Comparative genome analysis of strains of a pathogenic bacterial species can be a powerful tool to discover acquisition of mobile genetic elements related to virulence. Here, we compared 28 V. anguillarum strains that differed in virulence in fish larval models. By pan-genome analyses, we found that six of nine highly virulent strains had a unique core and accessory genome. In contrast, V. anguillarum strains that were medium to nonvirulent had low genomic diversity. Integration of genomic and phenotypic features provides insights into the evolution of V. anguillarum and can also be important for survey and diagnostic purposes.
KEYWORDS: pan-genome, genomics, virulence factors
ABSTRACT
Vibrio anguillarum is a marine bacterium that can cause vibriosis in many fish and shellfish species, leading to high mortalities and economic losses in aquaculture. Although putative virulence factors have been identified, the mechanism of pathogenesis of V. anguillarum is not fully understood. Here, we analyzed whole-genome sequences of a collection of V. anguillarum strains and compared them to virulence of the strains as determined in larval challenge assays. Previously identified virulence factors were globally distributed among the strains, with some genetic diversity. However, the pan-genome revealed that six out of nine high-virulence strains possessed a unique accessory genome that was attributed to pathogenic genomic islands, prophage-like elements, virulence factors, and a new set of gene clusters involved in biosynthesis, modification, and transport of polysaccharides. In contrast, V. anguillarum strains that were medium to nonvirulent had a high degree of genomic homogeneity. Finally, we found that a phylogeny based on the core genomes clustered the strains with moderate to no virulence, while six out of nine high-virulence strains represented phylogenetically separate clusters. Hence, we suggest a link between genotype and virulence characteristics of Vibrio anguillarum, which can be used to unravel the molecular evolution of V. anguillarum and can also be important from survey and diagnostic perspectives.
IMPORTANCE Comparative genome analysis of strains of a pathogenic bacterial species can be a powerful tool to discover acquisition of mobile genetic elements related to virulence. Here, we compared 28 V. anguillarum strains that differed in virulence in fish larval models. By pan-genome analyses, we found that six of nine highly virulent strains had a unique core and accessory genome. In contrast, V. anguillarum strains that were medium to nonvirulent had low genomic diversity. Integration of genomic and phenotypic features provides insights into the evolution of V. anguillarum and can also be important for survey and diagnostic purposes.
INTRODUCTION
The genus Vibrio belongs to a family of heterotrophic marine bacteria that includes many facultative symbiotic or pathogenic strains (1). Human pathogens include Vibrio cholerae, the causative agent of cholera (2), and V. parahaemolyticus and V. vulnificus, which are responsible for most cases of seafood poisoning (3). However, Vibrio infections are also common in marine organisms, as demonstrated by reports of V. coralliilyticus being capable of killing coral tissue (4), and several species of Vibrio are also of major concern in the aquaculture industry (5, 6). Vibrio (Listonella) anguillarum is the causative agent of a fatal hemorrhagic septicemic disease (vibriosis) and is one of the most important pathogens in the aquaculture and larviculture industry, infecting ~50 species of fish, molluscans, and crustaceans (7). Twenty-three different serotypes (O1 to O23) have been described, with serotypes O1, O2, and to some extent O3 being associated with fish vibriosis (8, 9). The other V. anguillarum serotypes are mostly nonpathogenic and represent environmental strains isolated from seawater, plankton, and sediment.
Although the mechanism of pathogenesis of V. anguillarum is not completely understood, virulence-related factors have been identified and include chemotaxis and motility (10, 11), adhesion (12), invasion (13, 14), iron sequestration (15, 16), and secretion of extracellular enzymes (17, 18). Several putative virulence genes have been detected in the genome of V. anguillarum strain H775-3 (the pJM1-cured strain of 775), including genes encoding exotoxins, adherence/colonization factors, invasion, capsule and cell surface components, and an iron uptake system (19, 20). The first complete genome sequence of V. anguillarum strain 775 revealed several genomic features that could explain the pathogenicity of the organism, including the presence of the virulence plasmid pJM1, 10 genomic islands (GIs), potential virulence factors, toxins, and genes evolved in biofilm formation (21). However, genome comparison analyses have demonstrated strain-specific toxins in other virulent V. anguillarum strains linked with the absence of the plasmid pJM1, suggesting that V. anguillarum strains have evolved different potential virulence mechanisms (21). In contrast, comparative genome analysis of 15 V. anguillarum isolates of serotypes O1, O2, and O3 revealed low genetic diversity, and the distribution of putative virulence factors was similar to that in strain 775, suggesting that virulence in V. anguillarum is multifactorial (22). The genotypes were compared in a subsequent study to virulence of 15 V. anguillarum strains using gnotobiotic European sea bass larvae as the model host (23). No clear correlation between virulence and genotypic was found, and more detailed analyses of whole-genome sequences in comparison with standardized virulence data are required to elucidate the evolution, physiology, and pathogenesis of this bacterium (23). This has been the purpose of the present study.
The microbial genome is divided into core and accessory elements, which combined constitute the pan-genome (24). The core genome includes the pool of genes shared by all the strains of the same bacterial species and typically contains genes required for the essential housekeeping functions of the cell. In contrast, the accessory genome comprises genes found in only some strains and includes genes acquired by horizontal gene transfer events (25). This strain-specific genome could be involved in functions related to pathogenicity, such as niche adaptation (26), antibiotic resistance (27), or production of strain-specific virulence factors (28), which are known to reside within genomic islands (29). Here, we whole-genome sequenced 26 V. anguillarum strains isolated from different geographic localities and from different years and hosts. We also included the genomes of two previously sequenced strains, 775 (21) and NB10 (30), and investigated the core genome and the accessory genome to identify virulence genes and explain differences in virulence potential among V. anguillarum strains. Our genomic analyses were compared to virulence of the strains as determined in cod, turbot, and halibut larval models (31).
RESULTS
Larval mortality caused by V. anguillarum strains.
Infection trials with cod, turbot, and halibut larvae divided the 28 strains into four groups of high, medium, low, or no mortality (31) (see Table S1 in the supplemental material).
Average mortality of fish larvae following infection with Vibrio anguillarum at high dose (HD) and low dose (LD). Download TABLE S1, DOCX file, 0.1 MB (17.3KB, docx) .
Copyright © 2017 Castillo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genome features of V. anguillarum strains.
The chromosome sizes of the 28 V. anguillarum strains ranged from 3.06 to 3.34 and 0.99 to 1.12 Mb for chromosomes I and II, respectively. The GC contents ranged from 44.0 to 44.8% and 43.6 to 44.1% for chromosomes I and II, respectively. The plasmid pJM1 was found in 17 of the 28 strains (Table 1; see Table S2 in the supplemental material). A total of 3,334 to 3,767 coding sequences (CDS) were predicted per strain (both chromosomes) (Table S2).
TABLE 1 .
Overview of the V. anguillarum strains analyzed in this study
| Strain | Origin | Yr of isolation | Isolate host | Serotype | Plasmid pJM1 | Accession no. |
|
|---|---|---|---|---|---|---|---|
| Chromosome I/II | Plasmid | ||||||
| 4299 | Norway | Unknown | Unknown | O2b | − | CP011458/CP011459 | |
| 87-9-116 | Finland | 1987 | Atlantic salmon | O1 | − | CP010044/CP010045 | |
| 87-9-117 | Finland | 1987 | Rainbow trout | O1 | + | CP010046/CP010047 | CP016253 |
| 90-11-286 | Denmark | 1990 | Rainbow trout | O1 | − | CP011460/CP011461 | |
| 90-11-287 | Denmark | 1990 | Rainbow trout | O1 | + | CP011475/CP011476 | CP016254 |
| 91-7-154 | Denmark | 1991 | Turbot | O1 | + | CP010082/CP010083 | CP016255 |
| 178/90 | Italy | Unknown | Sea bass | O1 | + | CP011470/CP011471 | CP016257 |
| 601/90 | Italy | Unknown | Sea bass | O1 | + | CP010076/CP010077 | CP016259 |
| 775 | United States | Unknown | Coho salmon | O1 | + | CP002284/CP002285 | AY312585 |
| 9014/8 | Denmark | 1990 | Rainbow trout | O1 | + | CP010038/CP010039 | CP016262 |
| DMS21597 | Norway | Unknown | Atlantic cod | O2 | − | CP010084/CP010085 | |
| H610 | Norway | Unknown | Atlantic cod | O2a | − | CP011462/CP011463 | |
| NB10 | Sweeden | Unknown | Unknown | O1 | + | LK021130/LK021129 | LK021128 |
| PF4 | Chile | 2004 | Salmon salar | O3 | − | CP010080/CP010081 | |
| PF7 | Chile | 2004 | Salmon salar | O3 | − | CP011464/CP011465 | |
| PF430-3 | Chile | 2013 | Unknown | O3 | − | CP011466/CP011467 | |
| S2 2/9 | Denmark | Unknown | Rainbow trout | O1 | − | CP011472/CP011473 | |
| VA1 | Greece | 2014 | Sea bass | O1 | + | CP010078/CP010079 | CP016265 |
| 6018/1 | Denmark | Unknown | Rainbow trout | O1 | + | CP010291/CP010292 | CP016260 |
| VIB18 | Denmark | Unknown | Rainbow trout | O1 | + | CP011436/CP011437 | CP016266 |
| 261/91 | Italy | Unknown | Sea bass | O1 | + | CP010032/CP010033 | CP016258 |
| A023 | Spain | Unknown | Turbot | O1 | − | CP010036/CP010037 | |
| LMG12010 | Unknown | Unknown | Unknown | O1 | + | CP011468/CP011469 | CP016263 |
| T265 | United Kingdom | Unknown | Atlantic salmon | O1/VaNT1 | + | CP010040/CP010041 | CP016263 |
| 51/82/2 | Germany | Unknown | Rainbow trout | O1 | − | CP010042/CP010043 | |
| VIB93 | Denmark | 1985 | Rainbow trout | O1 | + | CP011438/CP011439 | CP016267 |
| 91-8-178 | Norway | 1991 | Turbot | O1 | + | CP010034/CP010035 | CP016256 |
| Ba35 | United States | Unknown | Sockeye salmon | O1/VaNT1 | + | CP010030/CP010031 | CP016261 |
Genomic overview of the V. anguillarum strains analyzed in this study. Download TABLE S2, DOCX file, 0.1 MB (18KB, docx) .
