Abstract
Ascorbate is a critical compound in plants and animals. Humans are unable to synthesize ascorbate, and their main source of this essential vitamin are plants. However, the pathway of synthesis in plants is yet to be established, and several unknown enzymes are only postulated to exist. We describe a specific l-galactose-1-phosphate (l-gal-1-P) phosphatase that we partially purified from young kiwifruit (Actinidia deliciosa) berries. The enzyme had a native molecular mass of ≈65 kDa, was completely dependent on Mg2+ for activity and was very specific in its ability to hydrolyze l-gal-1-P. The activity had a pH optimum of 7.0, a KM(l-gal-1-P) of 20–40 μM and a Ka(Mg2+) of 0.2 mM. The activity was inhibited by Mg2+ at concentrations >2 mM. The enzyme from Arabidopsis thaliana shoots showed similar properties to the kiwifruit enzyme. The Arabidopsis thaliana enzyme preparation was digested with trypsin, and proteins present were identified by using liquid chromatography–MS. One of 24 proteins present in our preparation was an Arabidopsis thaliana protein, At3g02870, annotated myo-inositol-1-phosphate phosphatase in GenBank, that matched the characteristics of the purified l-gal-1-phosphate phosphatase. We then expressed a kiwifruit homologue of this gene in Escherichia coli and found that it showed 14-fold higher maximum velocity for l-gal-1-P than myo-inositol-1-P. The expressed enzyme showed very similar properties to the enzyme purified from kiwifruit and Arabidopsis, except that its KM(l-gal-1-P) and Ka(Mg2+) were higher in the expressed enzyme. The data are discussed in terms of the pathway to ascorbate biosynthesis in plants
Keywords: ascorbic acid, biosynthesis, vitamin C, myo-inositol-1-phosphate
Ascorbic acid is an essential compound in plants and animals, with a range of functions including scavenging reactive oxygen species (1) and maintaining human health (2). Humans cannot synthesize their own ascorbic acid (or vitamin C) (2) and are dependent on external sources of this important vitamin, mainly from plants. However, the pathway of ascorbic acid biosynthesis in plants has not been firmly established, and many enzymes have yet to be discovered (3, 4).
Three pathways of biosynthesis of ascorbic acid have been proposed in plants, one through l-galactose (l-gal) (5), another from myo-inositol (6–8), and a third through galacturonic acid (9). The l-gal pathway proceeds through l-gal to galactono-1,4-lactone and, thence, to ascorbate (5). The ascorbate can then be broken down to end products including oxalate (10). From mutant and biochemical studies (11–13), it is likely that the l-gal is derived from GDP-d-mannose through GDP-l-galactose to l-gal-1-phosphate (l-gal-1-P), which has been proposed to be converted to l-gal through the action of two yet-to-be-identified enzymes including an unknown specific phosphatase (14–18). In the myo-inositol pathway, inositol-1-P is derived from glucose-6-P. It is then converted to myo-inositol by a phosphatase (19–21) and, thence, to glucuronic acid by inositol oxygenase.
The enzyme activities and genes for several specific plant phosphatases have recently been identified, such as phosphoglycolate phosphatase (22), phosphoserine phosphatase (23), myo-inositol monophosphatase (21), sucrose-6-phosphate phosphatase (24), and trehalose-6-phosphate phosphatase (25), and enzyme activity for other specific phosphatases has been identified but not yet cloned; e.g., sorbitol-6-phosphate phosphatase (26) and 2-carboxy-d-arabinitol 1-phosphate phosphatase (27). There is also a wide range of information on nonspecific acid phosphatases (28–30), which would be expected to dephosphorylate l-gal-1-P among other substrates, but there appears to be no published information on a plant phosphatase that would catalyze the dephosphorylation of l-gal-1-P in a specific manner.
It is very likely that kiwifruit makes ascorbate through the galactose pathway as both leaves and fruit have been shown to express the gene for the committed enzyme l-gal dehydrogenase (31), and fruit shows high levels of this enzyme (W.A.L., unpublished data). However, the preceding phosphatase step in ascorbic acid biosynthesis has not been identified or described. In this work, we describe the partial purification of a specific l-gal-1-phosphate phosphatase (l-gal-1-Pase), which would act in the l-gal pathway of ascorbic acid biosynthesis characterize the enzyme, and identify the gene. The gene has been previously identified as a myo-inositol-1-phosphate phosphatase (myo-inositol-1-Pase), raising possibilities of linkage between two different related pathways of ascorbate biosynthesis.
Materials and Methods
Materials. Kiwifruit [Actinidia deliciosa (A. Chev.) C. F. Liang and A.R. Ferguson deliciosa cv. “Hayward”] berries were picked at Te Puke, New Zealand. For biochemical studies, 1-cm-diameter fruit were used and picked 4 weeks after anthesis. Arabidopsis thaliana (Columbia-1) plants were grown for ≈5 weeks at 20–25°C and 200 μmol·m-2·sec-1 photosynthetically active radiation. Tissues were stored frozen at –80°C until use.
Most chemicals were obtained from Sigma, except the protease inhibitor tablets (Roche Applied Sciences) and l-gal-1-P (Glycoteam, Hamburg, Germany). The l-gal-1-P appeared as a single peak by HPLC and had a mass of 260 Da. Column chromatography media were obtained from Amersham Biosciences. Bradford's reagent was provided by Bio-Rad.
