Abstract
The restriction endonuclease (REase) R.KpnI is an orthodox Type IIP enzyme, which binds to DNA in the absence of metal ions and cleaves the DNA sequence 5′-GGTAC^C-3′ in the presence of Mg2+ as shown generating 3′ four base overhangs. Bioinformatics analysis reveals that R.KpnI contains a ββα-Me-finger fold, which is characteristic of many HNH-superfamily endonucleases, including homing endonuclease I-HmuI, structure-specific T4 endonuclease VII, colicin E9, sequence non-specific Serratia nuclease and sequence-specific homing endonuclease I-PpoI. According to our homology model of R.KpnI, D148, H149 and Q175 correspond to the critical D, H and N or H residues of the HNH nucleases. Substitutions of these three conserved residues lead to the loss of the DNA cleavage activity by R.KpnI, confirming their importance. The mutant Q175E fails to bind DNA at the standard conditions, although the DNA binding and cleavage can be rescued at pH 6.0, indicating a role for Q175 in DNA binding and cleavage. Our study provides the first experimental evidence for a Type IIP REase that does not belong to the PD…D/EXK superfamily of nucleases, instead is a member of the HNH superfamily.
INTRODUCTION
The mechanism of phosphodiester cleavage comprises several important steps, including activation of a nucleophile, stabilization of the phosphoanion transition state and protonation of the leaving group (1). An underlying feature for a majority of the nucleases is the use of a metal ion as a cofactor for the activation of a nucleophilic water molecule and stabilization of the transition state. From the mechanistic point of view, the best-characterized prokaryotic endonucleases are Type II restriction enzymes (REases) (2,3). Type II REases bind and cleave DNA in presence of Mg2+ with remarkable specificity (4). Comparative studies of crystal structures and protein sequences followed by mutational analyses of Type II REases revealed the presence of a partially conserved motif ‘PD…D/EXK’. Two acidic residues (D and D/E) are important for the metal ion binding, and K is involved in catalysis (5). Substitutions of K and the two acidic residues in the active site typically lead to a drastic reduction of the cleavage activity (5). The PD…D/EXK motif has been found in numerous Type II REases as well as in other nucleases such as phage λ exonuclease (2), DNA repair enzyme MutH (6) and transposase TnsA (7). Together, these proteins form a PD…D/EXK superfamily of nucleases (8,9).
R.KpnI is a Type IIP REase (10), which recognizes a palindromic DNA sequence 5′-GGTAC^C-3′, and cleaves at indicated position [^], generating four base long 3′-overhangs. R.KpnI forms a restriction–modification system together with M.KpnI, a DNA methyltransferase (MTase), which transfers a methyl group from the cofactor AdoMet onto the N6-position of the adenine in both strands within the same sequence (11–13).
Bioinformatics analyses suggested that R.KpnI does not belong to the PD…D/EXK superfamily but rather exhibits an active site with the ββα-Me-finger fold, characteristic for the HNH-superfamily of nucleases. In order to identify the catalytic residues in the REase, we carried out homology modeling and used the models to identify targets for site-directed mutagenesis and to interpret the experimental data. Our results indicate that R.KpnI indeed belongs to the HNH superfamily and the architecture of its active site is completely different from the active sites of all Type II REases characterized to date.
MATERIALS AND METHODS
Enzymes and DNA
Plasmid pETRK contains the wild-type (wt) R.KpnI gene cloned into the pET11d expression vector (14). R.KpnI and its mutants were purified as described previously (14). The enzymes were diluted in binding buffer [20 mM Tris–HCl (pH 7.4), 25 mM NaCl and 5 mM 2-mercaptoethanol] for all the studies. The concentration of the proteins was estimated by the method of Bradford (15). T4 polynucleotide kinase, Pfu DNA polymerase and DpnI were purchased from New England Biolabs, USA. Oligonucleotides (Microsynth Inc, Switzerland) were purified on 18% urea-polyacrylamide gel (16). The purified oligonucleotides were end-labeled with T4 polynucleotide kinase and [γ-32P]ATP (6000 Ci/mmol).
Protein sequence analysis and structure prediction
Sequence searches of the non-redundant (nr) database were carried out with PSI-BLAST (17) via the NCBI website (http://www.ncbi.nlm.nih.gov), using the sequence of T4 Endonuclease VII (EndoVII) as a query. A relatively stringent cutoff of expectation (e)-value <10−3 was used to identify HNH-superfamily members related to endonuclease VII and build a multiple sequence alignment, subsequently converted to a position-specific scoring matrix (PSSM). The searches were iterated with the PSSM as a query until no more homologs with e-value <10−3 could be identified. Then, the cutoff was lowered to 0.1 and searches were continued with an additional criterion that potential HNH-superfamily members had to exhibit the residues of the common active site (otherwise, the sequences were not included in the PSSM).