Copyright © 2017 Castillo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The Vibrio anguillarum pan-genome.
To determine an overall approximation of the total gene pool for V. anguillarum based on the sequenced genomes, we calculated the pan-genome using the EDGAR software platform. The gene repertoire of the V. anguillarum pan-genome increased with each addition of a new genome and had at least 3,973 and 1,932 genes for chromosomes I and II, respectively (see Fig. S1 in the supplemental material). In contrast to this increase, the V. anguillarum core genome decreased with the addition of each new genome, as expected (Fig. S1). The V. anguillarum average gene contents were 1,891 and 479 genes for chromosomes I and II, respectively (Fig. 1; Fig. S1). These open reading frames (ORFs) belonging to the core genome were assigned to putative functional categories using the Clusters of Orthologous Groups of Proteins (COG) database (Fig. S1B). Approximately 55.4 and 17.3% of the predicted genes in the core genome were dedicated to metabolic functions for chromosomes I and II, respectively. Of these genes, 36.2 and 13.7% were split between cellular process/signaling functions and information storage/processing functions for chromosomes I and II, respectively. Finally, functions of the predicted genes in the remaining 8.4 and 69% of the core genome were assigned as uncharacterized proteins for chromosomes I and II, respectively (Fig. S1B).
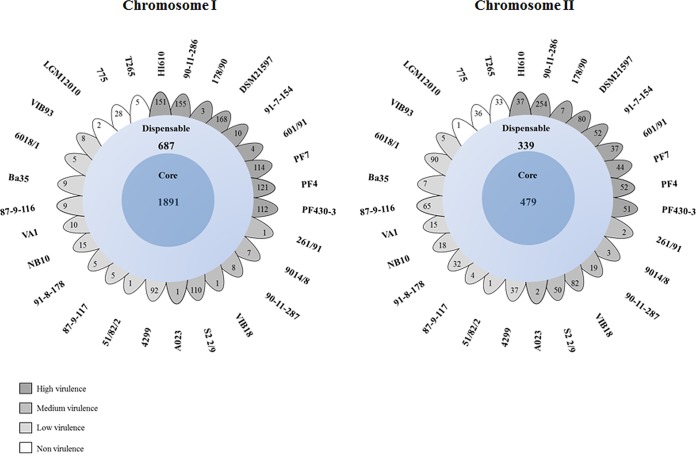
FIG 1 .
The pan-genome of V. anguillarum. The flower plots represent the number of shared (core) and specific (accessory/dispensable) genes based on cluster orthologs for each chromosome. Petals display numbers of strain-specific genes found in each genome of V. anguillarum strains with core gene numbers in the center. The gray colors indicate the virulence category as found in three fish larva model systems (31).
V. anguillarum pan-genome and core and accessory genome evolution according to the number of sequenced genomes. (A) Total numbers of genes (pan-genome), shared genes (core genome), and unique genes (accessory genome) for a given number of genomes sequentially added. (B) COG subcategories of predicted genes within the core genomes of V. anguillarum for chromosomes I and II. Each category or subcategory is graphed as a percentage of the total number of genes in the core genome. Download FIG S1, DOCX file, 0.5 MB (546.7KB, docx) .
Copyright © 2017 Castillo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The remaining 1,153 ORFs (chromosome I) and 1,117 ORFs (chromosome II) were defined as the V. anguillarum accessory genome (Fig. 1). The number of nonduplicated unique genes in each V. anguillarum strain varied from 1 to 168 for chromosome I and from 1 to 254 for chromosome II. The V. anguillarum strains 90-11-286, DSM21597, HI610, PF4, PF430-3, PF7, and S2 2/9 had the largest numbers of accessory genes (1,499 for both chromosomes).
Distribution of virulence factors.
Several virulence-associated genes have been described in V. anguillarum strain 775 (21). More than 90% of these virulence genes were present in all strains (see Table S3 in the supplemental material). Genes involved in iron transport, metalloproteases, motility, chemotaxis, type IV pilus, and quorum sensing were found in all strains (Table S3). However, V. anguillarum strains PF4, PF430-3, PF7, and S2 2/9 lacked the type VI secretion system present on chromosome I. Moreover, none of the V. anguillarum strains had the transcriptional regulator hylU or the unknown protein related to catechol siderophore metabolism positioned in chromosome II. Several strains did not have genes for secreted lipase and collagenase (Table S3). All strains in which the plasmid pJM1 was found carried genes related to anguibactin and iron metabolism (Table S3).
Distribution of virulence-related genes in V. anguillarum strains. Download TABLE S3, XLSX file, 0.1 MB (70.5KB, xlsx) .
Copyright © 2017 Castillo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
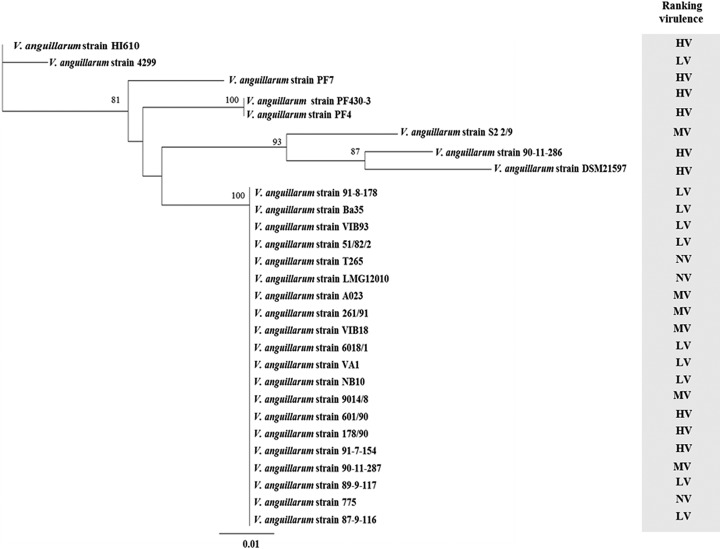
Although our results showed a global distribution of homologous virulence factors, nucleotide sequence dissimilarities could be translated to changes in amino acid level, which affect the dynamic functions or activities of these pathogenicity factors, leading to the development of a more invasive infection (32–34). To reveal the evolution of the virulence, we inferred the genetic diversity of 163 representative virulence factors shared by the V. anguillarum strains using the maximum likelihood algorithm (Fig. 2). V. anguillarum strains with medium to low virulence tended to cluster in a homogenous group, and the high-virulence strains 90-11-286, PF4, PF430-3, PF7, DSM21597, and HI610 clustered as separate groups (Fig. 2).
FIG 2 .
Phylogenetic tree of 163 concatenated virulence factors shared by all V. anguillarum strains. The phylogenetic tree was constructed based on the maximum likelihood algorithm, using a concatenated alignment of 163 amino acid sequences inferred from putative virulence factors identified in V. anguillarum strain 775 (20) (Table S2). Bootstrap values <80% were removed from the tree. The horizontal bar at the base of the figure represents 0.01 substitutions per amino acid site. The virulence ranking of the strains is based on three fish larva models (31). HV, high virulence; LV, low virulence; MV, medium virulence.
Phylogenetic relationship.
