Abstract
Considerable studies have investigated the associations between MDM4 gene polymorphisms and cancer risk recently, but with contradictory results. The aim of this meta-analysis was to evaluate the associations between MDM4 gene polymorphisms and cancer risk. Relevant studies were identified by a systematic search of PubMed, Embase, and CNKI databases. Crude odds ratios (ORs) and 95% confidence intervals (CIs) were used to describe the strength of the associations. Fifty-six studies published in 11 publications involving 18,910 cases and 51,609 controls were included in this meta-analysis. Five MDM4 gene polymorphisms were evaluated: rs4245739, rs1563828, rs11801299, rs10900598, and rs1380576. Our analyses suggested that the rs4245739 polymorphism was significantly associated with overall cancer risk. Furthermore, stratification analyses of ethnicity indicated that rs4245739 decreased the risk of cancer among the Asian population, and stratification analyses of smoking status indicated that rs4245739 decreased the risk of cancer among nonsmokers. However, stratification analyses of cancer type and sex suggested that rs4245739 was not related to cancer risk. There were no associations of rs1563828, rs11801299, rs10900598, or rs1380576 with overall cancer risk. In conclusion, our analyses indicated that rs4245739 polymorphism in the MDM4 gene may play an important role in the etiology of cancer.
Keywords: MDM4, single nucleotide polymorphisms, meta-analysis, cancer
INTRODUCTION
Cancer is a worldwide health problem and the second leading cause of morbidity and mortality in human diseases [1]. Cancer metastasis to other parts of the body is the major cause of death [2]. Chen et al. reported that approximately 4,292,000 new cancer cases and 2,814,000 cancer deaths would occur in China in 2015 [3]. It is obvious that cancer is a major threat to human health. However, the exact mechanism of carcinogenesis is still poorly understood. An increasing number of studies have reported that cancer is a complex disease, resulting from environmental and genetic factors and their interactions [4, 5]. In addition, genetic factors play important roles in the pathogenesis of cancer, and many genes have been identified as cancer-susceptible genes [6].
The tumor suppressor p53 plays an important role in many physiological processes, such as maintenance of genomic stability and control of cell growth and apoptosis [7, 8]. Inactivation of the p53 tumor suppressor is critical for tumorigenesis. The activity of p53 is strictly regulated, predominantly by MDM2 and its homolog MDM4 [9]. MDM4 shares an N-terminal p53-binding domain with MDM2 and can inhibit p53 activity during various malignancies [10–12]. Overexpressed MDM4 in human tumors may contribute to reduced p53 activity and tumorigenesis [13]. Many studies [14, 15] have demonstrated that mouse embryos lacking MDM4 die during embryogenesis. Migliorini et al. found MDM4 regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development [14]. Furthermore, transgenic mice overexpressing MDM4 showed spontaneous carcinogenesis and accelerated tumorigenesis [16]. Additionally, MDM4 is reported to be highly expressed in a significant percentage of human cancers, including 65% of retinoblastomas, 80% of adult pre-B lymphoblastic leukemia, 39% of head and neck squamous carcinomas, 19% of colon cancers, 19% of breast cancers, and 18% of lung cancers [17–20]. All above studies indicated that MDM4 gene may be significantly associated with cancer susceptibility.
Recently, many studies [13, 21–30] have described the associations between MDM4 gene polymorphisms and risks of various cancers, including colon cancer, lung cancer, breast cancer, gastric cancer, squamous cell carcinoma of the head and neck, non-Hodgkin lymphoma (NHL), and esophageal squamous cell carcinoma (ESCC). However, the results of these studies were conflicting and inconclusive. The clinical heterogeneity, different ethnic populations, and small sample sizes of previous studies may have contributed to these disparities. To overcome these limitations, we performed a meta-analysis of the contradictory results from these relevant studies to clarify the possible associations between MDM4 gene polymorphisms and cancer risk.
RESULTS
Characteristics of the included publications
A total of 149 publications were identified after our initial search. After removing duplicates and screening the titles and abstracts, 128 publications were removed. Finally, 21 publications were selected for further full text review. The following publications were excluded: four [31–34] in which the experimental designs were not case control studies; two publications [35, 36] that did not describe MDM4 polymorphisms (rs4245739, rs1563828, rs11801299, rs10900598, and rs1380576) and cancer risk, three [37–39] that not provide detailed genotyping data, and two [28, 40] that might have described partially overlapping populations, so we included the study [28] with the larger sample size. We finally identified 11 eligible publications [13, 21–30] including 56 studies (18,910 cases and 51,609 controls) in this meta-analysis. Selection for eligible publications included in this meta-analysis was presented in Figure 1. The characteristics of these included studies are summarized in Table 1. These publications were published from 2011 to 2015. Five single nucleotide polymorphisms (SNPs) (rs4245739, rs1563828, rs11801299, rs10900598 and rs1380576) of MDM4 gene were investigated. Genotype distributions of the controls about rs1380576 in one study [26] did not conform to HWE (P < 0.001). The NOS scores of all included studies ranged from 5 to 7 stars, suggesting that they were studies of high methodological quality. Four papers [13, 22, 27, 28] were carried out in Caucasian populations, and seven [21, 23–26, 29, 30] in Asian populations. The research [27] conducted by Garcia-Closaset et al. consisted of three genome-wide association studies (GWASs) involving 40 studies among Caucasian populations.
Figure 1. Selection for eligible publications included in this meta-analysis.
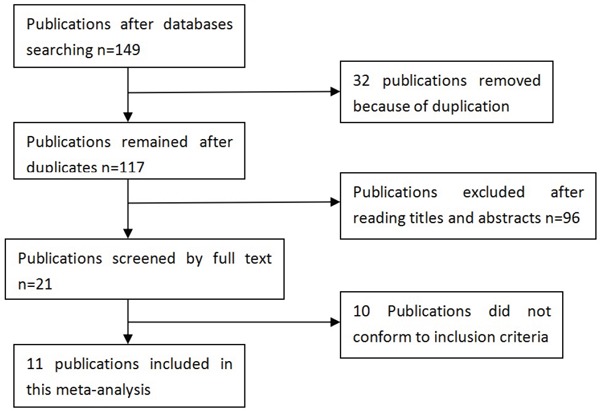
Table 1. Characteristics of included studies.
