Abstract
Aromatase inhibitor (AI) resistance during breast cancer treatment is mimicked by MCF-7:5C (5C) and MCF-7:2A (2A) cell lines that grow spontaneously. Survival signaling is reconfigured but cells are vulnerable to estradiol (E2)-inducible apoptosis. These model systems have alterations of stress related pathways including the accumulation of endoplasmic reticulum, oxidative, and inflammatory stress that occur prior to E2-induced apoptosis. We investigated miRNA expression profiles of 5C and 2A to characterize their AI resistance phenotypes. Affymetrix GeneChip miRNA2.0 arrays identified 184 miRNAs differentially expressed in 2A and 5C compared to E2-free wild-type MCF-7:WS8. In 5C, 34 miRNAs of the DLK1-DIO3 locus and miR-31 were overexpressed, whereas miR-222 was low. TCGA data revealed poor and favorable overall survival for low miR-31 and miR-222 levels, respectively (HR=3.0, 95% CI:1.9-4.8; HR=0.3, 95% CI:0.1-0.6). Targets of deregulated miRNAs were identified using CLIP-confirmed TargetScan predictions. KEGG enrichment analyses for 5C- and 2A-specific target gene sets revealed pathways associated with cell proliferation including insulin, mTOR, and ErbB signaling as well as immune response and metabolism. Key genes overrepresented in 5C- and 2A-specific pathway interaction networks including EGFR, IGF1R and PIK3R1 had lower protein levels in 5C compared to 2A and were found to be differentially modulated by respective miRNA sets. Distinct up-regulated miRNAs from the DLK1-DIO3 locus may cause these attenuative effects as they are predicted to interact with corresponding 3′ untranslated regions. These new miRNA profiles become an important regulatory database to explore E2-induced apoptotic mechanisms of clinical relevance for the treatment of resistant breast cancer.
Keywords: breast cancer, AI resistance, microRNA profiling, pathway enrichment, E2-inducible apoptosis
INTRODUCTION
Long-term estrogen deprivation is the standard of care in patients with estrogen receptor (ER)-positive breast cancer. Two proven treatment options exist: tamoxifen, a selective ER modulator which blocks 17ß-estradiol (E2) binding to ER to stop tumor growth, and aromatase inhibitors (AI), which block the aromatase enzyme that prevents the conversion of androgens to estrogens. Despite their well-established effectiveness [1, 2], patients frequently display de novo or acquired resistance which ultimately leads to disease progression and death.
Laboratory models of long-term E2-deprived breast cancer cells provide a valuable surrogate approach to study clinical AI resistance. The goal is to discover biomarkers and new therapeutic targets to subvert AI resistance. Long-term E2-deprivation selects for tumor cells that can grow spontaneously, but eventually, through clonal selection, become vulnerable to E2-inducible apoptosis [3]. Knowledge of the mechanisms of endocrine resistance originally evolved from in vivo studies in which MCF-7 cells were inoculated into athymic mice treated with E2 for tumor growth and tamoxifen for growth inhibition. Endocrine resistance evolves over time and manifests in phase I and phase II of which the latter refers to the cellular reprogramming towards E2-inducible apoptosis, for which clinical evidence exists.
Phase I-resistance occurs after one year of tamoxifen treatment in vivo in that growth becomes dependent on tamoxifen or estradiol [4, 5]. Clinical correlates are disease progression during tamoxifen therapy with breast cancer recurrence and development of metastasis. Phase II-resistance develops during 5 years of continuous passage of MCF-7 tumors in tamoxifen-treated athymic mice and refers to the reconfiguration of survival signaling [5]. While tumors still grow in response to tamoxifen treatment, they now rapidly regress with E2, referred to as E2-induced apoptosis. Clinical correlates are the successful treatment of metastatic and endocrine refractory breast cancer with high and/or low dose E2. Although estrogen was the first chemotherapy applied to cancer more than 70 years ago, it is recently being re-investigated with clinical trials [3]. To this end, 15 mg daily diethylstilbestrol (DES) showed a 30% response rate including some complete responses when given to patients with metastatic breast cancer exhaustively treated with anti-hormones [6]. Similarly, patients initially responding but then failing AI treatment benefitted from low (6 mg daily) and high dose E2 (30 mg daily) treatment with fewer toxic effects observed at low dose [7]. Notably, the antitumor effect is most effective when preceded by long-term E2-deprivation as is naturally the case in women 10 years beyond menopause. Additional supportive evidence comes from the Women's Health Initiative (WHI) trial on the health benefits of postmenopausal hormone replacement therapy where treatment of low concentrations of estrogen alone controlled the onset of breast cancer in epithelia with prior E2-deprivation, an effect that was most evident in postmenopausal hysterectomized women in their sixties [8].
The antihormone-resistant breast cancer models MCF-7:5C (5C) and MCF-7:2A (2A) were previously generated through clonal selection following long-term E2-deprivation of ER-positive MCF-7 cells [9, 10]. Their adaptation to E2-deficiency resulted in a wide range of alterations of stress-related pathways which became evident from global gene expression profiles [11]. As an example, both 5C and 2A cells have elevated basal expression levels of JNK (MAPK8), but not p38 (MAPK14) [12]. Moreover, many genes associated with response to stress, including inflammation (e.g. TNFRSF11B, CXCR4, TNF), oxidative stress (e.g. APOE, GPX2, SOD2), endoplasmic reticulum stress (e.g. EIF2AK3, ATF6, ERN1), as well as other stress-related kinases (e.g. SGK) have been altered after E2-deprivation in both cell lines [12]. The 5C cells undergo E2-induced apoptosis within seven days of E2-treatment, whereas 2A cells require two weeks. As 2A cells employ stronger antioxidant defense mechanisms compared to 5C cells, they ultimately require oxidative stress in order to die later in the response to E2-treatment [13]. At the cellular level, ERα is the target site for E2 to induce apoptosis, which can be completely blocked by 4-hydroxytamoxifen (4-OHT) or ERα knockdown [14, 15]. The accumulation of endoplasmic reticulum stress, oxidative stress, and inflammatory stress prior to E2-induced apoptosis [11] can be blocked by glucocorticoids and inhibition of c-Src [14, 16]. In particular, the endoplasmic reticulum functions as a key regulatory site for the cell fate decision after E2-treatment in both E2-deprived cell lines [12]. E2 first induces an unfolded protein response [13, 14] which is followed by reduced protein translation via protein kinase-like endoplasmic reticulum kinase (PERK) attenuation [14]. Simultaneously, the folding capacity of the endoplasmic reticulum is increased by activating transcription factor (ATF)-6 and inositol-requiring kinase-1 (IRE1) via up-regulation of endoplasmic reticulum chaperones and the endoplasmic reticulum-associated protein degradation (ERAD) machinery [12].
It is important to expand the known biology of AI resistance and E2-induced apoptosis to the level of gene regulation. MicroRNAs (miRNAs) have emerged as master regulators of gene expression and already have been proven to be of clinical use [17, 18]. MiRNAs are a family of short (22-24 nucleotides), non-coding, single-stranded RNA molecules that among many different biological processes regulate apoptosis and oncogenesis [19, 20]. In breast cancer numerous miRNAs regulate the ERα and vice versa. Examples are reviewed in Klinge et al. [21] and include the ERα regulators miR-22, miR-206, miR-221/−222, miR-18a, miR-18b, miR-193b and miR-302c. miR-221 and miR-222 show low expression in ERα-positive cell lines but are overexpressed in 4-OHT-resistant MCF-7 derivative cells as well as in ER-negative breast cancer [22]. Other miRNAs potentially involved in the development of endocrine resistance include miR-15a and miR-16 which suppress the antiapoptotic Bcl2 [23], miR-342 which re-sensitizes MCF-7 cell derivatives to tamoxifen [24], and miR-301, the blockade of which increases the tamoxifen sensitivity of MCF-7 cells [25]. Despite a growing body of literature on miRNAs in tamoxifen resistance little is known on the relevance of miRNAs in AI resistance [26]. Recently, 78 miRNAs were identified to be differentially expressed between MCF-7 and 2A cells including the ER-regulated let-7c, miR-99a, and miR-125b supporting a putative role in endocrine resistance [27].
Here we use AI-resistant breast cancer cell models and report miRNA candidates and their targeted pathways for ‘E2-independent growth’ and ‘E2-inducible apoptosis’ phenotypes.
RESULTS
miRNAs involved in AI resistance and E2-inducible apoptosis phenotypes
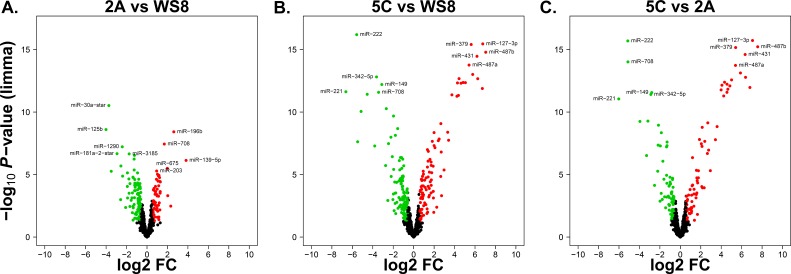
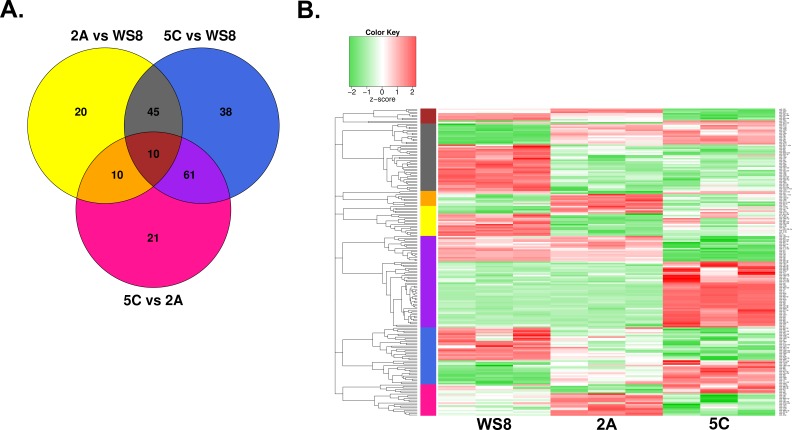
Top up- and down-regulated miRNA candidates were obtained from the pairwise comparisons 2A versus WS8, 5C versus WS8, and 5C versus 2A (Figure 1). We identified 85 relevantly differentially and significantly expressed miRNAs when comparing 2A versus WS8 (FC > 1.5 or < 1/1.5, P < 0.05) and 154 miRNAs for 5C versus WS8 (Supplementary Tables 1 and 2). Here, 2A cells revealed 31 upregulated miRNAs (top candidates: miR-196b, -708, -139-5p, -675, and -203), and 54 down-regulated miRNAs (top candidates: miR-30a-star, -125b, -1290, -181a-2-star, and -3185) (Figure 1A). 5C cells revealed 76 upregulated miRNAs (top candidates: miR-127-3p, -379, -487b, -431, and -487a), and 78 down-regulated miRNAs (top candidates: miR-222, -342-5p, -149, -221, and -708) (Figure 1B). The 5C versus 2A comparison revealed 102 significantly and relevantly differentially expressed miRNAs (Supplementary Table 3) of which 54 are up- and 48 down-regulated matching the top five 5C versus WS8 candidates (Figure 1C). The overlap between the 2A versus WS8 comparison and the 5C versus WS8 comparison disclosed a set of 55 miRNAs that could be relevant for E2-independent growth (Figure 2A). Overall, 30 miRNAs are specifically deregulated in 2A versus WS8, 99 in 5C versus WS8 and 21 additional candidates resulted from the direct comparison (Figure 2A and 2B). In total, 205 miRNAs were significantly and relevantly differentially expressed in at least one of the three pairwise comparisons (2A vs WS8, 5C vs WS8, 5C vs 2A). Figure 2B illustrates the relative expression levels (z-scores) for each of these miRNAs, ordered according to the seven subsets displayed in Figure 2A.
