Abstract
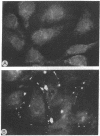
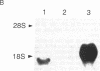
The gene family encoding gap junction proteins (connexins) consists of several known members, and multiple connexins are frequently coexpressed by coupled cells. To characterize the channel properties of the major rat liver gap junction protein (connexin 32) in isolation from other gap junction proteins, we have introduced the cDNA encoding it into a human hepatoma cell line (SKHep1) in which we have identified a gap junction deficiency. In this cell line, dye coupling was absent and junctional conductance was near zero. Connexins and connexin 32 mRNA were not detectable by immunocytochemistry and Northern blot analysis. After transfection and selection, cells were strongly coupled with regard to dye and electrical current, and connexin 32 mRNA and punctate connexin 32-immunoreactive membrane contacts were abundant. Functional gap junction channels were still expressed after 19 passages of the cells, indicating stable transfection. When junctional conductance was rendered reversibly low by exposing the cells to agents that uncouple other cell types, currents through single gap junction channels could be observed. The unitary conductance of these expressed channels was about 120-150 pS, a value that is distinctly larger than in heart cells, which express a different gap junction protein.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beyer E. C., Paul D. L., Goodenough D. A. Connexin43: a protein from rat heart homologous to a gap junction protein from liver. J Cell Biol. 1987 Dec;105(6 Pt 1):2621–2629. doi: 10.1083/jcb.105.6.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt J. M., Spray D. C. Single-channel events and gating behavior of the cardiac gap junction channel. Proc Natl Acad Sci U S A. 1988 May;85(10):3431–3434. doi: 10.1073/pnas.85.10.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt J. M., Spray D. C. Volatile anesthetics block intercellular communication between neonatal rat myocardial cells. Circ Res. 1989 Sep;65(3):829–837. doi: 10.1161/01.res.65.3.829. [DOI] [PubMed] [Google Scholar]
- Claudio T., Green W. N., Hartman D. S., Hayden D., Paulson H. L., Sigworth F. J., Sine S. M., Swedlund A. Genetic reconstitution of functional acetylcholine receptor channels in mouse fibroblasts. Science. 1987 Dec 18;238(4834):1688–1694. doi: 10.1126/science.3686008. [DOI] [PubMed] [Google Scholar]
- Dahl G., Miller T., Paul D., Voellmy R., Werner R. Expression of functional cell-cell channels from cloned rat liver gap junction complementary DNA. Science. 1987 Jun 5;236(4806):1290–1293. doi: 10.1126/science.3035715. [DOI] [PubMed] [Google Scholar]
- Dermietzel R., Traub O., Hwang T. K., Beyer E., Bennett M. V., Spray D. C., Willecke K. Differential expression of three gap junction proteins in developing and mature brain tissues. Proc Natl Acad Sci U S A. 1989 Dec;86(24):10148–10152. doi: 10.1073/pnas.86.24.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerr R., Zvibel I., Chiuten D., D'Olimpio J., Reid L. M. Clonal growth of tumors on tissue-specific biomatrices and correlation with organ site specificity of metastases. Cancer Res. 1989 Jan 15;49(2):384–392. [PubMed] [Google Scholar]
- Ebihara L., Beyer E. C., Swenson K. I., Paul D. L., Goodenough D. A. Cloning and expression of a Xenopus embryonic gap junction protein. Science. 1989 Mar 3;243(4895):1194–1195. doi: 10.1126/science.2466337. [DOI] [PubMed] [Google Scholar]
- Gimlich R. L., Kumar N. M., Gilula N. B. Sequence and developmental expression of mRNA coding for a gap junction protein in Xenopus. J Cell Biol. 1988 Sep;107(3):1065–1073. doi: 10.1083/jcb.107.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Kistler J., Christie D., Bullivant S. Homologies between gap junction proteins in lens, heart and liver. Nature. 1988 Feb 25;331(6158):721–723. doi: 10.1038/331721a0. [DOI] [PubMed] [Google Scholar]
