Abstract
Red clover (Trifolium pratense L.) is one of the most important legume forage species in temperate livestock agriculture. Tetraploid red clover cultivars are generally producing less seed than diploid cultivars. Improving the seed setting potential of tetraploid cultivars is necessary to utilize the high forage quality and environmentally sustainable nitrogen fixation ability of red clover. In the current study, our aim was to identify candidate genes involved in seed setting. Two genotypes, ‘Tripo’ with weak seed setting and ‘Lasang’ with strong seed setting were selected for transcriptome analysis. De novo and reference based analyses of transcriptome assemblies were conducted to study the global transcriptome changes from early to late developmental stages of flower development of the two contrasting red clover genotypes. Transcript profiles, gene ontology enrichment and KEGG pathway analysis indicate that genes related to flower development, pollen pistil interactions, photosynthesis and embryo development are differentially expressed between these two genotypes. A significant number of genes related to pollination were overrepresented in ‘Lasang’, which might be a reason for its good seed setting ability. The candidate genes detected in this study might be used to develop molecular tools for breeding tetraploid red clover varieties with improved seed yield potentials.
Red clover (Trifolium pratense L.) is a perennial forage legume species. It is outcrossing with a gametophytic self-incompatibility system, and it is cultivated mostly in temperate regions. Due to its nitrogen fixation ability, high protein content and digestibility, red clover is one of the most important forage legumes. Naturally, red clover is diploid (2n = 2X = 14); however, artificially induced tetraploid varieties (2n = 4X = 28) are also in commercial use. Tetraploid plants were first developed in 1939 by treating germinating seeds, young seedlings or apical meristem of diploids with the mitosis-inhibiting chemical colchicine1,2. New tetraploid plants can also be developed by treating diploid plants with nitrous oxide (N2O) and by gametic non-reduction2,3,4. However, red clover breeders develop new tetraploid varieties mainly by crossing plants from two or more tetraploid varieties or breeding lines.
The main advantages of cultivating tetraploid compared to diploid red clover are its higher forage yield, better persistency and tolerance to some diseases like Sclerotinia trifoliorum Eriks1,4,5,6. However, lower seed yield of tetraploid varieties is the major disadvantage compared to diploid cultivars1,7,8.
Seed yield of red clover, especially tetraploids, has not been improved in Scandinavia for a long time. The reasons for this are probably complex. A main reason is that several studies indicate that forage and seed yield are negatively correlated making seed yield improvement difficult9,10,11,12,13. Red clover is primarily grown for forage and forage yield is the main breeding goal; however, seed yield is crucial for the commercial value of new varieties14,15. The outcrossing nature and strong self-incompatibility system of red clover prevent the development of inbred lines and hybrids, thus only a proportion of the potential heterosis for seed yield can be captured in the usual synthetic varieties13,15.
Genomic resources related to seed yield are scarce in red clover compared to other model legume species. Currently, four genetic linkage maps for identification of markers linked to important traits have been developed in red clover12,16,17,18. Several QTL studies of seed yield and seed yield components have been conducted in species like white clover (Trifolium repens L.), soybean (Glycine max L.) and perennial ryegrass (Lolium perenne L.)19,20,21. However, so far, only one QTL study of seed yield has been performed in red clover, and this study identified 38 QTL12.
Rapid advancements in next generation sequencing (NGS) technology allow characterization and quantification of RNA through cDNA sequencing at massive scale22. A draft assembly of the red clover genome based on 16 different genotypes of red clover was recently published23. Furthermore, Yates et al.24 performed de novo transcriptome studies in red clover and provided insights into the drought response. De Vega et al.25 recently assembled a red clover genome to the chromosome level, estimating its size to be ~309 Mb. The group annotated 40,868 genes and identified clusters involved in forage quality and livestock nutrition.
With the availability of new genomic resources in red clover and the advancements in RNA-seq technologies, we performed both de novo and reference (red clover genome) based transcriptome analysis of the global transcriptome response during flower and seed development in two red clover genotypes with contrasting seed setting ability. The aim of this study was to identify molecular responses and to elucidate genes determining seed setting ability in red clover.
Materials and Methods
Plant material
In 2011, nine single plants of each of the low seed yielding variety ‘Tripo’, and the high seed yielding variety ‘Lasang’ were scored for the following seed yield components: number of flower heads per plant, number of florets per flower head, number of seed per flower head, fertility, seed weight per flower head, and length of the corolla tube26. The two lowest ranking plants of ‘Tripo’ and the two highest ranking plants of ‘Lasang’, for the majority of registered seed yield components, were selected for further analysis.
RNA sampling
A total number of 12 flower buds, one bud from each of three flower development periods (early – 12th of July, middle – 21st of July and late – 27th of July) from each of the four selected plants, were picked, flash frozen in liquid N2 and stored at −80 °C until RNA extraction. The frozen flower bud samples were crushed with a pestle and mortar. Using SIGMA SPECTRUM PLANT TOTAL RNA KIT (Sigma Life Science), total RNA was extracted from the 12 flower buds. On-Column DNAse Kit (Sigma Life Science) was used to remove DNA contamination. The quality and concentration of RNA were measured using the NANODROP (Nanodrop Technologies, Wilmington, DE, USA) and BIOANALYZER (Agilent Technologies, Palo Alto, CA, USA) equipment.
RNA-seq library preparation and Illumina sequencing
Twelve flower bud RNA samples with RIN (RNA Integrity Number) values above 7 were used to construct separate cDNA libraries with fragment lengths of 200 bp (±25 bp). Single-end sequencing was performed at the Norwegian Sequencing Centre (NSC), University of Oslo, using the Illumina sequencing platform (HISEQ 2000) generating single-end reads with a length of 50 bp. The FastQC program (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to analyse the quality of the raw sequencing reads.
De novo transcriptome analysis
The de novo assembly was performed in a similar manner as described by Kovi et al.27. Briefly, adapter sequences and low quality reads were removed using the sickle program (https://github.com/najoshi/sickle/blob/master/README.md). The clean reads derived from the four individual genotypes named Tripo42 and Tripo55, Lasang77 and Lasang108, were used to construct separate de novo assemblies for each genotype using the Trinity assembler (release 2013-02-25)28. The de novo assembled transcriptome was then used as a reference to map the individual reads using the Bowtie program29. Transcript abundance was measured for each genotype and time point combination as the expected number of fragments per kilobase (kb) of transcript sequence per million mapped reads (FPKM)30 using RSEM version 1.1.1131.