In order to examine potential associations between the core genome composition and the virulence properties, the phylogeny of V. anguillarum strains was inferred by constructing a genome-relatedness maximum likelihood tree using orthologous alignment of 2,370 protein-coding genes (both chromosomes) of the core genome (Fig. 1). The evolutionary tree displayed different cluster patterns, which varied in the levels of diversity. Interestingly, 20 of the 28 strains that were medium to nonvirulent in the larval assays grouped with a very low genetic diversity (Fig. 3), with the exception of strains 601/90, 178/90, and 91-7-154, which were highly virulent in the larval systems (30) (Table S1). In contrast, V. anguillarum strains 90-11-286, PF4, PF430-3, PF7, DSM21597, and HI610, all of which were highly virulent, each constituted a separate cluster (Fig. 3). V. anguillarum strain DSM21597 was the most distant lineage in the phylogenetic tree. However, strains S2 2/9 and 4299, which were medium to low virulence, grouped with strains 90-11-286 and HI610, respectively (Fig. 3). These findings suggest an association between virulence and shared gene content. Moreover, the core phylogenetic tree indicated a geographic association among the most genetically diverse V. anguillarum strains. For example, the V. anguillarum strains from Chile, PF4, PF430-3, and PF7, shared a common ancestor. Similarly, strains HI610 and 4299, isolated in Norway, and strains S2 2/9 and 90-11-286, isolated in Denmark, clustered according to the geographic locality of isolation (Fig. 3).
FIG 3 .
Core genome phylogeny of V. anguillarum strains. The maximum likelihood tree was obtained from a concatenated nucleotide sequence alignment of the orthologous core genes (1,723 genes for both chromosomes) for the 28 V. anguillarum strains. The virulence properties of the strains and geographical places of isolation were added to improve comparison. Bootstrap values of <80% were removed from the tree. The horizontal bar at the base of the figure represents 0.6 substitution per nucleotide site. The virulence ranking of the strains is based on three fish larva models (31). HV, high virulence; LV, low virulence; MV, medium virulence.
GIs, prophages, and strain-specific genes.
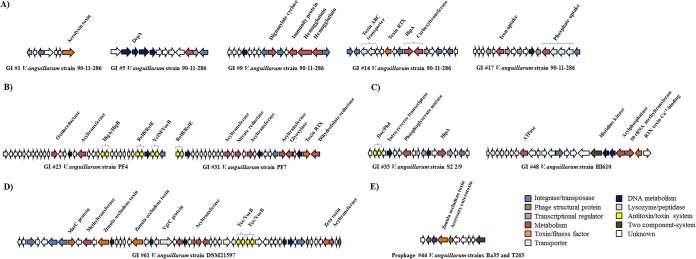
Ten genomic islands (GIs) have been described in V. anguillarum strain 775 (21), and their distribution in our collection was determined (see Fig. S2 in the supplemental material). All GIs (GIs 1 through 10) were found only in strains T265 and 775. Interestingly, the specific GIs 4, 6, 7, 8, and 10 were present in 82, 68, 64, 57, and 55% of the strains in the V. anguillarum collection, respectively (Fig. S2). For the purpose of this study, a GI was defined as a specific genomic region containing five or more ORFs (>5 kb). We detected a total of 64 strain-specific GIs between 6 and 132.1 kb for strains 90-11-286, PF4, PF430-3, PF7, HI610, DSM21597, 4299, and S2 2/9, all associated with transposases or integrases (see Table S4 in the supplemental material). Six of these nine strains were all highly virulent in cod, turbot, and halibut larva systems (31) (Table S1). A total of 1,067 strain-specific ORFs were found in these GIs. The G+C contents ranged from 26.2 to 44.9% for GIs in chromosome I and 32.0 to 52.9% for GIs in chromosome II (Table S4). Genes related to toxins, fitness factors, modification-restriction systems, antitoxin-toxin systems, transport, and metabolism were found in the GIs. V. anguillarum strain 90-11-286 had 21 specific GIs: one of them had an aerolysin toxin (GI 1), one contained diaguanylate cyclase and hemagglutinin genes (GI 9), and two harbored genes related to toxin RTX and toxin ABC transporter (GI 14) and iron and phosphate uptake systems (GI 17) (Fig. 4A). This strain contained a GI (GI 5) encoding a DprA protein, which has been associated with DNA transport and natural transformation competence (Fig. 4A).
FIG 4 .
Schematic representation of accessory elements carrying virulence or fitness factors in the V. anguillarum strains. (A) Genomic islands in strain 90-11-286. (B) Genomic islands in strains PF4 and PF7. (C) Genomic islands in strains S2 2/9 and HI610. (D) Genomic island in strain DSM21597. (E) Prophage-related elements in V. anguillarum strains T265 and Ba35 that contain a gene related to zonula occludens toxin (Zot). The positions of GIs and prophage-like elements are shown in Tables S4 and S7. The colors were assigned according to the possible role of each ORF as shown in the figure.
Distribution of genomic islands (GIs) previously identified in V. anguillarum strain 775. Shown is a graphic representation of the distribution of 10 GIs (GIs 1 to 10) in the V. anguillarum collection. Black and white squares represent the presence and absence of GI, respectively. Download FIG S2, DOCX file, 0.3 MB (362.2KB, docx) .
Copyright © 2017 Castillo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Features of the 64 strain-specific genomic islands in the V. anguillarum genomes. Download TABLE S4, DOCX file, 0.1 MB (24KB, docx) .
Copyright © 2017 Castillo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
V. anguillarum strain PF4 had a GI of 24.8 kb that encoded three antitoxin-toxin systems and one oxidoreductase gene (GI 23) (Fig. 4B). Also, strain PF7 had a GI that encoded many acyltransferases, RTX toxin, nitrate reductase, and one glyoxalase gene related to antibiotic resistance (GI 31) (Fig. 4B). One GI of strain S2 2/9 carried genes coding for HipA protein and an antitoxin-toxin system (GI 35) (Fig. 4C). Strain HI610 harbored a GI with the presence of 50 rRNA methyltransferase, which is related to antibiotic resistance and RTX toxin Ca2+-binding protein (GI 48) (Fig. 4C). The GI of strain DSM21597 had genes encoding zonula occludens toxins (Zots) and the protein MarC related to resistance to antibiotics (Fig. 4D). Many other GIs harbored genes of ecological interest, but specific details for each of these are out of the scope of this article.
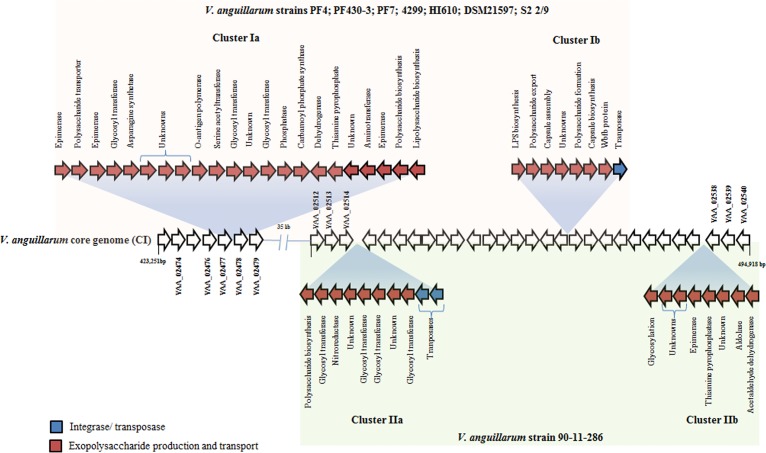
Strains that grouped in different phylogenetic lineages possessed a new set of gene clusters participating in the biosynthesis and transport of exopolysaccharides (Fig. 5). Strains PF4, FP430-3, PF7, HI610, 4299, S2 2/9, and DMS21597 had two clusters of 18.6 kb and 9.1 kb (one of them linked to transposases), encoding polysaccharide transports, glycosyltransferases, capsule assembly, and O-antigen polymerase (Fig. 5). Also, V. anguillarum strain 90-11-286 contained two gene clusters of 7.5 kb and 8.5 kb at chromosome I that contain genes related to glycosyltransferases, epimerases, and polysaccharide biosynthesis (Fig. 5).
FIG 5 .
Schematic representation of accessory gene clusters in V. anguillarum strains PF4, PF430-3, PF7, 4299, HI610, DSM21597, S2 2/9, and 90-11-286. The position of the core genome was assigned according to V. anguillarum strain 775.
Finally, a new set of specific putative virulence factors were identified in strains 90-11-286 and 91-7-154 (see Table S5 in the supplemental material). V. anguillarum strain 90-11-286 had three additional hemagglutinin proteins, one toxin Fic protein, and one cytotoxic necrotizing factor 2 protein (Table S5). Strain 91-7-154 had a Zot. Interestingly, noticeable amino acid similarities (>74%) of these virulence factors were found to those of other Vibrio species (V. cholerae, V. ordalii, and V. harveyi), indicating that the genes coding for these proteins may have extrachromosomal origin (Table S5).
Distribution of virulence factors not associated with prophages related sequences or genomic islands in V. anguillarum strains. Download TABLE S5, DOCX file, 0.1 MB (15.7KB, docx) .
Copyright © 2017 Castillo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Prophages.
