Abstract
E2F transcription factors play a critical role in the control of cell cycle progression, regulating the expression of genes involved in DNA replication, DNA repair, mitosis, and cell fate. This involves both positive-acting and negative-acting E2F proteins, the latter group including the E2F6 protein, which has been shown to function as an Rb-independent repressor of E2F-target gene transcription. In an effort to better delineate the context of E2F6 function, including the mechanisms of E2F6 functional specificity, we used chromatin immunoprecipitation assays to assess when and with what genes E2F6 associates during a cell cycle. We find that E2F6 associates specifically with the E2F target genes that are activated at G1/S; this interaction occurs during S phase of the cell cycle. In sharp contrast, E2F6 does not bind to E2F-regulated genes activated at G2/M. In the absence of E2F6, E2F4 can bind to the G1/S-regulated promoters and compensate for loss of E2F6 function. Indeed, inhibition of both E2F4 and E2F6 activity results in specific derepression of these genes during S phase. We conclude that E2F6 functions as a repressor of E2F-dependent transcription during S phase and given the specificity for the G1/S-regulated genes, we propose that E2F6 functions to distinguish G1/S and G2/M transcription during the cell cycle.
Keywords: E2F6, transcriptional regulations, G1/S, G2/M, cell cycle control
Numerous studies have demonstrated the role of E2F proteins in the control of genes whose products are essential for DNA replication, differentiation, and cell cycle progression (Dyson 1998; Nevins 1998). In particular, these studies have detailed the importance of E2F proteins in controlling gene expression at the G1/S transition, involving the activation of genes important for S-phase events including DHFR, thymidine kinase, and DNA polymerase. Specifically, overexpression of the E2F1 product was shown to drive quiescent cells into S phase (Johnson et al. 1993; Qin et al. 1994; Shan and Lee 1994) and, in cooperation with an activated ras oncogene, lead to oncogenic transformation of primary rat embryo cells (Johnson et al. 1994). In addition, to this role for E2Fs in controlling S phase, more recent work has also demonstrated a role for E2F activity during G2/M transition (Ishida et al. 2001; Polager et al. 2002; Ren et al. 2002; Zhu et al. 2004).
E2F is a family of structurally related DNA-binding proteins comprised of seven distinct gene products (E2Fs 1-7) (Adams et al. 2000; He et al. 2000; Leone et al. 2000; Dahme et al. 2002; Trimarchi and Lees 2002; Cam and Dynlacht 2003; de Bruin et al. 2003; Di Stefano et al. 2003). The size and complexity of the E2F family of proteins has led to complexity in function with individual E2Fs performing both distinct and overlapping roles in proliferation, apoptosis, and development (Muller et al. 2001; Trimarchi et al. 2001; DeGregori 2002). In general, however, the E2F family of proteins can be divided into three subclasses based on shared functional properties and sequence homologies. E2F1, E2F2, and E2F3a make up one of these subsets. Each of these E2Fs is a strong transcriptional activator that can induce quiescent cells to enter S phase (Lukas et al. 1996; DeGregori et al. 1997; Verona et al. 1997). The accumulation of these E2F proteins is tightly regulated, with essentially no expression in quiescent cells and then a large induction as cells are stimulated to grow. As cells enter mid-to-late G1 many E2F-responsive promoters are bound by E2F1, E2F2, and E2F3a coincident with histone acetylation and gene activation (Takahashi et al. 2000; Giangrande et al. 2004). E2F4, E2F5, and the alternative version of E2F3 termed E2F3b (Adams et al. 2000; Leone et al. 2000), constitute the second subset of E2F family members. They are not regulated by cell growth but instead can be found at nearly equivalent levels in both quiescent and proliferating cells (DeGregori 2002). In contrast to the activating E2Fs, E2F4, E2F5, and E2F3b are mainly involved in the repression of growth promoting E2F-responsive genes. E2F4 and E2F5 complexes recruit histone deacetylase (Dyson 1998; Nevins 1998) or the CtBP corepressor (Meloni et al. 1999) to E2F-responsive promoter elements. Specifically, repression of promoters in quiescent cells has been shown to be associated with recruitment of E2F4 and p130 and low histone acetylation (Takahashi et al. 2000).
E2F6 and E2F7 form a third class of E2Fs. These proteins lack the N-terminal sequences of E2F1-3 as well as the C-terminal domain common to all the other E2F proteins (Morkel et al. 1997; Cartwright et al. 1998; Gaubatz et al. 1998; Trimarchi et al. 1998; de Bruin et al. 2003; Di Stefano et al. 2003). Because of the absence of the activation and Rb-binding domains, E2F6 and E2F7 appear to function as Rb-independent transcriptional repressors (Trimarchi et al. 2001; de Bruin et al. 2003; Di Stefano et al. 2003). While the mechanism of E2F7 mediated repression is still not well understood, it is thought that the biological properties of E2F6 may be mediated through its ability to recruit the polycomb transcriptional repressor complex and histone methyltransferases (Trimarchi et al. 2001; Ogawa et al. 2002). Recent evidence also suggests that E2F6 can repress transcription of target genes independent of histone methylation (Oberley et al. 2003). Moreover, E2F6 appears to form complexes with Mga and Max in cells in G0, suggesting that E2F6 complexes might repress Myc- and Brachyury-responsive genes as well as E2F-responsive ones (Ogawa et al. 2002). Furthermore, other studies by Cartwright et al. (1998) showed that ectopic expression of E2F6 leads to accumulation of cells in S phase, suggesting that E2F6 may play a role in delaying exit of cells out of S phase. To explore the specific role of E2F6 in the control of E2F-dependent transcription, we have used chromatin immunoprecipitation assays to identify the targets of action. We find that E2F6 binds to a subset of E2F target genes during S phase, suggesting a role for this E2F in controlling cell cycle progression by balancing the function of the activating E2Fs. Furthermore, we show that another E2F family member, E2F4, compensates for loss of E2F6 by binding to and repressing E2F target genes during S phase in E2F6-deficient cells.