Identification of differentially expressed genes (DEGs), annotation and gene ontology (GO) analysis
The edgeR package32 in R program language (https://www.r-project.org/) was employed to identify DEGs and a false discovery rate (FDR) of 0.05 was further used to determine the significant DEGs. Transcripts showing differential expression at any flower development time-point were clustered using a K-means clustering algorithm. The annotation of the DEGs were performed using the Blast2GO program33. Initially, BLASTx was performed with an E-value threshold of 10e-06, followed by annotation with a cut-off value of 55 and GO weight Hsp-hit value of 20. The GO enrichment analysis was performed with a p-value of 0.01. The GO classification of DEGs in the two genotypes were generated using the WEGO program34. KEGG Pathway analysis was performed with the Blast2GO program33.
Validation of de novo assembly by CEGMA
The CEGMA software (version 2.4)35 was used to evaluate the quality of the four transcriptome assembly datasets. Several genome and transcriptome assembly studies have used CEGMA for evaluating the quality of assemblies27. CEGMA detects the presence of 248 extremely conserved core eukaryotic genes (CEGs) and their coverage in transcriptome assemblies for evaluation of the completeness of the assembly.
Red clover reference based transcriptome analysis, detecting DEGs and functional annotation
Using a reference-based approach, we mapped all the clean reads from the two genotypes (‘Tripo’ and ‘Lasang’) and the three time points (early, middle and late flower development) to the red clover reference genome25 using STAR, an ultrafast universal RNA-seq. aligner program36. The Cufflinks program30 was used to assemble the transcriptomes and to estimate the transcript abundance, followed by the cuffmerge and cuffdiff programs, which is included with the Cufflinks package. The Cuffmerge program merged the transcriptome assemblies from the three flower development time- points of each genotype for performing differential expression analysis. The Cuffdiff program compared the expression levels of genes and transcripts between the three time-points for each genotype, and detected genes that are up- or down-regulated between the time-points. The merged GTF files obtained from the Cuffmerge program was used in the TransDecoder program37 to identify the coding regions within transcripts. The longest homology coding sequences obtained from TransDecoder were blasted against the Viridiplantae database extracted from NCBI to find the gene names for the coding sequences. Further annotation was performed using the SWISS-PROT database. GFF3 (generic feature format) annotation file describing genomic features, was generated using in-house developed python scripts.
Comparison of significant DEGs to seed yield related QTL
To compare the DEGs with the QTL for seed yield and seed yield traits described by Hermann et al.12, we identified flanking SSR markers associated with the QTL and downloaded the marker sequences from the NCBI database. The chromosome locations of markers and DEGs were identified using the BLAST program with the marker sequences and DEG sequences as the query and the red clover genome sequence25 as the subject. A physical map was created based on the physical location of the DEGs in the red clover genome by the MapDraw software38. Briefly, all the physical location (bp) of DEGs were converted to centimorgan (cM) by an average of 450 kb/cM in red clover and spanned 440 cM across seven linkage groups (LGs), approximately similar to 444 cM of Hermann et al.12.
Results
De novo assembly
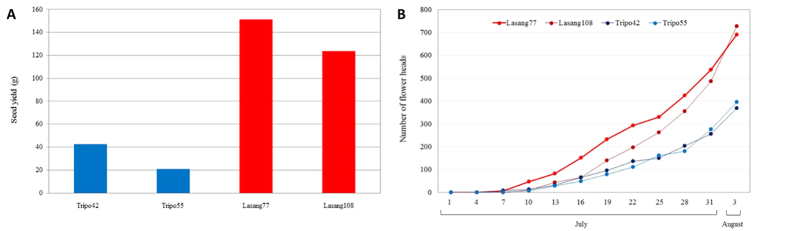
The low seed yielding ‘Tripo’ and the high seed yielding ‘Lasang’ genotypes (Fig. 1) were sequenced and characterized by the de novo transcriptome assembly (Table 1). A total number of 218 million reads of 50 bp were generated for the four genotypes (Tripo42, Tripo55, Lasang77 and Lasang108). 112 million reads were from the ‘Tripo’ genotypes and 106 million reads from the ‘Lasang’ genotypes. Individual transcriptome assemblies were generated for each genotype. The numbers of contigs observed in Lasang108 and Lasang77 were 80,328 (N50 of 930 bp) and 83,489 (N50 of 982 bp), respectively, while in Tripo42 and Tripo55, they were 84,545 (N50 of 1016 bp), and 84,442 (N50 of 982 bp), respectively. The longest contig sizes were 7469, 7295, 7447 and 7339 bp for Lasang108, Lasang77, Tripo42 and Tripo55, respectively. CEGMA analysis determined the complete CEGs (Core Eukaryotic Genes) in Lasang108, Lasang77, Tripo42 and Tripo55 transcriptome assemblies to be 89.11, 92.34, 92.34 and 92.34%, respectively, while the percentage of partially complete CEGs ranged from 97.18 to 97.98 (Table 2). The average number of orthologues per CEG in the four assemblies ranged from 3.18 to 3.30, while the percentage of CEGs that had more than one orthologue ranged from 89.59 to 95.20 (Table 2).
Figure 1.
(A) Cumulative curve for number of flower heads for four tetraploid red clover genotypes: two low seed yielding ‘Tripo’ genotypes and two high seed yielding ‘Lasang’ genotypes. The x-axis indicates the date when counting of flower heads was performed. (B) Seed yield per plant in low seed yielding tetraploid red clover cv. ‘Tripo’ and high seed yielding tetraploid red clover cv. ‘Lasang’.
Table 1. Characteristics of the de novo transcriptome assemblies.
| Genotypes | Total number of contigs | N50 (bp) | Maximum contig length (bp) | Total number of reads |
|---|---|---|---|---|
| Lasang108 | 80,328 | 930 | 7469 | 51,000,000 |
| Lasang77 | 83,489 | 982 | 7295 | 55,000,000 |
| Tripo42 | 84,545 | 1016 | 7447 | 57,000,000 |
| Tripo55 | 84,442 | 982 | 7339 | 55,000,000 |
Table 2. Results of CEGMA analysis for de novo assembly validation.
| Out of 248 CEGs1 | Lasang108 | Lasang77 | Tripo42 | Tripo55 |
|---|---|---|---|---|
| % of fully represented | 89.11 | 92.34 | 92.34 | 92.34 |
| % of at least partially represented | 97.98 | 97.58 | 97.18 | 97.98 |
| Average number of orthologues per CEG | 3.19 | 3.28 | 3.18 | 3.30 |
| % of detected CEGs with more than 1 orthologue | 89.59 | 95.20 | 90.39 | 89.96 |
1CEGs: Core Eukaryotic Genes.