Protein structure prediction was carried out via the GeneSilico metaserver gateway [http://genesilico.pl/meta/ (18)]. The sequence alignment between KpnI and structurally characterized HNH-superfamily nucleases obtained from PSI-BLAST (after 15 iterations) was used as starting points for homology modelling using the ‘FRrankenstein's monster’ approach, comprising cycles of model building, evaluation, realignment in poorly scored regions and merging of best-scoring fragments [see (19) for a detailed description]. The positions of predicted catalytic residues and secondary structure elements were used as spatial restraints.
Site-directed mutagenesis
R.KpnI mutants were generated by site-directed mutagenesis using the megaprimer inverse PCR method (20). Expression plasmid pETRK encoding the wt kpnR gene was used as a template. The oligonucleotide primers carrying the respective mutant amino acid codon substitutions (Table 1) were used as forward primers and T7 terminator primer was used as reverse primer (Table 1). The mega primers generated were used as complementary primers for the second round of PCR amplification. After confirming the mutation by sequencing, the mutant REases were expressed in Escherichia coli BL26 [F− omp T hsdSB (rB− mB−) gal dcm Δlac (DE3) nin5 lac UV5-T7 gene 1] expressing M.KpnI and purified as described previously (14).
Table 1. DNA oligonucleotides used in this study.
| Primers used for mutagenesis | |
|---|---|
| D148G | 5′-CTAACACCTGGCCATATGACAC-3′ |
| H149L | 5′-ACACCAGACCTTATGACACCTC-3′ |
| Q175E | 5′-TGTGGACGTCATGAAGTTATGAAA-3′ |
| D148A | 5′-AACTAACACCAGCCCATATGACA-3′ |
| H149A | 5′-ACACCAGACGCTATGACACCTC-3′ |
| Reverse primer (T7 terminator sequence) | 5′-GCTAGTTATTGTTCAGCGGTGGG-3′ |
| Duplex DNA used for EMSA | 5′-ATTGCGTGGTACCCGCTCTT-3′ |
| 3′-TAACGCACCATGGGCGAGAA-5′ |
Electrophoretic mobility shift assay
Different concentrations of the wt and mutant R.KpnI (0.1–128 nM) were incubated with 3.75 nM of end-labeled double-stranded oligonucleotides containing cognate site (Table 1) in the buffers [20 mM Tris–HCl (pH 7.4), 25 mM NaCl and 5 mM 2-mercaptoethanol or 20 mM HEPES (pH 6.0), 25 mM NaCl and 5 mM 2-mercaptoethanol] on ice for 15 min. The free DNA and the enzyme-bound complexes were then separated on 8% native polyacrylamide gels with running buffers containing 90 mM Tris-borate pH 8.0 and 1 mM EDTA or 50 mM HEPES, pH 6.0 and 1 mM EDTA. The gels were electrophoresed at 4°C for 1 h, dried and autoradiographed. The amounts of DNA in free and bound form were quantitated using Phosphor imager Image Gauge software version 2.54. The assays were repeated thrice and the average values were considered for Scatchard analysis to determine the affinity of the proteins.
In vitro DNA cleavage activity assay
Purified R.KpnI and its variants were incubated in 20 mM HEPES–KOH (pH 6.0) or 10 mM Tris–HCl (pH 7.0–9.0), 1 mM EDTA, 5 mM β-mercaptoethanol, 5 mM MgCl2 for 1 h at 37°C and 500 ng of plasmid DNA. The cleavage products were analyzed on 1% agarose gel.
