Abstract
Recurrent selection (RS) has been used in plant breeding to successively improve synthetic and other multiparental populations. Synthetics are generated from a limited number of parents but little is known about how affects genomic selection (GS) in RS, especially the persistency of prediction accuracy () and genetic gain. Synthetics were simulated by intermating = 2–32 parent lines from an ancestral population with short- or long-range linkage disequilibrium () and subjected to multiple cycles of GS. We determined and genetic gain across 30 cycles for different training set (TS) sizes, marker densities, and generations of recombination before model training. Contributions to and genetic gain from pedigree relationships, as well as from cosegregation and between QTL and markers, were analyzed via four scenarios differing in (i) the relatedness between TS and selection candidates and (ii) whether selection was based on markers or pedigree records. Persistency of was high for small where predominantly cosegregation contributed to , but also for large where replaced cosegregation as the dominant information source. Together with increasing genetic variance, this compensation resulted in relatively constant long- and short-term genetic gain for increasing > 4, given long-range LDA in the ancestral population. Although our scenarios suggest that information from pedigree relationships contributed to for only very few generations in GS, we expect a longer contribution than in pedigree BLUP, because capturing Mendelian sampling by markers reduces selective pressure on pedigree relationships. Larger TS size () and higher marker density improved persistency of and hence genetic gain, but additional recombinations could not increase genetic gain.
Keywords: genomic prediction, recurrent selection, synthetic populations, prediction accuracy, genetic gain, GenPred, Shared Data Resources, Genomic Selection
RS is an integral tool in plant breeding that targets the systematic improvement of quantitative traits in broad-based populations by increasing the frequency of favorable alleles, while maintaining genetic variability (Hallauer and Carena 2012). Source materials in allogamous crops include open-pollinated and synthetic populations (synthetics, Hallauer 1992). Synthetics are created by intermating a limited number of parental components and cross-pollinating the progeny for one or several generations (Falconer and Mackay 1996). A prominent example is the Iowa Stiff Stalk Synthetic (BSSS), which was developed from 16 inbred lines in the 1930s and has since been subjected to two long-term RS programs (Hallauer 2008), which have contributed a large proportion of today’s commercial maize germplasm (Mikel and Dudley 2006).
GS is a novel statistical method (Meuwissen et al. 2001) with the capability to accelerate future genetic progress in plant breeding (Heffner et al. 2010). Several studies indicate a potential superiority of GS over phenotypic selection (Bernardo 2009; Wong and Bernardo 2009; Jannink 2010; Yabe et al. 2013), marker-assisted selection (Bernardo and Yu 2007; Wong and Bernardo 2009; Heffner et al. 2010, Yabe et al. 2013), as well as pedigree-based selection (Muir 2007; Wolc et al. 2011a, 2016; Bastiaansen et al. 2012; Van Grevenhof et al. 2012). Although the usefulness of GS across two selection cycles has empirically been demonstrated in biparental maize families (Massman et al. 2013; Beyene et al. 2015), experimental results on long-term GS are still missing.
GS has further been proposed as a particularly suitable tool for RS in synthetics (Windhausen et al. 2012; Gorjanc et al. 2016). In this context, an established prediction equation could be used repeatedly for multiple cycles of selection without retraining. Combined with the use of off-season nurseries, this promises to increase genetic gain per unit time and to reduce costs for phenotyping (Bernardo and Yu 2007). The success of this strategy largely depends on persistency of the of estimated breeding values (EBV) across selection cycles to ensure satisfactory genetic gain when selection candidates are separated by one or more cycles from the model training generation. Although formulas for forecasting in a single cycle were derived (Daetwyler et al. 2008; Hayes et al. 2009; Goddard 2009; Goddard et al. 2011), no closed analytical solutions are available for calculating the additive genetic variance () and the cumulative genetic gain () across several selection cycles. This is because changes in the LD pattern, allele frequencies, and loss of polymorphisms are unpredictable (Jannink 2010).
While empirical results on persistency of in actual plant breeding programs are scarce to date, several simulation studies across multiple generations investigated of GS, assuming random mating of the whole population between generations (Meuwissen et al. 2001; Habier et al. 2007; Nielsen et al. 2009; Solberg et al. 2009). Others assumed selection and were therefore able to evaluate potential genetic gain using GS (Muir 2007; Sonesson and Meuwissen 2009; Jannink 2010; Bastiaansen et al. 2012; Yabe et al. 2013, 2016; Liu et al. 2015). However, these studies generally considered fairly large effective population sizes which are unrealistic for synthetics in plant breeding. In synthetics, the number of parents is usually relatively small and parents are often related, leading to small of the population. It is yet unclear how such a small influences the persistency of in genomic RS.
Initially, LD between QTL and molecular markers (commonly SNPs) of high density maps was considered as the only source of information exploited in GS (Meuwissen et al. 2001). In synthetics, LD between QTL and SNPs is attributable to (i) in the population from which the parents were taken, and (ii) sample LD, randomly generated by using a restricted number of parents (Schopp et al. 2017). Sample LD is conserved from parents to progeny between cosegregating loci, and has therefore been termed cosegregation. However, it was also demonstrated that SNPs contribute to by capturing pedigree relationships between individuals (Habier et al. 2007). Research in a companion paper (Schopp et al. 2017) showed that the choice of in synthetics crucially affects the relative importance of and cosegregation as well as the contribution of pedigree relationships in a single cycle of GS in synthetics. However, no study systematically investigated the importance of these information sources for the persistency of and in recurrent GS.
Besides the choice of an important question is how often the source material should be recombined before starting RS. Additional recombination might release genetic variability useful for long-term genetic gain (Schnable et al. 1996). For instance, Bernardo (2009) recommended the use of F2 instead of F1 plants in the production of maize doubled haploids. However, additional recombination might also adversely affect the three information sources in GS, and so far studies have not addressed whether this can outweigh the potential increase in long-term genetic gain.
In the present study, we applied fully stochastic forward-in-time simulations and generated two ancestral populations differing substantially in From these, we sampled different numbers of parents to create synthetics that were subjected to multiple cycles of recurrent GS, either directly or after additional generations of recombination. Our objectives were to (i) analyze and in recurrent GS, depending on the number of parents and the number of recombination generations and (ii) determine the importance of the three information sources, considering also and SNP density. Finally, we discuss implications for practical decisions in breeding programs employing recurrent GS.
