Abstract
MicroRNAs (MIRs) are a novel group of conserved short ∼22 nucleotide-long RNAs with important roles in regulating gene expression. We have established a MIR-specific oligonucleotide microarray system that enables efficient analysis of the expression of the human MIRs identified so far. We show that the 60-mer oligonucleotide probes on the microarrays hybridize with labeled cRNA of MIRs, but not with their precursor hairpin RNAs, derived from amplified, size-fractionated, total RNA of human origin. Signal intensity is related to the location of the MIR sequences within the 60-mer probes, with location at the 5′ region giving the highest signals, and at the 3′ end, giving the lowest signals. Accordingly, 60-mer probes harboring one MIR copy at the 5′ end gave signals of similar intensity to probes containing two or three MIR copies. Mismatch analysis shows that mutations within the MIR sequence significantly reduce or eliminate the signal, suggesting that the observed signals faithfully reflect the abundance of matching MIRs in the labeled cRNA. Expression profiling of 150 MIRs in five human tissues and in HeLa cells revealed a good overall concordance with previously published results, but also with some differences. We present novel data on MIR expression in thymus, testes, and placenta, and have identified MIRs highly enriched in these tissues. Taken together, these results highlight the increased sensitivity of the DNA microarray over other methods for the detection and study of MIRs, and the immense potential in applying such microarrays for the study of MIRs in health and disease.
The study of noncoding RNAs, and especially of small noncoding RNAs, has gained increasing attention in recent years. Among the noncoding RNAs, microRNAs (MIRs) comprise an evolutionary conserved group with important regulatory roles (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001; for review, see Bartel 2004). MIRs are ∼22 nucleotide long single-stranded RNAs that regulate the expression of target genes by interacting with complementary sites in the 3' UTR of the target mRNAs (Bartel 2004).
In the nucleus, DNA-encoding microRNAs is first transcribed as long primary transcripts (pri-MIR), probably by RNA-pol II (Lee et al. 2002; Houbaviy et al. 2003), and are subsequently processed into 60–120 nucleotide long precursors with hairpin structures (pre-MIR) by the RNase III enzyme Drosha (Lee et al. 2003). The hairpin pre-MIRs are then exported to the cytoplasm, where they are processed into mature MIRs by another RNase III enzyme, Dicer (Hutvagner et al. 2001; Ketting et al 2001). Mature MIRs are found in RNP complexes (Mourelatos et al. 2002), and exert their regulatory roles through the interaction of the RNP complex with the target mRNA, resulting in either suppression of translation or cleavage (Hutvagner and Zamore 2002; Doench et al. 2003; Zeng et al. 2003). In most cases, the target sites in the 3′UTR are not perfectly matched to the MIR, and the MIR causes suppression of the translation of the mRNA. In a few cases in which a perfect, or near perfect match has been found between the MIR and its target site, it was shown that the MIR mediates cleavage of the target mRNA (Pfeffer et al. 2004; Yekta et al. 2004).
Studies on the regulatory roles of MIRs in a variety of organisms revealed that they have critical roles in central biological processes. In Caenorhabditis elegans, MIRs were shown to regulate various developmental processes, such as the timing of early (Wightman et al. 1993; Moss et al. 1997) and late (Reinhart et al. 2000; Slack et al. 2000; Lin et al. 2003) larval developmental transition and left/right asymmetry of chemoreceptor expression (Johnson and Hobert 2003). In plants, MIRs were shown to regulate flowering development (Aukerman and Sakai 2003; Chen 2003), leaf development, and embryonic patterning (Palatnik et al. 2003). In insects, MIRs were shown to regulate apoptosis, growth control, and fat metabolism (Brennecke et al. 2003; Xu et al. 2003). Finally, in mammals, MIRs were shown to be involved in the regulation of lymphoid subset lineage development (Chen et al. 2004). In addition, MIRs found within the homeobox clusters were suggested to have roles in regulating the highly ordered expression pattern of homeobox genes, which are involved in many aspects of embryonic development (Yekta et al. 2004).
The wide variety of MIR regulatory functions in many different species is an indication of the important role they play in development and disease. To shed additional light on MIR functions, it is important to define the cell and tissue specificity, as well as the spatial and temporal expression pattern of MIRs. However, the detection of MIR expression has met with significant difficulties, mostly due to their small size, and the relative poor sensitivity and low comparative power of the current methods used for their detection, most notably, Northern blot analysis (Sempere et al. 2004), cloning (e.g., Lagos-Quintana et al. 2002), and membrane arrays using radioactive detection methods (Krichevsky et al. 2003). The application of DNA microarray technology to MIR expression profiling may offer significant advantages over the other methods, including higher sensitivity, high-throughput, and higher comparative capabilities. In the present study, we have successfully applied the DNA microarray technology for the detection of MIR expression in cells and tissues, representing a significant advance in the study of MIR expression. We have studied several aspects of the methodology in order to standardize it and define the parameters needed to achieve efficient hybridization and reliable results. We then utilized this methodology to profile the expression of 150 known human MIRs in HeLa cells and five human tissues.
Results
To explore the ability of DNA microarrays (DNA chips) to detect microRNAs or their hairpin precursors, we have designed two DNA chips, prepared by Agilent using their SurePrint technology, containing the known human MIR sequences (Sanger database; http://www.sanger.ac.uk/Software/Rfam/mirna/index.shtml) in various settings of 60-mer oligonucleotides. The first chip, MIRChip1, served as an exploratory chip and was based on MIR sequences contained within their hairpin precursor sequences. For each hairpin precursor, we included its antisense, two tiled 60-mer probes from the region just upstream, and two from the region just downstream to the precursor sequence on the genome, as well as probes with mismatches, varying the number and position of the mismatches along and outside of the MIR sequences. In addition, MIRChip1 included probes for tRNAs, snoRNAs, and for a variety of ubiquitously expressed mRNAs.
The material for hybridization onto the chip was derived from adaptor-ligated, size-fractionated RNA (Elbashir et al. 2001). Following amplification, the double-stranded cDNA, carrying a T7 RNA polymerase promoter on the 3′ adaptor, was used for the labeling reaction. Labeled cRNA (lcRNA) incorporating either Cy3 or Cy5 was purified through a G-50 column and hybridized under standard conditions. In all experiments, a Cy5 lcRNA, derived from a mix of HeLa and HEK293 cell RNA, was used as a common control.