Fifty-five different prophage-related elements were detected in the V. anguillarum genome sequences: of these, 9% were intact prophages, and the rest were defined as a “cryptic” or incomplete prophages. Both types of prophages were found in chromosomes I and II (see Tables S6 and S7 in the supplemental material). Specifically, 40 (72%) unique phage-related sequences between 5.3 and 49.2 kb were specific in 17 of the 28 V. anguillarum strains (Table S6). On other hand, 15 phage-related sequences (28%) were shared in 24 out of 28 V. anguillarum strains independently of locality and year of isolation (Table S7). Investigation of the presence of virulence or fitness factors encoded inside these sequences showed that V. anguillarum strains T265 and Ba35 carried a prophage-like element of 9.2 kb linked to a Zot-like toxin (Fig. 4E; Table S7 [prophage 43]).
Unique prophage-related sequences distributed in V. anguillarum strains. Download TABLE S6, DOCX file, 0.1 MB (17.8KB, docx) .
Copyright © 2017 Castillo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Shared prophage-related sequences in V. anguillarum strains. Download TABLE S7, DOCX file, 0.1 MB (16.6KB, docx) .
Copyright © 2017 Castillo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
The 28 V. anguillarum strains analyzed in the present study represent the largest collection of genome-sequenced strains for this fish-pathogenic bacterium. The multiscale comparative approach used in this work provides insights into the diversity of V. anguillarum strains (Fig. 1 to 4). Identification and characterization of accessory genome included genes that confer resistance to antibiotics and encode toxins and/or genes that improve the fitness of the organism, which may have been acquired via lateral gene transfer (35) (Fig. 4). In addition, core genome diversity indicated that the most virulent strains grouped in different genetic clusters (Table S1; Fig. 3). Thus, altogether our data indicate that virulence is multifactorial in V. anguillarum and that both the core and accessory genomes affect the pathogenicity of this Vibrio species. Similarly, the accessory and core genomes are significant sources of virulence-associated genes in Klebsiella pneumoniae (36), Escherichia coli (37), Staphylococcus aureus (38), and Pseudomona aeruginosa (39).
The plasmid pJM1 has been described as an important virulence factor in V. anguillarum (21, 40). However, V. anguillarum strains 90-11-286, PF4, PF7, and HI610, which were highly virulent against fish larvae, did not contain the plasmid pJM1 (Table 1; Table S1), but they had a functional vanchrobactin locus (not interrupted by a transposon) in the bacterial chromosome (31). This observation is in accordance with a previous study in which virulent pJM1-deficient strains did not carry the anguibactin system but produced the chromosomally encoded siderophore vanchrobactin, which is potentially a virulence factor (23, 41, 42). Thus, our results suggest that the presence or absence of the pJM1 plasmid is not an essential factor for V. anguillarum to cause disease in fish larvae.
The pan-genome analysis revealed that V. anguillarum contained a core genome of 2,370 nonduplicated ORFs for both chromosomes (Fig. 1; Fig. S1). This level of core gene content is higher than that reported for V. mimicus (43) but lower than those in V. parahaemolyticus (44) and V. cholerae (45). When the V. anguillarum core genome was used to determine phylogeny, the 28 V. anguillarum strains clustered in 5 groups, and six of the nine most virulent strains were found in separate clusters (Fig. 3). In contrast, most of the strains that showed moderate to no virulence in our larval systems shared a very similar backbone, and hence probably all originated from a common ancestor (Fig. 3). These characteristics could be relevant for understanding the relationship between the core genome diversity and the influence of acquired mobile elements on pathogenicity in V. anguillarum. We speculate that these phylogenetically distant bacteria could occupy different niches in fish farms and that the acquisition of GIs may vary in these aquatic systems and be influenced by the genetic background. Interestingly, three V. anguillarum strains (91-7-154, 601/90, and 178/90) that displayed high-virulence properties in the larval systems clustered with the strains that showed medium- or low-virulence properties (Fig. 3; Table S1). This observation allows us to suggest that specific mutations in the core genome may be linked to their high-virulence phenotypes.
Both chromosomes contained strain-specific elements and new virulence genes as a result of insertion of different genomic islands, prophages, and/or acquisition of other mobile genetic elements (described below) (Tables S4, S6, and S7). These results are in contrast to previous analysis in V. antiquaries (46), or V. mimicus (43), where chromosome II represents a collection of accessory elements and likely participate in the adaptation to different niches, having a critical role in the speciation and evolution of the genus Vibrio (47). However, our results indicated that chromosomes I and II both have genome plasticity (Fig. 1; Fig. S1), leading us to suggest that both chromosomes are involved in and driving the evolution in V. anguillarum.
To capture the dynamic nature of virulence gene repertoires across V. anguillarum, we screened for >200 virulence related genes (21) (Table S3). A total of 163 genes were present in all of the strains (belonging to core genome), and phylogenetic relationships displayed a functional divergence in six out of nine of the most virulent strains (Fig. 2). This diversification leads us to suggest that virulence activity is under strong selection, affecting the dynamic functions or activities of these proteins, leading to the development of a stronger interaction with the host in the different steps of infection, as has been proposed for Pseudomonas syringae (32) and P. aeruginosa (48).
Genomic islands contribute to the evolution and diversification of microbial communities (49). We found 64 GIs that belonged to the accessory genome among six out of the nine most virulent V. anguillarum strains (Table S4). V. anguillarum strain 90-11-286 was highly virulent in the larval models (Table S1) and harbored GIs carrying a diversity of virulence factors (Fig. 4A). For example, one GI encoded the channel-forming toxin aerolysin, which has been associated with diarrheal diseases and deep wound infections, by interacting with eukaryotic cells and aggregating to form pores, leading to the destruction of the membrane permeability barrier and osmotic lysis (50). A second GI had a diguanylate cyclase gene, which affects the adhesive and invasive capabilities of the human pathogen Porphyromonas gingivalis (51). Finally, a third GI contained a hemolysin toxin, RTX, associated with toxin ABC transporter (52). Interestingly, this strain harbored the highest number of accessory genes in both chromosomes (Fig. 1; Fig. S1), and this feature could be associated with the presence of the protein DprA (Fig. 4A), which participates in uptake, transport, and protection of DNA in the natural transformation process (53).
Strains PF4, PF7, and S2 2/9 harbored GIs that encoded several acyltransferases and toxin-antitoxin systems (Fig. 4B and C). Acyltransferases are enzymes that transfers acyl groups to specific targets and may be an important factor regulating quorum-sensing virulence-related phenotypes, including the production of virulence factors, motility, and biofilm formation (54). Toxin-antitoxin systems, which were originally linked to the plasmid maintenance and stabilization of the bacterial chromosome, are now known to be involved in general stress response (55), persistence (56), biofilm formation (57), and virulence capacity of pathogenic bacteria (58). Finally, strains HI610 and DSM21597 exhibited GIs which encode toxins and resistance to reactive oxygen species (Fig. 4C and D). Genomic island in strain HI610 had a 50 rRNA methyltransferase, which contribute to the virulence in Staphylococcus aureus by conferring resistance to oxidative stress (59) and hemolysin toxin RTX Ca2+ binding (52). Strain DSM21597 had two zonula occludens toxins (Zot), described previously in V. cholerae, whose function is to increase intestinal permeability by interacting with a mammalian cell receptor, with subsequent activation of intracellular signaling leading to the disassembly of the intercellular tight junctions (60, 61).
A set of prophage-related elements were identified in all 28 V. anguillarum strains (Table S6 and S7). Two prophage-like elements in the strains T265 and Ba35 contained Zot genes, which shared homology with V. cholerae (Fig. 4E). The presence of this toxin has also been documented in a prophage genome in V. coralliilyticus (62). However, recent studies have indicated that prophage elements may provide a benefit on virulence or fitness evolution, even if they do not carry virulence factors (63). Thus, this finding could be a starting point for future experimental studies on the role of bacteriophages as a potential central driver of pathogenicity in V. anguillarum.
Comparative genome analysis also revealed that most virulent strains 90-11-286, PF4, PF430, PF7, HI610, and DSM21597, and the low-virulence strain 4299 carried a new set of gene clusters related to biosynthesis, modification, and transport of exopolysaccharides (Fig. 5). In contrast to the limited diversity observed in this region among the 21 remaining strains, it clearly indicated that these clusters have been horizontally transferred. The existence of these accessory genes is regarded as essential virulence factor in Burkholderia pseudomallei (64) and V. cholerae (45). More importantly, the presence of these genes has been associated with the modification of capsule polysaccharide content and evasion of immune response (65).
