Abstract
Lysosomes, the major membrane-bound degradative organelles, have a multitude of functions in eukaryotic cells. Lysosomes are the terminal compartments in the endocytic pathway, though they display highly dynamic behaviors, fusing with each other and with late endosomes in the endocytic pathway, and with the plasma membrane during regulated exocytosis and for wound repair. After fusing with late endosomes, lysosomes are reformed from the resulting hybrid organelles through a process that involves budding of a nascent lysosome, extension of the nascent lysosome from the hybrid organelle, while remaining connected by a membrane bridge, and scission of the membrane bridge to release the newly formed lysosome. The newly formed lysosomes undergo cycles of homotypic fusion and fission reactions to form mature lysosomes. In this study, we used a forward genetic screen in Caenorhabditis elegans to identify six regulators of lysosome biology. We show that these proteins function in different steps of lysosome biology, regulating lysosome formation, lysosome fusion, and lysosome degradation.
Keywords: lysosome, Caenorhabditis elegans, coelomocyte
Lysosomes are membrane-bound organelles that serve as the major degradative compartments for endocytic, phagocytic, and autophagic materials targeted for destruction in eukaryotic cells. This degradation is critical to many physiological processes, including processing endocytosed nutrients, downregulating signaling receptors, presenting antigens, killing pathogenic organisms, and degrading normal and abnormal cellular proteins (Dunn 1994; Mullins and Bonifacino 2001; Luzio et al. 2005). In addition, lysosomes mediate some cell death pathways and repair damage to the plasma membrane (Rodriguez et al. 1997; Reddy et al. 2001; Yamashima et al. 2003; Fehrenbacher and Jaattela 2005; Gyrd-Hansen et al. 2006). Given these diverse activities, it is perhaps not surprising that lysosomal dysfunction has been linked to the progression of many diseases, including a group of ∼50 that are referred to as lysosomal storage disorders (Vellodi 2005; Ballabio and Gieselmann 2009).
Lysosomes are dynamic structures that undergo cycles of fusion and reformation with endocytic, phagocytic, and autophagic organelles. In the endocytic pathway, mature lysosomes fuse with late endosomes to form hybrid organelles using a molecular machinery that includes Rab7, HOPS complex, and SNARES (Luzio et al. 2005; Balderhaar and Ungermann 2013; Yasuda et al. 2016). Lysosomes are formed, or reformed, from hybrid organelles in complex eukaryotes by the budding of a small nascent lysosome from the hybrid organelle, movement of the nascent lysosome away from the hybrid organelle while maintaining a membrane bridge, and scission of the membrane bridge to release a discrete primary lysosome; molecules destined for lysosomes are concentrated in the nascent lysosome during lysosome formation/reformation (Treusch et al. 2004; Bright et al. 2005; Thompson et al. 2007; Miller et al. 2015). Autophagic lysosome reformation following fusion of autophagosomes with lysosomes is similar, perhaps identical, to lysosome formation in the endocytic pathway (Yu et al. 2010; Rong et al. 2011, 2012). Lysosome formation requires the Mucolipidosis type IV protein TRPML1 (CUP-5 in Caenorhabditis elegans), Ca2+ efflux from hybrid organelles, and the C. elegans small Rab GTPase 2-like UNC-108 protein (Pryor et al. 2000; Chun et al. 2008; Lu et al. 2008; Miller et al. 2015); autophagic lysosome reformation is activated by mTOR signaling, and requires clathrin and phosphatidylinositol-4,5-bisphosphate kinases (Yu et al. 2010; Rong et al. 2012). Primary lysosomes that are formed are thought to undergo cycles of homotypic fusion with each other using the HOPS complex and SNARES, and fission reactions that yield mature lysosomes that can fuse with late endosomes (Ward et al. 2000; Wang et al. 2003). In addition to all of these membrane dynamics, lysosomal enzymes are targeted from the biosynthetic pathway to lysosomes, a process referred to as lysosome biogenesis (Mullins and Bonifacino 2001; Luzio et al. 2003). Much remains to be discovered about molecular regulation of these complex fusion, fission, and reformation reactions in the late endocytic pathway.
We had made Pmyo-3::ssGFP transgenic worms in which GFP is secreted into the body cavity when it is attached to a signal sequence and expressed in body wall muscles. The GFP is endocytosed primarily by six scavenger cells called coelomocytes in the body cavities of hermaphrodite worms, and subsequently degraded in lysosomes; Pmyo-3::ssGFP worms display diffuse GFP fluorescence in the body cavity and GFP-filled endolysosomal compartments in coelomocytes (Fares and Greenwald 2001a,b; Treusch et al. 2004). Mutations in “cup” (coelomocyte uptake) genes that reduce endocytic uptake by coelomocytes lead to increased levels of GFP in the body cavity, while mutations that decrease lysosomal function result in increased compartmental GFP levels in coelomocytes (Fares and Greenwald 2001a). A previous pilot mutational screen identified 10 cup genes required for uptake of GFP from the body cavity, and a single gene, cup-5, which encodes the worm ortholog of mammalian TRPML1, that is required for lysosome function and that we showed is required for lysosome formation/reformation (Fares and Greenwald 2001a,b). In this study, we carried out a forward genetic screen focused on lysosomal phenotypes and identified six additional cup genes that function in different aspects of lysosome biology.
Materials and Methods
C. elegans strains and methods
Standard methods were used for the growth and manipulation of C. elegans (Brenner 1974). Ethyl methane sulfonate mutagenesis was performed on the wild type var. “Bristol” strain GS1912: dpy-20(e1282); arIs37[Pmyo-3::ssGFP; dpy-20] as previously described (Fares and Greenwald 2001a). Briefly, five mutagenized P0 parents were placed on a single NGM (nematode growth medium) plate that had been previously seeded with Escherichia coli OP50 (Brenner 1974). The F2 hermaphrodites from each plate were visually screened using a modified dissection microscope equipped with fluorescence optics (Zeis, Thornwood, NY) for a defect in lysosome function by coelomocytes evidenced by brighter GFP in coelomocytes. Independently isolated mutant F2 worms (only one from each plate) were picked onto individual fresh NGM plate (with OP50), and those that yielded viable progeny were kept for further analysis. Strain GS1912 was also used to backcross mutant strains. The wild type var. “Hawaiian” strain was used for single nucleotide polymorphism mapping, as previously described (Davis et al. 2005). RNAi was done by the feeding method; control RNAi was done using bacteria carrying the double-stranded RNA generating vector L4440/pPD129.36 (Timmons and Fire 1998). The C. elegans strains used in this study are listed in Supplemental Material, Table S1.
Microscopy
Microinjection:
The body cavities of young adult hermaphrodites were microinjected with 1 mg/ml BSA-Alexa Fluor 594 in 1× PBS at room temperature, as previously described (Dang et al. 2004; Treusch et al. 2004; Schaheen et al. 2009). Microinjected worms were allowed to recover on NGM + OP50 plates, and imaging was done after 30 min or 24 hr on worms placed in 6 μl of 9 mM levamisole/1× PBS on a 2.2% agarose pad.
Marker analysis:
Young adult wild type and mutant hermaphrodites were placed in 6 μl of 9 mM levamisole/1× PBS on a 2.2% agarose pad for imaging.