DEGs identified by de novo and reference based methods
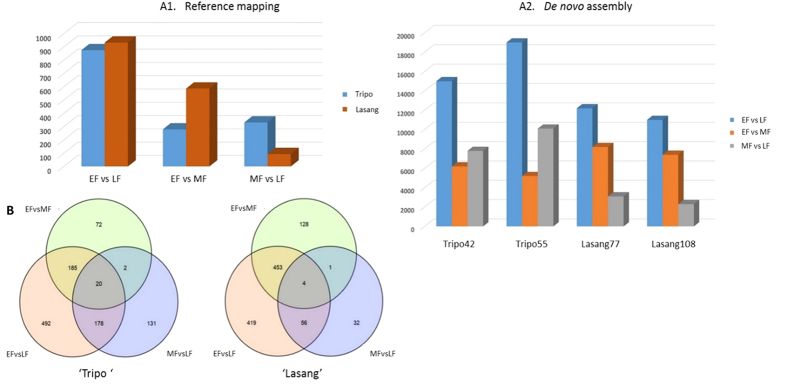
Clean reads from each sample were mapped onto their respective genotype specific de novo assemblies and to the reference genome (red clover genome sequence) to estimate the expression levels of transcripts at different flower development time-points, early (EF), middle (MF) and late (LF) flower development. The DEGs identified in a series of pairwise comparisons between the three flower development time-points EF-LF, EF-MF and LF-MF were 15,000, 7,204 and 7,903, respectively, in Tripo42; 18,105, 6,050 and 10,100, respectively, in Tripo55; 12,040, 8,426 and 2,304, respectively, in Lasang77; and 10,986, 7,492 and 2,430, respectively, in Lasang108 with a false discovery rate (FDR) <0.05 (Fig. 2B). In the reference-based analysis, 875 and 932 DEGs were observed between EF-LF samples; 279 and 586 between EF-MF samples and 331 and 93 between MF-LF samples in the ‘Tripo’ and ‘Lasang’ genotypes, respectively, including up- and down-regulated transcripts (Fig. 2A).
Figure 2.
The number of differentially expressed transcripts identified using reference based assembly (A1) and de novo based assembly (A2). (B) Venn diagrams represents the number of up- and down-regulated transcripts that were common and specific for the pairwise comparisons using the reference based assembly in pairwise comparisons between EF vs LF (early vs. late flowering), EF vs MF (early vs. mid flowering), and MF vs LF (mid vs. late flowering) in genotypes ‘Tripo’ and ‘Lasang’ using a false discovery rate (FDR) of <0.05.
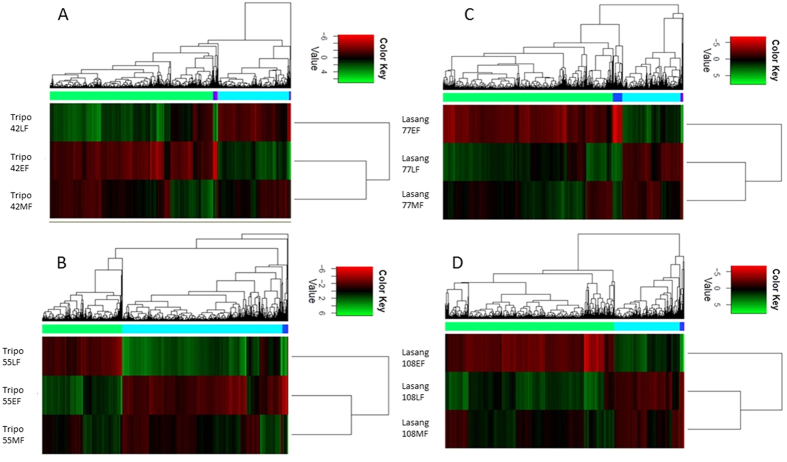
To determine the sample relations, differential expression data from the edgeR program were used to generate heat maps (Fig. 3). EF and MF grouped together in the low seed yielding ‘Tripo’ genotypes, while MF and LF grouped together in the high seed yielding ‘Lasang’ genotypes, indicating that unique genes expressed during late flower development (LF) in ‘Tripo’ and early flower development (EF) in ‘Lasang’ were playing major roles in their flowering and seed setting abilities.
Figure 3. Heat maps of differentially expressed genes detected using de novo assemblies for each genotype and grouped according to their expression patterns.
Y-axis represents the experimental conditions. (A) Tripo42, (B) Tripo55, (C) Lasang77, and (D) Lasang108. EF; early flowering, MF; middle flowering. LF; late flowering.
Blast, annotation and GO of differentially expressed genes
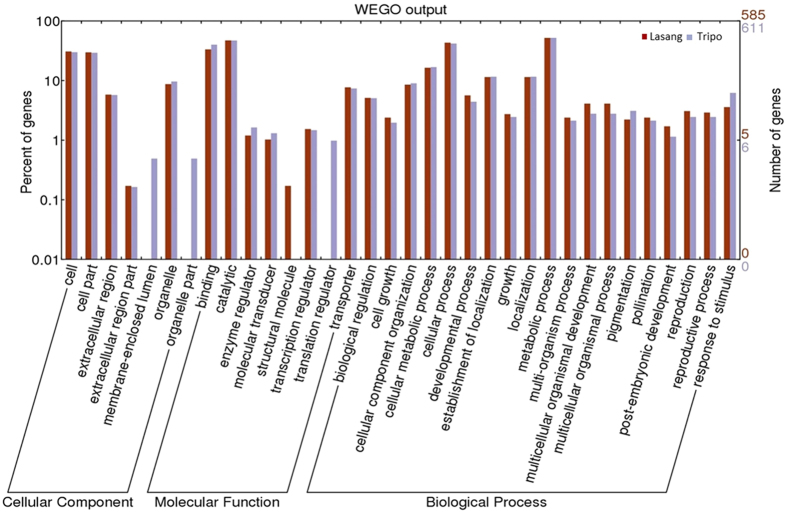
BLASTx was performed for all the DEGs against the Viridiplantae database derived from NCBI. Approximately 80% of the DEGs had blast hits and 60% were annotated using the Blast2GO program33. The top blast hit species were Trifolium subterraneum, followed by Medicago truncatula. Bboth species are closely related to red clover. Gene ontology (GO) classification of DEGs of ‘Tripo’ and ‘Lasang’ were represented as three main GO categories, i.e. cellular component, molecular function and biological process in a histogram (Fig. 4) using the WEGO (Web Gene Ontology Annotation Plot) graphical tool34. GO comparisons between ‘Tripo’ and ‘Lasang’ showed some differences regarding the cellular component and molecular function categories, while relatively small differences were observed for the biological process category. DEGs involved in membrane-enclosed lumen and translation regulator were present only in ‘Tripo’, while DEGs involved in structural molecule were present only in ‘Lasang’.
Figure 4. Gene ontology classifications of differentially expressed genes observed during pairwise comparisons of ‘Tripo’ and ‘Lasang’ genotypes generated by the WEGO tool (http://wego.genomics.org.cn/cgi-bin/wego/index.pl) using the newest GO archive provided.