To examine the ability of MIRs or pre-MIRs in the lcRNA to hybridize to MIRChip1, we examined hybridizations with 5, 17, or 50 μg of lcRNA derived from HeLa cells. The results have shown that the 17-μg hybridization gave the optimal outcome (Supplemental Fig. 1 and Supplemental Table 1). In general, signal intensity of MIR-containing probes followed their known abundance in HeLa cells. In contrast, the antisense and our tiled probes gave no signal above background. Of the other controls, signals of tRNA probes were, at most, similar to those of the most abundant MIRs, whereas probes for abundant mRNAs gave only background signals. Hybridizations of MIRChip1 with total RNA oligo-dT-derived lcRNA resulted in the expected pattern of signals from the mRNA probes, but no signals above background were observed from the MIR-containing probes.
The design of MIRChip2 was based on the results obtained from MIRChip1, and was aimed to examine several variables, including MIR sequence settings, MIR location on the 60-mer probes, and mismatch effects. The complete design is detailed in the Methods section, and probe sequences are provided in the Supplemental material. Briefly, the chip included a total of 150 distinct human MIRs and their precursors, taken from the Sanger database. The MIRs were placed in various settings on the 60-mer probes. This included single MIRs, duplex MIRs, and triplex MIRs. The ∼22 nucleotides of the single MIRs were surrounded by sequences not present in the human genome (NHG-sequences). For the single MIR probes, we have varied the location of the MIRs along the 60-mer probes and included additional probes in which the MIR sequences included mismatches. The set of duplex MIRs included a subset of probes with mismatches in the first, second, or both MIR copies. Similarly, the set of triplex MIRs included subsets with mismatches in the first, second, or all three MIR copies. Control probe groups included ubiquitous mRNA, various small RNAs, NHG-sequences, and randomly chosen 26-mers from the human genome.
MIRChip2 was hybridized with 17 μg of lcRNA derived from HeLa cells, and various variables of the hybridization results were examined. Comparison of 60-mer probes containing MIRs within their precursor sequence to those in which the MIRs were embedded in NHG-sequences show that both give similar signal levels (Fig. 1A). In contrast, probes containing precursor sequences without MIRs or with truncated MIRs gave low or background signals (Fig. 1A). Moreover, a similar hybridization on MIRChip1, which included mismatches either in the MIR or in the non-MIR precursor regions, have shown that mismatches within the MIR sequence result in significant reduction in signal intensity, whereas no change is observed in mismatches outside of the MIR (Fig. 1B). Control 60-mer probes composed only of the NHG-sequences gave only background signals. We conclude that MIRs, and not their hairpin precursors, are responsible for the observed signals.
Figure 1.
MIRs are responsible for signals from probes of precursor sequences. Signals from hybridization of 60-mer probes with HeLa lcRNA are presented. (A) Three probes were considered for each MIR as follows: (1) precursor sequence containing the full MIR (hairpin + MIR); (2) MIR in the 5′ end of the probe, followed by nonhuman genome sequences (MIR in 5′); (3) Precursor sequence containing no more than 16 nucleotides of the MIR sequence (hairpin–MIR). Only probe sets in which the microarray signal of the hairpin + MIR probe was >2000 were considered. For each probe set, the signal intensities for the three types of probes were calculated as a ratio of the signal intensity of the hairpin + MIR probe. The MIR in 5′ and hairpin–MIR bars show the average computed for 23 sets of probes. (B) A detailed example of a mismatch analysis of pre-MIR-125b. The pre-MIR-125b probe and its respective mismatch (mm) probes from MirChip1 are presented. Wild-type (wt) probe contains MIR-125b at position 2 to 23 (boxed). Probes with mismatches are listed below the wild-type probe with the location of the substituted nucleotides specified. The signal column presents the microarray signal intensity observed for each probe. Similar results were obtained for let-7a and miR-30c.
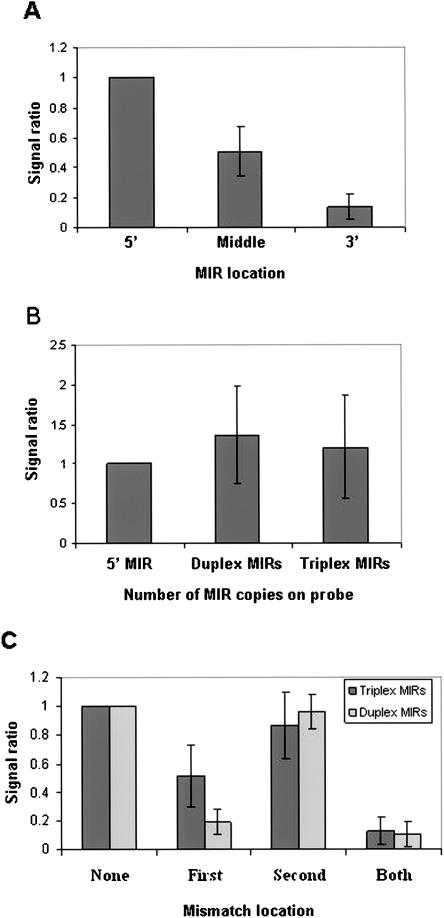
MIRChip2 included MIRs in three locations along the 60-mer probes to examine the importance of MIR location. Figure 2A shows that MIRs located at the 5′-end of the 60-mer probes result in significantly higher signals than MIRs located in the middle, with MIRs located at the 3′-end giving the lowest signals. Comparison of the 60-mer probes containing a single MIR to the duplex and triplex 60-mer probes show that the inclusion of additional MIR copies in the 60-mer probes results, at most, in a minor increase in signal intensity (Fig. 2B). Moreover, analysis of duplex and triplex 60-mer probes containing mismatches revealed that, in general, mismatches within the 5′ MIR cause a significant reduction in signal levels, whereas mismatches in MIRs located in the middle or in the 3′ ends had a significantly lower effect on signal intensity (Fig. 2C). We conclude that a single MIR located at the 5′ end of the probe, furthest from the surface of the chip, is sufficient to obtain high signals.
Figure 2.
