measured by ORs with 95% CIs based on five genetic models: allelic model, homozygous model, heterozygous model, dominant model, and recessive model. Statistical heterogeneity across studies was assessed using Cochran's Q test and I2 statistics, for which P < 0.1 and/or I2 > 50% indicated the presence of significant heterogeneity. The DerSimonian and Laird random-effects methods were used to calculate the OR if significant heterogeneity was present; otherwise the Mantel-Haenszel fixed-effects model was applied. Meta-regression analysis was undertaken to explore potential sources of heterogeneity across studies when statistical heterogeneity was detected. The covariates included publication year, ethnicity, cancer type, genotyping methods, and source of controls. In order to avoid false-positive results, the Bonferroni method was used to adjust the significance level of each covariate. Subgroup analyses were performed based on ethnicity cancer type, and source of controls.
Leave-one-out sensitivity analysis was conducted by sequentially excluding individual studies one at a time and recalculating ORs to evaluate the stability of the results. We also performed a sensitivity analysis by excluding the low-quality studies and computing the ORs for high-quality studies only. Publication bias was assessed using the funnel plot, Begg's and Egger's linear regression test, and the trim-and-fill method [36]. The fail-safe number of negative studies that would be required to nullify the effect size (i.e., to make P > 0.05) was also calculated [37].
A P value of < 0.05 (two-sided) was considered statistically significant. All statistical analyses were performed using Stata software (version 12.0, Stata Corporation, College Station, TX).
SUPPLEMENTARY MATERIALS TABLES
Acknowledgments
J.L., Y.J.Z. and Z.J.D. designed and wrote this article. Y.J.Z. and Z.J.D. screened out relevant articles and extracted data. H.R.H., F.G., L.H.Y., and Y.L.D. analyzed data. All authors approved the final version.
Footnotes
CONFLICTS OF INTEREST
All authors declare no conflicts of interest.
GRANT SUPPORT
This work was supported by the National Social Science Foundation of China (no. 16BGL183), the Natural Science Foundation of Shaanxi Province (no. 2015JM8415) and the Fundamental Research Funds for the Central Universities of China (no. 2011jdhz55).
REFERENCES
- 1.Marine JC, Dyer MA, Jochemsen AG. MDMX: from bench to bedside. J Cell Sci. 2007;120:371–8. doi: 10.1242/jcs.03362. [DOI] [PubMed] [Google Scholar]
- 2.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–23. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 3.Wasylishen AR, Lozano G. Attenuating the p53 Pathway in Human Cancers: Many Means to the Same End. Cold Spring Harb Perspect Med. 2016. [DOI] [PMC free article] [PubMed]
- 4.Stegeman S, Moya L, Selth LA, Spurdle AB, Clements JA, Batra J. A genetic variant of MDM4 influences regulation by multiple microRNAs in prostate cancer. Endocr Relat Cancer. 2015;22:265–76. doi: 10.1530/ERC-15-0013. [DOI] [PubMed] [Google Scholar]
- 5.Wynendaele J, Bohnke A, Leucci E, Nielsen SJ, Lambertz I, Hammer S, Sbrzesny N, Kubitza D, Wolf A, Gradhand E, Balschun K, Braicu I, Sehouli J, et al. An illegitimate microRNA target site within the 3′ UTR of MDM4 affects ovarian cancer progression and chemosensitivity. Cancer Res. 2010;70:9641–9. doi: 10.1158/0008-5472.CAN-10-0527. [DOI] [PubMed] [Google Scholar]
- 6.Fan C, Wei J, Yuan C, Wang X, Jiang C, Zhou C, Yang M. The functional TP53 rs1042522 and MDM4 rs4245739 genetic variants contribute to non-hodgkin lymphoma risk. PLoS ONE. 2014. p. 9. [DOI] [PMC free article] [PubMed]