Figure 1. Volcano plots illustrating the differentially expressed miRNAs of breast cancer AI resistance cell models.
X-axes: log2 FC (fold change); Y-axes: -log10 P-value from limma analysis. miRNAs with P-value < 0.05 and FC > 1.5 are marked in red, with P-value < 0.05 and FC < 1/1.5 in green, all others in black. The top five significantly up- down-regulated miRNAs are labeled. Pairwise comparisons: A. 2A versus WS8. B. 5C versus WS8. C. 5C versus 2A.
Figure 2. miRNA subsets associated with the phenotypes of AI resistance and E-inducible apoptosis.
Overall, 205 miRNAs were significantly and relevantly (Holm-adjusted P-value < 0.05 and FC > 1.5 or < 1/1.5) differentially expressed in at least one of the three pairwise comparisons (2A vs WS8, 5C vs WS8, 5C vs 2A). A. Venn diagram of the three comparisons with numbers of significantly and relevantly differentially expressed miRNAs. There are 55 candidates defining the miRNA set relevant for E2-independent growth (grey, red), 99 miRNAs are 5C-specific (blue, purple), and 30 miRNAs are 2A-specific (yellow, orange). The direct comparison of 5C versus 2A highlights 21 miRNAs (pink). B. Heatmap of the 205 miRNAs comparing WS8, 2A, and 5C with three replicates per cell line. Left color bar matches with colored subsets in A. Within each of the seven color-coded subsets, miRNAs were grouped by complete-linkage hierarchical clustering based on a Pearson correlation distance metric. Corresponding dendrograms are shown.
A summary of the miRNAs, which were differentially expressed in at least one of the three pairwise comparisons is given in Table 1A and 1B; for the purpose of reduction, only miRNAs with a mean normalized log2 signal of > 4 and FC > 2 and < ½ are enlisted. The overlapping candidates of both AI resistance models include miR-18a, -18b and -301a (upregulated) and miR-125b, -99a, -27b*, -27b, -342-5p, -708, -181a-2*, -30b, -30b*, -30d, -30e, and -34a* (downregulated), (Table 1A). The 2A-specific miRNAs include miR-139-5p, and -671-5p (upregulated), and miR-29a, -374b, -181a*, and -30a (downregulated), (Table 1B). The 5C-specific miRNAs include miR-18a*, -31, -505, -505*, -375, and -181d (upregulated), and miR-221/222, -218, -149, -342-3p, -23b, -574-3p, -574-5p, -328, -210, -200a*, and -200b (downregulated) (Table 1B). Other important 5C candidates include numerous up-regulated miRNAs from the DLK1-DIO3 locus on Chr. 14q32.31 (Table 1B, underlined).
Table 1A. miRNAs sets differentially expressed in AI-resistant cell lines.
| miRNA | Fold change>a | Holm-adj. Pb | Mean log2 signalc (5C) | Mean log2 signalc (2A) | Mean log2 signalc (WS8) |
|---|---|---|---|---|---|
| Common elements in 5C vs WS8 and 2A vs WS8 | |||||
| hsa-miR-3074 | 3.82 | 1.63E-02 | 4.64 | 4.74 | 2.71 |
| hsa-miR-1226 | 3.28 | 1.24E-04 | 6.04 | 5.62 | 4.32 |
| hsa-miR-503 | 3.26 | 6.88E-05 | 7.48 | 6.92 | 5.78 |
| hsa-miR-18a | 3.03 | 4.71E-05 | 10.42 | 9.95 | 8.83 |
| hsa-miR-18b | 2.90 | 3.51E-04 | 7.80 | 7.25 | 6.26 |
| hsa-miR-301a | 2.71 | 1.30E-05 | 9.45 | 8.94 | 8.02 |
| hsa-miR-152 | 2.66 | 6.01E-05 | 9.77 | 9.47 | 8.36 |
| hsa-miR-1979 | 2.33 | 8.28E-04 | 7.96 | 7.96 | 6.74 |
| hsa-miR-148a | 2.12 | 1.13E-03 | 8.03 | 8.12 | 6.94 |
| hsa-miR-125b | 1/35.27 | 1.64E-08 | 0.93 | 2.06 | 6.07 |
| hsa-miR-1246 | 1/5.77 | 2.36E-02 | 5.33 | 5.28 | 7.86 |
| hsa-miR-3185 | 1/4.68 | 1.33E-06 | 4.07 | 4.55 | 6.30 |
| hsa-miR-181a-2* | 1/3.62 | 1.47E-03 | 2.99 | 1.92 | 4.85 |
| hsa-miR-1296 | 1/3.36 | 7.47E-03 | 2.29 | 1.65 | 4.04 |
| hsa-miR-628-3p | 1/2.36 | 1.90E-03 | 4.86 | 4.94 | 6.10 |
| hsa-miR-324-3p | 1/2.31 | 1.81E-03 | 6.07 | 6.41 | 7.28 |
| hsa-miR-361-5p | 1/2.28 | 3.63E-05 | 10.26 | 10.22 | 11.45 |
| hsa-miR-30b | 1/2.24 | 1.96E-04 | 9.14 | 9.40 | 10.30 |
| hsa-miR-193a-5p | 1/2.17 | 6.64E-04 | 7.07 | 7.23 | 8.19 |
| hsa-miR-663 | 1/2.16 | 2.96E-03 | 6.50 | 6.43 | 7.61 |
| hsa-miR-1908 | 1/2.13 | 8.83E-04 | 6.89 | 6.73 | 7.98 |
| hsa-miR-30e | 1/2.13 | 4.41E-03 | 6.14 | 5.88 | 7.23 |
| hsa-miR-1287 | 1/2.12 | 1.21E-03 | 5.19 | 5.35 | 6.27 |
| hsa-miR-34a* | 1/2.05 | 6.42E-04 | 4.08 | 4.20 | 5.11 |
| hsa-miR-30d | 1/2.02 | 6.64E-04 | 8.73 | 9.10 | 9.74 |
| hsa-miR-1975 | 1/1.69 | 3.33E-02 | 10.02 | 9.57 | 10.77 |
| Common elements in 5C vs WS8, 2A vs WS8 and 5C vs 2A | |||||
| hsa-miR-1972 | 5.64 | 1.81E-06 | 7.28 | 6.07 | 4.79 |
| hsa-miR-30a* | 1.88 | 1.32E-02 | 5.42 | 0.78 | 4.51 |
| hsa-miR-99a | 1/44.62 | 2.70E-06 | 0.82 | 2.80 | 6.30 |
| hsa-miR-27b* | 1/23.35 | 9.09E-10 | 1.75 | 4.92 | 6.29 |
| hsa-miR-342-5p | 1/12.43 | 8.82E-11 | 5.41 | 8.30 | 9.05 |
| hsa-miR-708 | 1/10.79 | 6.63E-10 | 1.77 | 6.89 | 5.20 |
| hsa-miR-30b* | 1/5.50 | 3.74E-06 | 3.74 | 5.05 | 6.20 |
| hsa-miR-27b | 1/3.91 | 3.64E-08 | 8.62 | 9.92 | 10.59 |
| hsa-miR-675 | 1/3.88 | 2.30E-04 | 1.28 | 5.23 | 3.24 |
| hsa-miR-422a | 1/1.58 | 5.09E-03 | 6.87 | 8.22 | 7.53 |
| Elements only in 5C vs 2A | |||||
| hsa-miR-3065-5p | 1/4.63 | 4.63E-04 | 4.31 | 6.52 | 5.32 |
| hsa-miR-3065-3p | 1/3.65 | 1.39E-02 | 3.71 | 5.57 | 4.32 |
| hsa-miR-378c | 1/2.10 | 4.25E-03 | 7.87 | 8.94 | 8.20 |
Listed are all miRNAs which fulfill the following criteria for at least one of the three pairwise comparisons (5C vs WS8, 2A vs WS8, 5C vs 2A): (a) fold change > 2 or < ½, (b) Holm-adjusted P < 0.05, and (c) Mean log2 signal > 4 in at least one of the two cell lines being compared, bold: comparison for which respective FC and P-values are given.
Table 1B. miRNAs sets differentially expressed in AI-resistant cell lines.
| miRNA | Fold changea | Holm-adj. Pb | Mean log2 signalc (5C) | Mean log2 signalc (2A) | Mean log2 signalc (WS8) |
|---|---|---|---|---|---|
| Elements only in 2A vs WS8 | |||||
| hsa-miR-1273d | 2.43 | 1.79E-02 | 3.37 | 4.02 | 2.74 |
| hsa-miR-671-5p | 2.18 | 2.65E-02 | 7.80 | 8.11 | 6.98 |
| hsa-miR-374b | 1/2.77 | 3.29E-02 | 5.49 | 5.17 | 6.64 |
| hsa-miR-29a | 1/2.31 | 6.76E-03 | 8.17 | 7.50 | 8.70 |
| Common elements in 2A vs WS8 and 5C vs 2A | |||||
| hsa-miR-139-5p | 14.17 | 3.05E-04 | 3.06 | 5.59 | 1.76 |
| hsa-miR-1910 | 2.11 | 4.15E-03 | 4.44 | 5.31 | 4.23 |
| hsa-miR-181a-star | 1/3.78 | 1.67E-03 | 5.34 | 3.58 | 5.49 |
| hsa-miR-30a | 1/3.73 | 3.29E-02 | 7.95 | 5.61 | 7.51 |
| Elements only in 5C vs WS8 | |||||
| hsa-miR-1274a | 6.62 | 1.16E-02 | 4.77 | 3.10 | 2.05 |
| hsa-miR-18a* | 2.18 | 3.27E-03 | 5.70 | 5.34 | 4.57 |
| hsa-miR-328 | 1/4.59 | 6.64E-04 | 2.29 | 3.23 | 4.49 |
| hsa-miR-1231 | 1/4.07 | 2.05E-02 | 2.70 | 3.29 | 4.72 |
| hsa-miR-200a* | 1/2.51 | 9.84E-04 | 5.04 | 5.52 | 6.37 |
| hsa-miR-1285 | 1/2.36 | 1.87E-02 | 4.23 | 5.01 | 5.47 |
| hsa-miR-210 | 1/2.14 | 2.95E-03 | 6.93 | 7.29 | 8.03 |
| hsa-miR-550 | 1/2.13 | 2.14E-02 | 3.76 | 4.34 | 4.85 |
| hsa-miR-200b | 1/2.12 | 6.49E-04 | 8.43 | 8.89 | 9.51 |
| Common elements in 5C vs WS8 and 5C vs2A | |||||
| hsa-miR-487b | 132.73 | 1.86E-12 | 8.04 | 0.47 | 0.99 |
| hsa-miR-127-3p | 108.15 | 6.00E-13 | 7.74 | 0.68 | 0.99 |
| hsa-miR-432 | 105.24 | 3.72E-10 | 7.74 | 0.93 | 1.02 |
| hsa-miR-409-3p | 76.90 | 9.79E-11 | 7.28 | 0.90 | 1.02 |
| hsa-miR-431 | 73.35 | 3.15E-12 | 6.81 | 0.47 | 0.61 |