Unlike Salmonella enterica serovar Typhi (66), Bacillus anthracis (67) and V. parahaemolyticus O3:K6 (68), which showed low genetic diversity, V. anguillarum offered an example of how lateral gene transfer has an important role in accessorizing the genome, providing genes essential for pathogenicity or fitness (Fig. 4; Table S5). Taken altogether, we propose a hypothetical model of evolution in V. anguillarum occurring in distinct phylogenetic groups, which shows that the high-virulence properties of some strains were obtained mainly via acquisition of pathogenic genomic islands occurring in the natural environment (see Fig. S3 in the supplemental material). It should be noted that the stability and transmission of these GIs were speculative. For example, GIs 4 and 6 detected previously in strain 775 were present in 82 and 68% of the strains, respectively, independently of the geographic localities of isolation (Fig. S2). Thus, we assumed a vertical transmission and loss of these specific GIs in some phylogenetic clusters. In contrast, GIs 3 and 5 were presented in 7% and 14%, respectively (Fig. S2), and their transmission could be horizontal for those strains. Similarly, we hypothesized that temperate bacteriophages infected putative V. anguillarum genetic ancestors (e.g., Pp41) (Table S7) and consequently the prophage-related elements were transmitted vertically, while other bacteriophages could be strain specific (e.g., Pp 16). Altogether, this study proposes that GIs and prophage-related elements outside the core genome may be a driving force in diversity and pathogenicity of V. anguillarum (Fig. S3).
Hypothetical evolution pathway in V. anguillarum. The model of V. anguillarum evolution suggests insertions and deletion of genomic islands (Table S4) and infection by bacteriophages (Tables S6 and S7). The graphic representation of the genetic diversity is based on the core genome phylogeny. Putative ancestral strains are indicated as open circles. The virulence ranking of the strains is based on three fish larval models (1). Download FIG S3, DOCX file, 0.4 MB (450.4KB, docx) .
Copyright © 2017 Castillo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
V. anguillarum is an important part of the autochthonous marine microbial communities with a specific ecological niche, such as fish, where selective pressure may allow acquisition of genetic traits that could increase fitness and virulence potential (69). Data presented here clearly support this view, where genomic islands carrying a suite of virulence genes and other mobile elements are probably driving the pathogenic and/or fitness evolution of V. anguillarum (Fig. 4; Tables S4 and S5). It has been suggested that virulence factors have a dual function and are used by pathogens both during the host infection and in environmental adaptation (70). For example, the toxin hemagglutinin in V. cholerae has a role in intestinal colonization, but has also recently been implicated in biofilm formation on chitin-containing surfaces in aquatic environment (71). In the same way, V. antiquarius, isolated in a deep sea hydrothermal vent, exhibited Zot and RTX toxins (46), indicating a multifaceted role outside the host. Thus, the presence of these genes in the V. anguillarum strains (Fig. 4; Tables S4 and S5) suggests a dual role in non-host environments; however, clearly a new outlook is needed for inferring the putative secondary role of pathogenic genes in this bacteria.
Conclusions.
Using a comparative pan-genomic analysis of V. anguillarum, we identified new pathogenic genomic islands, prophages, and virulence factors, suggesting that independent acquisition of these mobile genetic elements could play an important role in the evolution and virulence of V. anguillarum. The phylogenetic relationship based on core genome and shared virulence factors revealed different cluster groups, which suggested a possible link with the virulence properties and supported the idea that pathogenicity is also driven by core genome content within this bacterial species. Altogether, the genome sequences analyzed could serve as a reference point for studies of pathogenicity in aquaculture when V. anguillarum is present.
MATERIALS AND METHODS
Strain selection, medium composition, and growth conditions.
Twenty-eight V. anguillarum strains isolated from different geographic localities (>13,000 km), temporal scales (>25 years), and hosts (Table 1) were included in the analyses. The strains were stored at −80°C in LB broth (12106; Mo-Bio) with 15% glycerol. Strains were grown in LB broth and incubated at 22°C with agitation for 24 h (72).
DNA extraction.
Bacterial DNA from V. anguillarum strains was extracted from cells harvested by centrifugation (5000 × g, 10 min) using the NucleoSpin tissue kit (Macherey-Nagel). The amount of genomic DNA was measured using a Nanodrop2000 UV-visible light (UV-Vis) spectrophotometer (Thermo Scientific).
Genome sequencing, assembly, and annotation.
The genomes of 25 V. anguillarum strains were sequenced using Illumina HiSeq platform (BGI, China) with paired-end read sizes of 100 bp. Library construction, sequencing, and data pipelining were performed in accordance with the manufacturer’s protocols. The Illumina data were assembled into contiguous sequences using Geneious software (version 9.1.4) (73), and short- and low-coverage contigs were filtered out. The remaining contigs were aligned using chromosomes I and II and plasmid pJM1 of V. anguillarum strain 775 as references (GenBank accession no. CP002284.1 [chromosome I], CP002285.1 [chromosome II], and AY312585 [plasmid pJM1]; December 2014). The genome assembly process was performed using the Geneious software version 9.1.4 and assembled into two scaffolds of 35 to 71 contigs with an average coverage of >88× for each isolate. The genome of V. anguillarum strain 90-11-286 was already fully sequenced and previously described (74). Annotation of the genomes was done by the NCBI Prokaryotic Genome Automatic Annotation Pipeline (PGAAP) (75). Alternatively, genomic annotation was done by RAST (76) and BaSys (77).
Identification of genomic islands, prophage-like elements, and virulence factors.
IslandViewer and MAUVE v2.3.1 were used to predict the putative genomic islands (GIs) (78, 79). IslandViewer integrated sequence composition-based genomic island prediction programs, including IslandPath-DIMOB, SIGI-HMM, and the comparative genome-based program IslandPick. The MAUVE alignment procedure allows the detection of unique regions using a comparative genomics approach. Putative virulence genes were predicted using the virulence database MvirDB (80). All predicted genes of the 28 V. anguillarum strains were searched against the MvirDB by BLASTP with loose criteria (E value, ≥1E−5; identity, ≥35%; coverage, ≥80%). Also, VirulenceFinder 1.2 (81) was used to screen for putative virulence factors using selected databases from Escherichia coli, Enterococcus, and Streptococcus aureus. Prophage-related sequences were identified and selected by running bacterial genomes in Phage_Finder v2.1 (82) and PHAST (83).
Virulence-related genes and genomic islands (GIs) of V. anguillarum strain 775 were used to identify homologs in the V. anguillarum genomes by BLAST analyses using the tBLASTn 2/2/25+ tool and an E value threshold of ≤10−10 (84). These DNA sequences were verified as reciprocal best hits.
Pan-genome analysis.
To predict the possible genomic dynamic changes at V. anguillarum, EDGAR (85) was used to predict the pan-genome: i.e., to determine the accessory genome (specific genes found in only one genome) and core genome (common genes mutually conserved). Comparative analyses at the protein level were done by an all-against-all comparison of the annotated genomes. The algorithm used was BLASTP with a standard scoring matrix, BLOMSUM62, and an E value cutoff of 10−4. All BLAST hits were normalized according to the best score (84). The score ratio value (SRV), which shows the quality of the hit, was calculated by dividing the scores of further hits by the best hit (86). Two genes were considered orthologous when revealing a bidirectional best BLAST hit with single SRV exceeding the predetermined cutoff of 76 (85).
Functional annotation of genes and transposase identification was accomplished by BLASTp alignment of annotated ORFs against the COG database (87) using BLAST+ v2.2.24 (88).
Phylogenomic tree reconstruction.
To reveal the phylogenetic relationship among V. anguillarum strains based on virulence factors, we selected 163 putative pathogenicity genes from V. anguillarum strain 775 (21). For each gene, protein sequences were aligned using ClustalW version 2.0 (89), and individual proteins were concatenated to infer phylogeny using maximum likelihood in Geneious version 9.1.4 (73). Similarly, to determine the core genome phylogenetic relationship among V. anguillarum strains based on genomic data, we selected a set of orthologous genes shared by all 28 strains and V. parahaemolyticus strain RIMD 2210633 (outgroup to root the tree) (1,723 genes present in a single copy, with paralogs not included) using OrthoMCL with an E value cutoff of 10−10 (90). The sets of 1,723 single core genes were first aligned at the amino acid level using ClustalW version 2.0 (89) and then back-translated to DNA sequences using PAL2NAL (91). The alignment of all orthologous sequences was concatenated using FASconCAT (92). The gene tree was constructed using PhyML (93).
Accession number(s).
Accession numbers for chromosomes and plasmids are listed in Table 1.
ACKNOWLEDGMENTS
This work was supported by a grant from The Danish Directorate for Food, Fisheries and Agri Business (ProAqua, project no. 09-072829).
The assistance of Jette Melchiorsen in preparing genomic DNA is acknowledged.