Effect of MIR location and number within the 60-mer probes on signal intensity. Signals from hybridization of 60-mer probes with HeLa lcRNA are presented. (A) A set of three probes was considered for each MIR as follows: (1) MIR in the 5′; (2) MIR in the middle; (3) MIR in the 3′. For all three probes, the rest of the 60-mer sequence was composed of nonhuman genome sequences. Only probe sets for which the microarray signal of the MIR in the 5′ probe was >2000 were considered. For each probe set, the signal intensities for the three types of probes were calculated as a ratio of the signal intensity of the MIR in the 5′ probe. (B) Probes containing one, two (duplex), or three (triplex) copies of a MIR were analyzed. For each probe set, the signal intensities for the three types of probes were calculated as a ratio of the signal intensity of the probe containing one MIR copy. (C) Probes containing two or three MIR copies were analyzed. (None) Probes containing duplex or triplex MIRS, in which none of the MIR copies contain mismatches. (First) Duplex or triplex probes in which the 5′ MIR copy contains two mismatches. (Second) Duplex or triplex probes in which the 3′ or middle MIR copy, respectively, contains two mismatches. (Both) Duplex or triplex probes in which both the 5′ and the 3′ or middle MIR copies, respectively, contain two mismatches. For each probe set, the signal intensities for the three types of probes were calculated as a ratio of the signal intensity of the “None probe”. Data are presented as mean signal ratio ± SEM for 23 sets of probes, except for the triplex “First” set in C, for which two probes were eliminated from the calculation due to sequence matches to adaptors.
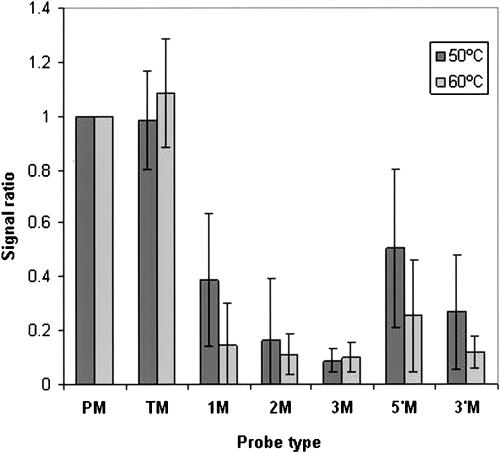
An important control of hybridization specificity is the effect of mismatches on observed signals. Figure 3 shows the results of hybridizations at temperatures of 50 and 60°C for a subset of 32 MIRs, each in two different settings of NHG-sequences for which mismatch probes were included. Whereas mismatches outside of the MIR sequences did not change signal levels, one, two, or three mismatches within the MIR significantly reduced the signal. In the 60°C hybridization, even a single mismatch reduced the signal close to background levels compared with a significantly lower reduction of signal intensity in the 50°C hybridization, in accordance with the lower stringency of these conditions. Similarly, mismatches in either the 5′ or 3′ regions of the MIR significantly reduced the signal intensity with higher effects at the 60°C hybridization temperature. Thus, under the standard conditions using hybridizations at 60°C, specificity was high.
Figure 3.
Effect of mismatches within MIRs on microarray signal intensity. Signals from hybridization of 60-mer probes with HeLa lcRNA at either 50 or 60°C were compared. Probes containing MIR at the 5′ end followed by nonhuman genome sequences were examined. Each probe containing a wild-type MIR (PM) was compared with a set of probes with the following mismatches: (TM) A probe with a wild-type MIR and six mismatches in the adjacent nonhuman genome sequence region; (1M) a probe with one mismatch at position 10 of the MIR sequence; (2M) a probe with two mismatches at positions 8 and 17 of the MIR; (3M) a probe with three mismatches at positions 6, 12, and 18 of the MIR. (5′M) a probe with a block of four mismatches at the 5′ end of the MIR; (3′M) a probe with a block of six mismatches at the 3′ end of the MIR. Data are presented as mean signal ratio ± SEM for 42 sets of probes.
We next hybridized MIRChip2 with lcRNAs derived from human brain, liver, thymus, testes, and placenta, and examined the tissue specificity of the various MIRs. The results obtained from the HeLa-cell hybridization mentioned above were included in the analysis. The full set of results can be found in the Supplemental material. A comparison was made mainly to results obtained by Sempere et al. (2004) that examined the expression of 119 MIRs by Northern blots in brain and liver, as well as other tissues not examined in the present study. Comparison was also made, when relevant, to the oligonucleotide array results of Krichevsky et al. (2003) and to the cloning data of Lagos-Quintana et al. (2002). MicroRNAs showing distinct brain (e.g., MIR-9 and MIR-124A) or liver (MIR-122A and MIR-194) tissue specificity gave identical results on our MIRChip hybridizations (Table 1). Also, the findings that certain MIRs, such as let-7A, let-7B, and MIR-30C (Sempere et al. 2004), are expressed at high levels in many tissues were confirmed using our microarrays, extending the results to the thymus, testes and placenta (Supplemental Table 1). An overall correlation of ∼0.6 was found between our results and those of Sempere et al. (2004). However, we also found distinct differences between our study and those of others. For example, we found very high expression of MIR-149 in the brain and high expression in the liver, whereas Sempere et al. (2004) found low levels in the brain and no signal in the liver. Similarly, we detected significant expression levels of MIR-20 in both brain and liver, compared with no signals on the Northern blots reported by Sempere et al. (2004). On the other hand, MIR-203 and MIR-137 showed only background signals in our study, compared with high levels of expression in both brain and liver or in the brain, respectively, observed by Sempere et al. (2004). In general, the main differences seem to be for MIRs expressed at relatively low levels, where the higher sensitivity of the DNA microarray had a clear advantage.
Table 1.