Results
E2F6 binds to E2F-target promoters during S phase
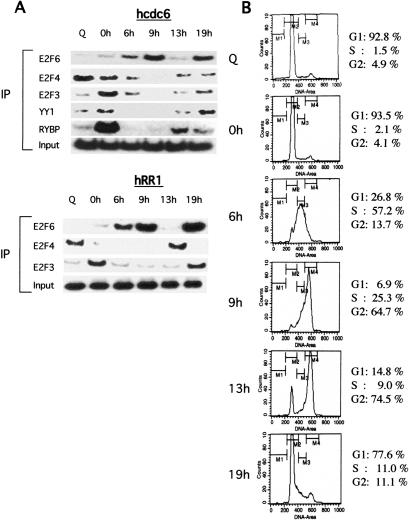
Because our previous work has demonstrated specificity in the interaction of various E2F proteins with E2F-target promoters during the cell cycle, we have carried out chromatin immunoprecipitation (ChIP) assays to measure the interaction of E2F6 with the cdc6 and RR1 promoters as cells progress through the cell cycle. Human neuroblastoma (T98G) cells were harvested either at quiescence or at 0, 6, 13, or 19 h following release from a hydroxy urea (HU) block (Fig. 1). The cell cycle profile was monitored by FACs analysis of propidium-iodide-stained cells (Fig. 1B). As previously demonstrated, E2F3, YY1, and RYBP are seen to associate with the cdc6 promoter during the G1/S phase transition (0 h), coincident with the time of activation of the cdc6 promoter (Fig. 1A, right panel; Schlisio et al. 2002). Similarly, E2F3 could be seen to interact with the RR1 promoter with kinetics similar to those for the interaction with the cdc6 promoter. E2F4 is bound to the promoters in the quiescent cell (Q) and then leaves the promoter as cells transition through G1/S (cf. 0-h, 6-h samples and Q sample), consistent with a role for E2F4 as a repressor of E2F target genes during quiescence. In sharp contrast to these other E2F interactions, E2F6 was observed to interact with both promoters during S phase and G2 of the cell cycle, a time when the transcription of these genes is turned off. Given the previous evidence implicating E2F6 as a transcriptional repressor, these results suggest a role for E2F6 as a specific repressor of E2F-mediated transcription at the S/G2 phase of the cell cycle. We do note that the lack of overlap in RYBP and E2F6 chromatin interaction is in contrast to the previous observation of a physical interaction between the two proteins (Trimarchi et al. 2001). The basis for this disconnect is unclear but may suggest that RYBP does not participate in the E2F6 chromatin interactions, at least as a function of the cell cycle.
Figure 1.
E2F6 binds to E2F-target promoters during G2/M. (A) Chromatin immunoprecipitation assays for interaction of E2F6, E2F4, E2F3, YY1, or RYBP with the hcdc6 promoter (left panel) and hRR1 promoter (right panel) as cells progress through the cell cycle. Human neuroblastoma T98G cells were harvested either at quiescence (Q), or 0, 6, 9, 13, and 24 h following release from a hydroxy urea (HU) block and cross-linked with addition of formaldehyde as described previously (Takahashi et al. 2000). (B) FACS analysis of T98G cells that were synchronized by hydroxyurea (HU) block, and then released into the cell cycle. (Q) Quiescent cells.
E2F4 compensates for loss of E2F6 in E2F6-/- cells
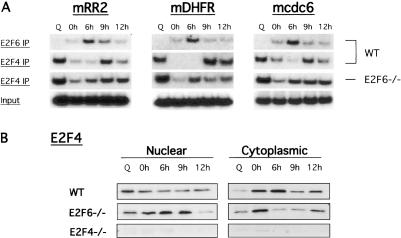
Consistent with a role for E2F6 in repression of genes during S phase, studies by Cartwright et al. (1998) have shown that ectopic expression of E2F6 leads to accumulation of cells in S phase. However, studies of E2F6-/- mouse embryo fibroblasts (MEFs) have revealed no defect in the proliferation ability of these cells, implying that E2F6 is dispensable for proliferation and suggesting that additional E2Fs may compensate for E2F6 deficiency in the E2F6-/- cells (Storre et al. 2002). A candidate for this compensation is E2F4, given its role as a transcriptional repressor. To address this possibility, we carried out ChIP assays for E2F6 and E2F4 in both wild-type and E2F6-/- MEFs. Consistent with the data in Figure 1, E2F6 was bound to the cdc6, RR2, and DHFR promoters during the S/G2 phase of the cell cycle (6 h) in wild-type cells. Again, no E2F6 was found to bind these promoter sequences during quiescence (Q) or in E2F6-/- cells (data not shown). As observed previously in Figure 1, E2F4 bound these promoters during quiescence (Q) and G1 (0 and 9 h-next cell cycle) in wild-type cells (Fig. 2A). However, an immunoprecipitate for E2F4 protein in E2F6-/- cells revealed E2F4 binding to the RR2, DHFR, and cdc6 promoters at quiescence (Q), as well as in S phase (Fig. 2A). The S-phase binding of E2F4 to these promoters in the absence of E2F6 is reminiscent of the binding of E2F6 to these promoters during S phase in wild-type cells. Furthermore, the presence of E2F4 on the RR2, DHFR, and cdc6 promoters during S phase coincides with retention of E2F4 protein in the nucleus in E2F6-/- cells (Fig. 2B, cf. wild-type [WT] and E2F6-/- cells). Together, these results show that in the absence of E2F6 protein, E2F4 is bound to E2F-target promoters in S phase similar to those of E2F6 in wild-type cells, suggesting that E2F4 may be compensating for loss of E2F6.
Figure 2.
E2F4 compensates for loss of E2F6 in E2F6-/- cells. (A) Chromatin immunoprecipitation assays for interaction of E2F6 and E2F4 with the mouse RR2, DHFR, and cdc6 promoters as cells progress through the cell cycle. Mouse embryo fibroblasts (wild-type [WT] or E2F6-/-) were harvested either at quiescence (Q) or 0, 6, 9, and 12 h following release from a hydroxyurea (HU) block and cross-linked with addition of formaldehyde as in Figure 1. (B) Nuclear and cytoplasmic protein extracts of wild-type (WT), E2F6-/-, or E2F4-/- cells. Cells were harvested either at quiescence (Q) or 0, 6, 9, and 12 h following a hydroxyurea block (HU). Cell extracts were resolved in an SDS acrylamide gel and assayed for the presence of mouse E2F4 protein by Western blotting with specific antibodies (see Materials and Methods).
Role for E2F6 in regulating a subset of E2F-target genes
The ChIP experiments in Figures 1 and 2 confirm that E2F6 binds to promoters during S phase of the cell cycle. We reasoned that E2F6 may be acting to specifically repress E2F-target genes that were activated at G1/S. Given the role for E2F proteins in positively regulating gene expression at both G1/S and G2 of the cell cycle, we next asked if there was a specificity in repression of gene expression by E2F6 (Ishida et al. 2001; Polager et al. 2002; Ren et al. 2002; Zhu et al. 2004). ChIP assays were performed for a large series of genes known to be regulated at G1/S or G2/M. Consistent with the data in Figure 2, E2F6 binds to the cdc6 promoter during S phase of the cell cycle (6-9 h). E2F6 was also found to be associated with additional G1/S promoters regulated by E2F (RR1, p68, RR2, PCNA, TS, TK-1) during the S phase (Fig. 3A, top and bottom panels). In contrast, we found no evidence for an association of E2F6 with any of the E2F-regulated G2/M genes (cdc2, cycA2, cycB2, Bub1, Kpna2, cdc20, cycB1) under these conditions (Fig. 3B). Again, consistent with the data in Figure 2, we observe that in the absence of E2F6 protein, E2F4 binds to the E2F-regulated G1/S promoters during S phase of the cell cycle (6-9 h) (Fig. 3A, top panel, cf. 6KO MEFs and wild-type [WT] MEFS), suggesting that E2F4 may be compensating for loss of E2F6 protein to repress genes during S phase. In contrast, no E2F6 protein is bound to these promoters during quiescence in E2F4-/- MEFs (Fig. 3A,B, top panels, cf. 4KO and wild-type [WT] MEFs), suggesting a role for E2F6 as a specific repressor of E2F-target G1/S genes during S phase of the cell cycle.