| hsa-miR-382 | 54.71 | 6.40E-11 | 6.29 | 0.40 | 0.51 |
| hsa-miR-379 | 49.44 | 6.00E-13 | 6.19 | 0.78 | 0.57 |
| hsa-miR-487a | 42.78 | 1.32E-11 | 5.93 | 0.53 | 0.51 |
| hsa-miR-433 | 33.39 | 1.60E-10 | 5.63 | 0.78 | 0.57 |
| hsa-miR-31 | 28.67 | 1.60E-10 | 5.98 | 0.97 | 1.14 |
| hsa-miR-134 | 24.79 | 1.60E-10 | 5.66 | 1.09 | 1.02 |
| hsa-miR-409-5p | 21.69 | 9.79E-11 | 4.82 | 0.75 | 0.38 |
| hsa-miR-495 | 20.78 | 1.01E-09 | 5.20 | 0.64 | 0.83 |
| hsa-miR-543 | 19.49 | 1.60E-10 | 4.77 | 0.45 | 0.49 |
| hsa-miR-329 | 18.76 | 1.12E-09 | 4.69 | 0.45 | 0.46 |
| hsa-miR-493 | 13.35 | 9.54E-10 | 4.92 | 0.93 | 1.18 |
| hsa-miR-376c | 10.65 | 2.22E-06 | 4.05 | 0.61 | 0.64 |
| hsa-miR-370 | 10.03 | 6.48E-07 | 4.25 | 0.65 | 0.92 |
| hsa-miR-505* | 6.32 | 1.41E-07 | 8.96 | 6.27 | 6.30 |
| hsa-miR-494 | 6.27 | 6.64E-04 | 7.53 | 5.37 | 4.88 |
| hsa-miR-505 | 5.32 | 2.33E-06 | 7.96 | 5.57 | 5.54 |
| hsa-miR-1308 | 3.99 | 6.67E-07 | 10.22 | 8.08 | 8.23 |
| hsa-miR-4284 | 3.75 | 1.36E-04 | 7.48 | 5.71 | 5.57 |
| hsa-miR-375 | 3.33 | 1.08E-03 | 4.36 | 1.71 | 2.62 |
| hsa-miR-330-3p | 2.54 | 1.56E-04 | 8.31 | 7.50 | 6.97 |
| hsa-miR-181d | 2.54 | 9.66E-06 | 8.09 | 6.90 | 6.75 |
| hsa-miR-221 | 1/99.01 | 6.36E-10 | 2.06 | 8.08 | 8.69 |
| hsa-miR-222 | 1/47.64 | 2.92E-13 | 3.16 | 8.30 | 8.73 |
| hsa-miR-218 | 1/14.02 | 5.31E-06 | 0.56 | 3.87 | 4.37 |
| hsa-miR-149 | 1/8.66 | 1.90E-10 | 6.00 | 8.80 | 9.11 |
| hsa-miR-342-3p | 1/6.39 | 1.01E-08 | 9.87 | 12.01 | 12.55 |
| hsa-miR-574-3p | 1/3.45 | 1.04E-06 | 7.12 | 8.98 | 8.91 |
| hsa-miR-23b | 1/2.95 | 3.41E-07 | 10.76 | 12.05 | 12.32 |
| hsa-miR-497 | 1/2.65 | 5.54E-05 | 5.83 | 6.79 | 7.24 |
| hsa-miR-489 | 1/2.56 | 2.44E-04 | 5.58 | 7.53 | 6.93 |
| hsa-miR-574-5p | 1/2.44 | 9.05E-04 | 4.96 | 6.67 | 6.25 |
| hsa-miR-27a* | 1/2.32 | 1.27E-03 | 3.62 | 4.96 | 4.83 |
| hsa-miR-7-1* | 1/2.10 | 2.75E-03 | 4.52 | 5.35 | 5.59 |
| hsa-miR-195 | 1/2.09 | 4.59E-05 | 9.15 | 9.91 | 10.21 |
Listed are all miRNAs which fulfill the following criteria for at least one of the three pairwise comparisons (5C vs WS8, 2A vs WS8, 5C vs 2A): (a) fold change > 2 or < ½, (b) Holm-adjusted P < 0.05, and (c) Mean log2 signal > 4 in at least one of the two cell lines being compared, bold: comparison for which respective FC and P-values are given; miRNAs of the DLK1-DIO3 cluster are underlined.
To distinguish miRNAs associated with E2-dependent growth that are altered upon short-term E2 deprivation from those relevant for the evolution of long-term changes associated to AI resistance and vulnerability to E2-induced apoptosis, we compared miRNA expression profiles of WS8 cells under 72h E2-treatment versus non-treated WS8 reference control. A total of 131 miRNAs were relevantly differentially and significantly expressed (FC > 1.5 or < 1/1.5, P < 0.05) (Supplementary Table 4). In addition, the 184 differentially expressed miRNAs between 5C versus WS8 and 2A versus WS8 contain 71 miRNAs that are altered during E2-stimulated growth (Supplementary Figure 1).
We confirmed the findings from our global miRNA profilings in that we compared relative miRNA expression levels of a selection of top candidates including miR-31, -221/222, -127-3p, -409-3p and miR-432-5p between 5C, 2A and WS8 using qRT-PCR (Supplementary Figure 2).
AI resistance-related miRNAs of chromosomal regions 14q32.31 and 21q21.1 are highly expressed in Luminal A breast cancers
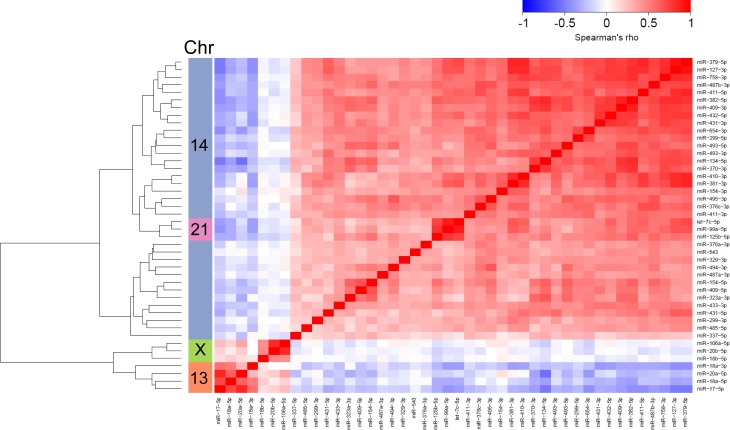
We used TCGA miRNA-Seq expression data of 746 breast cancer patients in order to interrogate the putative clinical relevance of miRNAs differentially expressed in the 2A and 5C AI resistance models. Pairwise comparisons of the 205 miRNAs revealed higher correlations between miRNAs located in chromosomal clusters compared to non-clustered miRNAs. Here we focus on those miRNAs that cluster at chromosomal regions 14q32.31, 21q21.1, and 13q31.3. The 14q32.31 mega-cluster comprises among others 34 positively correlated miRNAs overexpressed in 5C (Figure 3). The 21q21.1 cluster includes let-7c, miR-99a and miR-125b, the expression of which is decreased in 2A. In 5C cells, with the exception of let-7c similar changes were observed. Noteworthy, the three miRNAs also showed a strong relationship to miRNAs of the 14q32.31 locus (Figure 3). Furthermore, miRNAs of the 17-92 cluster on chromosome 13q31.3 (miR-17, -18a, -18a* and -20a) and the paralogous 106a-363 cluster on chromosome X (miR-20b, -18b, and -106a) upregulated in 5C show high intra-, but no positive inter-cluster correlations to miRNAs from the 14q32.31 locus, respectively (Figure 3).
Figure 3. miRNA expression correlations based on TCGA breast cancer data (primary tumors).
Hierarchical clustering illustrates intra- and inter-cluster relationships of pairwise comparisons of miRNAs located on chromosomes 14, 21, X and 13. Chromosomal assignments (left color bar) and color-coded Spearman's rho correlation coefficients (blue: low, red: high) are indicated.
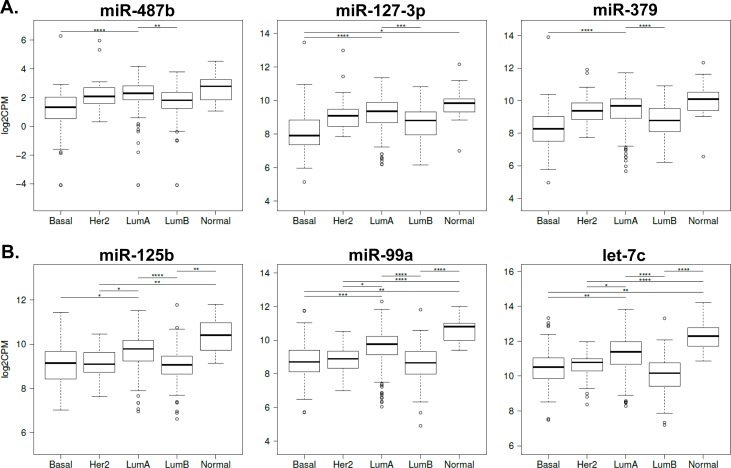
Next, we used the miRNA-Seq data of 513 patients with PAM50 annotation to test for a potential clinical relevance of dysregulated miRNAs (Luminal A, n = 253; Luminal B, n = 115; Basal-like, n = 86; HER2-Enriched, n = 43; and Normal breast-like tumors, n = 16). Differences between miRNA expression levels in the subtypes were identified for 19 out of the 34 miRNAs at 14q32.31 (Supplementary Figure 3A). For example, the expression of miR-487b, -127-3p and -379 was lowest in Basal-like and Luminal B tumors, however, the Luminal A subgroup shows significantly higher expressions (Figure 4A). Notably, a higher expression of miR-125b, -99a and let-7c (Chr. 21q21.1) was also observed for Luminal A tumors (Figure 4B), a finding that is in accordance with their high correlation to Chr. 14q32.31 miRNAs (Figure 3). AI resistance-related miRNAs at other chromosomal locations also showed differential expression between breast cancer subtypes. These include miR-30a, -375, -342-5p with low expression in Basal-like but high expression in Luminal A tumors. In contrast, we observed high expression of miR-20a, -222, and -18a in Basal-like tumors (Supplementary Figure 3B).
Figure 4. miRNA expression in PAM50-defined breast tumor subtypes from TCGA.
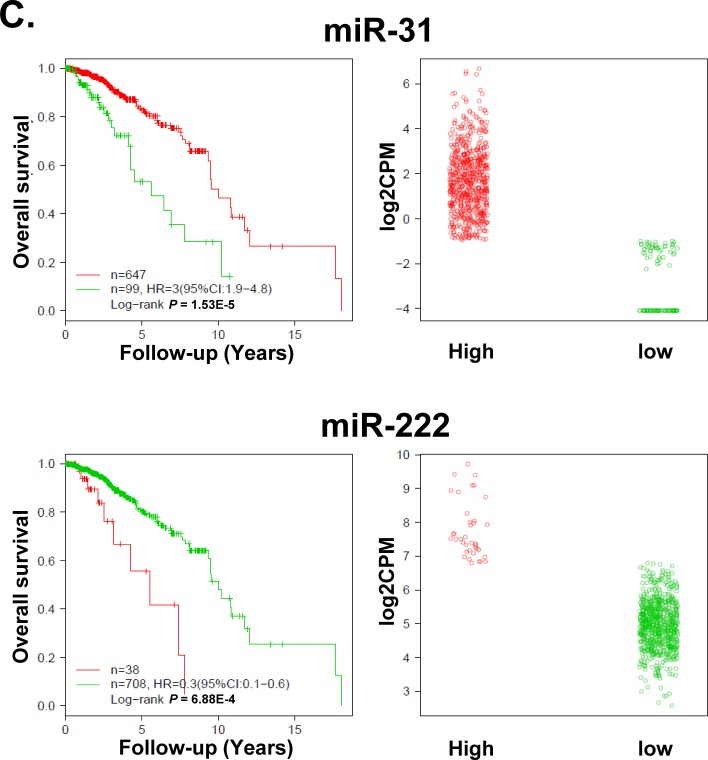
Box-Whisker plots indicate expression differences between the subgroups Basal-like (Basal, n = 86), HER2 positive (Her2, n = 43), Luminal A (LumA, n = 253), Luminal B (LumB, n = 115) and Normal breast-like (Normal, n = 16). A. Chromosome 14q32.31 miRNAs: miR-487b, miR-127-3p and miR-379. B. Chromosome 21q21.1 miRNAs: miR-125b, miR-99a and let-7c. Y-axes: log2CPM (CPM: counts per million). *: P < 0.05, **: P < 0.01, ***: P < 0.001 and ****: P < 0.0001. C. Kaplan-Meier curves for overall survival stratified by miRNA expression (n = 746). Favorable survival is associated with high miR-31 and low miR-222 expression. miRNA expression cut-offs were determined by conditional inference tree models. High (red) and low (green) expression are indicated on the log2CPM scale (CPM: counts per million).