Enzyme Assays. Two assays were used to measure the activity of the l-gal-1-Pase enzyme. The first assay measured the phosphate formed by the hydrolysis of l-gal-1-P by using a microplate-based colorimetric phosphate assay (32) and was used during purification and to characterize the enzyme. The standard conditions for this assay were 0.1 M Bis-Tris propane (pH 7.0), 2 mM MgCl2, and 0.2 mM l-gal-1-P in 100 μl. Between 1 and 20 μl of extract was added to start the reaction, incubated for 15 to 30 min at 30°C, and terminated by the addition of 100 μl of 3% ascorbate and 0.5% (both wt/vol) ammonium molybdate in 0.5 M HCl. Background assays were obtained by adding the enzyme after the reaction was terminated. The second assay, used during purification and to verify products, coupled the production of l-gal to l-gal dehydrogenase (5, 33) and measured the reduction of NAD+. The basic assay in 100 μl was the same as the first assay but additionally contained 0.2 mM NAD+ as well as an excess of l-gal dehydrogenase either partially purified from kiwifruit berries (31) or with the cloned enzyme expressed in Escherichia coli (S.B. and W.A.L., unpublished data). Time courses were followed up to 30 min at 20°C by using either a Victor fluorescence plate reader (Wallac, Gaithersburg, MD) to measure NADH fluorescence (excitation, 340 nm; emission, 460 nm) or absorbance spectroscopy at 340 nm.
Enzyme kinetic analyses for Km and Vmax measurements, [Mg2+] response, alternative substrates, and pH optimum were replicated three times within the experiment in a standard manner. Data are presented as means and SEs as appropriate. Experiments were repeated at least twice, and a representative experiment is presented.
l-Gal dehydrogenase activity was assayed in 0.1 M Tris (pH 8.0)/0.25 mM NAD/0.25 mM l-gal in 100 μl at 20°C by using the Victor fluorimeter to measure NADH fluorescence.
l-Gal-1-Pase Purification. Kiwifruit berries or Arabidopsis thaliana leaves were ground to a fine powder under liquid nitrogen, and ≈70 g of frozen tissue was extracted in 100 ml (kiwifruit) or 200 ml (Arabidopsis thaliana) of 0.18 M Mops buffer adjusted to pH 7.0 and containing 1.0% Triton X-100, 4% polyvinylpyrrolidone, 2.5 mM DTT, 10% (vol/vol) glycerol, 10 μM E64 (kiwifruit), and one proteinase inhibitor tablet per 50 ml. This extract was then filtered through a 20-μm nylon mesh and centrifuged (20 min at 20,000 × g), and the supernatant was decanted. This supernatant was applied to a 5 × 60-cm G75 Superdex column (GE Healthcare) run at 5 ml·min-1 equilibrated with 0.1 M Tris (pH 8.0)/2 mM EDTA/5 mM 2-mercaptoethanol/10% glycerol. Active fractions were pooled, diluted with water and glycerol to 50 mM Tris·Cl/10% glycerol, and applied to two 5-ml HiTrap Q FF columns (GE Healthcare) in series equilibrated with 20 mM Tris·Cl (pH 8.0)/2 mM EDTA/5 mM 2-mercaptoethanol/10% glycerol. The HiTrap Q column was eluted by using a gradient of 0–0.5 M NaCl in 20 mM Tris·Cl (pH 8.0) containing EDTA, 2-mercaptoethanol, and glycerol.
Active fractions were combined, and ammonium sulfate was added to 1 M before application of the sample to a 5-ml HiTrap Butyl FF column. Proteins bound to the column were eluted with a reverse gradient of ammonium sulfate from 1 M to 0 M. Active fractions were then concentrated to 1 ml by using a 10,000 molecular weight cutoff centrifugal concentrator and applied to a 40 × 1.6-cm G75 Superdex column equilibrated with the same buffer as the large G75 Superdex column used earlier. Active fractions were combined and used to characterize the enzyme.
SDS/PAGE was performed by using the Bio-Rad Miniprotean II system with a 4% (wt/vol) acrylamide stacking gel and a 10% (wt/vol) acrylamide separating gel by using the Tricine-based buffer (34). A broad molecular mass range of Bio-Rad prestained marker proteins were used to determine molecular mass. Gels were stained with SYPRO Ruby (Molecular Probes) followed by Coomassie blue by using standard techniques.
Liquid Chromatography–MS Analysis of Peptides. A partially purified l-gal-1-Pase preparation was digested with trypsin [0.5 μg of modified sequencing grade trypsin (Roche) in 0.2 M NH4HCO3 and 0.5 mM CaCl2], and gel slices containing proteins from an SDS/PAGE separation of the same preparation were subjected to in-gel digestion (35). Digested peptides were analyzed by liquid chromatography–MS by using an LCQ Deca ion-trap mass spectrometer fitted with a nanospray electrospray ionization interface (ThermoQuest, Finnigan, San Jose, CA) and coupled to a Surveyor HPLC (ThermoQuest, Finnigan). Five microliters of the digested protein sample was injected onto a reversed phase column (PepMap C18, 75 μm × 15 cm, 3 μm; LC Packings, San Francisco). Tryptic peptides were separated at a flow rate of 400 nl/min with a linear gradient from 2% to 80% B (acetonitrile plus 0.1% formic acid) over 50 min. Solvent A was 0.1% aqueous formic acid.
MS/MS data were analyzed by using turbosequest (36, 37). Spectra were searched against an Arabidopsis thaliana protein database. The criteria used for a positive peptide identification for a doubly charged peptide were a correlation factor >2.0, a δ cross-correlation factor >0.1, and a high preliminary scoring. For triply charged peptides, the correlation factor was set at 2.5. All matched peptides were confirmed by visual examination of the spectra.
