Abstract
To identify biologically relevant and drug-like protein ligands for medicinal chemistry and chemical biology research the grouping of proteins according to evolutionary relationships and conservation of molecular recognition is an established method. We propose to employ structure similarity clustering of the ligand-sensing cores of protein domains (PSSC) in conjunction with natural product guided compound library development as a synergistic approach for the identification of biologically prevalidated ligands with high fidelity. This is supported by the concepts that (i) in nature spatial structure is more conserved than amino acid sequence, (ii) the number of fold types characteristic for all protein domains is limited, and (iii) the underlying frameworks of natural product classes with multiple biological activities provide evolutionarily selected starting points in structural space. On the basis of domain core similarity considerations and irrespective of sequence similarity, Cdc25A phosphatase, acetylcholinesterase, and 11β-hydroxysteroid dehydrogenases type 1 and type 2 were grouped into a similarity cluster. A 147-member compound collection derived from the naturally occurring Cdc25A inhibitor dysidiolide yielded potent and selective inhibitors of the other members of the similarity cluster with a hit rate of 2–3%. Protein structure similarity clustering may provide an experimental opportunity to identify supersites in proteins.
Keywords: combinatorial chemistry, enzyme inhibition, bioinformatics, chemical biology, ligand-sensing core
The most crucial criteria to be met in compound library development for medicinal chemistry and chemical biology research are biological relevance (1, 2), drug likeness (3–7), and diversity (8–10). To meet these criteria usually potential target proteins are clustered into target families on the basis of functional relatedness and amino acid sequence homology (11). Also, recently a grouping into families based on ligand structure–activity relationship homology has been introduced (12, 13).
The major limitation of these concepts focusing on evolution and conserved molecular recognition is that they usually consider close sequence homologs (14). However, for proteins, spatial structure is more conserved in evolution than amino acid sequence (15). In addition, for certain protein folds a general tendency to bind substrates at so-called supersites, i.e., at similar locations in given domains, and independent of amino acid sequence, because of principles of protein structure and chemical constraints has been observed (16).
Inspired by these observations and the prediction that the number of fold types characteristic for all domains occurring in nature is fairly limited [probably ≈1,000 (17)], we recently proposed to employ this conservation in spatial structure as an alternative criterion for clustering in the context of ligand identification and compound library design (1, 2). In this approach, the ligand-sensing cores of individual protein domains are grouped on the basis of structural similarity and irrespective of sequence similarity to generate a protein structure similarity cluster (PSSC). The structures of ligands that bind to one member of this cluster may be used for the development of novel ligands for other members of the cluster.
To identify ligands that are biologically relevant and prevalidated with high fidelity we have proposed to employ the underlying structural frameworks characteristic for evolutionarily selected natural product classes with multiple biological activities that naturally have to bind to multiple proteins as additional guiding principle for compound library development (“natural product-guided combinatorial chemistry”) (1, 2). Natural product-derived compound libraries are expected to yield comparatively high hit rates at small library size.
Here we report on the successful application of the concepts to cluster according to structural similarity and to employ natural product structure as guiding principle for the development of compound libraries.
On the basis of domain core structure similarity considerations, enzymes carrying out different reactions and having different biological functions are grouped into one similarity cluster.
A small library (147 compounds) derived from a naturally occurring inhibitor of one of these enzymes yielded potent and selective inhibitors for the other enzymes of the similarity cluster at a hit rate of 2–3%. PSSC may provide an experimental opportunity to identify supersites in proteins.
Materials and Methods
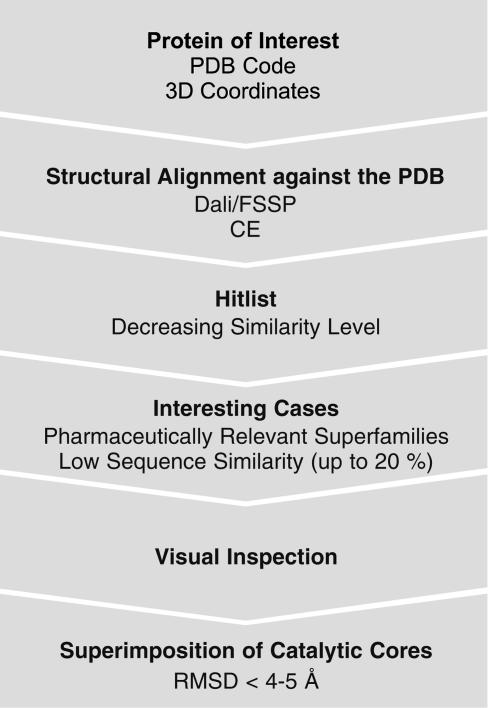
Data Mining and Structure Similarity Clustering. To identify structural similarity between a given protein of interest and other proteins the search strategy outlined in Fig. 1 was developed.