AI resistance-related miRNAs predict breast cancer outcome
Kaplan-Meier analyses of 746 patients from TCGA identified differences in breast cancer overall survival based on low versus high miRNA expression. Altogether, 35 top miRNA candidates (FC > 5 or < 1/5, Table 1A and 1B) and the remaining 16 miRNAs of the 14q32.31 cluster (Supplementary Tables 2 and 3) were analyzed. We identified low expression of miR-31 to be associated with a poor prognosis (HR = 3, 95% CI: 1.9-4.8; P = 1.53E-5; Figure 4C), and a low miR-222 expression being associated with a good prognosis (HR = 0.3, 95% CI: 0.1-0.6; P = 6.88E-4; Figure 4C). High expression of 7 miRNAs of the 14q32.31 locus (miR-410, -381, -485-5p, -487a, -376c, -411, and -127-3p) indicated a good prognosis, and four others, miR-431, -505*, -493* and -654-3p are associated with the outcome in patients with a Luminal A tumor (n = 253) (Supplementary Figure 4A and 4B).
AI resistance-related genes and key pathways enriched by miRNA sets
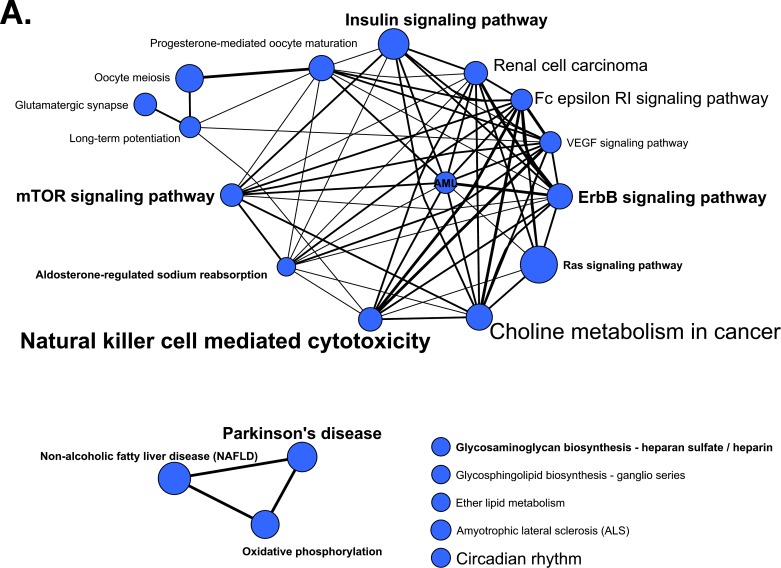
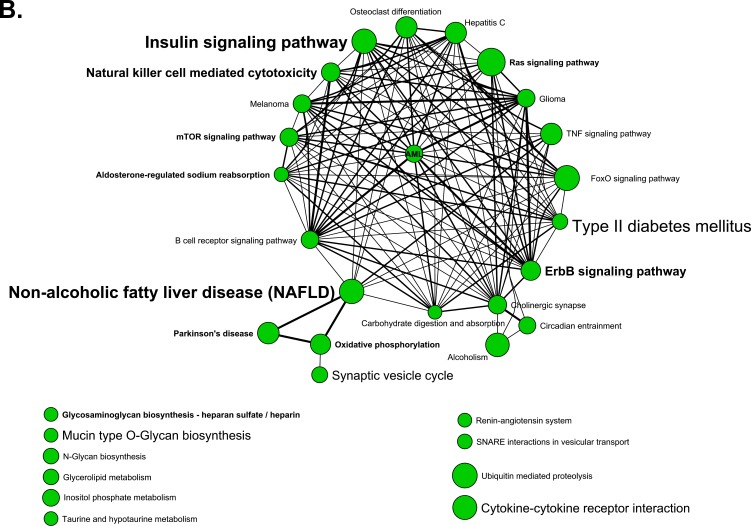
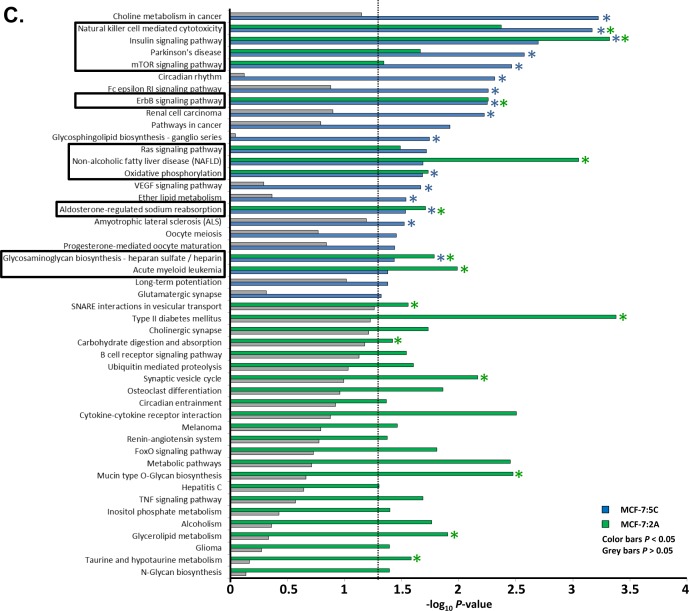
To identify pathways relevant in AI resistance, we performed miRNA functional enrichment analyses. First, we ascertained targets of miRNAs that are altered in 5C and 2A generating miRNA-mRNA interaction maps based on CLIP (cross-linking immunoprecipitation)-confirmed TargetScan predictions. Then, miRNAs were linked with KEGG pathways via their predicted targets in order to elucidate their functional context. We identified 24 and 34 enriched KEGG pathways for the 5C and 2A-specific miRNA sets, respectively. The networks in Figure 5A and 5B illustrate the overlaps between pathways based on their corresponding gene sets. Eleven pathways are enriched in both 5C and 2A and therefore portraying potential relevance in AI resistance (Figure 5C). These particularly affect, albeit to different degrees, immune response such as the ‘Natural killer cell mediated cytotoxicity’, and growth related pathways such as ‘Insulin signaling’, ‘mTOR signaling’, and ‘ErbB signaling’ (Figure 5A, 5B and 5C). Others affect metabolism such as ‘oxidative phosphorylation’ and ‘glycosaminoglycan biosynthesis-heparan sulfate/heparin’ and disease-related terms including ‘Parkinson's disease’, ‘non-alcoholic fatty liver disease’ and ‘acute myeloid leukemia’ (Figure 5A, 5B and 5C). To account for bias frequently associated with the hypergeometric distribution of miRNA enrichment analyses (standard method) we tested for enrichment using permutation tests (Figure 5C, blue and green asterisks).
Figure 5. KEGG pathways identified in miRNA functional enrichment analyses.
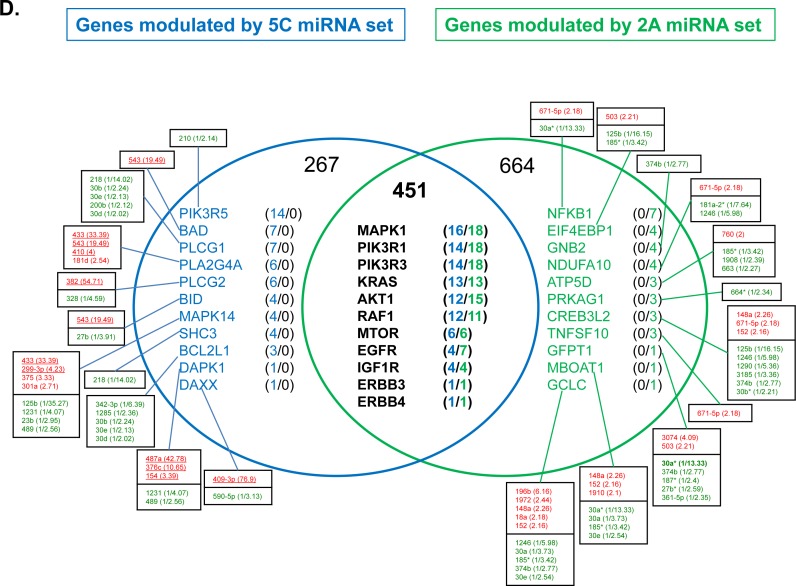
Specifically enriched pathways from the entire 5C- and 2A-specific miRNA sets (Tables S1 and S2): A. 5C (blue nodes) and B. 2A (green nodes). The node size reflects the number of pathway-related genes regulated by the specific miRNA set. The edge weight illustrates the degree of gene overlap between two pathways as measured by the Jaccard index. Pathways highlighted in bold are enriched in both AI resistant cell models. The font size reflects the unadjusted Fisher test P-values from enrichment analysis (P < 0.001, large (24 pts); 0.001 ≤ P < 0.01, intermediate (18 pts); 0.01 ≤ P < 0.05, small (12 pts)). C. Enriched pathways of 5C- and 2A-specific miRNA sets. X-axes represent -log10 P-Value, Y-axes: enriched pathways sorted according to specific pathways enriched in 5C with boxed pathways being enriched in both 5C and 2A models. P-value threshold (−log10 0.05) is indicated by the dotted line. Blue and green asterisks mark enriched pathways identified with 5C- and 2A-specific miRNA sets, respectively, based on analyses using permutation tests. P-values (Fisher and permutation test) are not corrected for multiple testing. D. Venn diagram of the 5C- and 2A- specific pathway-related gene sets extracted from the respective networks (A, B). The intersection represents the total number of genes (451) modulated by both 5C-and 2A-specific miRNA sets. Sections indicate the 5C- and 2A-specific miRNA-modulated genes (5C: 267 genes, 2A: 664 genes). Examples for each subgroup are given in descending order of the number of pathways a gene was detected in both networks. miRNAs with their respective FC (Table 1A and 1B) that interact with and modulate genes in 5C- and 2A cell models are depicted in red (up-regulated) and green (down-regulated). miRNA-mRNA interaction thresholds were defined as CLIP confirmed TargetScan 7.0 in silico predictions (> 50 percentile). miRNAs of the DLK-DIO3 cluster of chromosome 14 are underlined.
Stratification into up- and downregulated miRNA subsets of the 5C and 2A models as well as their pathway overlaps confirms those identified for the entire miRNA sets and enable their assignment (Supplementary Figure 5A, 5B, 5C and 5D). Multiple different metabolic pathways were enriched for the down-regulated miRNA subsets of both 5C and 2A. Proliferation-related pathways appeared to be contributed by both up- and down-regulated miRNA subsets (Supplementary Figure 5A, 5B and 5C).