Microscopy:
Confocal images were taken on a Zeiss LSM510 (Carl Zeiss, Oberkochen, Germany) laser scanning confocal, using LSM imaging software. Image analysis was done using Metamorph software (Molecular Devices, Sunnyvale, CA).
Molecular methods
Standard methods were used for the manipulation of recombinant DNA (Sambrook et al. 1989). Polymerase chain reaction was done using the Expand Long Template PCR System (Roche, Indianapolis, IN) according to the manufacturer’s instructions. All other enzymes were from New England Biolabs (Beverly, MA).
Whole-genome sequencing
Mutants were first backcrossed four times to the wild-type GS1912 strain. The backcrossed mutants were crossed one more time to GS1912, and 10 independently isolated homozygous mutants were picked from the F2 generation. These 10 hermaphrodites were mixed together for genomic DNA isolation for sequencing; this allowed us to identify background mutations that would not be homozygous in all 10 isolates (unless they are closely linked to the cup mutation).
C. elegans genomic DNA was sequenced by the University of Arizona Genomics Core using shotgun/de novo sequencing on an Illumina HiSequation 2000. Bowtie2 was used to map reads from each sample to the C. elegans reference WBcel215. samtools mpileup was then used to identify all of the variations from WBcel215. cup mutant sequences were compared to the whole genome sequence of GS1912; unique variants each had an average of 147 reads. This identified homozygous mutations in cup strains, relative to the parent strain GS1912.
Protein domain identification
SMART (Simple Modular Architectural Tool) was used for the identification of protein domains (Schultz et al. 1998; Letunic et al. 2015).
Statistical methods
The Student’s t-test was used to compare average measurements from two samples using a two-tailed distribution (Tails = 2), and a two-sample unequal variance (Type = 2).
Data availability
All data and reagents are available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Identification of novel cup mutants with lysosomal defects
We mutagenized ∼3000 P0 Pmyo-3::ssGFP hermaphrodites, and screened ∼70,000 F2 progeny for worms that had coelomocytes with brighter GFP, indicative of a delay in the lysosomal degradation of endocytosed GFP in coelomocytes. We identified 20 independent homozygous mutants that gave viable progeny (Table 1). All of the mutations were recessive, since crossing them to the parent wild-type Pmyo-3::ssGFP males yielded F1 hermaphrodite progeny that did not have a coelomocyte defect.
Table 1. List of cup genes and homologs.
| Gene | Worm ORF | Alleles | Mammalian homolog | S. cerevisiae homolog |
|---|---|---|---|---|
| cup-5 | R13A5.1 | cd9, cd10, cd12, cd18, cd19, cd20, cd21 | TRPML1 | None |
| cup-12/clh-6 | R07B7.1 | cd7, cd14, cd15, cd34, cd39 | CIC-7 | GEF1 |
| cup-13/aexr-2 | C25B8.7 | cd16, cd17 | Prokineticin Receptor | None |
| cup-14 | Y51H1A.2 | cd31, cd32 | PLEKHM1/3 | None |
| cup-15 | F42A8.3 | cd33, cd46 | OSTM1 | None |
| cup-16 | C34C6.7 | cd50 | None | None |
| cup-17/ctns-1 | C41C4.7 | cd49 | Cystinosin | ERS1 |
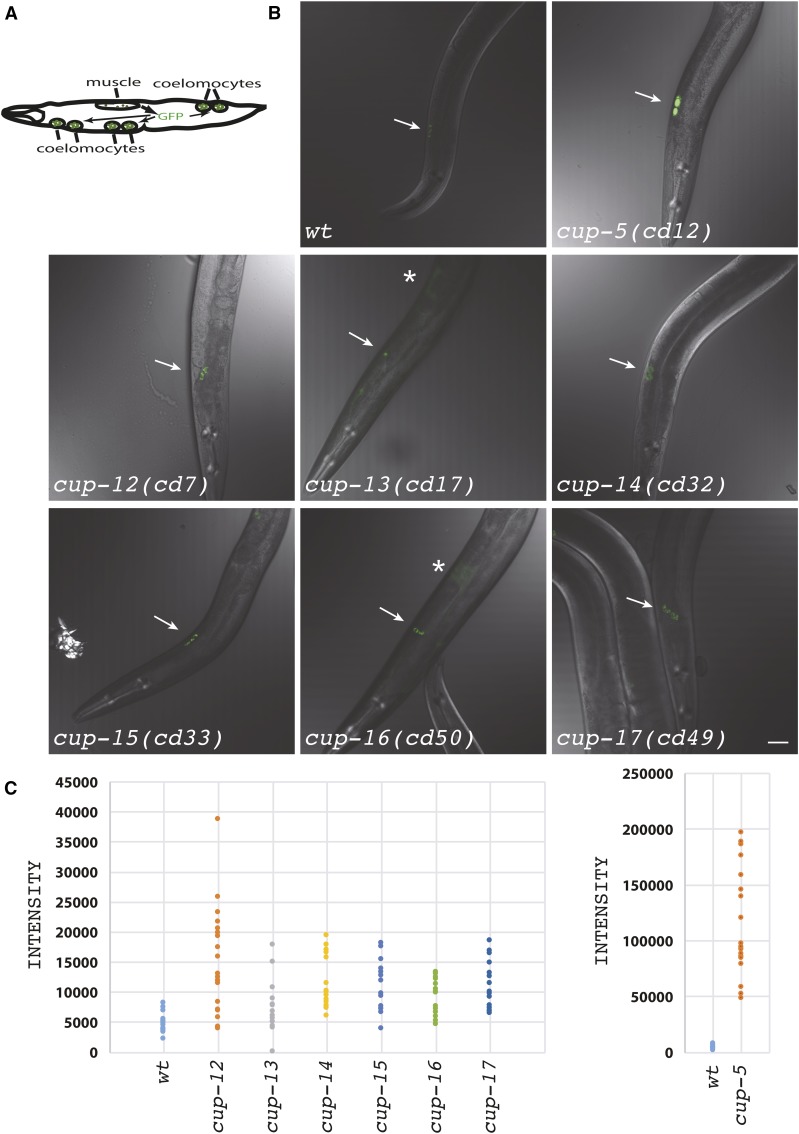
We used complementation analysis to determine the number of genes that correspond to the 20 mutations. We included the hypomorphic cup-5(ar465) allele in this analysis because we had originally identified this mutant on the basis of a similar phenotype in Pmyo-3::ssGFP worms (Fares and Greenwald 2001a). Our analysis indicates that we indeed identified several additional viable mutations in cup-5 (Table 1); these are hypomorphic mutations since null alleles in cup-5 result in embryonic lethality (Hersh et al. 2002; Schaheen et al. 2006). In addition to cup-5, we identified mutations in six other genes that we referred to provisionally as cup-12 through cup-17 (Table 1). cup-12, cup-14, cup-15, and cup-17 mutants have coelomocytes with brighter GFP than wild-type Pmyo-3::ssGFP hermaphrodites; in addition to this brighter GFP phenotype, cup-13 and cup-16 hermaphrodites also accumulate GFP in the body cavity, indicating a defect in the uptake of GFP from the body cavity (Figure 1) (Fares and Greenwald 2001a).
Figure 1.
