Abstract
Background: Recently, we analyzed genome-wide protein binding data for the Drosophila cell lines S2, Kc, BG3 and Cl.8 (modENCODE Consortium) and identified a set of 12 proteins enriched in the regions corresponding to interbands of salivary gland polytene chromosomes. Using these data, we developed a bioinformatic pipeline that partitioned the Drosophila genome into four chromatin types that we hereby refer to as aquamarine, lazurite, malachite and ruby.
Results: Here, we describe the properties of these chromatin types across different cell lines. We show that aquamarine chromatin tends to harbor transcription start sites (TSSs) and 5’ untranslated regions (5’UTRs) of the genes, is enriched in diverse “open” chromatin proteins, histone modifications, nucleosome remodeling complexes and transcription factors. It encompasses most of the tRNA genes and shows enrichment for non-coding RNAs and miRNA genes. Lazurite chromatin typically encompasses gene bodies. It is rich in proteins involved in transcription elongation. Frequency of both point mutations and natural deletion breakpoints is elevated within lazurite chromatin. Malachite chromatin shows higher frequency of insertions of natural transposons. Finally, ruby chromatin is enriched for proteins and histone modifications typical for the “closed” chromatin. Ruby chromatin has a relatively low frequency of point mutations and is essentially devoid of miRNA and tRNA genes. Aquamarine and ruby chromatin types are highly stable across cell lines and have contrasting properties. Lazurite and malachite chromatin types also display characteristic protein composition, as well as enrichment for specific genomic features. We found that two types of chromatin, aquamarine and ruby, retain their complementary protein patterns in four Drosophila cell lines.
Keywords: Drosophila, Cell lines, Chromatin types, Polytene chromosomes, Interbands, Genome-wide protein binding data
INTRODUCTION
Chromatin structure, genetic organization and local activity are known to be intimately coordinated. Yet, many facets and mechanisms of this interplay are still poorly explored. Polytene chromosomes serve as a convenient tool to analyze chromatin organization of a selected region of the genome in the context of an interphase nucleus. According to different estimates, salivary gland polytene chromosomes of Drosophila melanogaster contain about 3500 bands as much as interbands, which account for about 95% and 5% of the total length of euchromatic DNA, respectively [1, 2]. Compared to bands, interbands are formed by a decompacted chromatin, and compaction of bands has long been considered to reflect their genetic inactivity [3]. Large bands of intercalary heterochromatin (IH) constitute a special class of structures found in polytene chromosomes. They encompass multiple genes, span up to several hundred kbs, contain low levels of origin recognition complexes (ORCs) and are scattered along chromosome arms (for more details, see [2, 4, 5]). Clusters of single-copy tissue-specific genes with shared expression patterns are frequently found in IH regions; notably gene density is significantly decreased in IH [6, 7]. Besides IH bands, polytene chromosomes contain many “regular” bands that do not show attributes of IH mentioned above. Study of genetic and protein organization of bands and interbands was impossible until recently due to the lack of the knowledge on their genomic coordinates.
In our earlier reports, we used 12 insertions of P element-based transposons into interbands as tags to define a set of twelve non-histone proteins that are characteristic of these interbands. This set included RNA polymerase II (RNA pol II), CHRIZ (CHRO), dMi-2, NURF301, WDS, ISWI, JIL1, MLE, MOF, MRG15, MSL-1 and MBD-R2 [8-10]. Next, we demonstrated that in different cell lines these proteins tend to be significantly enriched in the same regions of the genome, which nicely correspond to 12 reference interbands of salivary gland polytene chromosomes [11]. We concluded then, that interbands correspond to the regions of permanently “open” chromatin in the Drosophila genome [10, 12]. Based on the genome-wide profiling data for these twelve proteins in four cell lines – S2, Kc, BG3 and Cl.8 [13], we developed a bioinformatic approach that partitions the molecular map of the euchromatic portion of the Drosophila genome into fragments corresponding to bands and interbands of polytene chromosomes. In so doing, we uncovered four chromatin types, which were originally color-coded as: cyan, blue, green and magenta [10], but are henceforth respectively referred to as aquamarine, lazurite, malachite and ruby to avoid confusion with the five principal chromatin types (BLUE, GREEN, BLACK, RED and YELLOW) previously described in Kc cells [14]. The aquamarine chromatin is enriched for the twelve interband-specific proteins described above; the lazurite type shows significant association with only three proteins: RNA pol II, JIL1 and MRG15; the malachite chromatin shows a borderline (non-significant) enrichment of the twelve proteins, whereas the fourth chromatin type, ruby, is significantly depleted for all of these proteins. Next, we expanded our set of the studied reference interbands to 32, all of which coincided with aquamarine chromatin. We found that in S2, Kc, BG3 and Cl.8 cells, this chromatin type is rich in “open” chromatin markers such as DNase I hypersensitive sites (DHSs), nucleosome remodeling complexes and ORCs [10]. Our analysis suggested that aquamarine chromatin is composed of 5748 domains, yet literature estimates indicate that there are much fewer interbands in the polytene chromosomes, about 3500 [2]. Therefore, we investigated whether all the aquamarine domains identified share the same organization and properties and which of them really correspond to interbands. We also wanted to understand how the rest of the chromatin types (lazurite, malachite and ruby) that correspond to different types of bands in polytene chromosomes [10] relate to genes and other genomic features. Thus, our present study has four goals: (i) to analyze the genetic composition of each chromatin type; (ii) to describe in detail the protein composition of each chromatin type; (iii) to analyze the distribution of genomic features in chromatin types; (iv) to explore the organization of various subtypes of aquamarine chromatin.
MATERIALS AND METHODS
Smoothing of the Whole-genome Map of the Four Chromatin Types
Four chromatin types used in the current study were defined earlier [10] and are based on the Release 5 Drosophila melanogaster genome assembly. Each chromatin type is composed of 200 bp-long non-overlapping fragments belonging to euchromatic portions of five large chromosome arms 2L, 2R, 3L, 3R and X. Chromatin types were not assigned to a total of about 5% of DNA sequence. Whenever such sequence (gap) was at most 400 bp and was flanked by the same chromatin types on both flanks, the gap was annotated as having the same type as these flanks. Whenever the flanking sequences belonged to distinct chromatin types or the gap exceeded 400 bp, the gap annotation was preserved. Series of consecutive fragments of the same chromatin type were merged together to define the positions of chromatin domains (Supplementary File 1). Numbers of domains within each chromatin type are summarized in the (Table 1).
Table 1.
Numbers of aquamarine, lazurite, malachite and ruby domains across euchromatic arms of D. melanogaster chromosomes.
| chr2L | chr2R | chr3L | chr3R | chrX | Total | |
|---|---|---|---|---|---|---|
| aquamarine | 991 | 1156 | 1145 | 1326 | 1130 | 5748 |
| lazurite | 705 | 856 | 768 | 970 | 840 | 4139 |
| malachite | 1687 | 1785 | 1930 | 2006 | 1781 | 9189 |
| ruby | 1150 | 1068 | 1354 | 1254 | 1199 | 6025 |
Data Sources
FlyBase database 5.50 was used to retrieve chromosomal positions of the majority of genetic features analyzed in the study (ftp://ftp.flybase.net/genomes/Drosophila_melanogaster/dmel_r5.50_FB2013_02/gff/dmel-all-no-analysisr5.50.gff.gz). Feature tags “mRNA”, “pseudogene”, “miRNA”, “tRNA”, “ncRNA”, “transposable_element” and “point_mutation” were used to get chromosomal positions of the transcripts, pseudogenes, microRNAs, transfer RNAs, non-coding RNAs, transposons and point mutations, respectively. As for the point mutations, we used data only for missense/nonsense mutations, which were marked by the “reported_pr_change” attribute. Our classification of natural transposon was based on the FlyBase symbols extracted by FlyBase QueryBuilder r5.57 for all “Natural Transposons”. Also, using FlyBase QueryBuilder r5.57, we retrieved all “Aberrations” in Drosophila melanogaster genome, and then we extracted information for 83 natural deletions by applying the following set of FlyBase controlled vocabulary (Fbcv) terms: 0000469 (caused by a spontaneous event), 0000470 (induced by recombination), 0000492 (induced by male recombination), 0000503 (induced by exposure to radiation), 0000506 (induced by gamma ray irradiation), 0000516 (induced by exposure to neutron radiation), 0000518 (induced by exposure to X rays), 0000530 (induced by ethyl methanesulfonate exposure), 0000532 (induced by hycanthon methanesulfonate exposure), 0000542 (induced by mitomycin C exposure) and 0000559 (induced by diepoxybutane exposure).
