Abstract
Myeloproliferative Neoplasms with myelofibrosis (MPN-MF) demonstrate constitutive activation of JAK-STAT signaling, which responds to treatment with the JAK1 & 2 kinase inhibitor (JAKi) ruxolitinib. However, MPN-MF often progresses (~20%) to secondary AML (sAML), where standard induction chemotherapy or ruxolitinib is relatively ineffective, necessitating the development of novel therapeutic approaches. In the present studies, we demonstrate that treatment with BET (bromodomain and extra terminal) protein inhibitor (BETi), e.g., JQ1, inhibits growth and induces apoptosis of cultured and primary, patient-derived (PD), post-MPN sAML blast progenitor cells. Reverse-phase protein array, mass-cytometry and Western analyses revealed that BETi treatment attenuated the protein expressions of c-MYC, p-STAT5, Bcl-xL, CDK4/6, PIM1 and IL-7R, while concomitantly inducing the levels of HEXIM1, p21 and BIM in the sAML cells. Co-treatment with BETi and ruxolitinib synergistically induced apoptosis of cultured and PD sAML cells, as well as significantly improved survival of immune-depleted mice engrafted with human sAML cells. While BETi or heat shock protein (HSP) 90 inhibitor alone exerted lethal activity, co-treatment with BETi and HSP90i was synergistically lethal against the ruxolitinib-persister or ruxolitinib-resistant sAML cells. Collectively, these findings further support in vivo testing of BETi-based combinations with JAKi and HSP90i against post-MPN sAML cells.
Keywords: secondary AML, BRD4, bromodomain antagonist, JAK2
INTRODUCTION
Myeloproliferative neoplasms with myelofibrosis (MPN-MF) express mutation in JAK2 (JAKV617F and exon 12 mutations), the thrombopoietin receptor c-MPL, or calreticulin (CALR) gene and exhibit constitutive activation of JAK-STAT signaling1,2. Ruxolitinib is a type I, ATP-competitive, wild-type and mutated JAK1 & 2 inhibitor (JAK-I) currently used as therapy for MPN-MF3,4. As a single agent, ruxolitinib confers notable clinical benefit by reducing the disease-related symptoms, splenomegaly, and improving patient survival in MPN-MF4-6. Ruxolitinib-induced responses and survival improvement occur independent of co-mutations in the genes other than JAK2, MPL and CALR7. However, continuous exposure to ruxolitinib only modestly reduces the allelic burden of the mutant JAK23. Prolonged exposure to ruxolitinib may also lead to a loss of response, causing the emergence of drug-tolerant and persistent (DTP) cells, or JAK inhibitor-resistant (JIR) cells8-10. Although lacking in additional mutations in JAK2, JIR cells exhibit reactivation of JAK-STAT signaling due to transphosphorylation of JAK2 by JAK1 or TYK2 tyrosine kinases (TK) 10,11. One-third of patients with MPN-MF exhibit recurrent mutations in genes encoding for chromatin modifiers (e.g., TET2 and IDH1 & 2) and splicing factors (e.g., SRSF2) 12,13. Co-mutations in ASXL1, TET2 and SRSF2 are associated with poorer spleen response, treatment discontinuation and adverse outcome in ruxolitinib-treated patients with MF13-15. Recurrent SRSF2 mutations are especially associated with shorter leukemia free survival14,15. The presence of 2 or more somatic mutations is strongly associated with the risk of AML transformation (sAML)12,13. Transformation to AML occurs in up to 20% of patients with MPN-MF13,16. Ruxolitinib exhibits modest activity and does not significantly impact the clinical outcome in sAML, where standard anthracycline and Ara-C-based chemotherapy is also mostly ineffective and may be associated with hematologic toxicity16-18. In sAML versus de novo AML, the recurrent, ‘driver’, somatic mutations are appreciably different, e.g. NPM1 and FLT3 mutations are rarely observed16,19,20. Sequential genomic assessments in pre- and post-sAML transformation have revealed mutations in TET2, ASXL1, IDH1 & 2, SRSF2, RUNX1, MYC, PTPN11, NRAS, SETBP1 and TP53 genes16,19. A co-occurrence of JAK2 V617F and mutant TP53 was documented in the dominant clones of sAML19,20. Since treatment with JAKi is ineffective, it is important to identify and elucidate the activity of novel agents for the therapy of the post-MPN sAML16,18.
The family of BET (bromodomain and extraterminal) proteins, including BRD4, are chromatin reader proteins that contain the N-terminal, double-tandem bromodomains, which bind to the acetylated lysine on the nucleosomal histones and transcription factors21. BET proteins also contain an extra-terminal (ET) domain in the C-terminus, through which they interact and recruit co-regulatory chromatin modifying enzymes, remodeling factors and the mediator elements to the chromatin for regulating gene transcription21, 22. The C-terminal domain (CTD) of BRD4 also interacts with pTEFb (positive transcription elongation factor b), the heterodimer composed of cyclin dependent kinase 9 (CDK9) and its regulatory subunit cyclin T 23. After recruitment to the gene promoters, the kinase activity of CDK9 in pTEFb phosphorylates serine 2 of the heptad repeats in the C-terminal domain (CTD) of RNA pol II (RNAP2), enabling it to mediate mRNA transcript elongation21, 24. Thus, BRD4 couples histone acetylation to transcript elongation, especially at the enhancers and promoters of oncogenes, including c-MYC, BCL-2, PIM1 and CDK4/6 that are regulated by clustered or ‘super’ enhancers and are important for cell growth and survival of AML cells21, 25. Although sustained inducible genetic knockdown of BRD4 causes multiple (but reversible) organ toxicities, pertinent to therapy, an RNAi screen identified BRD4 as an effective and promising target in AML cells26, 27. Several structure/activity-based BET protein small-molecule, acetyl-lysine-mimetic inhibitors (BET-I) have been developed, including JQ1, OTX-015 and GSK525762 28, 29. These agents displace BET proteins, along with the associated transcript initiation and elongation factors, from the chromatin, causing transcriptional repression of BCL-2, c-MYC, CDK4/628,29. Recently, we and others have demonstrated that BETis, e.g., JQ1 and I-BET-151, inhibit in vitro and in vivo growth and induce apoptosis of cultured and primary AML cells, especially those expressing MLL fusion-oncoproteins, mutant NPM1 and/or FLT3-ITD30,31. Additionally, co-treatment with BETi and FLT3-TKI exhibited synergistic lethality against AML expressing FLT3-ITD32. BETi was also shown to retain lethal activity against AML cells resistant to FLT3-TKIs32. However, none of these reports determined the pre-clinical activity of BET-I alone, or in combination with JAKi, against cultured and patient-derived (PD), CD34+ sAML blast progenitor cells. In the present studies, we determined that treatment with BETi attenuates the expression of c-MYC, p-STAT5, PIM1 and CDK4/6, inhibits growth and induces apoptosis of JAKi-sensitive or -resistant sAML cells, including those that co-express JAK2V617F and mutant TP53, e.g., HEL92.1.7 and SET2 cells. Additionally, we demonstrate that co-treatment with BETi and JAKi (ruxolitinib, pacritinib or SAR302503) is synergistically lethal against sAML cells sensitive to JAKi. Based on our previously reported observation that treatment with the heat shock protein 90 inhibitor (HSP90i) depletes mutant JAK2, AKT and c-RAF, as well as induces apoptosis of JAKi-resistant sAML cells8, here we also show that combined therapy with BETi and HSP90i exerts synergistic lethality against JAKi-resistant sAML cells.
Materials and Methods
Cell lines and cell culture
Human erythroleukemia HEL 92.1.7 (HEL) and SET2 cells expressing mutant JAK2-V617F and TP53 were obtained from ATCC (Manassas, VA) and the DSMZ (Braunschweig, Germany), respectively. All experiments with the cell lines were performed within 6 months after thawing or obtaining from ATCC or DSMZ. HEL92.1.7 ruxolitinib-persister cells were generated by exposing the cells to 1.0 μM of ruxolitinib for 48 hours. Dead cells were removed by Ficoll Hypaque density gradient centrifugation. Live cells were expanded in drug-free media and then treated with two additional rounds of identical ruxolitinib treatments and recovery. After this the viable, expanded cells were treated with JQ1 and/or AUY922 for the described studies.