Gene Cloning. The full-length cDNA was obtained from Hort-Research's (Auckland) kiwifruit EST resource cloned into Blue-script SK(-). The 880-bp fragment was excised by using BamHI and HindIII, cloned in frame into pET30c(+) (Novagen), and transformed into E. coli DH5α cells. A correct clone (verified by PCR amplification with pET primers and by restriction enzyme digestion) was transformed into E. coli BL21 RIL cells (Novagen). The construct contains an N-terminal His6-tag. A single colony was grown in 50 ml of Terrific broth to an OD600 of 0.6 at 37°C, then protein expression was induced by addition of isopropyl β-d-thiogalactoside to 0.3 mM. The induced culture was grown for 20 h at 20°C. Cells were harvested by centrifugation at 8,000 × g at 4°C for 5 min, resuspended in the His column binding buffer (Novagen) containing one protease inhibitor tablet per 50 ml, and broken by a high-pressure emulsifier (Emulsiflex, Avestin, Ottawa). The lysate was centrifuged at 12,000 × g for 30 min at 4°C, and the filtered (0.45 μm) supernatant was purified through a 5-ml HiTrap Nickel column. Fractions containing protein were dialyzed at 4°C against three changes of 10 mM Bis-Tris Propane (pH 7.1), 1 mM EDTA, 5 mM 2-mercaptoethanol, and 30 mM NaCl.
Results
Purification of a Specific l-Gal-1-Pase from Kiwifruit. A large peak of kiwifruit l-gal-1-Pase activity was eluted from the large G75 Superdex column after the void volume (Fig. 5, which is published as supporting information on the PNAS web site). Further analysis of this peak showed that the earlier eluting fractions consisted of a neutral pH, Mg2+, and l-gal dependent phosphatase activity, whereas activity in the later eluting fractions had a more acid pH optimum and was independent of MgCl2 (data not shown). Consequently, only the earlier fractions were applied to the anion exchange column, where a clear separation of the remaining acid phosphatase and the Mg2+-dependent phosphatase was achieved (Fig. 5). The first major peak of phosphatase activity was unaffected by MgCl2, whereas the second peak was stimulated 9-fold by MgCl2 (data not shown). The first peak, which had an optimum below pH 6 (data not shown), was most likely a nonspecific acid phosphatase with activity against l-gal-1-P.
Further purification of the second HiTrap Q peak was gained through fractionation on a hydrophobic interaction column and through rechromatography on a G75 Superdex column (Fig. 6, which is published as supporting information on the PNAS web site). The peak activity on this latter column eluted the fraction after BSA, suggesting a native molecular mass close to 65,000 Da.
Further purification (including chromatofocusing on a MonoP column and native gel electrophoresis) resulted in increases in protein purity but catastrophic losses in activity. The enzyme would not bind to a HiTrap SP column at pH 5.2. Consequently, characterization of the enzyme was carried out on combined fractions from the small G75 Superdex column. At this stage of the purification, at least 10 proteins with molecular masses between 30,000 and 100,000 Da were apparent by SDS/PAGE (data not shown).
Characterization of the Kiwifruit Phosphatase Activity. l-Gal-1-Pase activity was linear with the amount of enzyme until at least 10 nmol of phosphate had been generated in the assay, which represented ≈20% of the substrate present. The activity was also linear with time for at least 30 min.
Substrate Specificity of the Kiwifruit Enzyme. We verified that the product of the reaction was l-gal by coupling the phosphatase activity to the reduction of NAD by using semipurified l-gal dehydrogenase from kiwifruit. This enzyme has been shown to be active with l-gal as the substrate with some activity with l-sorbosone (Arabidopsis), l-fucose (Arabidopsis, but not kiwifruit), and l-gulose but no activity against a wide range of other sugars (31, 33). In all steps of purification, the coupled reaction paralleled the direct phosphate assay (data not shown).
The ability of the l-gal-1-Pase to hydrolyze a range of phosphorylated compounds was measured by using the standard phosphate assay. The results are the means of two experiments, and no substrate other than l-gal-1-P was significantly and reproducibly hydrolyzed by the partially purified enzyme (Table 1). Several substrates (fructose-2,6-bisP, ribulose-1,5-bisP, and dihydroxyacetone phosphate) were partially hydrolyzed (≈20% of the available substrate) by the acid stop mix and showed high backgrounds, explaining an apparent ability of the enzyme to hydrolyze fructose-2,6-bisP. However, this substrate showed a near-zero rate in other assays. Where there was slight activity, the enzyme showed a preference for phosphate in the C1 position.
Table 1. Substrate specifity for the partially purified l-gal-1-Pase activity.