REFERENCES
- 1.Lipp EK, Rose JB. 1997. The role of seafood in foodborne diseases in the United States of America. Rev Sci Tech 16:620–640. doi: 10.20506/rst.16.2.1048. [DOI] [PubMed] [Google Scholar]
- 2.Reidl J, Klose KE. 2002. Vibrio cholerae and cholera: out of the water and into the host. FEMS Microbiol Rev 26:125–139. doi: 10.1111/j.1574-6976.2002.tb00605.x. [DOI] [PubMed] [Google Scholar]
- 3.Baker-Austin C, Stockley L, Rangdale R, Martinez-Urtaza J. 2010. Environmental occurrence and clinical impact of Vibrio vulnificus and Vibrio parahaemolyticus: an European perspective. Environ Microbiol Rep 2:7–18. doi: 10.1111/j.1758-2229.2009.00096.x. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Haim Y, Thompson FL, Thompson CC, Cnockaert MC, Hoste B, Swings J, Rosenberg E. 2003. Vibrio coralliilyticus sp. nov., a temperature-dependent pathogen of the coral Pocillopora damicornis. Int J Syst Evol Microbiol 53:309–315. doi: 10.1099/ijs.0.02402-0. [DOI] [PubMed] [Google Scholar]
- 5.Soto-Rodriguez SA, Gomez-Gil B, Lozano-Olvera R, Betancourt-Lozano M, Morales-Covarrubias MS. 2015. Field and experimental evidence of Vibrio parahaemolyticus as the causative agent of acute hepatopancreatic necrosis disease of cultured shrimp (Litopenaeus vannamei) in northwestern Mexico. Appl Environ Microbiol 81:1689–1699. doi: 10.1128/AEM.03610-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pujalte MJ, Sitjà-Bobadilla A, Macián MC, Belloch C, Alvarez-Pellitero P, Pérez-Sánchez J, Uruburu F, Garay E. 2003. Virulence and molecular typing of Vibrio harveyi strains isolated from cultured dentex, gilthead sea bream and European sea bass. Syst Appl Microbiol 26:284–292. doi: 10.1078/072320203322346146. [DOI] [PubMed] [Google Scholar]
- 7.Frans I, Michiels CW, Bossier P, Willems KA, Lievens B, Rediers H. 2011. Vibrio anguillarum as a fish pathogen: virulence factors, diagnosis and prevention. J Fish Dis 34:643–661. doi: 10.1111/j.1365-2761.2011.01279.x. [DOI] [PubMed] [Google Scholar]
- 8.Sørensen UB, Larsen JL. 1986. Serotyping of Vibrio anguillarum. Appl Environ Microbiol 51:593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toranzo AE, Barja JL. 1990. A review of the taxonomy and seroepizootiology of Vibrio anguillarum, with special reference to aquaculture in the northwest Spain. Dis Aquat Org 9:73–82. doi: 10.3354/dao009073. [DOI] [Google Scholar]
- 10.O’Toole R, Milton DL, Wolf-Watz H. 1996. Chemotactic motility is required for invasion of the host by the fish pathogen Vibrio anguillarum. Mol Microbiol 19:625–637. doi: 10.1046/j.1365-2958.1996.412927.x. [DOI] [PubMed] [Google Scholar]
- 11.Ormonde P, Hörstedt P, O’Toole R, Milton DL. 2000. Role of motility in adherence to and invasion of a fish cell line by Vibrio anguillarum. J Bacteriol 182:2326–2328. doi: 10.1128/JB.182.8.2326-2328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang XH, Leung KY. 2000. Biochemical characterization of different types of adherence of Vibrio species to fish epithelial cells. Microbiology 146:989–998. doi: 10.1099/00221287-146-4-989. [DOI] [PubMed] [Google Scholar]
- 13.Croxatto A, Lauritz J, Chen C, Milton DL. 2007. Vibrio anguillarum colonization of rainbow trout integument requires a DNA locus involved in exopolysaccharide transport and biosynthesis. Environ Microbiol 9:370–382. doi: 10.1111/j.1462-2920.2006.01147.x. [DOI] [PubMed] [Google Scholar]
- 14.Rodkhum C, Hirono I, Crosa JH, Aoki T. 2005. Four novel hemolysin genes of Vibrio anguillarum and their virulence to rainbow trout. Microb Pathog 39:109–119. doi: 10.1016/j.micpath.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Di Lorenzo M, Stork M. 2014. Plasmid-encoded iron uptake systems. Microbiol Spectr 2:PLAS-0030-2014. doi: 10.1128/microbiolspec.PLAS-0030-2014. [DOI] [PubMed] [Google Scholar]
- 16.Naka H, Liu M, Actis LA, Crosa JH. 2013. Plasmid- and chromosome-encoded siderophore anguibactin systems found in marine vibrios: biosynthesis, transport and evolution. Biometals 26:537–547. doi: 10.1007/s10534-013-9629-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denkin SM, Nelson DR. 2004. Regulation of Vibrio anguillarum empA metalloprotease expression and its role in virulence. Appl Environ Microbiol 70:4193–4204. doi: 10.1128/AEM.70.7.4193-4204.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rock JL, Nelson DR. 2006. Identification and characterization of a hemolysin gene cluster in Vibrio anguillarum. Infect Immun 74:2777–2786. doi: 10.1128/IAI.74.5.2777-2786.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodkhum C, Hirono I, Stork M, Di Lorenzo M, Crosa JH, Aoki T. 2006. Putative virulence-related genes in Vibrio anguillarum identified by random genome sequencing. J Fish Dis 29:157–166. doi: 10.1111/j.1365-2761.2006.00692.x. [DOI] [PubMed] [Google Scholar]
- 20.Lemos ML, Osorio CR. 2007. Heme, an iron supply for vibrios pathogenic for fish. Biometals 20:615–626. doi: 10.1007/s10534-006-9053-8. [DOI] [PubMed] [Google Scholar]
- 21.Naka H, Dias GM, Thompson CC, Dubay C, Thompson FL, Crosa JH. 2011. Complete genome sequence of the marine fish pathogen Vibrio anguillarum harboring the pJM1 virulence plasmid and genomic comparison with other virulent strains of V. anguillarum and V. ordalii. Infect Immun 79:2889–2900. doi: 10.1128/IAI.05138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Busschaert P, Frans I, Crauwels S, Zhu B, Willems K, Bossier P, Michiels C, Verstrepen K, Lievens B, Rediers H. 2015. Comparative genome sequencing to assess the genetic diversity and virulence attributes of 15 Vibrio anguillarum isolates. J Fish Dis 38:795–807. doi: 10.1111/jfd.12290. [DOI] [PubMed] [Google Scholar]
- 23.Frans I, Dierckens K, Crauwels S, Van Assche A, Leisner JJ, Larsen MH, Michiels CW, Willems KA, Lievens B, Bossier P, Rediers H. 2013. Does virulence assessment of Vibrio anguillarum using sea bass (Dicentrarchus labrax) larvae correspond with genotypic and phenotypic characterization? PLoS One 8:e70477. doi: 10.1371/journal.pone.0070477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tettelin H, Riley D, Cattuto C, Medini D. 2008. Comparative genomics: the bacterial pan-genome. Curr Opin Microbiol 11:472–477. doi: 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 25.Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. 2005. The microbial pan-genome. Curr Opin Genet Dev 15:589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 26.Grim CJ, Kotewicz ML, Power KA, Gopinath G, Franco AA, Jarvis KG, Yan QQ, Jackson SA, Sathyamoorthy V, Hu L, Pagotto F, Iversen C, Lehner A, Stephan R, Fanning S, Tall BD. 2013. Pan-genome analysis of the emerging foodborne pathogen Cronobacter spp. suggests a species-level bidirectional divergence driven by niche adaptation. BMC Genomics 14:366. doi: 10.1186/1471-2164-14-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu P, Yang M, Zhang A, Wu J, Chen B, Hua Y, Yu J, Chen H, Xiao J, Jin M. 2011. Comparative genomics study of multi-drug-resistance mechanisms in the antibiotic-resistant Streptococcus suis R61 strain. PLoS One 6:e24988. doi: 10.1371/journal.pone.0024988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rasko DA, Rosovitz MJ, Myers GS, Mongodin EF, Fricke WF, Gajer P, Crabtree J, Sebaihia M, Thomson NR, Chaudhuri R, Henderson IR, Sperandio V, Ravel J. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol 190:6881–6893. doi: 10.1128/JB.00619-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castillo D, Christiansen RH, Dalsgaard I, Madsen L, Espejo R, Middelboe M. 2016. Comparative genome analysis provides insights into the pathogenicity of Flavobacterium psychrophilum. PLoS One 11:e0152515. doi: 10.1371/journal.pone.0152515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holm KO, Nilsson K, Hjerde E, Willassen NP, Milton DL. 2015. Complete genome sequence of Vibrio anguillarum strain NB10, a virulent isolate from the Gulf of Bothnia. Stand Genomic Sci 10:60. doi: 10.1186/s40793-015-0060-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rønneseth A, Castillo D, D’Alvise P, Tønnesen Ø, Haugland G, Grotkjær T, Engell-Sørensen K, Nørremark L, Bergh Ø, Wergeland HI, Gram L. Comparative assessment of Vibrio virulence in marine fish larvae. J Fish Dis, in press. doi: 10.1111/jfd.12612. [DOI] [PubMed] [Google Scholar]