Expression of tissue-specific or highly enriched MIRs in the five human tissues
| HeLa | Brain | Liver | Thymus | Testes | Placenta | |
|---|---|---|---|---|---|---|
| HSA-MIR-9 | 642 | a42659 | 3504 | 4455 | 4485 | 2313 |
| HSA-MIR-124A | 1879 | 65517 | 7025 | 3099 | 2672 | 2498 |
| HSA-MIR-128A | 2015 | 27701 | 4940 | 4876 | 5166 | 2495 |
| HSA-MIR-128B | 1168 | 21969 | 3954 | 4819 | 5383 | 2027 |
| HSA-MIR-129 | 503 | 22573 | 1175 | 2213 | 5364 | 2017 |
| HSA-MIR-194 | 501 | 910 | 65518 | 4737 | 2342 | 7952 |
| HSA-MIR-122A | 1051 | 447 | 65518 | 2644 | 617 | 570 |
| HSA-MIR-148 | 413 | 620 | 38436 | 5250 | 6204 | 2711 |
| HSA-MIR-192 | 452 | 606 | 20650 | 1628 | 1263 | 2607 |
| HSA-MIR-96 | 887 | 3100 | 1477 | 44800 | 2266 | 5466 |
| HSA-MIR-182 | 662 | 1944 | 1091 | 25771 | 2034 | 3683 |
| HSA-MIR-183 | 1026 | 1123 | 1286 | 8754 | 1681 | 2138 |
| HSA-MIR-200A | 415 | 429 | 547 | 2708 | 540 | 675 |
| HSA-MIR-10B | 410 | 433 | 477 | 3871 | 23083 | 738 |
| HSA-LET-7E | 557 | 2723 | 1367 | 4805 | 10147 | 4293 |
| HSA-MIR-212 | 608 | 1410 | 1414 | 3849 | 6425 | 1478 |
| HSA-MIR-187 | 510 | 752 | 851 | 962 | 3641 | 827 |
| HSA-MIR-134 | 448 | 617 | 698 | 763 | 2250 | 997 |
| HSA-MIR-141 | 696 | 805 | 1220 | 4063 | 2000 | 46845 |
| HSA-MIR-23A | 1312 | 3492 | 2990 | 6021 | 11173 | 40076 |
| HSA-MIR-136 | 465 | 725 | 709 | 776 | 3100 | 8840 |
Signal values are in bold in the tissue in which the MIR was found to be expressed at the highest level
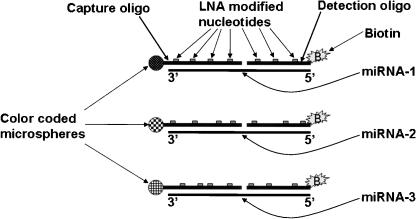
To validate the MIR expression detected by the MIRChip, we used the mirMASA technology (http://gene.genaco.com/miRNA.htm), which is a fluorescence-based solution hybridization method based on the xMAP technology developed by Luminex (Yang et al. 2001; http://www.luminexcorp.com/01_xMAPTechnology/index.html). The mirMASA technology uses a specific capture-oligo for each targeted MIR that is covalently coupled onto color-coded microspheres (beads), and a detection-oligo that is labeled with biotin (Fig. 4). Both capture and detection oligos are spiked with Locked Nucleic Acid (LNA) nucleotides to increase specificity and sensitivity (Petersen and Wengel 2003). Following hybridization of the capture and detection oligos with the RNA, streptavidin-phycoerythrin is added. The fluorescence associated with the color-coded beads provides a measure for MIR expression level. We have focused the mir-MASA validation study on the expression of MIRs, showing distinct differences between our MIRChip and the published Northern blot data. As shown in Table 2, the expression of MIR-20 was detected in brain and liver, as well as in the other three tissues, compared with no signals observed in the Northern blot analysis of Sempere et al. (2004). On the other hand, no expression is observed in any of the tissues for MIR-137 and MIR-203, compared with expression of MIR-137 in the brain and of MIR-203 in both brain and liver observed by Sempere et al. (2004). In addition, a good correlation was observed between the mirMASA and the MIRChip results in the expression of MIR-141 and let-7B in all five tissues. These results validate the expression patterns observed in the MIRChip experiments. Examination of the expression of MIR-127 and MIR-129 show no signals (Table 2), compared with clear expression predicted from the MIRChip experiments. These results are in agreement with the Northern blot data of Sempere et al. (2004). Possible reasons for these apparent false-positive signals in the MIRChip are discussed below.
Figure 4.
The mirMASA technology. A specific 10–12 nucleotide-long capture oligonucleotide (oligo) and a specific 8–10 nucleotide-long detection oligo are synthesized for each MIR. The capture oligo is covalently linked to color-coded microspheres (beads), whereas the detection oligo is labeled with biotin. For each MIR, the capture oligo is linked to a unique color-coded bead. Both capture and detection oligos are spiked with Locked Nucleic Acid (LNA) nucleotides to increase specificity and sensitivity. The biotin is used for detection following addition of streptavidin-phycoerythrin and reading the fluorescence associated with each color-coded bead.
Table 2.
Validation of MIR expression in the five human tissues by the mirMASA method
|
Blanka
|
Brain
|
Matchd
|
Liver
|
Match
|
Thymus
|
Me
|
Testes
|
M
|
Placenta
|
M
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample | Signalb | Signal | Ratioc | NB | CH | Ratio | NB | CH | Ratio | CH | Ratio | CH | Ratio | CH | ||||
| Let-7b | 83 | 5102 | 62.5 | + | + | 1436 | 17.3 | + | + | 3342 | 40.3 | + | 3292 | 39.7 | + | 1175 | 14.2 | + |
| mir-20 | 67 | 554 | 8.3 | – | + | 515 | 7.7 | – | + | 1193 | 17.9 | + | 970 | 14.6 | + | 751 | 11.3 | + |
| mir-127 | 89 | 179 | 2.0 | + | – | 128 | 1.4 | + | – | 102 | 1.2 | – | 125 | 1.4 | – | 127 | 1.4 | – |
| mir-129 | 84 | 118 | 1.4 | + | – | 96 | 1.1 | + | + | 93 | 1.1 | + | 104 | 1.2 | – | 120 | 1.4 | + |
| mir-137 | 73 | 85 | 1.2 | – | + | 81 | 1.1 | + | + | 68 | 0.9 | + | 63 | 0.9 | + | 75 | 1.0 | + |
| mir-141 | 76 | 115 | 1.5 | + | + | 102 | 1.3 | + | + | 154 | 2.0 | + | 165 | 2.2 | + | 664 | 8.8 | + |
| mir-199a | 103 | 187 | 1.8 | – | + | 454 | 4.4 | + | + | 979 | 9.5 | + | 1671 | 16.2 | + | 1552 | 15.0 | + |
| mir-203 | 56 | 78 | 1.4 | – | + | 81 | 1.4 | – | + | 99 | 1.8 | + | 73 | 1.3 | + | 79 | 1.4 | + |
| 5sRNA-1 | 111 | 3630 | 32.8 | 4059 | 36.7 | 3450 | 31.2 | 3758 | 33.9 | 4256 | 38.4 | |||||||
| 5sRNA-2 | 81 | 3016 | 37.2 | 3180 | 39.3 | 2878 | 35.5 | 3078 | 38.0 | 3317 | 41.0 | |||||||
Signal obtained from incubation of the capture and detection oligo's without RNA
The expression of each MIR in each tissue was measured in duplicates. Difference between duplicates was ∼5%–10%. The average of the duplicates is presented
The ratio between the signal value and the Blank value
Match between the mirMASA, Northern blot (NB), and DNA chip (CH) data. (+) A match (–) no match
M is short for match. For the thymus, testes, and placenta, no Northern blot data is available, and comparison was done only to DNA chip data
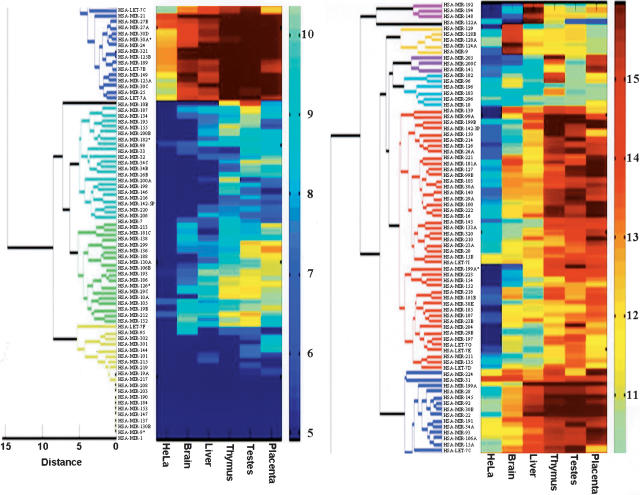