Methods
Genome properties and simulation of ancestral populations
Properties of the genome, construction of the genetic map, and simulation of ancestral populations are detailed in Schopp et al. (2017). In brief, we selected maize (Zea mays L.) as a model species using genetic map positions for 37,286 SNPs distributed over 10 chromosomes with 1913 in total. Using the software QMSim (Sargolzaei and Schenkel 2009), we simulated two ancestral populations with either short-range LDA (SR) or extensive long-range LDA (LR). First, we generated an initial population of 1500 diploid individuals by sampling alleles at each (biallelic) locus independently from a Bernoulli distribution with probability 0.5. Second, 5000 loci were randomly sampled from all SNPs and henceforth interpreted as QTL; all remaining loci were considered as SNP markers. Third, these individuals were randomly mated for 3000 generations with a constant population size of 1500 and a mutation rate of until mutation-drift-equilibrium was reached. Fourth, a strong population bottleneck was imposed by reducing the population size to 30 arbitrarily selected individuals, followed by 15 additional generations of random mating to generate extensive long-range LDA. Lastly, the population was expanded to individuals and randomly mated three times more to establish ancestral population LR. Ancestral population SR was derived from LR by continuing random mating for 100 generations with constant population size of to break down long-range LDA. Due to this large population size, genetic drift had only a negligible influence and hence allele frequencies were nearly identical in both ancestral populations. The heterozygous ancestral populations (LR and SR) were considered as unrelated and were used as reference bases for the pedigree of all subsequently derived individuals.
Simulation of synthetic populations
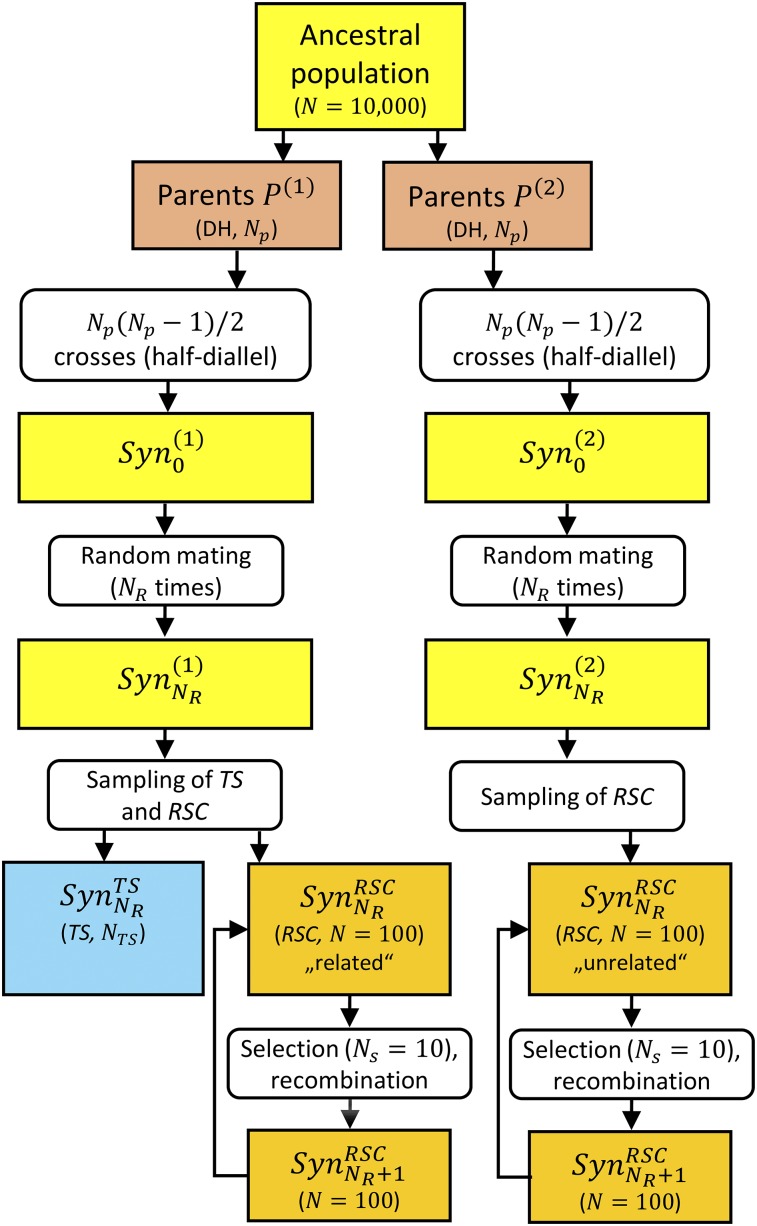
The RS breeding scheme applied is shown in Figure 1 and factors analyzed are listed in Table 1. The simulation of the synthetics varied, depending on whether the parents of the TS and the recurrent selection candidates (RSC) were identical () or disjoint For a single synthetic was simulated from which both the TS and the RSC were sampled, whereas for TS and RSC were taken from two synthetics having no parents in common. In both cases, parental gametes were randomly drawn from the same ancestral population and chromosomes were doubled in silico to generate fully homozygous parent lines. These were intermated to obtain all possible single crosses, denoted as generation Subsequently, single crosses were randomly mated times (allowing for selfings) to obtain generation from which the TS () and RSC () were later drawn. Here, counts the number of recombination generations conducted prior to initiating RS. For the special case of the corresponded to a F1 cross and to a F2 family.
Figure 1.
Schematic representation of the breeding program applied in this study. Two synthetic populations and were separately created by using recombination generations from parental gametes drawn from one ancestral population [with short- (SR) or long-range linkage disequilibrium (LR)]. If the training set (TS) and the recurrent selection candidates (RSC) were related, TS and RSC were sampled from the same synthetic and if they were unrelated, they were drawn from separate synthetics and In each cycle of recurrent selection, individuals were selected and recombined to establish the next generation.
Table 1. Overview of the factors analyzed in our simulation study.
| Factors | Levels | |
|---|---|---|
| Primary factors | ||
| Ancestral population | SR, LR | |
| Information scenario | ||
| Number of parents () | 2, 3, 4, 6, 8, 12, 16, 32 | |
| Secondary factors | ||
| Selection scenario | EBV, TBV, RBV | |
| Number of recombination generations () | 1, 2, 3, 4, 5 | |
| Marker density | 0.125, 2.5 | |
| Training set size () | 250, 1000 | |
For secondary factors, bold face type factor levels indicate the default simulation setting. SR, short-range; LR, long-range; Re, related; LDA, ancestral linkage disequilibrium; SNP, single nucleotide polymorphism; Ped, pedigree; LEA, ancestral linkage equilibrium; Un, unrelated; EBV, estimated breeding values; TBV, true breeding values; RBV, random breeding values.