- 32.Baltrus DA, Nishimura MT, Romanchuk A, Chang JH, Mukhtar MS, Cherkis K, Roach J, Grant SR, Jones CD, Dangl JL. 2011. Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog 7:e1002132. doi: 10.1371/journal.ppat.1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nunes A, Borrego MJ, Nunes B, Florindo C, Gomes JP. 2009. Evolutionary dynamics of ompA, the gene encoding the Chlamydia trachomatis key antigen. J Bacteriol 191:7182–7192. doi: 10.1128/JB.00895-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayfield JA, Liang Z, Agrahari G, Lee SW, Donahue DL, Ploplis VA, Castellino FJ. 2014. Mutations in the control of virulence sensor gene from Streptococcus pyogenes after infection in mice lead to clonal bacterial variants with altered gene regulatory activity and virulence. PLoS One 9:e100698. doi: 10.1371/journal.pone.0100698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dobrindt U, Hochhut B, Hentschel U, Hacker J. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol 2:414–424. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- 36.Marcoleta AE, Berríos-Pastén C, Nuñez G, Monasterio O, Lagos R. 2016. Klebsiella pneumoniae asparagine tDNAs are integration hotspots for different genomic islands encoding microcin E492 production determinants and other putative virulence factors present in hypervirulent strains. Front Microbiol 7:849. doi: 10.3389/fmicb.2016.00849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lloyd AL, Henderson TA, Vigil PD, Mobley HL. 2009. Genomic islands of uropathogenic Escherichia coli contribute to virulence. J Bacteriol 191:3469–3481. doi: 10.1128/JB.01717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moon BY, Park JY, Robinson DA, Thomas JC, Park YH, Thornton JA, Seo KS. 2016. Mobilization of genomic islands of Staphylococcus aureus by temperate bacteriophage. PLoS One 11:e0151409. doi: 10.1371/journal.pone.0151409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bianconi I, Jeukens J, Freschi L, Alcalá-Franco B, Facchini M, Boyle B, Molinaro A, Kukavica-Ibrulj I, Tümmler B, Levesque RC, Bragonzi A. 2015. Comparative genomics and biological characterization of sequential Pseudomonas aeruginosa isolates from persistent airways infection. BMC Genomics 16:1105. doi: 10.1186/s12864-015-2276-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crosa JH. 1980. A plasmid associated with virulence in the marine fish pathogen V. anguillarum specifies an iron-sequestering system. Nature 284:566–568. doi: 10.1038/284566a0. [DOI] [PubMed] [Google Scholar]
- 41.Balado M, Osorio CR, Lemos ML. 2006. A gene cluster involved in the biosynthesis of vanchrobactin, a chromosome-encoded siderophore produced by Vibrio anguillarum. Microbiology 152:3517–3528. doi: 10.1099/mic.0.29298-0. [DOI] [PubMed] [Google Scholar]
- 42.Lemos ML, Salinas P, Toranzo AE, Barja JL, Crosa JH. 1988. Chromosome-mediated iron uptake system in pathogenic strains of Vibrio anguillarum. J Bacteriol 170:1920–1925. doi: 10.1128/jb.170.4.1920-1925.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hasan NA, Grim CJ, Haley BJ, Chun J, Alam M, Taviani E, Hoq M, Munk AC, Saunders E, Brettin TS, Bruce DC, Challacombe JF, Detter JC, Han CS, Xie G, Nair GB, Huq A, Colwell RR. 2010. Comparative genomics of clinical and environmental Vibrio mimicus. Proc Natl Acad Sci U S A 107:21134–21139. doi: 10.1073/pnas.1013825107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li L, Wong HC, Nong W, Cheung MK, Law PT, Kam KM, Kwan HS. 2014. Comparative genomic analysis of clinical and environmental strains provides insight into the pathogenicity and evolution of Vibrio parahaemolyticus. BMC Genomics 15:1135. doi: 10.1186/1471-2164-15-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chun J, Grim CJ, Hasan NA, Lee JH, Choi SY, Haley BJ, Taviani E, Jeon YS, Kim DW, Lee JH, Brettin TS, Bruce DC, Challacombe JF, Detter JC, Han CS, Munk AC, Chertkov O, Meincke L, Saunders E, Walters RA, Huq A, Nair GB, Colwell RR. 2009. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci U S A 106:15442–15447. doi: 10.1073/pnas.0907787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hasan NA, Grim CJ, Lipp EK, Rivera IN, Chun J, Haley BJ, Taviani E, Choi SY, Hoq M, Munk AC, Brettin TS, Bruce D, Challacombe JF, Detter JC, Han CS, Eisen JA, Huq A, Colwell RR. 2015. Deep-sea hydrothermal vent bacteria related to human pathogenic Vibrio species. Proc Natl Acad Sci U S A 112:E2813–E2819. doi: 10.1073/pnas.1503928112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kirkup BC Jr, Chang L, Chang S, Gevers D, Polz MF. 2010. Vibrio chromosomes share common history. BMC Microbiol 10:137. doi: 10.1186/1471-2180-10-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winstanley C, O’Brien S, Brockhurst MA. 2016. Pseudomonas aeruginosa evolutionary adaptation and diversification in cystic fibrosis chronic lung infections. Trends Microbiol 24:327–337. doi: 10.1016/j.tim.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langille MG, Hsiao WW, Brinkman FS. 2010. Detecting genomic islands using bioinformatics approaches. Nat Rev Microbiol 8:373–382. doi: 10.1038/nrmicro2350. [DOI] [PubMed] [Google Scholar]
- 50.Szczesny P, Iacovache I, Muszewska A, Ginalski K, van der Goot FG, Grynberg M. 2011. Extending the aerolysin family: from bacteria to vertebrates. PLoS One 6:e20349. doi: 10.1371/journal.pone.0020349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chaudhuri S, Pratap S, Paromov V, Li Z, Mantri CK, Xie H. 2014. Identification of a diguanylate cyclase and its role in Porphyromonas gingivalis virulence. Infect Immun 82:2728–2735. doi: 10.1128/IAI.00084-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Linhartová I, Bumba L, Mašín J, Basler M, Osička R, Kamanová J, Procházková K, Adkins I, Hejnová-Holubová J, Sadílková L, Morová J, Sebo P. 2010. RTX proteins: a highly diverse family secreted by a common mechanism. FEMS Microbiol Rev 34:1076–1112. doi: 10.1111/j.1574-6976.2010.00231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smeets LC, Becker SC, Barcak GJ, Vandenbroucke-Grauls CM, Bitter W, Goosen N. 2006. Functional characterization of the competence protein DprA/Smf in Escherichia coli. FEMS Microbiol Lett 263:223–228. doi: 10.1111/j.1574-6968.2006.00423.x. [DOI] [PubMed] [Google Scholar]
- 54.Yeom DH, Kim SK, Lee MN, Lee JH. 2013. Pleiotropic effects of acyltransferases on various virulence-related phenotypes of Pseudomonas aeruginosa. Genes Cells 18:682–693. doi: 10.1111/gtc.12076. [DOI] [PubMed] [Google Scholar]
- 55.Ramage HR, Connolly LE, Cox JS. 2009. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet 5:e1000767. doi: 10.1371/journal.pgen.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim Y, Wood TK. 2010. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem Biophys Res Commun 391:209–213. doi: 10.1016/j.bbrc.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kasari V, Kurg K, Margus T, Tenson T, Kaldalu N. 2010. The Escherichia coli mqsR and ygiT genes encode a new toxin-antitoxin pair. J Bacteriol 192:2908–2919. doi: 10.1128/JB.01266-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Georgiades K, Raoult D. 2011. Genomes of the most dangerous epidemic bacteria have a virulence repertoire characterized by fewer genes but more toxin-antitoxin modules. PLoS One 6:e17962. doi: 10.1371/journal.pone.0017962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kyuma T, Kimura S, Hanada Y, Suzuki T, Sekimizu K, Kaito C. 2015. Ribosomal RNA methyltransferases contribute to Staphylococcus aureus virulence. FEBS J 282:2570–2584. doi: 10.1111/febs.13302. [DOI] [PubMed] [Google Scholar]
- 60.Di Pierro M, Lu R, Uzzau S, Wang W, Margaretten K, Pazzani C, Maimone F, Fasano A. 2001. Zonula occludens toxin structure-function analysis. Identification of the fragment biologically active on tight junctions and of the zonulin receptor binding domain. J Biol Chem 276:19160–19165. doi: 10.1074/jbc.M009674200. [DOI] [PubMed] [Google Scholar]