Lysosomal degradation defect of endocytosed GFP. (A) The top left panel is a schematic of an arIs37[Pmyo-3::ssGFP; dpy-20] hermaphrodite showing GFP being secreted from body wall muscle cells into the body cavity, and the endocytosis of this GFP by coelomocytes. Anterior is to the left. (B) Confocal images of the indicated genotypes also carrying the arIs37[Pmyo-3::ssGFP; dpy-20] transgene. All images were taken using the same microscopy settings. Images are shown as an overlay of the GFP fluorescence (green) and DIC (gray) of anterior coelomocytes. Arrows indicate coelomocytes. Asterisks indicate accumulation of GFP in the body cavity. Bar = 20 µm. (C) Quantitation of the GFP intensity in whole coelomocytes of the indicated genotypes also carrying the arIs37[Pmyo-3::ssGFP; dpy-20] transgene. cup-5 mutant worms are shown separately to adjust for scale.
We backcrossed alleles of each gene at least four times to the wild-type Pmyo-3::ssGFP strain. Imaging of the backcrossed strains confirmed that cup-12 to cup-17 mutants had brighter GFP in coelomocytes, indicating a defect in lysosomal transport and/or function in these mutants (Figure 1, A and B). The brighter GFP in coelomocytes of cup-13 and cup-16 mutants is an underestimate because of the reduced uptake of GFP into coelomocytes from the body cavity in these mutants (Figure 1, A and B). However, none of the new mutants exhibited as severe a defect as cup-5 mutants, in terms of the accumulation of GFP in coelomocytes (Figure 1, B and C). Having isolated these new mutants with lysosomal defects, we proceeded to identify the mutated genes.
Cloning cup-12 to cup-17
We used a similar approach to clone each gene. We first used rapid single-nucleotide mapping to map one mutation in each gene to a chromosome. We then did whole-genome sequencing to identify homozygous mutations that affected predicted protein sequences that mapped to the relevant chromosome for each gene; mutations identified by whole genome sequencing were reconfirmed by regular sequencing, which resulted in a list of candidate genes for each cup mutant. To identify the relevant gene/mutation, we used complementation analysis against previously identified alleles of the genes, and/or RNAi of the candidate genes in Pmyo-3::ssGFP hermaphrodites, to test for the relevant phenotypes.
cup-12:
We isolated five independent alleles of cup-12 (Table 1). We mapped cup-12(cd7) to chromosome V. Whole-genome sequence analysis of cup-12(cd7) and of cup-12(cd15) worms identified protein-altering mutations that were common in both alleles in three genes on chromosome V, Y108G3AL.3, ZC412.2/gcy-14, and R07B7.1/clh-6. We did complementation analysis by crossing cup-12(cd7); Pmyo-3::ssGFP males to existing null alleles of Y108G3AL.3 and gcy-14, and to an existing missense allele of clh-6 (Table S1): only the clh-6 cross gave F1 worms with the cup-12 mutant bright coelomocyte GFP defect. Indeed, a clh-6(gk248753); Pmyo-3::ssGFP strain showed the same Cup phenotype as cup-12; Pmyo-3::ssGFP. Furthermore, RNAi of clh-6, but not of Y108G3AL.3 or gcy-14, yielded the cup-12 mutant bright-coelomocyte defect in Pmyo-3::ssGFP worms. We identified homozygous mutations in clh-6 in all five cup-12 alleles; cd15 introduces a stop codon, while the rest of the alleles have amino acid substitutions in the predicted wild-type protein (Figure S1). Thus, CUP-12 is CLH-6, which is homologous to the lysosomal chloride channel CIC-7 in mammals and to Saccharomyces cerevisiae GEF1 (Borsani et al. 1995; Bianchi et al. 2001; Braun et al. 2010).
cup-13:
We isolated two independent alleles of cup-13 (Table 1). We mapped both cup-13 alleles to the X-chromosome. We had previously identified mutations in a gene we called cup-11 that mapped to the X-chromosome, and that showed similar defects as cup-13 (Fares and Greenwald 2001a). Complementation analysis indicated that cup-11 and cup-13 were identical. Whole-genome sequence analysis of cup-13(cd17) worms identified homozygous, protein-altering mutations in three genes on chromosome X: col-168, aexr-2, and F14B8.5. We did complementation analysis by crossing cup-13(cd16); Pmyo-3::ssGFP and cup-13(cd17); Pmyo-3::ssGFP males to existing null alleles of col-168 and aexr-2, and to existing missense alleles of F14B8.5 (Table S1): only the aexr-2 crosses gave F1 worms with the cup-13 mutant bright-coelomocyte GFP and body cavity GFP defects. Indeed, an aexr-2(gk533021); Pmyo-3::ssGFP strain showed the same Cup phenotypes as cup-13; Pmyo-3::ssGFP. In addition, we identified protein-altering changes in the two independently isolated cup-13 alleles, and in eight of the nine independently isolated cup-11 alleles (Figure S2). Thus, CUP-13 is CUP-11, which is AEXR-2, a predicted seven-transmembrane domain, G protein-coupled receptor with homology to Prokineticin and Galanin Receptors in mammals (Floren et al. 2000; Ngan and Tam 2008; Lau et al. 2015).
cup-14:
We isolated two independent alleles of cup-14 (Table 1). We mapped cup-14(cd32) to chromosome II. Whole-genome sequence analysis of cup-14(cd32) worms identified homozygous, protein-altering mutations in two genes on chromosome II, Y51H1A.2 and sre-46. RNAi of Y51H1A.2, but not of sre-46, yielded the cup-14 mutant bright coelomocyte GFP defect in Pmyo-3::ssGFP worms. Furthermore, cup-14(cd32); Pmyo-3::ssGFP did not complement an existing null allele of Y51H1A.2 (Table S1). Indeed, a Y51H1A.2(gk595958); Pmyo-3::ssGFP strain showed the same Cup phenotype as cup-14; Pmyo-3::ssGFP. We identified homozygous mutations in Y51H1A.2 in both cup-14 alleles; cd31 is a missense allele, and cd32 creates a stop codon that truncates the carboxyl terminus (Figure S3). Thus, CUP-14 is Y51H1A.2, which is homologous to the endolysosomal PLEKHM1 protein in mammals (McEwan et al. 2015a,b).
cup-15:
We isolated two independent alleles of cup-15 (Table 1). We mapped cup-15(cd33) to chromosome II. Whole-genome sequence analysis of cup-15(cd33) worms identified a homozygous, nonsense mutation in F42A8.3, and homozygous, missense mutations in bmy-1, cpna-2, T01H3.2, efl-3, and F14E5.1 on chromosome II. cup-15(cd33); Pmyo-3::ssGFP complemented existing null alleles of cpna-2 and T01H3.2, but did not complement an existing null allele of F42A8.3 (Table S1). Indeed, an F42A8.3(gk150714); Pmyo-3::ssGFP strain showed the same Cup phenotype as cup-15; Pmyo-3::ssGFP. Furthermore, RNAi of F42A8.3 yielded the cup-15 mutant bright coelomocyte GFP defect in Pmyo-3::ssGFP worms. We confirmed the presence of homozygous mutations in F42A8.3 in both cup-15 alleles; cd33 is a nonsense mutation that truncates the carboxyl terminus, including a transmembrane domain, and cd46 is a deletion that removes from upstream of the predicted start codon to the middle of the second intron (Figure S4). Thus, CUP-15 is F42A8.3, which is homologous to mammalian OSTM1 that forms the Cl−/H+ exchange channel with mammalian CIC-7 (worm CLH-6/CUP-12) on lysosomes (Lange et al. 2006; Leisle et al. 2011).