Genetic model
We assumed a quantitative trait based on 1000 biallelic QTL with purely additive gene action and absence of QTL × year interactions. For each simulation replicate, QTL were randomly sampled from the 37,286 SNPs present in the ancestral population. Following Meuwissen et al. (2001), absolute values of QTL effects were drawn from a gamma distribution with scale and shape parameter of 0.4 and 1.66, respectively. Signs of QTL effects were sampled from a Bernoulli distribution with probability 0.5. Although we assumed biallelic QTL, the alleles of neighboring QTL are strongly correlated due to and linkage, effectively leading to haploblocks that could be considered as higher-level multi-allelic QTL. The true breeding value (TBV) for any individual (either from the synthetics or from the ancestral populations) was computed as where counts the number of minor alleles at the -th QTL centered by the respective ancestral allele frequency in LR, and is the associated QTL effect. Phenotypes were simulated as where is an environmental noise variable. The error variance was assumed to be constant throughout all simulations and was determined as follows: for all individuals in the ancestral population LR, TBVs were calculated according to the above procedure under replicated sampling of 1000 QTL together with their associated effects. The variance of the noise variable was then set equal to the mean additive genetic variance . As the allele frequencies in both ancestral populations were virtually identical, was also the mean additive genetic variance in ancestral population SR. This approach implies that the heritability in ancestral populations LR and SR was, on average, 0.5. Heritability was lower in the synthetics due to the finite sample of parents and, on average, for
Information source scenarios
We employed four distinct scenarios to evaluate the contributions of the three information sources used in Genomic Best Linear Unbiased Prediction (GBLUP) for estimating actual relationships at causal loci by SNPs (cf. Habier et al. 2013). These scenarios can be distinguished by (i) the relatedness of the TS and RSC and (ii) the type of data employed for calculating the relationship matrix used as a kernel in GBLUP (Supplemental Material, Table S1).
Our standard scenario was where the TS and RSC were related () as their parents were identical The kernel in GBLUP was calculated based on SNPs (excluding QTL) and thus contained genomic relationships. As a consequence, this scenario harnesses all three sources of information, namely: (i) pedigree relationships captured by SNPs, (ii) cosegregation between QTL and SNPs by virtue of the parents being identical, and (iii) between QTL and SNPs due to the presence of in the ancestral population, which was carried over to the synthetics. is a realistic scenario and is perhaps the most frequent scenario encountered in applications of GS.
Scenario was artificial and was derived from Here, for each of the 10 chromosomes, the multi-locus genotypes of QTL and SNPs were regarded as separate units and were reshuffled among the parents prior to intermating. This procedure broke up historical associations between QTL and SNPs due to while conserving the LD structure among QTL and among SNPs as well as their allele frequencies. Hence, information from cannot contribute to and any LD between QTL and SNPs is exclusively due to sampling a limited number of parental gametes from the ancestral population, i.e., sample LD.
Scenario was identical to except that the kernel of GBLUP was the numerator relationship matrix calculated from pedigree records of all individuals (pedigree BLUP). This scenario provided a reference for and its dynamics across cycles that can be obtained exclusively from known pedigree relationships between TS and RSC.
In scenario , the TS and RSC were unrelated because their parents were distinct Thus, the influence of pedigree relationships captured by SNPs and cosegregation between QTL and SNPs is eliminated, and the only remaining connection between the TS and RSC is the LD shared due to their common ancestral population, i.e.,
Genomic prediction model
We used GBLUP to predict breeding values according to the model equation
where and are the phenotypic and breeding values, respectively, of the -th individual, is the overall population mean, and the associated model residual. Standard assumptions about the distribution of the random effects were , and stochastic independence of and Variance component estimates for and , as well as predicted breeding values were calculated using the R-package rrBLUP (Endelman 2011). The matrix describes the variance–covariance structure of the breeding values of all individuals ( and ) and was computed based on different types of data, depending on the information scenario. For , and SNP-based genomic relationship coefficients between individuals and were computed following VanRaden (2008) as
where are the genotypic SNP scores and is the frequency at the -th SNP marker in the ancestral populations. In scenario pedigree relationships were computed from the complete pedigree records of all individuals using the R-package pedigree (Coster 2013).
Recurrent genomic selection scheme
The TS was sampled once from synthetic (Figure 1) and thereupon was used to predict breeding values in all of 30 selection cycles. The initial 100 RS candidates were sampled from the remaining individuals of if or from the second synthetic if In each cycle the top individuals were selected (before flowering) either based on (i) EBV calculated by GBLUP or pedigree BLUP (scenario ), (ii) TBV, corresponding to phenotypic selection with or (iii) “random breeding values” (RBV), being chosen at random. While EBV shows the realistic decay of (taking into account that in earlier cycles influences in later cycles), TBV provides an identical and constant selection accuracy of one, independent of for all scenarios. RBV shows the decay of without directional selection, i.e., the decay that is caused by recombination and genetic drift alone. The selected fraction of 10% is realistic for practical applications and has been used in other simulation studies (e.g., Jannink 2010). The selected candidates were subsequently recombined by random mating to create 100 new progeny, serving as in the next selection cycle. The effects of and of SNP density 0.125, 2.5 SNPs per were examined in independent simulations, with default values of and SNPs. For each combination of factors, we conducted 500 independent simulation replicates. Here, one replicate encompasses: (i) sampling of parents from the ancestral population; (ii) sampling of 1000 QTL together with their QTL effects and an appropriate number of SNPs to reach the desired marker density; (iii) creation of the synthetics assuming different numbers of generations of random mating, and sampling of the TS and the initial RSC; (iv) simulation of phenotypes for TS individuals; and (v) conduction of recurrent GS without retraining for selection cycles. All simulations were performed with the R statistical language (R Core Team 2015) and code is provided in File S2.
Cumulative genetic gain, additive genetic variance, and prediction accuracy
In each selection cycle, the cumulative genetic gain () was computed as the average of all 100 TBVs of the RSC relative to the average in . The of the RSC was computed as the variance of values. The was expressed in units of and in units of was calculated as the Pearson correlation coefficient between TBVs and predicted breeding values of the RSC.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Dynamics of genetic gain, prediction accuracy, and additive genetic variance
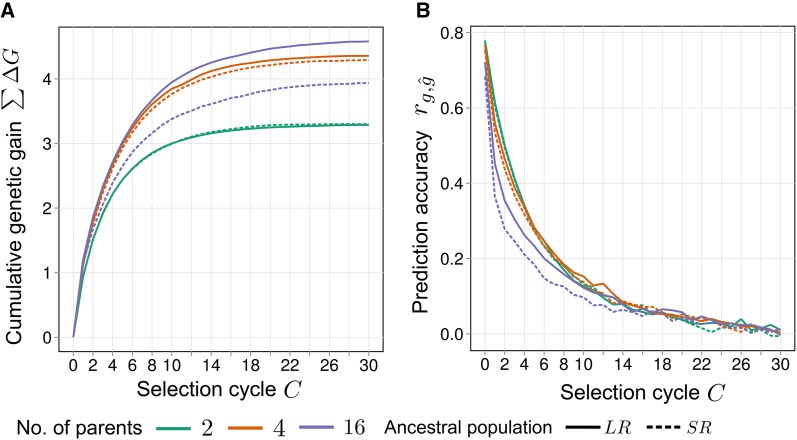
An overview of the dynamics of cumulative genetic gain and prediction accuracy under recurrent GS for the standard scenario is given in Figure 2. Across selection cycles, increased concavely, approaching a plateau. Regardless of the number of parents was higher in LR compared to SR. For LR, increased together with whereas for SR, was lowest for highest for , and intermediate for In the model training generation ranged between 0.7 and 0.8 and was higher for smaller After the first round of selection, there was a substantial decline in that was strongest for large generally approached an asymptotic value of ∼0.1 in cycle The overall level of (Figure S1) in the RSC was higher for larger and strongly declined during selection, especially after the first cycle. In was nearly identical for LR and SR, and showed a slightly steeper decline in LR.