| Author and year | Country | Ethnicity | Case | Control | Cancer type | HWE | NOS | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs4245739 | AA | AC | CC | AA | AC | CC | |||||
| Gansmo2015 | Norway | Caucasian | 823 | 600 | 108 | 2042 | 1439 | 266 | Colon cancer | 0.566 | 6 |
| Gansmo2015 | Norway | Caucasian | 715 | 515 | 101 | 2042 | 1439 | 266 | Lung cancer | 0.566 | 6 |
| Gansmo2015 | Norway | Caucasian | 1412 | 927 | 161 | 2042 | 1439 | 266 | Prostate cancer | 0.566 | 6 |
| Gansmo2015 | Norway | Caucasian | 966 | 643 | 108 | 2042 | 1439 | 266 | Breast cancer | 0.566 | 6 |
| Gao2015 | China | Asian | 297 | 22 | 1 | 548 | 90 | 2 | SCLC | 0.399 | 5 |
| Gao2015 | China | Asian | 183 | 17 | 0 | 321 | 77 | 2 | SCLC | 0.248 | 7 |
| Fan2014 | China | Asian | 187 | 13 | 0 | 346 | 53 | 1 | NHL | 0.487 | 6 |
| Liu2013 | China | Asian | 733 | 67 | 0 | 686 | 111 | 3 | Breast cancer | 0.505 | 6 |
| Liu2013 | China | Asian | 278 | 22 | 0 | 501 | 96 | 3 | Breast cancer | 0.484 | 6 |
| Zhou2013 | China | Asian | 501 | 37 | 2 | 478 | 70 | 2 | ESCC | 0.740 | 6 |
| Zhou2013 | China | Asian | 529 | 56 | 3 | 510 | 88 | 2 | ESCC | 0.379 | 6 |
| Garcia-Closas2013 | Mixed | Caucasian | 3318 | 2637 | 557 | 22825 | 15798 | 2828 | Breast cancer | 0.183 | 5 |
| rs1563828 | CC | CT | TT | CC | CT | TT | |||||
| Zhang2012 | China | Asian | 98 | 91 | 21 | 90 | 88 | 22 | NPC | 0.944 | 7 |
| Song2012 | China | Asian | 53 | 57 | 14 | 44 | 43 | 14 | Breast cancer | 0.506 | 5 |
| rs11801299 | GG | AG | AA | GG | AG | AA | |||||
| Wang2012 | America | Caucasian | 195 | Na | Na | 201 | Na | Na | Oral cancer | Na | 6 |
| Yu2011 | America | Caucasian | 684 | 351 | 40 | 665 | 376 | 38 | SCCHN | 0.086 | 6 |
| rs10900598 | GG | GT | TT | GG | GT | TT | |||||
| Wang2012 | America | Caucasian | 107 | Na | Na | 94 | Na | Na | Oral cancer | Na | 6 |
| Yu2011 | America | Caucasian | 307 | 545 | 223 | 296 | 552 | 231 | SCCHN | 0.377 | 6 |
| rs1380576 | CC | CG | GG | CC | CG | GG | |||||
| Wu2015 | China | Asian | 188 | 281 | 173 | 212 | 290 | 218 | Gastric cancer | <0.001 | 5 |
| Wang2012 | America | Caucasian | 141 | Na | Na | 149 | Na | Na | Oral cancer | Na | 6 |
| Yu2011 | America | Caucasian | 487 | 477 | 111 | 518 | 455 | 106 | SCCHN | 0.677 | 6 |
HWE, Hardy–Weinberg equilibrium; NOS, Newcastle–Ottawa scale; NPC, Nasopharyngeal carcinoma; SCCHN, squamous cell carcinoma of the head and neck; SCLC, Small Cell Lung Cancer; NHL, Non-Hodgkin lymphoma; ESCC, esophageal squamous cell carcinoma; Na, not available.
Meta-analysis of rs4245739
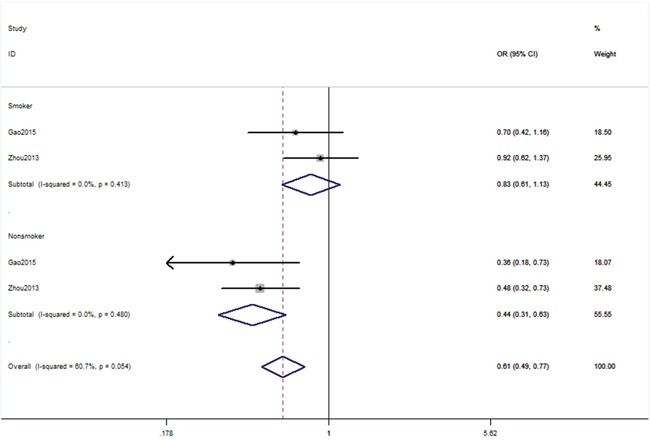
Six publications [13, 21, 24, 25, 27, 29] including 51 studies with 16,539 cases and 49,188 controls examined rs4245739 polymorphism. As shown in Table 2, rs4245739 polymorphism was significantly associated with a decreased risk of overall cancer risk in three models (C vs. A: OR, 0.78; 95% CI, 0.68–0.89, P < 0.001; CC+AC vs. AA: OR, 0.75; 95% CI, 0.64–0.87, P < 0.001; AC vs. AA: OR, 0.75; 95% CI, 0.64–0.87, P < 0.001, Figure 2). Stratification analyses were conducted according to ethnicity, cancer type, sex and smoking status. Our data indicated that rs4245739 polymorphism was also significantly associated with a decreased risk of cancer among Asian populations in three models (C vs. A: OR, 0.53; 95% CI, 0.45–0.62, P < 0.001; CC+AC vs. AA: OR, 0.51; 95% CI, 0.43–0.60, P < 0.001; AC vs. AA: OR, 0.51; 95% CI, 0.43–0.60, P < 0.001, Figure 3). Stratification analyses of smoking status indicated rs4245739 decreased the risk of nonsmokers (CC+AC vs. AA: OR, 0.44; 95% CI, 0.31–0.63, P < 0.001, Figure 4). However, stratification analyses of cancer type suggested rs4245739 was not related with the risks of lung cancer, breast cancer, gastrointestinal cancer and other cancers. Stratification analyses of sex also indicated this SNP was not associated with cancer risk. All included studies conform to HWE, indicating their controls subjects were representative of the general population.