Purification and next-generation sequencing of PD post MPN-MF secondary AML cells
Patient-derived peripheral blood and/or bone marrow aspirate samples were obtained with informed consent from patients with secondary AML transformed from high risk (≥ 3) MF (according to the International Prognostic Scoring System, IPSS) and processed as previously described8. Detailed methods for targeted next-generation sequencing (NGS) of DNA from 23 sAML samples are in the Supplemental Methods.
Assessment of apoptosis by annexin-V staining
Untreated or drug-treated cells were stained with Annexin-V (Pharmingen, San Diego, CA) and TO-PRO-3 iodide and the percentages of apoptotic cells were determined by flow cytometry as previously described8, 32. For patient-derived sAML cells, loss of viability following drug treatment was determined as previously described8, 32.
Reverse phase protein array (RPPA) and single cell mass cytometry (CyTOF) analysis
Detailed methods for the processing and analysis of sAML cells by RPPA and CyTOF are provided in the Supplemental Methods.
In vivo model of secondary acute myeloid leukemia
Detailed methods for the generation and treatment of mice engrafted with secondary AML cells are provided in the Supplemental Methods.
Statistical Analysis
Significant differences between values obtained in a population of sAML cells treated with different experimental conditions were determined using a two-tailed, unpaired t-test. P values of less than 0.05 were assigned significance.
RESULTS
BETi treatment attenuates the levels of IL-7R, JAK2-pSTAT5 and c-MYC while inducing caspases 3 and 7 in post-MPN sAML cells
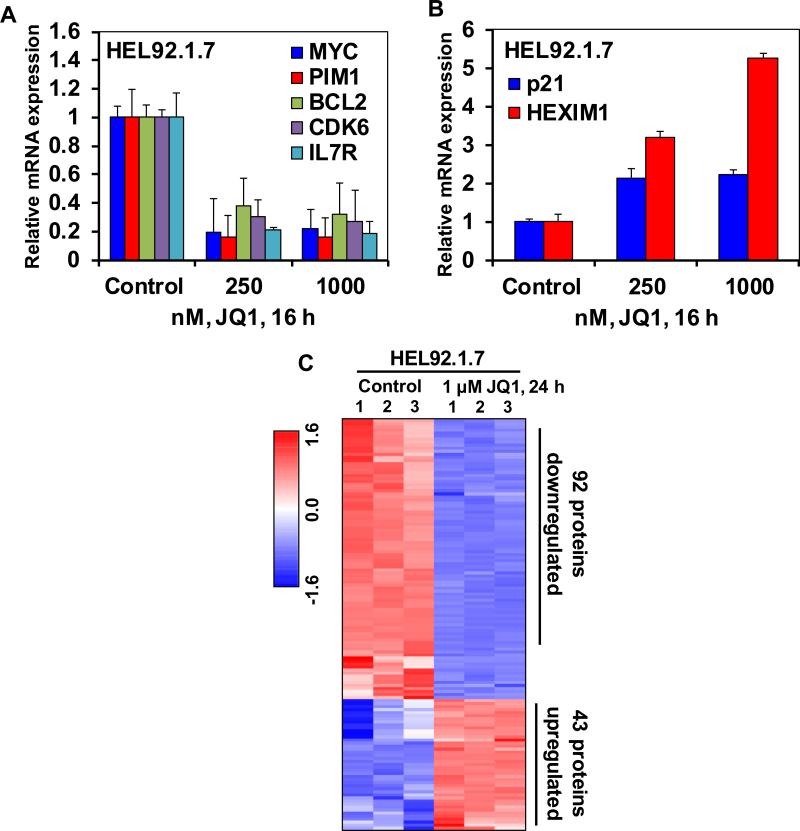
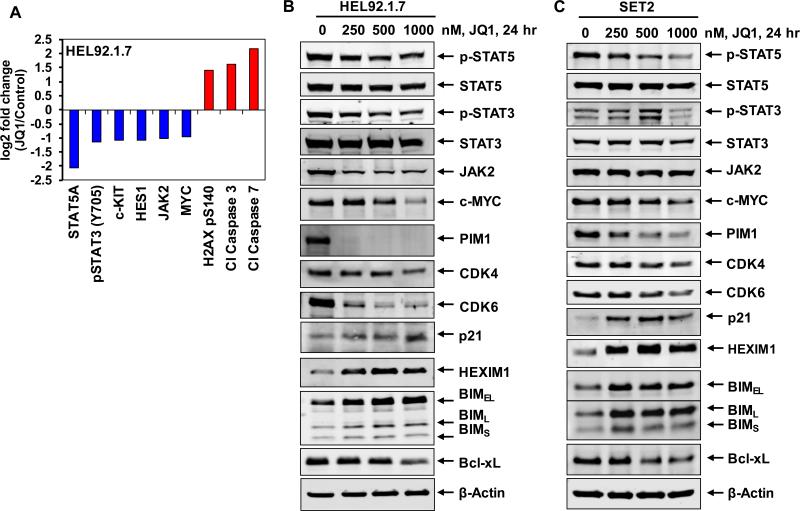
In a previous report we had demonstrated that treatment with the BETi JQ1 reduces the binding and occupancy of BRD4 on the chromatin associated with c-MYC, BCL-2 and CDK6 in the cultured and PD primary de novo AML cells30. Consistent with this, in the present studies the chromatin immunoprecipitation (ChIP) analyses showed that treatment with JQ1 also decreased the BRD4 occupancy on the enhancer and promoter of c-MYC, as well as on the promoters of BCL2 and CDK6 in the sAML HEL92.1.7 cells (Supplemental Figure S1A). Notably, qPCR analyses showed that JQ1 treatment dose-dependently attenuated the mRNA expression of c-MYC, BCL2 and CDK6, as well as of PIM1 and IL-7R in HEL92.1.7 and SET2 cells (Figure 1A and Supplemental Figure S1B). Concomitantly, in contrast, JQ1 treatment induced the mRNA expression of p21 and HEXIM1 in HEL92.1.7 cells (Figure 1B). HEXIM1, along with the small nuclear RNA 7SK, sequesters pTEFb in an inhibitory complex and induction of HEXIM1 has been shown to mechanistically mediate the inhibitory effects of BETi on RNAP2-mediated mRNA transcript elongation33. Treatment with a different BETi, I-BET151, also had a similar effect on the mRNA levels of c-MYC, BCL2, CDK6, PIM1 and p21 in HEL92.1.7 cells (Supplemental Figure S1C and S1D). We next determined the effects of JQ1 treatment on protein expressions, utilizing specific and validated antibodies coupled to a reverse phase protein array (RPPA). Figure 1C shows the heat map of the changes in expression (in triplicate) of those proteins that exhibited a ≥ 1.25 fold increase or decrease in expression and p <0.05 (relative to the untreated cells) following treatment with JQ1 (1.0 μM for 24 hours). More than twice as many proteins (92 proteins) were down regulated as were up regulated (43 proteins) (Figure 1C and Supplemental Table 1). Of these, the notable examples of the down regulated proteins were p-STAT5, p-STAT3, c-KIT, HES1, JAK2 and c-MYC, whereas protein levels of DNA damage-associated γ-H2AX (H2AX pS140) as well as of the apoptosis-inducing cleaved caspase 3 and 7 were simultaneously up regulated (Figure 2A). Western analyses were conducted to further confirm the effects of JQ1 on the protein levels in HEL92.1.7 and SET2 cells. Figure 2B and 2C show that exposure to JQ1 dose-dependently reduced the levels of c-MYC, p-STAT5, p-STAT3, JAK2, PIM1, CDK4 and CDK6. Concomitantly, JQ1 treatment increased the levels of HEXIM1, BIM and p21 (Figure 2B and 2C). Consistent with JQ1-mediated depletion of p-STAT5 levels, the protein expression of its target gene Bcl-xL was reduced in HEL92.1.7 and SET2 cells (Figure 2B and 2C) 34. Since a previous report had demonstrated that JQ1 treatment reduces p-STAT5 levels by also causing attenuation of the IL-7 receptor (IL-7R) expression35, we next determined the effects of JQ1 treatment on the mRNA and protein expression of IL-7R in sAML cells. QPCR and flow cytometric analysis, respectively, showed that treatment with JQ1 attenuated the mRNA expression and cell-surface protein expression of IL-7R in HEL92.1.7 and SET2 cells (Figure 1A and Supplemental Figure S2).
Figure 1. Treatment with the BET protein bromodomain inhibitor, JQ1 depletes the mRNA expression and protein expression of BRD4 target genes including c-MYC in cultured sAML cells.