Figure 2.
(A) Average cumulative genetic gain and (B) average prediction accuracy in scenario under recurrent genomic selection across selection cycles for synthetics produced from parents taken from ancestral populations SR or LR. Values of are expressed in units of LDA, ancestral linkage disequilibrium; LR, long-range linkage disequilibrium; Re, related; SNP, single nucleotide polymorphism; SR, short-range linkage disequilibrium.
Cumulative genetic gain
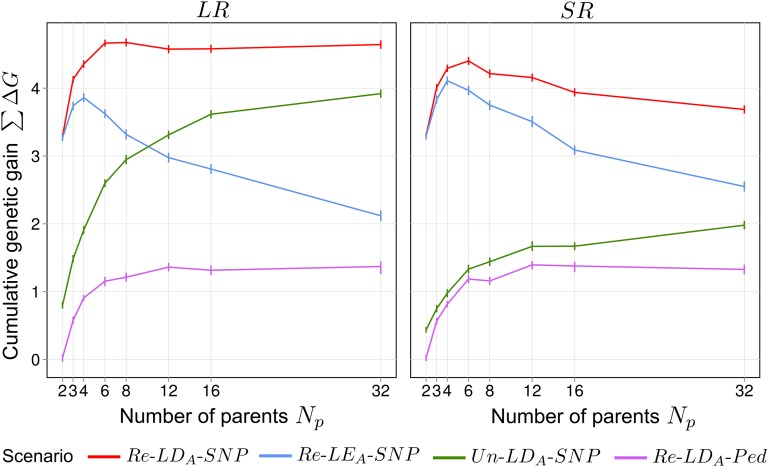
To explore in greater detail in and the information sources primarily exploited, we varied between 2 and 32 (Figure 3). Here, the relationship between and in scenario was strongly affected by the level of For LR, initially increased between and and then remained nearly constant for larger For SR, also increased initially, but then strongly decreased for larger In scenario was much lower than in and monotonically increased with growing This increase and the overall level of was much higher in LR than SR. In scenario was zero for and strongly increased with plateauing at For scenario virtually no further genetic gain could be realized after (Figure S2).
Figure 3.
Average cumulative genetic gain under recurrent genomic selection in selection cycle for synthetics produced from different numbers of parents taken from ancestral populations SR or LR. All values are expressed in units of , mean additive genetic variance; LDA, ancestral linkage disequilibrium; LEA, ; LR, long-range linkage disequilibrium; Ped, pedigree; Re, related; SNP, single nucleotide polymorphism; SR, short-range linkage disequilibrium.
Persistency of prediction accuracy
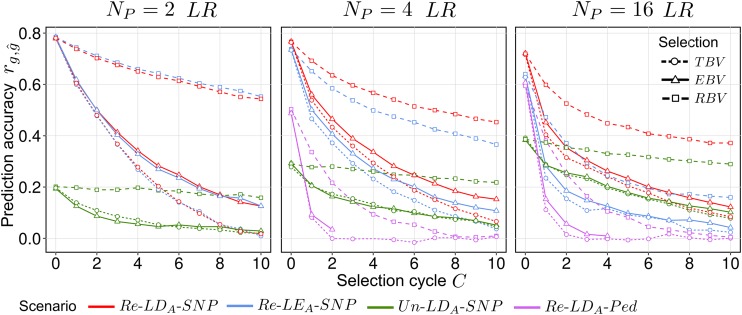
The persistency of for selection regimes EBV, TBV, and RBV under LR is shown in Figure 4. For scenarios and the overall level of declined with growing whereas it increased for scenario (compare Figure S3). In scenario the decay of was strongest in the first selection cycle, especially for large values of In scenario could not be calculated for and as discussed in File S1; for started in at intermediate values of 0.5 for and 0.6 for but declined to zero within a few cycles if the selection was based on either EBV or TBV. With selection based on RBV, approached zero only for Scenarios and showed identical for For , decreased faster in than in and more so with increasing When ancestral long-range LDA was absent (SR), the differences between and were generally much smaller, but otherwise trends were similar (results not shown). Scenario showed an overall low level of especially for SR, where it was close to zero. However, the decline of across cycles was attenuated compared to the other scenarios. When selection was exercised based on TBV, the decay of was similar to selection based on EBV, but much stronger compared with selection based on RBV.
Figure 4.
Average prediction accuracy under recurrent genomic selection across selection cycles for synthetics produced from parents taken from ancestral population LR. Selection of candidates was based on either true breeding values (TBV), random breeding values (RBV), or estimated breeding values (EBV). LDA, ancestral linkage disequilibrium; LEA, ; LR, long-range linkage disequilibrium; Ped, pedigree; Re, related; SNP, single nucleotide polymorphism.
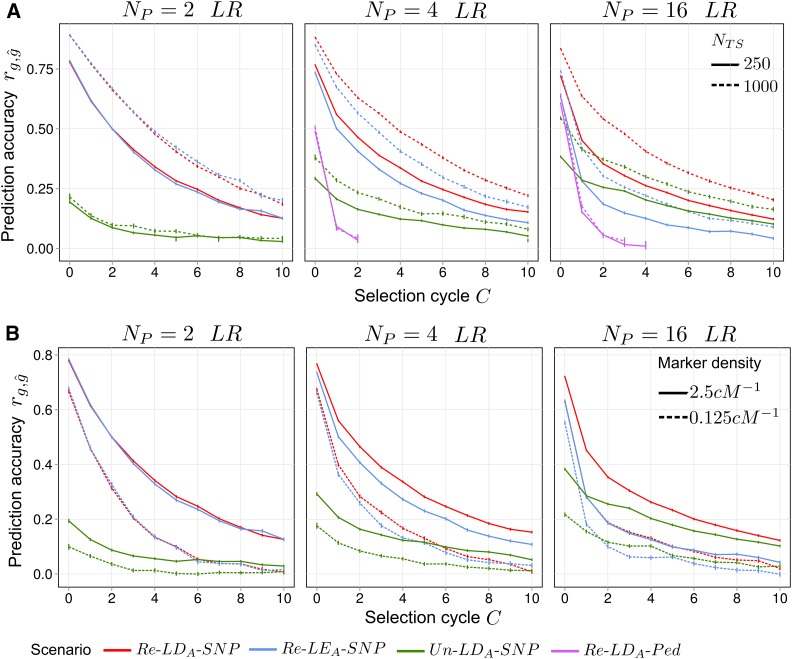
TS size and SNP density
The influence of and SNP density on under selection based on EBV is shown in Figure 5. For all scenarios, increasing elevated the level of across cycles. Specifically, for scenarios assuming increasing reduced the drop in after the first selection cycle, which was not observed for scenario Increasing marker density from 0.125 to 2.5 notably increased the level of for all SNP-based scenarios and led to higher persistency of for SNP-based scenarios with identical parents Scenario did not show an increased persistency with higher marker density.