Fig. 1.
Database search strategy. Dali/Fold Classification Based on Structure–Structure Alignment of Proteins (FSSP) searches or Combinatorial Extension (CE) searches are performed by using the Protein Data Bank (PDB) code of the protein of interest. Alternatively, coordinates of a query protein structure may be submitted, and Dali/FSSP or CE compares them against those in the PDB. The FSSP and CE databases are based on exhaustive all-against-all 3D structure comparison of protein structures currently included in the PDB. Hits are listed with decreasing similarity level (3D and sequence similarity). From this list proteins belonging to pharmaceutically relevant families/superfamilies with low sequence identity (SI up to 20%) are chosen and visually inspected. The relevant part of the protein with respect to the delineated concept, i.e., the catalytic core, the conserved part of the domain where the active site is located, must be structurally similar and superimposed. RMSD, rms deviation for aligned Cα positions. When protein structures become too large most superimposition algorithms might fail so that smaller subsets of such big domains containing the interesting catalytic core have to be superimposed. Here, according to our experience, the CE algorithm (see Supporting Materials and Methods) delivers the best results.
The following databases were used: (i) the Structural Classification of Proteins (SCOP) database [http://scop.mrclmb.cam.ac.uk/scop (18)], (ii) the Dali/Fold Classification Based on Structure–Structure Alignment of Proteins (FSSP) database [www.ebi.ac.uk/dali (19)], and (iii) the Database for 3D Protein Structure Comparison and Alignment using the Combinatorial Extension (CE) method [http://cl.sdsc.edu/ce.html (20)]. For manual superimposition of domain cores the CE superimposition algorithm was used (ref. 20; see Supporting Materials and Methods, which is published as supporting information on the PNAS web site). Figures were prepared by using molscript (www.avatar.se/molscript) and rendered with povray (www.povray.org).
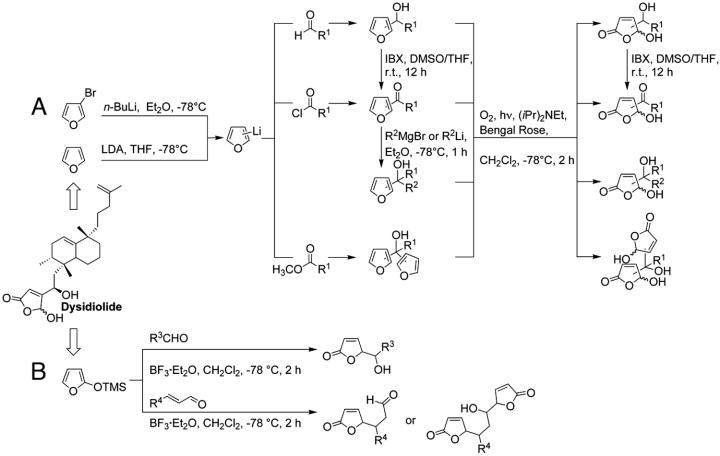
Synthesis. A library of γ-hydroxybutenolides and closely related α,β-unsaturated five-membered lactones based on the structure of the natural product dysidiolide (see Fig. 2) was synthesized by following established procedures (Fig. 2) (21–24).
Fig. 2.
Synthesis of 2- and 3-substituted furans and γ-hydroxybutenolides (A) and 5-substituted butenolides and bisbutenolides (B). LDA, lithium diisopropylamide; THF, tetrahydrofuran; IBX, 1-hydroxy-1,2-benziodoxol-3(1H)-one 1-oxide; r.t., room temperature.
Briefly, either 3-bromofuran or furan itself was selectively lithiated in the 3 or 2 position, respectively, and the furyl lithium intermediates were trapped with different electrophiles to generate secondary and tertiary alcohols or furyl ketones as intermediates. The ketones and the secondary alcohols subsequently were further transformed by addition of Grignard or aryl lithium reagents. Conversion of the furans into hydroxybutenolides was effected by means of cycloaddition with photochemically generated singlet oxygen and regioselective opening of the intermediate cycloadducts (25, 26). α,β-Unsaturated lactones were readily obtained by treatment of different aldehydes with 2-(trimethylsilyloxy)furan in the presence of a Lewis acid (27). A few thiophene derivatives were prepared similarly (not shown). For more details concerning general synthetic procedures and selected analytical data see Supporting Materials and Methods.
Inhibition of Cdc25A Phosphatase (Cdc25A). The clone pET9d/His-Cdc25A was expressed in the Escherichia coli strain BL21-DE3 and purified in the presence of 8 M urea. The inhibition assay was adapted from known procedures (28, 29), using p-nitrophenyl phosphate as colorimetric substrate. Data represent mean ± SD of at least three independent experiments (see Supporting Materials and Methods).