Figure 3.
Role of E2F6 in regulating a subset of E2F-target genes. (A, top) Chromatin immunoprecipitation assays for the interaction of E2F3, E2F4, or E2F6 with the mouse cdc6, RR1, and p68 promoters as cells progress through the cell cycle. (Bottom) Chromatin immunoprecipitation assays for interaction of E2F3, E2F4, or E2F6 with the mouse RR2, PCNA, TS, and TK-1 promoters as cells progress through the cell cycle. Mouse embryo fibroblasts (wild-type [WT], E2F4-/-, or E2F6-/-) were harvested either at quiescence (Q) or 0, 6, 9, and 12 h following release from a hydroxyurea block and cross-linked with addition of formaldehyde as in Figure 1. (B, top) Chromatin immunoprecipitation assays for the interaction of E2F3, E2F4, or E2F6 with the mouse cdc2 and cycA2 promoters as cells progress through the cell cycle. (Bottom) Chromatin immunoprecipitation assays for interaction of E2F3, E2F4, or E2F6 with the mouse cycB2, Bub1, kpna2, cdc20, and cycB1 promoters as cells progress through the cell cycle. Mouse embryo fibroblasts (wild-type [WT], E2F4-/-, or E2F6-/-) were harvested as above.
E2F6 binds to the same E2F-repressor site as E2F4
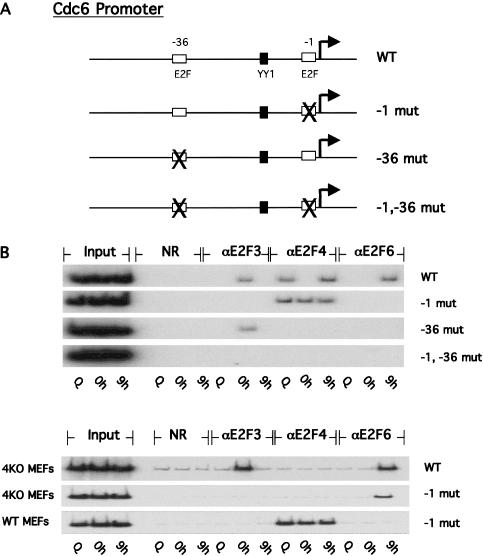
Our recent work has demonstrated a role for both positive and negative E2F-regulatory elements within the cdc6 promoter sequences (Schlisio et al. 2002). Specifically, mutation of the -36 E2F element resulted in a derepression of cdc6 promoter activity in quiescent cells, consistent with a role for E2F in negative control of transcription, particularly in quiescent cells. In contrast, mutation of the -1 E2F element reduced the activation of the promoter following growth stimulation, suggesting a role for the -1 E2F element in positive control of the promoter (Fig. 4A; Schlisio et al. 2002). Based on these results, we concluded that E2F activity, probably an E2F4-Rb complex acting through the -36 E2F element, functions to repress transcription in quiescent cells. In contrast, both E2F3 and YY1 contribute to positive activation of the promoter by binding to the -1 and YY1 sites, respectively (Schlisio et al. 2002). In light of the functional role of the individual E2F elements in the cdc6 promoter, we investigated the relationship between this function and the nature of the E2F proteins that interact with these sites during various stages of the cell cycle. To do so, we have made use of ChIP assays that measure protein-DNA interactions with transfected plasmid sequences rather than with endogenous promoter sequences (Giangrande et al. 2004). Using this modified ChIP assay, we can measure the effect of mutation of a particular element on the protein-DNA interaction profile. Specifically, we used the cdc6 reporter plasmid for these assays and measured the interaction of E2F3, E2F4, and E2F6 with the reporter in either quiescent or G1/S-arrested MEFs that were released into the cell cycle. The promoter mutants used in these assays, which include disruptions in either the -36, -1, or both E2F elements, are shown in Figure 4A.
Figure 4.
E2F6 binds to the same E2F-repressor site as E2F4. (A) Schematic of cdc6 luciferase reporter constructs assayed in the reporter chromatin immunoprecipitation assay. (B) Chromatin immunoprecipitation assay for interactions of E2F3, E2F4, and E2F6 with the cdc6 wild-type (WT) and mutant reporter constructs harboring mutations within the E2F sites at the -1 and -36 positions from the transcription start site (indicated by arrow). (Top panel) wild-type (WT) mouse embryo fibroblasts. (Bottom panel) E2F4-/- mouse embryo fibroblasts.
Samples of wild-type MEFs transfected with either wild-type or mutant cdc6 promoter constructs were assayed for interaction of E2F3, E2F4, or E2F6 proteins with reporter DNA (Fig. 4B). E2F3 bound to the wild-type promoter in G1/S-arrested cells (0 h) as was the case for the endogenous ChIP assays above. Similarly, E2F4 bound to the wild-type reporter plasmid at quiescence and at G2 (9 h) of the cell cycle, but not during G1/S (0 h), while binding of E2F6 to the promoter was again observed only at G2, but not in quiescence. Assays of the mutant promoters revealed that E2F3 bound to the -1 E2F site while the repressor E2Fs (E2F4 and E2F6) bound to the -36 site. Interestingly, the interaction of E2F4 with the cdc6 reporter persisted on the promoter containing the distal site mutation that blocked binding of both E2F3 and E2F6 to DNA (Fig. 4B, top panel, -1 mut). To assess whether binding of E2F6 to the -1 mutant cdc6 reporter was restored in the absence of E2F4 protein, we assayed for interaction of E2F6 protein with the reporter in E2F4-/- MEFs (Fig. 4B, bottom panel). As shown in Figure 4B, E2F6 binds to the wild-type and -1 mutant reporters in the absence of E2F4 protein. Together, these results demonstrate that the activator and repressor E2Fs bind to distinct E2F functional elements within the cdc6 promoter. Furthermore, the results suggest that the activator E2Fs (E2F3) displace the repressor E2F complexes and that distinct repressor E2Fs will bind to the same E2F element on the promoter but during different cell cycle stages.