A-B. HEL92.1.7 cells were treated with the indicated concentrations of JQ1 for 16 hours. At the end of treatment, RNA was isolated and reverse transcribed. The resulting cDNA was used for real-time, quantitative PCR analysis. The relative mRNA expression of each target was normalized to GAPDH and compared to the untreated cells. C. Heatmap of HEL92.1.7 treated with JQ1 for 24 hours, then analyzed by reverse phase protein analysis. Each column represents an independent sample. Samples were analyzed in biologic triplicates. Proteins shown were altered by at least 1.25-fold up or down relative to the untreated cells and had a p-value of less than 0.05.
Figure 2. Treatment with JQ1 depletes the expression levels of pSTAT5, c-MYC, CDK4 and CDK6 with concomitant induction of HEXIM1 and BIM expression levels in sAML cells.
A. HEL92.1.7 cells were treated with JQ1 for 24 hours, and then analyzed by reverse phase protein analysis. Figure shows log2 fold change of selected targets in JQ1-treated cells relative to the untreated control cells. All fold changes shown are significant (p<0.05). B-C. HEL92.1.7 and SET-2 cells were treated with the indicated concentrations of JQ1 for 24 hours. Total cell lysates were prepared and immunoblot analyses were conducted as indicated. The expression levels of β-Actin in the lysates served as the loading control.
BET-I treatment inhibits growth and induces apoptosis of post-MPN sAML cells
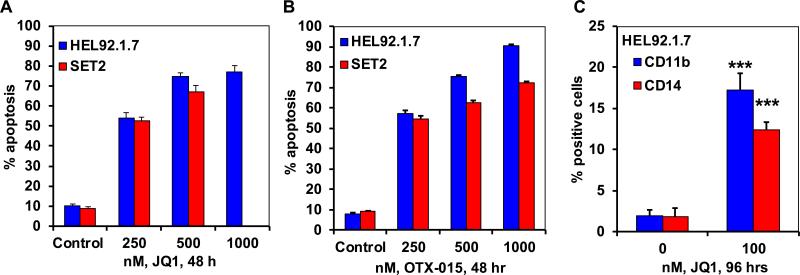
We next determined the effects of JQ1 treatment on the growth and apoptosis of the post-MPN sAML HEL92.1.7 and SET2 cells. Figure 3A demonstrates that consistent with its inhibitory effects on the levels of JAK2, p-STAT3, STAT5, c-MYC, PIM1 and Bcl-xL, as well as its induction of the levels of BIM, JQ1 treatment for 48 hours dose-dependently induced apoptosis of HEL92.1.7 and SET2 cells. Exposure to other BETi such as OTX-015 and I-BET-151 also dose- dependently induced apoptosis of HEL92.1.7 and SET2 cells (Figure 3B, and data not shown). We have previously reported and show here in the Supplemental Figure 3A to 3C that BETi JQ1 and OTX-015 treatment did not inhibit p-STAT5 and STAT5 levels, while it reduced c-MYC and CDK4/6, but induced HEXIM1, p21 and BIM, as well as induced apoptosis of non-JAK2 mutated cultured and primary de novo AML cells (30,32). Notably, a longer exposure (96 hours) to considerably lower concentrations of JQ1 (100 nM) inhibited the suspension culture growth of HEL92.1.7 and SET2 cells (Supplemental Figure 4A). This was associated with a significant increase in the cell surface expression of the differentiation-associated myeloid markers CD11b and CD14 on the HEL92.1.7 and SET2 cells (Figure 3C). Following a 96-hour exposure to JQ1, a significant increase in the % of non-viable cells was also observed (Supplemental Figure 4B). Collectively, these findings suggest that treatment with BETi inhibits growth, as well as induces differentiation and apoptosis of sAML cells.
Figure 3. Treatment with BET bromodomain inhibitors induces differentiation and apoptosis in sAML cells.
A-B. HEL92.1.7 and SET-2 cells were treated with the indicated concentrations of JQ1 or OTX-015 for 48 hours. The % apoptotic cells were determined by flow cytometry. Columns, mean apoptosis from three experiments; Bars, standard error of the mean. C. HEL92.1.7 cells were treated with 100 nM of JQ1 for 96 hours. At the end of treatment, the % CD11b and CD14-positive cells in HEL92.1.7 cells were determined. *** indicates significantly increased CD11b or CD14 expression levels in JQ1 treated cells compared to control cells (p<0.005).
Co-treatment with BETi and ruxolitinb exerts synergistic lethality and improves survival of mice engrafted with sAML cells
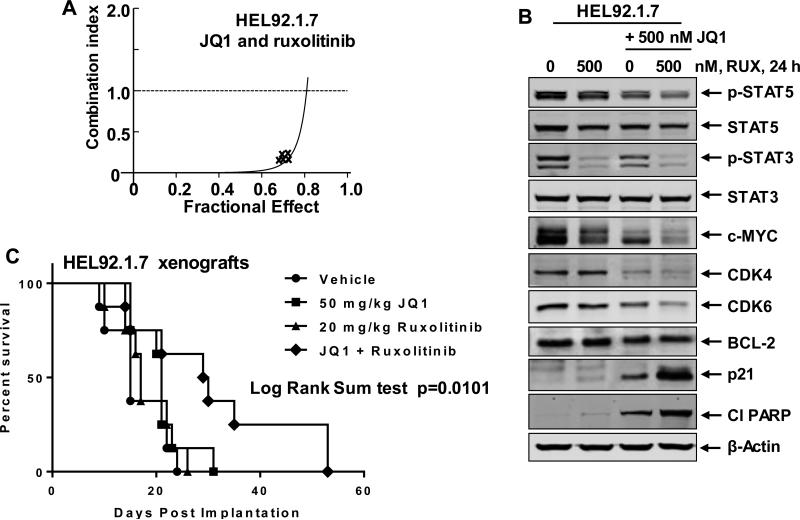
We had previously reported that treatment with a combination of BETi and tyrosine kinase inhibitor (TKI), which inhibits the activity of the growth and survival-promoting mutant and/or activated TKs, exerts synergistic lethality against AML and Mantle Cell Lymphoma cells32,36. Here, first, we utilized RPPA analyses to determine the effects of JAKi ruxolitinib or pacritinib (SB1518) on the signaling proteins in sAML cells37. As shown, both JAKis attenuated the levels of p-PRAS40, p-S6, p-4E-BP1, and p-STAT3, while also increasing γ-H2AX and cleaved caspase 3 & 7 levels (Supplemental Figure S5A and 5B and Supplemental Table 2 & 3). Notably, treatment with pacritinib caused greater inhibition of p-AKT. Next, we determined the activity of co-treatment with JQ1 with JAKis ruxolitinib, pacritinib or SAR against cultured post-MPN sAML cells. Figure 4A demonstrates that co-treatment with JQ1 and ruxolitinib synergistically induced apoptosis of HEL92.1.7, with combination indices below 1.0, utilizing the isobologram analyses (dose and fractional effect table is presented in Supplemental Figure S5A). Similar synergy between JQ1 and ruxolitinib was observed in SET2 cells (Supplemental Figure 6B). As compared to treatment with each agent alone, co-treatment with JQ1 and ruxolitinib caused greater attenuation of p-STAT5, CDK4, CDK6, c-MYC and p-STAT3 in HEL92.1.7 cells (Figure 4B). Whereas JQ1 treatment reduced BCL2 levels, co-treatment with ruxolitinib did not further reduce the levels of BCL2 in HEL92.1.7 cells. However, co-treatment with ruxolitinib further enhanced JQ1-mediated increase in the levels of p21 and cleaved PARP (Figure 4B). Co-treatment with JQ1 and ruxolitinib exerted similar effects on the protein levels of pSTAT5, CDK4, CDK6, MYC and p-STAT3 in SET2 cells (Supplemental Figure S6C). Combination of the BETi OTX-015 with ruxolitinib also synergistically induced apoptosis of SET2 cells (Supplemental Figure S6D). Notably, co-treatment of JQ1 with the other JAKis, pacritinib and SAR, also induced synergistic lethality against HEL92.1.7 and SET2 cells (Supplemental Figures S6E-S6I). Consistent with this, co-treatment with pacritinib also enhanced JQ1-mediated perturbations in the protein levels of c-MYC, CDK4, CDK6, BCL-2, Bcl-xL, p-STAT3, p-STAT5, PIM1 and cleaved PARP (Supplemental Figure S6G). We next determined the in vivo anti-sAML activity of JQ1 and/or ruxolitinib against the xenograft of HEL92.1.7 cells in NSG mice. Treatment with vehicle alone or JQ1 and/or ruxolitinib was initiated seven days after the tail-vein infusion and engraftment of HEL92.1.7 cells. JQ1 and ruxolitinib were administered by intraperitoneal injection and oral gavage, respectively, daily for 5 days/week for 3 weeks. The Kaplan Meier plot in Figure 4C demonstrates that, as compared to the treatment with vehicle control, JQ1 or ruxolitinib treatment alone, co-treatment with JQ1 and ruxolitinib significantly improved the median survival of the mice (Log Rank Sum test, p= 0.0101).