- 61.Marinaro M, Fasano A, De Magistris MT. 2003. Zonula occludens toxin acts as an adjuvant through different mucosal routes and induces protective immune responses. Infect Immun 71:1897–1902. doi: 10.1128/IAI.71.4.1897-1902.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weynberg KD, Voolstra CR, Neave MJ, Buerger P, van Oppen MJ. 2015. From cholera to corals: viruses as drivers of virulence in a major coral bacterial pathogen. Sci Rep 5:17889. doi: 10.1038/srep17889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Govind R, Vediyappan G, Rolfe RD, Dupuy B, Fralick JA. 2009. Bacteriophage-mediated toxin gene regulation in Clostridium difficile. J Virol 83:12037–12045. doi: 10.1128/JVI.01256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reckseidler SL, DeShazer D, Sokol PA, Woods DE. 2001. Detection of bacterial virulence genes by subtractive hybridization: identification of capsular polysaccharide of Burkholderia pseudomallei as a major virulence determinant. Infect Immun 69:34–44. doi: 10.1128/IAI.69.1.34-44.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sim BM, Chantratita N, Ooi WF, Nandi T, Tewhey R, Wuthiekanun V, Thaipadungpanit J, Tumapa S, Ariyaratne P, Sung WK, Sem XH, Chua HH, Ramnarayanan K, Lin CH, Liu Y, Feil EJ, Glass MB, Tan G, Peacock SJ, Tan P. 2010. Genomic acquisition of a capsular polysaccharide virulence cluster by non-pathogenic Burkholderia isolates. Genome Biol 11:R89. doi: 10.1186/gb-2010-11-8-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Holt KE, Parkhill J, Mazzoni CJ, Roumagnac P, Weill FX, Goodhead I, Rance R, Baker S, Maskell DJ, Wain J, Dolecek C, Achtman M, Dougan G. 2008. High-throughput sequencing provides insights into genome variation and evolution in Salmonella typhi. Nat Genet 40:987–993. doi: 10.1038/ng.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Keim P, Gruendike JM, Klevytska AM, Schupp JM, Challacombe J, Okinaka R. 2009. The genome and variation of Bacillus anthracis. Mol Aspects Med 30:397–405. doi: 10.1016/j.mam.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Loyola DE, Navarro C, Uribe P, García K, Mella C, Díaz D, Valdes N, Martínez-Urtaza J, Espejo RT. 2015. Genome diversification within a clonal population of pandemic Vibrio parahaemolyticus seems to depend on the life circumstances of each individual bacteria. BMC Genomics 16:176. doi: 10.1186/s12864-015-1385-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alcaide E, Blasco MD, Esteve C. 2005. Occurrence of drug resistant bacteria in two European eel farms. Appl Environ Microbiol 71:3348–3350. doi: 10.1128/AEM.71.6.3348-3350.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vezzulli L, Guzmán CA, Colwell RR, Pruzzo C. 2008. Dual role colonization factors connecting Vibrio cholerae’s lifestyles in human and aquatic environments open new perspectives for combating infectious diseases. Curr Opin Biotechnol 19:254–259. doi: 10.1016/j.copbio.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 71.Reguera G, Kolter R. 2005. Virulence and the environment: a novel role for Vibrio cholerae toxin-coregulated pili in biofilm formation on chitin. J Bacteriol 187:3551–3555. doi: 10.1128/JB.187.10.3551-3555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tan D, Gram L, Middelboe M. 2014. Vibriophages and their interactions with the fish pathogen Vibrio anguillarum. Appl Environ Microbiol 80:3128–3140. doi: 10.1128/AEM.03544-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rasmussen BB, Grotkjær T, D’Alvise PW, Yin G, Zhang F, Bunk B, Spröer C, Bentzon-Tilia M, Gram L. 2016. Vibrio anguillarum is genetically and phenotypically unaffected by long-term continuous exposure to the antibacterial compound tropodithietic acid. Appl Environ Microbiol 82:4802–4810. doi: 10.1128/AEM.01047-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res 44:6614–6624 doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Van Domselaar GH, Stothard P, Shrivastava S, Cruz JA, Guo A, Dong X, Lu P, Szafron D, Greiner R, Wishart DS. 2005. BASys: a web server for automated bacterial genome annotation. Nucleic Acids Res 33:W455–W459. doi: 10.1093/nar/gki593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dhillon BK, Laird MR, Shay JA, Winsor GL, Lo R, Nizam F, Pereira SK, Waglechner N, McArthur AG, Langille MGI, Brinkman FSL. 2015. IslandViewer 3: more flexible, interactive genomic island discovery, visualization and analysis. Nucleic Acids Res 43:W104–W108. doi: 10.1093/nar/gkv401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhou CE, Smith J, Lam M, Zemla A, Dyer MD, Slezak T. 2007. MvirDB—a microbial database of protein toxins, virulence factors and antibiotic resistance genes for bio-defence applications. Nucleic Acids Res 35:D391–D394. doi: 10.1093/nar/gkl791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, Aarestrup FM. 2014. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol 52:1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fouts DE. 2006. Phage_Finder: automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res 34:5839–5851. doi: 10.1093/nar/gkl732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res 39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gertz EM, Yu YK, Agarwala R, Schäffer AA, Altschul SF. 2006. Composition-based statistics and translated nucleotide searches: improving the tBLASTn module of BLAST. BMC Biol 4:41. doi: 10.1186/1741-7007-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Blom J, Albaum SP, Doppmeier D, Pühler A, Vorhölter FJ, Zakrzewski M, Goesmann A. 2009. EDGAR: a software framework for the comparative analysis of prokaryotic genomes. BMC Bioinformatics 10:154. doi: 10.1186/1471-2105-10-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lerat E, Daubin V, Moran NA. 2003. From gene trees to organismal phylogeny in prokaryotes: the case of the gamma-proteobacteria. PLoS Biol 1:E19. doi: 10.1371/journal.pbio.0000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, Kiryutin B, Galperin MY, Fedorova ND, Koonin EV. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res 29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. Blast+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 90.Chen F, Mackey AJ, Stoeckert CJ Jr, Roos DS. 2006. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res 34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Suyama M, Torrents D, Bork P. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kück P, Meusemann K. 2010. FASconCAT: convenient handling of data matrices. Mol Phylogenet Evol 56:1115–1118. doi: 10.1016/j.ympev.2010.04.024. [DOI] [PubMed] [Google Scholar]
- 93.Guindon S, Delsuc F, Dufayard JF, Gascuel O. 2009. Estimating maximum likelihood phylogenies with PhyML. Methods Mol Biol 537:113–137. doi: 10.1007/978-1-59745-251-9_6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Average mortality of fish larvae following infection with Vibrio anguillarum at high dose (HD) and low dose (LD). Download TABLE S1, DOCX file, 0.1 MB (17.3KB, docx) .
Copyright © 2017 Castillo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genomic overview of the V. anguillarum strains analyzed in this study. Download TABLE S2, DOCX file, 0.1 MB (18KB, docx) .
Copyright © 2017 Castillo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
V. anguillarum pan-genome and core and accessory genome evolution according to the number of sequenced genomes. (A) Total numbers of genes (pan-genome), shared genes (core genome), and unique genes (accessory genome) for a given number of genomes sequentially added. (B) COG subcategories of predicted genes within the core genomes of V. anguillarum for chromosomes I and II. Each category or subcategory is graphed as a percentage of the total number of genes in the core genome. Download FIG S1, DOCX file, 0.5 MB (546.7KB, docx) .
Copyright © 2017 Castillo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Distribution of virulence-related genes in V. anguillarum strains. Download TABLE S3, XLSX file, 0.1 MB (70.5KB, xlsx) .
Copyright © 2017 Castillo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Distribution of genomic islands (GIs) previously identified in V. anguillarum strain 775. Shown is a graphic representation of the distribution of 10 GIs (GIs 1 to 10) in the V. anguillarum collection. Black and white squares represent the presence and absence of GI, respectively. Download FIG S2, DOCX file, 0.3 MB (362.2KB, docx) .
Copyright © 2017 Castillo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Features of the 64 strain-specific genomic islands in the V. anguillarum genomes. Download TABLE S4, DOCX file, 0.1 MB (24KB, docx) .
Copyright © 2017 Castillo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Distribution of virulence factors not associated with prophages related sequences or genomic islands in V. anguillarum strains. Download TABLE S5, DOCX file, 0.1 MB (15.7KB, docx) .
Copyright © 2017 Castillo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Unique prophage-related sequences distributed in V. anguillarum strains. Download TABLE S6, DOCX file, 0.1 MB (17.8KB, docx) .
Copyright © 2017 Castillo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Shared prophage-related sequences in V. anguillarum strains. Download TABLE S7, DOCX file, 0.1 MB (16.6KB, docx) .
Copyright © 2017 Castillo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Hypothetical evolution pathway in V. anguillarum. The model of V. anguillarum evolution suggests insertions and deletion of genomic islands (Table S4) and infection by bacteriophages (Tables S6 and S7). The graphic representation of the genetic diversity is based on the core genome phylogeny. Putative ancestral strains are indicated as open circles. The virulence ranking of the strains is based on three fish larval models (1). Download FIG S3, DOCX file, 0.4 MB (450.4KB, docx) .
Copyright © 2017 Castillo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.