The modENCODE ChIP-chip data sets [13] were extracted via the InterMine [15]. DamID data sets were extracted from the GEO database (GSE22069 and GSE36175) [14, 16].
Each pairwise combination “chromatin type – localization class” shown was subjected to random permutation analysis and the probability of the occurrence by chance for the observed overlap between chromatin type and localization class was computed. The permutation process guaranteed that random (expected) distribution of chromatin domains did not have any relationship to the position of genes on chromosomes (localization class). For instance, the combination “aquamarine-TSS” shows the P value of 10-2908. This value reflects the probability that the overlap between random distribution of aquamarine domains and TSSs exceeds (↑) or matches that for the observed distribution. Similarly, the “lazurite-GENE” combination has the P value of 10-1256. This value indicates chances of higher (↑) or equal overlap between lazurite domains and gene parts distinct from TSSs or introns for the simulated distribution in comparison with the observed one. The down arrows (↓) indicate chances of lower overlap between certain types of domains and localization classes. P values below 10-6 were obtained using normal distribution as an approximation for the distribution of random chromatin domains. Asterisks mark statistically significant “chromatin type – localization class” combinations (P value < 0.05).
Monte Carlo Testing
To obtain an estimate of non-randomness of the observed distribution of chromatin types relatively to the four localization classes (TSS, GENE, INTRON and INTERGENIC REGION), random permutation test was performed for each of the five chromosome arms. To do so, length distributions of domains and inter-domain spacers were calculated for each chromatin type. Domains were named as An = {1, 2, … N} and spacers were named as Sn = {1 (region from the chromosome start to the start of the first domain), 2, … N, N+1 (region from the end of the ultimate domain to the chromosome end)}. Index shuffling for An and Sn arrays was used to obtain random distribution of domains on the chromosome. Thus, the expected distribution of domains is generated, wherein overlap with the localization classes is totally random. In each iteration, the number of domains belonging to each localization class is counted within the confines of each chromatin type (see RESULTS). In total, we generated M ~ 106 expected distributions and so we calculated the probability that the count of the localization class in expected domain distribution {Randm} is above or below than that for observed one {Real}. These probabilities are estimates for the enrichment/depletion of the chromatin type within domains belonging to a chosen structural class. When calculated P values were equal to zero or one (i.e. fewer/greater counts observed for all expected distributions, respectively), the exact P value was calculated using normal distribution as an approximation of the expected one {Randm}. Specifically, the average Av(Rand) and the standard deviation SD(Rand) for the expected distribution were computed, followed by establishing the Z-score: z = {Real-Av(Rand)}/SD(Rand). Area under the left (Real<Av(Rand), z<0) or right (Real>Av(Rand), z>0) tail of the normal distribution were estimated by the asymptotic expansion of the (complementary) error function. For instance, the right tail area of the normal distribution (z>0) is described by the complementary error function erfc(t) as follow:
.The asymptotic expansion of erfc(t) function for large absolute z values computed as following convergent series:
.To perform the transition from individual chromosomes to the whole genome, we ran permutation test simultaneously on all chromosomes. First, in a single iteration, we generated the distributions of domains and inter-domain spacers for the whole genome, i.e. we computed five distributions for five chromosome arms at once. Second, these five distributions of domains and the whole-genome map of genes allowed distinguishing the four localization classes (TSS, GENE, INTRON and INTERGENIC REGION) of the domains. Third, multiple iterations allowed estimating the probabilities of occurrence by the chance for all localization classes across the entire genome. Table 2 summarizes the results of resampling tests. The source code used for the Monte Carlo simulations and examples of its input and output files are provided in the Supplementary File 2.
Table 2. Probabilities of occurrence by chance of the observed distribution of the four chromatin types relatively to the gene structure.
| Chromatin Type | Localization Class | |||
|---|---|---|---|---|
| TSS | GENE | INTRON |
INTERGENIC
REGION |
|
| aquamarine | 10-2908 ↑ * | 10-142 ↓ * | 10-120 ↓ * | 10-107 ↓ * |
| lazurite | 10-148 ↓ * | 10-1256 ↑ * | 10-140 ↓ * | 10-125 ↓ * |
| malachite | 10-5 ↓ * | 10-7 ↑ * | 0.032 ↓ * | 0.450 ↓ |
| ruby | 10-63 ↓ * | 0.025 ↓ * | 10-16 ↑ * | 10-8 ↑ * |
Statistical Significance of Overlap Between the Four Chromatin Types and Distribution of Chromatin Proteins and Genomic Features
Pearson’s chi-square goodness of fit test was used to estimate the significance of chromatin enrichment with proteins and genomic features. The null hypothesis was that centers of the regions enriched with proteins, histone modifications, as well as centers of genomic features (with the exception of point mutations) are distributed across the genome proportionally to the coverage by the four chromatin types. In the analysis of point mutations, null hypothesis was that they are distributed across the chromatin types proportionally to the total length of exons overlapping with each chromatin type. Results having P values below 0.02 and 0.001 were considered significant and highly significant, respectively. Enrichment value was calculated as log2 (observed/expected). The scripts used for the data analysis as well as the processed data sets are provided in the Supplementary File 3.
R programming and Bioconductor libraries were used to analyze and visualize the data [17-20]. For the purpose of visualization, we transformed log2(observed/expected) values for proteins into Euclidean distances and proceeded with hierarchical complete linkage clusterization to group similar proteins on the heat maps.
Electron Microscopy
Salivary gland polytene chromosome squashes were prepared for electron microscopy analysis and examined as described earlier [21]. The sections (120–150 nm) were cut using an LKB-IV ultratome (LKB Bromma, Sweden) and examined under a JEM-100C (JEOL, Japan) electron microscope at 80 kV.
Fluorescence In Situ Hybridization
Salivary glands were dissected in Ephrussi-Beadle solution and then fixed in a 3:1 mixture of ethanol and acetic acid for 30 minutes at –20°C, squashed in 45% acetic acid, snap-frozen in liquid nitrogen and stored in 70% ethanol at –20°С. Fluorescence in situ hybridization (FISH) on polytene chromosomes was performed as described [22]. DNA probes were labeled with biotin-16-dUTP or digoxigenin-11-dUTP (Roche) in random-primed polymerase reaction with a Klenow fragment. Primers used in this study were: Cul-3 (5’–ACTGCAAAGCGAGTAAGGGGCTT–3’, 5’–CACCCTAC GCGGATTGTAGCGT–3’), lace (5’–GCTCATGTGAGC GGTGGAGC–3’, 5'–CCCGGAACACTGCAGATGGGT–3’), CycE (5’–AGGTCCGTCGACTGATCGGC–3’, 5'–ATG ACGCGTGTGCGGTGACA–3’).
RESULTS
1. Correspondence Between the Four Chromatin Types and Gene Structure
To address the question of whether the four chromatin types may correspond to distinct parts of the genes, we compared positions of the chromatin domains with those of 27979 unique protein-coding transcripts encoded in euchromatic arms of chromosomes (transcript annotations were obtained from FlyBase; see MATERIALS AND METHODS). We adopted the following classification of how chromatin domains are positioned relatively to the gene structure: domains overlapping with at least one of the gene transcription start site (TSS) were classified as “TSS”; domains entirely embedded within introns were termed “INTRON”; all other domains overlapping with the gene bodies were defined as “GENE”; finally, the domains falling outside of any of the genes were classified as “INTERGENIC REGION” (Fig. 1). When a chromatin domain overlapped with more than one transcript, and was classified as belonging to several distinct localization classes, each of those classes was assigned equal weight (Supplementary Fig. 1).
Fig. (1).
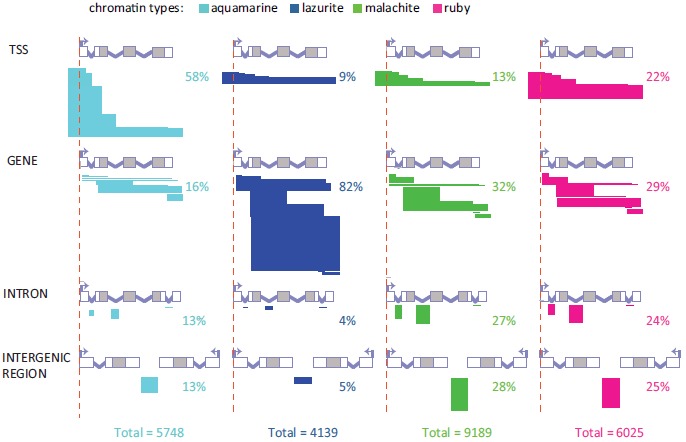
Detailed classification of the relative positioning of aquamarine, lazurite, malachite and ruby chromatin domains versus gene structure. Bent arrows and red vertical dashed lines indicate the positions of TSSs. Grey boxes represent coding parts of exons and white boxes correspond to 5’ and 3’ UTRs. Introns are shown as broken lines. Horizontal bars depict different possible overlaps between chromatin domains and genes for the all defined localization classes: TSS, GENE, INTRON and INTERGENIC REGION. The width of the bars reflects the percentage of each localization subclass in each chromatin type. For each localization class, fragments with borders mapped to various structural parts of a gene are shown separately.