Inhibition of 11β-Hydroxysteroid Dehydrogenases (11βHSDs). 11βHSD1-dependent oxoreduction of cortisone and 11βHSD2-dependent oxidation of cortisol were measured in lysates of stably transfected HEK-293 cells as described previously (30, 31). The rate of conversion of cortisone to cortisol or the reverse reaction was determined by using 1,2,6,7-3H-labeled substrate at a final concentration of 200 nM cortisone or 25 nM cortisol, respectively, in the presence of inhibitor (0–200 μM). Data represent mean ± SD of at least four independent experiments (see Supporting Materials and Methods).
Inhibition of Acetylcholinesterase (AChE). AChE inhibitory activity was measured by using the spectrophotometric method of Ellman et al. (32, 33). Acetylthiocholine iodide was used as the substrate of the enzymatic reaction and 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) was used for the measurement of cholinesterase activity. Data represent mean ± SD of at least three independent experiments (see Supporting Materials and Methods).
Results
Cdc25A is a dual-specificity phosphatase that regulates progression of cell division at the G1 → S checkpoint by dephosphorylating Cdk2/cyclin complexes (34). On the basis of the crystal structure of the fragment of its catalytic domain (residues 336–523) it is assigned, according to the SCOP database (18), to the rhodanese/cell cycle control phosphatase fold. It employs Cys-430, which points into the center of the CX5R phosphate-binding loop, and Glu-431 as important amino acids that act as nucleophile and general acid in the course of phosphate hydrolysis (34, 35). Cdc25A is considered a viable target in the development of new antitumor drugs (36).
Initially, searches in the Dali/FSSP (19) and CE (20) databases were performed that used the structure of Cdc25A (PDB 1C25) as search motif.
This search yielded Hevea brasiliensis hydroxynitrile lyase (PDB 3YAS) as a similar enzyme sharing 17% sequence identity (SI) and a rms deviation for aligned Cα positions of 2.6 Å over an alignment length of 76 residues. This enzyme belongs to the superfamily of α/β-hydrolases from which subsequently AChE was chosen as a pharmaceutically relevant member.
AChE hydrolyzes the neurotransmitter acetylcholine and thereby terminates impulse transmission at cholinergic synapses (37). The Torpedo californica enzyme (PDB 2ACE), which is structurally homologous to the related AChEs occurring in vertebrate nerve and muscle, adopts the α/β-hydrolase fold. Ser-200, His-440, and Glu-327 form the catalytic triad residing in the esteratic subsite of the enzyme's active site. AChE inhibitors are used in the treatment of various disorders such as myasthenia gravis, glaucoma, and Alzheimer's disease (38).
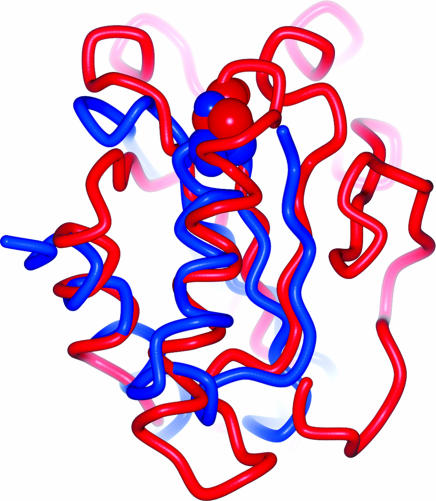
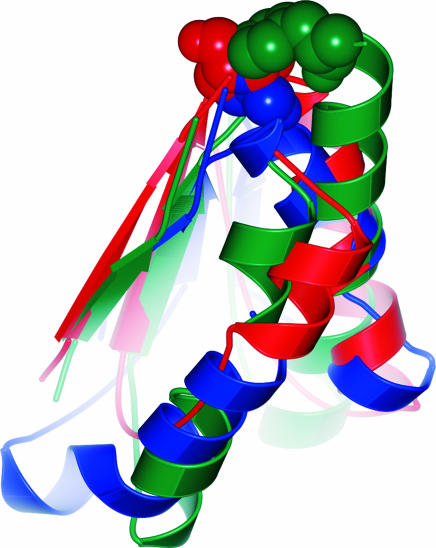
Searches in the Dali/FSSP database may be insensitive for local similarity when structures of substantially different size are being compared. Therefore, in the next step of the analysis, a careful comparative structural exploration of the catalytic cores (i.e., the parts of the catalytic domains where the active site is located) was carried out by means of superimposition using protein structure alignment by incremental combinatorial extension of the optimal path (20) and subsequent visual inspection. As shown in Fig. 3, the two cores of Cdc25A and AChE exhibit significant 3D similarity.