| Partially purified kiwifruit enzyme
|
Partially purified Arabidopsis enzyme
|
E. coli expressed kiwifruit enzyme EST 233909
|
||||
|---|---|---|---|---|---|---|
| Substrate | nmol/min | % | nmol/min | % | nmol/min | % |
| l-Gal-1-P | 0.220 | 100.0 | 0.163 | 100.0 | 1.61* | 100.0 |
| d-Gal-1-P | 0.011 | 4.6 | 0.066 | 4.1 | ||
| d-Mannose-1-P | 0.008 | 3.3 | 0.059 | 3.7 | ||
| Dihydroxyacetone phosphate | -0.001 | -0.3 | ||||
| Fructose-1, 6-bisP | -0.001 | -0.4 | ||||
| Fructose-1-P | -0.003 | -1.3 | ||||
| Fructose-6-P | 0.000 | -0.2 | ||||
| Fructose-2, 6-bisP | 0.020 | 9.8 | ||||
| Guanosine diphosphate | 0.007 | 3.6 | ||||
| Glucose-6-P | 0.000 | -0.1 | -0.001 | 0.0 | ||
| Glucose-1-P | 0.016 | 6.7 | ||||
| Glucose-1-6-bisP | 0.002 | 1.0 | ||||
| Guanosine triphosphate | -0.004 | -2.1 | ||||
| Mannose-6-P | -0.001 | -0.3 | ||||
| p-Nitrophenyl phosphate | 0.009 | 4.2 | 0.001 | 0.4 | -0.014 | -0.9 |
| Phosphoglyceric acid | 0.001 | 0.3 | 0.000 | 0.0 | ||
| Phosphoglycolate | 0.008 | 3.6 | 0.009 | 5.9 | ||
| Ribose-5-P | 0.003 | 1.5 | 0.014 | 0.9 | ||
| Ribulose-1, 5-bisP | -0.003 | -1.2 | ||||
| Sucrose-6-P | 0.006 | 2.7 | 0.003 | 1.9 | -0.001 | -0.1 |
| Trehalose-6-P | -0.001 | -0.6 | 0.003 | 1.7 | -0.003 | -0.2 |
| Myo-inositol-1-P | 0.353 | 21.9* | ||||
Reactions were run in 100 mM Bis-Tris Propane buffer (pH 7.0), with 2 mM MgCl2, and 0.5 mM phosphorylated substrate for 30 min (kiwifruit and Arabidopsis) or 11 min (E. coli) at 30°C. SEs were ≈2% of the mean for these assays. Empty positions indicate that the substrate was not assayed with that enzyme.
The test for alternative substrates used excess E. coli expressed enzyme in this assay to maximize sensitivity for alternative substrates, and the control rate was beyond the linear portion of the activity versus the amount of enzyme curve (data not shown). The true level of l-Gal-1-Pase activity was about three times this value. In separate measurements, we established that under saturating substrate concentration and 2 mM MgCl2 at pH 7.0, myo-inositol-1-P was hydrolysed at 7.1% the rate of l-gal-1-P
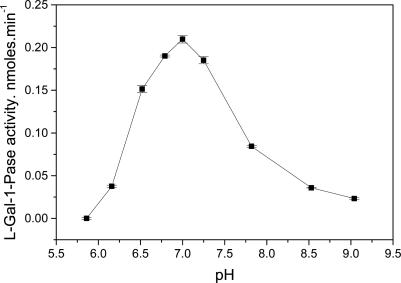
The pH optimum showed a well defined maximum at pH 7 with a falloff to half of the maximum rate within ±0.6 pH of the maximum (Fig. 1).
Fig. 1.
pH response of l-gal-1-Pase. Assays were run in 0.2 M Bis-Tris propane buffer (pH as specified on the graph), 2 mM MgCl2, and 0.5 mM l-gal-1-P for 30 min at 30°C. Data are the mean ± SE of three replicates. Separate experiments showed very similar responses.
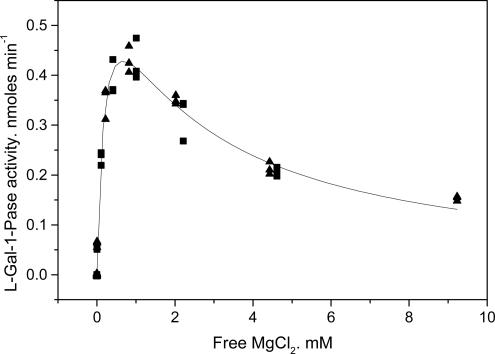
The response of the l-gal-1-Pase to MgCl2 showed a Ka(Mg2+) of 0.2 ± 0.04 mM, but Mg2+ was inhibitory at concentrations >2 mM (Fig. 2). The apparent Ki(Mg2+) for substrate inhibition was 0.46 ± 0.07 mM. No activity was found in the presence of 0.2 mM EDTA without added Mg2+.
Fig. 2.
Response of l-gal-Pase activity to MgCl2. The reaction mix contained either 0.4 mM (▴) or 0.2 mM (▪) EDTA in 0.2 M Bis-Tris propane buffer (pH 7.0) and 0.5 mM gal-1-P. Assays (30 min at 30°C) were initiated with enzyme. Free Mg2+ was calculated by using cheaqs (a program for calculating chemical equilibria in aquatic systems) written by Wilko Verweij (http://home.tiscali.nl/cheaqs/index.html).
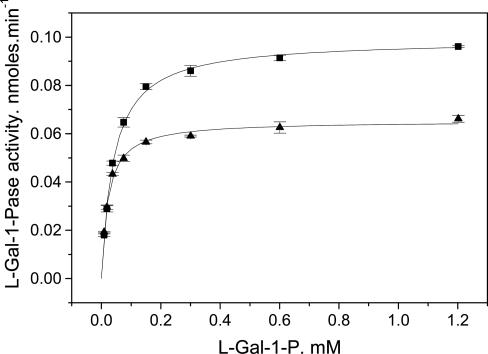
The KM(gal-1-P) depended on MgCl2 being 0.041 ± 0.002 mM at 1.8 mM MgCl2 and 0.022 ± 0.001 mM at 4.8 mM MgCl2 (Fig. 3). In some experiments, there was evidence of mild substrate inhibition, but the Ki was in the range of 3–9 mM. As is the case for the Mg2+ response, the Vmax was halved when the Mg2+ was raised from close to the optimum to inhibitory concentrations (e.g., 4.8 mM).
Fig. 3.
Response of l-gal-1-Pase activity to l-gal-1-P concentration. Reactions were carried out in 100 mM Bis-Tris propane (pH 7.0) at 30°C for 30 min. Reactions were run at either 1.9 mM (▪) or 4.8 mM (▴) MgCl2. The curves are the best fits of a Menten–Michaelis equation to the data.