Table 2. Meta-analysis of associations between the rs4245739 polymorphism and cancer risk.
| Comparison | Overall and Stratification analyses | Studies | OR (95% CI) | P-value | Random/Fixed effect model | P for heterogeneity | I2 (%) |
|---|---|---|---|---|---|---|---|
| C vs. A | Overall | 51 | 0.78(0.68,0.89) | <0.001 | Random | <0.001 | 90.6 |
| Caucasian | 44 | 1.02(0.92,1.12) | 0.301 | Random | <0.001 | 88.5 | |
| Asian | 7 | 0.53 (0.45,0.62) | <0.001 | Random | 0.584 | 0.0 | |
| Gastrointestinal cancer | 3 | 0.74(0.49,1.12) | 0.157 | Random | 0.001 | 85.6 | |
| Lung cancer | 3 | 0.61(0.31,1.19) | 0.148 | Random | <0.001 | 90.4 | |
| Breast cancer | 43 | 0.78 (0.59,1.02) | 0.070 | Random | <0.001 | 94.4 | |
| Other cancers | 2 | 0.70(0.35,1.39) | 0.307 | Random | 0.024 | 80.5 | |
| CC vs. AA+AC | Overall | 47 | 1.04(0.88,1.21) | 0.666 | Random | 0.016 | 59.5 |
| Caucasian | 44 | 1.03 (0.86,1.23) | 0.748 | Random | 0.584 | 0.0 | |
| Asian | 3 | 1.20(0.38,3.84) | 0.754 | Random | 0.032 | 78.1 | |
| Gastrointestinal cancer | 3 | 1.00 (0.80,1.26) | 0.996 | Random | 0.895 | 0.0 | |
| Lung cancer | 2 | 1.07(0.85,1.36) | 0.556 | Random | 0.953 | 0.0 | |
| Breast cancer | 41 | 1.08(0.75,1.55) | 0.696 | Random | 0.003 | 88.4 | |
| Other cancers | 1 | 0.90 (0.74,1.10) | 0.312 | Na | Na | Na | |
| CC+AC vs. AA | Overall | 51 | 0.75(0.64,0.87) | <0.001 | Random | <0.001 | 90.1 |
| Caucasian | 44 | 1.02(0.91,1.14) | 0.740 | Random | <0.001 | 85.5 | |
| Asian | 7 | 0.51(0.43,0.60) | <0.001 | Random | 0.662 | 0.0 | |
| Gastrointestinal cancer | 3 | 0.72 (0.45,1.14) | 0.157 | Random | <0.001 | 87.0 | |
| Lung cancer | 3 | 0.59(0.29,1.20) | 0.142 | Random | <0.001 | 90.5 | |
| Breast cancer | 43 | 0.76(0.56,1.04) | 0.089 | Random | <0.001 | 94.0 | |
| Other cancers | 2 | 0.69 (0.34,1.39) | 0.294 | Random | 0.025 | 80.0 | |
| Male | 5 | 0.72 (0.49,1.07) | 0.107 | Random | 0.008 | 70.8 | |
| Female | 5 | 0.62 (0.32,1.19) | 0.150 | Random | <0.001 | 83.4 | |
| Smoker | 4 | 0.83 (0.61,1.13) | 0.237 | Fixed | 0.413 | 0.0 | |
| Nonsmoker | 4 | 0.44 (0.31,0.63) | <0.001 | Fixed | 0.480 | 0.0 | |
| CC vs. AA | Overall | 47 | 1.04(0.85,1.26) | 0.360 | Random | 0.001 | 71.8 |
| Caucasian | 44 | 1.03(0.83,1.28) | 0.769 | Random | <0.001 | 83.8 | |
| Asian | 3 | 1.13 (0.35,3.59) | 0.840 | Random | 0.938 | 0.0 | |
| Gastrointestinal cancer | 3 | 1.01(0.80,1.28) | 0.915 | Random | <0.001 | 90.6 | |
| Lung cancer | 2 | 1.08(0.85,1.38) | 0.522 | Random | <0.001 | 88.5 | |
| Breast cancer | 41 | 1.09 (0.70,1.71) | 0.698 | Random | 0.584 | 0.0 | |
| Other cancers | 1 | 0.88(0.71,1.08) | 0.208 | Random | 0.032 | 78.1 | |
| AC vs. AA | Overall | 51 | 0.75 (0.64,0.87) | <0.001 | Random | <0.001 | 88.6 |
| Caucasian | 44 | 1.02 (0.93,1.12) | 0.694 | Random | 0.001 | 77.3 | |
| Asian | 7 | 0.51(0.43,0.60) | <0.001 | Random | 0.747 | 0.0 | |
| Gastrointestinal cancer | 3 | 0.70(0.43,1.14) | 0.157 | Random | <0.001 | 87.7 | |
| Lung cancer | 3 | 0.58(0.29,1.19) | 0.136 | Random | <0.001 | 90.0 | |
| Breast cancer | 43 | 0.78(0.58,1.04) | 0.091 | Random | <0.001 | 92.6 | |
| Other cancers | 2 | 0.70(0.35,1.39) | 0.307 | Random | 0.028 | 79.3 |
*Bold values are statistically significant (P < 0.05). OR, odds ratio; 95% CI, 95% confidence interval; Na, not available.
Figure 2. Forest plot shows odds ratio for the associations between rs4245739 and cancer risk (a: C vs. A; b: CC+AC vs. AA; c: AC vs. AA).
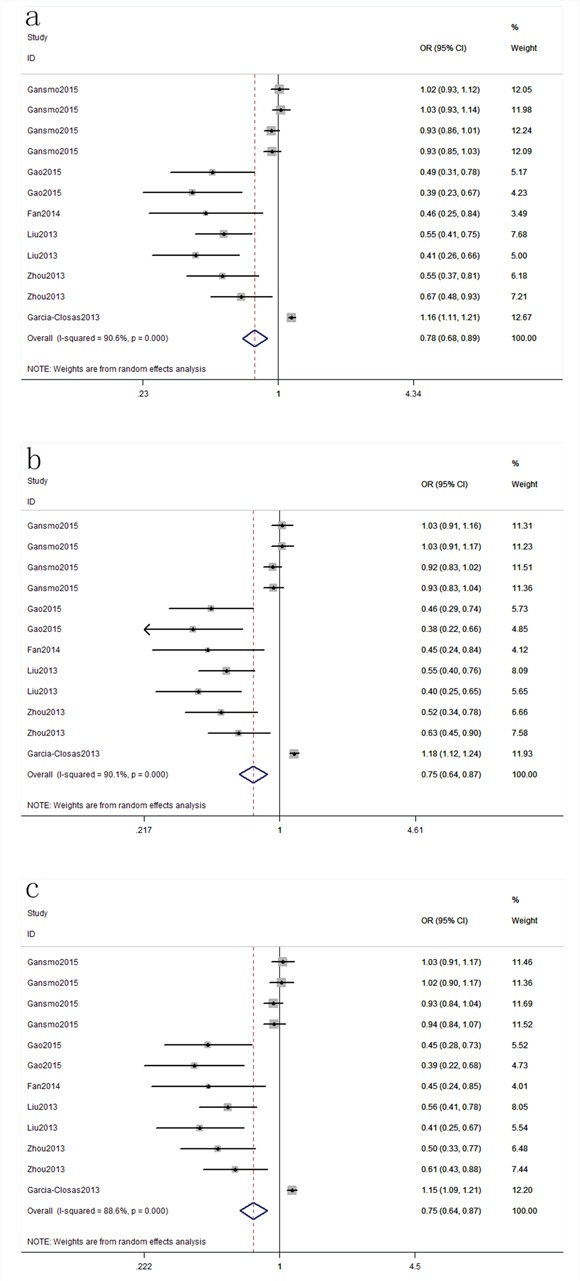
Figure 3. Stratification analyses of ethnicity between rs4245739 and cancer risk (a: C vs. A; b: CC+AC vs. AA; c: AC vs. AA).