RESULTS
Sequence alignment and homology modelling of R.KpnI
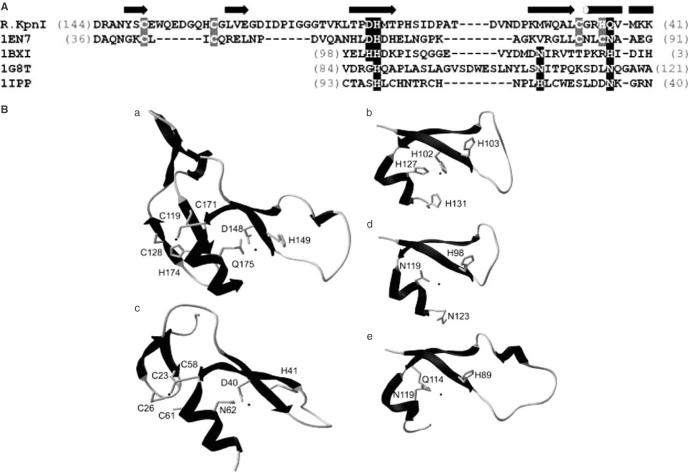
PSI-BLAST sequence searches of the nr database (see Materials and Methods) revealed remote homology between EndoVII and KpnI [(21); this work]. Our analyses resulted in a sequence alignment slightly different from that published in the earlier work (21); we confirmed the earlier prediction of the catalytic core, but also identified a potential Zn-finger conserved between KpnI and EndoVII, which was partially misaligned previously (21). In order to facilitate the interpretation of experimental data in the sequence–structure–function context, we built a theoretical model of the KpnI catalytic domain (residues 97–190) in complex with the target DNA. The homology model of the KpnI monomer was generated based on the sequence alignment with known structures of HNH-superfamily members (see Materials and Methods). The mutual orientation of the two KpnI monomers as well as the coordinates of the DNA molecule and the Mg2+ ions were based on superposition onto the subunits A and B in the I-PpoI co-crystal structure (22). The GGTACC site recognized by KpnI was generated by ‘mutating’ bases in the original I-PpoI target (CTTAAG) and optimizing their geometry using HyperChem 7.1 (Hypercube, Inc.). The final model of the KpnI dimer complexed with the GGTACC sequence (available for download from ftp://genesilico.pl/iamb/models/KpnI/) was obtained after steric clashes between a few residues from both monomers and the DNA were removed by selecting different rotamers of the respective side chains.
Generation of site-directed mutants in R.KpnI
The homology model of R.KpnI shown in Figure 1B reveals the ββα-Me finger fold composed of two antiparallel β-strands, an α-helix and a metal ion, between the first strand and the helix. Based on the bioinformatics analysis, amino acid residues D148, H149 and Q175 of R.KpnI are predicted to be functionally equivalent to the catalytic/metal-binding residues D40, H41 and N62 of T4 endonuclease VII (23), and the corresponding residues of other HNH nucleases. Thus, D148, H149 and Q175 were targeted for mutagenesis. The mutations were confirmed by restriction digestion and sequencing analysis (data not shown). The mutant kpnIR genes present in the pET11d vector were expressed in E.coli BL26 and the mutant proteins were purified (as described in Materials and Methods). During purification steps, all the mutants exhibited properties similar to that of wt enzyme, suggesting that they were properly folded.
Figure 1.
(A) Multiple sequence alignment of R.KpnI and the experimentally solved structures of HNH-superfamily nucleases used as modeling templates. The secondary structure of T4 Endonuclease VII is shown above the alignment as tubes (helices) and arrows (strands). Amino acid residues of the active site are highlighted in black, residues involved in formation of a Zn-finger in Endonuclease VII and R.KpnI are highlighted in grey. The numbers in parentheses indicate the size of terminal extensions of the conserved ‘ββα-finger’, which do not share either the sequence or the structure similarity. The arrow and the cylinder represent the β-strand and the α-helix, respectively. (B) Comparison of the modeled structure of the catalytic domain of KpnI (a) and the experimentally solved structures of HNH-superfamily nucleases used as modeling templates (b) T4 Endonuclease VII (1en7), (c) colicin E9 (1bxi), (d) intron-encoded homing endonuclease I-PpoI (1ipp) and (e) Serratia marcenscens endonuclease (1g8t). Amino acid residues of the active site and those involved in formation of a Zn-finger in Endonuclease VII and KpnI are indicated and labeled.
DNA cleavage properties of R.KpnI mutants
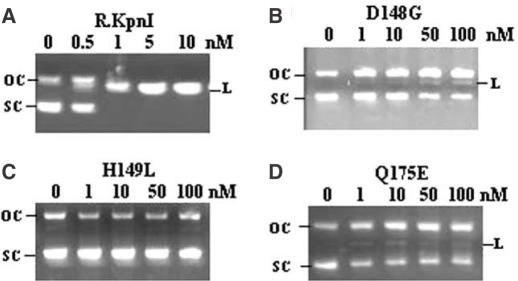
Wild-type R.KpnI and its mutants were assayed for the ability to cleave DNA. As shown in Figure 2, pUC18 DNA was completely linearized by 1 nM wt enzyme. Under the same assay conditions, no cleavage product was detected with the mutant H149L even at 100-fold excess protein concentrations (Figure 2C). Mutant H149A also behaved in the same fashion (See Supplementary Material). Mutants D148G and Q175E showed only traces of the DNA cleavage activity when used in large excess (Figure 2B and D). Similar results were observed with D148A (See Supplementary Material). In order to measure the residual activity more precisely, reactions were performed with higher concentrations of D148G and Q175E. No complete linearization of the DNA substrate could be observed even at 2 μM (2000-fold excess) of the enzyme concentration. However, prolonged incubation for 12 h resulted in near-complete DNA cleavage with D148G, while the pattern with H149L and Q175E did not change significantly under standard assay conditions (data not shown). In contrast, incubation with very low amounts of the wt enzyme (0.05 units) for prolonged incubation leads to complete DNA cleavage. Thus, all three residues of the predicted HNH motif were found to be essential for the REase activity.