Identification of the Gene from Arabidopsis. Because highly purified preparations of the kiwifruit enzyme were very unstable, we prepared the enzyme from Arabidopsis thaliana shoots by using the same procedure as kiwifruit and identified the protein from this source. The Arabidopsis thaliana shoot preparation of the l-gal-1-Pase showed similar properties to the kiwifruit enzyme, showing absolute Mg2+ dependency with a Ka(Mg2+) of 0.016 ± 0.004 mM and a Ki(Mg2+) of 0.620 ± 0.17 mM. The Km(l-gal-1-P) was 0.044 ± 0.003 mM (2 mM MgCl2), and the pH optimum was between 6.8 and 7 at 2 mM MgCl2 and 0.5 mM l-gal-1-P (data not shown). For those substrates assayed, the Arabidopsis enzyme showed a similar spectrum of activity to the kiwifruit enzyme (Table 1).
To identify the gene, our strategy was to make as pure a preparation as possible, digest that mixture with trypsin, and identify the proteins in the mixture by liquid chromatography–MS by using the complete Arabidopsis predicted protein databases available. In parallel, we separated the mixture by using SDS/PAGE (data not shown) and excised nine protein-stained gel bands, in-gel digested these with trypsin, and analyzed the mixture by liquid chromatography–MS. We identified 24 proteins based on MS analysis, one of which was annotated as myo-inositol phosphatase. Nineteen proteins were identified from the analysis of the partially pure preparation, and five extra proteins were identified from the SDS bands. A protein annotated as myo-inositol phosphatase was found both from the mixture and from a gel band. None of the other proteins was identified as a phosphatase. The putative phosphatase protein was Arabidopsis protein At3g02870, a protein annotated in The Arabidopsis Information Resource as a putative inositol-1 (or 4) monophosphatase with a predicted mass of 29,121 Da and a predicted pI of 5.01.
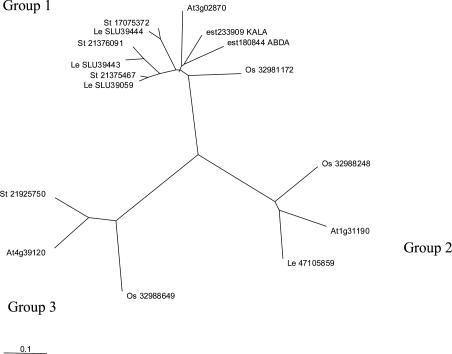
Expression of the Kiwifruit l-Gal-1-Pase. We identified a range of genes from public databases, as well as kiwifruit and apple full-length ESTs from the HortResearch EST database resource, that showed homology to AT3g02870, and we used clustalx (38) to align these proteins. The translated genes formed into three main clusters (Fig. 4), with the tomato myo-inositol phosphatases (21) all in one group with At3g02870, a kiwifruit EST, and an apple EST. The two other clusters also included kiwifruit, apple, and Arabidopsis sequences.
Fig. 4.
Cluster analysis of purported myo-inositol phosphate phosphatases. The alignment was based on translated GenBank sequences or The Arabidopsis Information Resource (At, Arabidopsis thaliana) peptide sequences. Tomato sequences (Le) represent isoforms 1 (LeIMP1 and SLU39444), 2 (LeIMP2 and SLU39443), and 3 (LeIMP3 and SLU39059). Rice sequences are coded Os, potato sequences are coded St, and they are followed by their GenBank ID. The ESTs from the HortResearch fruit database are coded ABDA (apple) or KALA (kiwifruit), after their EST number. EST 233909 is the EST shown to posses l-gal-1-Pase activity in this paper.
We cloned and expressed EST 233909, a putative kiwifruit myo-inositol-1-Pase that fell in group 1 (Fig. 4), in E. coli. This protein showed some myo-inositol-1-Pase activity but had ≈14 times more gal-1-Pase activity and very little activity against a wide range of other phosphorylated compounds (Table 1). The expressed enzyme had a maximum specific activity against l-gal-1-P of 16.4 ± 1.0 μmol π-generated per min-1 per mg of protein (as calculated from the l-gal-1-P response curve), whereas an extract from E. coli expressing the empty vector after His-column purification showed no phosphatase activity against l-gal-1-P, myo-inositol-1-P, or other substrates tested. Coupled assays with E. coli expressed kiwifruit l-gal dehydrogenase, which uses l-gal to reduce NAD+, showed that the products of the reaction were l-gal as well as inorganic phosphate (data not shown).
Kinetic analysis of the cloned and expressed kiwifruit enzyme showed it had a Km(l-gal-1-P) of 0.15 ± 0.02 mM, a Km(myo-inositol-1-P) of 0.33 ± 0.02 mM, and a Ka(Mg2+) of 0.47 ± 0.16 mM with a Ki(Mg2+) of 13.4 ± 5.9 mM (data not shown). The high SEs for Mg inhibition reflect the much lower inhibition by Mg2+ seen with this enzyme compared with the enzymes extracted from plants. The enzyme showed a pH optimum of 7.0 (data not shown), very similar to the Arabidopsis and kiwifruit enzymes. LiCl inhibited the l-gal-1-Pase activity, although with a Ki of 3.7 ± 0.4 mM (data not shown), which is considerably higher than that observed with the expressed tomato myo-inositol-1-Pase (21). The enzyme also appeared to be a dimer by gel filtration. The predicted subunit molecular mass of the expressed enzyme, including the His-tag, is ≈34,800 Da. On a G75 Sephadex column, the enzyme eluted (data not shown) just before a BSA standard (68,000 Da), as would be expected for a dimer (≈70,000 Da).