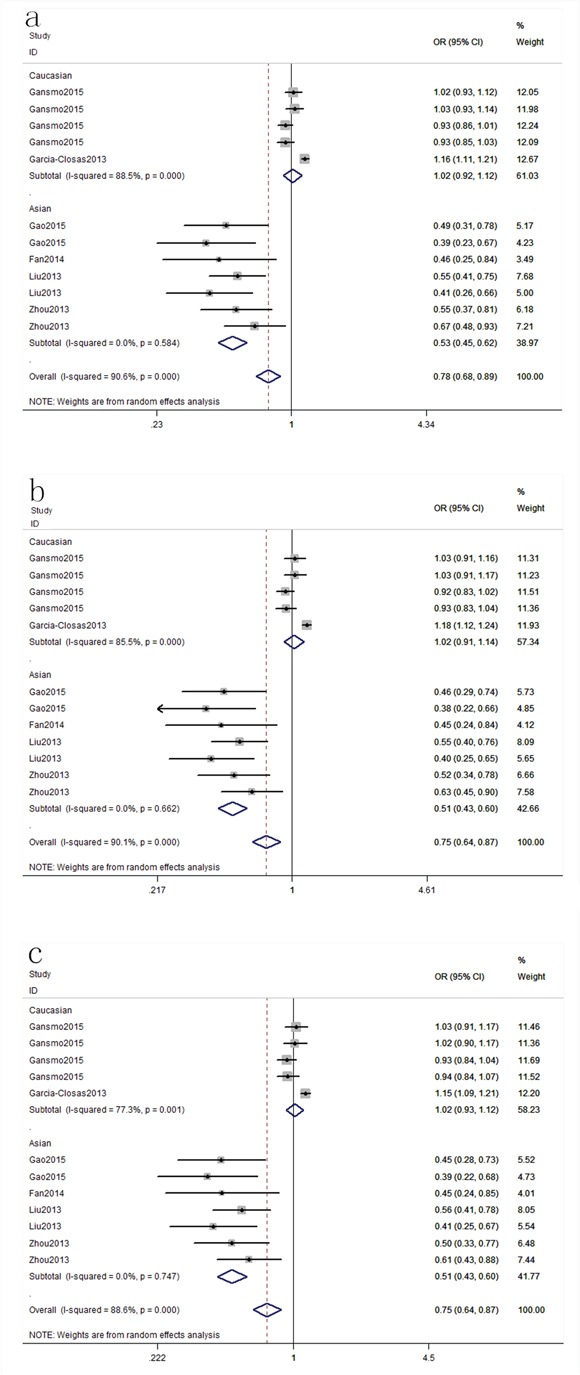
Figure 4. Stratification analyses of smoke status between rs4245739 and cancer risk (CC+AC vs. AA).
Meta-analysis of rs1563828, rs11801299, rs10900598 and rs1380576
Two publications [23, 30] with 334 cases and 301 controls examined rs1563828 polymorphism; two publications [22, 28] with 1,395 cases and 1,400 controls studied rs11801299 polymorphism; two publications [22, 28] with 1,395 cases and 1,400 controls studied rs10900598 polymorphism; three publications [22, 26, 28] with 2,037 cases and 2,120 controls studied rs1380576 polymorphism. With regard to rs11801299, rs10900598 and rs1380576, we could only calculate the data in dominant model as they did not provide enough data. As shown in Table 3, we found these four SNPs were not associated with cancer risk. For these four SNPs, we did not perform stratification analyses due to limited data. Genotype distributions of the controls about rs1380576 in one study [26] did not conform to HWE (P < 0.001). By exclusion of this study, the pooled estimates of the remaining studies showed that rs1380576 polymorphism was also not associated with cancer risk (CG+GG vs. CC: OR, 1.11; 95% CI, 0.96–1.29, P = 0.164), suggesting that the results of this SNP was stable.
Table 3. Meta-analysis of associations between rs1563828, rs11801299, rs10900598 and rs1380576 polymorphisms and cancer risk.
| Comparison | OR(95%CI) | P-value | Random/Fixed effect model | P for heterogeneity | I2 (%) |
|---|---|---|---|---|---|
| rs1563828 | |||||
| T vs. C | 0.95(0.75,1.20) | 0.658 | Fixed | 0.928 | 0.0% |
| TT+CT vs. CC | 0.96(0.70,1.32) | 0.814 | Fixed | 0.744 | 0.0% |
| TT vs. CC+CT | 0.86(0.52,1.40) | 0.536 | Fixed | 0.804 | 0.0% |
| CT vs. CC | 1.00(0.72,1.39) | 0.997 | Fixed | 0.678 | 0.0% |
| TT vs. CC | 0.86(0.51,1.45) | 0.566 | Fixed | 0.921 | 0.0% |
| rs11801299 | |||||
| AG+AA vs. GG | 0.95(0.82,1.11) | 0.529 | Fixed | 0.399 | 0.0% |
| rs10900598 | |||||
| GT+TT vs. GG | 0.91(0.78,1.08) | 0.288 | Fixed | 0.483 | 0.0% |
| rs1380576 | |||||
| CG+GG vs. CC | 1.08(0.95,1.22) | 0.227 | Fixed | 0.785 | 0.0% |
*Bold values are statistically significant (P < 0.05).