Figure 2.
Restriction digestion of pUC18 DNA by R.KpnI and its mutants. Purified enzymes (A) R.KpnI (B) D148G (C) H149L and (D) Q175E mutants were incubated with 500 ng of pUC18 DNA and the products were electrophoresed on 1% agarose gel. The concentration of the enzyme used is depicted in each panel. SC, L and OC indicate the respective position of the supercoiled, linear and open circular forms of plasmid.
DNA-binding ability of R.KpnI mutants
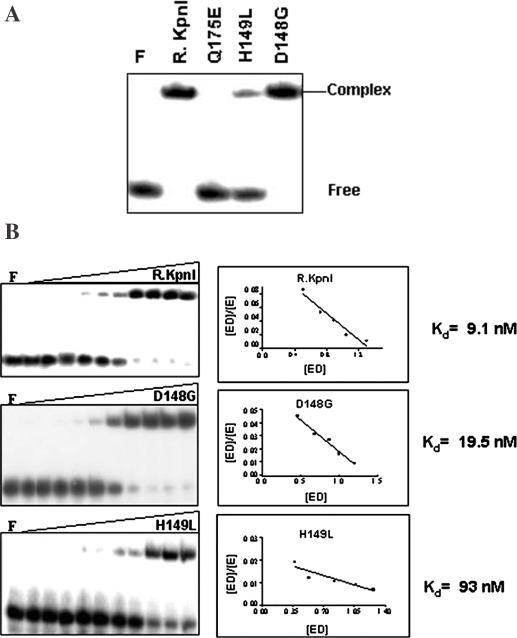
To analyze the cause for decreased or abolished DNA cleavage observed with mutant REases, the DNA binding property was assayed by the electrophoretic mobility shift assays (EMSA) using 32P-labeled 20mer oligonucleotide containing KpnI recognition site. As shown in Figure 3A, D148G and H149L bind DNA. However, the H149L mutant appears to bind DNA poorly in this qualitative assay. Increasing concentrations of mutant proteins were used to calculate the dissociation constant (Kd) for DNA binding. Mutants D148G and H149L bind DNA with 2 and 10 times reduced affinities, respectively, compared to the wt protein (Figure 3B). In electrophoretic mobility shift assays, R.KpnI does not bind to nonspecific DNA substrates (13) and the mutants behaved in similar fashion.
Figure 3.
DNA binding properties of mutant proteins. (A) EMSA analysis: 50 nM of wt and mutant enzymes were incubated with 3.75 nM of 32P-labeled 20mer duplex oligonucleotides containing recognition sequence (Table 1) under standard assay conditions as described in Materials and Methods. (B) Determination of DNA binding affinities of R.KpnI, D148G and H149L. The assays were carried out as above with the increasing amounts of proteins (0, 0.1, 0.5, 1, 2, 4, 8, 16, 32, 64 and 128 nM) to carry out Scatchard analysis. Lane F in each panel refers to oligonucleotides incubated only with EMSA buffer in the absence of protein.