cup-16:
We isolated one allele of cup-16 (Table 1). We mapped cup-16(cd50) to chromosome II. Whole-genome sequence analysis of cup-16(cd50) worms identified homozygous, protein-altering mutations in several genes on chromosome II: cpb-2, srh-60, W10G11.17, F19B10.1, ani-2, F18A1.1, cpna-2, C34C6.7, cct-2, C18E9.9, T15H9.6, and let-23. cup-16(cd50); Pmyo-3::ssGFP did not complement an existing null allele of C34C6.7, but complemented missense (srh-60, W10G11.17, C18E9.9) and nonsense (cpna-2, F19B10.1, ani-2, F18A1.1, C34C6.7, cct-2, T15H9.6, let-23) alleles in the other genes (Table S1). Indeed, a C34C6.7(gk358192); Pmyo-3::ssGFP strain showed the same Cup phenotypes as cup-16; Pmyo-3::ssGFP. Furthermore, RNAi of C34C6.7 yielded the cup-16 mutant bright coelomocyte GFP and body cavity GFP defects in Pmyo-3::ssGFP worms. cd50 is a missense mutation (Figure S5). Thus, CUP-16 is C34C6.7, which, based on primary sequence blast searches, appears to be found only in the Caenorhabditis genus.
cup-17:
We isolated one allele of cup-17 (Table 1). We mapped cup-17(cd49) to chromosome II. Whole-genome sequence analysis of cup-17(cd49) worms identified homozygous, nonsense mutations in C41C4.7/ctns-1 and T19H5.2/chil-26, and missense mutations in ZK1127.4, ZK1290.13, cec-3, C18E9.7, T24H10.1, R09D1.4, mrps-22, kin-6, F13D12.8, and F44E5.3 on chromosome II. cup-17(cd49); Pmyo-3::ssGFP did not complement an existing null allele of ctns-1 (Table S1). Indeed, a ctns-1(ok813); Pmyo-3::ssGFP strain showed the same Cup phenotype as cup-17; Pmyo-3::ssGFP. Furthermore, RNAi of ctns-1 yielded the cup-17 mutant bright coelomocyte GFP defect in Pmyo-3::ssGFP worms. cd49 is a nonsense mutation that truncates the carboxyl terminus, including a transmembrane domain (Figure S6). Thus, CUP-17 is CTNS-1, which is homologous to the lysosomal cysteine transporter Cystinosin in mammals, and to S. cerevisiae ERS1 (Kalatzis et al. 2001; Gao et al. 2004; Liu et al. 2012).
Having identified the genes from our mutational screen, we analyzed endolysosomal transport to identify which aspects of lysosome biology they regulate.
Determining organelle sizes to evaluate membrane traffic
The sizes of endosomes and lysosomes increase with membrane that they receive from other compartments through fusion reactions, and decrease with membrane that they lose through fission reactions (for example, vesicular or tubular transport). To determine where the CUP proteins functioned in the endocytic pathway, we used established markers for organelles to evaluate sizes of endosomes and of lysosomes. We included the cup-5 mutant in this analysis for comparative purposes.
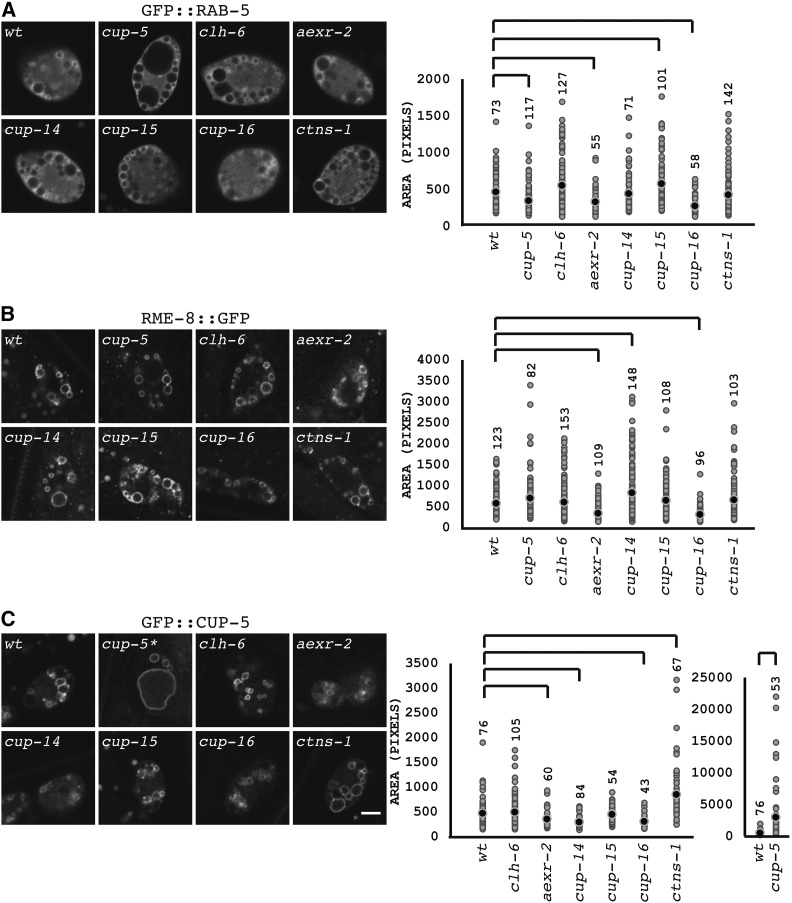
We first used GFP::RAB-5 as a marker of early endosomes (Dang et al. 2004; Patton et al. 2005; Schaheen et al. 2009). The cup-5 (ar465) mutant showed early endosomes that were 70% the size of early endosomes in wild-type coelomocytes (P 4.3 × 10−5); however, the large vacuoles that push against other compartments in the cup-5 mutant make determination of early endosome sizes inaccurate (Figure 2, A and C). The cup-15 mutant showed slightly larger early endosomes than wild-type cells (1.24-times larger; P 0.02), suggesting increased endocytic uptake, increased early endosome fusion, or decreased membrane transport from early endosomes in the absence of CUP-15 (Figure 2A). The most significant differences were decreased early endosome sizes compared to wild-type coelomocytes in the cup-13/aexr-2 mutant (70% of wild type; P 2.6 × 10−4) and the cup-16 mutant (58% of wild type; P 1.5 × 10−8); these are consistent with the decreased endocytic uptake of GFP by coelomocytes, and the accumulation of GFP in the body cavities of cup-13/aexr-2 and cup-16 mutants (Figure 1B and Figure 2A).
Figure 2.
Sizes of endolysosomal compartments. (A) Confocal images of coelomocytes in young adult hermaphrodites of the indicated genotypes that also carry the cdIs131[Pcc1::GFP::rab-5; unc-119-myo-2::GFP] transgene. The scatter plot shows the sizes of individual GFP::RAB-5-labeled compartments in the coelomocytes. (B) Confocal images of coelomocytes in young adult hermaphrodites of the indicated genotypes that also carry the cdIs153[rme-8::GFP; unc-119-myo-2::GFP] transgene. The scatter plot shows the sizes of individual RME-8::GFP-labeled compartments in the coelomocytes. (C) Confocal images of coelomocytes in young adult hermaphrodites of the indicated genotypes that also carry the cdIs40[Pcc1::GFP::cup-5; unc-119-myo-2::GFP] transgene; the cup-5 mutant carries the pwIs50[lmp-1::GFP; unc-119] transgene. The left scatter plot shows the sizes of individual GFP::CUP-5-labeled compartments in the coelomocytes; the cup-5 mutant is shown on a separate chart to adjust for scale. The black circle indicates the average size of organelles. The number above each column indicates the number of organelles measured for each strain. Inverted brackets indicate P < 0.05 of mutants compared to wild type. Bar = 5 µm.