Domains of different chromatin types are non-randomly distributed relatively to the gene structure (Fig. 1; Supplementary Fig. 1). In particular, 58.2% of aquamarine domains encompass TSSs. Overlap of TSSs with the other three chromatin types is substantially lower. Lazurite domains largely fall into the GENE localization class (82.4%), so they mainly correspond to the transcribed portions of the genes.
To estimate the significance of differences in enrichment of the localization classes in the four chromatin types, we performed Monte Carlo simulation (random permutation) using DNA sequences covering euchromatic portions of the genome. As it follows from the data presented in the (Table 2) (see MATERIALS AND METHODS), aquamarine and lazurite chromatin types are significantly enriched for TSSs and gene bodies, respectively. At the same time, aquamarine chromatin is significantly depleted for genes bodies, introns and intergenic regions, whereas lazurite chromatin is significantly depleted for TSSs, introns and intergenic regions. Malachite chromatin is also significantly depleted for TSSs and is enriched for gene bodies. Ruby chromatin is significantly depleted for TSSs, yet it is significantly enriched with introns and intergenic regions.
2. Protein Composition of the Four Chromatin Types
To explore in more detail the protein composition of the four chromatin types, we analyzed genome-wide ChIP-chip data for 64 chromatin proteins and histone modifications mapped in S2, Kc, BG3 and Cl.8 cells [13] and DamID-chip data for 104 chromatin proteins in Kc cells [14, 16]. Regions significantly enriched with these proteins were overlaid with the map of the four chromatin types to measure the abundance of each protein in each chromatin type. These data were normalized to the genomic baseline (see MATERIALS AND METHODS). Note that insulator proteins were not included in this study and their behavior will be described elsewhere. Below we describe the properties of these chromatin types across S2, Kc, BG3 and Cl.8 cells.
Most of the proteins and histone modifications demonstrate consistent enrichment/depletion values in the four chromatin types across the cell lines (Supplementary Fig. 2). We found that aquamarine and ruby chromatin types appear to be reciprocal in the composition: the proteins highly enriched in aquamarine are virtually absent in ruby, and vice versa, aquamarine chromatin is depleted for those few proteins that appear enriched in ruby (Fig. 2; Supplementary Fig. 2). Although less pronounced, reciprocity in chromatin composition is also observed for the lazurite and malachite chromatin types. For example, lazurite chromatin is enriched, whereas malachite chromatin is depleted, for JIL1, MRG15, H2B-ubiq, H3K36me3 and H3K79me3, all of which have been implicated in the elongation step of transcription [23-25]. On the contrary, GAF, LSD1, PCL, Polycomb (PC), DNA topoisomerase II (Top2), H3K18ac, H3K23ac, H3K27me3 and H3K36me1 tend to map to malachite but not lazurite chromatin (Fig. 2). Notably, histone modifications H3K4me1 and H2Av moderately (1.5-2 fold) enriched in both lazurite and malachite chromatin types are also enriched in aquamarine chromatin.
Fig. (2).
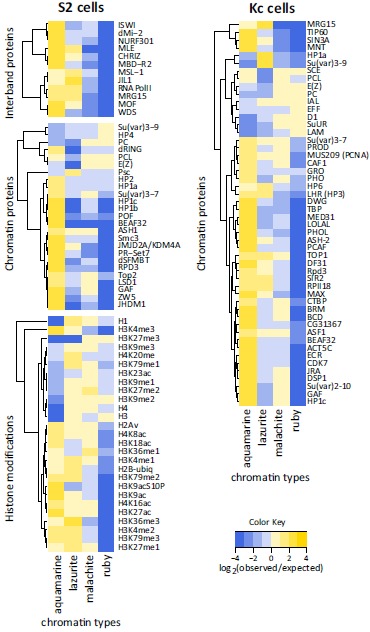
Protein composition of the four chromatin types in S2 and Kc cells. Heat maps of the enrichment (Grey) and depletion (black) for proteins and histone modifications as mapped in S2 cells by ChIP-chip [13] and in Kc cells by DamID [14]. Data are presented as a log2-transformed ratio of the observed and expected by chance overlap between the protein-bound regions and each of four chromatin types.
Aquamarine chromatin is highly enriched (3-7-fold) for histone modifications H3K4me3, H3K27ac and H3K9ac associated with active and decondensed regions of the genome [26, 27], as well as for the H4K16ac mark reported to be coupled to activity of housekeeping genes [28-30]. Aquamarine chromatin is also rich in histone-modifying proteins: methyltransferase ASH1 [31], histone demethylases JHDM1 (Kdm2) and JMJD2A/KDM4A [32, 33], histone deacetylase RPD3 [34], histone methyltransferase PR-Set7 and ubiquitin transferase dRING, the latter two being involved in transcription regulation [35-37]. This chromatin type is also characterized by binding of components of nucleosome remodeling complexes: dMi-2 [38], WDS [39, 40], NURF301 and ISWI [41-43]. Our observation that aquamarine chromatin tends to map to promoter regions is in line with its high (5-fold) enrichment with HP1b, HP1c and GAF, as these proteins are associated with paused RNA pol II and H3K4me3 at TSSs [44-46]. Aquamarine chromatin is also enriched for the MBD-R2, which is found at active gene promoters and is involved in transcription [40, 47].
Next, aquamarine chromatin displays 6-7 fold enrichment for subunit of PhoRC (Pho-repressive complex) dSFMBT [48, 49], cohesin complex subunit Smc3 [50], dMi-2, a chromatin remodeling factor and a regulator of cohesin binding [38]. Enrichment within aquamarine chromatin is less pronounced (2-3 fold) for Top2 [51], HP1a and HP2 likewise associated with chromatin remodeling complexes [52, 53].
Several proteins and histone modifications turned out to be significantly underrepresented in aquamarine chromatin. These include histones H1, H3 and H4 (20-25 fold), as well as an epigenetic transcription regulator PC and H3K27me3, a key PC-dependent silencing mark [54]. Aquamarine chromatin also shows several-fold depletion in histone modification H3K9me2 and heterochromatin proteins affecting position effect variegation (PEV), HP4 and Su(var)3-9 [55, 56] (Fig. 2; Supplementary Fig. 2).
Analysis of DamID data sets for a number of additional chromatin proteins [14, 16] indicated that aquamarine chromatin is enriched for many proteins in Kc cells (Fig. 2; Supplementary Fig. 2) including Su(var)2-10, a protein implicated in the maintenance of chromosome structure and known to interact with HP1a [57]. On the other hand, aquamarine chromatin is depleted for SuUR, D1 and lamin Dm0 (LAM), all of which co-localize in inactive chromatin of Kc cells [14, 58].
Lazurite chromatin is associated with transcription-related histone modifications and is poor in silent chromatin marks. We observed at least 2-fold enrichment of lazurite chromatin in H3K79me1/2/3, H4K20me, H2B-ubiq, H2Av,
H3K27me1, H3K4me1/2/3 and H3K36me3 [59-63]. The latter histone modification is an established mark associated with elongating RNA pol II and it is predominantly found in gene exons [14, 24]. Lazurite chromatin displays notable depletion (5-fold) for H3K27me3. Other histone marks do not show consistent enrichment or depletion across the cell lines analyzed. There is significant enrichment (3-4 fold) of lazurite chromatin with JIL1 and MRG15, both of which bind methylated H3K36 and H3K4 and participate in the maintenance of a decompacted state of chromatin and regulate gene transcription [14, 25, 64]. Lazurite chromatin is also moderately enriched in histone demethylase JMJD2A/KDM4A (1.2-1.6 fold), which was reported to interact with HP1a and to control the methylation status of H3K36 [65, 66]. The rest of the studied chromatin proteins appear depleted in lazurite chromatin (Fig. 2; Supplementary Fig. 2).
Malachite chromatin shows binding of Top2, histone H3K4 demethylase LSD1 (1.5-1.9 fold) [67], GAF, transcription regulator PC [68] and Su(var)3-7 that interacts genetically with JIL1 [69]. Malachite chromatin shows about 4-fold enrichment with a histone mark H3K36me1. According to DamID data sets for Kc cell line [14], malachite chromatin appears slightly associated with the prominent components of inactive chromatin, SuUR [70] and D1 [71] (Fig. 2; Supplementary Fig. 2).