The results are distributed in three main GO categories: cellular component, molecular function and biological process. The right y-axis indicates the number of genes in a GO category for each genotype. The left y-axis indicates the percentage of a specific category of genes in the respective main categories for each genotype.
Over- or underrepresented GO terms were determined using Fischer’s exact test in the Blast2GO program, and the REVIGO tool for reducing and visualising gene ontologies39. Six GO terms were enriched when compared ‘Tripo’ and ‘Lasang’ genotypes. Out of these, four GO terms, i.e. plasma membrane, pollination, transport and Golgi apparatus were overrepresented in the high seed yielding ‘Lasang’ genotypes. Transcripts assigned to DNA metabolic processes and nucleic acid binding were overrepresented in the low seed yielding ‘Tripo’ genotypes (Fig. 5, Supplementary Figure 1).
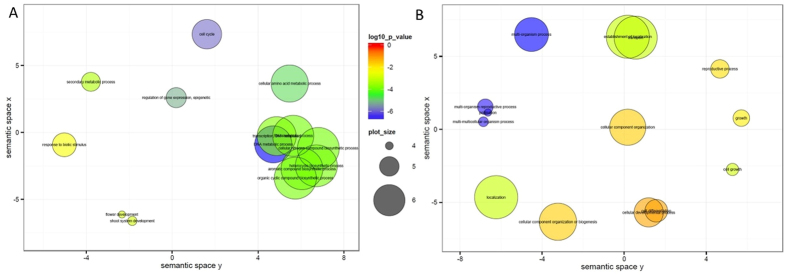
Figure 5. Gene ontology (GO) enrichment analysis by Fischer’s exact test.
The scatterplot of GO terms which are associated with differentially expressed genes in ‘Tripo’ (A) and in ‘Lasang’ (B) shows the cluster representatives (i.e. terms remaining after the redundancy reduction) in a two dimensional space derived by applying multidimensional scaling to a matrix of the GO terms’ semantic similarities. Bubble color indicates p-value (-log10 p-value); size indicates the frequency of the GO term in the underlying GOA database (bubbles of more general terms are larger). In ‘Tripo’ (low seed yield) (A) has higher representation of GO of flower and shoot system development. In ‘Lasang’ (high seed yield) (B) has higher representation of GO for pollination or pollen-pistel interactions (multi-organism process).
Several genes, putatively involved in flower and seed development were detected in these studies, e.g. walls are thin related protein (WAT1), tubby-like F-box protein, gibberellin (GA) 2–beta-dioxygenase, putative aquaporin NIP4-1, zinc finger protein 4, which all were significantly upregulated from the EF to the MF stage, and significantly downregulated from MF to LF (Table 3, Fig. 2). Ethylene-responsive transcription factor (ERF106), probable inorganic phosphate transporter 1–4 (OsPht1;4) were significantly downregulated from the EF to the MF stage, while they were upregulated from MF to LF stage (Table 3, Fig. 2). Furthermore, the Kyoto encyclopedia of genes and genomes (KEGG) database detected different pathways between ‘Tripo’ and ‘Lasang’ at EF-MF and MF-LF stages. In total, 1196 DEGs were involved in 87 pathways (Supplementary Table 1). Pathways with highest representation among the genes were involved in starch and sucrose metabolism (4.84%, 58 genes), pentose and glucoronate interconversions (2.84%, 34 genes), phenylpropanoid biosynthesis (2.75%, 33 genes), purine metabolism (2.34%, 28 genes) and thiamine metabolism (2%, 24 genes).
Table 3. List of differentially expressed genes that can be considered as potential candidate genes involved in seed setting in two red clover genotypes, ‘Tripo’ and ‘Lasang’.
| Sequence ID | Description | Chromosome position | LogFC EF/MF | LogFC MF/LF |
|---|---|---|---|---|
| XLOC_000673 | Probable inorganic phosphate transporter 1–4 | LG1:16251287–16253813 | −100.00 | 100.00 |
| XLOC_001313 | Polyadenylate-binding protein 8 | LG1:2381924–2385791 | −4.37 | −2.69 |
| XLOC_001371 | Polyadenylate-binding protein 2 | LG1:3705560–3712487 | 4.44 | −1.06 |
| XLOC_002319 | Peroxidase 15 | LG1:27442419–27444838 | −4.42 | −4.35 |
| XLOC_002704 | Ethylene-responsive transcription factor ERF106 | LG1:5667372–5667944 | −100.00 | 100.00 |
| XLOC_005163 | Uncharacterized protein | LG2:31978172–31979622 | −4.12 | −0.79 |
| XLOC_008531 | Long chain acyl-CoA synthetase 3 | LG3:4387754–4394714 | 4.78 | −0.45 |
| XLOC_009167 | Type III polyketide synthase A | LG3:18921285–18925387 | −5.42 | −3.29 |
| XLOC_009463 | Gibberellic acid methyltransferase 2 | LG3:26837375–26842634 | −4.62 | −2.65 |
| XLOC_009550 | Protein DETOXIFICATION 45, chloroplastic | LG3:29591163–29602019 | 5.69 | −2.03 |
| XLOC_013980 | Tubby-like F-box protein 13 | LG4:5131256–5132175 | 100.00 | −2.95 |
| XLOC_015206 | Tubby-like F-box protein 11 | LG4:5130358–5131150 | 100.00 | −2.86 |
| XLOC_016540 | Non-specific lipid-transfer protein C6 | LG5:486967–488591 | −6.78 | −5.40 |
| XLOC_016934 | Potassium transporter 19 | LG5:12411777–12412424 | 100.00 | −2.14 |
| XLOC_017036 | Inorganic phosphate transporter 1–1 | LG5:725199–729206 | 4.13 | −0.70 |
| XLOC_019592 | Cucumisin | LG6:5874355–5878558 | 6.48 | −2.67 |
| XLOC_020680 | Alpha-L-fucosidase 3 | LG6:8011079–8011470 | 100.00 | −0.23 |
| XLOC_022367 | Elongation factor TuA, chloroplastic | LG7:23203090–23207956 | 5.30 | −2.26 |
| XLOC_022717 | Uncharacterized protein | LG7:1020381–1022267 | 100.00 | −1.77 |
| XLOC_024498 | Zinc finger protein 4 | LG7:11459754–11460460 | 100.00 | −0.20 |
| XLOC_025967 | Putative aquaporin NIP4-1 | scaf_10543:0–1249 | 100.00 | −2.40 |
| XLOC_028325 | Probable E3 ubiquitin-protein ligase ARI10 | scaf_1364:21411–23094 | 100.00 | −5.16 |
| XLOC_028939 | Glycerophosphodiester phosphodiesterase GDPDL4 | scaf_14591:517–949 | 100.00 | −1.10 |
| XLOC_029741 | Cystinosin homolog | scaf_15837:737–851 | 100.00 | −2.98 |
| XLOC_029976 | Nudix hydrolase 1 | scaf_1621:8252–8547 | 100.00 | −1.89 |
| XLOC_034559 | Probable magnesium transporter NIPA2 | scaf_25880:5–754 | 100.00 | −1.62 |
| XLOC_037847 | 1-aminocyclopropane-1-carboxylate oxidase homolog 1 | scaf_34610:27–594 | 100.00 | −0.94 |
| XLOC_039083 | Probable 2-oxoglutarate/Fe(II)-dependent dioxygenase | scaf_3804:1914–2262 | 100.00 | −3.64 |
| XLOC_039720 | 17.2 kDa class II heat shock protein | scaf_4026:0–243 | 100.00 | −5.90 |
| XLOC_040357 | B3 domain-containing protein | scaf_440:86915–89742 | 100.00 | −3.55 |
| XLOC_041752 | Cytochrome P450 94B1 | scaf_521:81827–84098 | 5.26 | −1.65 |
| XLOC_043901 | L-type lectin-domain containing receptor kinase VIII.2 | scaf_6561:0–1615 | −100.00 | 100.00 |