Sensitivity analysis and publication bias
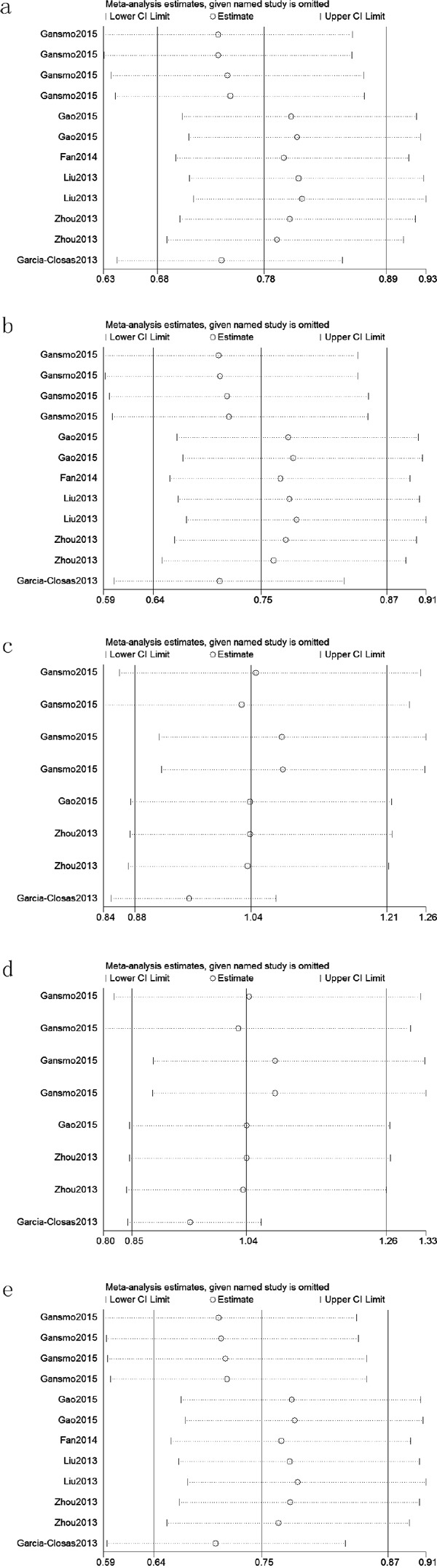
We assessed sensitivity by omitting each study once at a time in every genetic model for rs4245739. The pooled ORs for the effects of rs4245739 on the risk for cancer risk indicated that our data were stable and trustworthy about this SNP (Figure 5). We did not perform sensitivity analyses about rs1563828, rs11801299, rs10900598 and rs1380576 due to limited data. Both Egger's and Begg's tests were used to evaluated the publication bias of this meta-analysis. Our data revealed that there was obvious publication bias in three models for rs4245739 (C vs. A, Pbegg = 0.075 and Pegger < 0.001; CC+AC vs. AA, Pbegg = 0.055 and Pegger< 0.001; AC vs. AA, Pbegg = 0.040 and Pegger < 0.001). Due to limited studies, we did not conduct Egger's and Begg's tests about rs1563828, rs11801299, rs10900598 and rs1380576.
Figure 5. Sensitivity analyses between rs4245739 and cancer risk in five models (a: allele model; b: dominant model; c: recessive model; d: homozygous model; e: heterozygous model).
DISCUSSION
To the best of our knowledge, this is the first meta-analysis emphasizing the associations between MDM4 gene polymorphisms and cancer risk. Our data indicated that the rs4245739 polymorphism was significantly associated with a decreased risk of cancer overall. In addition, stratification analyses of ethnicity indicated that rs4245739 decreased the risk of cancer in the Asian population, and stratification analyses of smoking status indicated that rs4245739 decreased the risk of cancer among nonsmokers. However, stratification analyses of cancer type suggested rs4245739 was not related to the risks of lung cancer, breast cancer, gastrointestinal cancer, and other cancers. Stratification analyses of sex also indicated this SNP was not associated with cancer risk in male or female populations. With regard to the remaining four SNPs, no associations were found between rs1563828, rs11801299, rs10900598, or rs1380576and overall cancer risk.
One of most important tumor suppressors in human cells is p53. MDM2, a key regulator of the p53 tumor suppressor signaling pathway, can induce degradation of p53 through the ubiquitin-proteasome pathway [41]. MDM4 is structurally homologous to MDM2, and MDM4 can cooperate with MDM2 to inhibit p53 activities [10]. Furthermore, MDM4 can interact with MDM2 via the Really Interesting New Gene finger domain that inhibits the degradation of MDM2 protein [12, 42]. In light of these findings, we hypothesize that MDM4 may play pivotal roles in the pathogenesis of cancer, and that MDM4 is a candidate susceptibility gene for cancer. Currently, many studies [13, 21–30] have investigated the associations between MDM4 gene polymorphisms and cancer risk. However, these studies showed inconsistent results, because they had inadequate statistical power due to relatively small samples.
To provide a comprehensive and reliable conclusion, we conducted a meta-analysis to assess the associations between MDM4 gene polymorphisms and cancer risk. Our data indicated that the rs4245739 polymorphism was significantly associated with a decreased risk of overall cancer. Six publications [13, 21, 24, 25, 27, 29] including 51 studies involving 16,539 cases and 49, 188 controls investigated this SNP. Among these included publications, four [21, 24, 25, 29] from China assessed this variant in ESCC, small cell lung cancer (SCLC), and NHL and breast cancer and concluded that rs4245739 was associated with a reduced risk of cancer. Studies from Norway [13] suggested that rs4245739 is associated with a reduced risk of breast cancer but is not associated with lung cancer, colon cancer, or prostate cancer. Studies by Garcia-Closaset et al. [27], consisting of three GWAS studies among Caucasian populations, indicated that rs4245739 increased the risk of breast cancer. As mentioned previously, we found that the associations between rs4245739 and breast cancer risk were inconsistent. Two studies [13, 24] reported that rs4245739 decreased the risk of breast cancer, while the study by Garcia-Closaset et al. [27] reported that it increased the risk of breast cancer. It is noteworthy that Garcia-Closaset et al. reported that rs4245739 increased the risk of estrogen receptor (ER)-negative but not ER-positive breast cancer, which was consistent with the conclusion of two other GWAS studies [37, 38]. There are important differences in genetic susceptibility with these two types (ER-negative and ER-positive) of breast cancer. Garcia-Closaset et al. reported that rs4245739 is located in an ER-negative-specific breast cancer risk locus. It is therefore reasonable to hypothesize that this SNP may specifically affect susceptibility to ER-negative breast cancer. Stratification analyses of cancer type in this meta-analysis concluded that rs4245739 is not associated with overall breast cancer. The reasons why the results of Garcia-Closaset et al. differed from the results of those two studies [13, 24] and this meta-analysis are unclear, but it may be partially explained by differences in the genetic susceptibility of different types of breast cancer. Due to limited data, we could not conduct stratification analyses of ER status. Larger studies are therefore needed to identify the possible association between rs4245739 and ER-negative and ER-positive breast cancer.