Clustering analysis revealed that MIRs are expressed in almost any conceivable pattern (Fig. 5). This includes MIRs expressed in all tissues, MIRs expressed in some tissues, tissue-specific MIRs, and MIRs undetectable in any of the tissues examined. The analysis revealed distinct clusters of MIRs specifically expressed in brain, liver, and thymus, whereas clusters of MIRs that are specifically expressed in testes and placenta are more obscure. A thorough analysis of the testes and placenta hybridization data revealed MIRs that are specific, or highly enriched in these tissues. Table 1 shows, in addition to the brain and liver data, the MIRs that are tissue specific or highly enriched in the three tissues not examined before by others, that is, MIR-96, MIR-182, and MIR 183 in the thymus, MIR-10b, MIR-212, and MIR let-7E in the testes, and MIR-141, MIR-23A, and MIR-136 in the placenta. Some MIRs were expressed in two of these tissues (e.g., MIR-197 and MIR-205), and others were expressed in all three tissues (e.g., MIR-26a, MIR-100, and MIR-222). Interestingly, we observed an overall low expression of MIRs in HeLa cells. Only 44 of the MIRs show signal levels above background, compared with 86 to 119 in the five tissues. In addition, none of the MIRs was found to be specifically enriched in HeLa cells, and the vast majority of the MIRs showing significant signals were expressed at lower levels than in the five tissues. These results are compatible with other reports observing lower expression levels of various MIRs in cancer cells (Michael et al. 2003; Calin et al. 2004).
Figure 5.
Clustering analysis of the expression of 150 human MIRs in placenta, testes, thymus, liver, brain, and HeLa cells. The clustering procedure used to derive the dendrogram is described in the Methods section. The level of expression of each MIR in each of the samples is indicated by the color shown next to the MIR name. The relationship between the color and the expression levels is defined by the color key on the right side of the figure. The color code numbers on the color key are log2 of the signal intensity. The original figure was cut in half due to space considerations. HSA-LET-7c, found in the bottom, right, and at top, left, links the two halves.
Discussion
In this study, we have established an oligonucleotide microarray platform for the analysis of microRNA expression. Our results provide strong evidence for the high specificity and sensitivity of the MIR-specific microarray and present an initial comparative study of tissue distribution of MIRs in several human tissues.
We have shown that signals, obtained from lcRNA prepared from size-fractionated RNA smaller than 140 nucleotides, were derived from MIRs and not from their hairpin precursors. It is possible that the hairpin structure of the precursors makes them less available for hybridization. It is also possible that their fleeting existence and low abundance make them undetectable. Similarly, the oligonucleotide 60-mer probes matching the hairpin precursors are also expected to fold into hairpin structures. Our finding that MIRs are able to effectively hybridize to these probes suggests that MIRs are able to efficiently replace any self-annealed strands. It is possible that the nonpaired G-T in the DNA renders the probes to form less-stable hairpins.
Mismatch analysis is critical for examining the specificity of the hybridization. Our findings that single mismatches in the middle of the MIR sequence, or two mismatches in either side, reduce the signal to background levels, suggest that the signals are specific. Moreover, MIRs that are different by a few nucleotides from each other often show different expression patterns. For example, MIRs let-7A and let-7B, which differ from each other in only 2 nucleotides, have a very similar pattern, whereas let-7c, which is one nucleotide different from both let-7A and let-7B, has a different expression pattern with significantly lower expression in placenta and brain, but not in the other tissues. The similar expression pattern of let-7A and let-7B is most likely related to the fact that they are clustered on chromosome 22. Taken together, our data strongly support the specificity of the signals observed in our MIRChip experiments.
The location of a MIR in the 5′ end of the 60-mer oligonucleotide probes gave significantly higher signals, compared with locations in the middle or at the 3′ end. Although the interpretation of this observation is not straightforward, it may suggest that locations toward the 3′ terminus, which is closer to the chip surface, are less accessible to the lcRNA molecules. It also indicates that probe synthesis quality, expected to be lower at the 5′ end, is not a significant factor.
The inclusion of 60-mer oligonucleotide probes composed of MIR duplexes and triplexes allowed us to compare our results with those described by Krichevsky et al. (2003), which used triplex MIR oligonucleotides printed on nylon membranes coupled with hybridization with radioactively labeled RNA. We found that both MIR duplex and triplex 60-mer probes gave comparable signals to those obtained from 60-mers containing a single MIR at the 5′ terminus. This result is compatible with the notion that 60-mer probe molecules are in excess over the matching MIRs in the lcRNA. It would be interesting to investigate whether shorter oligonucleotides probes harboring MIRs at their 5′ can be used to obtain the same signal levels seen with the 60-mer probes.
Compared with hybridizations with poly(A) RNA-derived probes, requiring 1μg or less of lcRNA, the optimal detection of expression of a wide variety of MIRs required 17 μg of lcRNA. Under these conditions, abundant MIRs result in saturated signals. However, for obtaining significant signals for MIRs expressed at lower levels, such high lcRNA amounts are required. Additional reasons for the difference may be the smaller amount of the MIR-containing lcRNA molecules compared with poly(A)-derived lcRNA molecules, and the lower efficiency of hybridization of the short cRNA molecules to the 60-mer probes. Whereas the background signals in these experiments were ∼500, we have used a conservative approach and used a signal of 1500 as the threshold for positive expression. Our experiments suggest that signals close to this threshold are specific, as mismatches within the MIRs on the probes reduce the signals to background levels. Moreover, several MIRs showing low levels of expression were shown to be present in the relevant cells by cloning and sequencing. Current methods, such as Northern blotting and RNase protection, are not sensitive enough to examine such low expression levels. Thus, a more rigorous test of the specificity of these results depends on the availability of methods with sensitivities approaching that of the DNA microarray. Using 17 μg of lcRNA, many abundant MIRs result in signals at the upper limit of detection. For a more accurate profiling of the expression of these MIRs, lower amounts of lcRNA are required. Thus, to extensively follow the expression of rare and abundant MIRs, several hybridizations using varying amounts of lcRNA are needed.