Figure 4. Co-treatment with JQ1 and JAK inhibitor ruxolitinib synergistically induces apoptosis of cultured sAML cells and improves the survival of mice bearing HEL92.1.7 cell xenografts.
A. HEL92.1.7 were treated with JQ1 (dose range: 200-1000 nM) and ruxolitinib (dose range: 200-1000 nM) at a constant ratio for 48 hours. Then, the % of annexin V- and TO-PRO-3-iodide positive, apoptotic cells was determined by flow cytometry. Median dose effect and isobologram analyses were performed utilizing CompuSyn, assuming mutual exclusivity. Combination index (CI) values less than 1.0 indicate a synergistic interaction of the two agents in the combination. B. HEL92.1.7 cells were treated with the indicated concentrations of JQ1 and/or ruxolitinib for 24 hours. Following this, the cells were harvested and total cell lysates were prepared. Immunoblot analyses were conducted as indicated. The expression levels of β-actin in the lysates served as the loading control. C. HEL92.1.7 cells were injected into the lateral tail vein of NSG mice (n=8) that had been pre-conditioned with 2.5 Gy of gamma irradiation. Seven days post cell implantation, mice were treated with 50 mg/kg of JQ1 (daily × 5 days, by IP injection) and/or 20 mg/kg ruxolitinib (daily × 5 days, P.O.) for 3 weeks. The survival of the mice is represented by a Kaplan Meier plot.
Effects of BETi and/or JAKi in patient-derived (PD) sAML stem or blast progenitor cells expressing JAK2 V617F mutation
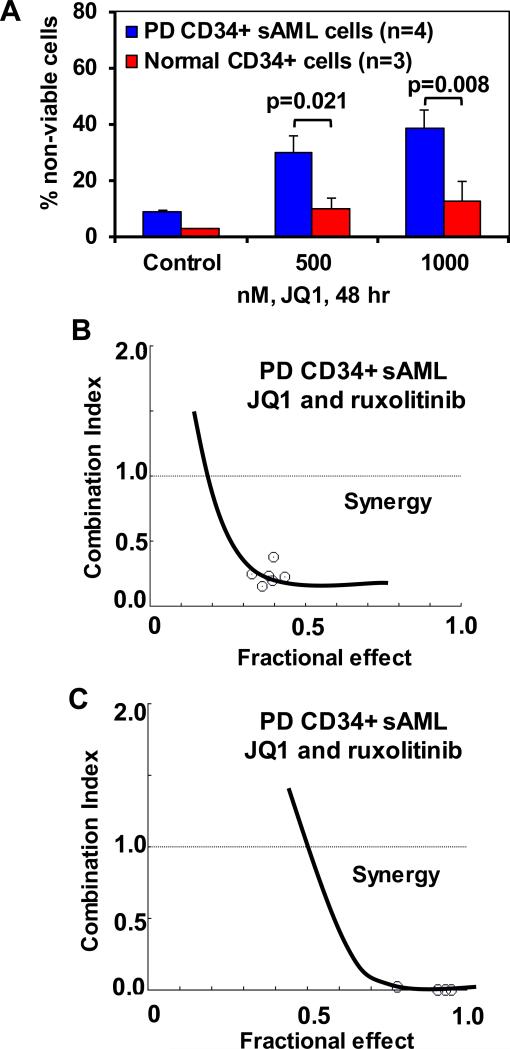
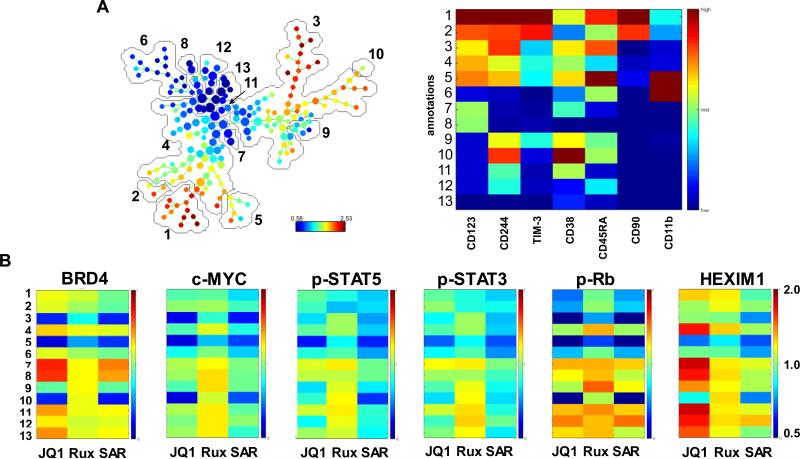
We next compared the lethal effects of JQ1 in PD CD34+ sAML versus normal progenitor cells. As shown in Figure 5A, treatment with JQ1 dose-dependently induced significantly more lethality in four samples of PD CD34+ sAML, as compared to the normal CD34+ progenitor cells (3 samples) (p < 0.01). Utilizing NextGen sequencing, we next determined the genetic alterations in 23 PD sAML samples, including the 4 samples utilized above (sample 2, 4, 6 and 7), as well as 19 additional sAML samples (Supplemental Figure S7). Similar to previous reports, we also identified genetic alterations in JAK2, c-MPL, CALR, KRAS, NRAS, ASXL1, TET2, IDH1 & 2, EZH2, RUNX1 and DNMT3A. In this cohort, only one sample each exhibited mutant TP53 (sample 4) or CALR (sample 8). We next determined the lethal effects of co-treatment with JQ1 and ruxolitinib or pacritinib in PD sAML cells. As shown, co-treatment with ruxolitinib synergistically induced apoptosis in two representative samples of PD CD34+ sAML cells (Figure 5B and 5C) and Supplemental Figures S8A and 8B). Co-treatment with JQ1 and pacritinib also synergistically induced apoptosis in the two samples of PD CD34+ sAML cells (Supplemental Figures 8C to 8F). We next determined the effects of JQ1, as well as of ruxolitinib and SAR on the immunophenotypic and intracellular protein expressions in PD CD34+ sAML cells, utilizing a recently described mass cytometry approach38, 39. A bioinformatics algorithm, SPADE (Spanning-tree Progression Analysis of Density-normalized Events), was utilized on mass cytometry data, using 7 cell surface markers, to yield 13 cell groupings or clusters which represent the various immunophenotypic cell subsets (Figure 6A). Among these, clusters 1 and 2 phenotypically represented stem/progenitor cells, based on high expression of CD90 and CD244 (also high CD123+, TIM3Fc+ and CD45RA+ but low expression of CD38+ and CD11b), whereas cluster 6 represented the more differentiated granulocyte-macrophage progenitor (GMP) cells with low expression of CD90 and CD244 (also low CD123+, TIM3Fc+ and CD45RA+ but high expression of CD11b and CD38+ (expressions ranging between 0.58 and 2.53)). Clusters 7 and 8 were phenotypically closer to the stem/progenitor cells but with lower expression of CD123. Next, among the immuno-phenotypically distinct clusters, especially clusters 1 and 2 versus cluster 6, we compared the alterations in the intracellular protein expressions of 13 proteins in JQ1-, ruxolitinib- or SAR-treated cells (Figure 6B). The range of expression was between 0.5 and 2.0, with the untreated control at 1.0 in the range. Results of this PD sAML sample (sample 4 in Supplemental Figure S6) are shown in Figure 6B and Supplemental Figure 9. They demonstrate that treatment with JQ1 reduced the expression of c-MYC, p-STAT5, p-STAT3, p-Rb, AKT, p-AKT, p-ERK, p-S6, BCL2 and Bcl-xL in clusters 1 and 2 (Figure 6B and Supplemental Figure 9). In contrast, JQ1 treatment increased the expression of HEXIM1 and BRD4 in clusters 7 and 8 more so than in clusters 1 and 2 (Figure 6B). Exposure to ruxolitinib exerted a greater inhibitory effect than JQ1, but less than SAR, on the expression of the signaling proteins in clusters 1 and 2. In contrast, overall, JAKi treatment exerted lesser effects than JQ1 on BRD4 and HEXIM1 in clusters 1, 2, 7 and 8 (Figure 6B).