| XLOC_044539 | Gibberellin 2-beta-dioxygenase | scaf_702:17734–18233 | 100.00 | −0.65 |
| XLOC_045146 | WAT1-related protein | scaf_743:105103–107134 | 100.00 | −2.56 |
DEGs compared to the seed yield QTL
The DEGs identified in this study were compared to seed yield related QTL in order to see if any of the genes identified are co-located with the seed yield QTL as described by Hermann et al.12. Out of 15 SSR markers flanking the seed yield QTL, six SSR markers are located in the corresponding regions as six DEGs detected in this study positioned on four linkage groups (Fig. 6). The six DEGs are myb-related protein MYBAS2, 4-coumarate–CoA ligase-like 2, protein cornichon homolog 3, ethylene-responsive transcription factor ERF113, protein DETOXIFICATION 45, and UDP-glucuronate 4-epimerase 4.
Figure 6. Comparative mapping of significant differentially expressed genes (DEGs) detected in this study to the red clover seed yield related QTL (Hermann et al., 2006).
The distribution of DEGs on the seven linkage groups (LG) of red clover. The SSR markers (denoted in brackets) (Hermann et al., 2006), linked to QTL (denoted in bars) which are co-located to the DEGs are highlighted. In LG1, MYBA2 mapped to the QTL for thousand seed weight (TSW). In LG2, 4CLL2 and CNIH3 mapped to QTL for time of flowering (TOF) and TSW. In LG3, EF113 mapped to QTL for seed yield per plant (SYP). In LG6, GAE4, mapped to the QTL for seed yield per head (SYH).
Discussion
Comparative analysis between de novo and reference based transcriptome assays
When a reference genome is available, reference-based approaches have been considered more effective than de novo assembly (Martin and Wang, 2011), but very few studies have compared the two strategies27,40. Moreover, it is important to see whether the de novo assembly can detect the same genes and the molecular responses even in the absence of a reference genome. In the present transcriptome analysis of red clover, we compared both strategies. The CEGMA analysis showed that the de novo assemblies were very complete in terms of gene content since they captured high percentages of ultra-conserved CEGs in all assemblies of the ‘Tripo’ and ‘Lasang’ genotypes. De novo and reference-based (red clover reference genome) gene expression data indicated that genes expressed during the early flower development stage (EF) in ‘Lasang’ and during the late flower development stage (LF) in ‘Tripo’ might play key roles in their differential seed setting abilities. In both de novo and the reference-based mapping, the pattern of the differentially expressed transcripts was similar. A larger number of differentially expressed transcripts was observed in early vs late flower development stage than in middle vs late and middle vs early stage (Figs 2 and 3). This might be due to the presence of several differentially expressed transcripts at all three stages. In addition, there was a larger number of differentially expressed transcripts at the early vs middle flower development stage in ‘Lasang’ than in ‘Tripo’, whereas there was more differentially expressed transcripts at the early vs late stage in ‘Tripo’ compared to ‘Lasang’ (Fig. 2). Furthermore, we found the proportion of differentially expressed transcripts to be higher in de novo compared to the reference based mapping, which is similar to the findings of Kovi et al.27. The trinity de novo assembler yield more transcripts due to the lack of strand-specific information. However, most of the differentially expressed genes identified in both these methods were related to the Medicago truncatula (Supplementary Figure 2), which is the most closely related species to red clover. Furthermore, both methods identified many similar candidate genes putatively involved in flower and seed development (Table 3), thus demonstrating the potential of the de novo method of capturing genes even in the absence of a reference genome. This comparative analysis study might be very useful for the researchers working on orphan species with no reference genome.
Potential candidate genes involved in flower and seed development
Several genes putatively involved in flower development were detected in this study (Table 3). WAT1 related protein is a cell wall protein mainly responsible for transmembrane transporter activity (http://www.uniprot.org/uniprot/Q94AP3). Ranocha et al.41 reported that stem apices in the mutant wat1 produced significantly lower seed yields in Arabidopsis thaliana compared to wild type stem apices. It might be that the downregulated expression of this gene in ‘Tripo’ flower buds in the early and middle flower development periods, negatively affected its seed setting ability and thus seed yield.
Tubby-like proteins are involved in abscisic acid (ABA) signaling pathways and plays a key role in seed germination and early seed growth42. In a recent study, Verma et al.43 identified a tubby-like F-box protein as a potential candidate gene for the seed weight QTL qSW in chickpea (Cicer arietinum L.). Gibberellin 2–beta-dioxygenase was highly expressed in EF and MF. According to Xue et al.44, genes that encodes gibberellin 2-beta-dioxygenase 1 were highly expressed in rice embryo.
NIP4-1 belongs to the aquaporin gene family, which are small integral membrane proteins that facilitate water and solute movement across different tissues throughout development and growth45. Regulation of water and nutrient state is very relevant for pollen development, pollen tube growth and germination46. Recently Di Giorgio et al.47 showed that NIP4-1 and NIP4-2 are required for pollen development and pollination in Arabidopsis thaliana. Furthermore, single nip4;1 mutant plants showed a significantly higher frequency of abnormal, stunted siliques and fewer seeds when compared with the wild type47. This indicate that NIP4-1 plays a prominent role in determining seed yield. In our studies, the significant upregulation of this gene in ‘Lasang’ during the EF and MF stages might play a key role in determining the better seed yielding capacity of this cultivar.