- Kumar N. M., Gilula N. B. Cloning and characterization of human and rat liver cDNAs coding for a gap junction protein. J Cell Biol. 1986 Sep;103(3):767–776. doi: 10.1083/jcb.103.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins L. A., Gruss P., Pozzatti R., Khoury G. Characterization of enhancer elements in the long terminal repeat of Moloney murine sarcoma virus. J Virol. 1984 Jan;49(1):183–189. doi: 10.1128/jvi.49.1.183-189.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J., Malek T. R., Leonard W. J., Greene W. C., Shevach E. M., Germain R. N. Nucleotide sequence and expression of a mouse interleukin 2 receptor cDNA. J Immunol. 1985 Jun;134(6):4212–4217. [PubMed] [Google Scholar]
- Miller T., Dahl G., Werner R. Structure of a gap junction gene: rat connexin-32. Biosci Rep. 1988 Oct;8(5):455–464. doi: 10.1007/BF01121644. [DOI] [PubMed] [Google Scholar]
- Neyton J., Trautmann A. Single-channel currents of an intercellular junction. 1985 Sep 26-Oct 2Nature. 317(6035):331–335. doi: 10.1038/317331a0. [DOI] [PubMed] [Google Scholar]
- Nicholson B., Dermietzel R., Teplow D., Traub O., Willecke K., Revel J. P. Two homologous protein components of hepatic gap junctions. Nature. 1987 Oct 22;329(6141):732–734. doi: 10.1038/329732a0. [DOI] [PubMed] [Google Scholar]
- Okayama H., Berg P. A cDNA cloning vector that permits expression of cDNA inserts in mammalian cells. Mol Cell Biol. 1983 Feb;3(2):280–289. doi: 10.1128/mcb.3.2.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D. L. Molecular cloning of cDNA for rat liver gap junction protein. J Cell Biol. 1986 Jul;103(1):123–134. doi: 10.1083/jcb.103.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somogyi R., Kolb H. A. Cell-to-cell channel conductance during loss of gap junctional coupling in pairs of pancreatic acinar and Chinese hamster ovary cells. Pflugers Arch. 1988 Jul;412(1-2):54–65. doi: 10.1007/BF00583731. [DOI] [PubMed] [Google Scholar]
- Spray D. C., Bennett M. V. Physiology and pharmacology of gap junctions. Annu Rev Physiol. 1985;47:281–303. doi: 10.1146/annurev.ph.47.030185.001433. [DOI] [PubMed] [Google Scholar]
- Spray D. C., Ginzberg R. D., Morales E. A., Gatmaitan Z., Arias I. M. Electrophysiological properties of gap junctions between dissociated pairs of rat hepatocytes. J Cell Biol. 1986 Jul;103(1):135–144. doi: 10.1083/jcb.103.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spray D. C., Saez J. C., Brosius D., Bennett M. V., Hertzberg E. L. Isolated liver gap junctions: gating of transjunctional currents is similar to that in intact pairs of rat hepatocytes. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5494–5497. doi: 10.1073/pnas.83.15.5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson B. R., Siliciano J. D., Mooseker M. S., Goodenough D. A. Identification of ZO-1: a high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J Cell Biol. 1986 Sep;103(3):755–766. doi: 10.1083/jcb.103.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson K. I., Jordan J. R., Beyer E. C., Paul D. L. Formation of gap junctions by expression of connexins in Xenopus oocyte pairs. Cell. 1989 Apr 7;57(1):145–155. doi: 10.1016/0092-8674(89)90180-3. [DOI] [PubMed] [Google Scholar]
- Sáez J. C., Gregory W. A., Watanabe T., Dermietzel R., Hertzberg E. L., Reid L., Bennett M. V., Spray D. C. cAMP delays disappearance of gap junctions between pairs of rat hepatocytes in primary culture. Am J Physiol. 1989 Jul;257(1 Pt 1):C1–11. doi: 10.1152/ajpcell.1989.257.1.1-a. [DOI] [PubMed] [Google Scholar]
- Traub O., Look J., Dermietzel R., Brümmer F., Hülser D., Willecke K. Comparative characterization of the 21-kD and 26-kD gap junction proteins in murine liver and cultured hepatocytes. J Cell Biol. 1989 Mar;108(3):1039–1051. doi: 10.1083/jcb.108.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner R., Miller T., Azarnia R., Dahl G. Translation and functional expression of cell-cell channel mRNA in Xenopus oocytes. J Membr Biol. 1985;87(3):253–268. doi: 10.1007/BF01871226. [DOI] [PubMed] [Google Scholar]
- Young J. D., Cohn Z. A., Gilula N. B. Functional assembly of gap junction conductance in lipid bilayers: demonstration that the major 27 kd protein forms the junctional channel. Cell. 1987 Mar 13;48(5):733–743. doi: 10.1016/0092-8674(87)90071-7. [DOI] [PubMed] [Google Scholar]