Role for E2F6 in E2F-target gene expression
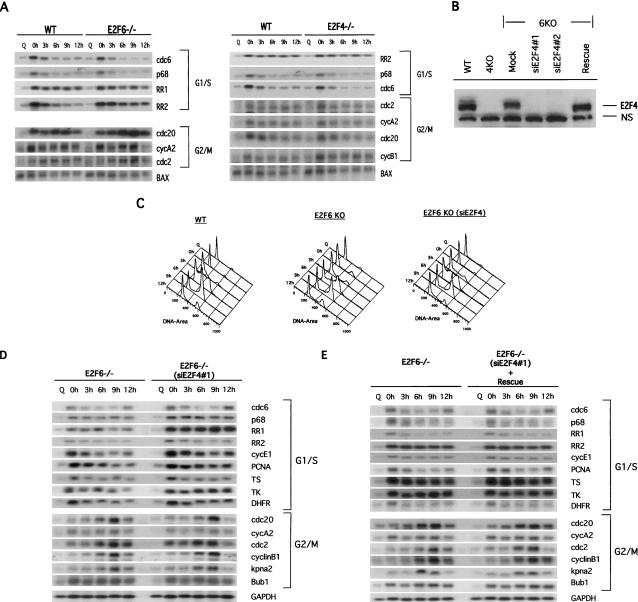
The chromatin binding studies suggest a role for E2F6 as a specific repressor of E2F-target G1/S genes during S phase of the cell cycle. To explore the extent to which the specificity of promoter interactions reflects specificity of transcription control of these genes, we compared expression of a subset of G1/S and G2/M E2F-responsive genes in wild-type MEFs and cells deficient for either E2F4 or E2F6 (Fig. 5A). Quiescent primary MEFs or MEFs blocked at G1/S were stimulated to grow by serum addition, and samples were taken at the indicated times to generate RNA for Northern blot analysis (Fig. 5A). The patterns of expression of these E2F-regulated genes were compared to the levels of BAX, a protein whose expression does not vary dramatically during the cell cycle. FACs analysis of propidium-iodide-stained cells from the same experiment demonstrated no significant difference in the cell cycle profile of wild-type, E2F4-/-, and E2F6-/- MEFs (data not shown). Each of the G1/S genes assayed, exhibited a cell cycle-regulated pattern of expression with peak accumulation occurring at the time of release from a hydroxy urea (HU) block (G1/S) and declined as cells exited S phase (3 h). In contrast, the expression of the G2/M genes tested peaked either at 0 or 6-9 h following release from HU and in all cases remained elevated as cells moved through S/G2 of the cell cycle (Fig. 5A). The expression of the G1/S and G2/M genes was not affected by loss of E2F6. Likewise, the expression patterns were also not altered by the absence of E2F4. The fact that ChIP assays revealed a potential role for E2F4 in compensating for absence of E2F6 function (see Figs. 2, 3) raised the possibility that the lack of an effect on gene expression from loss of either E2F4 or E2F6 alone might be due to compensation.
Figure 5.
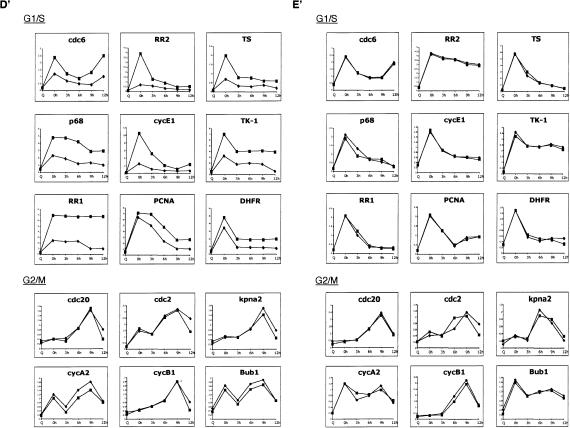
Role of E2F6 in E2F-target gene expression. (A) Wild-type (WT), E2F4-null, and E2F6-null MEFs were synchronized by serum starvation (Q, quiescence) and then stimulated by the addition of serum and 0.5 mM hydroxyurea (HU) to block cells at the G1/S boundary. Cells were then released from the HU block using media containing 15% FBS and allowed to progress through the cell cycle. Cells were harvested for RNA at 0, 3, 6, 9, and 12 h after HU release. Equal amounts of mRNA were subjected to Northern blot analysis to determine the pattern of expression of a number of E2F-responsive genes.(B) Extracts of E2F6-/- cells infected with either Retrovirus alone (Mock), Retrovirus encoding siRNAs against E2F4 (siE2F4#1 and siE2F4#2), or siE2F4#1 along with pCDNA3-E2F4m (Rescue) were resolved on SDS acrylamide gels and assessed for presence of E2F4 protein by Western blotting with specific antibodies. (N.S.) Nonspecific protein. Extracts from wild-type (WT) and E2F4-/- cells (4KO) were included as controls. (C) FACS analysis of wild-type (WT), E2F6-/- (E2F6 KO), and E2F6-/- cells infected with an siE2F4-expressing retrovirus [E2F6 KO(siE2F4)]. Cells were synchronized by a hydroxyurea (HU) block, and then released into the cell cycle. (Q) Quiescent cells. Time points were collected at 0, 3, 6, 9, and 12 h after release from HU block. (D) Northern blot analysis of E2F6-/- cells transfected with either a pool of four nonspecific siRNAs (control) or an siRNA specific to E2F4 (siE2F4#1). Cells were synchronized by serum starvation (Q, quiescence) and then stimulated by the addition of serum and 0.5 mM hydroxyurea (HU) to block cells at the G1/S boundary. Cells were then released from the HU block using media containing 15% FBS and allowed to progress through the cell cycle. Cells were harvested for RNA at 0, 3, 6, 9, and 12 h after HU release. Equal amounts of mRNA were subjected to Northern blot analysis to determine the pattern of expression of a number of E2F-responsive genes. (D′) The expression level of the genes shown in D was quantitated by PhosphorImager analysis and normalized to the GAPDH control. E2F6-/- (diamond); siE2F4#1 (square). (E) Northern blot analysis of E2F6-/- cells transfected with either a pool of four nonspecific siRNAs (control), or siE2F4 plus an expression plasmid for human E2F4 (Rescue). The human E2F4 cDNA was mutated to prevent degradation of exogenous E2F4 mRNA and therefore depletion of E2F4 protein levels. Cells were treated as above and harvested for RNA at 0, 3, 6, 9, and 12 h after HU release. Equal amounts of mRNA were subjected to Northern blot analysis to determine the pattern of expression of several E2F-responsive genes. (E′) The expression level of the genes shown in E was quantitated by PhosphorImager analysis and normalized to the GAPDH control. (Diamond) E2F6-/-; (square) Rescue.