Proceeding along the endolysosomal pathway, we next used RME-8::GFP as a marker of early and late endosomes (Zhang et al. 2001; Treusch et al. 2004; Patton et al. 2005). Both the cup-13/aexr-2 mutant (58% of wild type; P 2.3 × 10−9), and the cup-16 mutant (52% of wild type; P 10−11), showed decreased sizes consistent with the decreased amount of membrane transported to endosomes and lysosomes in these mutants (Figure 2B). In addition, the cup-14 mutant showed slightly larger early/late endosomes than wild-type cells (1.4-times larger; P 1.5 × 10−4); the normal sized GFP::RAB-5-marked early endosomes in the cup-14 mutant suggest a trafficking defect from late endosomes in the absence of CUP-14 (Figure 2B).
We next analyzed lysosomes using a GFP::CUP-5 marker for these organelles; for the cup-5 mutant, we used an LMP-1::GFP fusion to assay the sizes of the terminal compartments, since GFP::CUP-5 rescues the cup-5 mutation (Treusch et al. 2004). Consistent with the reduced sizes of early and late endosomes, both the cup-13/aexr-2 mutant (74% of wild type; P 3.5 × 10−3) and the cup-16 mutant (52% of wild type; P 1.9 × 10−4) showed decreased sizes of lysosomes (Figure 2C). In addition, the cup-14 mutant showed severely reduced lysosome sizes relative to wild type (61% of wild type; P 7.4 × 10−8), suggestive of a defect in lysosome homotypic fusion or increased homotypic lysosome fission (Figure 2C). The cup-17/ctns-1 mutant showed significantly increased sizes of terminal compartments compared to wild type (1.8 times larger, P 3.6 × 10−7), though that increase was much more pronounced in the cup-5 mutant that showed terminal compartments that were, on average, 6.4 times larger than wild type (Figure 2C).
The analysis of endolysosomal compartment sizes revealed significant defects in the cup-13/aexr-2, cup-14, cup-16, and cup-17/ctns-1 mutants. Given the defect in GFP degradation in all of the mutants, this suggested that some mutants had milder transport defects that caused a delay in the degradation of endocytosed GFP without grossly affecting sizes of late endosomes and lysosomes, at least at this resolution. We therefore followed the membrane transport of an endocytosed substrate to assay the efficiency of trafficking to lysosomes in the cup mutants.
Pulse-chase analysis of endocytosed substrate
In Pmyo-3::ssGFP worms, the GFP that is endocytosed by coelomocytes from the body cavity is visible primarily in lysosomes, where it accumulates and is eventually degraded (Fares and Greenwald 2001a; Treusch et al. 2004). We used pulse-chase analysis of a fluorescent substrate injected into the body cavities of worms to visualize the rate of flow of endocytosed substrate through endosomal compartments to lysosomes in coelomocytes. BSA-Alexa Fluor 594 microinjected into the body cavity appears by 5 min in endosomal compartments, and is concentrated in small nascent lysosomes budding from hybrid organelles; by 30 min the BSA-Alexa Fluor 594 is fully localized to lysosomes (Figure S7) (Zhang et al. 2001; Dang et al. 2004; Treusch et al. 2004; Schaheen et al. 2009).
We microinjected BSA-Alexa Fluor 594 into the body cavity of Pmyo-3::ssGFP worms and imaged two time points, 30 min and 24 hr; at these time points, the BSA-Alexa Fluor 594 is completely found in lysosomes of wild type worms, where it colocalizes with endocytosed GFP (Figure 3 and Figure S7). Pulse-chase analysis with microinjected BSA-Alexa Fluor 594 into the body cavities of the mutants revealed three types of behavior:
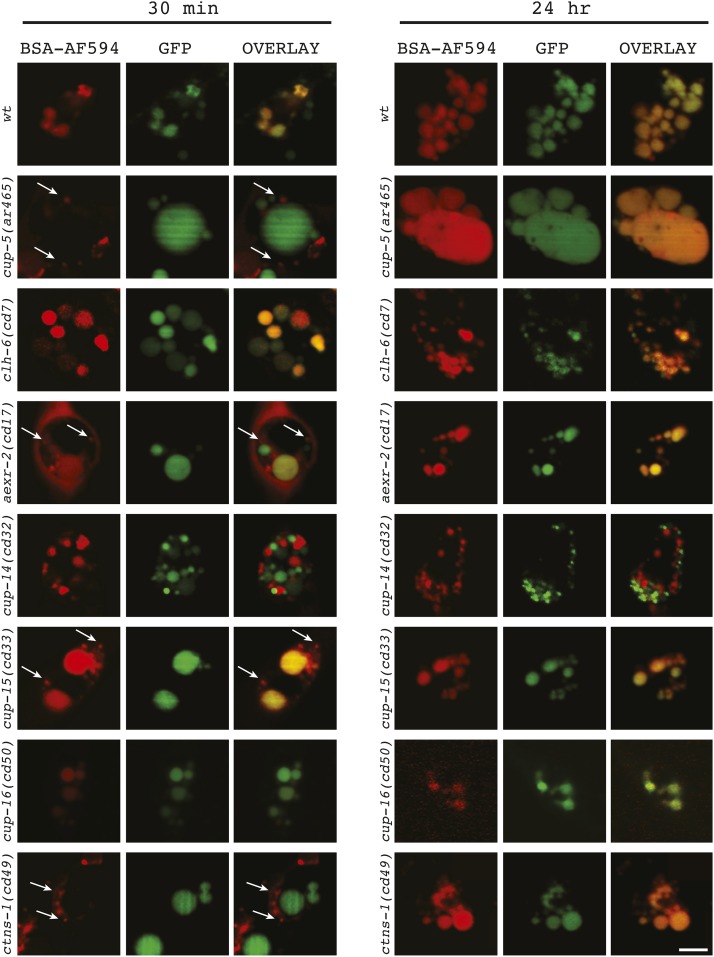
Figure 3.
Endolysosomal transport of endocytosed BSA-Alexa Fluor 594. Single optical section confocal images of the indicated genotypes also carrying the arIs37[Pmyo-3::ssGFP; dpy-20] transgene at the indicated times after microinjection of BSA-Alexa Fluor 594 into their body cavities. Arrows indicate nascent lysosomes containing concentrated BSA-Alexa Fluor 594. Bar = 5 µm.
cup-12/clh-6 and cup-16 mutants showed normal trafficking of BSA-Alexa Fluor 594, such that all of the BSA-Alexa Fluor 594 was colocalized with GFP-filled compartments by 30 min (Figure 3). Thus, there is no delay in the transport of endocytosed BSA-Alexa Fluor 594 to lysosomes in cup-12/clh-6 and cup-16 mutants.