Discussion
Ascorbic acid is a critical chemical in plants, for both plant and human health, and understanding the path of biosynthesis is important in achieving the significant goal of increasing plant ascorbic acid levels. In this work, by discovering a phosphatase highly specific to the l-gal pathway of ascorbic acid biosynthesis, we further establish the significance of this pathway in higher plants, as well as open new possibilities to manipulate ascorbic acid levels in plants. In addition, we raise the possibility that two pathways of ascorbic acid biosynthesis, namely the l-gal and myo-inositol pathways, may be linked through the action of a common phosphatase enzyme.
We have partially purified enzymes from kiwifruit berries and Arabidopsis thaliana shoots that show significant specifity in hydrolyzing l-gal-1-P to l-gal and inorganic phosphate as shown by direct assays of both products. The activities show a low Km for both l-gal-1-P and Mg2+ and a pH optimum of 7.0. These properties make them very viable candidates to be the phosphatase in the pathway to ascorbate biosynthesis because there appears to be no other use for l-gal in plants other than ascorbate biosynthesis (15, 18).
l-Gal-1-Pase Gene. We have also identified a kiwifruit cDNA as coding the protein l-gal-1-Pase. This clone shows significant homology to an Arabidopsis thaliana gene (At3g02870), identified in GenBank as a putative myo-inositol-1-Pase protein, in a partially purified protein preparation from Arabidopsis shoots. The E. coli expressed enzyme had very similar properties and highly specific ability to hydrolyze l-gal-1-P as found in both the Arabidopsis and kiwifruit partially purified enzymes. These properties of the partially purified enzymes included a native mass close to 65,000 Da (presumably a homodimer), a low pI (excluded by SP column at pH 5.2), an absolute Mg2+ dependency with inhibition at higher MgCl2 concentrations, a very strong preference for l-gal-1-P as substrate, and a pH optimum close to 7.0. The main differences between the recombinantly expressed and recombinantly extracted enzymes were that the recombinant enzyme had a higher Ka for Mg2+ and a higher Km for l-gal-1-P than the extracted enzymes, but this may reflect the bacterial folding environment or the presence of the His-tag and other sequence contributed by the pET vector as well as another 14 residues from the 5′ UTR of the EST in front of the N-terminal of the expressed enzyme. The Arabidopsis enzyme does not have a targeting sequence nor does the Kiwifruit enzyme. The next enzyme in the pathway, l-gal dehydrogenase, has no targeting sequence either (31).
Putative myo-Inositol-1-Pase Genes in Apple and Kiwifruit. We observed that apple, kiwifruit, and Arabidopsis thaliana putative myo-inositol-1-Pase sequences partition into three clusters (groups 1–3 in Fig. 4). We have analyzed cDNA sequences that belong to the three clusters. Within apple, there were 23 ESTs belonging to the myo-inositol phosphatase family in total, of which 18 belong to group 1, the clade with demonstrated gal-1-Pase activity. Of these group-1 ESTs, 13 ESTs were from libraries made from fruit tissue, whereas five ESTs came from shoot and xylem libraries. Because HortResearch has sequenced ≈160,000 apple ESTs, these 23 ESTs represent a frequency of 0.014%. The group-2 and group-3 apple ESTs came from fruit (3 ESTs) and shoot (5 ESTs) tissues. In the case of kiwifruit, there were also 23 ESTs identified (a frequency of 0.017%). Group 1 ESTs came from fruit (15) and buds (4). Group-2 and group-3 ESTs also came from fruit (3) and buds (1). Thus, overall, group-1 (l-gal-1-Pase-like) ESTs were the most commonly found ESTs (total 37 of 46), predominantly from fruit (28) and, to a lesser extent, shoot (9) tissues. This result is compatible with ascorbic acid biosynthesis occurring via the l-gal pathway in fruit tissue.
The Inositol-1-Monophosphatase Gene Family. This inositol-1-monophosphatase family is a widely distributed family (39, 40) and includes the E. coli phosphomonoesterases such as SuhB (41). SuhB preferentially hydrolyses myo-inositol-1-P but can also hydrolyze glycerophosphate, myo-inositol-2-P, and adenosine-2′-monophosphate. The myo-inositol family also includes members of the N-amidino-scyllo-inosamine-4-phosphate phosphatase and fructose bisphosphatase families, and enzymes with the ability to cleave d-gal-1-P and fructose-1–6-bisphosphate are described in refs. 42–44.
In plants, two genomes have now been fully sequenced: rice and the model plant Arabidopsis thaliana. Both have one putative myo-inositol phosphatase gene belonging to group 1 in our phylogenetic tree and one myo-inositol phosphatase-like gene in each of the other groups. Tomato (21, 45) and potato have three putative myo-inositol phosphatase genes in group 1 (Fig. 4), the same cluster in which we found an EST with l-gal-1-Pase activity (EST 233909, Fig. 4). There is a GenBank Solanaceae sequence that belong to each of the other two groups.