To determine whether E2F4 was, indeed, responsible for repressing E2F-responsive genes during S phase in the absence of E2F6, we performed Northern blot analysis using E2F6-null cells that had been depleted of E2F4 protein using siRNAs specific to E2F4 (Fig. 5B-E′). As shown in Figure 5B, two different siRNAs against E2F4, siE2F4#1 and siE2F4#2, both down-regulate E2F4 protein levels, whereas, a control siRNA (Mock) has no effect on E2F4 protein. Moreover, E2F4 protein levels were restored in cells transfected with an E2F4 expression plasmid harboring mutations that conferred resistance to the siE2F4s (Rescue) (Fig. 5B). MEFs deficient for either E2F6 alone or both E2F6 and E2F4 proteins were treated as in Figure 5A to generate RNA for Northern blot analysis (Fig. 5D). As a control for these experiments, MEFs deficient for both E2F6 and E2F4 proteins were transfected with a mutant E2F4 construct to rescue E2F4 deficiency (Fig. 5E). The pattern of expression of both E2F-responsive G1/S and G2/M genes were normalized to GAPDH, a protein whose expression does not vary dramatically during the cell cycle (Fig. 5D′,E′). FACs analysis of propidium-iodide-stained cells from the same experiment showed no significant difference in the cell cycle profile of wild-type, E2F6-/-, and E2F6-/- (siE2F4#1) MEFs (Fig. 5C). As observed previously in Figure 5A, the E2F-responsive G1/S genes exhibit a pattern of expression with peak accumulation at 0 h following release from HU, in E2F6-/- cells (Fig. 5D,E). The expression of these genes was deregulated in cells lacking both E2F6 and E2F4 proteins, with genes being derepressed at G1/S (0 h) and remaining elevated during S/G2 transition (6-9 h). In contrast, there was no change in the expression of G2/M genes under these same conditions. The deregulated expression of the E2F-responsive G1/S genes was restored by addition of the rescue plasmid encoding an E2F4 mutant transcript that is resistant to the siE2F4s (Fig. 5E,E′). Together, these data confirm that E2F6 represses E2F-responsive G1/S genes during S-phase exit and that E2F4 is the key factor that compensates for E2F6 deficiency to repress G1/S genes.
Although it is true that in most cases the overall pattern of accumulation of the G1/S-regulated genes is maintained in the double deficient cells, it is also true that the level in S phase is consistently elevated over the level at G1/S in the wild-type cells. The fact that there was no evidence for deregulation in either the E2F4-null or E2F6-null cells, but that there was deregulation in the double null cells, says that both E2F4 and E2F6 are contributing to this deregulation. We would attribute the elevated expression at G1/S to E2F4 given previous work pointing to a role for E2F4 in the transition from G0 to G1/S leaving the elevation in expression during S phase to the effects of E2F6. The overlap in function and compensation that is evident for E2F4 and E2F6 precludes a separation of these events to allow a focus on the specific role of E2F6. We also note that the recently described role for E2F7 activity in S phase (Di Stefano et al. 2003) further complicates the situation and may be partially responsible for the lack of a dramatic effect in S phase.
Discussion
A variety of studies have now documented the central role for E2F activities in regulating gene expression in relation to cell growth. This involves not only the control of transcription as cells transition out of a quiescent state and into the growth cycle but also the control of transcription as cells cycle during a growth state. These studies have defined roles for E2F proteins, together with Rb proteins, in the repression of transcription in the quiescent cell, in the activation of transcription of a large number of genes at G1/S to generate the activities for DNA replication and further cell cycle progression, and the activation of transcription at G2/M to generate activities necessary for mitosis (Dyson 1998; Nevins 1998; DeGregori 2002). The experiments we report here now point to a novel role for the E2F6 protein in this process, acting to repress transcription of the G1/S-regulated genes as cells move through S phase. By so doing, it would appear that E2F6 plays an important role in defining the distinction in E2F-dependent gene activation at G1/S and G2/M, thus establishing part of the mechanistic basis that generates the temporal control of gene expression during a cell cycle.
We do note previous work that suggested a different role for E2F6 in the control of cell growth-regulated transcription. Ogawa et al. (2002) reported binding of E2F6 to a subset of E2F-regulated promoters during G0 of the cell cycle. We also note that in the study of Ogawa et al. (2002), neither E2F4 nor p130 was found to be promoter-associated in G0 cells but then were bound at G1. This result is in contrast to both our findings as well as the work reported by Dynlacht and colleagues that describe a G0-specific interaction for E2F4 and p130 that is then reduced as cells enter the cell cycle (Takahashi et al. 2000). While the precise explanation for this distinction is unclear, the drastic difference in the nature of the interactions suggests that cell-type distinctions or different conditions for G0 induction could be responsible.
Specificity of E2F6 function
The fact that E2F6 shows specificity in the interaction with G1/S-regulated promoters, despite the presence of E2F-binding sites in both G1/S and G2/M promoters, raises the question of how this specificity is achieved. Our previous work has provided evidence for protein-protein interactions, involving other promoter-specific factors, as underlying the ability of particular E2F proteins to recognize specific promoters. These include the interaction of E2F3 with the E-box-binding TFE3 factor to activate the DNA pol α p68 promoter (Giangrande et al. 2003, 2004) and E2F2 or E2F3 acting together with the YY1 transcription factor, via an interaction with the RYBP protein, to activate cdc6 transcription (Schlisio et al. 2002). In addition, our more recent work has now extended this concept to the G2/M-regulated genes by demonstrating that the ability of each of the activator E2Fs to interact with the cdc2 promoter is dependent on the CCAAT element that binds the NF-Y transcription factor (Zhu et al. 2004). We have further shown that the interaction of the E2F4 protein with the cdc2 promoter, which coincides with a repression of cdc2 promoter activity, is dependent on an adjacent CHR element, also shown to be critical for repression (Zhu et al. 2004). As such, the ability of an activator or a repressor E2F to interact with a specific element would be dictated by the presence of the partner protein. Taken together, these previous studies would suggest that the mechanism by which E2F6 is specifically recruited to G1/S promoters may involve an interaction with one or more additional factors that allows specific complex formation and binding to these promoters.