Figure 5.
Average prediction accuracy under recurrent genomic selection across selection cycles depending on (A) training set size and (B) marker density for synthetics produced from parents taken from ancestral population LR. LDA, ancestral linkage disequilibrium; LEA, ; LR, long-range linkage disequilibrium; Ped, pedigree; Re, related; SNP, single nucleotide polymorphism.
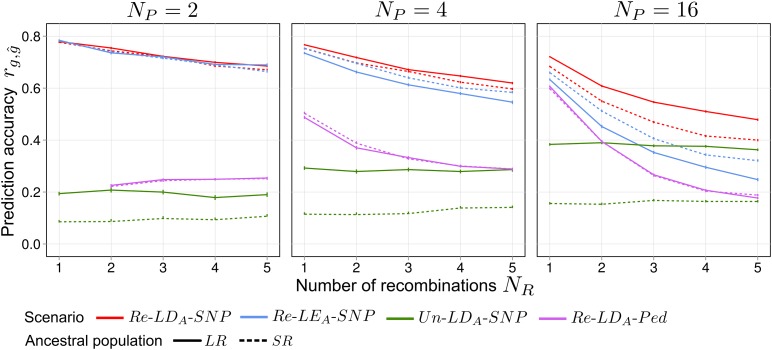
Number of recombinations
In general, increasing the number of recombinations resulted in a decrease of ( Figure 6), except for scenario where stayed nearly constant. Increasing in scenario resulted in the strongest decline in of all scenarios, except if where it remained constant. For scenario increasing from 1 to 5 slightly increased long-term in for selection based on TBV, but not notably for selection based on EBV (Figure 7). The in was not affected by (Figure S4A).
Figure 6.
Average prediction accuracy in selection cycle for different numbers of recombination generations used for production of synthetics from parents taken from ancestral populations SR or LR. LDA, ancestral linkage disequilibrium; LEA, ; LR, long-range linkage disequilibrium; Ped, pedigree; Re, related; SNP, single nucleotide polymorphism; SR, short-range linkage disequilibrium.
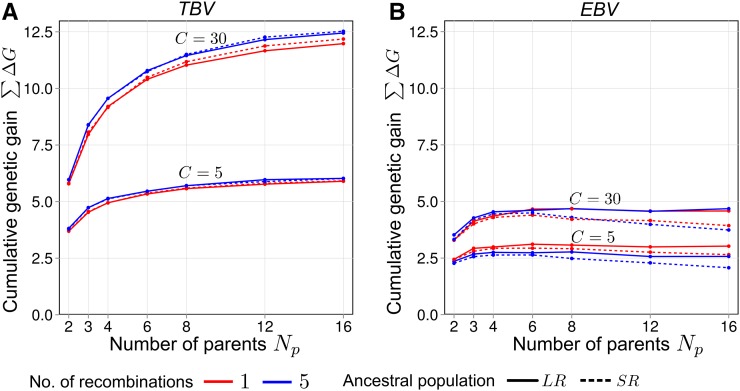
Figure 7.
Average cumulative genetic gain under recurrent genomic selection in selection cycle and for synthetics produced from different numbers of parents taken from ancestral populations SR or LR for and recombination generations. (A) Selection based on true breeding values (TBV), averages across all information scenarios (because values are expected to be identical). (B) Selection based on estimated breeding values (EBV) for scenario All values are expressed in units of , mean additive genetic variance; LDA, ancestral linkage disequilibrium; LR, long-range linkage disequilibrium; Re, related; SNP, single nucleotide polymorphism; SR, short-range linkage disequilibrium.
Discussion
In plant breeding, small effective population sizes that result from a small number of population parents crucially influence the information sources contributing to in a single cycle of GS. For a large number of parents, and pedigree relationships are the driving forces of accuracy, whereas for few parents, cosegregation between QTL and SNPs dominates. While exploitation of information from cosegregation leads to high accuracy, it is unclear how this affects persistency of across selection cycles. Moreover, genetic gain depends on the available genetic variance, which is expected to be reduced for a small number of parents, as opposed to the trend expected for . Although persistency and genetic gain in GS have been previously studied, the important situation of the very small effective population sizes in plant breeding, where cosegregation plays a central role, has not been addressed. Hence, the purpose of the present study was to investigate the contributions of the information sources to persistency of and genetic gain across multiple cycles of recurrent GS in synthetic populations, depending on the number of parents.
Persistency of prediction accuracy across cycles
The persistency of in GS is of crucial importance for practical breeding, because it determines the number of generations that can be employed until retraining of the prediction equation becomes necessary. Thus, it affects the optimum design of a breeding program using recurrent GS and its costs and efficiency compared to phenotypic RS. In agreement with previous studies, we observed a substantial drop in in scenario especially after the first cycle (Figure 4). It was hypothesized that this decline is due to a loss of information from pedigree relationships captured by SNPs (Habier et al. 2007; Wolc et al. 2011b, 2016). In support of this explanation, we observed to plummet after the first cycle in scenario and this can be attributed to two reasons. First, even without directional selection, the variation in pedigree relationships between the TS and RSC erodes as the number of generations between both increases (Figure S5C, selection based on RBV). Second, selection based on pedigree relationships favors the choice of candidates closely related to one another (Quinton et al. 1992; Daetwyler et al. 2007), as verified by the substantial increase in inbreeding and the reduced variation in pedigree relationships (Figure S5, A and C), making the breeding population already genetically narrow after only one selection cycle. This causes EBVs to be more similar to each other and hence, also is severely reduced, although the top pedigree relationships between the TS and RSC individuals increase (Figure S5B). Conversely, selection on TBV (corresponding to phenotypic selection with ) imposes less inbreeding (Figure S5A), because candidates can have equally high breeding values without necessarily being closely related, which results in the selection of clusters of closely related candidates (Figure S8).
The strong drop of in scenario for selection based on EBV might suggest that pedigree relationships only contribute for one or at least very few generations to of scenario However, it has to be taken into account that cosegregation of SNPs and QTL allows capturing of Mendelian sampling (Daetwyler et al. 2007), which reduces the selection pressure on pedigree relationships and in turn increases persistency of in scenario The effect of reduced selection pressure on pedigree relationships can be inferred from scenario under selection based on RBV, where essentially all selection pressure was removed and individuals were selected irrespective of their ancestry. Here, showed a much slower decay compared to selection based on EBV (Figure 4). This suggests that in scenario with selection based on EBV, pedigree relationships probably contribute longer to than indicated by (selection based on EBV).