We have performed a comparative expression profiling using RNA from human brain, liver, thymus, testes, placenta, and Hela cells. Comparison of the results from brain and liver to those reported by others (Sempere et al. 2004; Logos-Quintana et al. 2002; Krichevsky et al. 2003) reveals an overall agreement. Our results validate the brain-specific expression of MIR-9, MIR-124A, and MIR-128, and the liver-specific expression of MIR-122a/b and MIR-148. In both tissues, we identified additional tissue-specific MIRs (MIR-129 in the brain and MIR-192 and MIR-194 in the liver). Major differences consisting of no expression detected in our chips compared with high signals on Northern blots were very few (e.g., MIR-137 and MIR-203) and can be accounted for by the higher specificity of the chips and occurrence of cross-hybridization in the Northern blot method. The higher stringency conditions used in the MIRChip hybridizations (60°C compared with 42°C) support this interpretation. The possibility of failure of specific probes on our microarrays is unlikely, as results obtained from miRChip1 and miRChip2, which were independently prepared, were highly similar with a correlation coefficient of 0.98.
We have applied the mirMASA methodology to further validate the results of our study. The results obtained from this fundamentally different methodology have lent further support to our interpretation of the higher sensitivity and specificity of the microarray in comparison with the currently used methods for detection and characterization of MIRs. Thus, the absence of detectable expression of MIR-137 and MIR-203 in our study was validated by the mirMASA method. On the other hand, the distinct expression levels of MIR-20 detected by the microarray, but showing no signals in the Northern blots (Sempere et al. 2004), were validated by the mirMASA analysis. However, the mirMASA results for MIR-127 and MIR-129, indicating undetectable levels of these MIRs in the brain tissue, support the Northern blot data and disagree with the microarray results. It is possible that this difference is due to the large size and high complexity of the human brain, and the fact that RNA preparations from total human brain do not actually use the entire brain. Thus, different RNA preparations are likely to result in variability of the RNA population. This is further supported by the observation that MIR-127 is expressed in mouse brain (Sempere et al. 2004) at levels similar to those observed in our MIRChip experiments, and by preliminary studies showing variability in expression of MIR-129 in different brain preparation. In this regard, we note that we obtained total brain RNA from Ambion, whereas Sempere et al. (2004) obtained it from the Biochain Institute. As for MIR-127, the mirMASA study did not validate its expression in placenta, thymus, and testes. Although differences in sensitivity and specificity between the various methodologies may explain this result, it is also possible that the MIR-127 signals observed in the microarray are false positive. In general, the microarray platform is prone to infrequent false-negative and false-positive data-points (e.g., Cole et al. 2003). Thus, as is common in the general applications of microarray expression analysis, the move from studying general expression patterns into interest in specific genes requires specific validation experiments.
Most recently, data from microarray analysis of microRNA expression have been published by others (Liu et al. 2004). Comparing these published results of miRNA expression to the Northern blot data of Sempere et al. (2004) show much more disagreements then agreements. We estimate the correlation between these two data sets to be 0.008 for brain and 0.28 for liver. By comparison, the correlation between our data and Sempere et al. (2004) show a highly significant correlation of 0.71 for brain and 0.6 for liver. As expected, the correlation between our data and that of Liu et al. (2004) is very low (0.11 for brain and 0.15 for liver). The main difference between our methodology and that of Liu et al. (2004) is in the nature of the labeled material. Liu et al. (2004) used random hexameres to label the entire population of RNAs found in total RNA, whereas our approach included a highly enriched population of short RNAs produced in the cells by cleavage with RNaseIII enzymes. It is, therefore, not surprising that in the Liu et al. (2004) study, mostly the highly expressed MIRs match the published Northern blot data.
The clustering analysis of MIR expression revealed a wide variety of expression patterns. Overlaying the expression pattern with MIR clustering on the genome has shown that, in most cases, clustered MIRs have the same expression pattern with occasional variability in the expression level. For example, MIR-194 and MIR-192 are clustered on chromosome 11 and have a similar liver-specific expression. Interestingly, MIR-194 is clustered with MIR-215 on chromosome 1, but the latter is not expressed in any of the tissues examined. One can conclude that the liver-specific expression of MIR-194 derives from transcription of its locus on chromosome 11. In a similar manner, MIR-96, MIR-182, and MIR-183 are clustered on chromosome 7, and all show thymus-specific expression. In some cases, major differences in expression levels of MIRs found in the same cluster were observed. For example, MIR-23B is expressed at much lower levels in all tissues, compared with MIR-27B, MIR-189, and MIR-24, all residing in the same cluster on chromosome 9. Another example of a cluster showing variability in expression pattern is the cluster of MIR-25, MIR-93, and MIR-106B on chromosome 7. Whereas MIR-25 is expressed at high levels in the five tissues, MIR-93 is expressed at much lower levels in the brain and liver, and MIR-106B is expressed at low levels in all five tissues. Possible explanations for these differences may involve a variety of post-trancriptional regulation modes.
Taken together, this study has clearly demonstrated the feasibility of using the microarray technology for the detection of MIRs in human tissues. It opens the door for the wide application of this technology for the study of the physiological role of MIRs in many species, as well as for their role in disease. The MIR microarray is a further step toward a better understanding of this fast-evolving field of biology.
Methods
Microarrays
Custom microarrays comprised of oligonucleotides synthesized in situ were manufactured by Agilent Technologies using SurePrint technology, as described (Hughes et. al. 2001). Sequences of the oligonucleotides are detailed in the Supplemental material.
MIRChip2 design
The full list of the 60-mer oligonucleotide sequences used in the microarray are given in the Supplemental material. The MIRs and their respective precursor sequences were taken from the Sanger Database, yielding a total of 150 distinct MIRs and 186 precursor sequences (some MIRs appear in more than one precursor). The following different probe groups were constructed.
Single MIR probes
From each precursor, we took 26-mer containing the MIR, then assigned three probes for each extended MIR sequence as follows: (1) the 26-mer at the 5′ of the 60-mer probe; (2) the 26-mer at the 3′ of the 60-mer probe; (3) the 26-mer in the middle of the 60-mer probe.
One of two different 34-mer words, which do not appear in the human genome (NHG-sequences), were attached to the 26-mer to complete a 60-mer probe. The 34-mer words are a combination of 10-mer words, which are very rare in the human genome. Each potential 34-mer word was compared with the human genome by the BLAST program, and we ended up with two different rare words that have a homology of no more than 40% and have no 15-mer subsequences with >80% homology.