Mechanisms of action
A common theme found from simple to complex organisms is the presence of multiple E2Fs that carry out distinct functions of transcription activation and transcription repression. As the organism becomes more complex, so does the complexity of the E2F proteins performing these roles. In some instances, this complexity appears to reflect tissue-specific roles for the proteins such as the case for E2F5 in the choroid plexus (Lindeman et al. 1998) and E2F2 and E2F7 in lymphoid tissue (de Bruin et al. 2003). The evolution of E2F6 and E2F7 may define yet another class—E2F repressors that function independently of Rb. The fact that E2F6 and E2F7 lack C-terminal sequences responsible for activation of transcription and interaction with Rb family members, has suggested a role for these E2Fs as Rb-independent transcriptional repressor. Indeed, functional assays have provided evidence for an ability of E2F6 to repress transcription in an Rb-independent manner (Morkel et al. 1997; Cartwright et al. 1998; Gaubatz et al. 1998; Trimarchi et al. 1998). While a mechanism for E2F6-mediated repression appears to involve recruitment of polycomb protein-containing complexes, the manner in which E2F7 represses is still poorly understood since E2F7 lacks the domain involved in binding RYBP/polycomb proteins. Nevertheless, both E2F6 and E2F7 do appear to play a role in S phase and, by extension to the work we present here, may both be involved in defining the distinction between G1/S- and G2/M-regulated transcription.
Distinguishing G1/S and G2/M transcription
Various experiments have documented the role of E2F proteins in the control of G2/M transcription (Ishida et al. 2001; Polager et al. 2002; Ren et al. 2002) as well as the previously documented role in controlling genes at G1/S. Our recent work has provided insight into the mechanisms that define this distinction and that establish a temporal relationship between events at G1/S and G2/M. Analysis of the promoters of the G2-regulated cdc2 and cyclin B1 genes has demonstrated the presence of both positive- and negative-acting E2F elements as well as additional promoter elements that provide both positive and negative control (Zhu et al. 2004). One of the positive elements is a binding site for the Myb transcription factor. Myb is expressed at G1/S in an E2F-dependent manner and then subsequently associates with and contributes to the activation of G2/M genes. Importantly, the ability of Myb to interact with the cdc2 promoter requires E2F, thus linking the control of gene expression at G1/S with control at G2/M. The results we present here, which define E2F6 as a specific repressor of E2F-regulated G1/S genes, now provide yet a further dimension to the distinction in G1/S and G2/M control involving E2F activities. A role for E2F6 can be seen as ensuring that the G1/S transcription is efficiently extinguished following the activation of these genes but, at the same time, allowing the G2/M group of genes to be activated. By so doing, the action of E2F6 would help to further define the distinction in expression at these two points in the cell cycle.
Materials and methods
Cell culture and FACs analysis
Human glioblastoma T98G cells (ATCC) were maintained in DMEM containing 10% fetal bovine serum (FBS). E2F6-/- MEFs were a gift from Stefan Gaubatz (Philipps University, Marburg, Germany). Primary and established mouse embryo fibroblasts (MEFs) were maintained in DMEM containing 15% heat-inactivated FBS. Null MEFs, of a given genotype, were compared to MEFs generate from wild-type 13.5-embryonic-day-old (E13.5) littermates. Cell samples for FACS analysis were harvested by trypsinization, and then resuspended in 1 mL of nuclear isolation buffer (1× PBS, 0.5% BSA, 0.1% NP-40) with 10 μL of propidium iodide (0.5 mg/mL) and 10 μL of RNase A (10 mg/mL). Flow cytometry (FACS) analysis was carried out to confirm the cell cycle synchrony (Duke Cancer Center flow cytometry facility).
Transfection assays
Transient transfection assays were performed using Superfect Reagent (QIAGEN) as previously described (Giangrande et al. 2003). All transient transfections were carried out in triplicate. Passage 23 E2F6-/- MEFs were plated at a density of 6 × 105 on 60-mm plates 24 h before transfection with the siRNAs. The siRNAs to E2F4 were a gift from B. Sullenger (Duke University, Durham, NC). The nonspecific siRNA pool (M-006368-00), purchased from Dharmacon, was used as a negative control for these experiments (Mock). The pCDNA3-HAE2F4 expression plasmid for human E2F4 was mutated to generate a mutant E2F4 transcript (E2F4m) that would not be targeted and degraded by siE2F4#1 using site-directed mutagenesis (GeneEditor Kit; Promega). E2F4m was cotransfected into E2F6-/- MEFs previously infected with the siE2F4#1 retrovirus in order to reconstitute E2F4 protein levels. Transfection assays were performed with Superfect Reagent as detailed above. Transfected MEFs were then lysed as detailed below, and extracts were resolved on SDS-PAGE gels for immunoblotting.
siE2F4#1 retrovirus
The retrovirus vectors for siRNA expression were derived from pSuper.retro (OligoEngine). This hairpin produces an shRNA that is analogous to the siE2F4#1 duplex described above. Retroviruses were packaged using Plat-E cells (Morita et al. 2000). The supernatants were collected 36 h post-transfection and used for the infection of E2F6-/- MEFs. Infected MEFs were selected and maintained in DMEM/15% FBS containing 3 μg/mL puromycin.
Western blot assay
Nuclear and cytoplasmic extracts of wild-type, E2F4-/-, or E2F6-/- MEFs containing equal amounts of total protein were boiled for 5 min in protein sample buffer and subjected to SDS-PAGE on 8.5% polyacrylamide gels. Proteins were subsequently transferred onto a PVDF membrane and blocked in PBS containing 5% skim milk for 2 h. Blots were then incubated with an antibody to E2F4 (1:1000 dilution; Santa Cruz; SC-1082) in PBS containing 5% milk overnight at 4°C, washed with PBS plus 0.05% NP-40, and incubated in PBS plus 0.05% NP-40 and a rabbit secondary antibody (1:5000 dilution) for 1 h at room temperature. Blots were processed with Amersham's ECL system as described by the manufacturer.
Northern blot analysis
Passage 4 wild-type, E2F4-/-, and E2F6-/- null MEFs, derived from E13.5 littermates, as well as passage 20 E2F6-/- MEFs infected with either Retrovirus alone (Mock), Retrovirus encoding an siRNA against E2F4 (siE2F4#1), or Retrovirus encoding an siRNA against E2F4 (siE2F4#1) plus a mammalian expression plasmid for E2F4m (Rescue) were brought to quiescence by contact inhibition for 24 h following serum deprivation for an additional 24 h. Cells were then blocked at the G1/S boundary by the addition of medium containing 0.5 mM hydroxy urea (HU) for 18 h. Cells were subsequently released from the HU block by addition of fresh medium. Total RNA was isolated using RNeasy Kit (QIAGEN). The Oligotex mRNA Kit (QIAGEN) was then used to purify mRNA from total RNA. Approximately 2 μg of mRNA for each time point was separated by agarose gel electrophoresis under denaturing conditions, transferred to the GeneScreen membrane (NEN Life Science), and probed with various probes labeled with 32P as described previously (DeGregori et al. 1995). Mouse cDNA fragments used as probes were either made from Image Clones purchased from ResearchGenetics, Inc. or obtained by restriction digests of full-length cDNAs: RR1, mouse EcoRI fragment; RR2, 600-bp mouse EcoRI fragment; TK-1, human BamHI fragment; E2F1, 1.3-kb mouse EcoRI/XhoI fragment; E2F2, human ScaI/PstI fragment; DHFR, hamster BamHI fragment; GAPDH, 780-bp PstI/XbaI fragment.