It was previously shown that information from is highly persistent across generations (Habier et al. 2007). In synthetics, the observed LD largely corresponds to only if is large, which implies that mainly contributes to for large (Schopp et al. 2017). Consistent with these findings, for large (e.g., 16) was the dominant information source across selection cycles, as verified by the strong reduction in when was artificially removed from scenario as in (Figure 4). Conversely, for small , the representation of in the synthetics is hampered by randomly created sample LD when selecting the parents, which raises the question how this influences persistency of for small Our results show that for the persistency of in scenario was even higher than compared with where it decreased more strongly, even though the contribution of was markedly reduced (the drop of in scenario was larger for than ) compared to This implies that sample LD and therefore information from cosegregation behaves similarly to regarding the decay of information across selection cycles. The strong conservation of can be directly assessed from scenario where TS and RSC are unrelated and was the only information source (Figure 4). Here, the decay of was generally small, and if selection was based on RBV it was even diminutive, indicating that recombination between QTL and SNPs only marginally drives ancestral LD structures of the TS and the RSC apart. Even if cosegregation information dominates over in the case of small (e.g., 4), still substantially contributes to especially in later selection cycles (Figure 4, vs. ).
The genomic prediction methodology used can also have a bearing on the exploitation of the sources of information, which was not considered in this study. Previous research indicated that (Bayesian) variable selection methods are better suited to capture information from compared to GBLUP, especially if traits are oligogenic and individual QTL have strong effects (Habier et al. 2007, 2013; Zhong et al. 2009). Therefore, we expect that such methods are advantageous in situations where heavily relies on information from as is the case for large or if TS and RSC are unrelated.
Steady state cumulative genetic gain
In any population advanced by RS, the cumulative increase in overall performance is of central interest to breeders. Here, we continued RS until cycle where further increases in were only marginal because either was depleted (Figure S6) and/or was near zero (Figure 4). This approach allowed for direct comparisons between for different scenarios and conclusions were not contingent on the amount of left.
Increasing leads to an asymptotic increase in the initially available which was independent of the ancestral population in our simulation (Figure S7). According to the breeder’s equation, increasing results in higher genetic gain, which partially explains the increase in for larger However, besides higher differential contributions of the three sources of information to play a major role. In scenario was relatively constant from medium on (Figure 3), which is presumably the result of the counterbalancing effects of a slight increase in and a moderate decrease in with increasing As pointed out by Schopp et al. (2017), increasing from medium to large values decreases the frequency of close relatives between TS and RSC and hence, reduces (Figure S3). The contribution of pedigree relationships to long-term genetic gain in scenario should therefore be relatively constant for medium to large As the contribution of cosegregation to decreases with larger of scenario strongly declined. Conversely, of scenario strongly increased with larger due to more information from Given that there is sufficient present in the ancestral population (LR), both effects largely compensate for each other and hence, in scenario appears to be insensitive to changes in beyond four parents for LR (Figure 3). When there is not sufficient as applies to SR, increasing information due to can no longer compensate for the loss in cosegregation information and therefore in decreased for higher Although we considered close to its steady state, it is important to note that the essential trends in are already apparent for as few as two selection cycles (Figure S2), which implies that our observations do not only apply to the situation of extreme long-term selection without retraining, but also to few selection cycles.
Influence of TS size and SNP density
We found that increasing leads to higher persistency of in early selection cycles for scenarios with pedigree relationship between TS and RSC ( Figure 5). This is because, for a given increasing enhances the probability of obtaining TS individuals that share an exceptionally large portion of their genome with the RSC individuals due to Mendelian sampling and because of similarities between individuals due to Hence, for small there is a higher reliance on information from pedigree relationships (Jannink et al. 2010; Schopp et al. 2017) that quickly erodes under directional selection. For large there is a higher weight on information from cosegregation and which in turn increases the persistency of This shift in emphasis also entails reduced inbreeding, especially in early selection cycles (results not shown), in agreement with the findings of Jannink (2010). Therefore, if a prediction equation is to be used for multiple cycles, should be chosen large enough to not only guarantee high initial but also high persistency of and reduced inbreeding in order to improve genetic gain from GS. Increasing SNP density from 0.125 to 2.5 corresponding to ∼250 and 5000 SNPs in the case of maize, led to an increase in the persistency of (Figure 5), which is in concordance with previous studies (Solberg et al. 2009; Sonesson and Meuwissen 2009). Higher SNP density theoretically affects all three sources of information, but its influence should be strongest on and cosegregation because they rely on physical proximity of SNPs and QTL. If the SNP density is extremely low (e.g., ), it is unlikely that SNPs and QTL are tightly linked and hence, SNPs mainly capture pedigree relationships, whereas and cosegregation play only subordinate roles. Therefore, high SNP density improves persistency of over generations, because information from both (Figure 5, ) and cosegregation (Figure 5, ) are less prone to decay, compared to pedigree relationships. The highest SNP density we investigated was 2.5 which is relatively low compared to what is nowadays available in many plant species. However, because of the strong influence of cosegregation in synthetics that are produced from a low to intermediate number of parents, we would expect that little can be gained by further increasing SNP density, especially if long-range LDA is present, as can be assumed for elite germplasm in practical applications. However, the situation can be quite different for large and if there is only short-range LDA in the ancestral population, which rapidly increases the need for higher SNP densities.
Influence of the number of recombination generations
We hypothesized that larger might lead to enhanced long-term by virtue of a stronger fragmentation of chromosomes in the synthetic. Actually, the average length of chromosomal segments of unique parental origin decreased from ∼66 cM for to 30 cM () and 20 cM () for (Figure S4B). However, as information from pedigree relationships strongly declined with increasing (Figure 6, scenario ), in generally decreased in scenario Conversely, the decline of information contributed by with increasing was negligible (scenario). Decreasing selection accuracy reduces which can conceal the positive effect of higher genome fragmentation. Analysis of the latter factor alone is possible with selection regime TBV, where selection accuracy was always constant and equal to one, regardless of Here, we found higher for compared to (Figure 7) because finer fragmentation promotes occurrence of genotypes with favorable allele combinations for selection. This is accompanied by a reduced coselection of QTL, such that more QTL stay polymorphic and therefore remains higher in advanced selection cycles. The positive effect of on under selection on TBV increased with increasing presumably because larger results in even finer genome fragmentation (Figure S4B). For selection regime EBV, in was not higher for than for suggesting that positive and negative effects of recombination cancelled out each other. For ancestral population SR, was even slightly lower for because compared to LR, stochastic dependency between QTL is relatively low from the beginning and hence, higher fragmentation has only a minor effect. A special situation existed for which is explained in File S1.