Effect of pH on DNA cleavage of R.KpnI mutants
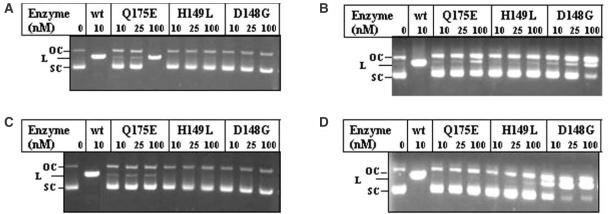
Experiments described above (Figure 2) showed that the mutants D148G and Q175E retained residual DNA cleavage activity. This prompted us to carry out DNA digestion at varied pH conditions to further evaluate their activity at different ionization conditions. R.KpnI exhibits cleavage activity at wide pH range; cleavage of pUC18 was observed from pH 6.0 to 9.0 (Figure 4 and See Supplementary Material). Under these conditions, the mutant proteins displayed differential activities. H149L was not able to cleave the DNA under any of these conditions. D148G showed slightly improved DNA cleavage under alkaline pH, yet the cleavage was not complete even at a 100-fold excess concentration of the enzyme. Surprisingly, the Q175E mutant showed the ability to completely cleave the DNA substrate at pH 6.0 at higher concentrations of the enzyme, while its activity under neutral and alkaline pH was very low (Figure 4). We consider this behavior to be similar to the Q115E mutant of EcoRI (24). It is noteworthy that the deprotonated Q115E mutant of EcoRI was defective in DNA binding at neutral pH. Under acidic pH, however, Q115E is protonated and bound to DNA to carry out efficient cleavage. Similarly, deprotonation of the Glu residue in the Q175E variant of R.KpnI could occur near neutral pH resulting in poor binding and rendering it catalytically inefficient. To verify this hypothesis, we have carried out DNA binding experiment at pH 6.0. The mutant REase Q175E binds to DNA in acidic conditions (Figure 5), unlike its behavior at near-neutral pH. These results confirm that Q175 has an important role in DNA binding coupled to catalysis.
Figure 4.
Effect of pH on DNA cleavage activities of ββα-Me motif mutants of KpnI REase. Increasing amounts of wt, Q175E, H149L and D148G were incubated with pUC18 in buffer with different pH (A) pH 6.0 (B) pH 7.0 (C) pH 8.0 and pH 9.0 (D) at 37°C for 1 h. The samples were electrophoresed through 1% agarose. SC, L and OC indicate the respective positions of the supercoiled, linear and open circular forms of pUC18.
Figure 5.
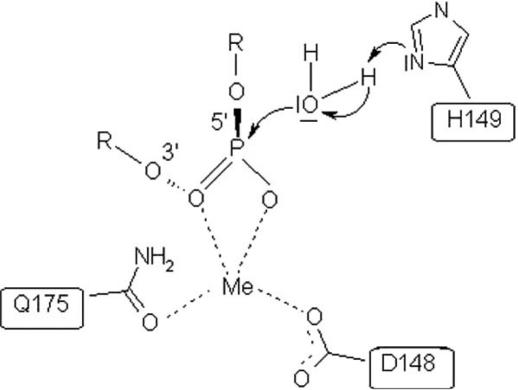
Proposed catalytic mechanism of R.KpnI. Residues D148 and Q175 are shown in this scheme to be involved in metal coordination and H149 acting as a general base. H149 activates the nucleophilic water molecule to facilitate the phosphodiester bond cleavage.
DISCUSSION
To date, a large number of mutational and structural studies have shown that most REases possess the PD…D/EXK motif for metal binding and catalysis (9,25,26). A single exception described for which experimental evidence have been provided, is the Type IIS REase BfiI, which does not contain the PD…DE/XK motif. Instead, it belongs to the phospholipase D superfamily and carries out the DNA cleavage in the absence of divalent metal ions (27). Moreover, bioinformatic analyses suggested that a Type IV, m5C-specific REase McrA, and few Type II REases of different subclasses, including KpnI, HpyI, NlaIII, SphI, SapI, NspHI, NspI and MboII are related to the HNH superfamily (21,28,29), while Eco29kI and its close relatives are members of the GIY-YIG superfamily (28). Thus, motifs other than PD…D/EXK are found in REases, although these predictions remained untested.
We report here the first biochemical evidence for a Type II REase that belongs to the HNH superfamily and exhibits a new type of the catalytic motif based on the ββα-Me finger fold. The HNH nucleases include heterogeneous group of enzymes with varied sequence or structure specificity exhibiting different biological functions. For example, mitochontrial EndoG is a DNA/RNA non-specific nuclease, which employs the mechanism of DNA cleavage similar to that of sequence-specific homing nuclease I-PpoI and non-specific Serratia nuclease (30–32). A typical HNH nuclease, I-HmuI is encoded by group I intron present in the DNA polymerase gene of SPO1 bacteriophage of Bacillus subtilis. The recently solved structure of this single-strand-cleaving enzyme reveal its reaction mechanism to be similar to that of I-PpoI and Serratia nuclease (33). Likewise, the caspase-activated Dnase, another member belonging to ββα-Me finger fold, follows the mechanism of structure-specific T4 endonuclease VII and non-specific colicin E9 (23,34).