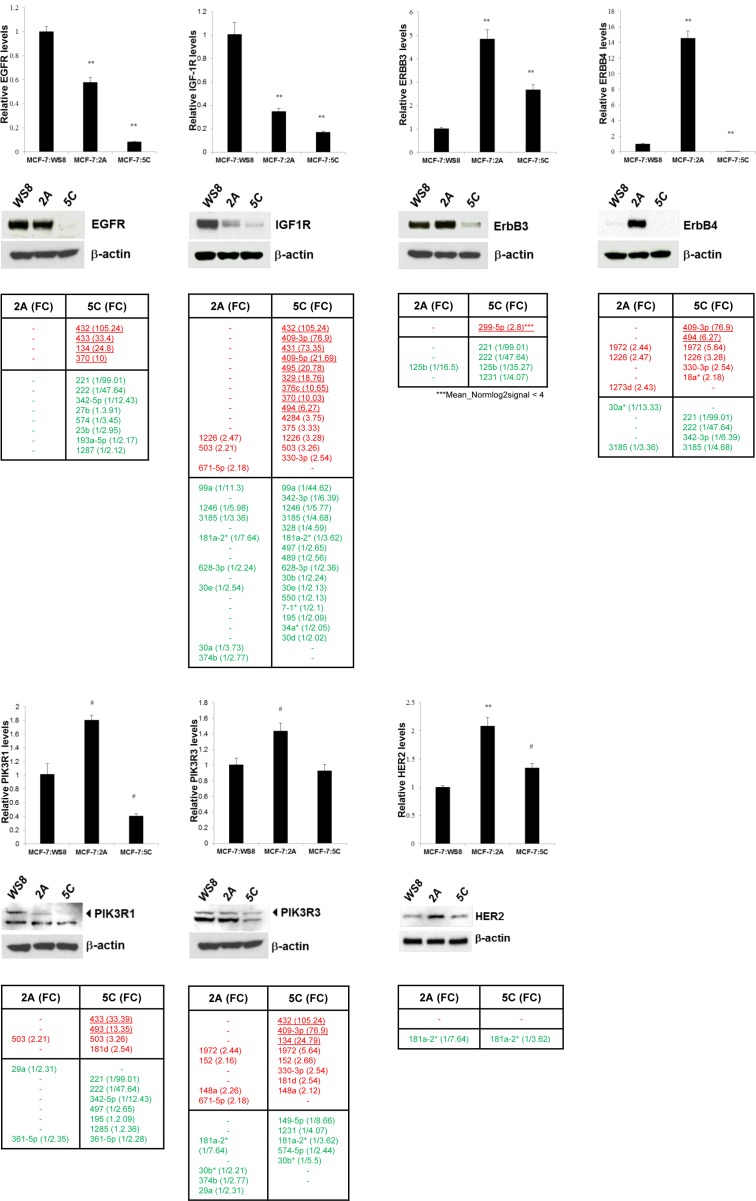
To identify AI resistance-relevant genes modulated by both 5C- and 2A-specific miRNA sets we compared their corresponding gene sets obtained from the 5C- and 2A-specific KEGG pathway networks (Figure 5A and 5B). The overlay identifies 451 genes that are modulated by either the 5C- or the 2A-specific miRNA sets. Accordingly, the targeting of 664 genes by the 2A-specific miRNA set and the targeting of 267 genes targeted by the 5C-specific miRNA set may therefore influence the 2A and 5C phenotypes (Figure 5D). Important genes triggered by 5C- and 2A-specific miRNAs include growth factor receptors such as IGF1R, EGFR, INSR, and respective downstream key signal transducers including serine/threonine protein kinases AKT1, MAPK1, MTOR, RAF1, as well as other mediators such as SOS1, SOS2, JUN, PIK3R1, KRAS (Figure 5D). The modulatory impact of miRNAs acting as posttranscriptional regulators cannot be solely captured by RT-PCR of the targeted genes. To identify putative modulatory effects, we therefore investigated the protein levels of key growth factor signal transduction mediators by Western blotting. A strong influence of the 5C-specific miRNA set indicates inhibitory effects on protein expression levels of growth factor receptors. For example, multiple strongly up- and downregulated miRNAs point to a role in the reduction of EGFR, IGF1R, ErbB3 and ErbB4 expression in 5C versus 2A cells, respectively (Figure 6). The dominance of the strongly upregulated miRNAs mainly originating from the DLK1-DIO3 cluster (Chr. 14q32.31) underlines their modulatory role on the difference in protein expression which is substantiated by our in silico finding of the presence of various dinstinct 3′ UTR CLIP-confirmed interaction sites. Similar findings but with a lesser effect on the protein level were obtained for PIK3R1, PIK3R3 and HER2 (Figure 6) as well as AKT1, MAPK1, KRAS and RAF-1 (Supplementary Figure 6). Minor opposite effects, i.e increased protein expression, have been observed for MTOR (Supplementary Figure 6). miRNAs with postulated major effects on the protein expression of multiple growth related genes are miR-432-5p (e. g. EGFR, IGF1R and PIK3R3) and miR-409-3p (e. g. IGF1R, ERBB4, PIK3R3, AKT1, MAPK1, and KRAS) (Figure 6 and Supplementary Figure 6).
Figure 6. Basal levels of gene and protein expression of key growth regulators in AI resistance models 2A and 5C and their modulatory miRNAs.
Gene expression levels were quantified by RT-PCR and protein expression levels examined by Western blotting with ß-actin as loading control (WS8: E2 growth sensitive control). Up (red) and down (green) regulating miRNAs were obtained from specific miRNA sets given in Table 1A and 1B. miRNA-mRNA interaction thresholds were defined as CLIP confirmed TargetScan 7.0 in silico predictions (> 50 percentile). miRNAs of the DLK-DIO3 cluster of chromosome 14 are underlined. Modulatory effects towards lower protein expression in 5C as compared to 2A are shown for EGFR, ErbB3, ErbB4, IGF1R, PIK3R1, PIK3R3 and HER2. *: P < 0.05, #: P < 0.01, **: P < 0.001 (comparisons to WS8).
Additional GO term enrichment analyses have been performed with respective gene sets (Figure 5D) to better understand their underlying biological processes. Gene set enrichment analyses (GORILLA) resulted in the identification of major (superordinate) and minor (subordinate) GO terms (Supplementary Figures 7A, 7B, 7C and 8). The gene set targeted by 5C specific miRNAs revealed GO terms referring to signal transduction, protein localization, apoptosis, metabolic processes, and regulation of cell differentiation including Wnt signaling (Supplementary Figure 8). Hence, we observed a dominant role of the regulation of protein phosphorylation and kinase activity including MAPK. Notably, some of the 267 genes targeted by the 5C-specific miRNA set (Figure 5D) enrich for pathways that trigger apoptosis. They include pro-apoptotic genes encoding BAD, BID, DAPK1 and DAXX postulated to be particularly attenuated by miRNAs from the DLK1-DIO3 cluster (e.g. miR-543, miR-487a, miR-409-3p). In contrast, BCL2L1 may be modulated via disinhibitory effects of miRNAs such as miR-342-3p and/or members of the miR-30 family (Figure 5D). MAPK14 (p38) is targeted by numerous up- and down-regulated miRNAs and therefore the protein levels may not be affected. The 2A-specific phenotype with 664 specifically targeted genes is characterized by numerous pathways related to metabolism including lipid and amino acids (e.g. glutamine) metabolism. Representative genes include GFPT1, MBOAT1 and GCLC (Figure 5D). We postulate that the 2A-specific down-regulated miRNAs such as miR-125b, miR-30a*, miR-374b, miR-181a-2* and miR-1246 may have a dominant effect on the posttranscriptional regulation towards their higher protein expression (Figure 5D). A similar trend pertains to CREB3L2 (Figure 5D), a transcriptional activator involved in the UPR (unfolded protein response) and known to counteract ER stress-induced cell death.
EGFR protein expression is modulated by miRNAs of the chromosomal region 14q32.31
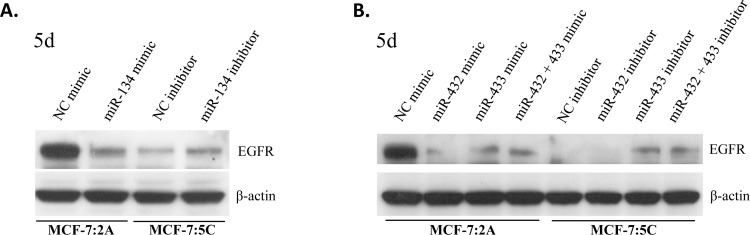
Our in silico findings identified distinct CLIP-confirmed interaction sites of the growth factor receptor EGFR 3′UTR sequence to be targeted by miRNAs from both the 5C- and 2A-specific miRNA sets with a dominant role of miRNAs deregulated in 5C (Figure 6). The distinct difference in miRNA panels of 2A and 5C point to a possible role of EGFR and its downstream pathways in the mediation of AI resistance in the 2A model. To determine the impact of upregulated miRNAs from the DLK1-DIO3 cluster on EGFR expression we mimicked the expression of the three top miRNA candidates, miR-432-5p, miR-433-3p and miR-134-5p in 2A cells and inhibited these miRNAs in 5C cells. First data show particularly an influence of miR-134-5p and miR-433-3p on EGFR protein expression. Figure 7A shows a significant downregulation of EGFR protein level by miR-134-5p mimic in 2A cells (not at mRNA level, Supplementary Figure 9A), and a significant upregulation via miR-134-5p inhibition in 5C cells after 5 days. Corresponding qRT-PCR controls of miR-134-5p relative levels as well as mRNA expression levels of EGFR are shown in Supplememtary Figure 9A, B. Mimic experiments with miR432-5p, and miR-433-3p as well as simultaneous mimic with both miRNAs in 2A cells showed a clear downregulation of the EGFR protein level (Figure 7B). miRNA inhibiton experiments with single miR-433-3p and simultaneous miR-432-5p and miR-433-3p inhibition in 5C cells show a clear increase of EGFR protein, which appears to be mainly influenced by miR-433-3p (Figure 7B).
Figure 7. miRNAs of the DLK1-DIO3 cluster downregulate EGFR protein levels in MCF-7:2A (mimics) and upregulate EGFR protein levels in MCF-7:5C (inhibition).
A. Western blot following mimic and inhibition of miR-134 at day 5. B. Western blot following mimic and inhibition of miR-432 and miR-433 as well as simultaneous miR-432 and miR-433 mimic and inhibition at day 5. ß-actin was used as loading control; NC: negative control.
DISCUSSION
We identified more than two hundred up- and down regulated miRNAs relevant to the E2-independent growth phenotype inherent to Phase I/II endocrine resistance (Figure 2). We distinguished miRNAs common to both AI resistance models from those specific to 5C and 2A, and investigated their clinical relevance. Thirty-four 5C-specific miRNAs are located at Chr. 14q32.31 (Supplementary Table 2 and 3), a chromosomal region that hosts the largest microRNA cluster in the genome known as the DLK1-DIO3 domain-containing cluster of 54 miRNAs. Together with numerous imprinted genes, antisense and neighbouring long intergenic RNAs they are differentially expressed in several pathologic processes including various cancers [28]. For 34 miRNAs of this cluster, we identified a positive correlation of expression which stresses the existence of common regulatory elements and the dominance of co-transcribed polycistronic primary transcripts (Figure 3). Our finding of high intra-cluster correlations is in accordance with similar findings reported from a large-scale study on miRNA landscapes in breast cancer [29], and highlights their putative implication for AI resistance and vulnerability for E2-inducible apoptosis. Notably, we observed high correlations of numerous Chr. 14q32.31 miRNAs with let-7c/miR-99a/miR-125b that are located at another chromosomal cluster 21q21.1 (Figure 3), suggesting their potential cooperation in common regulatory pathways.