It is noteworthy that in our simulations the initial () was unaffected by although strong sample LD between QTL was broken up. In reality, ancestral populations (corresponding to source germplasms in breeding) generally underwent some sort of directional selection, which can theoretically cause a reduction in due to the Bulmer effect (Bulmer 1971; Long et al. 2011). This hidden part of attributable to negative LD between causal loci can be recovered by recombination, which might lead to an increase in for
Implications for practical applications
At the start of any breeding program employing GS with the goal of improving quantitative traits, breeders have to make a number of crucial decisions, including the source germplasm, parents, and mating scheme used to develop the breeding population. Further decisions specific to GS concern the and marker density. All of these factors influence the importance of the three information sources in GS and thereby have ramifications on the success of the breeding program.
The choice of the source germplasm crucially determines the improvement potential for the target trait (Fountain and Hallauer 1996), because it determines the genetic diversity and linkage disequilibrium (i.e., ), which are both of central importance for the success of GS. Our study demonstrates that information from generally offers high persistency across selection cycles in synthetics, irrespective of Hence, is particularly important for ensuring sustained genetic progress during the breeding program. However, the contribution of to genetic gain is itself highly dependent on Whereas for large LD in synthetics adequately represents small generates sample LD and, in turn, cosegregation that dominates LD in synthetics. Cosegregation has a similarly high persistency as but it can only contribute to genetic gain if TS and selection candidates are related by having parents in common. However, it must be taken into account that reducing also reduces the initially available genetic variance for breeding, thereby impairing . In essence, high persistency of and thereby prolonged genetic progress may be achieved irrespective of but if is large, substantial is required.
Pedigree relationships also contribute to predictive information for and harnessing pedigree information has been recommended to achieve high in GS (e.g., Wolc et al. 2011a). Frequent retraining of the prediction equation, at best in every generation, would be required to optimally exploit pedigree relationships because information from them rapidly erodes over generations, especially under directional selection. In addition, selection using pedigree relationships increases the rate of inbreeding due to intraclass correlation of EBV for members of the same family and their coselection (Daetwyler et al. 2007), a result that is well known in animal breeding (Belonsky and Kennedy 1988) and was confirmed in our study for synthetics in plant breeding (Figure S5A). A high rate of inbreeding is undesirable in long-term selection, because genetic diversity is rapidly depleted and eventually is compromised. In GS, it was shown that molecular markers not only capture deviations of genomic relationships from pedigree relationships, but also the pedigree relationships themselves (Habier et al. 2007), i.e., the latent family structure in the case of synthetics. Therefore, the same concerns as for pedigree-based selection partially apply to GS, so that GS is also prone to selection of close relatives and inbreeding (Jannink 2010). If the breeding objective is long-term as classically targeted by RS in genetically broad-based populations (Hallauer and Carena 2012), corresponding to large in our study, deliberate avoidance of using pedigree relationships might be desirable for maximizing long-term
There are different possibilities to reduce the influence of pedigree relationships. Increasing both and marker density leads to an improved capturing of Mendelian sampling and similarities between individuals due to which reduces the reliance on pedigree relationships and in turn reduces inbreeding. Another possibility could be modeling information from cosegregation (Calus et al. 2008; Legarra and Fernando 2009), and pedigree relationships in a joint linear mixed model in an attempt to isolate information from pedigree relationships. Alternatively, one could modify the mating scheme used for generating the synthetic. Additional generations of recombination successfully decreased strong variation in pedigree relationships between individuals, but only up to where a baseline level was reached (Figure S4C). Mating schemes as employed for establishing the Multi-parent Advanced Generation Intercrosses (MAGIC) largely avoid population substructure and pedigree relationships, while they complement the favorable properties of synthetics such as high genetic diversity and elevated minor allele frequencies with a fine-grained mosaic of the genome (compare Dell’Acqua et al. 2015; Holland 2015). Thus, they potentially represent ideal candidates for long-term recurrent GS, but this warrants further research.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.036582/-/DC1.
Acknowledgments
This study was financially supported by the project “Climate Resilient Maize for ASIA (CRMA)” from the International Maize and Wheat Improvement Center, México and the Deutsche Gesellschaft für Internationale Zusammenarbeit, project no. 15.7860.8-001.00 (contract no. 81194991).
Footnotes
Communicating editor: J. B. Holland
Literature Cited
- Bastiaansen J. W. M., Coster A., Calus M. P. L., van Arendonk J. A. M., Bovenhuis H., 2012. Long-term response to genomic selection: effects of estimation method and reference population structure for different genetic architectures. Genet. Sel. Evol. 44: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belonsky G. M., Kennedy B. W., 1988. Selection on individual phenotype and best linear unbiased predictor of breeding value in a closed swine herd. J. Anim. Sci. 66: 1124–1131. [DOI] [PubMed] [Google Scholar]
- Bernardo R., 2009. Should maize doubled haploids be induced among F1 or F 2 plants? Theor. Appl. Genet. 119: 255–262. [DOI] [PubMed] [Google Scholar]
- Bernardo R., Yu J., 2007. Prospects for genomewide selection for quantitative traits in maize. Crop Sci. 47: 1082–1090. [Google Scholar]
- Beyene Y., Semagn K., Mugo S., Tarekegne A., Babu R., et al. , 2015. Genetic gains in grain yield through genomic selection in eight bi-parental maize populations under drought stress. Crop Sci. 55: 154–163. [Google Scholar]
- Bulmer M. G., 1971. The effect of selection on genetic variability. Am. Nat. 105: 201–211. [Google Scholar]
- Calus M. P. L., Meuwissen T. H. E., de Roos A. P. W., Veerkamp R. F., 2008. Accuracy of genomic selection using different methods to define haplotypes. Genetics 178: 553–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coster, A., 2013 Pedigree: Pedigree Functions. Available at: https://rdrr.io/cran/pedigree. Accessed: Month day, year.