Similar to cup-5 mutants, cup-13/aexr-2, cup-15, and cup-17/ctns-1 mutants showed a delay in the transport of BSA-Alexa Fluor 594 to lysosomes, such that there were still nascent lysosomes with concentrated BSA-Alexa Fluor 594 after 30 min; the BSA-Alexa Fluor 594 was fully localized to GFP-filled compartments by 24 hr (Figure 3) (Treusch et al. 2004). Between these three mutants, the cup-17/ctns-1 mutant had the most severe defect in transport to lysosomes, such that no BSA-Alexa Fluor 594 had reached GFP-filled lysosomes by 30 min; in contrast, cup-13/aexr-2 and cup-15 mutants had a milder defect, such that some BSA-Alexa Fluor 594 had reached GFP-filled compartments by 30 min (Figure 3). Thus, CUP-13, CUP-15, and CUP-17 likely function in the process of lysosome formation, that is, the efficient scission of nascent lysosomes budding from late endosomes/hybrid organelles.
The cup-14 mutant did not show a delay in transport to lysosomes since there were no BSA-Alexa Fluor 594 concentrations in nascent lysosomes by 30 min (Figure 3); however, the BSA-Alexa Fluor did not colocalize with GFP-filled lysosomes after 30 min, or even after 24 hr (Figure 3). This implicates CUP-14 in the fusion of lysosomes with each other (see Discussion).
Thus, the mutants we identified affect at least four different aspects of lysosome biology.
Discussion
Lysosomes are one of the major degradative organelles in eukaryotic cells that carry out diverse cellular functions. Lysosomes show highly dynamic behaviors, including homotypic and heterotypic fusions, fission, and formation/reformation, which itself involves budding, extension, and scission. We carried out an unbiased forward mutational screen to identify novel regulators of lysosome dynamics and/or function; this screen is based on the degradation of a substrate, GFP, that is endocytosed by scavenger cells in worms. We identified cup-5 and six additional proteins that have lysosomal functions in C. elegans coelomocytes. CUP-16 is only conserved in the genus Caenorhabditis, and likely functions in endocytic uptake at the plasma membrane and in lysosomal degradation. Besides CUP-16, five of the mammalian homologs of the other CUP proteins, CIC-7, OSTM1, PLEKHM1, Cystinosin, and TRPML1, had been previously implicated in lysosome biology, thus validating this approach (Bach 2001; Lange et al. 2006; Wartosch et al. 2009; Weinert et al. 2010; Leisle et al. 2011; Ivanova et al. 2015; McEwan et al. 2015a,b; Andrzejewska et al. 2016). There are certainly additional genes that function in lysosome biology, some of which are essential for embryonic development, and would require isolating rarer hypomorphic mutations that impair lysosomal function without affecting viability, as was the case for cup-5. In addition, we relied on a visual screen of whole worms, so more subtle defects in lysosome function or structure would have been missed. However, we have identified regulators and generated novel insights into at least four different steps in the endocytic pathway (Figure S8).
First, mutation of the G protein-coupled receptor CUP-13/AEXR-2 results in an early defect in endocytic uptake of a solute from the extracellular space. G protein-coupled receptors are known to function in signaling pathways at the plasma membrane through their coupling to heterotrimeric G proteins. The activities of G protein-coupled receptors is regulated by their internalization into early endosomes, followed by their recycling to the plasma membrane or their degradation in lysosomes (Seachrist and Ferguson 2003; Tsvetanova et al. 2015). Our study shows that, besides regulating their own transport and activities, G protein-coupled receptors can regulate general endocytic transport.
Second, we identified CUP-13/AEXR-2, CUP-15, and CUP-17/CTNS-1 as novel regulators of the poorly understood process of lysosome formation (Figure S8). This is based primarily on an apparently normal endocytic uptake of BSA-Alexa Fluor 594 in the mutants, and the persistence of nascent lysosomes with concentrated BSA-Alexa Fluor 594 at later time points of the pulse-chase analysis; however, this does not preclude additional roles of these proteins in other lysosomal trafficking events, for example, fusion of late endosomes with lysosomes. Based on the mutant phenotypes in C. elegans, and our quantitative live imaging studies in mammalian cells, CUP-5/TRPML1 plays a central role in lysosome formation (Treusch et al. 2004; Miller et al. 2015). The mutant phenotypes suggest that, though the activities of CUP-13/AEXR-2, CUP-15, and CUP-17/CTNS-1 may not be strictly essential for the completion of the budding, extension, and scission events during lysosome formation, these proteins modulate the efficiency of lysosome formation. It is currently unclear how G protein-coupled receptors (CUP-13/AEXR-2), OSTM1 (CUP-15), and Cystinosin (CUP-17/CTNS-1) regulate lysosome formation, and whether the defects in this process in C. elegans mutants are an indirect consequence of their reduced or absent function. OSTM1 and Cystinosin have roles in ion and cysteine transport across membranes, respectively, such that the lysosome formation defect in the cup-15 and cup-17/ctns-1 mutants may be an indirect consequence of altered ion/cysteine homeostasis (Kalatzis et al. 2001; Lange et al. 2006; Leisle et al. 2011; Liu et al. 2012). Alternatively, these proteins may have direct roles in lysosome formation, either dependent or independent of their channel functions, for example, Cystinosin has dual and independent roles as a cysteine transporter, and as a component of the mTORC1 pathway (Andrzejewska et al. 2016; Ivanova et al. 2016).
Third, our studies indicate a role for CUP-14 in promoting primary lysosomes fusing with each other (Figure S8). In the absence of CUP-14, endocytosed GFP and BSA-Alexa Fluor 594 localize to discrete lysosomes that are reduced in size. This is in agreement with our identification of scavenger receptors that are required for the endocytosis and for the transport from late endosomes to lysosomes of endocytosed GFP, but not of BSA (H.F. unpublished data). Our model is that scavenger receptors localize to nascent lysosomes where they concentrate certain solutes (like GFP or BSA). Following scission, CUP-14 and other proteins, yet to be identified, regulate the fusion of these primary lysosomes, resulting in content mixing (Figure S8). Our results further suggest that primary lysosomes are not all identical, or “homotypic”; primary lysosomes that carry GFP may be homotypic, primary lysosomes that carry BSA may also be homotypic, but the GFP-bearing lysosomes and the BSA-bearing lysosomes likely have different scavenger receptors and possibly other constituents, and may thus require a specific machinery, that includes CUP-14, for their fusion with each other. PLEKHM1, the mammalian homolog of CUP-14, functions in “heterotypic” lysosomal fusion events with Salmonella-containing vacuoles and autophagososmes (McEwan et al. 2015a,b). Our study extends PLEKHM1 functions to other lysosomal fusion events, and supports a model of receptor-mediated concentration of solutes in nascent lysosomes. CUP-14 may also have earlier roles in late endosomes, as evidenced by the slightly larger RME-8-labeled compartments in the cup-14 mutant. These larger late endosomes are not due to a defect in membrane transport to lysosomes, as determined by the BSA-Alexa Fluor 594 pulse-chase analysis. One possibility is that the lysosome fusion defect in the absence of CUP-14 makes these smaller lysosomes more fusogenic with late endosomes, thus increasing the sizes of the large endosomes; alternatively, the cup-14 mutant may have defects in membrane transport from late endosomes to other cellular organelles besides lysosomes, for example the Golgi apparatus.