Chromatin immunoprecipitation assays
We performed chromatin immunoprecipitations using a modification of a previously published method (Takahashi et al. 2000). Immunoprecipitates were incubated with 4 μg each of antibodies against E2F3 (Santa Cruz; SC-879), E2F4 (Santa Cruz; SC-1082), or E2F6 (Santa Cruz; SC-8175 and SC-8366) at 4°C overnight. Decross-linking of chromatin was performed as described, and samples were analyzed by semiquantitative PCR (Takahashi et al. 2000; Giangrande et al. 2004). Twenty-eight cycles of PCR were performed in 25 μL with 5 μL of immunoprecipitated material, 50 pmol of each primer set, 0.5 units of Taq Gold DNA polymerase (Applied Biosystems), and 0.01 mCi of 32P-dGTP or 32P-dCTP. We used EZ Retrieve (http://siriusb.umdnj.edu:18080/EZRetriev) to access human and mouse promoter sequences. To amplify E2F-responsive promoter regions, the following primer sets were designed to mouse promoter sequences between -500 and +50 for the following genes: p68 (Unigene ID: Mm.320); cdc6 (Unigene ID: Mm.20912); RR1 (Unigene ID: Mm.656); RR2 (Unigene ID: Mm.99); PCNA (Unigene ID: Mm.7141); TK-1 (Unigene ID: Mm.2661); DHFR (Unigene ID: Mm.23695); TS (Unigene ID: Mm.5879); cdc2 (Unigene ID: Mm.281367); cyclin A2 (Unigene ID: Mm.4189); cyclin B1 (Unigene ID: Mm.60114); cyclin B2 (Unigene ID: Mm.22592); Bub1 (Unigene ID: Mm.2185); Kpna2 (Unigene ID: Mm.12508); cdc20 (Unigene ID: Mm.289740). The cdc6 and RR1 primer sequences specific for the human cdc6 and RR1 promoters, respectively, have been described previously (Schlisio et al. 2002). The primer sequences for the above promoter regions are available upon request. PCR products were electrophoresed on 6% polyacrylamide gels. Each experiment was performed at least three independent times, and representative data are shown.
Reporter chromatin immunoprecipitation
Wild-type and E2F4-/- mouse embryo fibroblasts were transiently transfected with cdc6-reporter plasmids using Superfect Reagent (QIAGEN) as detailed above. The cdc6-reporter plasmids have been previously described (Schlisio et al. 2002). Briefly, for a 150-mm culture dish, 30 μg of cdc6-reporter plasmid, 5 μg of pRL-renilla, and 5 μg of pCMV-bgal were mixed with 60 μL of Superfect reagent and 700 μL of OPTI-MEM medium as previously described (Giangrande et al. 2004). The Superfect-DNA mix was then added to cells. After 24 h, the cells were split 1:3 in medium containing 0.2% FBS to render them quiescent. Cells were harvested either at quiescence, or at different time points after an HU block. Cells were cross-linked and prepared for chromatin immunoprecipitation as described previously (Giangrande et al. 2004). After cell nuclei were collected, soluble nuclear material was extracted by incubation in 1× RIPA (50 mM HEPES at pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxychlate) for 30 min on ice. Soluble nuclear material containing reporter plasmids was collected, sonicated for 20 sec., and then used for immunoprecipitation of protein associated with plasmid DNA. Immunoprecipitations were performed with 4 μg of anti-E2F3 (Santa Cruz; SC-879), anti-E2F4 (Santa Cruz; SC-1082), anti-E2F6 (Santa Cruz; SC-8175 and SC-8366), or normal rabbit serum (Pierce), and collected with 20 μL of 50% Protein A/G Agarose bead slurry (Oncogene). Associated plasmid DNA material was collected, cleaned, and subjected to analysis by semiquantitative PCR. PCR for pRL-renilla and pCMV-βgal plasmids served as internal controls for transfection efficiency and specificity of immunoprecipitation (data not shown). PCR primers specific for the cdc6-Luc reporters were developed and used to detect associated reporter plasmid in each immunoprecipitation. Primer sequences are as follows: Cdc6 (5′-GAAAGGCTCTGTGACTACAGCCA-3′, a pGL2-basic specific primer GLprimer2; Promega); Renilla (5′-GGAAACGGATGATAACTGGTC-3′, 5′-TGCCCATACCAATAAGGTCTGG-3′); β-gal (5′-ACTGGCAGATGCACGGTTACGATG-3′, 5′-CACATCTGAACTTCAGCCTCCAG-3′).
Acknowledgments
We thank Bruce Sullenger for his gift of reagents. We also thank Kaye Culler for help with the preparation of the manuscript and Laszlo Jakoi and Ginger Ehmann for technical assistance. J.R.N. is an Investigator of the Howard Hughes Medical Institute. P.H.G. and W.Z. were supported by the Howard Hughes Medical Institute.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1239304.