Ruby chromatin is depleted (at least 10-fold) for most of the proteins and histone modifications tested. Components of inactive chromatin E(Z) and PC show moderate (1.3-1.5 fold) enrichment. Histone marks associated with decondensed and active chromatin, namely H3K9ac, H3K27ac, H3K4me1/2/3, H4K16ac and H3K36me3, display the most pronounced depletion within ruby domains. Analysis of DamID data sets [14] indicates that in Kc cells ruby chromatin is specifically enriched in ubiquitin ligase Effete (EFF) [72] and LAM, and, as well as malachite chromatin, it is enriched in SuUR and D1 (Fig. 2; Supplementary Fig. 2).
Thus, our expanded analysis confirmed the 4-state model [10]: aquamarine and lazurite are active chromatin types; ruby is inactive chromatin type, whereas malachite chromatin is intermediate in terms of activity. Aquamarine and ruby chromatin types have complementary enrichment/depletion patterns for most of the proteins analyzed in S2, Kc, BG3 and Cl.8 cells (Supplementary Fig. 2). In contrast, lazurite and malachite chromatin types appear to be more dynamic in their protein composition across these cell lines.
3. Distribution of Genomic Features in the Four Chromatin Types
We proceeded to investigate the distribution of deletion breakpoints and missense/nonsense point mutations across the four chromatin types. We analyzed whether there is any preference in their localization in a given chromatin type using chi-square test. FlyBase annotations were available for 150 natural deletion breakpoints and 2882 missense/nonsense point mutations (Fig. 3). Appropriate fractions of the genome were taken into account in our analysis (for details, see MATERIALS AND METHODS). We found that lazurite chromatin is 1.7-fold enriched for natural deletion breakpoints and 1.4-fold enriched for point mutations (P value < 0.001). In total, this chromatin type harbors 28.6% and 55.5% of natural deletion breakpoints and point mutations, respectively. On the contrary, aquamarine and ruby chromatin types show up to 2-fold depletion in point mutations (P value < 0.001) and ruby chromatin is 1.4-fold depleted for natural deletion breakpoints (P value < 0.02). Distribution of point mutations in malachite chromatin is not significantly different from that expected by chance.
Fig. (3).
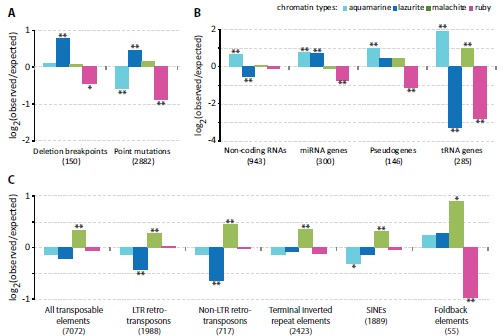
Enrichment and depletion of the four chromatin types with genomic features. Y axis shows log2-transformed ratio of the observed numbers of genomic features to the expected ones. Single and double asterisks indicate P < 0.02 and P < 0.001, respectively (Pearson’s chi-square goodness of fit test). A. Distribution of deletion breakpoints and missense/nonsense point mutations in the chromatin types. B. Distribution of non-coding RNAs, pseudogenes, miRNA and tRNA genes in the chromatin types. C. Relative enrichment and depletion of the four chromatin types with natural transposable elements (all) and individual classes thereof (LTR retrotransposons, non-LTR retrotransposons, terminal inverted repeat elements, SINEs, Foldback elements).
Next, we asked whether there is any correlation between the four chromatin types and localization of non-coding RNAs, pseudogenes, miRNA and tRNA genes. FlyBase annotations for 943 non-coding RNAs, 300 miRNA genes, 146 pseudogenes and 285 tRNA genes were used (Fig. 3). We found that aquamarine chromatin is significantly enriched, whereas lazurite chromatin is significantly depleted for non-coding RNAs (1.5-fold, P value < 0.001 for both chromatin types). miRNA genes are 1.7 times more frequent in aquamarine and lazurite chromatin types (P value < 0.001 for both chromatin types), with ruby chromatin showing 1.6-fold depletion (P value < 0.001). Aquamarine chromatin has 1.9 times higher occurrence in pseudogenes than it is expected by chance (P value < 0.001). Pseudogenes are 2.3 times less abundant in ruby chromatin (P value < 0.001). Half (47.7%) of tRNA genes map to aquamarine chromatin, which translates into 3.7-fold excess (P value < 0.001). Malachite domains host 39% of tRNA genes; this is about 2-fold enrichment over the baseline (P value < 0.001). Significant depletion for tRNA genes is observed for lazurite and ruby chromatin types (P value < 0.001 for both chromatin types), which encompass 1% and 7% of tRNA genes, respectively (Fig. 3).
Next, we investigated the relative distribution of natural transposons across the four chromatin types. Significance of co-localization in the distribution of transposons and the four chromatin types was estimated by chi-square test and corrected for the total span of each chromatin type in the genome. We used FlyBase annotations for all the transposon classes taken together, as well as the data for the individual classes: 1988 LTR retrotransposons, 717 non-LTR retrotransposons, 1889 SINEs, 2423 terminal inverted repeat elements and 55 Foldback-class transposons (Fig. 3). Overall, transposons were moderately (1.2-1.5-fold), yet significantly, enriched in malachite chromatin (P value < 0.001). At the same time, when individual classes of transposons were considered, lazurite chromatin displayed about 1.5-fold depletion for both retrotransposon classes (P value < 0.001). Depletion level for other transposon classes in lazurite chromatin failed to reach the significance level. Aquamarine chromatin is moderately, yet significantly, depleted for SINEs (1.2 fold, P value < 0.02). Foldback DNA transposons display distinct behavior: they are 1.8-fold enriched in malachite chromatin (P value < 0.02) and 2-fold depleted in ruby chromatin (P value < 0.001). The rest of the transposon classes were found in ruby chromatin at frequencies close to the genome baseline.
Thus, aquamarine chromatin is a preferred localization place for tRNA genes, non-coding RNAs, miRNA genes and pseudogenes. Lazurite chromatin is enriched with miRNA genes, deletion breakpoints and point mutations and is virtually devoid of tRNA genes. Transposons of different classes, particularly Foldback elements, are abundant in malachite chromatin. Ruby chromatin is rarely hit by natural deletion breakpoints and point mutations and is generally poor in pseudogenes, Foldback DNA transposons, tRNA and miRNA genes.
4. Specificity of Organization of Different Aquamarine Chromatin Subtypes
Not all the aquamarine domains identified using our bioinformatic pipeline correspond to interbands. We previously found that 32 reference interbands are formed by aquamarine chromatin enriched with TSSs, CHRIZ and GAF [10]. Here we studied the morphological organization of the cytological region 35D1-4 using FISH analysis, namely we precisely mapped the borders of the bands 35D1-2 and 35D3-4, as well as the intervening interband 35D1-2/35D3-4 (Fig. 4). This interband encompasses the TSS, 5’ UTR, the first exon and most of the first intron of the lace gene (Fig. 4). Electron microscopy image of this region clearly shows a single interband at 35D1-2/35D3-4 (Fig. 4). At the molecular level, there are two aquamarine domains near the FISH-mapped interband, one of which (the left one on Fig. 4) shows all the features of reference interbands; whereas the other one (the right one on Fig. 4) is devoid of CHRIZ, GAF and TSS, but hosts the 3’ UTR of lace (Fig. 4). Thus, aquamarine chromatin is an open chromatin type; however the extent of this “openness” may vary considerably, from the interband (TSS of lace gene) to a less decompacted chromatin (3’UTR of lace gene).
Fig. (4).
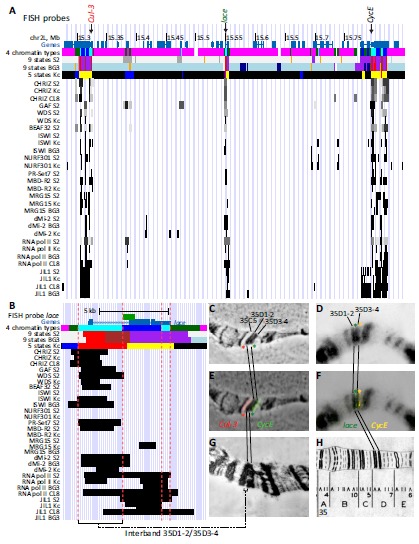
FISH mapping of 35D1-4 region of salivary gland polytene chromosomes on the molecular map. А. Physical map of the 35D1-4 region: genomic coordinates, distribution of chromatin types predicted by different models (“4 types” [10], “9 states S2” [73], “9 states BG3” [73] and “5 states” [14]) and regions of significant enrichment of interband-specific proteins [13]. Positions of the probes used for FISH are shown by arrows at the top. B. Physical map of the 35D1-2/35D3-4 interband encompassing the lace gene. Red dashed lines show borders of the two aquamarine regions in the 35D1-2/35D3-4. C-H. FISH mapping of DNA probes from the Cul-3 gene (marks the distal border of band 35D1-2), lace gene (marks interband 35D1-2/35D3-4) and CycE (marks the proximal border of the band 35D3-4). C. Phase-contrast photograph. D. DAPI signal (DNA, inverted image). E, F. Merged image of hybridization signals and phase-contrast or DAPI channel. G. Electron microphotograph of the region 35BE. F. Bridges’ map [85].