Figure 5. Co-treatment with JQ1 and JAK2 inhibitors synergistically induces cell death in patient-derived CD34+ sAML cells.
A. Patient-derived (PD) CD34+ sAML cells and normal CD34+ cells were treated with the indicated concentrations of JQ1 for 48 hours. At the end of treatment, the % of PI-positive, non-viable cells was determined by flow cytometry. Columns, mean loss of cell viability; Bars, standard error of the mean. B-C. Patient-derived CD34+ sAML cells (n=2) were treated with JQ1 and ruxolitinib for 48 hours. The % of non-viable cells was determined by propidium iodide (PI) staining and flow cytometry. Median dose effect and isobologram analyses were performed utilizing CompuSyn, assuming mutual exclusivity. Combination index (CI) values less than 1.0 indicate a synergistic interaction of the two agents in the combination.
Figure 6. Treatment with JQ1 or JAK inhibitors ruxolitinib or SAR302503 depletes JAK/STAT signaling and attenuates c-MYC, BCL-2 and Bcl-xL protein expression in primary CD34+ secondary AML cells as determined by mass cytometry analysis.
Primary CD34+ secondary AML cells were treated with JQ1 or ruxolitinib or SAR302503 for 16 hours. At the end of treatment, cells were fixed and labeled with a cocktail of primary antibodies conjugated to rare metal elements. Mass cytometry ‘CyTOF’ analysis was conducted and data were normalized and exported. Data were further analyzed utilizing the SPADE software. A. SPADE tree colored by c-MYC and a heatmap of the 13 annotated clusters on the SPADE tree based on the expression levels of CD markers and cellular markers associated with stem cells or differentiated cells. B. Heatmaps of the protein expression changes caused by treatment with JQ1, ruxolitinib, or SAR302503 relative to the untreated control cells (set to 1) are shown.
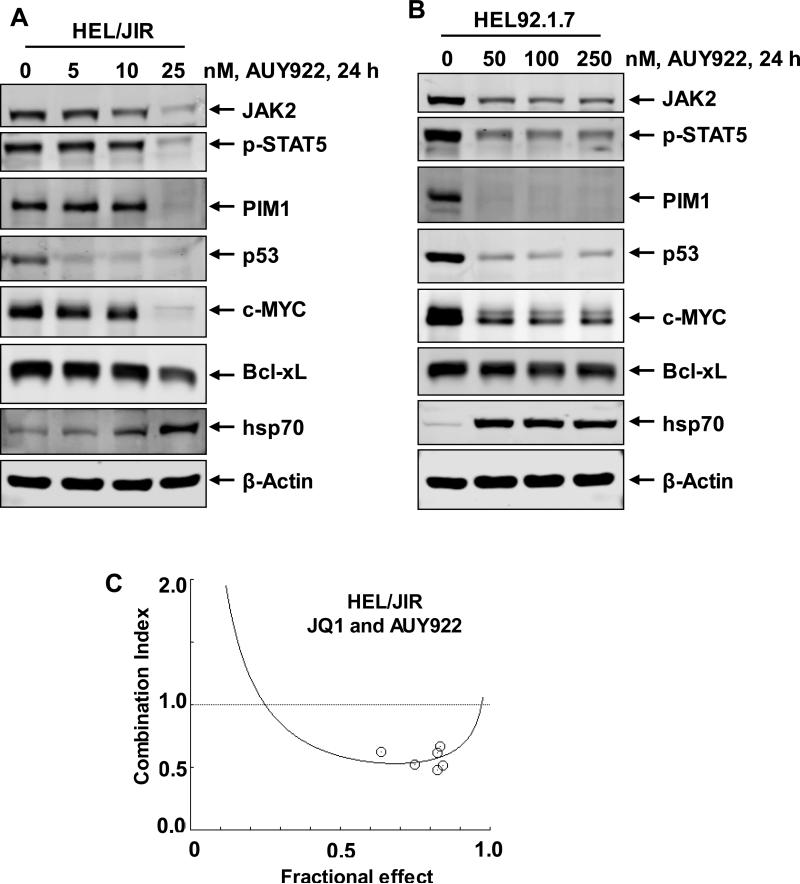
Co-treatment with BETi and HSP90i exerts synergistic lethality against in vitro generated ruxolitinib-persister and ruxolitinib-resistant cells
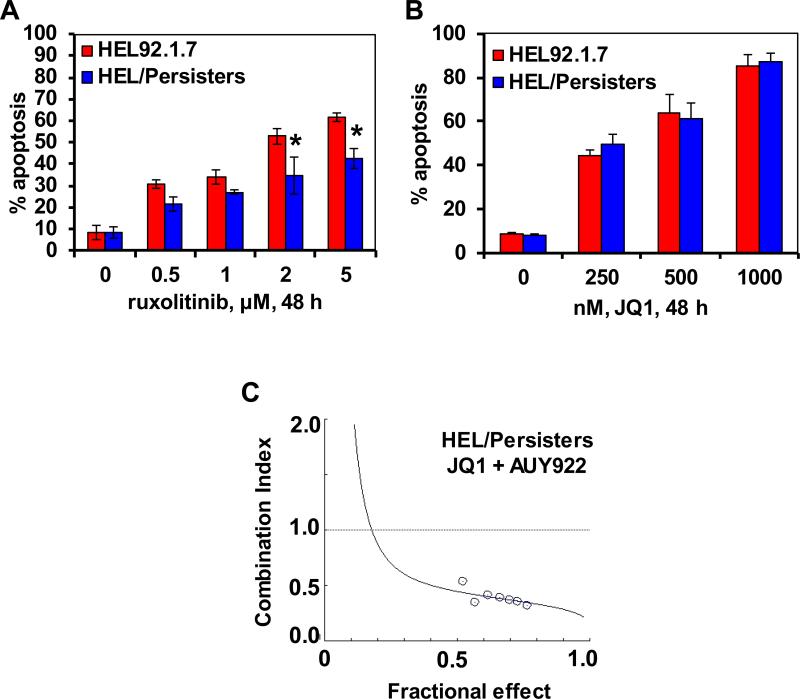
We had previously reported the isolation and characterization of JAKi-resistant (HEL/JIR) cells8. As shown in the Supplemental Figure S10A, unlike the parental HEL92.1.7, HEL/JIR cells are relatively resistant to ruxolitinib-induced apoptosis. It is notable that as compared to HEL92.1.7, UKE1 cells were endogenously less sensitive to ruxolitinib-induced apoptosis (Supplemental Figure 10B). In contrast, HEL/JIR cells are significantly more sensitive to apoptosis induced by HSP90i, e.g., AUY922 (p < 0.01) (Supplemental Figure S8C)40. Exposure to 20 nM of AUY922 for 48 hours induced apoptosis in a mean of 75.2% of HEL/JIR versus 27.7% of HEL92.1.7 cells. Consistent with this, exposure to 20 nM of AUY922 markedly reduced the protein levels of JAK2, PIM1, p-STAT5, c-MYC, Bcl-xL and p53, while simultaneously inducing the levels of HSP70, in HEL/JIR cells (Figure 7A). In contrast, markedly higher concentrations of AUY922 were necessary to achieve similar effects in HEL92.1.7 cells (Figure 7B, and data not shown). Notably, co-treatment with JQ1 and AUY922 was synergistically lethal against HEL/JIR cells (CI of < 1.0) (Figure 7C and Supplemental Figure S10D). Concomitantly, combined treatment with JQ1 and AUY922 led to greater attenuation of PIM1, c-MYC, CDK4, CDK6 and Bcl-xL in HEL/JIR cells (Supplemental Figure S10E). Synergistically lethal effects of this combination were also observed in the ruxolitinib-sensitive HEL92.1.7 and SET2 cells (data not shown). As previously reported for EGFRi-resistant-persisters9, we also developed ex vivo ruxolitinib-persisters, following repeated weekly exposure (1.0 μM for 48 hours) and recovery of the surviving HEL92.1.7 cells. Figure 8A demonstrates that, as compared to the parental HEL92.1.7, HEL/Persister cells are significantly less sensitive to ruxolitinib-induced apoptosis (p < 0.05). This was associated with less attenuation in the levels of p-STAT5 and Bcl-xL levels in HEL/Persister versus HEL92.1.7 cells (Supplemental Figure S11A). However, HEL/Persister cells are as sensitive to JQ1-induced apoptosis as HEL92.1.7 cells (Figure 8B). Notably, treatment with AUY922 induced significantly more apoptosis in HEL/Persister than in HEL92.1.7 cells (Supplemental Figure 11B). This correlated with greater AUY922-mediated attenuation of PIM1, c-MYC and Bcl-xL levels in HEL/Persister versus HEL92.1.7 cells (Supplemental Figure S11A). Furthermore, co-treatment with JQ1 and AUY922 was also synergistically lethal against HEL/Persister cells, with combination indices below 1.0) (Figure 8C and Supplemental Figure S11C). These findings highlight that treatment with an HSP90i is more effective, and combination of BETi and HSP90i exerts superior activity, against JAKi/Persister cells as well as JIR cells.