Zinc finger proteins (ZFP) play an important role in various biological functions, such as plant growth and development (flower, shoot, seed, pistil and leaf)48,49. Recently it was found that ZFP3, ZFP4 and the related ZFP subfamily of zinc finger factors regulate light and ABA responses during germination and early seedling development50. Higher expression of ZFP4 during the EF and MF stages indicate that it might be important for seed setting in our tetraploid red clover genotypes.
The gene ERF106 belongs to the APETALA2 (AP2) gene family, which controls seed weight (Ohto et al.)51, was overexpressed during the EF and MF stages in ‘Lasang’ flower buds. APETALA2 influences the development of embryo, endosperm and seed coat51. According to Xue et al.44, genes involved in ethylene mediated signaling were highly expressed in rice developing seeds.
The rice gene OsPht1,4 belongs to a group of genes that regulate phosphorus homeostasis in plant cells52. Jia et al.53 reported that suppression of OsPT4 in rice resulted in lower P content in unfilled rice grains, which again resulted in lower seed yields. This gene was overexpressed during the MF and LF stages in ‘Lasang’ flower buds indicating its positive effect on seed yield. This gene is also involved in the embryo development in rice54.
GO differences in ‘Tripo’ and ‘Lasang’
Gene ontology (GO) has provided a way of consistently describing genes and proteins to computationally process data at the functional level34,39. Gene ontology (GO) comparisons between the two genotypes showed differences regarding the cellular component and molecular function categories (Fig. 4). Six GO terms were enriched between these two genotypes. Out of these four GO terms, plasma membrane, pollination, transport and Golgi apparatus were overrepresented in the high seed yielding ‘Lasang’. Transcripts assigned to DNA metabolic processes and nucleic acid binding were overrepresented in the low seed yielding ‘Tripo’ (Fig. 5; Supplementary Figure 1). Genes, such as pollen-specific leucine-rich repeat extensin-like protein 1-like (PEX1), pollen profiling variant 1, phd finger protein male sterility 1–like (MS1-PHD), and polypyrimidine tract-binding protein, were observed in the pollination GO term. PEX1 reported to be involved in reproduction with in the pollen tube wall during its rapid growth55. Another gene, MS1-PHD encodes a PHD-type transcription factor and regulates pollen and tapetum development and pollen wall biosynthesis56. In the GO term plasma membrane, genes like aberrant pollen transmission (APT1), flotillin-like protein, sodium transporter hkt1-like were observed. Xu and Dooner57 showed that the APT1 protein is involved in membrane trafficking and is required for the high secretory demands of tip growth in pollen tubes. Most of the overrepresented genes are linked to pollen development, which is crucial for fertility and seed setting, thus likely involved in determining the higher seed yield capacity of ‘Lasang’ compared with ‘Tripo’.
Validation of the DEGs by comparing to previous red clover seed yield QTL
Comparative mapping studies are powerful tools to validate the detected DEGs by comparing the sequences of the genes to the sequences of markers located inside or flanking QTL58. The physical map of DEGs was created based on their physical locations (bp) in the red clover genome. However, there is no constant ratio to convert between bp and cM, as some regions of the genome with frequent recombination have fewer bp per cM than regions with low recombination. The best approach might be to pick the most detailed genetic map (in our case ref. 12), fetch the sequences for each SSR marker on the map, and BLAST these marker sequences against the genome sequence (red clover genome). From the BLAST analysis, we were able to translate each of these cM distances into a bp distance between the points of alignment from the markers to the chromosome, thus calculated as 450 kb/cM. A similar approach was carried out in Arabidopsis by estimating genetic distance as 250 kb/cM59. In this study, we detected six DEGs that mapped to the seed yield QTL regions identified by Hermann et al.12, and positioned on linkage groups LG1, LG2, LG3 and LG6 (Fig. 6). Among them, MYB transcription factors play a key role in plant development, pollen development60, pollen tube differentiation61, floral initiation and seed development62. The gene ‘protein cornichon homolog’ belongs to a conserved protein family found in eukaryotes demonstrated to participate in the selection of integral membrane proteins as cargo for their correct targeting63. Further, Man et al.58 detected protein cornichon homolog as a potential gene encoding for the yield related QTL in cotton. 4-coumarate–CoA ligase-like 2, belongs to a group of essential enzymes involved in the phenylpropanoid-derived compound (PDC) pathway, which generates various secondary compounds like lignin, anthocyanins, and isoflavonoids. Doughty et al.64 suggests that flavonoids may play a fundamental role in regulating communication between the seed coat and the endosperm also.
Conclusions
In this study, transcriptome analysis was conducted for cv. ‘Tripo’ with inferior seed setting ability and two from cv.‘Lasang’ with improved seed setting ability, and several DEGs were identified. Many genes related to pollination, flower and seed development were upregulated during the early to middle (EF-MF) flower development stage in the ‘Lasang’ and downregulated during the middle to late (MF-LF) flower development stage in the ‘Tripo’, indicating their major role in determining seed setting and potential seed yield. GO enrichment analysis further confirmed that plasma membrane, pollination, transport and Golgi apparatus related genes are overrepresented in the ‘Lasang’. Further, comparative mapping, co-located six seed yield related QTL to the six DEGs on the same linkage groups, thus validating the detected DEGs in this study. Putative candidate genes detected in this study might provide a basis for future functional genomics research in understanding the biology of seed yield in red clover. Loss-of-function techniques like RNA interference methods can further be used to understand the role of these genes in the seed setting.
Additional Information
Accession codes: The raw Illumina sequencing data generated in this study were deposited in the EMBL-EBI ArrayExpress Archive, under accession number E-MTAB-5117. De novo transcriptome assemblies of four red clover genotypes generated by trinity program are deposited in DRYAD Digital Repository along with the GFF3 annotation files and script. (http://datadryad.org/resource/doi:10.5061/dryad.0bk52).
How to cite this article: Kovi, M. R. et al. De novo and reference transcriptome assembly of transcripts expressed during flowering provide insight into seed setting in tetraploid red clover. Sci. Rep. 7, 44383; doi: 10.1038/srep44383 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by the Norwegian Research Council (NRC) grant No. 209702 (Industrial PhD program) and by Graminor Breeding AS. We are also grateful to Elena Gusakova for the help with RNA extraction. We sincerely acknowledge the efforts of Torben Asp from Arhus University and Tina Graceline Kirubakaran from CIGENE, NMBU for providing valuable suggestions of bioinformatics analysis.
Footnotes
The authors declare no competing financial interests.
Author Contributions M.A. and O.A.R. designed the study with inputs from H.A. and M.R.K. H.A. performed phenotype experiments and collected plant material. M.R.K. was responsible for RNA sequencing, bioinformatics and expression analysis. M.R.K. and H.A. drafted the manuscript with inputs from M.A. and O.A.R. All authors read and approved the final manuscript.