Genome-wide analysis of S2, Kc, BG3 and Cl.8 data sets revealed 3991 aquamarine domains (constituting 69.43% of all aquamarine domains) enriched for both CHRIZ and TSSs across all four cell types. Some of these aquamarine domains are also characterized by the presence of GAF (Fig. 5; Supplementary Table 1). This number is very close to the estimates of the number of interbands present in the salivary gland polytene chromosomes of Drosophila [2].
Fig. (5).
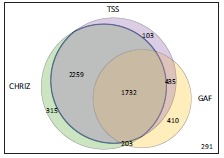
Venn diagram showing the numbers of aquamarine subdomains in the Drosophila genome that display features characteristic of the reference set of interbands: binding of CHRIZ [86] and GAF proteins and overlap with TSSs in all four cell types analyzed (S2, Kc, BG3 and Cl.8 cells). The total number of aquamarine domains in the genome is 5748. Only 291 aquamarine domains lack all of these features.
Other aquamarine domains (1757), as a rule, are enriched with DHSs, 3’UTRs of genes and particular combinations of some proteins used for partitioning the Drosophila genome into the four chromatin types (RNA pol II, dMi-2, NURF301, WDS, ISWI, JIL1, MLE, MOF, MRG15, MSL-1 and MBD-R2). These domains also tend to encompass ORCs although not in all the cell lines studied (data not shown). Interestingly, aquamarine domains lacking CHRIZ, TSS and GAF are relatively rare (291 cases, 5.06% of all the aquamarine domains). Thus, aquamarine chromatin is heterogeneous: some of the aquamarine domains correspond to open chromatin, yet they do not appear as interbands in the context of polytene chromosomes (the right aquamarine domain in Fig. 4).
5. Comparison of the Four Chromatin Types with Other Chromatin Models
Previous studies partitioned the Drosophila genome into 5- and 9-states [14, 73]. 5-state model was based on the DamID profiles of 53 broadly selected chromatin proteins in Kc cells [14], whereas 9-state model was a result of analysis of ChIP profiles of histone modifications in S2 and BG3 cells [73]. Our analysis was distinct from these models, since we pooled together the chromatin features from S2, Kc, BG3 and Cl.8 cell lines to identify basic protein composition of the regions corresponding to polytene chromosome interbands [10]. To characterize the four chromatin types in more details, we overlaid them with 5- and 9-states.
Expectedly, aquamarine chromatin mostly consists of active chromatin states RED and YELLOW in Kc cells as well as putative enhancer regions (State 3) and the regions where transcription initiation occurs (State 1) in S2 and BG3 cells (Fig. 6). Besides, 8-13% of aquamarine chromatin
Fig. (6).
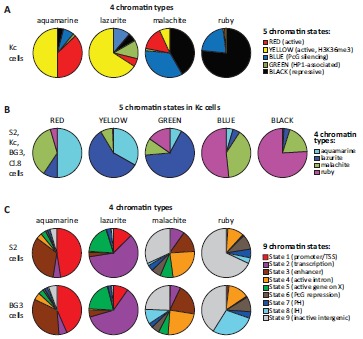
Overlap of the four chromatin types with other models of chromatin. A. Distribution of five principal chromatin states mapped in Kc cells [14] in each of the four chromatin types studied in this work [10]. B. Distribution of the four chromatin types [10] in five principal chromatin states mapped in Kc cells [14]. C. Distribution of nine chromatin states mapped in S2 and BG3 cells [73] in each of the four chromatin types studied in this work [10]. PH, pericentromeric heterochromatin; IH intercalary heterochromatin.
corresponds to the chromatin annotated as repressed in both 5- and 9-state models. This discrepancy might be partially attributable to the inadvertent errors of data smoothing and modeling. Chromatin plasticity between different cell lines is likely a contributing factor as well. Lazurite chromatin primarily consists of the following transcriptional states: YELLOW in Kc cells and State 2 (transcription elongation on autosomes) and State 5 (active genes on the X chromosome) in S2 and BG3 cells. Note that there is a moderate overlap of lazurite chromatin with inactive chromatin states in Kc, but not S2 and BG3 cells (Fig. 6). In Kc cells, malachite chromatin mostly intersects with closed chromatin states BLACK (SuUR-, LAM- and D1-associated repressive state) and BLUE (Polycomb repression). Yet, about one fifth of malachite chromatin (22%) overlaps with active chromatin states RED and YELLOW (Fig. 6). In S2 and BG3 cells, malachite chromatin corresponds to a wide range of poorly characterized active and inactive chromatin states (Fig. 6). Ruby chromatin overlaps almost exclusively with repressive chromatin states BLACK and BLUE in Kc cells (considering 0.5-1% overlap with the active chromatin states as a smoothing error) (Fig. 6). In S2 and BG3 cells, 80% of ruby chromatin coincides with inactive regions: State 8 (intercalary heterochromatin), State 9 (inactive intergenic areas) and State 6 (Polycomb-mediated repression). Notably, 11-14% of ruby chromatin intersects with State 4 characterized as active gene introns (Fig. 6).
DISCUSSION
Aquamarine, in all likelihood, is open chromatin, which is stable across different cell types studied. Our analysis suggests that aquamarine domains (~70% of which apparently match interbands of polytene chromosomes) are enriched with open chromatin proteins, as well as with components of transcription initiation complexes and chromatin remodelers [24, 30, 47]. In our earlier study, we demonstrated that in S2, Kc, BG3 and Cl.8 сells the regions corresponding to interbands of polytene chromosomes tend to be much more transcriptionally active than regions corresponding to bands [10]. Aquamarine chromatin was also shown to be enriched for broad-class promoters [10] with ubiquitous expression pattern [74-77] in S2, Kc, BG3 and Cl.8 сells. From these data one could conclude that aquamarine chromatin (and so the interbands of polytene chromosomes as well) is nothing but promoter regions of housekeeping genes, therefore interbands of polytene chromosomes may appear decompacted due to the permanent expression of such genes. However, this is not quite the case, as only ~60% of aquamarine domains overlap with TSSs of protein-coding genes (Fig. 1 ). Moreover, data are available indicating that open chromatin structure is not invariably coupled to gene expression. For instance, interbands in the regions 3C and 61C of salivary gland polytene chromosomes correspond to aquamarine chromatin, yet their morphologically decompacted state is unrelated to the transcription status of the underlying genes [78]. Thus, aquamarine is characterized as a universally open chromatin, which is not necessarily coupled to local gene expression. Understanding the functioning of a wide range of proteins enriched in aquamarine chromatin is of special interest. Many of these proteins can have dual functions, depending on the context of complexes they form with other proteins [79].
Roughly 40% of the euchromatic part of the genome is composed of the chromatin types having intermediate morphological levels of compaction (stronger than aquamarine, yet weaker than ruby) observed in polytene chromosomes – these are lazurite and malachite chromatin types [10]. In all four cell lines studied, lazurite chromatin is enriched with proteins and histone modifications associated with the elongation step of transcription (Supplementary Fig. 2). On average, lazurite chromatin corresponds to transcribed portions of protein-coding genes that include both exons and introns (GENE localization class) (Fig. 1 ). Thus, organization and protein composition of lazurite chromatin appear consistent with our view of this chromatin type as composed of actively and broadly transcribed sequences many of which are protein-coding [80-83]. Malachite chromatin type is rich in natural transposon insertions [84]. Transposons and expression thereof are known to have a regulatory role in the genome [80-82]. Ruby chromatin displays the features that are opposite to those of aquamarine chromatin. All the proteins typically found in open chromatin tend to be depleted in ruby across different cell types. This chromatin type frequently maps to densely packed bands and intercalary heterochromatin bands of polytene chromosomes [5, 10] that are known to encompass clusters of co-expressed stage- or tissue-specific genes [6, 83, 84]. Ruby chromatin is enriched for a set of proteins and histone modifications characteristic of inactive chromatin (Fig. 2), it is depleted for natural deletion breakpoints, point mutations, miRNA genes and pseudogenes, and is virtually devoid of tRNA genes (Fig. 3). Its average expression level is significantly lower than that of aquamarine chromatin [10].