Figure 7. Co-treatment with JQ1 and AUY922 induces synergistic apoptosis of sAML cells.
A-B. HEL/JIR and HEL92.1.7 cells were treated with the indicated concentrations of AUY922 for 24 hours. At the end of treatment, total cell lysates were prepared and immunoblot analyses were conducted. The expression levels of β-actin in the lysates served as the loading control. C. HEL/JIR cells were treated with JQ1 and AUY922 at a constant ratio for 48 hours. Then, the % of annexin V- and TO-PRO-3 iodide-positive, apoptotic cells was determined by flow cytometry. Median dose effect and isobologram analyses were performed utilizing CompuSyn, assuming mutual exclusivity. Combination index (CI) values less than 1.0 indicate a synergistic interaction of the two agents in the combination.
Figure 8. Co-treatment with JQ1 and AUY922 induces synergistic apoptosis of ruxolitinib persistent sAML cells.
A. HEL92.1.7 and HEL cells made persistent to ruxolitinib (HEL/Persisters) were treated with the indicated concentrations of ruxolitinib for 48 hours. Then, the % of annexin V- and TO-PRO-3-positive, apoptotic cells was determined by flow cytometry. Columns represent the mean of three experiments; Bars represent the standard error of the mean. * indicates apoptosis values significantly less in HEL/Persister cells than in parental cells (p<0.05). B. HEL92.1.7 and HEL/Persisters were treated with the indicated concentrations of JQ1 for 48 hours. Following this, the % of annexin V-positive, apoptotic cells was determined by flow cytometry. Columns represent the mean of three experiments; Bars represent the standard error of the mean. C. HEL/Persister cells were treated with JQ1 and AUY922 at a constant ratio for 24 hours. Then, the % of annexin V-positive apoptotic cells was determined by flow cytometry. The median dose effect and combination indices (isobologram) were determined utilizing Compusyn software assuming mutual exclusivity. Combination index (CI) values less than 1.0 indicate a synergistic interaction of the two agents in the combination.
DISCUSSION
Similar to their documented activity against de novo AML blast progenitor cells expressing MLL fusion proteins, mutant NPM1 and FLT3-ITD29,30,32, present studies demonstrate that treatment with BETi also exert lethal activity against cultured (HEL92.1.7 and SET2) and PD sAML progenitor cells, including those expressing JAK2-V617F, KRAS, TET2, ASXL1, IDH2, RUNX1, DNMT3A, EZH2 and/or TP53. Notably, as compared to the normal CD34+ progenitor cells, the lethal effects of JQ1 were significantly more pronounced against PD CD34+ sAML cells. The cytotoxic effects of BETi treatment were associated with attenuation of the mRNA and protein levels of important oncoproteins in sAML cells, including c-MYC, BCL2, CDK6 and PIM1. The serine–threonine kinase PIM1 is involved in phosphorylating and affecting the levels and/or activity of its target proteins, which include c-MYC, p21, p27, BAD and PRAS4041. Therefore, JQ1-mediated reduction in the levels of PIM1, along with attenuation of c-MYC and CDK4, could explain the inhibitory effects of JQ1 on the cell-growth, differentiation and cell-viability of sAML cells. Also, consistent with our previously reported findings, JQ1-mediated induction of p21 and HEXIM1 also likely contributed here to the inhibition of growth and induction of differentiation in sAML cells33. Mutations in JAK2, c-MPL or CALR constitutively activate JAK-STAT signaling, as well as promote survival and proliferation of MPN and sAML progenitor cells2,11,19. Additionally, signaling downstream of IL-7R may further accentuate JAK-STAT signaling in sAML progenitor cells42. Therefore, JQ1-mediated attenuation of the levels of IL-7R, JAK2 and p-STAT5, as well as of the STAT5 transcriptional target Bcl-xL, demonstrated by the RPPA findings presented here, further likely contributed to the anti-sAML activity of JQ1. RPPA findings also demonstrated that treatment with JQ1 attenuated the NOTCH pathway transcriptional regulator HES1, whose activity inhibits differentiation and promotes the stemness in the transformed progenitor cells43. Utilizing mass cytometry coupled to SPADE algorithm clustering of the data, we have also demonstrated for the first time that treatment with JQ1 reduces the expression of c-MYC, BCL-2 and Bcl-xL, as well as several other signaling proteins involved in growth and survival, including AKT, p-AKT, p-ERK, p-S6, p-STAT5, p-STAT3, in the immunophenotypically defined stem/progenitor versus the more differentiated GMP sAML cells. Mass cytometry also demonstrated induction of HEXIM1 by JQ1 treatment in the clusters representing stem/progenitor cells, which would contribute to JQ1-mediated growth inhibitory and apoptotic effects in sAML cells33. We also observed that exposure to SAR > ruxolitinib > JQ1 inhibited the levels of several signaling proteins in the sAML stem/progenitor cells. This could be because SAR has been recognized to exert both JAKi as well as BETi activities44. Recent studies have highlighted that prolonged exposure to JQ1 leads to accumulation of BRD4 levels45. This could limit the effects of JQ1 in attenuating the oncoprotein levels, as well reducing the growth-inhibitory and apoptotic effects of JQ145. A similar increase in BRD4 levels due to JQ1 treatment was also observed here in the stem/progenitor sAML cells. This was seen in clusters 7 and 8 more so than in clusters 1 and 2. This observation highlights that, as compared to BETi, treatment with the hetero-bifunctional PROTACs (proteolysis targeting chimera) may exert superior activity against sAML cells46. A PROTAC has been shown to bind and recruit the E3 ubiquitin ligase cereblon to ubiquitylate and proteasomally degrade BRD4, which is likely to exert a more pronounced inhibitory effect on the levels of the oncoproteins as well as on the growth and survival of sAML cells46.
Although it imparts significant clinical benefit and improves survival in patients with advanced MPN or primary MF, ruxolitinib has modest and short-term clinical efficacy in patients with sAML16,18. This may be due to a lack of efficacy of ruxolitinib against sAML stem/progenitor cells. Findings presented here show that co-treatment with a BETi (JQ1 or OTX-015) and JAKi (ruxolitinib, pacritinib or SAR302503) exerted synergistic lethality against cultured and PD sAML blast progenitor cells. Additionally, as compared to treatment with each agent alone, co-treatment with JQ1 and ruxolitinib induced significantly greater improvement in median survival of the immune-depleted mice engrafted with the sAML HEL92.1.7 cells. The synergistically lethal anti-sAML activity of the combination could potentially be due to greater inhibitory effects on the levels of the pro-growth and pro-survival oncoproteins, including p-STAT5, p-STAT3, c-MYC, BCL-2 and CDK4/6. This was accompanied with greater induction in the levels of p21 and cleaved PARP in sAML cells. A greater reduction in the PIM1 levels due to the combination was also observed in SET2 cells. These results are consistent with previous observations that combined therapy with BETi and TKIs that target STAT5 signaling could cooperatively extinguish the AML-associated deregulated transcriptome, leading to a synergistic lethality against the myeloid leukemia stem/progenitor cells32,47.