The higher expression of numerous miRNAs of the 14q32.31 and 21q21.1 clusters in Luminal A breast tumors compared to Luminal B and Basal like tumors supports the notion of a prominent role of ER as the key histopathological covariate affecting miRNA variability [29]. The high expression of miRNAs miR-210, -17, -18a, and -20a in Basal-like tumors together with our observation of their high expression in AI resistance models suggests their oncogenic potential. The low expression of miRNAs miR-30a, -149, and -342 in the AI resistance models contrasts with a high expression in Luminal A tumors thereby underscoring their role as tumor suppressors during the evolution of AI resistance. Notably, oncogenic and tumor suppressor capacities of these miRNAs have been previously discussed [30–32]. The tumor expression of some miRNAs described herein correlated with breast cancer outcome of patients recorded in TCGA. For example, low expression of miR-31 was associated with a 3-fold increased risk of death. This observation is consistant with the finding of miR-31 as a master regulator of the invasion-metastasis cascade [33]. Its known anti-metastatic function of inhibiting the multistep invasion-metastasis cascade via the repression of pro-metastatic targets such as WASF3 and ITGB1 [33, 34] aid in the characterization of AI resistance phenotypes. We demonstrated that both genes were only identified in 5C-enriched pathways and respective gene sets (Figure 5A and 5D) and therefore specifically characterize the 5C-specific phenotype. In addition to miR-31, also miR-493-5p from the DLK1-DIO3 locus targets their transcripts and may support attenuation of their protein expression in 5C. Moreover, the capacity of miR-31 to sensitize human breast cells to apoptosis by targeting protein kinase Cε (epsilon) [35] may assist in the understanding of the susceptibility to early E2-inducible apoptosis of breast cancer cells particularly in the light of its upregulation in the 5C model. Of note, our CLIP-confirmed TargetScan predictions point to a dominant role of multiple other up-regulated miRNAs in both models with the prevalence of miRNAs from the DLK1-DIO3 locus which may impact on PRKCE levels in 5C.
We showed, that a high expression of miR-222 clearly correlated with non-favorable overall survival of patients recorded in TCGA supporting its role in breast cancer aggressiveness. This finding is in line with recent in vitro evidence that miR-221/miR-222 promotes S-phase entry of the cell cycle and cellular migration, and that miR-221 and miR-222 negatively regulate the expression of the tumor suppressor genes, suppressor of cytokine signaling 1 (SOCS1) and cyclin-dependent kinase inhibitor 1B (CDKN1B) [36]. Stinson et al. [37] reported that miR-221/222 promote a Basal-like phenotype by acting downstream of the oncogenic RAS-RAF-MEK pathway and the triggering of epithelial-to-mesenchymal transition (EMT). Notably, they recognized miR-221/222 as part of the EMT signature as high miR-221/222 levels were correlated with high vimentim and low E-cadherin expression. Our findings of high vimentin (Supplementary Table 5) and low E-cadherin expression in 2A compared to 5C cells supports this role. Moreover, miR-221/222 levels are known to be triggered via the activation of the EGFR-(RAS-RAF-MEK) pathway axis [37]. We observed nearly complete loss of EGFR protein in 5C cells and showed that EGFR is the target of miRNAs mainly of the DLK1-DIO3 cluster. We therefore propose that the low EGFR levels in 5C compared to 2A may account for the lowering of miR-221/222 expression thereby suppressing EMT. This may explain the known phenotypic difference with regards to the 5C and 2A EMT status which may have its origin in attenuating effects of miRNAs of the DLK1-DIO3 cluster on EGFR protein expression. Importantly, miR-222 is associated with anti-estrogen resistance [31], and when transfected with miR-221- and/or miR-222, ER-positive cell lines (MCF-7, T47D) develop resistance to tamoxifen, yet their knockdown in ER-negative cells (MDA-MB-468) leads to tamoxifen-induced cell growth arrest and apoptosis. The lack of tamoxifen response in the former can be explained by the negative regulation of ERα protein expression as a consequence of the miR-221/222 targeting of ERα [22]. In this context, the differences in elevation of ERα levels with higher levels detected in 2A compared to 5C (data not shown) may originate from additional modulatory influences of up-regulated miRNAs in 5C. TargetScan predicts multiple high scoring 3′UTR binding sites for miRNAs from the DLK1-DIO3 locus pointing to attenuative functions on ERα protein level in 5C, although one has to consider the complex nature of its regulation via other non-posttranscriptional mechanisms. As the mediation of tamoxifen resistance has been further linked to miR-221/222 enclosed in exosomes that act as intercellular bio-messengers [38], we conclude that miR-221/222 is critical for the portrayal of the AI resistance phenotype and susceptibility to E2-inducible apoptosis particularly since miR-221/222 is low in 5C cells.
We could not confirm the prognostic evidence for the low expression of let-7c/miR-99a/miR-125b in Luminal A tumors that correlate with poor outcome as recently reported by Bailey et al. [27]. They described these miRNAs being present in MCF-7 but not 2A cells and suggest that let-7c and miR-125b inhibit HER2 protein expression in the latter. We observed low expression of these miRNAs in 2A cells, however we were not able to confirm their prognostic character albeit our TCGA sample size was much larger. Interestingly, findings from other AI resistance models demonstrated that miR-125b could be a novel marker of poor prognosis as its overexpression is sufficient to confer resistance to letrozole and anastrozole [39]. Whether or not miR-125b is prognostic in distinct breast cancer subsets remains elusive. In support of a putative clinical relevance of miRNAs of the DLK1-DIO3 cluster, it is noteworthy, that 7 miRNAs (miR-410, -381, -485-5p, -487a, -376c, -411, and -127-3p) were associated with overall survival, with high expression being favorable. Although these clustered miRNAs could be of interest within the context of prognosis, our findings may be preliminary as the median follow-up of 2.2 years in the TCGA breast cancer PAM50 dataset is short. From the patient-based findings, we conclude that the differentially expressed miRNAs identified by us (Table 1A and 1B) bear clinical relevance and should therefore be further investigated as putative treatment targets to subvert AI resistance. In the absence of clinical data linking miRNAs to the development of AI resistance it must be emphasized that miRNAs moving towards cancer therapeutic development are those with sufficient mechanistic data that allow a fairly accurate placement of the miRNA into the disease-related pathways [40].
To elucidate the underlying key cellular processes and pathways, it is important to explore the complex modulatory roles of miRNAs in determining specific AI resistance phenotypes. Aided by miRNA-mRNA networks we enriched for pathways common to the 5C and 2A models and for additional pathways intrinsic to their distinct biology. Both 5C- and 2A-specific miRNA sets enriched for pathways modulating cell growth and proliferation via cell surface tyrosine kinase receptors such as insulin-, EGFR-, and ErbB-signaling. Moreover, pathways related to immune response regulation including cytokine signaling via JAK/STAT and NFκB were enriched. Our findings support evidence demonstrating that growth factor receptors, such as insulin-like growth factor receptor (IGF-1R), EGFR, and ErbB2, play critical roles in the mediation of endocrine resistance in breast cancer [41–44]. Their functional dominance in the development of resistance is variable and depends on the context of cell lines [42–44]. This is particularly evident from our results in that the clonally selected 5C cells clearly differ from 2A cells in terms of ErbB signaling which we attribute to the distinct modulatory actions of respective miRNA sets on growth pathway-relevant transcipts. Our data show higher protein levels of all ErbB receptors in 2A compared to 5C, for the latter only HER2 is expressed at similar amounts as in the MCF-7:WS8 reference (Figure 6). Therefore we may speculate, that up-regulation of ErbB signaling is used as a compensatory mechanism to continue proliferation in the AI resistant 2A cells, whereas 5C cells seem not to depend on this for survival. These phenotypic differences may account for the stronger resistance to E2-inducible apoptosis in the 2A model [11] which is the focus of future studies. On the miRNome level, these differences may be attributed to modulatory effects of multiple miRNAs of the DLK1-DIO3 cluster on key signal transduction molecules in ErbB signaling. First functional data highlight the role of miR-134-5p, miR-432-5p and miR-433-3p in the modulation of EGFR protein levels in 2A and 5C AI resistance models, a finding that is in line with the recently reported direct targeting of the EGFR 3′UTR sequence by miR-134-5p in non-small cell lung cancer cell models [45].
Moreover, it is evident from our results that miRNA modulatory effects on phosphatidylinositol-mediated signaling, Ras signaling, and the MAPK cascade play a dominant role. This is also reflected by the enrichment for protein phosphorylation and kinase activity, in particular MAPK activity, by both 5C- and 2A-specific miRNA sets. Our own observations confirm that IGF-1R is a growth driver in 5C and 2A cells [12, 13]. IGF-1R is regulated by E2 in an ER-dependent manner [12] and therefore results in the reduction of total IGF-1R protein in resistance models [44, 46, 47]. However, the cross-talk between IGF-1R and other membrane-associated molecules keeps these resistant cells with higher levels of phosphorylated IGF-1R [44, 46, 47]. The higher levels of phosphorylated IGF-1R are possibly a consequence of an increased modulatory influence of miRNAs on the total amount of protein phosphorylation.
Our enrichment analysis further reveals that the mTOR signaling axis is a main target for miRNAs in both cell lines (Figure 5A and 5B). PI3K/mTOR is a common downstream pathway of growth factor receptors, which is often upregulated in endocrine-resistant breast cancer and proven clinically as a novel treatment target [48, 49]. Notably, the 5C-specific miRNA set additionally enriches for serine/threonine kinase activity and cellular response to stress strengthening our conclusion of an increased modulatory influence on mTOR signaling in 5C. The AKT/mTOR pathway is constitutively activated in AI resistant cell models [39], which is consistent with our pathway enrichment findings. Interestingly, Vilquin et al. showed that overexpression of miR-125b activates the AKT/mTOR pathway and increases the capacity to form stem cell-like properties. As miR-125b is one of the top downregulated miRNAs in both of our AI resistance models, we suggest that other miRNAs such as miR-99a [50] and miRNAs of the DLK1-DIO3 locus [51] may interfere with the mTOR pathway during the reprogramming of growth signaling. A suppressive role of the DLK1-DIO3 miRNA mega-cluster on the PI3K-mTOR pathway has been identified in hematopoetic stem cells resulting in a reduction of mitochondrial biosynthesis and metabolic activity and the protection of these cells from excessive production of reactive oxygene species [51]. As miRNAs of this cluster are up-regulated in 5C, future studies will decipher their role in the control of oxidative stress and apoptosis.
A major difference between the AI-resistant 5C and 2A phenotypes is the variable response to oxidative stress triggered by E2-treatment. Both cell models respond with endoplasmic reticulum stress and subsequent apoptosis, however at different time points. We attribute the delay of several days of the 2A cells to gluthatione as its basal expression is higher in 2A as compared to 5C cells [13]. Our identified miRNA-mRNA interaction maps revealed potential modulatory effects on the posttranscriptional regulation of enzymes involved in glutathione synthesis such as the γ- glutamylcysteine sythetase (GCLC), as the observed low miR-30a expression in 2A supports this view. Overall, our pathway enrichment analyses suggest a higher metabolic activity for 2A compared to 5C cells. As changes for example in amino acid metabolic pathways might contribute to the complex biology of the early vulnerability of 5C cells to E2-induced apoptosis this research topic requires further attention in the future. Importantly, both AI-resistant cell lines exhibit stem cell-like and EMT-like phenotypes, demonstrated by gene expression levels of the putative stem cell markers CD44high and CD24low, regression of the cytokeratin expression (KRT18), and activation of vimentin expression (Supplementary Table 5). Additionally, Twist, Notch, and Wnt signaling pathways are activated in 5C and 2A cells (Supplementary Table 5). All of these factors have been implicated to be involved in EMT- or stem cell-associated processes which contribute to endocrine resistance. Our pathway enrichment indicated that miRNAs frequently modulate these pathways. Therefore, targeting critical miRNAs to interfere with EMT and to modulate its associated pathways in stem cells has been considered as a potential approach to overcome endocrine resistance [52].
The primary aim of the study was to identify in well characterized cell models of AI resistance the global effects on the modulation of pathways and networks affected by their respective underlying miRNome phenotypes. We defined the intrinsic phenotypes of two AI resistance models, extend available data [27] and provide comprehensive miRNA sets and targeted pathways of 2A and 5C cells. We defined their common and specific features at the miRNA-mRNA interaction level, however changes in miRNA expression are the result of a complex interplay of genomic, transcriptional and post-transcriptional mechanisms which are frequently subtle. Notably, each miRNA can have several targets, some belonging to the same functional network or signaling pathway, and 3′UTR regions of a single gene are frequently targeted by several different miRNAs [39]. Enhancing E2-induced apoptosis in AI resistant breast cancer from 30% to 100% responsiveness is an important goal in therapeutics. Understanding the miRNA modulations necessary to trigger apoptosis is an essential first step in the broad application of low dose estrogen as a safe intervention for patients. This extensive database, cross-referenced with clinical databases or functional data, provides the conduit to identify the events that trigger or prevent E2-induced apoptosis in AI resistant breast cancer.