Based on the results of our analysis, we propose roles for active site residues of R.KpnI (Figure 6) relying mainly on the mechanism proposed for T4 endonuclease VII (23). The studies presented here indicate that the H149 of KpnI REase might be acting as a general base, which could activate a water molecule for an in-line nucleophilic attack on the scissile phosphate similar to H41 of T4 endonuclease VII. Residues D40 and N62 of T4 endonuclease VII (corresponding to D148 and Q175 in R.KpnI) are involved in metal ion binding (23). D148 of R.KpnI might be acting as a general acid involved in metal binding. Interestingly, Q175 appears to have roles both in metal coordination and DNA binding by R.KpnI. This conclusion is based on the ability of Q175E mutant to bind DNA under acidic condition when it is protonated (See Figure 5).
Figure 6.
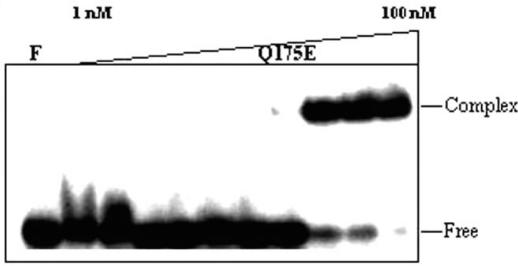
Role of Q175E in DNA binding activity. Different concentrations of Q175E (1, 2.5, 5, 7.5, 10, 15, 20, 25, 50 and 100 nM) were incubated with 3.75 nM of 32P-labeled 20 bp duplex DNA containing KpnI recognition sequence on ice for 15 min at pH 6.0. The products were analyzed on 8% PAGE as described in Materials and Methods.
Another interesting feature of HNH endonucleases is the generation of 3′-overhangs after DNA cleavage, a property shared by R.KpnI. A majority of the HNH nucleases including His–Cys box containing I-PpoI act by cutting both scissile phosphates across the minor groove. In so doing, they exhibit very low specificity at the central four base pairs that are located between these scissile phosphate groups, making specific contacts to bases present beyond the phosphates. Being a REase, R.KpnI should be able to recognize its cognate sequence with higher specificity compared to other diverse HNH nucleases. Accordingly, footprinting results showed that the enzyme makes base-specific contacts with the first three bases (5′-GGT-) of each strand of the recognition sequence 5′-GGTACC-3′ (13), a pattern different from that of I-PpoI. However, the data may not adequately reveal all the specific contacts of the enzyme until its structure with the DNA is solved.
It appears that the majority of REases belong to the PD…D/EXK superfamily, even though a few REases such as BfiI or KpnI, may exhibit alternative, unrelated three-dimensional folds and mechanisms for DNA cleavage (9,27,28). We speculate that the PD…D/EXK fold was more suitable for development of extremely stringent recognition of DNA sequences, characteristic for Type II REases, while other nuclease superfamilies predominantly gave rise to enzymes with more relaxed specificities or preference towards particular DNA structures. For instance T4 EndoVII, the closest homolog of R.KpnI, with known structure, primarily acts as a broad specificity repair enzyme. Other enzymes containing ββα-Me finger fold, such as periplasmic non-specific nuclease Vvn, Serratia nuclease, colicin nuclease E7 and E9 exhibit low sequence specificity (35–38). In support of this view, we have recently demonstrated the relaxed specificity exhibited by R.KpnI (39). Thus, we hypothesize that KpnI and other few members of the HNH superfamily predicted amongst REases developed more stringent sequence specificity only when they teamed up with highly specific DNA MTases and thereby appeared in the context of RM systems relatively late in the evolution.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank R. Kanagaraj and D. R. Radha for help in experiments and other members of the VN lab for discussions. This work was supported by a grant from Department of Science and Technology, Government of India. We thank Alfred Pingoud and International Quality Network programme for discussions. M.S. is a Senior Research Fellow of Council of Scientific and Industrial Research, Government of India. J.M.B. was supported by KBN (grant PBZ-KBN-088/P04/2003), EMBO/HHMI Young Investigator award, and the Fellowship for Young Researchers from the Foundation for Polish Science.