We propose that the observed stability of organization of the four chromatin types across cell lines relies on the common principles of chromosome organization into domains that are universal across somatic cells. Further analysis of such domains and their regulation should provide deeper insight into chromatin organization in the interphase nucleus.
ACKNOWLEDGeMENTS
We thank Sergey A. Demakov, Stepan N. Belyakin, Tatyana D. Kolesnikova, Evgeniya N. Andreyeva, Galina V. Pokholkova and Elena S. Belyaeva for helpful suggestions and critical reading of the manuscript. The computer tools for processing the genome annotation tracks were developed by VGL under support of the ICG SB RAS budget project 0324-2015-0003; the FISH mapping of 35D1-4 region was performed by LVB, OVD and VFS under support of fundamental scientific research on the project 0310-2014-0002. The work of AVP and IFZ at IMCB SB RAS was funded by the Russian Scientific Foundation grant #14-14-00934. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s web site along with the published article.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
REFERENCES
- 1.Beermann W. Chromomeres and genes. Results Probl. Cell Differ. 1972;4:1–33. doi: 10.1007/978-3-540-37164-9_1. [DOI] [PubMed] [Google Scholar]
- 2.Zhimulev I.F. Genetic organization of polytene chromosomes. Adv. Genet. 1999;39:1–589. doi: 10.1016/s0065-2660(08)60476-9. [DOI] [PubMed] [Google Scholar]
- 3.Gersh E.S. Sites of gene activity and of inactive genes in polytene chromosomes of diptera. J. Theor. Biol. 1975;50:413–428. doi: 10.1016/0022-5193(75)90090-9. [DOI] [PubMed] [Google Scholar]
- 4.Belyaeva E.S., Andreyeva E.N., Belyakin S.N., Volkova E.I., Zhimulev I.F. Intercalary heterochromatin in polytene chromosomes of Drosophila melanogaster. Chromosoma. 2008;117:411–418. doi: 10.1007/s00412-008-0163-7. [DOI] [PubMed] [Google Scholar]
- 5.Belyaeva E.S., Goncharov F.P., Demakova O.V., Kolesnikova T.D., Boldyreva L.V., Semeshin V.F., Zhimulev I.F. Late replication domains in polytene and non-polytene cells of Drosophila melanogaster. PLoS One. 2012;7:e30035. doi: 10.1371/journal.pone.0030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belyakin S.N., Christophides G.K., Alekseyenko A.A., Kriventseva E.V., Belyaeva E.S., Nanayev R.A., Makunin I.V., Kafatos F.C., Zhimulev I.F. Genomic analysis of Drosophila chromosome underreplication reveals a link between replication control and transcriptional territories. Proc. Natl. Acad. Sci. USA. 2005;102:8269–8274. doi: 10.1073/pnas.0502702102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belyakin S.N., Babenko V.N., Maksimov D.A., Shloma V.V., Kvon E.Z., Belyaeva E.S., Zhimulev I.F. Gene density profile reveals the marking of late replicated domains in the Drosophila melanogaster genome. Chromosoma. 2010;119:589–600. doi: 10.1007/s00412-010-0280-y. [DOI] [PubMed] [Google Scholar]
- 8.Demakov S.A., Vatolina T.Y., Babenko V.N., Semeshin V.F., Belyaeva E.S., Zhimulev I.F. Protein composition of interband regions in polytene and cell line chromosomes of Drosophila melanogaster. BMC Genomics. 2011;12:566. doi: 10.1186/1471-2164-12-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vatolina T.Y., Demakov S.A., Semeshin V.F., Makunin I.V., Babenko V.N., Belyaeva E.S., Zhimulev I.F. Identification and molecular genetic characterization of the polytene chromosome interbands in Drosophila melanogaster. Russ. J. Genet. 2011;47:521–532. [PubMed] [Google Scholar]
- 10.Zhimulev I.F., Zykova T.Y., Goncharov F.P., Khoroshko V.A., Demakova O.V., Semeshin V.F., Pokholkova G.V., Boldyreva L.V., Demidova D.S., Babenko V.N., Demakov S.A., Belyaeva E.S. Genetic organization of interphase chromosome bands and interbands in Drosophila melanogaster. PLoS One. 2014;9:e101631. doi: 10.1371/journal.pone.0101631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vatolina T.Y., Boldyreva L.V., Demakova O.V., Demakov S.A., Kokoza E.B., Semeshin V.F., Babenko V.N., Goncharov F.P., Belyaeva E.S., Zhimulev I.F. Identical functional organization of nonpolytene and polytene chromosomes in Drosophila melanogaster. PLoS One. 2011;6:e25960. doi: 10.1371/journal.pone.0025960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhimulev I.F., Belyaeva E.S., Vatolina T.Y., Demakov S.A. Banding patterns in Drosophila melanogaster polytene chromosomes correlate with DNA-binding protein occupancy. BioEssays. 2012;34:498–508. doi: 10.1002/bies.201100142. [DOI] [PubMed] [Google Scholar]
- 13.Roy S., Ernst J., Kharchenko P.V., Kheradpour P., Negre N., Eaton M.L., Landolin J.M., Bristow C.A., Ma L., Lin M.F., Washietl S., Arshinoff B.I., Ay F., Meyer P.E., Robine N., Washington N.L., Di Stefano L., Berezikov E., Brown C.D., Candeias R., Carlson J.W., Carr A., Jungreis I., Marbach D., Sealfon R., Tolstorukov M.Y., Will S., Alekseyenko A.A., Artieri C., Booth B.W., Brooks A.N., Dai Q., Davis C.A., Duff M.O., Feng X., Gorchakov A.A., Gu T., Henikoff J.G., Kapranov P., Li R., MacAlpine H.K., Malone J., Minoda A., Nordman J., Okamura K., Perry M., Powell S.K., Riddle N.C., Sakai A., Samsonova A., Sandler J.E., Schwartz Y.B., Sher N., Spokony R., Sturgill D., van Baren M., Wan K.H., Yang L., Yu C., Feingold E., Good P., Guyer M., Lowdon R., Ahmad K., Andrews J., Berger B., Brenner S.E., Brent M.R., Cherbas L., Elgin S.C., Gingeras T.R., Grossman R., Hoskins R.A., Kaufman T.C., Kent W., Kuroda M.I., Orr-Weaver T., Perrimon N., Pirrotta V., Posakony J.W., Ren B., Russell S., Cherbas P., Graveley B.R., Lewis S., Micklem G., Oliver B., Park P.J., Celniker S.E., Henikoff S., Karpen G.H., Lai E.C., MacAlpine D.M., Stein L.D., White K.P., Kellis M. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Filion G.J., van Bemmel J.G., Braunschweig U., Talhout W., Kind J., Ward L.D., Brugman W., de Castro I.J., Kerkhoven R.M., Bussemaker H.J., van Steensel B. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith R.N., Aleksic J., Butano D., Carr A., Contrino S., Hu F., Lyne M., Lyne R., Kalderimis A., Rutherford K., Stepan R., Sullivan J., Wakeling M., Watkins X., Micklem G. InterMine: a flexible data warehouse system for the integration and analysis of heterogeneous biological data. Bioinformatics. 2012;28:3163–3165. doi: 10.1093/bioinformatics/bts577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Bemmel J.G., Filion G.J., Rosado A., Talhout W., de Haas M., van Welsem T., van Leeuwen F., van Steensel B. A network model of the molecular organization of chromatin in Drosophila. Mol. Cell. 2013;49:759–771. doi: 10.1016/j.molcel.2013.01.040. [DOI] [PubMed] [Google Scholar]
- 17.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Hornik K., Hothorn T., Huber W., Iacus S., Irizarry R., Leisch F., Li C., Maechler M., Rossini A.J., Sawitzki G., Smith C., Smyth G., Tierney L., Yang J.Y., Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrence M., Gentleman R., Carey V. rtracklayer: an R package for interfacing with genome browsers. Bioinformatics. 2009;25:1841–1842. doi: 10.1093/bioinformatics/btp328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawrence M., Huber W., Pages H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., Carey V.J. Software for computing and annotating genomic ranges. PLOS Comput. Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.R Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2015. [Google Scholar]
- 21.Semeshin V.F., Belyaeva E.S., Shloma V.V., Zhimulev I.F. Electron microscopy of polytene chromosomes. Methods Mol. Biol. 2004;247:305–324. doi: 10.1385/1-59259-665-7:305. [DOI] [PubMed] [Google Scholar]