Discovering myo-inositol-1-Pase activity by the expressed tomato genes was based on screening a phage library for 5-bromo-4-chloro-3-indoyl phosphate phosphatase activity at neutral pH (21). The one positive clone (LelMP) was then used to screen a young fruit cDNA library resulting in isolation of the two other clones (21). The three clones show greater than ≈80% similarity, whereas the three Arabidopsis thaliana clones only show 20–40% similarity. This finding may explain why putative myo-inositol-1-Pases belonging to groups 2 and 3 were not cloned by these authors from tomato. The activity reported for the tomato clones used a very sensitive assay (21), and as we show, our expressed l-gal-1-Pase can hydrolyze myo-inositol-1-P at ≈7% of the rate of l-gal-1-P hydrolysis. Gillespy et al. (21) did not test specificity of activity against other substrates and did not characterize the enzyme's properties except to suggest that it was a dimer, Mg2+-dependent, and Li+-sensitive (21). Curiously, whereas Gillespy et al. isolated LeIMP1 from a young fruit library, this gene is more highly expressed in mature fruit and flowers and increases in expression with exposure to light (21). Such an increase in expression with exposure to light could suggest that this gene may also have l-gal-1-Pase activity due to activation of ascorbate synthesis. Although we currently do not know the activity of the proteins encoded by genes found in the other two groups, it may be that the group-1 genes are actually all l-gal-1-Pases and the real myo-inositol-1-Pase has yet to be identified.
The Function of a Phosphatase in the Biosynthesis of Ascorbate. Two pathways of ascorbic acid biosynthesis involve a phosphatase step: the l-gal pathway (5) and the myo-inositol pathway (8, 21). Myo-inositol metabolism is also of significance in the widespread phosphatidylinositol signaling pathway (39). It is of note that the one enzyme we describe shows activity against both of these substrates, but it is much more effective against l-gal-1-P than myo-inositol-1-P. It is possible that both pathways involve the one gene and enzyme, or more likely, that the l-gal pathway has seconded the myo-inositol-1-Pase into a new role through gene duplication. This finding would imply that the group-2 or group-3 members of the myo-inositol-1-Pase family are the true phosphatases for myo-inositol-1-P, and that the group-1 phosphatases are the seconded enzyme.
In conclusion, we have partially purified and characterized a specific l-gal-1-Pase that we propose is significant in the biosynthesis of ascorbic acid, further supporting the importance of the l-gal route to ascorbate biosynthesis, and we have identified the gene that encodes this protein and discussed its relationship with other genes.
Note Added in Proof. Since submission of this paper, we have shown that the Arabidopsis thaliana group 1 clone At3g02870 expressed in E. coli shows ≈12 times as much l-gal-1-Pase activity as myo-inositol-1-Pase activity (data not shown).
Supplementary Material
Acknowledgments
We thank Dr. Andrew Gleave (HortResearch) for providing sequencing facilities; Dr. Ross Crowhurst (HortResearch), manager of the EST sequence database and bioinformatics suite; and Dr. Lesley Beuning (HortResearch), who oversaw tissue collection and library construction for kiwifruit and apple ESTs. This work was supported by Foundation for Science and Technology Grant CO6X0006.
Author contributions: W.A.L. designed research; W.A.L., S.B., M.W., J.C., D.J., and D.B. performed research; W.A.L. and S.B. analyzed data; J.C., D.J. and D.B. contributed new reagents/analytic tools; W.A.L., S.B., and E.M. wrote the paper; E.M. obtained funding; and E.M. provided direction for research.
Abbreviations: l-gal, l-galactose; l-gal-1-P, l-gal-1-phosphate; l-gal-1-Pase, l-gal-1-P phosphatase; myo-inositol-1-P, myo-inositol-1-phosphate; myo-inositol-1-Pase, myo-inositol-1-P phosphatase.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY787585 and AY787586).
References
- 1.Apel, K. & Hirt, H. (2004) Annu. Rev. Plant Biol. 55, 373-399. [DOI] [PubMed] [Google Scholar]
- 2.Naidu, K. A. (2003) Nutr. J. 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smirnoff, N., Conklin, P. L. & Loewus, F. A. (2001) Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 437-467. [DOI] [PubMed] [Google Scholar]
- 4.Smirnoff, N., Running, J. A. & Gatzek, S. (2004) in Vitamin C: Its Function and Biochemistry in Animals and Plants, eds. Asard, H., May, J. M. & Smirnoff, N. (Bios Scientific, London), pp. 7-29.
- 5.Wheeler, G. L., Jones, M. A. & Smirnoff, N. (1998) Nature 393, 365-369. [DOI] [PubMed] [Google Scholar]
- 6.Loewus, F. A. & Kelly, S. (1961) Arch. Biochem. Biophys. 95, 483-493. [DOI] [PubMed] [Google Scholar]
- 7.Isherwood, F. A. & Mapson, L. W. (1956) Biochem. J. 64, 13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorence, A., Chevone, B. I., Mendes, P. & Nessler, C. L. (2004) Plant Physiol. 134, 1200-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agius, F., Gonzalez-Lamothe, R., Caballero, J. L., Munoz-Blanco, J., Botella, M. A. & Valpuesta, V. (2003) Nat. Biotechnol. 21, 177-181. [DOI] [PubMed] [Google Scholar]
- 10.Banhegyi, G. & Loewus, F. A. (2004) in Vitamin C: Function and Biochemistry in Animals and Plants, eds. Asard, H., May, J. M. & Smirnoff, N. (Bios Scientific, London), pp. 31-48.