In post-MPN MF and sAML, resistance to ruxolitinb has been attributed to persistent JAK-STAT signaling due to transphosphorylation of JAK2 by either JAK1 or TYK210,19. However, this was associated with a collateral sensitivity of the sAML cells to HSP90i therapy8,10,19. Additional mechanisms of therapy resistance in the post-MPN sAML stem/progenitor cells may be due to the co-occurrence of mutations in TP53 and several of the chromatin regulators, coupled to the activity of the BRD4-dependent, super enhancer-driven, oncogenic deregulated transcriptome19,22, 25,48,49. Therefore, disrupting BRD4 activity at the super-enhancers and promoters by BETi represents a promising therapy, which we also harnessed here in the synergistic combination with HSP90i against sAML cells, sensitive or resistant to ruxolitinib. Since BETi also exert single agent activity against AML cells that express mutant TP53, as well as express mutant versions of chromatin regulators, this also supports the rationale to utilize BETi in combination with HSP90i against JAKi-resistant cells and JAKi-Persisters. Also, since TYK2, p-STAT5 and mutant TP53 are chaperoned by HSP90, and treatment with HSP90i has been shown to induce proteasomal degradation of TYK2, p-STAT5 and mutant TP53, this also explains the synergy of the combination against the mutant TP53-expressing JAKi-resistant and JAK-Persisters50-52. Taken together, these observations strongly support the rationale for further evaluation of the combined therapy with BETi and HSP90i against sAML cells. Recent reports have demonstrated that the type-II JAK2i CHZ868 may reverse type-I JAK inhibitor (e.g., ruxolitinib, pacritinib and SAR302503) resistance, demonstrating efficacy against post-MPN cells53. Therefore, in the future, it would also be important to determine the activity of co-treatment with a BETi and type II JAK2i against sAML, resistant to type I JAKi.
Supplementary Material
Acknowledgements
The authors would like to thank the Functional Proteomics RPPA Core facility which is supported by MD Anderson Cancer Center Support Grant 5P30 CA016672-40. The initial heatmaps were developed by the MD Anderson Cancer Center Department of Bioinformatics and Computational Biology, In Silico Solutions, Santeon and SRA International. This work was supported in part by U.S. National Cancer Institute (NCI; MD Anderson TCGA Genome Data Analysis Center) grant numbers CA143883 and CA083639, the Mary K. Chapman Foundation and the Michael & Susan Dell Foundation (honoring Lorraine Dell). Additional support was also provided by CPRIT Metabolomics Core Facility Support Award RP120092 (CC), a pilot grant from Alkek Center for Molecular Discovery (CC). This research is supported in part by the MD Anderson Cancer Center Support Grant (P30 CA016672) and the MD Anderson Cancer Center Leukemia SPORE (P50 CA100632).
Footnotes
Conflict of Interest: Dr. Pemmaraju serves on the scientific advisory board of Incyte Pharmaceuticals. He has also served as a consultant and received research funding from Novartis Pharmaceuticals. All other authors state that they have no conflict of interest to declare.
Supplementary information is available at Leukemia's website.
REFERENCES
- 1.Vainchenker W, Delhommeau F, Constantinescu SN, Bernard OA. New mutations and pathogenesis of myeloproliferative neoplasms. Blood. 2011;118:1723–1735. doi: 10.1182/blood-2011-02-292102. [DOI] [PubMed] [Google Scholar]
- 2.Rampal R, Al-Shahrour F, Abdel-Wahab O, Patel JP, Brunel JP, Mermel CH, et al. Integrated genomic analysis illustrates the central role of JAK-STAT pathway activation in myeloproliferative neoplasm pathogenesis. Blood. 2014;123:e123–133. doi: 10.1182/blood-2014-02-554634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keohane C, Mesa R, Harrison C. The role of JAK1/2 inhibitors in the treatment of chronic myeloproliferative neoplasms. Am Soc Clin Oncol Educ Book. 2013:301–5. doi: 10.14694/EdBook_AM.2013.33.301. [DOI] [PubMed] [Google Scholar]
- 4.Vannucchi AM, Kiladjian JJ, Griesshammer M, Masszi T, Durrant S, Passamonti F, et al. Ruxolitinib versus standard therapy for the treatment of polycythemia vera. N Engl J Med. 2015;372:426–435. doi: 10.1056/NEJMoa1409002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vannucchi AM, Kantarjian HM, Kiladjian JJ, Gotlib J, Cervantes F, Mesa RA, et al. A pooled analysis of overall survival in COMFORT-I and COMFORT-II, 2 randomized phase III trials of ruxolitinib for the treatment of myelofibrosis. Haematologica. 2015;100:1139–1145. doi: 10.3324/haematol.2014.119545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Passamonti F, Maffioli M, Cervantes F, Vannucchi AM, Morra E, Barbui T, et al. Impact of ruxolitinib on the natural history of primary myelofibrosis: a comparison of the DIPSS and the COMFORT-2 cohorts. Blood. 2014;123:1833–1835. doi: 10.1182/blood-2013-12-544411. [DOI] [PubMed] [Google Scholar]
- 7.Guglielmelli P, Biamonte F, Rotunno G, Artusi V, Artuso L, Bernardis I, et al. Impact of mutational status on outcomes in myelofibrosis patients treated with ruxolitinib in the COMFORT-II study. Blood. 2014;123:2157–2160. doi: 10.1182/blood-2013-11-536557. [DOI] [PubMed] [Google Scholar]
- 8.Fiskus W, Verstovsek S, Manshouri T, Rao R, Balusu R, Venkannagari S, et al. Heat shock protein 90 inhibitor is synergistic with JAK2 inhibitor and overcomes resistance to JAK2-TKI in human myeloproliferative neoplasm cells. Clin Cancer Res. 2011;17:7347–7358. doi: 10.1158/1078-0432.CCR-11-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koppikar P, Bhagwat N, Kilpivaara O, Manshouri T, Adli M, Hricik T, et al. Heterodimeric JAK STAT activation as a mechanism of persistence to JAK2 inhibitor therapy. Nature. 2012;489:155–159. doi: 10.1038/nature11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meyer SC, Levine RL. Molecular pathways: molecular basis for sensitivity and resistance to JAK kinase inhibitors. Clin Cancer Res. 2014;20:2051–2059. doi: 10.1158/1078-0432.CCR-13-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lundberg P, Karow A, Nienhold R, Looser R, Hao-Shen H, Nissen I, et al. Clonal evolution and clinical correlates of somatic mutations in myeloproliferative neoplasms. Blood. 2014;123:2220–2228. doi: 10.1182/blood-2013-11-537167. [DOI] [PubMed] [Google Scholar]
- 13.Hobbs GS, Rampal RK. Clinical and molecular genetic characterization of myelofibrosis. Curr Opin Hematol. 2015;22:177–183. doi: 10.1097/MOH.0000000000000122. [DOI] [PubMed] [Google Scholar]
- 14.Lasho TL, Jimma T, Finke CM, Patnaik M, Hanson CA, Ketterling RP, et al. SRSF2 mutations in primary myelofibrosis: significant clustering with IDH mutations and independent association with inferior overall and leukemia-free survival. Blood. 2012;120:4168–4171. doi: 10.1182/blood-2012-05-429696. [DOI] [PubMed] [Google Scholar]
- 15.Zhang SJ, Rampal R, Manshouri T, Patel J, Mensah N, Kayserian A, et al. Genetic analysis of patients with leukemic transformation of myeloproliferative neoplasms shows recurrent SRSF2 mutations that are associated with adverse outcome. Blood. 2012;119:4480–4485. doi: 10.1182/blood-2011-11-390252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rampal R, Mascarenhas J. Pathogenesis and management of acute myeloid leukemia that has evolved from a myeloproliferative neoplasm. Curr Opin Hematol. 2014;21:65–71. doi: 10.1097/MOH.0000000000000017. [DOI] [PubMed] [Google Scholar]
- 17.Eghtedar A, Verstovsek S, Estrov Z, Burger J, Cortes J, Bivins C, et al. Phase 2 study of the JAK kinase inhibitor ruxolitinib in patients with refractory leukemias, including postmyeloproliferative neoplasm acute myeloid leukemia. Blood. 2012;119:4614–4618. doi: 10.1182/blood-2011-12-400051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kundranda MN, Tibes R, Mesa RA. Transformation of a chronic myeloproliferative neoplasm to acute myelogenous leukemia: does anything work? Curr Hematol Malig Rep. 2012;7:78–86. doi: 10.1007/s11899-011-0107-9. [DOI] [PubMed] [Google Scholar]