References
- Sjödin J. & Ellerström S. In Research and Results in Plant Breeding (eds Olsson G.) 102–113 (1986). [Google Scholar]
- Boller B., Schubiger F. X. & Kölliker R. In Fodder Crops and Amenity Grasses (eds Beat B. K., Ulrich P. & Fabio V.) 211–260 (Springer New York, 2010). [Google Scholar]
- Meglic V. & Smith R. R. Self-Incompatibility and Seed Set in Colchicine-, Nitrous Oxide-, and Sexually Derived Tetraploid Red Clover. Crop Sci. 32, 1133–1137, doi: 10.2135/cropsci1992.0011183X003200050013x (1992). [DOI] [Google Scholar]
- Taylor N. L. & Quesenberry K. H. In Current Plant Science and Biotechnology in Agriculture Vol. 28 (Red Clover Science, Kluwer Academic Publishers, Dordrecht, 1996). [Google Scholar]
- Vestad R. In Present status and future prospects of Norwegian plant breeding (ed. Rognli O. A.) Meld. Norg.: Landbr, 165–172 (1990). [Google Scholar]
- Vleugels T., Cnops G. & van Bockstaele E. Screening for resistance to clover rot (Sclerotinia spp.) among a diverse collection of red clover populations (Trifolium pratense L.). Euphytica 194, 371–382, doi: 10.1007/s10681-013-0949-4 (2013). [DOI] [Google Scholar]
- Wexelsen H. & Vestad R. Observations on pollination and seed setting in diploid and tetraploid red clover. 64–68 (European Grassland Conference, Paris, 1954). [Google Scholar]
- Valle O. In Precedings of the Symposium on fertility in tetraploid red clover (ed Ellerström S.) 28–33 (Eucarpia, Section “Fodder crops” group, Svalof, Sweden, 1961). [Google Scholar]
- Clifford P. T. P. & Baird I. J. In Proceedings of the XVII International Grassland Congress 1678–1679 (Palmerston North, New Zealand, 1993). [Google Scholar]
- Steiner J. J., Smith R. R. & Alderman S. C. Red Clover Seed Production: IV. Root Rot Resistance under Forage and Seed Production Systems. Crop Sci. 37, 1278–1282, doi: 10.2135/cropsci1997.0011183X003700040042x (1997). [DOI] [Google Scholar]
- Vasiljević S. et al. Mutual relationships among green forage and seed yield components in genotypes of red clover (Trifolium pratense L.). Genetika 32, 188–191 (2000). [Google Scholar]
- Herrmann D., Boller B., Studer B., Widmer F. & Kölliker R. QTL analysis of seed yield components in red clover (Trifolium pratense L.). Theor. Appl. Genet. 112, 536–545, doi: 10.1007/s00122-005-0158-1 (2006). [DOI] [PubMed] [Google Scholar]
- Sleper D. A. & Poehlman J. M. In Breeding field crops (Blackwell publishing, 2006). [Google Scholar]
- Ravagnani A., Abberton M. T. & Skøt L. Development of Genomic Resources in the Species of Trifolium L. and Its Application in Forage Legume Breeding. Agronomy 2, 116 (2012). [Google Scholar]
- Annicchiarico P., Barrett B., Brummer E. C., Julier B. & Marshall A. H. Achievements and Challenges in Improving Temperate Perennial Forage Legumes. Crit. Rev. Plant Sci. 34, 327–380, doi: 10.1080/07352689.2014.898462 (2015). [DOI] [Google Scholar]
- Isobe S., Klimenko I., Ivashuta S., Gau M. & Kozlov N. N. First RFLP linkage map of red clover (Trifolium pratense L.) based on cDNA probes and its transferability to other red clover germplasm. Theor. Appl. Genet. 108, 105–112, doi: 10.1007/s00122-003-1412-z (2003). [DOI] [PubMed] [Google Scholar]
- Sato S. et al. Comprehensive structural analysis of the genome of red clover (Trifolium pratense L.). DNA Res. 12, 301–364, doi: 10.1093/dnares/dsi018 (2005). [DOI] [PubMed] [Google Scholar]
- Isobe S. et al. Construction of a consensus linkage map for red clover (Trifolium pratense L.). BMC Plant Biol. 9, 1–11, doi: 10.1186/1471-2229-9-57 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett B. A., Baird I. J. & Woodfield D. R. A QTL Analysis of White Clover Seed Production. Crop Sci. 45, 1844–1850, doi: 10.2135/cropsci2004.0679 (2005). [DOI] [Google Scholar]
- Mansur L. M. et al. Genetic Mapping of Agronomic Traits Using Recombinant Inbred Lines of Soybean. Crop Sci. 36, 1327–1336, doi: 10.2135/cropsci1996.0011183X003600050042x (1996). [DOI] [Google Scholar]
- Cogan N. O. I. et al. QTL analysis and comparative genomics of herbage quality traits in perennial ryegrass (Lolium perenne L.). Theor. Appl. Genet. 110, 364–380, doi: 10.1007/s00122-004-1848-9 (2005). [DOI] [PubMed] [Google Scholar]
- Shendure J. & Ji H. Next-generation DNA sequencing. Nat. Biotechnol. 26, 1135–1145, doi: 10.1038/nbt1486 (2008). [DOI] [PubMed] [Google Scholar]
- Istvanek J., Jaros M., Krenek A. & Repkova J. Genome assembly and annotation for red clover (Trifolium pratense; Fabaceae). Am. J. Bot. 101, 327–337, doi: 10.3732/ajb.1300340 (2014). [DOI] [PubMed] [Google Scholar]
- Yates S. A. et al. De novo assembly of red clover transcriptome based on RNA-Seq data provides insight into drought response, gene discovery and marker identification. BMC Genomics 15, 1–15, doi: 10.1186/1471-2164-15-453 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vega J. J. et al. Red clover (Trifolium pratense L.) draft genome provides a platform for trait improvement. Sci. Rep. 5, 17394, doi: 10.1038/srep17394 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amdahl H. et al. Seed Yield Components in Single Plants of Diverse Scandinavian Tetraploid Red Clover Populations (Trifolium pratense L.). Crop Sci., doi: 10.2135/cropsci2016.05.0321 (2016). [DOI] [Google Scholar]
- Kovi M. R. et al. Global transcriptome changes in perennial ryegrass during early infection by pink snow mould. Sci. Rep. 6, 28702, doi: 10.1038/srep28702 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652, doi: 10.1038/nbt.1883 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M. & Salzberg S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, 1–10, doi: 10.1186/gb-2009-10-3-r25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotech. 28, 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B. & Dewey C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323, doi: 10.1186/1471-2105-12-323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J. & Smyth G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140, doi: 10.1093/bioinformatics/btp616 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A. & Gotz S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics 2008, 619832, doi: 10.1155/2008/619832 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J. et al. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 34, W293–297, doi: 10.1093/nar/gkl031 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G., Bradnam K. & Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23, 1061–1067, doi: 10.1093/bioinformatics/btm071 (2007). [DOI] [PubMed] [Google Scholar]
- Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. J. et al. De novo transcript sequence reconstruction from RNA-Seq: reference generation and analysis with Trinity. Nature protocols 8, 10.1038/nprot.2013.1084, doi: 10.1038/nprot.2013.084 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R. H. & Meng J. L. [MapDraw: a microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data]. Yi chuan = Hereditas/Zhongguo yi chuan xue hui bian ji 25, 317–321 (2003). [PubMed] [Google Scholar]
- Supek F., Bošnjak M., Škunca N. & Šmuc T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 6, e21800, doi: 10.1371/journal.pone.0021800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. A., Ponnala L. & Weber C. A. Strategies for transcriptome analysis in nonmodel plants. Am. J. Bot. 99, 267–276, doi: 10.3732/ajb.1100334 (2012). [DOI] [PubMed] [Google Scholar]
- Ranocha P. et al. Arabidopsis WAT1 is a vacuolar auxin transport facilitator required for auxin homoeostasis. Nat. Commun. 4, 2625, doi: 10.1038/ncomms3625 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y. et al. Characterization of Arabidopsis Tubby-like proteins and redundant function of AtTLP3 and AtTLP9 in plant response to ABA and osmotic stress. Plant Mol. Biol. 86, 471–483, doi: 10.1007/s11103-014-0241-6 (2014). [DOI] [PubMed] [Google Scholar]
- Verma S. et al. High-density linkage map construction and mapping of seed trait QTLs in chickpea (Cicer arietinum L.) using Genotyping-by-Sequencing (GBS). Sci. Rep. 5, 17512, doi: 10.1038/srep17512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L. J., Zhang J. J. & Xue H. W. Genome-Wide Analysis of the Complex Transcriptional Networks of Rice Developing Seeds. PLoS ONE 7, e31081, doi: 10.1371/journal.pone.0031081 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan K., Agarwal P., Shanware A. & Sane V. A. Heterologous Expression of Two Jatropha Aquaporins Imparts Drought and Salt Tolerance and Improves Seed Viability in Transgenic Arabidopsis thaliana. PLoS ONE 10, e0128866, doi: 10.1371/journal.pone.0128866 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firon N., Nepi M. & Pacini E. Water status and associated processes mark critical stages in pollen development and functioning. Ann. Bot. 109, 1201–1214, doi: 10.1093/aob/mcs070 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giorgio J. A. et al. Pollen-Specific Aquaporins NIP4;1 and NIP4;2 Are Required for Pollen Development and Pollination in Arabidopsis thaliana. Plant Cell 28, 1053–1077, doi: 10.1105/tpc.15.00776 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M. et al. Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96, 296–301 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo K. I., Kanno Y., Nishino T. & Takatsuji H. Zinc-Finger Genes That Specifically Express in Pistil Secretory Tissues of Petunia. Plant Cell Physiol. 41, 377–382, doi: 10.1093/pcp/41.3.377 (2000). [DOI] [PubMed] [Google Scholar]
- Joseph M. P. et al. The Arabidopsis ZINC FINGER PROTEIN3 Interferes with Abscisic Acid and Light Signaling in Seed Germination and Plant Development. Plant Physiol. 165, 1203–1220, doi: 10.1104/pp.113.234294 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto M. A., Floyd S. K., Fischer R. L., Goldberg R. B. & Harada J. J. Effects of APETALA2 on embryo, endosperm, and seed coat development determine seed size in Arabidopsis. Sex. Plant Reprod. 22, 277–289, doi: 10.1007/s00497-009-0116-1 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y. et al. The Phosphate Transporter Gene OsPht1;4 Is Involved in Phosphate Homeostasis in Rice. PLoS ONE 10, e0126186, doi: 10.1371/journal.pone.0126186 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H. et al. The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant Physiol. 156, 1164–1175, doi: 10.1104/pp.111.175240 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F. et al. Involvement of OsPht1;4 in phosphate acquisition and mobilization facilitates embryo development in rice. Plant J. 82, 556–569, doi: 10.1111/tpj.12804 (2015). [DOI] [PubMed] [Google Scholar]
- Rubinstein A. L., Broadwater A. H., Lowrey K. B. & Bedinger P. A. Pex1, a pollen-specific gene with an extensin-like domain. Proc. Natl. Acad. Sci. USA (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Vizcay-Barrena G., Conner K. & Wilson Z. A. MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell 19, 3530–3548, doi: 10.1105/tpc.107.054981 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z. & Dooner H. K. The Maize aberrant pollen transmission 1 Gene Is a SABRE/KIP Homolog Required for Pollen Tube Growth. Genetics 172, 1251–1261, doi: 10.1534/genetics.105.050237 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man W. et al. A comparative transcriptome analysis of two sets of backcross inbred lines differing in lint-yield derived from a Gossypium hirsutum x Gossypium barbadense population. Mol. Genet. Genomics 291, 1749–1767, doi: 10.1007/s00438-016-1216-x (2016). [DOI] [PubMed] [Google Scholar]
- Lukowitz W., Gillmor C. S. & Scheible W.-R. Positional Cloning in Arabidopsis. Why It Feels Good to Have a Genome Initiative Working for You. Plant Physiol. 123, 795–806, doi: 10.1104/pp.123.3.795 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan H. A., Iacuone S., Li S. F. & Parish R. W. The MYB80 Transcription Factor Is Required for Pollen Development and the Regulation of Tapetal Programmed Cell Death in Arabidopsis thaliana. Plant Cell 23, 2209–2224, doi: 10.1105/tpc.110.082651 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leydon A. R. et al. Three MYB transcription factors control pollen tube differentiation required for sperm release. Curr. Biol. 23, 1209–1214, doi: 10.1016/j.cub.2013.05.021 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodger F. J., Gubler F., Pogson B. J. & Jacobsen J. V. A Mak-like kinase is a repressor of GAMYB in barley aleurone. Plant J. 33, 707–717 (2003). [DOI] [PubMed] [Google Scholar]
- Rosas-Santiago P. et al. Identification of rice cornichon as a possible cargo receptor for the Golgi-localized sodium transporter OsHKT1;3. J. Exp. Bot. 66, 2733–2748, doi: 10.1093/jxb/erv069 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doughty J., Aljabri M. & Scott R. J. Flavonoids and the regulation of seed size in Arabidopsis. Biochem. Soc. Trans. 42, 364–369, doi: 10.1042/bst20140040 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.