References
- Adams M.R., Sears, R., Nuckolls, F., Leone, G., and Nevins, J.R. 2000. Complex transcriptional regulatory mechanisms control expression of the E2F3 locus. Mol. Cell. Biol. 20: 3633-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam H. and Dynlacht, B.D. 2003. Emerging roles for E2F: Beyond the G1/S transition and DNA replication. Cancer Cell 3: 311-316. [DOI] [PubMed] [Google Scholar]
- Cartwright P., Muller, H., Wagener, C., Holm, K., and Helin, K. 1998. E2F-6: A novel member of the E2F family is an inhibitor of E2F-dependent transcription. Oncogene 17: 611-623. [DOI] [PubMed] [Google Scholar]
- Dahme T., Wood, J., Livingston, D.M., and Gaubatz, S. 2002. Two different E2F6 proteins generated by alternative splicing and internal translation initiation. Eur. J. Biochem. 269: 5030-5036. [DOI] [PubMed] [Google Scholar]
- de Bruin A., Maiti, B., Jakoi, L., Timmers, C., Buerki, R., and Leone, G. 2003. Identification and characterization of E2F7, a novel mammalian E2F family member capable of blocking cellular proliferation. J. Biol. Chem. 278: 42041-42049. [DOI] [PubMed] [Google Scholar]
- DeGregori J. 2002. The genetics of the E2F family of transcription factors: Shared functions and unique roles. Biochem. Biophys. Acta 1602: 131-150. [DOI] [PubMed] [Google Scholar]
- DeGregori J., Leone, G., Ohtani, K., Miron, A., and Nevins, J.R. 1995. E2F1 accumulation bypasses a G1 arrest resulting from the inhibition of G1 cyclin-dependent kinase activity. Genes & Dev. 9: 2873-2887. [DOI] [PubMed] [Google Scholar]
- DeGregori J., Leone, G., Miron, A., Jakoi, L., and Nevins, J.R. 1997. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc. Natl. Acad. Sci. 94: 7245-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano L., Jensen, M.R., and Helin, K. 2003. E2F7, a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. EMBO J. 22: 6289-6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson N. 1998. The regulation of E2F by pRB-family proteins. Genes & Dev. 12: 2245-2262. [DOI] [PubMed] [Google Scholar]
- Gaubatz S., Wood, J.G., and Livingston, D.M. 1998. Unusual proliferation arrest and transcriptional control properties of a newly discovered E2F family member, E2F-6. Proc. Natl. Acad. Sci. 95: 9190-9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giangrande P.H., Hallstrom, T.C., Tunyaplin, C., Calame, K., and Nevins, J.R. 2003. Identification of the E box factor TFE3 as a functional partner for the E2F3 transcription factor. Mol. Cell. Biol. 23: 3707-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giangrande P., Zhu, W., Rempel, R.E., Laakso, N., and Nevins, J.R. 2004. Combinatorial gene control involving E2F and E box family members. EMBO J. 23: 1336-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Armanious, M.K., Thomas, M.J., and Cress, W.D. 2000. Identification of E2F-3B, an alternative form of E2F-3 l acking a conserved N-terminal region. Oncogene 19: 3422-3433. [DOI] [PubMed] [Google Scholar]
- Ishida S., Huang, E., Zuzan, H., Spang, R., Leone, G., West, M., and Nevins, J.R. 2001. Role for E2F in the control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 21: 4684-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.G., Schwarz, J.K., Cress, W.D., and Nevins, J.R. 1993. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature 365: 349-352. [DOI] [PubMed] [Google Scholar]
- Johnson D.G., Cress, W.D., Jakoi, L., and Nevins, J.R. 1994. Oncogenic capacity of the E2F1 gene. Proc. Natl. Acad. Sci. 91: 12823-12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone G., Nuckolls, F., Ishida, S., Adams, M., Sears, R., Jakoi, L., Miron, A., and Nevins, J.R. 2000. Identification of a novel E2F3 product suggests a mechanism for determining specificity of repression by Rb proteins. Mol. Cell. Biol. 20: 3626-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeman G.J., Dagnino, L., Gaubatz, S., Xu, Y., Bronson, R.T., Warren, H.B., and Livingston, D.M. 1998. A specific, nonproliferative role for E2F-5 in choroid plexus function revealed by gene targeting. 12: 1092-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas J., Petersen, B.O., Holm, K., Bartek, J., and Helin, K. 1996. Deregulated expression of E2F family members induces S-phase entry and overcomes p16INK4A-mediated growth suppression. Mol. Cell. Biol. 16: 1047-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meloni A.R., Smith, E.J., and Nevins, J.R. 1999. A mechanism for Rb/p130-mediated transcription repression involving recruitment of the CtBP corepressor. Proc. Natl. Acad. Sci. 96: 9574-9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S., Kojima, T., and Kitamura, T. 2000. Plat-E: An efficient and stable system for transient packaging of retroviruses. Gene Ther. 7: 1063-1066. [DOI] [PubMed] [Google Scholar]
- Morkel M., Wenkel, J., Bannister, A.J., Kouzarides, T., and Hagemeier, C. 1997. An E2F-like repressor of transcription. Nature 390: 567-568. [DOI] [PubMed] [Google Scholar]
- Muller H., Bracken, A.P., Vernell, R., Moroni, M.C., Christians, F., Grassilli, E., Prosperini, E., Vigo, E., Oliner, J.D., and Helin, K. 2001. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes & Dev. 15: 267-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins J.R. 1998. Toward an understanding of the functional complexity of the E2F and Retinoblastoma families. Cell Growth & Diff. 9: 585-593. [PubMed] [Google Scholar]
- Oberley M.J., Inman, D.R., and Farnham, P.J. 2003. E2F6 negatively regulates BRCA1 in human cancer cells without methylation of histone H3 on lysine 9. J. Biol. Chem. 278: 42466-42476. [DOI] [PubMed] [Google Scholar]
- Ogawa H., Ishiguro, K., Gaubatz, S., Livingston, D.M., and Nakatani, Y. 2002. A complex with chromatin modifiers that occupies E2F and Myc responsive genes in G0. Science 296: 1132-1136. [DOI] [PubMed] [Google Scholar]
- Polager S., Kalma, Y., Berkovich, E., and Ginsberg, D. 2002. E2Fs up-regulate expression of genes involved in DNA replication, DNA repair and mitosis. Oncogene 21: 437-446. [DOI] [PubMed] [Google Scholar]
- Qin X.-Q., Livingston, D.M., Kaelin, W.G., and Adams, P.D. 1994. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc. Natl. Acad. Sci. 91: 10918-10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B., Cam, H., Takahashi, Y., Volkert, T., Terragni, J., Young, R.A., and Dynlacht, B.D. 2002. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes & Dev. 16: 245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlisio S., Halperin, T., Vidal, M., and Nevins, J.R. 2002. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 21: 5775-5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan B. and Lee, W.-H. 1994. Deregulated expression of E2F-1 induces S-phase entry and leads to apoptosis. Mol. Cell. Biol. 14: 8166-8173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storre J., Elsasser, H.P., Fuchs, M., Ullmann, D., Livingston, D.M., and Gaubatz, S. 2002. Homeotic transformations of the axial skeleton that accompany a targeted deletion of E2f6. EMBO Rep. 3: 695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Rayman, J.B., and Dynlacht, B.D. 2000. Analysis of promoter binding by the E2F and Rb families in vivo: Distinct E2F proteins mediate activation and repression. Genes & Dev. 14: 804-816. [PMC free article] [PubMed] [Google Scholar]
- Trimarchi J.M. and Lees, J.A. 2002. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell. Biol. 3: 11-20. [DOI] [PubMed] [Google Scholar]
- Trimarchi J.M., Fairchild, B., Verona, R., Moberg, K., Andon, N., and Lees, J.A. 1998. E2F-6, a member of the E2F family that can behave as a transcriptional repressor. Proc. Natl. Acad. Sci. 95: 2850-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi J.M., Fairchild, B., Wen, J., and Lees, J.A. 2001. The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc. Natl. Acad. Sci. 98: 1519-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verona R., Moberg, K., Estes, S., Starz, M., Vernon, J.P., and Lees, J.A. 1997. E2F activity is regulated by cell cycle-dependent changes in subcellular localization. Mol. Cell. Biol. 17: 7268-7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Giangrande, P.H., and Nevins, J.R. 2004. E2Fs link the control of G1/S and G2/M transcription. EMBO J. (in press). [DOI] [PMC free article] [PubMed]