For a subset of 32 of Single MIR probes, we designed an additional six mismatch mutation probes as follows: (1) a block of four mismatches at the 5′ end of the MIR; (2) a block of six mismatches at the 3′ end of the MIR; (3) one mismatch at position 10 of the MIR; (4) two mismatches at positions 8 and 17 of the MIR; (5) three mismatches at positions 6, 12, and 18 of the MIR; (6) six mismatches at different positions outside of the MIR.
Duplex MIR probes
From each precursor, we took a 30-mer containing the MIR, and then duplicated it to obtain a 60-mer probe. For a subset of 32 MIRs, we designed an additional three mismatch mutation probes as follows: (1) two mismatches on the first MIR; (2) two mismatches on the second MIR; (3) two mismatches on each of the MIRs.
Triplex MIR probes
Following Krichevsky et al. (2003), we attached head-to-tail MIR sequences of ∼22 nucleotides to obtain 60-mer probes containing up to three times the same MIR sequence. For a subset of 32 of probes, we designed an additional three mismatch mutation probes as follows: (1) two mismatches on the first MIR; (2) two mismatches on the second MIR; (3) two mismatches on each of the MIR copies.
Precursor with MIR probes
For each precursor, we took a 60-mer sequence containing the entire MIR.
Precursor without MIR probes
For each precursor, we took a 60-mer sequence containing no more than 16 nucleotides of the MIR. For a subset of 32 of probes, we designed additional mismatch probes containing four mismatches.
General control Groups included 100 probes for mRNAs, representing mostly genes expressed in a wide variety of cell types, 85 representative tRNAs, and 19 representative snoRNA probes.
Negative controls included one group composed of 294 randomly chosen 26-mer sequences from the human genome, not contained in published precursors sequences, placed at the 5′ end and complemented with 34-mer rare word as described above. A second group was composed of 182 different 60-mer probes containing different combinations of 10-mer rare words.
Preparation of cDNA
The cDNA preparation procedure was based on the method described by Elbashir et al. (2001). Total RNA was size-fractionated using an YM-100 column (Millipore Corporation) and 3 μg of the size-fractionated RNA was used for the ligation of adaptor sequences.
A 5′ adaptor (5′-AAAGGAGGAGCTCTAGaua-3′) and 3′ adaptor containing a T7 RNA polymerase promoter [5′-(P)uggCCTATAGTGAGTCGTATTA(3InvdT)-3′] were ligated to the size-fractionated RNA with subsequent gel-fractionation steps (Elbashir et al. 2001). Uppercase letters denote deoxyribonucleotides and lowercase letters denote ribonucleotides. Following ligation, the material was converted to cDNA using a primer complementary to the 3′ adaptor (5′-TAATACGACTCACTATAGGCCA-3′).
Next, the cDNA was amplified by PCR using the above mentioned oligonucleotide as a reverse primer and a forward primer matching the adaptor (5′-AAAGGAGGAGCTCTAGATA-3′).
The amplified DNA was digested with XbaI to remove the majority of the 5′ adaptor sequence.
Labeled material and hybridization
cRNA labeled with either cyanine 3-CTP (Cy3-CTP) or cyanine 5-CTP (Cy5-CTP) was generated from each cDNA source using the Low-Input Linear Amplification Kit (Agilent) according to the manufacturer's protocol, except that synthesis was initiated at the in vitro transcription step using 1 μg of cDNA as starting material. Hybridization solutions that contained the indicated amount of each of labeled cRNA from either the control or the test samples were prepared using the in situ Hybridization Reagent Kit (Agilent). Hybridized microarrays were scanned using the Agilent LP2 DNA Microarray Scanner at 10-μm resolution. Microarray images were visually inspected for defects.
Data analysis
Microarray images were analyzed using Feature Extraction Software (Version 7.1.1, Agilent). We set the signal of each probe as its median intensity. We observed a nearly constant background intensity signal of 430. Using NHG-sequence negative control probes, the threshold for reliable probe signals was set at 1500. No NHG-sequence probes with signals higher than 1500 were observed in HeLa, brain, liver, and thymus, and <0.5% of these probes gave signals higher than 1500 in testes and placenta.
In all hybridization experiments, a high correlation of 0.96 to 0.98 was observed between the Cy5-labeled common control lcRNA. In addition, lcRNAs derived from the same RNA source and hybridized to MIRChip1 and MIRChip2 gave a correlation coefficient of 0.98 when identical probes on the two chips were compared.
Clustering analysis
For all 150 MIRs used for clustering, the background signal of 500 was first subtracted from the values observed in all six different tissues. A threshold of 30 was set as a minimal value. A log2 transformation was applied, and the Euclidian distance matrix was calculated. A hierarchical clustering using Average Linkage algorithm was performed with an output of a dendrogram. A distance threshold of six was used to distinguish between the most significant clusters.
Analysis of MIR expression by the mirMASA technology
Details on the mirMASA technology can be found in http://gene.genaco.com/miRNA.htm. The analysis was done by multiplexing in two groups. One group included let-7b, MIR-127, MIR-129, MIR-137, MIR-203, and 5sRNA control. The second group included MIR-20, MIR-199a, MIR-141, and 5sRNA control. The analysis of each group was done on 1 μg of total RNA. A negative bead-control was performed for each group, shown as “blank” in Table 2.