Fourth, mutation of CUP-12/CLH-6 does not affect membrane transport steps, but likely reduces lysosomal degradative functions (Figure S8). This is in agreement with studies on the mammalian homolog CIC-7, which functions as a Cl−/H+ exchange protein that is required for lysosome acidification (Wartosch et al. 2009; Weinert et al. 2010). Mammalian studies identified OSTM1 (CUP-15) as a beta subunit of the CIC-7 channel that is required for the lysosomal transport and the channel activity of CIC-7 (Lange et al. 2006; Wartosch et al. 2009; Leisle et al. 2011). Our studies suggest additional functions for OSTM1 (CUP-15) in the regulation of lysosome formation that are independent of its roles with CIC-7 (CUP-12/CLH-6).
CIC-7, OSTM1, Cystinosin, and TRPML1 are all associated with human diseases (Bach 2001; Kalatzis et al. 2001; Lange et al. 2006; Weinert et al. 2010). Our study has identified potentially novel roles of these and other proteins in lysosome biology; this allows more accurate investigations into the basis for lysosomal defects in human disorders.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.037515/-/DC1.
Acknowledgments
We thank Susan Miller and Muhammad Noon for whole-genome sequencing and analysis at the University of Arizona. Several strains were acquired from the Caenorhabditis Genetics Center (CGC). This work was funded by National Science Foundation (NSF) grant MCB-1243037 to H.F.
Footnotes
Communicating editor: B. J. Andrews
Literature Cited
- Andrzejewska Z., Nevo N., Thomas L., Chhuon C., Bailleux A., et al. , 2016. Cystinosin is a component of the vacuolar H+-ATPase-Ragulator-Rag complex controlling mammalian target of rapamycin complex 1 signaling. J. Am. Soc. Nephrol. 27: 1678–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach G., 2001. Mucolipidosis type IV. Mol. Genet. Metab. 73: 197–203. [DOI] [PubMed] [Google Scholar]
- Balderhaar H. J., Ungermann C., 2013. CORVET and HOPS tethering complexes—coordinators of endosome and lysosome fusion. J. Cell Sci. 126: 1307–1316. [DOI] [PubMed] [Google Scholar]
- Ballabio A., Gieselmann V., 2009. Lysosomal disorders: from storage to cellular damage. Biochim. Biophys. Acta 1793: 684–696. [DOI] [PubMed] [Google Scholar]
- Bianchi L., Miller D. M., III, George A. L., Jr, 2001. Expression of a CIC chloride channel in Caenorhabditis elegans gamma-aminobutyric acid-ergic neurons. Neurosci. Lett. 299: 177–180. [DOI] [PubMed] [Google Scholar]
- Borsani G., Rugarli E. I., Taglialatela M., Wong C., Ballabio A., 1995. Characterization of a human and murine gene (CLCN3) sharing similarities to voltage-gated chloride channels and to a yeast integral membrane protein. Genomics 27: 131–141. [DOI] [PubMed] [Google Scholar]
- Braun N. A., Morgan B., Dick T. P., Schwappach B., 2010. The yeast CLC protein counteracts vesicular acidification during iron starvation. J. Cell Sci. 123: 2342–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright N. A., Gratian M. J., Luzio J. P., 2005. Endocytic delivery to lysosomes mediated by concurrent fusion and kissing events in living cells. Curr. Biol. 15: 360–365. [DOI] [PubMed] [Google Scholar]
- Chun D. K., McEwen J. M., Burbea M., Kaplan J. M., 2008. UNC-108/Rab2 regulates postendocytic trafficking in Caenorhabditis elegans. Mol. Biol. Cell 19: 2682–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang H., Li Z., Skolnik E. Y., Fares H., 2004. Disease-related myotubularins function in endocytic traffic in Caenorhabditis elegans. Mol. Biol. Cell 15: 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. W., Hammarlund M., Harrach T., Hullett P., Olsen S., et al. , 2005. Rapid single nucleotide polymorphism mapping in C. elegans. BMC Genomics 6: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn W. A., Jr, 1994. Autophagy and related mechanisms of lysosome-mediated protein degradation. Trends Cell Biol. 4: 139–143. [DOI] [PubMed] [Google Scholar]
- Fares H., Greenwald I., 2001a Genetic analysis of endocytosis in Caenorhabditis elegans: coelomocyte uptake defective mutants. Genetics 159: 133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares H., Greenwald I., 2001b Regulation of endocytosis by CUP-5, the Caenorhabditis elegans mucolipin-1 homolog. Nat. Genet. 28: 64–68. [DOI] [PubMed] [Google Scholar]
- Fehrenbacher N., Jaattela M., 2005. Lysosomes as targets for cancer therapy. Cancer Res. 65: 2993–2995. [DOI] [PubMed] [Google Scholar]
- Floren A., Land T., Langel U., 2000. Galanin receptor subtypes and ligand binding. Neuropeptides 34: 331–337. [DOI] [PubMed] [Google Scholar]
- Gao Y., Dickerson J. B., Guo F., Zheng J., Zheng Y., 2004. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc. Natl. Acad. Sci. USA 101: 7618–7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyrd-Hansen M., Farkas T., Fehrenbacher N., Bastholm L., Hoyer-Hansen M., et al. , 2006. Apoptosome-independent activation of the lysosomal cell death pathway by caspase-9. Mol. Cell. Biol. 26: 7880–7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersh B. M., Hartwieg E., Horvitz H. R., 2002. The Caenorhabditis elegans mucolipin-like gene cup-5 is essential for viability and regulates lysosomes in multiple cell types. Proc. Natl. Acad. Sci. USA 99: 4355–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova E. A., De Leo M. G., Van Den Heuvel L., Pastore A., Dijkman H., et al. , 2015. Endo-lysosomal dysfunction in human proximal tubular epithelial cells deficient for lysosomal cystine transporter cystinosin. PLoS One 10: e0120998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova E. A., van den Heuvel L. P., Elmonem M. A., De Smedt H., Missiaen L., et al. , 2016. Altered mTOR signalling in nephropathic cystinosis. J. Inherit. Metab. Dis. 39: 457–464. [DOI] [PubMed] [Google Scholar]
- Kalatzis V., Cherqui S., Antignac C., Gasnier B., 2001. Cystinosin, the protein defective in cystinosis, is a H(+)-driven lysosomal cystine transporter. EMBO J. 20: 5940–5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange P. F., Wartosch L., Jentsch T. J., Fuhrmann J. C., 2006. ClC-7 requires Ostm1 as a beta-subunit to support bone resorption and lysosomal function. Nature 440: 220–223. [DOI] [PubMed] [Google Scholar]
- Lau S. T., Hansford L. M., Chan W. K., Chan G. C., Wan T. S., et al. , 2015. Prokineticin signaling is required for the maintenance of a de novo population of c-KIT(+) cells to sustain neuroblastoma progression. Oncogene 34: 1019–1034. [DOI] [PubMed] [Google Scholar]