- 22.Ashburner M., Golic K.G., Hawley R.S. Drosophila: A Laboratory Handbook. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- 23.Wu L., Zee B.M., Wang Y., Garcia B.A., Dou Y. The RING finger protein MSL2 in the MOF complex is an E3 ubiquitin ligase for H2B K34 and is involved in crosstalk with H3 K4 and K79 methylation. Mol. Cell. 2011;43:132–144. doi: 10.1016/j.molcel.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwak H., Lis J.T. Control of transcriptional elongation. Annu. Rev. Genet. 2013;47:483–508. doi: 10.1146/annurev-genet-110711-155440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Regnard C., Straub T., Mitterweger A., Dahlsveen I.K., Fabian V., Becker P.B. Global analysis of the relationship between JIL-1 kinase and transcription. PLoS Genet. 2011;7:e1001327. doi: 10.1371/journal.pgen.1001327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernstein B.E., Humphrey E.L., Erlich R.L., Schneider R., Bouman P., Liu J.S., Kouzarides T., Schreiber S.L. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. USA. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schübeler D., MacAlpine D.M., Scalzo D., Wirbelauer C., Kooperberg C., van Leeuwen F., Gottschling D.E., O'Neill L.P., Turner B.M., Delrow J., Bell S.P., Groudine M. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conrad T., Cavalli F.M., Holz H., Hallacli E., Kind J., Ilik I., Vaquerizas J.M., Luscombe N.M., Akhtar A. The MOF chromobarrel domain controls genome-wide H4K16 acetylation and spreading of the MSL complex. Dev. Cell. 2012;22:610–624. doi: 10.1016/j.devcel.2011.12.016. [DOI] [PubMed] [Google Scholar]
- 29.Kind J., Vaquerizas J.M., Gebhardt P., Gentzel M., Luscombe N.M., Bertone P., Akhtar A. Genome-wide analysis reveals MOF as a key regulator of dosage compensation and gene expression in Drosophila. Cell. 2008;133:813–828. doi: 10.1016/j.cell.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 30.Lam K.C., Muhlpfordt F., Vaquerizas J.M., Raja S.J., Holz H., Luscombe N.M., Manke T., Akhtar A. The NSL complex regulates housekeeping genes in Drosophila. PLoS Genet. 2012;8:e1002736. doi: 10.1371/journal.pgen.1002736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dorighi K.M., Tamkun J.W. The trithorax group proteins Kismet and ASH1 promote H3K36 dimethylation to counteract Polycomb group repression in Drosophila. Development. 2013;140:4182–4192. doi: 10.1242/dev.095786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klose R.J., Kallin E.M., Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 33.Lorbeck M.T., Singh N., Zervos A., Dhatta M., Lapchenko M., Yang C., Elefant F. The histone demethylase Dmel\Kdm4A controls genes required for life span and male-specific sex determination in Drosophila. Gene. 2010;450:8–17. doi: 10.1016/j.gene.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Rubertis F., Kadosh D., Henchoz S., Pauli D., Reuter G., Struhl K., Spierer P. The histone deacetylase RPD3 counteracts genomic silencing in Drosophila and yeast. Nature. 1996;384:589–591. doi: 10.1038/384589a0. [DOI] [PubMed] [Google Scholar]
- 35.Karachentsev D., Sarma K., Reinberg D., Steward R. PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes Dev. 2005;19:431–435. doi: 10.1101/gad.1263005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gutiérrez L., Oktaba K., Scheuermann J.C., Gambetta M.C., Ly-Hartig N., Muller J. The role of the histone H2A ubiquitinase Sce in Polycomb repression. Development. 2012;139:117–127. doi: 10.1242/dev.074450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishioka K., Rice J.C., Sarma K., Erdjument-Bromage H., Werner J., Wang Y., Chuikov S., Valenzuela P., Tempst P., Steward R., Lis J.T., Allis C.D., Reinberg D. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol. Cell. 2002;9:1201–1213. doi: 10.1016/s1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- 38.Fasulo B., Deuring R., Murawska M., Gause M., Dorighi K.M., Schaaf C.A., Dorsett D., Brehm A., Tamkun J.W. The Drosophila MI-2 chromatin-remodeling factor regulates higher-order chromatin structure and cohesin dynamics in vivo. PLoS Genet. 2012;8:e1002878. doi: 10.1371/journal.pgen.1002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hollmann M., Simmerl E., Schafer U., Schafer M.A. The essential Drosophila melanogaster gene wds (will die slowly) codes for a WD-repeat protein with seven repeats. Mol. Genet. Genomics. 2002;268:425–433. doi: 10.1007/s00438-002-0768-0. [DOI] [PubMed] [Google Scholar]
- 40.Raja S.J., Charapitsa I., Conrad T., Vaquerizas J.M., Gebhardt P., Holz H., Kadlec J., Fraterman S., Luscombe N.M., Akhtar A. The nonspecific lethal complex is a transcriptional regulator in Drosophila. Mol. Cell. 2010;38:827–841. doi: 10.1016/j.molcel.2010.05.021. [DOI] [PubMed] [Google Scholar]
- 41.Bouazoune K., Brehm A. ATP-dependent chromatin remodeling complexes in Drosophila. Chromosome Res. 2006;14:433–449. doi: 10.1007/s10577-006-1067-0. [DOI] [PubMed] [Google Scholar]
- 42.Carre C., Ciurciu A., Komonyi O., Jacquier C., Fagegaltier D., Pidoux J., Tricoire H., Tora L., Boros I.M., Antoniewski C. The Drosophila NURF remodelling and the ATAC histone acetylase complexes functionally interact and are required for global chromosome organization. EMBO Rep. 2008;9:187–192. doi: 10.1038/sj.embor.7401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deuring R., Fanti L., Armstrong J.A., Sarte M., Papoulas O., Prestel M., Daubresse G., Verardo M., Moseley S.L., Berloco M., Tsukiyama T., Wu C., Pimpinelli S., Tamkun J.W. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol. Cell. 2000;5:355–365. doi: 10.1016/s1097-2765(00)80430-x. [DOI] [PubMed] [Google Scholar]
- 44.Font-Burgada J., Rossell D., Auer H., Azorin F. Drosophila HP1c isoform interacts with the zinc-finger proteins WOC and Relative-of-WOC to regulate gene expression. Genes Dev. 2008;22:3007–3023. doi: 10.1101/gad.481408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fuda N.J., Guertin M.J., Sharma S., Danko C.G., Martins A.L., Siepel A., Lis J.T. GAGA factor maintains nucleosome-free regions and has a role in RNA polymerase II recruitment to promoters. PLoS Genet. 2015;11:e1005108. doi: 10.1371/journal.pgen.1005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwon S.H., Florens L., Swanson S.K., Washburn M.P., Abmayr S.M., Workman J.L. Heterochromatin protein 1 (HP1) connects the FACT histone chaperone complex to the phosphorylated CTD of RNA polymerase II. Genes Dev. 2010;24:2133–2145. doi: 10.1101/gad.1959110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feller C., Prestel M., Hartmann H., Straub T., Soding J., Becker P.B. The MOF-containing NSL complex associates globally with housekeeping genes, but activates only a defined subset. Nucleic Acids Res. 2012;40:1509–1522. doi: 10.1093/nar/gkr869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kahn T.G., Stenberg P., Pirrotta V., Schwartz Y.B. Combinatorial interactions are required for the efficient recruitment of pho repressive complex (PhoRC) to polycomb response elements. PLoS Genet. 2014;10:e1004495. doi: 10.1371/journal.pgen.1004495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klymenko T., Papp B., Fischle W., Kocher T., Schelder M., Fritsch C., Wild B., Wilm M., Muller J. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006;20:1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eichinger C.S., Kurze A., Oliveira R.A., Nasmyth K. Disengaging the Smc3/kleisin interface releases cohesin from Drosophila chromosomes during interphase and mitosis. EMBO J. 2013;32:656–665. doi: 10.1038/emboj.2012.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lupo R., Breiling A., Bianchi M.E., Orlando V. Drosophila chromosome condensation proteins Topoisomerase II and Barren colocalize with Polycomb and maintain Fab-7 PRE silencing. Mol. Cell. 2001;7:127–136. doi: 10.1016/s1097-2765(01)00161-7. [DOI] [PubMed] [Google Scholar]
- 52.Alekseyenko A.A., Gorchakov A.A., Zee B.M., Fuchs S.M., Kharchenko P.V., Kuroda M.I. Heterochromatin-associated interactions of Drosophila HP1a with dADD1, HIPP1, and repetitive RNAs. Genes Dev. 2014;28:1445–1460. doi: 10.1101/gad.241950.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eissenberg J.C., Elgin S.C. HP1a: a structural chromosomal protein regulating transcription. Trends Genet. 2014;30:103–110. doi: 10.1016/j.tig.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schuettengruber B., Oded Elkayam N., Sexton T., Entrevan M., Stern S., Thomas A., Yaffe E., Parrinello H., Tanay A., Cavalli G. Cooperativity, specificity, and evolutionary stability of Polycomb targeting in Drosophila. Cell Reports. 2014;9:219–233. doi: 10.1016/j.celrep.2014.08.072. [DOI] [PubMed] [Google Scholar]