Acknowledgments
The following authors made inventive contributions to this study: O.B. was involved in the microarray design, data analysis, and interpretation; E.M. conceived and prepared the RNA samples for the preparation of the labeled cRNA; I.B. and A.A. conceived and designed the methodology of detecting microRNAs using microarrays and directed the work of the other authors in its implementation. Other authors made the following significant noninventive contributions to this study: R.A., Y.K., E.S., A.B., and U.E. were involved in various bioinformatics and algorithmic aspects of microarray design; S.G., M.S., and Y.M.S. were involved in the preparation of RNA for microarray analysis; P.H. and E.K.L. performed the microarray work; Z.B. provided scientific guidance and edited the manuscript; P.E. assisted in the microarray design, work, data analysis, interpretation, and wrote the manuscript. We are grateful to all members of Rosetta Genomics for their invaluable contributions to this work. We thank Inbal Halperin, Renata Kabiljo, Liron Sivan, and Micha Tugentman for their assistance in microarray design, and Yaffa Schindler for helpful comments on the manuscript.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2845604.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Aukerman, M.J. and Sakai, H. 2003. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15: 2730-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281-297. [DOI] [PubMed] [Google Scholar]
- Brennecke, J., Hipfner, D.R., Stark, A., Russell, R.B., and Cohen, S.M. 2003. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113: 25-36. [DOI] [PubMed] [Google Scholar]
- Calin, G.A., Sevignan, I.C., Dumitru, C.D., Hyslop, T., Noch, E., Yendamuri, S., Shimizu, M., Rattan, S., Bullrich, F., Negrini, M., et al. 2004. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. 101: 2999-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. 2003. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.Z., Li, L., Lodish, H.F., and Bartel, D.P. 2004. MicroRNAs modulate hematopoietic lineage differentiation. Science 303: 83-86. [DOI] [PubMed] [Google Scholar]
- Cole, S.W., Galic, Z., and Zack, J.A. 2003. Controlling false-negative errors in microarray differential expression analysis: A PRIM approach. Bioinformatics 19: 1808-1816. [DOI] [PubMed] [Google Scholar]
- Doench, J.G., Petersen, C.P., and Sharp, P.A. 2003. siRNAs can function as MIRNAs. Genes & Dev. 17: 438-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Lendeckel, W., and Tuschl, T. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 15: 188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaviy, H.B., Murray, M.F., and Sharp, P.A. 2003. Embryonic stem cell-specific MicroRNAs. Dev. Cell 5: 351-358. [DOI] [PubMed] [Google Scholar]
- Hughes, T.R., Mao, M., Jones, A.R., Burchard, J., Marton, M.J., Shannon, K.W., Lefkowitz, S.M., Ziman, M., Schelter, J.M., Meyer, M.R., et al. 2001. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 19: 342-347. [DOI] [PubMed] [Google Scholar]
- Hutvagner, G. and Zamore, P.D. 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297: 2002-2003. [DOI] [PubMed] [Google Scholar]
- Hutvagner, G., McLachlan, J., Pasquinelli, A.E., Balint, E., Tuschl, T., and Zamore, P.D. 2001. A cellular function for the RNA interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science 293: 834-838. [DOI] [PubMed] [Google Scholar]
- Johnston, R.J. and Hobert, O. 2003. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426: 845-849. [DOI] [PubMed] [Google Scholar]
- Ketting, R.F., Fischer, S.E., Bernstein, E., Sijen, T., Hannon, G.J., and Plasterk, R.H. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & Dev. 15: 2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky, A.M., King, K.S., Donahue, C.P., Khrapko, K., and Kosik, K.S. 2003. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 9: 1274-1281.Erratum in: RNA. 10: 551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Lendeckel, W., and Tuschl, T. 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853-858. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., and Tuschl, T. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12: 735-739. [DOI] [PubMed] [Google Scholar]
- Lau, N.C., Lim, L.P., Weinstein, E.G., and Bartel, D.P. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858-862. [DOI] [PubMed] [Google Scholar]
- Lee, R.C. and Ambros, V. 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862-864. [DOI] [PubMed] [Google Scholar]
- Lee, Y., Jeon, K., Lee, J.T., Kim, S., and Kim, V.N. 2002. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 21: 4663-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Radmark, O., Kim, S., et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415-419. [DOI] [PubMed] [Google Scholar]
- Lin, S.Y., Johnson, S.M., Abraham, M., Vella, M.C., Pasquinelli, A., Gamberi, C., Gottlieb, E., and Slack, F.J. 2003. The C. elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev. Cell 4: 639-650. [DOI] [PubMed] [Google Scholar]
- Liu, C.-G., Calin, G.A., Meloon, B., Gamliel, N., Sevignani, C., Ferracin, M., Dumitru, C.D., Shimizu, M., Zupo, S., Dono, M., et al. 2004. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc. Nat. Acad. Sci. 101: 9740-9744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael, M.Z., O'Connor, S.M., van Holst Pellekaan, N.G., Young, G.P., and James, R.J. 2003. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol. Cancer Res. 1: 882-891. [PubMed] [Google Scholar]
- Moss, E.G., Lee, R.C., and Ambros, V. 1997. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88: 637-646. [DOI] [PubMed] [Google Scholar]
- Mourelatos, Z., Dostie, J., Paushkin, S., Sharma, A., Charroux, B., Abel, L., Rappsilber, J., Mann, M., and Dreyfuss, G. 2002. MIRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes & Dev. 16: 720-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik, J.F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J.C., and Weigel, D. 2003. Control of leaf morphogenesis by microRNAs. Nature 425: 257-263. [DOI] [PubMed] [Google Scholar]
- Petersen, M. and Wengel, J. 2003. LNA: A versatile tool for therapeutics and genomics. Trends Biotechnol. 21: 74-81. [DOI] [PubMed] [Google Scholar]
- Pfeffer, S., Zavolan, M., Grasser, F.A., Chien, M., Russo, J.J., Ju, J., John, B., Enright, A.J., Marks, D., Sander, C., et al. 2004. Identification of virus-encoded microRNAs. Science 304: 734-736. [DOI] [PubMed] [Google Scholar]
- Reinhart, B.J., Slack, F.J., Basson, M., PasquinellI, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R., and Ruvkun, G. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901-906. [DOI] [PubMed] [Google Scholar]
- Sempere, L.F., Freemantle, S., Pitha-Rowe, I., Moss, E., Dmitrovsky, E., and Ambros, V. 2004. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 5: R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack, F.J., Basson, M., Liu, Z., Ambros, V., Horvitz, H.R., and Ruvkun, G. 2000. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 5: 659-669. [DOI] [PubMed] [Google Scholar]
- Wightman, B., Ha, I., and Ruvkun, G. 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75: 855-862. [DOI] [PubMed] [Google Scholar]
- Xu, P., Vernooy, S.Y., Guo, M., and Hay, B.A. 2003. The Drosophila microRNA MIR-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 13: 790-795. [DOI] [PubMed] [Google Scholar]
- Yang, L., Tran, D.K., and Wang, X. 2001. BADGE, beads array for the detection of gene expression, a high-throughput diagnostic bioassay. Genome Res. 11: 1888-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta, S., Shih, I.H., and Bartel, D.P. 2004. MicroRNA-directed cleavage of HOXB8 mRNA. Science 304: 594-596. [DOI] [PubMed] [Google Scholar]
- Zeng, Y., Yi, R., and Cullen, B.R. 2003. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl. Acad. Sci.. 100: 9779-9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
Web site references
- http://www.sanger.ac.uk/Software/Rfam/mirna/index.shtml;
- http://gene.genaco.com/miRNA.htm; description of the mirMASA technology.
- http://www.luminexcorp.com/01_xMAPTechnology/index.html; description of the xMAP technology.