Stratification analyses of ethnicity suggested rs4245739 decreased the risk of cancer in Asian population. The genetic background of cancer may vary among different ethnicities. We did not find that rs4245739 was associated with the risk of cancer in Caucasians, although three GWAS studies [27, 37, 38] reported that this SNP increased the risk of ER-negative or triple negative breast cancer (defined by the absence of ER, progesterone receptor and human epidermal growth factor receptor-2) in the Caucasian population. In this meta-analysis, a large difference was found in the distribution of rs4245739 between Caucasians and Asians, with minor allele frequencies of 26.2% and 6.2%, respectively, and this was possibly affected by the power of the studies conducted in Asian populations and the final relationships between the rs4245739 polymorphism and cancer risks among different racial groups [13]. Furthermore, the etiology of ER-negative breast cancer is different from that of ER-positive breast cancer, including differences in genetic predisposition [43, 44]. Therefore, it is reasonable to assume that the genetic susceptibility of ER-negative or triple negative breast cancer is different than those of other breast cancer subtypes or overall breast cancer, which may explain why the results of these GWAS studies in Caucasians are different from the results of this meta-analysis in Caucasians. The reasons why rs4245739 decreased the risk of cancer among Asian population may be that the functional rs4245739 SNP A>C locating in the MDM43′-untranslated (3′-UTR) region creates a miR-191-5p or miR-887-3p targeting sites [23]. MiR-191-5p and miR-887-3p could bind to MDM43′-UTR with the rs4245739 C allele selectively [23]. These changes could result in decreased expression of oncogene MDM4, which could reduce the inhibition of p53 activities. Stratification analyses of smoking status indicated that rs4245739 decreased the risk of cancer among nonsmokers. Two studies [21, 29] investigating SCLC and ESCC were included in those stratification analyses. The studies also revealed significant multiple interactions between rs4245739 and smoking. Notably, SCLC and ESCC share similar environmental etiologies such as heavy smoking.
Regarding the remaining four SNPs, we failed to identify any associations between rs1563828, rs11801299, rs10900598, and rs1380576 and cancer risk. We cannot definitively conclude that these four SNPs are not associated with cancer risk, because this meta-analysis included only a few studies with limited sample sizes, and any associations between gene polymorphisms and disease are greatly affected by the number of participants. Given the limited sample size, the relationships between these four SNPs and cancer risk should be interpreted with caution. Genotype distributions of the controls in one study [26] on rs1380576 did not conform to HWE. After excluding this study, the conclusions of the remaining studies did not significantly change, suggesting that the result of rs1380576 was trustworthy.
Several potential limitations of this meta-analysis should be considered. First, our data indicated that publication bias existed with respect to studies regarding rs4245739, and potential language bias may have resulted from the inclusion of published studies in English or Chinese only. Second, the number of studies on rs1563828, rs11801299, rs10900598, and rs1380576 included in the meta-analysis was small, and the sample size was limited, which prevented further stratification analyses of other potential factors. Third, our results were based on unadjusted estimates for confounding factors, which might have affected the final results. Fourth, we could not assess potential gene-gene and gene-environment interactions because of the lack of original data. Fifth, this meta-analysis only included Asian and Caucasian populations; future studies on other ethnic groups are necessary because of ethnic differences in gene polymorphisms. Sixth, heterogeneity was considerable in this meta-analysis because the included studies involved different ethnicities and environments. Although sensitivity analyses indicated that our data were stable and trustworthy, we should interpret these data with caution.
In conclusion, this meta-analysis indicates that rs4245739 polymorphism of MDM4 gene plays important roles in cancer pathogenesis, especially among Asian populations. Stratification analyses also indicate that rs4245739 decreases the risk of cancer among nonsmokers. However, the other four SNPs are not associated with cancer risk. Larger well-designed studies are necessary to validate these findings.
MATERIALS AND METHODS
Literature search
We systematically searched the PubMed, Embase, and China Knowledge Resource Integrated Database to identify studies through January 1, 2016. The following search terms were used: “cancer,” “carcinoma,” “neoplasm,” “tumor,” “MDM4,” “MDMX,” “HDMX,” “polymorphism,” “SNP” and “polymorphisms”. Two independent authors conducted the search. No language or other restrictions were placed on the search. Additional initially omitted studies have been identified by hand screening.
Criteria of inclusion and exclusion
The included studies conformed to the following criteria: (1) studies that evaluated the associations between cancer risk and MDM4 gene polymorphisms (at least one of the five polymorphisms), (2) studied on human beings, (3) study provided sufficient data to calculate the odds ratios (ORs) and 95% confidence intervals (CIs), and P value, and (4) case-control study. Exclusion criteria were: (1) duplication of previous publications; (2) case reports or review articles; (3) studies without detailed genotype data.
Data extraction and quality assessment
Relevant information was carefully extracted from all eligible studies. The extracted information including: name of first author, publication year, country of origin, ethnicity, numbers of cases and controls, and cancer type. Two authors independently performed the extraction of data and assessed the study quality based on the Newcastle-Ottawa Scale (NOS) [45]. Total NOS scores ranged from 0 to 9. A score ranging 5 to 9 stars is considered to be a generally high methodological quality whereas a score ranging 0 to 4 is regarded as a relatively poor quality. All disagreements were discussed and resolved with consensus.
Statistical analysis
All statistical analyses were performed using the Stata 11.0 software (StataCorp, College Station, TX, USA). The strength of associations between MDM4 gene polymorphisms and cancer risk were estimated for each study by crude ORs and 95% CIs. Stratification analyses were carried out by ethnicity, cancer type, sex and smoking status. P < 0.05 was considered statistically significant. When a significant Q test (P < 0.1) or I2 < 50% indicated heterogeneity across studies, a fixed-effect model was used. Otherwise, the fixed-effects model was applied [46]. Pooled ORs were calculated for allele model, dominant model, recessive model, homozygous model, and heterozygous model. We performed sensitivity analyses by omitting each study in turn to determine the effect on the test of heterogeneity and evaluated the stability of the overall results. Hardy–Weinberg equilibrium (HWE) was assessed in the controls using Pearson's χ2 test. Potential publication bias was assessed by Begger's and Egger's linear regression test [47]; P < 0.05 was considered to indicate statistically significant.