MATERIALS AND METHODS
Materials
Sources for antibodies for Western blotting are as follows: EGFR antibody was from EMD Millipore (Temecula, CA); IGF-1R antibody was purchased from Santa Cruz (Santa Cruz, CA); ERBB2, ERBB3, ERBB4, MTOR, AKT1, MAPK1, KRAS and RAF-1 antibodies were from cell signaling technology (Beverly, MA); PIK3R1 and PIK3R3 antibodies were obtained from ThermoFisher Scientific (Rockford, IL). mirVanaTM miRNA Mimics and Inhibitors for miR-134-3p, 432-5p and 433-3p and respective negative controls (NC) were purchased from ThermoFisher Scientific (Rockford, IL).
Cell culture
The E2-hypersensitive MCF-7:WS8 (WS8) cells were clonally selected from MCF-7 human breast cancer cells and used as the E2-dependent reference cell line [53]. The E2 deprivation resistant and refractory MCF-7:2A (2A) cells and the sensitive to E2-induced apoptosis MCF-7:5C (5C) cells were also clonally selected from MCF-7 cells for maximal growth under long-term estrogen-free conditions. Estrogen-dependent WS8 cells were maintained in fully estrogenized media (phenol red containing RPMI-1640 and 10% whole FBS supplemented with 6 ng/mL insulin, 2 mM glutamine, 100 μM nonessential amino acids, and 100 U penicillin and streptomycin per mL), whereas 5C and 2A cells were maintained in estrogen-free medium (phenol red-free RPMI-1640 plus 10% dextran-coated charcoal-stripped FBS and the same supplements as for fully estrogenized medium) as previously described [11]. Cells were maintained at 37°C in a humidified 5% CO2 atmosphere. E2-dependent WS8 cells were switched to E2-free media (10% SFS) for 3 days, subsequently treated with 0.1% EtOH or 10−9 M E2 for 3 days and harvested. The DNA content of cells, a measure of proliferation, was determined as previously described [11]. For total RNA extraction for the validation of miRNAs, 5C, 2A and WS8 cells were seeded at 3×105/well in triplicate and harvested after 1 day. WS8 cells were switched to E2-free media beforehand for 3 days.
For miRNA mimic and inhibition experiments, 2A and 5C cells were loaded in 6-well plates with a density of 2×105/well in triplicate. After one day, 2A cells were transfected with miR-134-3p, 432-5p, 433-3p mimics and the respective negative control (NC), whereas 5C cells were transfected with respective miRNA inhibitors and negative control (NC) at final concentrations of 30 nM according to the manufacturers specifications. After three days, cells were double transfected with mimics or inhibitors. Cells were harvested 5 days after transfection for qRT-PCR analyses and measurement of protein expression levels of EGFR by Western Blot.
Western blotting
Proteins were extracted in cell lysis buffer (Cell Signaling Technology, Beverly, MA), supplemented with Proteinase Inhibitor Cocktail Set I and Phosphatase Inhibitor Cocktail Set II (Calbiochem, San Diego, CA). Western blotting was perfomed as previously described [14].
Quantitative real-time PCR
Quantitative RT-PCR assays were done as previously described [14] using SYBR Green PCR Master Mix from Applied Biosystems (Foster City, CA) on a QuantStudio 6 Flex7900HT Real-time PCR System (Applied Biosystems). For the validation of differentially expressed candidate miRNAs, commercially available TaqMan MicroRNA assays and reagents from ThermoFisher Scientific were used as described in the manufacturers specifications (ThermoFisher Scientific, Rockford, IL, USA). Total RNA was extracted with mirVana™ miRNA Isolation Kit. Briefly, 10 ng total RNA were reverse transcribed and cDNAs were diluted 1:12. For normalization of miRNA qRT-PCR results, RNU44 was used. In parallel, 1μg total RNA was converted to first-strand cDNA using a high capacity cDNA reverse transcription kit (Applied Biosystems) to quantitate expression levels of EGFR.
Total RNA extraction
Total RNA was isolated using the mirVanaTM miRNA Isolation Kit (Applied Biosystems, AM1560) according to the manufacturer's specifications. Briefly, 300 μl Lysis/Binding solution was added to 106 harvested cells and vortexed to obtain a homogenous lysate. RNA samples were checked for purity and integrity using a Nanodrop 1,000 spectrophotometer (Thermo Fisher Scientific, Inc., Waltham, MA, USA) and Agilent 2100 Bioanalyzer (Agilent G2938B) and stored at −80°C until further use.
MicroRNA microarray
MicroRNA expression profiles were generated utilising GeneChip miRNA2.0 arrays (Affymetrix Inc., Santa Clara, CA, USA) with 1 μg of each total RNA sample according to the manufacturer's instructions. The profiling series included a total of 12 samples. Affymetrix software miRNA QC Tool (Version 1.1.1.0, Affymetrix, 2010) was used for quality control of microarrays and preprocessing of expression data by robust multichip average (RMA). Resulting data were on log2-scale and preprocessed data from all 12 samples (triplicates for WS8, WS8 E2 72h, 2A and 5C) were extracted for this study. The microarray data have been submitted to the GEO online data repository with the accession number GSE79326.
Statistical analyses and bioinformatics
R-3.2.0 [54] with additional package limma-3.24.10 [55, 56] were used to test the expression data of the three groups (2A, 5C, WS8) against each other. In this investigation, only the 4560 human probe sets were considered. The Holm correction procedure [57] was applied to adjust resulting P-values for multiple testing unless otherwise specified. All statistical tests were two-sided unless otherwise specified. Statistical significance was defined as P < 0.05. All other statistical analyses including KEGG pathway and GO term enrichment analyses are described in detail in Supplementary materials.
SUPPLEMENTARY MATERIALS FIGURES
Acknowledgments
The results shown here are in part based on data generated by The Cancer Genome Atlas (TCGA) Research Network. We would like to thank the TCGA initiative, all tissue donors, and the investigators who contributed to the acquisition and analyses of the samples used in this study. Information about TCGA and the investigators and institutions constituting the TCGA research network can be found online (http://cancergenome.nih.gov).
Footnotes
CONFLICTS OF INTEREST
All authors declare no potential conflict of interest.
GRANT SUPPORT
This work was supported by the Robert Bosch Foundation, Stuttgart (RH, SW, HB), the German Cancer Consortium (DKTK) and German Cancer Research Center (DKFZ), Heidelberg (HB, FB), the Interfaculty Centre for Pharmacogenomics and Drug Research (ICEPHA), University of Tübingen (HB), and the Federal Ministry for Education and Research (BMBF, 01ZP0502, HB), Germany, and the 7FP EU Marie Curie Initial Training Network “FightingDrugFailure” (GA238132, HB).
This work was further supported by the National Institutes of Health (NIH), MD Anderson's Cancer Center support grant CA016672 and Susan G. Komen for the Cure Foundation under award number SAC100009, and Cancer Prevention Research Institute of Texas (CPRIT) for the STARs and STARs Plus Awards. VCJ thanks the benefactors of the Dallas/Ft. Worth Living Legend Chair of Cancer Research for their generous support.
REFERENCES
- 1.Davies C, Godwin J, Gray R, Clarke M, Cutter D, Darby S, McGale P, Pan HC, Taylor C, Wang YC, Dowsett M, Ingle J, Peto R. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet. 2011;378:771–84. doi: 10.1016/S0140-6736(11)60993-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dowsett M, Cuzick J, Ingle J, Coates A, Forbes J, Bliss J, Buyse M, Baum M, Buzdar A, Colleoni M, Coombes C, Snowdon C, Gnant M, et al. Meta-analysis of breast cancer outcomes in adjuvant trials of aromatase inhibitors versus tamoxifen. J Clin Oncol. 2010;28:509–18. doi: 10.1200/JCO.2009.23.1274. [DOI] [PubMed] [Google Scholar]
- 3.Jordan VC. The new biology of estrogen-induced apoptosis applied to treat and prevent breast cancer. Endocr Relat Cancer. 2015;22:R1–31. doi: 10.1530/ERC-14-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottardis MM, Jordan VC. Development of tamoxifen-stimulated growth of MCF-7 tumors in athymic mice after long-term antiestrogen administration. Cancer Res. 1988;48:5183–7. [PubMed] [Google Scholar]
- 5.Yao K, Lee ES, Bentrem DJ, England G, Schafer JI, O'Regan RM, Jordan VC. Antitumor action of physiological estradiol on tamoxifen-stimulated breast tumors grown in athymic mice. Clin Cancer Res. 2000;6:2028–36. [PubMed] [Google Scholar]
- 6.Lonning PE, Taylor PD, Anker G, Iddon J, Wie L, Jorgensen LM, Mella O, Howell A. High-dose estrogen treatment in postmenopausal breast cancer patients heavily exposed to endocrine therapy. Breast Cancer Res Treat. 2001;67:111–6. doi: 10.1023/a:1010619225209. [DOI] [PubMed] [Google Scholar]
- 7.Ellis MJ, Gao F, Dehdashti F, Jeffe DB, Marcom PK, Carey LA, Dickler MN, Silverman P, Fleming GF, Kommareddy A, Jamalabadi-Majidi S, Crowder R, Siegel BA. Lower-dose vs high-dose oral estradiol therapy of hormone receptor-positive, aromatase inhibitor-resistant advanced breast cancer: a phase 2 randomized study. JAMA. 2009;302:774–80. doi: 10.1001/jama.2009.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson GL, Chlebowski RT, Aragaki AK, Kuller LH, Manson JE, Gass M, Bluhm E, Connelly S, Hubbell FA, Lane D, Martin L, Ockene J, Rohan T, et al. Conjugated equine oestrogen and breast cancer incidence and mortality in postmenopausal women with hysterectomy: extended follow-up of the Women's Health Initiative randomised placebo-controlled trial. Lancet Oncol. 2012;13:476–86. doi: 10.1016/S1470-2045(12)70075-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang SY, Wolf DM, Yingling JM, Chang C, Jordan VC. An estrogen receptor positive MCF-7 clone that is resistant to antiestrogens and estradiol. Mol Cell Endocrinol. 1992;90:77–86. doi: 10.1016/0303-7207(92)90104-e. [DOI] [PubMed] [Google Scholar]
- 10.Pink JJ, Jiang SY, Fritsch M, Jordan VC. An estrogen-independent MCF-7 breast cancer cell line which contains a novel 80-kilodalton estrogen receptor-related protein. Cancer Res. 1995;55:2583–90. [PubMed] [Google Scholar]
- 11.Ariazi EA, Cunliffe HE, Lewis-Wambi JS, Slifker MJ, Willis AL, Ramos P, Tapia C, Kim HR, Yerrum S, Sharma CG, Nicolas E, Balagurunathan Y, Ross EA, et al. Estrogen induces apoptosis in estrogen deprivation-resistant breast cancer through stress responses as identified by global gene expression across time. Proc Natl Acad Sci U S A. 2011;108:18879–86. doi: 10.1073/pnas.1115188108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan P, Cunliffe HE, Maximov PY, Agboke FA, McDaniel RE, Zou X, Ramos P, Russell ML, Jordan VC. Integration of Downstream Signals of Insulin-like Growth Factor-1 Receptor by Endoplasmic Reticulum Stress for Estrogen-Induced Growth or Apoptosis in Breast Cancer Cells. Mol Cancer Res. 2015;13:1367–76. doi: 10.1158/1541-7786.MCR-14-0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sweeney EE, Fan P, Jordan VC. Mechanisms underlying differential response to estrogen-induced apoptosis in long-term estrogen-deprived breast cancer cells. Int J Oncol. 2014;44:1529–38. doi: 10.3892/ijo.2014.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan P, Griffith OL, Agboke FA, Anur P, Zou X, McDaniel RE, Creswell K, Kim SH, Katzenellenbogen JA, Gray JW, Jordan VC. c-Src modulates estrogen-induced stress and apoptosis in estrogen-deprived breast cancer cells. Cancer Res. 2013;73:4510–20. doi: 10.1158/0008-5472.CAN-12-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Obiorah IE, Fan P, Jordan VC. Breast cancer cell apoptosis with phytoestrogens is dependent on an estrogen-deprived state. Cancer Prev Res (Phila) 2014;7:939–49. doi: 10.1158/1940-6207.CAPR-14-0061. [DOI] [PubMed] [Google Scholar]