- 55.Greil F., de Wit E., Bussemaker H.J., van Steensel B. HP1 controls genomic targeting of four novel heterochromatin proteins in Drosophila. EMBO J. 2007;26:741–751. doi: 10.1038/sj.emboj.7601527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schotta G., Ebert A., Krauss V., Fischer A., Hoffmann J., Rea S., Jenuwein T., Dorn R., Reuter G. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hari K.L., Cook K.R., Karpen G.H. The Drosophila Su(var)2-10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family. Genes Dev. 2001;15:1334–1348. doi: 10.1101/gad.877901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pindyurin A.V., Moorman C., de Wit E., Belyakin S.N., Belyaeva E.S., Christophides G.K., Kafatos F.C., van Steensel B., Zhimulev I.F. SUUR joins separate subsets of PcG, HP1 and B-type lamin targets in Drosophila. J. Cell Sci. 2007;120:2344–2351. doi: 10.1242/jcs.006007. [DOI] [PubMed] [Google Scholar]
- 59.Boros I.M. Histone modification in Drosophila. Brief. Funct. Genomics. 2012;11:319–331. doi: 10.1093/bfgp/els029. [DOI] [PubMed] [Google Scholar]
- 60.Eissenberg J.C., Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev. Biol. 2010;339:240–249. doi: 10.1016/j.ydbio.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mozzetta C., Boyarchuk E., Pontis J., Ait-Si-Ali S. Sound of silence: the properties and functions of repressive Lys methyltransferases. Nat. Rev. Mol. Cell Biol. 2015;16:499–513. doi: 10.1038/nrm4029. [DOI] [PubMed] [Google Scholar]
- 62.Swaminathan A., Gajan A., Pile L.A. Epigenetic regulation of transcription in Drosophila. Front. Biosci. (Landmark Ed.) 2012;17:909–937. doi: 10.2741/3964. [DOI] [PubMed] [Google Scholar]
- 63.Wright D.E., Wang C.Y., Kao C.F. Flickin' the ubiquitin switch: the role of H2B ubiquitylation in development. Epigenetics. 2011;6:1165–1175. doi: 10.4161/epi.6.10.17745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Joshi A.A., Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol. Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 65.Lin C.H., Li B., Swanson S., Zhang Y., Florens L., Washburn M.P., Abmayr S.M., Workman J.L. Heterochromatin protein 1a stimulates histone H3 lysine 36 demethylation by the Drosophila KDM4A demethylase. Mol. Cell. 2008;32:696–706. doi: 10.1016/j.molcel.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin C.H., Paulson A., Abmayr S.M., Workman J.L. HP1a targets the Drosophila KDM4A demethylase to a subset of heterochromatic genes to regulate H3K36me3 levels. PLoS One. 2012;7:e39758. doi: 10.1371/journal.pone.0039758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Di Stefano L., Ji J.Y., Moon N.S., Herr A., Dyson N. Mutation of Drosophila Lsd1 disrupts H3-K4 methylation, resulting in tissue-specific defects during development. Curr. Biol. 2007;17:808–812. doi: 10.1016/j.cub.2007.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schuettengruber B., Chourrout D., Vervoort M., Leblanc B., Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 69.Deng H., Cai W., Wang C., Lerach S., Delattre M., Girton J., Johansen J., Johansen K.M. JIL-1 and Su(var)3-7 interact genetically and counteract each other's effect on position-effect variegation in Drosophila. Genetics. 2010;185:1183–1192. doi: 10.1534/genetics.110.117150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Makunin I.V., Volkova E.I., Belyaeva E.S., Nabirochkina E.N., Pirrotta V., Zhimulev I.F. The Drosophila suppressor of underreplication protein binds to late-replicating regions of polytene chromosomes. Genetics. 2002;160:1023–1034. doi: 10.1093/genetics/160.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Levinger L., Varshavsky A. Selective arrangement of ubiquitinated and D1 protein-containing nucleosomes within the Drosophila genome. Cell. 1982;28:375–385. doi: 10.1016/0092-8674(82)90355-5. [DOI] [PubMed] [Google Scholar]
- 72.Cipressa F., Romano S., Centonze S., zur Lage P.I., Verni F., Dimitri P., Gatti M., Cenci G. Effete, a Drosophila chromatin-associated ubiquitin-conjugating enzyme that affects telomeric and heterochromatic position effect variegation. Genetics. 2013;195:147–158. doi: 10.1534/genetics.113.153320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kharchenko P.V., Alekseyenko A.A., Schwartz Y.B., Minoda A., Riddle N.C., Ernst J., Sabo P.J., Larschan E., Gorchakov A.A., Gu T., Linder-Basso D., Plachetka A., Shanower G., Tolstorukov M.Y., Luquette L.J., Xi R., Jung Y.L., Park R.W., Bishop E.P., Canfield T.K., Sandstrom R., Thurman R.E., MacAlpine D.M., Stamatoyannopoulos J.A., Kellis M., Elgin S.C., Kuroda M.I., Pirrotta V., Karpen G.H., Park P.J. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hoskins R.A., Landolin J.M., Brown J.B., Sandler J.E., Takahashi H., Lassmann T., Yu C., Booth B.W., Zhang D., Wan K.H., Yang L., Boley N., Andrews J., Kaufman T.C., Graveley B.R., Bickel P.J., Carninci P., Carlson J.W., Celniker S.E. Genome-wide analysis of promoter architecture in Drosophila melanogaster. Genome Res. 2011;21:182–192. doi: 10.1101/gr.112466.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Juven-Gershon T., Kadonaga J.T. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 2010;339:225–229. doi: 10.1016/j.ydbio.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rach E.A., Winter D.R., Benjamin A.M., Corcoran D.L., Ni T., Zhu J., Ohler U. Transcription initiation patterns indicate divergent strategies for gene regulation at the chromatin level. PLoS Genet. 2011;7:e1001274. doi: 10.1371/journal.pgen.1001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rach E.A., Yuan H.Y., Majoros W.H., Tomancak P., Ohler U. Motif composition, conservation and condition-specificity of single and alternative transcription start sites in the Drosophila genome. Genome Biol. 2009;10:R73. doi: 10.1186/gb-2009-10-7-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kvon E.Z., Demakov S.A., Zhimulev I.F. Chromatin decompaction in the interbands of Drosophila polytene chromosomes does not correlate with high transcription level. Russ. J. Genet. 2011;47:674–681. [PubMed] [Google Scholar]
- 79.Meier K., Brehm A. Chromatin regulation: how complex does it get? Epigenetics. 2014;9:1485–1495. doi: 10.4161/15592294.2014.971580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mani S.R., Juliano C.E. Untangling the web: the diverse functions of the PIWI/piRNA pathway. Mol. Reprod. Dev. 2013;80:632–664. doi: 10.1002/mrd.22195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McCue A.D., Slotkin R.K. Transposable element small RNAs as regulators of gene expression. Trends Genet. 2012;28:616–623. doi: 10.1016/j.tig.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 82.Gbadegesin M.A. Transposable elements in the genomes: parasites, junks or drivers of evolution? Afr. J. Med. Med. Sci. 2012;41(Suppl.):13–25. [PubMed] [Google Scholar]
- 83.Makunin I.V., Kolesnikova T.D., Andreyenkova N.G. Underreplicated regions in Drosophila melanogaster are enriched with fast-evolving genes and highly conserved noncoding sequences. Genome Biol. Evol. 2014;6:2050–2060. doi: 10.1093/gbe/evu156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ranz J.M., Diaz-Castillo C., Petersen R. Conserved gene order at the nuclear periphery in Drosophila. Mol. Biol. Evol. 2012;29:13–16. doi: 10.1093/molbev/msr178. [DOI] [PubMed] [Google Scholar]
- 85.Bridges P.N. A new map of the salivary gland 2L-chromosome of Drosophila melanogaster. J. Hered. 1942;33:403–408. [Google Scholar]
- 86.Gortchakov A.A., Eggert H., Gan M., Mattow J., Zhimulev I.F., Saumweber H. Chriz, a chromodomain protein specific for the interbands of Drosophila melanogaster polytene chromosomes. Chromosoma. 2005;114:54–66. doi: 10.1007/s00412-005-0339-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s web site along with the published article.