- 7.Liu J, Tang X, Li M, Lu C, Shi J, Zhou L, Yuan Q, Yang M. Functional MDM4 rs4245739 genetic variant, alone and in combination with P53 Arg72Pro polymorphism, contributes to breast cancer susceptibility. Breast Cancer Research and Treatment. 2013;140:151–7. doi: 10.1007/s10549-013-2615-x. [DOI] [PubMed] [Google Scholar]
- 8.Zhou L, Zhang X, Li Z, Zhou C, Li M, Tang X, Lu C, Li H, Yuan Q, Yang M. Association of a Genetic Variation in a miR-191 Binding Site in MDM4 with Risk of Esophageal Squamous Cell Carcinoma. PLoS ONE. 2013. p. 8. [DOI] [PMC free article] [PubMed]
- 9.Eeles RA, Olama AA, Benlloch S, Saunders EJ, Leongamornlert DA, Tymrakiewicz M, Ghoussaini M, Luccarini C, Dennis J, Jugurnauth-Little S, Dadaev T, Neal DE, Hamdy FC, et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet. 2013;45:385–91. 91e1–2. doi: 10.1038/ng.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atwal GS, Kirchhoff T, Bond EE, Montagna M, Menin C, Bertorelle R, Scaini MC, Bartel F, Bohnke A, Pempe C, Gradhand E, Hauptmann S, Offit K, et al. Altered tumor formation and evolutionary selection of genetic variants in the human MDM4 oncogene. Proc Natl Acad Sci U S A. 2009;106:10236–41. doi: 10.1073/pnas.0901298106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Oliveira Reis AH, de Carvalho IN, de Sousa Damasceno PB, Ferman SE, Lucena E, Lopez-Camelo JS, Seuanez HN, Vargas FR. Influence of MDM2 and MDM4 on development and survival in hereditary retinoblastoma. Pediatr Blood Cancer. 2012;59:39–43. doi: 10.1002/pbc.24014. [DOI] [PubMed] [Google Scholar]
- 12.Yu H, Wang LE, Liu Z, Wei S, Li G, Sturgis EM, Wei Q. Polymorphisms of MDM4 and risk of squamous cell carcinoma of the head and neck. Pharmacogenetics and Genomics. 2011;21:388–96. doi: 10.1097/FPC.0b013e32834632e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z, Sturgis EM, Zhang Y, Huang Z, Zhou Q, Wei Q, Li G. Combined p53-related genetic variants together with HPV infection increase oral cancer risk. International Journal of Cancer. 2012;131:E251–E8. doi: 10.1002/ijc.27335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu H, Sturgis EM, Liu Z, Wang LE, Wei Q, Li G. Modifying effect of MDM4 variants on risk of HPV16-associated squamous cell carcinoma of oropharynx. Cancer. 2012;118:1684–92. doi: 10.1002/cncr.26423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu GC, Zhang ZT. Genetic association of single nucleotide polymorphisms in P53 pathway with gastric cancer risk in a Chinese Han population. Medical Oncology. 2015;32:1–5. doi: 10.1007/s12032-014-0401-1. [DOI] [PubMed] [Google Scholar]
- 16.Song CG, Fu FM, Wu XY, Wang C, Shao ZM. [Correlation of polymorphism rs1563828 in MDM4 gene with breast cancer risk and onset age]. [Article in Chinese] Zhonghua Wai Ke Za Zhi. 2012;50:53–6. [PubMed] [Google Scholar]
- 17.Zhang YW, Guan J, Zhang Y, Qiu YR, Chen LH. Role of an MDM4 polymorphism in the early age of onset of nasopharyngeal carcinoma. Oncology Letters. 2012;3:1115–8. doi: 10.3892/ol.2012.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thunell LK, Bivik C, Waster P, Fredrikson M, Stjernstrom A, Synnerstad I, Rosdahl I, Enerback C. MDM2 SNP309 promoter polymorphism confers risk for hereditary melanoma. Melanoma Res. 2014;24:190–7. doi: 10.1097/CMR.0000000000000063. [DOI] [PubMed] [Google Scholar]
- 19.Feng JB, Nie YQ, Lin Y, Li YF, Du YL. Association between single nucleotide polymorphism at miR-191 target site and susceptibility to gastric cancer. The Journal of Practical Medicine. 2014. pp. 1217–20.