- 19.Rampal R, Ahn J, Abdel-Wahab O, Nahas M, Wang K, Lipson D, et al. Genomic and functional analysis of leukemic transformation of myeloproliferative neoplasms. Proc Natl Acad Sci. 2014;111:E5401–5410. doi: 10.1073/pnas.1407792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milosevic JD, Puda A, Malcovati L, Berg T, Hofbauer M, Stukalov A, et al. Clinical significance of genetic aberrations in secondary acute myeloid leukemia. Am J Hematol. 2012;87:1010–1016. doi: 10.1002/ajh.23309. [DOI] [PubMed] [Google Scholar]
- 21.Shi J, Vakoc CR. The mechanism behind the therapeutic activity of BET bromodomain inhibition. Mol Cell. 2014;54:728–36. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roe JS, Mercan F, Rivera K, Pappin DJ, Vakoc CR. BET Bromodomain inhibition suppresses the function of hematopoietic transcription factors in acute myeloid leukemia. Mol Cell. 2015;58:1028–39. doi: 10.1016/j.molcel.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Itzen F, Greifenberg AK, Bosken CA, Geyer M. Brd4 activates P-TEFB for RNA polymerase II CTD phosphorylation. Nucleic Acids Res. 2014;42:7577–90. doi: 10.1093/nar/gku449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nechaev S, Adelman K. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta. 2011;1809:34–45. doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–34. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolden JE, Tasdemir N, Dow LE, van Es JH, Wilkinson JE, Zhao Z, et al. Inducible in vivo silencing of Brd4 identifies potential toxicities of sustained BET protein inhibition. Cell Rep. 2014;8:1919–1929. doi: 10.1016/j.celrep.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Filippakapoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov. 2014;13:337–56. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- 29.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–33. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fiskus W, Sharma S, Qi J, Valenta JA, Schaub LJ, Shah B, et al. Highly active combination of BRD4 antagonist and histone deacetylase inhibitor against human acute myelogenous leukemia cells. Mol Cancer Ther. 2014;13:1142–1154. doi: 10.1158/1535-7163.MCT-13-0770. [DOI] [PubMed] [Google Scholar]
- 31.Dawson MA, Gudgin EJ, Horton SJ, Giotopoulos G, Meduri E, Robson S, et al. Recurrent mutations, including NPM1c, activate a BRD4-dependent core transcriptional program in acute myeloid leukemia. Leukemia. 2014;28:311–20. doi: 10.1038/leu.2013.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fiskus W, Sharma S, Qi J, Shah B, Devaraj SG, Leveque C, et al. BET protein antagonist JQ1 is synergistically lethal with FLT3 tyrosine kinase inhibitor (TKI) and overcomes resistance to FLT3-TKI in AML cells expressing FLT-ITD. Mol Cancer Ther. 2014;13:2315–2327. doi: 10.1158/1535-7163.MCT-14-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Devaraj SG, Fiskus W, Shah B, Qi J, Sun B, Iyer SP, et al. HEXIM1 induction is mechanistically involved in mediating anti-AML activity of BET protein bromodomain antagonist. Leukemia. 2016;30:504–508. doi: 10.1038/leu.2015.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sonkin D, Palmer M, Rong X, Horrigan K, Regnier CH, Fanton C, et al. The identification and characterization of a STAT5 gene signature in hematologic malignancies. Cancer Biomark. 2015;15:79–87. doi: 10.3233/CBM-140434. [DOI] [PubMed] [Google Scholar]
- 35.Ott CJ, Kopp N, Bird L, Paranal RM, Qi J, Bowman T, et al. BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia. Blood. 2012;120:2843–2852. doi: 10.1182/blood-2012-02-413021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun B, Shah B, Fiskus W, Qi J, Rajapakshe K, Coarfa C, et al. Synergistic activity of BET protein bromodomain antagonist-based combinations against Mantle Cell Lymphoma (MCL) cells sensitive or resistant to ibrutinib. Blood. 2015;126:1565–74. doi: 10.1182/blood-2015-04-639542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hart S, Goh KC, Novotny-Diermayr V, Hu CY, Hentze H, Tan YC, et al. SB1518, a novel macrocyclic pyrimidine-based JAK2 inhibitor for the treatment of myeloid and lymphoid malignancies. Leukemia. 2011;25:1751–9. doi: 10.1038/leu.2011.148. [DOI] [PubMed] [Google Scholar]
- 38.Han L, Qiu P, Zeng Z, Jorgensen JL, Mak DH, Burks JK, et al. Single-cell mass cytometry reveals intracellular survival/proliferative signaling in FLT3-ITD-mutated AML stem/progenitor cells. Cytometry. 2015;87:346–56. doi: 10.1002/cyto.a.22628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Behbehani GK, Samusik N, Bjornson ZB, Fantl WJ, Medeiros BC, Nolan GP. Mass Cytometric Functional Profiling of Acute Myeloid Leukemia Defines Cell-Cycle and Immunophenotypic Properties That Correlate with Known Responses to Therapy. Cancer Discov. 2015;5:988–1003. doi: 10.1158/2159-8290.CD-15-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sessa C, Shapiro GI, Bhalla KN, Britten C, Jacks KS, Mita M, et al. First-in-human phase I dose-escalation study of the HSP90 inhibitor AUY922 in patients with advanced solid tumors. Clin Cancer Res. 2013;19:3671–80. doi: 10.1158/1078-0432.CCR-12-3404. [DOI] [PubMed] [Google Scholar]
- 41.Blanco-Aparicio C, Carnero A. Pim kinases in cancer: diagnostic, prognostic and treatment opportunities. Biochem Pharmacol. 2013;85:629–43. doi: 10.1016/j.bcp.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 42.Ribeiro D, Melão A, Barata JT. IL-7R-mediated signaling in T-cell acute lymphoblastic leukemia. Adv Biol Regul. 2013;53:211–22. doi: 10.1016/j.jbior.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 43.Sang L, Roberts JM, Coller HA. Hijacking HES1: how tumors co-opt the anti-differentiation strategies of quiescent cells. Trends Mol Med. 2010;16:17–26. doi: 10.1016/j.molmed.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ciceri P, Müller S, O'Mahony A, Fedorov O, Filippakopoulos P, Hunt JP, et al. Dual kinase bromodomain inhibitors for rationally designed polypharmacology. Nat Chem Biol. 2014;10:305–12. doi: 10.1038/nchembio.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu J, Qian Y, Altieri M, Dong H, Wang J, Raina K, et al. Hijacking the E3 Ubiquitin Ligase Cereblon to Efficiently Target BRD4. Chem Biol. 2015;22:755–63. doi: 10.1016/j.chembiol.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toure M, Crews CM. Small-molecule PROTACS: New approaches to protein degradation. Agnew Chem Int Ed Engl. 2016;55:1966–73. doi: 10.1002/anie.201507978. [DOI] [PubMed] [Google Scholar]
- 47.Liu S, Walker SR, Nelson EA, Cerulli R, Xiang M, Toniolo PA, et al. Targeting STAT5 in hematologic malignancies through inhibition of the bromodomain and extra-terminal (BET) bromodomain protein BRD2. Mol Cancer Ther. 2014;13:1194–205. doi: 10.1158/1535-7163.MCT-13-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12:599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- 49.Hnisz D, Schuijers J, Lin CY, Weintraub AS, Abraham BJ, Lee TI, et al. Convergence of Developmental and Oncogenic Signaling Pathways at Transcriptional Super-Enhancers. Mol Cell. 2015;58:362–70. doi: 10.1016/j.molcel.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taldone T, Ochiana SO, Patel PD, Chiosis G. Selective targeting of the stress chaperome as a therapeutic strategy. Trends Pharmacol Sci. 2014;35:592–603. doi: 10.1016/j.tips.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zong H, Gozman A, Caldas-Lopes E, Taldone T, Sturgill E, Brennan S, et al. A Hyperactive Signalosome in Acute Myeloid Leukemia Drives Addiction to a Tumor-Specific Hsp90 Species. Cell Rep. 2015;13:2159–73. doi: 10.1016/j.celrep.2015.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Akahane K, Sanda T, Mansour MR, Radimerski T, DeAngelo DJ, Weinstock DM, et al. HSP90 inhibition leads to degradation of the TYK2 kinase and apoptotic cell death in T-cell acute lymphoblastic leukemia. Leukemia. 2016;30:219–228. doi: 10.1038/leu.2015.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meyer SC, Keller MD, Chiu S, Koppikar P, Guryanova OA, Rapaport F, et al. CHZ868, a Type II JAK2 Inhibitor, Reverses Type I JAK Inhibitor Persistence and Demonstrates Efficacy in Myeloproliferative Neoplasms. Cancer Cell. 2015;28:15–28. doi: 10.1016/j.ccell.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.