REFERENCES
- 1.Pingoud A. and Jeltsch,A. (1997) Recognition and cleavage of DNA by type II restriction endonucleases. Eur. J. Biochem., 246, 1–22. [DOI] [PubMed] [Google Scholar]
- 2.Kovall R.A. and Mathews,B.W. (1999) Type II restriction endonucleases: structural, functional and evolutionary relationships, Curr. Opin. Chem. Biol., 3, 578–583. [DOI] [PubMed] [Google Scholar]
- 3.Pingoud A. and Jeltsch,A. (2001) Structure and function of type II restriction endonucleases. Nucleic Acids Res., 29, 3705–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roberts R.J. and Halford,S.E. (1993) Type II restriction enzymes. In Linn,S.M., Lloyd,R.S. and Roberts,R.J. (eds), Nucleases. Cold Spring Harbor, NY, pp 35–88. [Google Scholar]
- 5.Grabowski G., Jeltsch,A., Wolfes,H., Maass,G. and Alves,J. (1995) Site-directed mutagenesis in the catalytic center of the restriction endonuclease. Gene, 157, 113–118. [DOI] [PubMed] [Google Scholar]
- 6.Ban C. and Yang,W. (1998) Structural basis for MutH activation in E.coli mismatch repair and relationship of MutH to restriction endonucleases. EMBO J., 17,1526–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hickman A.B., Li,Y., Mathew,S.V., May,E.W., Craig,N.L and Dyda,F. (2000) Unexpected structural diversity in DNA recombination: the restriction endonuclease connection. Mol. Cell., 5, 1025–1034. [DOI] [PubMed] [Google Scholar]
- 8.Bujnicki J.M. (2000) Phylogeny of the restriction endonuclease-like superfamily inferred from comparison of protein structures. J. Mol. Evol., 50, 39–54. [DOI] [PubMed] [Google Scholar]
- 9.Bujnicki J.M. (2004) Molecular phylogenetics of restriction endonucleases. In Pingoud,A. (Ed.) Restriction Endonucleases, Springer-Verlag, Berlin, Heidelberg, Vol. 114, pp. 63–93. [Google Scholar]
- 10.Roberts R.J., Belfort,M., Bestor,T., Bhagwat,A.S., Bickle,T.A., Bitinaite,J., Blumenthal,R.M., Degtyarev,S., Dryden,D.T., Dybvig,K. et al. (2003) A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res., 31, 1805–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomassini J, Roychoudhury,R, Wu,R. and Roberts,R.J. (1978) Recognition sequence of restriction endonuclease KpnI from Klebsiella pneumoniae. Nucleic Acids Res., 5, 4055–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiss A., Finta,C. and Venetianer,P. (1991) M. KpnI is an adenine-methyltransferase. Nucleic Acids Res., 19, 3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandrashekaran S., Manjunatha,U.H. and Nagaraja,V. (2004) KpnI restriction endonuclease and methyltransferase exhibit contrasting mode of sequence recognition. Nucleic Acids Res., 32, 3148–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandrashekaran S., Babu,P. and Nagaraja V. (1999) Characterization of DNA binding activities of over-expressed KpnI REase and modification methylase. J. Biosci., 24, 269–277. [Google Scholar]
- 15.Bradford M.M. (1976) A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 35–88. [Google Scholar]
- 17.Kurowski. M.A. and Bujnicki,J.M. (2003) GeneSilico protein structure prediction meta-server. Nucleic Acids Res., 31, 3305–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kosinski J, Cymerman,I.A., Feder,M., Kurowski,M.A., Sasin,J.M. and Bujnicki,J.M. (2003) A ‘FRankenstein's monster’ approach to comparative modeling: merging the finest fragments of Fold-Recognition models and iterative model refinement aided by 3D structure evaluation. Proteins, 53, 369–379. [DOI] [PubMed] [Google Scholar]
- 19.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirsch R.D. and Joly,E. (1998) An improved PCR mutagenesis strategy for two-site mutagenesis or sequence swapping between related genes. Nucleic Acids Res., 26, 1848–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aravind L., Makarova,K.S. and Koonin,E.V. (2000) Holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res., 28, 3417–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galburt E.A., Chevalier,B., Tang,W., Jurica,M.S., Flick,K.E., Monnat,R.J.,Jr and Stoddard,B.L. (1999) A novel endonuclease mechanism directly visualized for I-PpoI. Nature Struct Biol., 6, 1096–1099. [DOI] [PubMed] [Google Scholar]