Fig. 3.
Superimposed catalytic cores of AChE (blue) and Cdc25A (red). The best matching parts were aligned with an rms deviation of 2.74 Å at an alignment length of 49 residues. The sequence identities in this best matching part amount to 8.2%. Also shown, in Corey–Pauling–Koltun (CPK) representation, are the catalytic residues, Ser-200 and Cys-430. They share the same location.
Moreover, the structural alignment and the sequence alignment derived thereof (see Supporting Materials and Methods) reveal that the central catalytic residues Cys-430 (Cdc25A) and Ser-200 (AChE) are correlated.
The Dali/FSSP search employing the structure of Cdc25A as search structure also yielded Methylobacterium extorquens methylenetetrahydromethanopterin dehydrogenase (PDB 1LU9) as similarly folded protein with an rms deviation of 4.9 Å over an alignment length of 89 residues and a sequence identity of 7%. This enzyme displays the NAD(P)-binding Rossmann fold. An additional Dali/FSSP search employing the structure of AChE as search motif identified as similar to Cdc25A as described above, additionally yielded Datura stramonium tropinone reductase (PDB 1AE1) as similar protein with an rms deviation of 3.9 Å at an alignment length of 155 residues and 6% sequence identity. This protein also exhibits the Rossmann fold. Thus, both searches converged on proteins displaying the same fold. Consequently, as a third member of a potential similarity cluster, an enzyme belonging to the superfamily of Rossmann-fold domains was chosen. Interestingly, tropinone reductase belongs to the family of the short-chain dehydrogenases, which also contains the pharmaceutically highly relevant hydroxysteroid dehydrogenases. Thus, 11βHSDs type 1 and type 2 were chosen as further members of the cluster.
The two 11βHSD isoforms regulate the interconversion of biologically active 11β-hydroxyglucocorticoids (cortisol and corticosterone) and the inactive 11-ketosteroids (cortisone and 11-dehydrocorticosterone) (39). The enzymes have a catalytic tyrosine embedded in a YXXXK motif in common and they share a glycine-rich N-terminal cosubstrate-binding site (40). 11βHSD1 is essential for the local activation of glucocorticoid receptors because it catalyzes the oxoreduction of cortisone to cortisol. 11βHSD1 may be a promising therapeutic target for the antagonization of glucocorticoid actions (39, 41). Its inhibition is considered to be a promising approach to the treatment of obesity (30, 42), the metabolic syndrome (43, 44), diabetes type 2 (45, 46), and cognitive dysfunction (47). The 11βHSD2 isoform catalyzes exclusively the oxidation of cortisol, and inhibition of 11βHSD2 causes sodium retention resulting in hypertension (48). Therefore, isoenzyme-specific inhibitors of 11βHSD1 are actively sought.
Because crystal structures of the 11βHSDs were not available, homology models for both isoforms were constructed (see Supporting Materials and Methods).
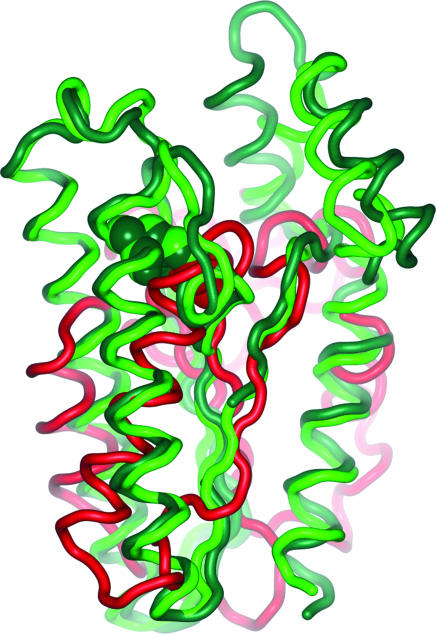
Structural comparison of the Cdc25A catalytic core with the substrate-binding catalytic cores identified from the homology models of the 11βHSDs revealed significant similarity (Fig. 4).
Fig. 4.
Superimposed catalytic cores of Cdc25A (red), 11βHSD1 (dark green), and 11βHSD2 (light green). Cdc25A and the pharmaceutically relevant 11βHSD1 exhibit an rms deviation of 4.13 Å at an alignment length of 80 residues. The sequence identities in this part amount to 5.0%. Also shown, in CPK representation, are the catalytic residues, Cys-430 (Cdc25A), Tyr-183 (11βHSD1), and Tyr-232 (11βHSD2). They share the same space although they derive from different locations when Cdc25A and 11βHSDs are compared.