Acknowledgments
This study was supported by National Natural Science Foundation of China, No. 81371927.
Abbreviations
- SCCHN
squamous cell carcinoma of the head and neck
- NHL
Non-Hodgkin lymphoma
- ESCC
esophageal squamous cell carcinoma
- SCLC
small cell lung cancer
- CI
confidence interval
- OR
odds ratio
- NOS
Newcastle-Ottawa Scale
- HWE
Hardy–Weinberg equilibrium
- SNP
single nucleotide polymorphism
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Pharoah PD, Dunning AM, Ponder BA, Easton DF. Association studies for finding cancer-susceptibility genetic variants. Nature reviews Cancer. 2004;4:850–860. doi: 10.1038/nrc1476. [DOI] [PubMed] [Google Scholar]
- 5.Hoover RN. Cancer--nature, nurture, or both. N Engl J Med. 2000;343:135–136. doi: 10.1056/NEJM200007133430210. [DOI] [PubMed] [Google Scholar]
- 6.Ponder BA. Cancer genetics. Nature. 2001;411:336–341. doi: 10.1038/35077207. [DOI] [PubMed] [Google Scholar]
- 7.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 8.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 9.Junttila MR, Evan GI. p53--a Jack of all trades but master of none. Nature reviews Cancer. 2009;9:821–829. doi: 10.1038/nrc2728. [DOI] [PubMed] [Google Scholar]
- 10.Hu Z, Wu C, Shi Y, Guo H, Zhao X, Yin Z, Yang L, Dai J, Hu L, Tan W, Li Z, Deng Q, Wang J, Wu W, Jin G, Jiang Y, et al. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nature genetics. 2011;43:792–796. doi: 10.1038/ng.875. [DOI] [PubMed] [Google Scholar]
- 11.Shvarts A, Steegenga WT, Riteco N, van Laar T, Dekker P, Bazuine M, van Ham RC, van der Houven van Oordt W, Hateboer G, van der Eb AJ, Jochemsen AG. MDMX: a novel p53-binding protein with some functional properties of MDM2. The EMBO journal. 1996;15:5349–5357. [PMC free article] [PubMed] [Google Scholar]
- 12.Wade M, Wang YV, Wahl GM. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends in cell biology. 2010;20:299–309. doi: 10.1016/j.tcb.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gansmo LB, Romundstad P, Birkeland E, Hveem K, Vatten L, Knappskog S, Lonning PE. MDM4 SNP34091 (rs4245739) and its effect on breast-, colon-, lung-, and prostate cancer risk. Cancer medicine. 2015;4:1901–1907. doi: 10.1002/cam4.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Migliorini D, Lazzerini Denchi E, Danovi D, Jochemsen A, Capillo M, Gobbi A, Helin K, Pelicci PG, Marine JC. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Molecular and cellular biology. 2002;22:5527–5538. doi: 10.1128/MCB.22.15.5527-5538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, Jochemsen AG, Lozano G. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nature genetics. 2001;29:92–95. doi: 10.1038/ng714. [DOI] [PubMed] [Google Scholar]
- 16.Xiong S, Pant V, Suh YA, Van Pelt CS, Wang Y, Valentin-Vega YA, Post SM, Lozano G. Spontaneous tumorigenesis in mice overexpressing the p53-negative regulator Mdm4. Cancer research. 2010;70:7148–7154. doi: 10.1158/0008-5472.CAN-10-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valentin-Vega YA, Barboza JA, Chau GP, El-Naggar AK, Lozano G. High levels of the p53 inhibitor MDM4 in head and neck squamous carcinomas. Human pathology. 2007;38:1553–1562. doi: 10.1016/j.humpath.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Danovi D, Meulmeester E, Pasini D, Migliorini D, Capra M, Frenk R, de Graaf P, Francoz S, Gasparini P, Gobbi A, Helin K, Pelicci PG, Jochemsen AG, Marine JC. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Molecular and cellular biology. 2004;24:5835–5843. doi: 10.1128/MCB.24.13.5835-5843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laurie NA, Donovan SL, Shih CS, Zhang J, Mills N, Fuller C, Teunisse A, Lam S, Ramos Y, Mohan A, Johnson D, Wilson M, Rodriguez-Galindo C, Quarto M, Francoz S, Mendrysa SM, et al. Inactivation of the p53 pathway in retinoblastoma. Nature. 2006;444:61–66. doi: 10.1038/nature05194. [DOI] [PubMed] [Google Scholar]
- 20.Han X, Garcia-Manero G, McDonnell TJ, Lozano G, Medeiros LJ, Xiao L, Rosner G, Nguyen M, Fernandez M, Valentin-Vega YA, Barboza J, Jones DM, Rassidakis GZ, Kantarjian HM, Bueso-Ramos CE. HDM4 (HDMX) is widely expressed in adult pre-B acute lymphoblastic leukemia and is a potential therapeutic target. Modern pathology. 2007;20:54–62. doi: 10.1038/modpathol.3800727. [DOI] [PubMed] [Google Scholar]
- 21.Zhou L, Zhang X, Li Z, Zhou C, Li M, Tang X, Lu C, Li H, Yuan Q, Yang M. Association of a genetic variation in a miR-191 binding site in MDM4 with risk of esophageal squamous cell carcinoma. PloS one. 2013;8:e64331. doi: 10.1371/journal.pone.0064331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, Sturgis EM, Zhang Y, Huang Z, Zhou Q, Wei Q, Li G. Combined p53-related genetic variants together with HPV infection increase oral cancer risk. International journal of cancer. 2012;131:E251–258. doi: 10.1002/ijc.27335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song CG, Fu FM, Wu XY, Wang C, Shao ZM. Correlation of polymorphism rs1563828 in MDM4 gene with breast cancer risk and onset age [Article in Chinese] Zhonghua wai ke za zhi. 2012;50:53–56. [PubMed] [Google Scholar]
- 24.Liu J, Tang X, Li M, Lu C, Shi J, Zhou L, Yuan Q, Yang M. Functional MDM4 rs4245739 genetic variant, alone and in combination with P53 Arg72Pro polymorphism, contributes to breast cancer susceptibility. Breast cancer research and treatment. 2013;140:151–157. doi: 10.1007/s10549-013-2615-x. [DOI] [PubMed] [Google Scholar]