- Daetwyler H. D., Villanueva B., Bijma P., Woolliams J. A., 2007. Inbreeding in genome-wide selection. J. Anim. Breed. Genet. 124: 369–376. [DOI] [PubMed] [Google Scholar]
- Daetwyler H. D., Villanueva B., Woolliams J. A., 2008. Accuracy of predicting the genetic risk of disease using a genome-wide approach. PLoS One 3: e3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Acqua M., Gatti D. M., Pea G., Cattonaro F., Coppens F., et al. , 2015. Genetic properties of the MAGIC maize population: a new platform for high definition QTL mapping in Zea mays. Genome Biol. 16: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endelman J. B., 2011. Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome J. 4: 250. [Google Scholar]
- Falconer D. S., Mackay T. F. C., 1996. Introduction to Quantitative Genetics. Benjamin Cummings, San Francisco. [Google Scholar]
- Fountain M. O., Hallauer A. R., 1996. Genetic variation within maize breeding populations. Crop Sci. 36: 26–32. [Google Scholar]
- Goddard M., 2009. Genomic selection: prediction of accuracy and maximisation of long term response. Genetica 136: 245–257. [DOI] [PubMed] [Google Scholar]
- Goddard M. E., Hayes B. J., Meuwissen T. H. E., 2011. Using the genomic relationship matrix to predict the accuracy of genomic selection. J. Anim. Breed. Genet. 128: 409–421. [DOI] [PubMed] [Google Scholar]
- Gorjanc G., Jenko J., Hearne S. J., Hickey J. M., 2016. Initiating maize pre-breeding programs using genomic selection to harness polygenic variation from landrace populations. BMC Genomics 17: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habier D., Fernando R. L., Dekkers J. C. M., 2007. The impact of genetic relationship information on genome-assisted breeding values. Genetics 177: 2389–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habier D., Tetens J., Seefried F.-R., Lichtner P., Thaller G., 2010. The impact of genetic relationship information on genomic breeding values in German Holstein cattle. Genet. Sel. Evol. 42: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habier D., Fernando R. L., Garrick D. J., 2013. Genomic BLUP decoded: a look into the black box of genomic prediction. Genetics 194: 597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallauer A. R., 1992. Recurrent selection in maize. Plant Breed. Rev. 9: 115–179. [Google Scholar]
- Hallauer A. R., Carena M. J., 2012. Recurrent selection methods to improve germplasm in maize. Maydica 57: 266–283. [Google Scholar]
- Hayes B. J., Visscher P. M., Goddard M. E., 2009. Increased accuracy of artificial selection by using the realized relationship matrix. Genet. Res. 91: 47–60. [DOI] [PubMed] [Google Scholar]
- Heffner E. L., Lorenz A. J., Jannink J. L., Sorrells M. E., 2010. Plant breeding with genomic selection: gain per unit time and cost. Crop Sci. 50: 1681–1690. [Google Scholar]
- Holland J. B., 2015. MAGIC maize: a new resource for plant genetics. Genome Biol. 16: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannink J.-L., 2010. Dynamics of long-term genomic selection. Genet. Sel. Evol. 42: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannink J.-L., Lorenz A. J., Iwata H., 2010. Genomic selection in plant breeding: from theory to practice. Brief. Funct. Genomics 9: 166–177. [DOI] [PubMed] [Google Scholar]
- Legarra A., Fernando R. L., 2009. Linear models for joint association and linkage QTL mapping. Genet. Sel. Evol. 41: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Meuwissen T., Sørensen A. C., Berg P., 2015. Upweighting rare favourable alleles increases long-term genetic gain in genomic selection programs. Genet. Sel. Evol. 47: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long N., Gianola D., Rosa G. J. M., Weigel K. A., 2011. Marker-assisted prediction of non-additive genetic values. Genetica 139: 843–854. [DOI] [PubMed] [Google Scholar]
- Massman J. M., Jung H. J. G., Bernardo R., 2013. Genomewide selection vs. marker-assisted recurrent selection to improve grain yield and stover-quality traits for cellulosic ethanol in maize. Crop Sci. 53: 58–66. [Google Scholar]
- Meuwissen T. H. E., Hayes B. J., Goddard M. E., 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157: 1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikel M. A., 2006. Availability and analysis of proprietary dent corn inbred lines with expired U.S. plant variety protection. Crop Sci. 46: 2555–2560. [Google Scholar]
- Mikel M. A., Dudley J. W., 2006. Evolution of North American dent corn from public to proprietary germplasm. Crop Sci. 46: 1193–1205. [Google Scholar]
- Muir W. M., 2007. Comparison of genomic and traditional BLUP-estimated breeding value accuracy and selection response under alternative trait and genomic parameters. J. Anim. Breed. Genet. 124: 342–355. [DOI] [PubMed] [Google Scholar]
- Nielsen H. M., Sonesson A. K., Yazdi H., Meuwissen T. H. E., 2009. Comparison of accuracy of genome-wide and BLUP breeding value estimates in sib based aquaculture breeding schemes. Aquaculture 289: 259–264. [Google Scholar]
- Quinton M., Smith C., Goddard M. E., 1992. Comparison of selection methods at the same level of inbreeding. J. Anim. Sci. 70: 1060–1067. [DOI] [PubMed] [Google Scholar]
- R Core Team , 2015. R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Sargolzaei M., Schenkel F. S., 2009. QMSim: a large-scale genome simulator for livestock. Bioinformatics 25: 680–681. [DOI] [PubMed] [Google Scholar]
- Schnable P. S., Xu X., Civardi L., Xia Y., Hsia A.-P., et al. , 1996. The role of meiotic recombination in generating novel genetic variability, pp. 103–110 in The Impact of Plant Molecular Genetics, edited by Sobral B. W. S. Birkhäuser, Boston, MA. [Google Scholar]
- Schopp P., Müller D., Technow F., Melchinger A. E., 2017. Accuracy of genomic prediction in synthetic populations depending on the number of parents, relatedness and ancestral linkage disequilibrium. Genetics 205: 441–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solberg T. R., Sonesson A. K., J. A. Woolliams, J. Odegard, and Meuwissen T. H. E., 2009. Persistence of accuracy of genome-wide breeding values over generations when including a polygenic effect. Genet. Sel. Evol. 41: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonesson A. K., Meuwissen T. H. E., 2009. Testing strategies for genomic selection in aquaculture breeding programs. Genet. Sel. Evol. 41: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Grevenhof E. M., Van Arendonk J. A., Bijma P., 2012. Response to genomic selection: the Bulmer effect and the potential of genomic selection when the number of phenotypic records is limiting. Genet. Sel. Evol. 44: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRaden P. M., 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91: 4414–4423. [DOI] [PubMed] [Google Scholar]
- Windhausen V. S., Atlin G. N., Hickey J. M., Crossa J., Jannink J.-L., et al. , 2012. Effectiveness of genomic prediction of maize hybrid performance in different breeding populations and environments. G3 2: 1427–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolc A., Arango J., Settar P., Fulton J. E., O’Sullivan N. P., et al. , 2011a Persistence of accuracy of genomic estimated breeding values over generations in layer chickens. Genet. Sel. Evol. 43: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolc A., Stricker C., Arango J., Settar P., Fulton J. E., et al. , 2011b Breeding value prediction for production traits in layer chickens using pedigree or genomic relationships in a reduced animal model. Genet. Sel. Evol. 43: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolc A., Arango J., Settar P., Fulton J. E., O’Sullivan N. P., et al. , 2016. Mixture models detect large effect QTL better than GBLUP and result in more accurate and persistent predictions. J. Anim. Sci. Biotechnol. 7: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe S., Ohsawa R., Iwata H., 2013. Potential of genomic selection for mass selection breeding in annual allogamous crops. Crop Sci. 53: 95–105. [Google Scholar]
- Yabe S., Yamasaki M., Ebana K., Hayashi T., Iwata H., 2016. Island-model genomic selection for long-term genetic improvement of autogamous crops. PLoS One 11(4): e0153945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Dekkers J. C. M., Fernando R. L., Jannink J.-L., 2009. Factors affecting accuracy from genomic selection in populations derived from multiple inbred lines: a Barley case study. Genetics 182: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.