- 23.Raaijmakers H., Toro,I., Birkenbihl,R., Kemper,B. and Suck,D. (2001) Conformational flexibility in T4 endonuclease VII revealed by crystallography: implications for substrate binding. J. Mol. Biol., 308, 311–323. [DOI] [PubMed] [Google Scholar]
- 24.Jeltsch A., Alves,J., Oelgeschlager,T., Wolfes,H., Maass,G. and Pingoud,A. (1993) Mutational analysis of the function of Gln115 in the EcoRI restriction endonuclease, a critical amino acid for recognition of the inner thymidine residue in the sequence -GAATTC- and coupling specific DNA binding to catalysis. J. Mol. Biol., 229, 221–234. [DOI] [PubMed] [Google Scholar]
- 25.Bujnicki J.M. (2001) Understanding the evolution of restriction-modification systems: clues from sequence and structure comparisons. Acta Biochim. Pol., 48, 935–967. [PubMed] [Google Scholar]
- 26.Bujnicki J.M. and Rychlewski,L. (2001) Grouping together highly diverged PD-(D/E)XK nucleases and identification of novel superfamily members using structure-guided alignment of sequence profiles. J. Mol. Microbiol. Biotechnol., 3, 69–72. [PubMed] [Google Scholar]
- 27.Sapranauskas R., Sanauskas,G., Lagunavicius,A., Vilkaitis,G., Lubys,A. and Siksnys,V. (2000) Novel subtype of type IIS restriction enzymes, J. Biol. Chem., 275, 30878–30885. [DOI] [PubMed] [Google Scholar]
- 28.Bujnicki J.M., Radlinska,M. and Rychlewski,L. (2001) Polyphyletic evolution of type II restriction enzymes revisited: two independent sources of second-hand folds revealed. Trends Biochem. Sci., 26, 9–11. [DOI] [PubMed] [Google Scholar]
- 29.Bujnicki J.M., Radlinska M. and Rychlewski,L. (2000) Atomic model of the 5-methylcytosine-specific restriction enzyme McrA reveals an atypical zinc-finger and structural similarity to beta-beta-alpha-Me endonucleases. Mol. Microbiol., 37, 1280–1281. [DOI] [PubMed] [Google Scholar]
- 30.Friedhoff P., Franke,I., Meiss,G., Wende,W., Krause,K.L. and Pingoud,A. (1999) A similar active site for non-specific and specific endonucleases. Nature Struct. Biol., 6, 112–113. [DOI] [PubMed] [Google Scholar]
- 31.Kowalski J.C. and Derbyshire,V. (2002) Characterization of homing endonucleases. Methods, 28, 356–373. [DOI] [PubMed] [Google Scholar]
- 32.Schafer P., Scholz., Gimadutdinow,O., Cymerman,I.A., Bujnicki,J.M., Carrillo,A.R., Pingoud,A. and Meiss,G. (2004) Structural and functional characterization of mitochondrial EndoG, a sugar non-specific nuclease which plays an important role during apoptosis. J. Mol. Biol., 338, 217–228. [DOI] [PubMed] [Google Scholar]
- 33.Shen B.W., Landthaler,M., Shub,D.A. and Stoddard,B.L. (2004) DNA binding and cleavage by the HNH homing endonuclease I-HmuI. J. Mol. Biol., 342, 43–56. [DOI] [PubMed] [Google Scholar]
- 34.Scholz S.R., Korn,C., Bujnicki,J.M., Gimadutdinow,O., Pingoud,A. and Meiss,G. (2003) Experimental evidence for a ββα-Me finger nuclease motif to represent the active site of the caspase-activated DNase. Biochemistry, 42, 9288–9294. [DOI] [PubMed] [Google Scholar]
- 35.Li C.L., Hor,L.I., Chang,Z.F., Tsai,L-C., Yan,W.Z. and Yuan,H.S. (2003). DNA binding and cleavage by the periplasmic nuclease Vvn: a novel structure with a known active site. EMBO J., 22, 4014–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolmes B., Franke,I., Friedhoff,P. and Pingoud,A. (1996) Analysis of the reaction mechanism of the non-specific endonuclease of Serratia marcescens using an artificial minimal substrate. FEBS Lett., 397, 343–346. [DOI] [PubMed] [Google Scholar]
- 37.Cheng Y.S., Hsia,K.C., Doudeva,L.G., Chak,K.F. and Yuan,H.S. (2002). The crystal structure of the nuclease domain of colicin E7 suggests a mechanism for binding to double-stranded DNA by the HNH endonucleases. J. Mol. Biol., 324, 227–236. [DOI] [PubMed] [Google Scholar]
- 38.Walker D.C., Georgiou,T., Pommer,A.J., Walker,D., Moore,G.R., Klenthous,C. and James,R. (2002). Mutagenic scan of the H-N-H motif of colicin E9: implications for the mechanistic enzymology of colicinns, homing enzymes and apoptotic endonucleases. Nucleic Acids Res., 30, 3225–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chandrashekaran S., Saravanan,M., Radha,D.R. and Nagaraja,V. (2004) Ca2+ mediated site-specific DNA cleavage and suppression of promiscuous activity of KpnI restriction endonuclease. J. Biol. Chem., in press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.