The catalytic residues Cys-430 (Cdc25A) and Tyr-183 (11βHSD1)/Tyr-232 (11βHSD2) could not be aligned (see the structure-based sequence alignment in Supporting Materials and Methods). The superimposition of the structures, however, clearly demonstrates that these crucial amino acids occupy the same space.
On the basis of this analysis (see Supporting Materials and Methods for a graphical summary) we group Cdc25A, AChE, and the 11βHSDs in a similarity cluster. This grouping is further supported by a similar analysis of the active sites of all three enzymes (Fig. 5), which reveals the overlapping location of the key catalytic residues in space at significant structural similarity in their immediate vicinity.
Fig. 5.
Top view of the catalytic sites of Cdc25A (red), 11βHSD1 (green), and AChE (blue). The key catalytic residues, Cys-430 (Cdc25A), Tyr-183 (11βHSD1), and Ser-200 (AChE), shown in CPK representation, are located similarly.
In the light of this structural similarity, a compound collection was synthesized that is based on a naturally occurring inhibitor of one of the enzymes.
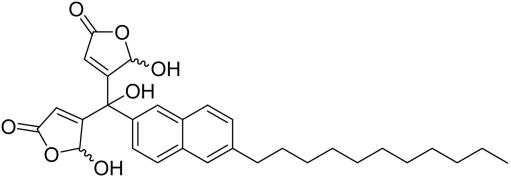
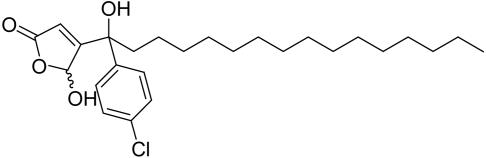
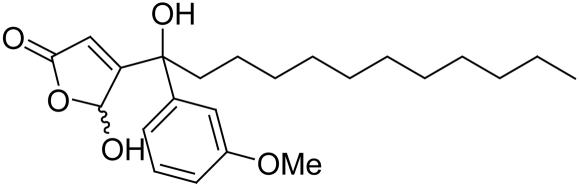
The sesterterpene dysidiolide (see Fig. 2) is an inhibitor of Cdc25A. On the basis of earlier investigations (21, 22) and literature reports on the phosphatase-inhibiting activity of related natural products (49) we hypothesized that the γ-hydroxybutenolide group incorporated into the natural product is a major determinant of its phosphatase-inhibiting activity. Consequently, a library of γ-hydroxybutenolides and closely related α,β-unsaturated five-membered lactones was synthesized (Fig. 2). In total, 147 compounds were prepared (see Supporting Materials and Methods for the structures).
This compound collection was then subjected to biochemical investigation for possible inhibition of Cdc25A, AChE, or 11βHSD1/2. Compounds displaying IC50 values ≤10 μM were considered as hits (see Table 1). Of the 147 compounds investigated, 42 qualified as hits in the Cdc25A assay. The most potent compound (1) had an IC50 value of 350 nM, which is significantly lower than the reported (50) IC50 value for dysidiolide (9.4 μM). Three compounds (2-4) inhibited AChE with IC50 values of 1.3–4.5 μM. The collection contained three 11βHSD1 inhibitors with IC50 values of 7.8–10 μM and four 11βHSD2 inhibitors with IC50 values of 2.4–6.7 μM. Thus, the hit rates for the enzymes identified as being similar to Cdc25A are ≈2–3%, which is a very acceptable value for an initial screen aimed exclusively at identifying hit classes. The results for the most relevant compounds are given in Table 1.
Table 1. Synopsis of inhibition data for selected compounds.
Gratifyingly, even at this small library size, the hits indicated a pronounced degree of selectivity for individual enzymes and also for the isoenzymes 11βHSD1 and 11βHSD2.
Thus, 1 was a significantly more potent inhibitor for Cdc25A than for the other enzymes and 5 showed a clear preference for 11βHSD2. Most remarkably, the α,β-unsaturated lactone 6 inhibited only the therapeutically relevant 11βHSD1 but not or only very weakly the other enzymes investigated. Also, a furan derivative (7) was identified as being an inhibitor for Cdc25A and for 11βHSD2. A selective inhibitor for AChE was not discovered. Promiscuous inhibition of the enzymes due to aggregate formation was ruled out by performing control experiments in the presence of either 0.01% or 0.001% Triton X-100 (ref. 51; see Supporting Materials and Methods).
Discussion
These results convincingly demonstrate the applicability of the PSSC approach delineated above. It provides a filter for the initial identification of compound classes that may yield ligands for multiple members of the similarity cluster.