- Leisle L., Ludwig C. F., Wagner F. A., Jentsch T. J., Stauber T., 2011. ClC-7 is a slowly voltage-gated 2Cl(-)/1H(+)-exchanger and requires Ostm1 for transport activity. EMBO J. 30: 2140–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Doerks T., Bork P., 2015. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 43: D257–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Du H., Rutkowski R., Gartner A., Wang X., 2012. LAAT-1 is the lysosomal lysine/arginine transporter that maintains amino acid homeostasis. Science 337: 351–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Zhang Y., Hu T., Guo P., Li W., et al. , 2008. C. elegans Rab GTPase 2 is required for the degradation of apoptotic cells. Development 135: 1069–1080. [DOI] [PubMed] [Google Scholar]
- Luzio J. P., Poupon V., Lindsay M. R., Mullock B. M., Piper R. C., et al. , 2003. Membrane dynamics and the biogenesis of lysosomes. Mol. Membr. Biol. 20: 141–154. [DOI] [PubMed] [Google Scholar]
- Luzio J. P., Pryor P. R., Gray S. R., Gratian M. J., Piper R. C., et al. , 2005. Membrane traffic to and from lysosomes. Biochem. Soc. Symp. 72: 77–86. [DOI] [PubMed] [Google Scholar]
- McEwan D. G., Popovic D., Gubas A., Terawaki S., Suzuki H., et al. , 2015a PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol. Cell 57: 39–54. [DOI] [PubMed] [Google Scholar]
- McEwan D. G., Richter B., Claudi B., Wigge C., Wild P., et al. , 2015b PLEKHM1 regulates Salmonella-containing vacuole biogenesis and infection. Cell Host Microbe 17: 58–71. [DOI] [PubMed] [Google Scholar]
- Miller A., Schafer J., Upchurch C., Spooner E., Huynh J., et al. , 2015. Mucolipidosis type IV protein TRPML1-dependent lysosome formation. Traffic 16: 284–297. [DOI] [PubMed] [Google Scholar]
- Mullins C., Bonifacino J. S., 2001. The molecular machinery for lysosome biogenesis. BioEssays 23: 333–343. [DOI] [PubMed] [Google Scholar]
- Ngan E. S., Tam P. K., 2008. Prokineticin-signaling pathway. Int. J. Biochem. Cell Biol. 40: 1679–1684. [DOI] [PubMed] [Google Scholar]
- Patton A., Knuth S., Schaheen B., Dang H., Greenwald I., et al. , 2005. Endocytosis function of a ligand-gated ion channel homolog in Caenorhabditis elegans. Curr. Biol. 15: 1045–1050. [DOI] [PubMed] [Google Scholar]
- Pryor P. R., Mullock B. M., Bright N. A., Gray S. R., Luzio J. P., 2000. The role of intraorganellar Ca(2+) in late endosome-lysosome heterotypic fusion and in the reformation of lysosomes from hybrid organelles. J. Cell Biol. 149: 1053–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A., Caler E. V., Andrews N. W., 2001. Plasma membrane repair is mediated by Ca(2+)-regulated exocytosis of lysosomes. Cell 106: 157–169. [DOI] [PubMed] [Google Scholar]
- Rodriguez A., Webster P., Ortego J., Andrews N. W., 1997. Lysosomes behave as Ca2+-regulated exocytic vesicles in fibroblasts and epithelial cells. J. Cell Biol. 137: 93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y., McPhee C. K., Deng S., Huang L., Chen L., et al. , 2011. Spinster is required for autophagic lysosome reformation and mTOR reactivation following starvation. Proc. Natl. Acad. Sci. USA 108: 7826–7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y., Liu M., Ma L., Du W., Zhang H., et al. , 2012. Clathrin and phosphatidylinositol-4,5-bisphosphate regulate autophagic lysosome reformation. Nat. Cell Biol. 14: 924–934. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritch E. F., Maniatis T., 1989. Molecular Cloning: A Laboratory Manual, Ed. 2 Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- Schaheen B., Dang H., Fares H., 2009. Derlin-dependent accumulation of integral membrane proteins at cell surfaces. J. Cell Sci. 122: 2228–2239. [DOI] [PubMed] [Google Scholar]
- Schaheen L., Dang H., Fares H., 2006. Basis of lethality in C. elegans lacking CUP-5, the Mucolipidosis Type IV orthologue. Dev. Biol. 293: 382–391. [DOI] [PubMed] [Google Scholar]
- Schultz J., Milpetz F., Bork P., Ponting C. P., 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95: 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seachrist J. L., Ferguson S. S., 2003. Regulation of G protein-coupled receptor endocytosis and trafficking by Rab GTPases. Life Sci. 74: 225–235. [DOI] [PubMed] [Google Scholar]
- Thompson E. G., Schaheen L., Dang H., Fares H., 2007. Lysosomal trafficking functions of mucolipin-1 in murine macrophages. BMC Cell Biol. 8: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., Fire A., 1998. Specific interference by ingested dsRNA. Nature 395: 854. [DOI] [PubMed] [Google Scholar]
- Treusch S., Knuth S., Slaugenhaupt S. A., Goldin E., Grant B. D., et al. , 2004. Caenorhabditis elegans functional orthologue of human protein h-mucolipin-1 is required for lysosome biogenesis. Proc. Natl. Acad. Sci. USA 101: 4483–4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetanova N. G., Irannejad R., von Zastrow M., 2015. G protein-coupled receptor (GPCR) signaling via heterotrimeric G proteins from endosomes. J. Biol. Chem. 290: 6689–6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellodi A., 2005. Lysosomal storage disorders. Br. J. Haematol. 128: 413–431. [DOI] [PubMed] [Google Scholar]
- Wang C. W., Stromhaug P. E., Kauffman E. J., Weisman L. S., Klionsky D. J., 2003. Yeast homotypic vacuole fusion requires the Ccz1-Mon1 complex during the tethering/docking stage. J. Cell Biol. 163: 973–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward D. M., Pevsner J., Scullion M. A., Vaughn M., Kaplan J., 2000. Syntaxin 7 and VAMP-7 are soluble N-ethylmaleimide-sensitive factor attachment protein receptors required for late endosome-lysosome and homotypic lysosome fusion in alveolar macrophages. Mol. Biol. Cell 11: 2327–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wartosch L., Fuhrmann J. C., Schweizer M., Stauber T., Jentsch T. J., 2009. Lysosomal degradation of endocytosed proteins depends on the chloride transport protein ClC-7. FASEB J. 23: 4056–4068. [DOI] [PubMed] [Google Scholar]
- Weinert S., Jabs S., Supanchart C., Schweizer M., Gimber N., et al. , 2010. Lysosomal pathology and osteopetrosis upon loss of H+-driven lysosomal Cl- accumulation. Science 328: 1401–1403. [DOI] [PubMed] [Google Scholar]
- Yamashima T., Tonchev A. B., Tsukada T., Saido T. C., Imajoh-Ohmi S., et al. , 2003. Sustained calpain activation associated with lysosomal rupture executes necrosis of the postischemic CA1 neurons in primates. Hippocampus 13: 791–800. [DOI] [PubMed] [Google Scholar]
- Yasuda S., Morishita S., Fujita A., Nanao T., Wada N., et al. , 2016. Mon1-Ccz1 activates Rab7 only on late endosomes and dissociates from the lysosome in mammalian cells. J. Cell Sci. 129: 329–340. [DOI] [PubMed] [Google Scholar]
- Yu L., McPhee C. K., Zheng L., Mardones G. A., Rong Y., et al. , 2010. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465: 942–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Grant B., Hirsh D., 2001. RME-8, a conserved J-domain protein, is required for endocytosis in Caenorhabditis elegans. Mol. Biol. Cell 12: 2011–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and reagents are available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.