- 25.Fan C, Wei J, Yuan C, Wang X, Jiang C, Zhou C, Yang M. The functional TP53 rs1042522 and MDM4 rs4245739 genetic variants contribute to Non-Hodgkin lymphoma risk. PloS one. 2014;9:e107047. doi: 10.1371/journal.pone.0107047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu GC, Zhang ZT. Genetic association of single nucleotide polymorphisms in P53 pathway with gastric cancer risk in a Chinese Han population. Medical oncology (Northwood, London, England) 2015;32:401. doi: 10.1007/s12032-014-0401-1. [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Closas M, Couch FJ, Lindstrom S, Michailidou K, Schmidt MK, Brook MN, Orr N, Rhie SK, Riboli E, Feigelson HS, Le Marchand L, Buring JE, Eccles D, Miron P, Fasching PA, Brauch H, et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nature genetics. 2013;45:392–398. doi: 10.1038/ng.2561. 398e391-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu H, Wang LE, Liu Z, Wei S, Li G, Sturgis EM, Wei Q. Polymorphisms of MDM4 and risk of squamous cell carcinoma of the head and neck. Pharmacogenetics and genomics. 2011;21:388–396. doi: 10.1097/FPC.0b013e32834632e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao F, Xiong X, Pan W, Yang X, Zhou C, Yuan Q, Zhou L, Yang M. A Regulatory MDM4 Genetic Variant Locating in the Binding Sequence of Multiple MicroRNAs Contributes to Susceptibility of Small Cell Lung Cancer. PloS one. 2015;10:e0135647. doi: 10.1371/journal.pone.0135647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang YW, Guan J, Zhang Y, Qiu YR, Chen LH. Role of an MDM4 polymorphism in the early age of onset of nasopharyngeal carcinoma. Oncology letters. 2012;3:1115–1118. doi: 10.3892/ol.2012.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salvi S, Calistri D, Gurioli G, Carretta E, Serra L, Gunelli R, Zoli W, Casadio V. Copy number analysis of 24 oncogenes: MDM4 identified as a putative marker for low recurrence risk in non muscle invasive bladder cancer. International journal of molecular sciences. 2014;15:12458–12468. doi: 10.3390/ijms150712458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stegeman S, Moya L, Selth LA, Spurdle AB, Clements JA, Batra J. A genetic variant of MDM4 influences regulation by multiple microRNAs in prostate cancer. Endocrine-related cancer. 2015;22:265–276. doi: 10.1530/ERC-15-0013. [DOI] [PubMed] [Google Scholar]
- 33.Kulkarni DA, Vazquez A, Haffty BG, Bandera EV, Hu W, Sun YY, Toppmeyer DL, Levine AJ, Hirshfield KM. A polymorphic variant in human MDM4 associates with accelerated age of onset of estrogen receptor negative breast cancer. Carcinogenesis. 2009;30:1910–1915. doi: 10.1093/carcin/bgp224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun T, Lee GS, Oh WK, Pomerantz M, Yang M, Xie W, Freedman ML, Kantoff PW. Single-nucleotide polymorphisms in p53 pathway and aggressiveness of prostate cancer in a Caucasian population. Clin Cancer Res. 2010;16:5244–5251. doi: 10.1158/1078-0432.CCR-10-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atwal GS, Kirchhoff T, Bond EE, Montagna M, Menin C, Bertorelle R, Scaini MC, Bartel F, Bohnke A, Pempe C, Gradhand E, Hauptmann S, Offit K, Levine AJ, Bond GL. Altered tumor formation and evolutionary selection of genetic variants in the human MDM4 oncogene. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:10236–10241. doi: 10.1073/pnas.0901298106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Oliveira Reis AH, de Carvalho IN, de Sousa Damasceno PB, Ferman SE, Lucena E, Lopez-Camelo JS, Seuanez HN, Vargas FR. Influence of MDM2 and MDM4 on development and survival in hereditary retinoblastoma. Pediatric blood & cancer. 2012;59:39–43. doi: 10.1002/pbc.24014. [DOI] [PubMed] [Google Scholar]
- 37.Stevens KN, Vachon CM, Couch FJ. Genetic susceptibility to triple-negative breast cancer. Cancer research. 2013;73:2025–2030. doi: 10.1158/0008-5472.CAN-12-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Purrington KS, Slager S, Eccles D, Yannoukakos D, Fasching PA, Miron P, Carpenter J, Chang-Claude J, Martin NG, Montgomery GW, Kristensen V, Anton-Culver H, Goodfellow P, Tapper WJ, Rafiq S, Gerty SM, et al. Genome-wide association study identifies 25 known breast cancer susceptibility loci as risk factors for triple-negative breast cancer. Carcinogenesis. 2014;35:1012–1019. doi: 10.1093/carcin/bgt404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reincke S, Govbakh L, Wilhelm B, Jin H, Bogdanova N, Bremer M, Karstens JH, Dork T. Mutation analysis of the MDM4 gene in German breast cancer patients. BMC cancer. 2008;8:52. doi: 10.1186/1471-2407-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu H, Sturgis EM, Liu Z, Wang LE, Wei Q, Li G. Modifying effect of MDM4 variants on risk of HPV16-associated squamous cell carcinoma of oropharynx. Cancer. 2012;118:1684–1692. doi: 10.1002/cncr.26423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landers JE, Cassel SL, George DL. Translational enhancement of mdm2 oncogene expression in human tumor cells containing a stabilized wild-type p53 protein. Cancer research. 1997;57:3562–3568. [PubMed] [Google Scholar]
- 42.Linares LK, Hengstermann A, Ciechanover A, Muller S, Scheffner M. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:12009–12014. doi: 10.1073/pnas.2030930100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang XR, Chang-Claude J, Goode EL, Couch FJ, Nevanlinna H, Milne RL, Gaudet M, Schmidt MK, Broeks A, Cox A, Fasching PA, Hein R, Spurdle AB, Blows F, Driver K, Flesch-Janys D, et al. Associations of breast cancer risk factors with tumor subtypes: a pooled analysis from the Breast Cancer Association Consortium studies. Journal of the National Cancer Institute. 2011;103:250–263. doi: 10.1093/jnci/djq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mavaddat N, Antoniou AC, Easton DF, Garcia-Closas M. Genetic susceptibility to breast cancer. Molecular oncology. 2010;4:174–191. doi: 10.1016/j.molonc.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. European journal of epidemiology. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 46.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Statistics in medicine. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 47.Peters JL, Sutton AJ, Jones DR, Abrams KR, Rushton L. Comparison of two methods to detect publication bias in meta-analysis. Jama. 2006;295:676–680. doi: 10.1001/jama.295.6.676. [DOI] [PubMed] [Google Scholar]