We point out and stress that ultimately the precise structural organization of the binding sites is the most important factor determining the binding of potential ligands to the proteins. Because in the context of a clustering as proposed above the different binding sites, although commonly located, may be diverse, the compound library to be synthesized must match this biological diversity with an appropriate chemical diversity. Thus, PSSC should be regarded only as an initial abstracting guiding principle. It yields biologically relevant frameworks of small molecules whose structures then have to be (!) refined and varied in a library approach.
Our results demonstrate that potency and selectivity among members of the similarity cluster can be achieved even at relatively small library size and lend proof to the idea that compound libraries derived from natural products may yield protein ligands with high fidelity.
Natural products are exposed to and must bind to multiple proteins in the course of their biosynthesis and when exerting their biological function (e.g., in chemical defense or communication in nature). Thus, if biological activity is enriched in a group of natural products, because of the conservation of structural elements in proteins it is to be expected that the underlying structural framework does confer the ability to bind to proteinaceous receptors to the entire compound class. Compound libraries based on such privileged evolutionarily selected structures (52) from nature should, therefore, yield high hit rates of biologically prevalidated protein ligands at comparably small library sizes.
PSSC also allows for the discovery of completely new and unexpected ligand types. Thus, although many inhibitor classes are known for AChE (53, 54) and the 11βHSDs (39) γ-hydroxybutenolides and α,β-unsaturated five-membered lactones were not described as AChE or 11βHSD inhibitors until now.
The investigation reported herein provides a successful example for the joint application of PSSC and natural product-guided compound library development. However, we stress that this approach is successful for other cases as well. Thus, we have analyzed literature data reported for the biological investigation of natural product-inspired compound collections. This analysis indicated that, for instance, the development of high-affinity agonists for the farnesoid X receptor and of potent inhibitors of leukotriene A4 hydrolase would have been possible by following our approach (2, 55).
Finally, we point out that PSSC may provide an experimental opportunity to identify supersites in proteins, a goal that usually is achieved only by bioinformatics tools. This possibility will need to be investigated in greater depth, in particular with respect to homology or analogy between the proteins involved.
Supplementary Material
Acknowledgments
We thank Dr. Ingrid R. Vetter (Max Planck Institute, Dortmund) for continuing stimulating discussions, Dr. Heino Prinz (Max Planck Institute, Dortmund) for advice with enzymatic assays, Dr. Ingrid Hoffmann (Deutsches Krebsforschungszentrum, Heidelberg) for providing the Cdc25A clone, and Sasikala Thavam, Heike Rimpel, Walburga Hecker, Heidi Jamin, and Regula Suter for excellent technical support. This research was supported by the Max-Planck-Gesellschaft, the Deutsche Forschungsgemeinschaft, and the Fonds der Chemischen Industrie. M.A.K. is the recipient of a doctoral scholarship from the Studienstiftung des Deutschen Volkes. A.O. was supported by grants from the Cloëtta Research Foundation and the Swiss Cancer League (OCS-01402-08-2003).
Author contributions: M.A.K. and H.W. designed research; M.A.K., L.-O.W., S.B., and D.A.J. performed research; M.A.K., L.-O.W., S.B., D.A.J., E.G., K.R., and A.O. contributed new reagents/analytical tools; M.A.K. analyzed data; and M.A.K. and H.W. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PSSC, protein structure similarity clustering; PDB, Protein Data Bank; FSSP, Fold Classification Based on Structure–Structure Alignment of Proteins; CE, Combinatorial Extension; 11βHSD, 11β-hydroxysteroid dehydrogenase; AChE, acetylcholinesterase; CPK, Corey–Pauling–Koltun.