- 20.Gansmo LB, Romundstad P, Birkeland E, Hveem K, Vatten L, Knappskog S, Lonning PE. MDM4 SNP34091 (rs4245739) and its effect on breast-, colon-, lung-, and prostate cancer risk. Cancer Med. 2015;4:1901–7. doi: 10.1002/cam4.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao F, Xiong X, Pan W, Yang X, Zhou C, Yuan Q, Zhou L, Yang M. A regulatory MDM4 genetic variant locating in the binding sequence of multiple MicroRNAs contributes to susceptibility of small cell lung cancer. PLoS ONE. 2015. p. 10. [DOI] [PMC free article] [PubMed]
- 22.Gansmo LB, Bjornslett M, Halle MK, Salvesen HB, Dorum A, Birkeland E, Hveem K, Romundstad P, Vatten L, Lonning PE, Knappskog S. The MDM4 SNP34091 (rs4245739) C-allele is associated with increased risk of ovarian-but not endometrial cancer. Tumour Biol. 2016. [DOI] [PMC free article] [PubMed]
- 23.Garcia-Closas M, Couch FJ, Lindstrom S, Michailidou K, Schmidt MK, Brook MN, Orr N, Rhie SK, Riboli E, Feigelson HS, Le Marchand L, Buring JE, Eccles D, et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat Genet. 2013;45:392–8. 8e1–2. doi: 10.1038/ng.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–9. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 25.Marshall H. Genetic and epigenetic factors in development of lung cancer. Lancet Oncol. 2012;13:1188. doi: 10.1016/s1470-2045(12)70523-5. [DOI] [PubMed] [Google Scholar]
- 26.Marine JC, Jochemsen AG. MDMX (MDM4), a Promising Target for p53 Reactivation Therapy and Beyond. Cold Spring Harb Perspect Med. 2016. p. 6. [DOI] [PMC free article] [PubMed]
- 27.Liu H, Zhou Y, Liu Q, Xiao G, Wang B, Li W, Ye D, Yu S. Association of miR-608 rs4919510 polymorphism and cancer risk: a meta-analysis based on 13,664 subjects. Oncotarget. 2016. [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 28.Kang HJ, Feng Z, Sun Y, Atwal G, Murphy ME, Rebbeck TR, Rosenwaks Z, Levine AJ, Hu W. Single-nucleotide polymorphisms in the p53 pathway regulate fertility in humans. Proc Natl Acad Sci U S A. 2009;106:9761–6. doi: 10.1073/pnas.0904280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kulkarni DA, Vazquez A, Haffty BG, Bandera EV, Hu W, Sun YY, Toppmeyer DL, Levine AJ, Hirshfield KM. A polymorphic variant in human MDM4 associates with accelerated age of onset of estrogen receptor negative breast cancer. Carcinogenesis. 2009;30:1910–5. doi: 10.1093/carcin/bgp224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson SG. Why sources of heterogeneity in meta-analysis should be investigated. BMJ. 1994;309:1351–5. doi: 10.1136/bmj.309.6965.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. 2009;339:b2535. doi: 10.1136/bmj.b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Wu XS, He J, Ma T, Lei W, Shen ZY. A novel TP53 variant (rs78378222 A > C) in the polyadenylation signal is associated with increased cancer susceptibility: Evidence from a meta-analysis. Oncotarget. 2016;7:32854–65. doi: 10.18632/oncotarget.9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niu YM, Du XY, Cai HX, Zhang C, Yuan RX, Zeng XT, Luo J. Increased risks between Interleukin-10 gene polymorphisms and haplotype and head and neck cancer: a meta-analysis. Sci Rep. 2015;5:17149. doi: 10.1038/srep17149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng X, Zhang Y, Kwong JS, Zhang C, Li S, Sun F, Niu Y, Du L. The methodological quality assessment tools for preclinical and clinical studies, systematic review and meta-analysis, and clinical practice guideline: a systematic review. J Evid Based Med. 2015;8:2–10. doi: 10.1111/jebm.12141. [DOI] [PubMed] [Google Scholar]
- 36.Zhang L, Chen LM, Wang MN, Chen XJ, Li N, Huang YD, Chen M. The G894t, T-786c and 4b/a polymorphisms in Enos gene and cancer risk: a meta-analysis. J Evid Based Med. 2014;7:263–9. doi: 10.1111/jebm.12126. [DOI] [PubMed] [Google Scholar]
- 37.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