- 16.Sweeney EE, Fan P, Jordan VC. Molecular modulation of estrogen-induced apoptosis by synthetic progestins in hormone replacement therapy: an insight into the women's health initiative study. Cancer Res. 2014;74:7060–8. doi: 10.1158/0008-5472.CAN-14-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nana-Sinkam SP, Croce CM. Clinical applications for microRNAs in cancer. Clin Pharmacol Ther. 2013;93:98–104. doi: 10.1038/clpt.2012.192. [DOI] [PubMed] [Google Scholar]
- 18.Tessitore A, Cicciarelli G, Mastroiaco V, Vecchio FD, Capece D, Verzella D, Fischietti M, Vecchiotti D, Zazzeroni F, Alesse E. Therapeutic Use of MicroRNAs in Cancer. Anticancer Agents Med Chem. 2015;16:7–19. doi: 10.2174/1871520615666150824153358. [DOI] [PubMed] [Google Scholar]
- 19.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Lee CG. MicroRNA and cancer—focus on apoptosis. J Cell Mol Med. 2009;13:12–23. doi: 10.1111/j.1582-4934.2008.00510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klinge CM. miRNAs and estrogen action. Trends Endocrinol Metab. 2012;23:223–33. doi: 10.1016/j.tem.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, Coppola D, Cheng JQ. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem. 2008;283:31079–86. doi: 10.1074/jbc.M806041200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Cittelly DM, Das PM, Salvo VA, Fonseca JP, Burow ME, Jones FE. Oncogenic HER2{Delta}16 suppresses miR-15a/16 and deregulates BCL-2 to promote endocrine resistance of breast tumors. Carcinogenesis. 2010;31:2049–57. doi: 10.1093/carcin/bgq192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cittelly DM, Das PM, Spoelstra NS, Edgerton SM, Richer JK, Thor AD, Jones FE. Downregulation of miR-342 is associated with tamoxifen resistant breast tumors. Mol Cancer. 2010;9:317. doi: 10.1186/1476-4598-9-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi W, Gerster K, Alajez NM, Tsang J, Waldron L, Pintilie M, Hui AB, Sykes J, P'ng C, Miller N, McCready D, Fyles A, Liu FF. MicroRNA-301 mediates proliferation and invasion in human breast cancer. Cancer Res. 2011;71:2926–37. doi: 10.1158/0008-5472.CAN-10-3369. [DOI] [PubMed] [Google Scholar]
- 26.Hayes EL, Lewis-Wambi JS. Mechanisms of endocrine resistance in breast cancer: an overview of the proposed roles of noncoding RNA. Breast Cancer Res. 2015;17:40. doi: 10.1186/s13058-015-0542-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bailey ST, Westerling T, Brown M. Loss of estrogen-regulated microRNA expression increases HER2 signaling and is prognostic of poor outcome in luminal breast cancer. Cancer Res. 2015;75:436–45. doi: 10.1158/0008-5472.CAN-14-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benetatos L, Hatzimichael E, Londin E, Vartholomatos G, Loher P, Rigoutsos I, Briasoulis E. The microRNAs within the DLK1-DIO3 genomic region: involvement in disease pathogenesis. Cell Mol Life Sci. 2013;70:795–814. doi: 10.1007/s00018-012-1080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dvinge H, Git A, Graf S, Salmon-Divon M, Curtis C, Sottoriva A, Zhao Y, Hirst M, Armisen J, Miska EA, Chin SF, Provenzano E, Turashvili G, et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature. 2013;497:378–82. doi: 10.1038/nature12108. [DOI] [PubMed] [Google Scholar]
- 30.Khuu C, Utheim TP, Sehic A. The Three Paralogous MicroRNA Clusters in Development and Disease, miR-17-92, miR-106a-363, and miR-106b-25. Scientifica (Cairo) 2016;2016:1379643. doi: 10.1155/2016/1379643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muluhngwi P, Klinge CM. Roles for miRNAs in endocrine resistance in breast cancer. Endocr Relat Cancer. 2015;22:R279–R300. doi: 10.1530/ERC-15-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rothe F, Ignatiadis M, Chaboteaux C, Haibe-Kains B, Kheddoumi N, Majjaj S, Badran B, Fayyad-Kazan H, Desmedt C, Harris AL, Piccart M, Sotiriou C. Global microRNA expression profiling identifies MiR-210 associated with tumor proliferation, invasion and poor clinical outcome in breast cancer. PLoS One. 2011;6:e20980. doi: 10.1371/journal.pone.0020980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sossey-Alaoui K, Downs-Kelly E, Das M, Izem L, Tubbs R, Plow EF. WAVE3, an actin remodeling protein, is regulated by the metastasis suppressor microRNA, miR-31, during the invasion-metastasis cascade. Int J Cancer. 2011;129:1331–43. doi: 10.1002/ijc.25793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Augoff K, Das M, Bialkowska K, McCue B, Plow EF, Sossey-Alaoui K. miR-31 is a broad regulator of beta1-integrin expression and function in cancer cells. Mol Cancer Res. 2011;9:1500–8. doi: 10.1158/1541-7786.MCR-11-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Körner C, Keklikoglou I, Bender C, Worner A, Munstermann E, Wiemann S. MicroRNA-31 sensitizes human breast cells to apoptosis by direct targeting of protein kinase C epsilon (PKCepsilon) J Biol Chem. 2013;288:8750–61. doi: 10.1074/jbc.M112.414128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y, Liang C, Ma H, Zhao Q, Lu Y, Xiang Z, Li L, Qin J, Chen Y, Cho WC, Pestell RG, Liang L, Yu Z. miR-221/222 promotes S-phase entry and cellular migration in control of basal-like breast cancer. Molecules. 2014;19:7122–37. doi: 10.3390/molecules19067122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stinson S, Lackner MR, Adai AT, Yu N, Kim HJ, O'Brien C, Spoerke J, Jhunjhunwala S, Boyd Z, Januario T, Newman RJ, Yue P, Bourgon R, et al. TRPS1 targeting by miR-221/222 promotes the epithelial-to-mesenchymal transition in breast cancer. Sci Signal. 2011;4:ra41. doi: 10.1126/scisignal.2001538. [DOI] [PubMed] [Google Scholar]
- 38.Wei Y, Lai X, Yu S, Chen S, Ma Y, Zhang Y, Li H, Zhu X, Yao L, Zhang J. Exosomal miR-221/222 enhances tamoxifen resistance in recipient ER-positive breast cancer cells. Breast Cancer Res Treat. 2014;147:423–31. doi: 10.1007/s10549-014-3037-0. [DOI] [PubMed] [Google Scholar]
- 39.Vilquin P, Donini CF, Villedieu M, Grisard E, Corbo L, Bachelot T, Vendrell JA, Cohen PA. MicroRNA-125b upregulation confers aromatase inhibitor resistance and is a novel marker of poor prognosis in breast cancer. Breast Cancer Res. 2015;17:13. doi: 10.1186/s13058-015-0515-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bader AG, Brown D, Winkler M. The promise of microRNA replacement therapy. Cancer Res. 2010;70:7027–30. doi: 10.1158/0008-5472.CAN-10-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma CX, Reinert T, Chmielewska I, Ellis MJ. Mechanisms of aromatase inhibitor resistance. Nat Rev Cancer. 2015;15:261–75. doi: 10.1038/nrc3920. [DOI] [PubMed] [Google Scholar]
- 42.Fan P, Wang J, Santen RJ, Yue W. Long-term treatment with tamoxifen facilitates translocation of estrogen receptor alpha out of the nucleus and enhances its interaction with EGFR in MCF-7 breast cancer cells. Cancer Res. 2007;67:1352–60. doi: 10.1158/0008-5472.CAN-06-1020. [DOI] [PubMed] [Google Scholar]
- 43.Gilani RA, Kazi AA, Shah P, Schech AJ, Chumsri S, Sabnis G, Jaiswal AK, Brodie AH. The importance of HER2 signaling in the tumor-initiating cell population in aromatase inhibitor-resistant breast cancer. Breast Cancer Res Treat. 2012;135:681–92. doi: 10.1007/s10549-012-2148-8. [DOI] [PubMed] [Google Scholar]
- 44.Fagan DH, Uselman RR, Sachdev D, Yee D. Acquired resistance to tamoxifen is associated with loss of the type I insulin-like growth factor receptor: implications for breast cancer treatment. Cancer Res. 2012;72:3372–80. doi: 10.1158/0008-5472.CAN-12-0684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qin Q, Wei F, Zhang J, Wang X, Li B. miR-134 inhibits non-small cell lung cancer growth by targeting the epidermal growth factor receptor. J Cell Mol Med. 2016 doi: 10.1111/jcmm.12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fan P, Agboke FA, Cunliffe HE, Ramos P, Jordan VC. A molecular model for the mechanism of acquired tamoxifen resistance in breast cancer. Eur J Cancer. 2014;50:2866–76. doi: 10.1016/j.ejca.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Massarweh S, Osborne CK, Creighton CJ, Qin L, Tsimelzon A, Huang S, Weiss H, Rimawi M, Schiff R. Tamoxifen resistance in breast tumors is driven by growth factor receptor signaling with repression of classic estrogen receptor genomic function. Cancer Res. 2008;68:826–33. doi: 10.1158/0008-5472.CAN-07-2707. [DOI] [PubMed] [Google Scholar]
- 48.Chumsri S, Sabnis G, Tkaczuk K, Brodie A. mTOR inhibitors: changing landscape of endocrine-resistant breast cancer. Future Oncol. 2014;10:443–56. doi: 10.2217/fon.13.178. [DOI] [PubMed] [Google Scholar]
- 49.Fruman DA, Rommel C. PI3K and cancer: lessons, challenges and opportunities. Nat Rev Drug Discov. 2014;13:140–56. doi: 10.1038/nrd4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Z, Han Y, Cheng K, Zhang G, Wang X. miR-99a directly targets the mTOR signalling pathway in breast cancer side population cells. Cell Prolif. 2014;47:587–95. doi: 10.1111/cpr.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qian P, He XC, Paulson A, Li Z, Tao F, Perry JM, Guo F, Zhao M, Zhi L, Venkatraman A, Haug JS, Parmely T, Li H, et al. The Dlk1-Gtl2 Locus Preserves LT-HSC Function by Inhibiting the PI3K-mTOR Pathway to Restrict Mitochondrial Metabolism. Cell Stem Cell. 2015 doi: 10.1016/j.stem.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garg M. Targeting microRNAs in epithelial-to-mesenchymal transition-induced cancer stem cells: therapeutic approaches in cancer. Expert Opin Ther Targets. 2015;19:285–97. doi: 10.1517/14728222.2014.975794. [DOI] [PubMed] [Google Scholar]
- 53.Pink JJ, Jordan VC. Models of estrogen receptor regulation by estrogens and antiestrogens in breast cancer cell lines. Cancer Res. 1996;56:2321–30. [PubMed] [Google Scholar]
- 54.R Core Team . R Foundation for Statistical Computing. Vienna, Austria: 2015. R: A language and environment for statistical computing. [Google Scholar]
- 55.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 57.Holm S. A simple sequentially rejective multiple test procedure. Scandinavian Journal of Statistics. 1979;6:65–70. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.