References
- 1.Breinbauer, R., Vetter, I. R. & Waldmann, H. (2002) Angew. Chem. Int. Ed. 41, 2878–2890. [DOI] [PubMed] [Google Scholar]
- 2.Koch, M. A., Breinbauer, R. & Waldmann, H. (2003) Biol. Chem. 384, 1265–1272. [DOI] [PubMed] [Google Scholar]
- 3.Walters, W. P., Ajay & Murcko, M. A. (1999) Curr. Opin. Chem. Biol. 3, 384–387. [DOI] [PubMed] [Google Scholar]
- 4.Ajay, Walters, W. P. & Murcko, M. A. (1998) J. Med. Chem. 41, 3314–3324. [DOI] [PubMed] [Google Scholar]
- 5.Sadowski, J. & Kubinyi, H. (1998) J. Med. Chem. 41, 3325–3329. [DOI] [PubMed] [Google Scholar]
- 6.Ghose, A. K., Viswanadhan, V. N. & Wendoloski, J. J. (1999) J. Comb. Chem. 1, 55–68. [DOI] [PubMed] [Google Scholar]
- 7.Lee, M.-L. & Schneider, G. (2001) J. Comb. Chem. 3, 284–289. [DOI] [PubMed] [Google Scholar]
- 8.Golebiowski, A., Klopfenstein, S. R. & Portlock, D. E. (2001) Curr. Opin. Chem. Biol. 5, 273–284. [DOI] [PubMed] [Google Scholar]
- 9.Mason, J. S. & Hermsmeier, M. A. (1999) Curr. Opin. Chem. Biol. 3, 342–349. [DOI] [PubMed] [Google Scholar]
- 10.Schreiber, S. L. (2000) Science 287, 1964–1969. [DOI] [PubMed] [Google Scholar]
- 11.Schuffenhauer, A., Floersheim, P., Acklin, P. & Jacoby, E. (2003) J. Chem. Inf. Comput. Sci. 43, 391–405. [DOI] [PubMed] [Google Scholar]
- 12.Frye, S. V. (1999) Chem. Biol. 6, R3–R7. [DOI] [PubMed] [Google Scholar]
- 13.Jacoby, E., Schuffenhauer, A. & Floersheim, P. (2003) Drug News Perspect. 16, 93–102. [DOI] [PubMed] [Google Scholar]
- 14.Gerlt, J. A. & Babbitt, P. C. (2001) Annu. Rev. Biochem. 70, 209–246. [DOI] [PubMed] [Google Scholar]
- 15.Grishin, N. V. (2001) J. Struct. Biol. 134, 167–185. [DOI] [PubMed] [Google Scholar]
- 16.Russell, R. B., Sasieni, P. D. & Sternberg, M. J. E. (1998) J. Mol. Biol. 282, 903–918. [DOI] [PubMed] [Google Scholar]
- 17.Koonin, E. V., Wolf, Y. I. & Karev, G. P. (2002) Nature 420, 218–223. [DOI] [PubMed] [Google Scholar]
- 18.Murzin, A. G., Brenner, S. E., Hubbard, T. & Chothia, C. (1995) J. Mol. Biol. 247, 536–540. [DOI] [PubMed] [Google Scholar]
- 19.Holm, L. & Sander, C. (1996) Nucleic Acids Res. 24, 206–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shindyalov, I. N. & Bourne, P. E. (1998) Protein Eng. 11, 739–747. [DOI] [PubMed] [Google Scholar]
- 21.Brohm, D., Metzger, S., Bhargava, A., Müller, O., Lieb, F. & Waldmann, H. (2002) Angew. Chem. Int. Ed. 41, 307–311. [DOI] [PubMed] [Google Scholar]
- 22.Brohm, D., Philippe, N., Metzger, S., Bhargava, A., Müller, O., Lieb, F. & Waldmann, H. (2002) J. Am. Chem. Soc. 124, 13171–13178. [DOI] [PubMed] [Google Scholar]
- 23.Ihara, M., Suzuki, S., Taniguchi, N. & Fukumoto, K. (1993) J. Chem. Soc. Perkin Trans. 1, 2251–2258.
- 24.Frigerio, M., Santagostino, M., Sputore, S. & Palmisano, G. (1995) J. Org. Chem. 60, 7272–7276. [Google Scholar]
- 25.Corey, E. J. & Roberts, B. E. (1997) J. Am. Chem. Soc. 119, 12425–12431. [Google Scholar]
- 26.Kernan, M. R. & Faulkner, D. J. (1988) J. Org. Chem. 53, 2773–2776. [Google Scholar]
- 27.Brown, S. P., Goodwin, N. C. & MacMillan, D. W. C. (2003) J. Am. Chem. Soc. 125, 1192–1194. [DOI] [PubMed] [Google Scholar]
- 28.Blomberg, I. & Hoffmann, I. (1999) Mol. Cell. Biol. 19, 6183–6194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baratte, B., Meijer, L., Galaktionov, K. & Beach, D. (1992) Anticancer Res. 12, 873–880. [PubMed] [Google Scholar]
- 30.Schweizer, R. A. S., Atanasov, A. G., Frey, B. M. & Odermatt, A. (2003) Mol. Cell. Endocrinol. 212, 41–49. [DOI] [PubMed] [Google Scholar]
- 31.Frick, C., Atanasov, A. G., Arnold, P., Ozols, J. & Odermatt, A. (2004) J. Biol. Chem. 279, 31131–31138. [DOI] [PubMed] [Google Scholar]