- 11.Hancock, R. D., McRae, D., Haupt, S. & Viola, R. (2003) BMC Plant Biol. 3, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conklin, P. L., Norris, S. R., Wheeler, G. L., Williams, E. H., Smirnoff, N. & Last, R. L. (1999) Proc. Natl. Acad. Sci. USA 96, 4198-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolucka, B. A., Persiau, G., Van Doorsselaere, J., Davey, M. W., Demol, H., Vandekerckhove, J., Van Montagu, M., Zabeau, M. & Boerjan, W. (2001) Proc. Natl. Acad. Sci. USA 98, 14843-14848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conklin, P. L. (2001) Plant Cell Environ. 24, 383-394. [Google Scholar]
- 15.Davey, M. W., Van Montagu, M., Inze, D., Sanmartin, M., Kanellis, A., Smirnoff, N., Benzie, I. J. J., Strain, J. J., Favell, D. & Fletcher, J. (2000) J. Sci. Food Agric. 80, 825-860. [Google Scholar]
- 16.Smirnoff, N. (2000) Curr. Opin. Plant Biol. 3, 229-235. [PubMed] [Google Scholar]
- 17.Smirnoff, N. & Wheeler, G. L. (2000) Crit. Rev. Biochem. Mol. Biol. 35, 291-314. [DOI] [PubMed] [Google Scholar]
- 18.Smirnoff, N. (2001) Vitam. Horm. 61, 241-266. [DOI] [PubMed] [Google Scholar]
- 19.Gumber, S. C., Loewus, M. W. & Loewus, F. A. (1984) Plant Physiol. 76, 40-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loewus, M. W. & Loewus, F. A. (1982) Plant Physiol. 70, 765-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gillaspy, G. E., Keddie, J. S., Oda, K. & Gruissem, W. (1995) Plant Cell 7, 2175-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mamedov, T. G., Suzuki, K., Miura, K., Kucho Ki, K. & Fukuzawa, H. (2001) J. Biol. Chem. 276, 45573-45579. [DOI] [PubMed] [Google Scholar]
- 23.Ho, C. L., Noji, M. & Saito, K. (1999) J. Biol. Chem. 274, 11007-11012. [DOI] [PubMed] [Google Scholar]
- 24.Lunn, J. E., Ashton, A. R., Hatch, M. D. & Heldt, H. W. (2000) Proc. Natl. Acad. Sci. USA 97, 12914-12919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogel, G., Aeschbacher, R. A., Muller, J., Boller, T. & Wiemken, A. (1998) Plant J. 13, 673-683. [DOI] [PubMed] [Google Scholar]
- 26.Zhou, R., Cheng, L. & Wayne, R. (2003) Plant Sci. 165, 227-232. [Google Scholar]
- 27.Kingston-Smith, A. H., Major, I., Parry, M. A. & Keys, A. J. (1992) Biochem. J. 287, 821-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turner, W. L. & Plaxton, W. C. (2001) Planta 214, 243-249. [DOI] [PubMed] [Google Scholar]
- 29.Schenk, G., Guddat, L. W., Ge, Y., Carrington, L. E., Hume, D. A., Hamilton, S. & de Jersey, J. (2000) Gene 250, 117-125. [DOI] [PubMed] [Google Scholar]
- 30.Bozzo, G. G., Raghothama, K. G. & Plaxton, W. C. (2002) Eur. J. Biochem. 269, 6278-6286. [DOI] [PubMed] [Google Scholar]
- 31.Laing, W. A., Frearson, N., Bulley, S. & MacRae, E. (2004) Funct. Plant Biol. 31, 1015-1025. [DOI] [PubMed] [Google Scholar]
- 32.Chifflet, S., Torriglia, A., Chiesa, R. & Tolosa, S. (1988) Anal. Biochem. 168, 1-4. [DOI] [PubMed] [Google Scholar]
- 33.Gatzek, S., Wheeler, G. L. & Smirnoff, N. (2002) Plant J. 30, 541-553. [DOI] [PubMed] [Google Scholar]
- 34.Schagger, H. & von Jagow, G. (1987) Anal. Biochem. 166, 368-379. [DOI] [PubMed] [Google Scholar]
- 35.Jimenez, C. R., Huang, L., Qiu, Y. & Burlingame, A. L. (2003) in Current Protocols in Protein Science, eds. Coligan, J. E., Dunn, B. M., Ploegh, H. L., Speicher, D. W. & Wingfield, P. T. (Wiley, Hoboken, NJ), pp. 16.4.1-16.4.5.
- 36.Eng, J., McCormack, A. L. & Yates, J. R. (1994) J. Am. Soc. Mass Spectrom. 5, 976-989. [DOI] [PubMed] [Google Scholar]
- 37.Yates, J. R., Eng, J. K., McCormack, A. L. & Schieltz, D. (1995) Anal. Chem. 67, 1426-1436. [DOI] [PubMed] [Google Scholar]
- 38.Jeanmougin, F., Thompson, J. D., Gouy, M., Higgins, D. G. & Gibson, T. J. (1998) Trends Biochem. Sci. 23, 403-405. [DOI] [PubMed] [Google Scholar]
- 39.Majerus, P. W., Kisseleva, M. V. & Norris, F. A. (1999) J. Biol. Chem. 274, 10669-10672. [DOI] [PubMed] [Google Scholar]
- 40.York, J. D., Ponder, J. W. & Majerus, P. W. (1995) Proc. Natl. Acad. Sci. USA 92, 5149-5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsuhisa, A., Suzuki, N., Noda, T. & Shiba, K. (1995) J. Bacteriol. 177, 200-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parthasarathy, R., Parthasarathy, L. & Vadnal, R. (1997) Brain Res. 778, 99-106. [DOI] [PubMed] [Google Scholar]
- 43.Bhat, P. J. (2003) Med. Hypotheses 60, 123-128. [DOI] [PubMed] [Google Scholar]
- 44.Stec, B., Yang, H., Johnson, K. A., Chen, L. & Roberts, M. F. (2000) Nat. Struct. Biol. 7, 1046-1050. [DOI] [PubMed] [Google Scholar]
- 45.Styer, J. C., Keddie, J., Spence, J. & Gillaspy, G. E. (2004) Gene 326, 35-41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.