- 32.Ellman, G. L., Courtney, K. D., Andres, V., Jr., & Featherstone, R. M. (1961) Biochem. Pharmacol. 7, 88–95. [DOI] [PubMed] [Google Scholar]
- 33.Contreras, J.-M., Parrot, I., Sippl, W., Rival, Y. M. & Wermuth, C. G. (2001) J. Med. Chem. 44, 2707–2718. [DOI] [PubMed] [Google Scholar]
- 34.Fauman, E. B., Cogswell, J. P., Lovejoy, B., Rocque, W. J., Holmes, W., Montana, V. G., Piwnica-Worms, H., Rink, M. J. & Saper, M. A. (1998) Cell 93, 617–625. [DOI] [PubMed] [Google Scholar]
- 35.McCain, D. F., Catrina, I. E., Hengge, A. C. & Zhang, Z.-Y. (2002) J. Biol. Chem. 277, 11190–11200. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, Z.-Y. (2001) Curr. Opin. Chem. Biol. 5, 416–423. [DOI] [PubMed] [Google Scholar]
- 37.Sussman, J. L., Harel, M., Frolow, F., Oefner, C., Goldman, A., Toker, L. & Silman, I. (1991) Science 253, 872–879. [DOI] [PubMed] [Google Scholar]
- 38.Ibach, B. & Haen, E. (2004) Curr. Pharm. Des. 10, 231–251. [DOI] [PubMed] [Google Scholar]
- 39.Walker, B. R. & Seckl, J. R. (2003) Expert Opin. Ther. Targets 7, 771–783. [DOI] [PubMed] [Google Scholar]
- 40.Jörnvall, H., Persson, B., Krook, M., Atrian, S., Gonzàlez-Duarte, R., Jeffery, J. & Ghosh, D. (1995) Biochemistry 34, 6003–6013. [DOI] [PubMed] [Google Scholar]
- 41.Chrousos, G. P. (2004) Proc. Natl. Acad. Sci. USA 101, 6329–6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Masuzaki, H., Paterson, J., Shinyama, H., Morton, N. M., Mullins, J. J., Seckl, J. R. & Flier, J. S. (2001) Science 294, 2166–2170. [DOI] [PubMed] [Google Scholar]
- 43.Masuzaki, H. & Flier, J. S. (2003) Curr. Drug Targets Immune Endocr. Metab. Disord. 3, 255–262. [DOI] [PubMed] [Google Scholar]
- 44.Paterson, J. M., Morton, N. M., Fievet, C., Kenyon, C. J., Holmes, M. C., Staels, B., Seckl, J. R. & Mullins, J. J. (2004) Proc. Natl. Acad. Sci. USA 101, 7088–7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alberts, P., Engblom, L., Edling, N., Forsgren, M., Klingström, G., Larsson, C., Rönquist-Nii, Y., Öhman, B. & Abrahmsén, L. (2002) Diabetologia 45, 1528–1532. [DOI] [PubMed] [Google Scholar]
- 46.Ross, S. A., Gulve, E. A. & Wang, M. (2004) Chem. Rev. 104, 1255–1282. [DOI] [PubMed] [Google Scholar]
- 47.Sandeep, T. C., Yau, J. L. W., MacLullich, A. M. J., Noble, J., Deary, I. J., Walker, B. R. & Seckl, J. R. (2004) Proc. Natl. Acad. Sci. USA 101, 6734–6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.New, M. I. & Wilson, R. C. (1999) Proc. Natl. Acad. Sci. USA 96, 12790–12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lyon, M. A., Ducruet, A. P., Wipf, P. & Lazo, J. S. (2002) Nat. Rev. Drug Discov. 1, 961–976. [DOI] [PubMed] [Google Scholar]
- 50.Gunasekera, S. P., McCarthy, P. J., Kelly-Borges, M., Lobkovsky, E. & Clardy, J. (1996) J. Am. Chem. Soc. 118, 8759–8760. [Google Scholar]
- 51.McGovern, S. L., Helfand, B. T., Feng, B. & Shoichet, B. K. (2003) J. Med. Chem. 46, 4265–4272. [DOI] [PubMed] [Google Scholar]
- 52.Evans, B. E., Rittle, K. E., Bock, M. G., DiPardo, R. M., Freidinger, R. M., Whitter, W. L., Lundell, G. F., Veber, D. F., Anderson, P. S., Chang, R. S. L., et al. (1988) J. Med. Chem. 31, 2235–2246. [DOI] [PubMed] [Google Scholar]
- 53.Maelicke, A. & Albuquerque, E. X. (1996) Drug Discov. Today 1, 53–59. [Google Scholar]
- 54.Kaur, J. & Zhang, M.-Q. (2000) Curr. Med. Chem. 7, 273–294. [DOI] [PubMed] [Google Scholar]
- 55.Koch, M. A. & Waldmann, H. (2004) in Chemogenomics in Drug Discovery: A Medicinal Chemistry Perspective, eds. Kubinyi, H. & Müller, G. (Wiley-VCH, Weinheim, Germany), pp. 377–403.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.