Abstract
The causal agents of Lyme disease in North America, Borrelia burgdorferi and Borrelia mayonii, are transmitted primarily by Ixodes scapularis ticks. Due to their limited metabolic capacity, spirochetes rely on the tick blood meal for nutrients and metabolic intermediates while residing in the tick vector, competing with the tick for nutrients in the blood meal. Metabolomics is an effective methodology to explore dynamics of spirochete survival and multiplication in tick vectors before transmission to a vertebrate host via tick saliva. Using gas chromatography coupled to mass spectrometry, we identified statistically significant differences in the metabolic profile among uninfected I. scapularis nymphal ticks, B. burgdorferi-infected nymphal ticks and B. mayonii-infected nymphal ticks by measuring metabolism every 24 hours over the course of their up to 96 hour blood meals. Specifically, differences in the abundance of purines, amino acids, carbohydrates, and fatty acids during the blood meal among the three groups of nymphal ticks suggest that B. mayonii and B. burgdorferi may have different metabolic capabilities, especially during later stages of nymphal feeding. Understanding mechanisms underlying variable metabolic requirements of different Lyme disease spirochetes within tick vectors could potentially aid development of novel methods to control spirochete transmission.
Lyme disease is the most commonly reported vector-borne disease in the United States, with more than 30,000 cases reported each year and indirect information sources indicating that the true numbers of annual Lyme disease cases are 10-fold higher1,2,3. Most of these cases occur in the Northeast and Upper Midwest, caused by the spirochete Borrelia burgdorferi and transmitted to humans primarily by the nymphal stage of the blacklegged tick, Ixodes scapularis4,5. In the Upper Midwest, the same tick also transmits another recently described Lyme disease spirochete, Borrelia mayonii6,7,8. In the continued absence of a human vaccine against these Lyme disease spirochetes, new approaches are needed to suppress vector ticks and disrupt the natural transmission of the spirochetes in their tick vectors and vertebrate reservoir hosts.
Various metabolomic methods, used to measure the abundance or flux of metabolites, or small molecule metabolites in a biological system, have been employed to provide insight into the complex molecular interactions that underpin the proliferation of vector-borne pathogens9,10,11,12,13, but similar efforts have been scarce for Lyme disease spirochetes14,15. A wide variety of instruments are employed for metabolomics projects, often utilizing a particular metabolite extraction solvent system16 followed by a chromatographic separation scheme (gas chromatography (GC), liquid chromatography (LC), hydrophilic interaction (HILIC)) coupled with mass spectrometry (MS)17. Non-targeted metabolomics studies provide global, unbiased coverage of metabolites18, closely reflecting the phenotype of an organism19. Analysis of metabolites involved in complex vector-pathogen interactions provides opportunities to discover innovative control methods for arthropod-borne pathogens9.
Pathogens that are maintained exclusively in arthropod vector-vertebrate reservoir host transmission chains often have considerably limited metabolic capacity, requiring exploitation of major vector/reservoir host cell functions for the acquisition of metabolic intermediates for development10,20. This is the case for B. burgdorferi, which in the eastern United States is maintained in a transmission chain involving Ixodes ticks, particularly I. scapularis, and various vertebrate reservoir hosts21, and presumably also for B. mayonii. Because B. burgdorferi spirochetes lack pathways for de novo biosynthesis of nucleotides, amino acids, fatty acids, and enzyme cofactors, they depend on the blood meal for metabolic intermediates and for intra-tick migration signals when residing within the tick vector15,22,23. Knowledge of Borrelia metabolism within the tick and the metabolic impact of Borrelia colonization of I. scapularis is limited. However, a metabolic cascade in the tick gut is initiated upon ingestion of blood, which also affects the phenotype of the colonizing pathogen24,25. Within unfed nymphs, nutrient deprived spirochetes remain in a metabolic state that has been largely uncharacterized24,26. The number of spirochetes within the tick gut remain at a low level during the first 24 hours of nymphal feeding27,28, but increases exponentially as the feeding progresses29, placing an increased metabolic demand on the tick.
By studying metabolic processes of Borrelia-infected ticks, in our case B. burgdorferi and B. mayonii, we can gain a more detailed understanding of intricate vector-pathogen interactions as well as explore variation in metabolic requirements among different Lyme disease spirochetes as they are exposed in the tick midgut to blood ingested from a vertebrate blood meal host, start to multiply and penetrate the tick midgut to reach the salivary glands, and are transmitted to the vertebrate tick host. Such knowledge can facilitate discovery of metabolites with potential utility for novel methods to control the spirochetes within the vector tick. While the roles of specific metabolites have previously been investigated21,30,31,32,33,34, this is the first account of the global metabolome of Borrelia-infected ticks during the period of nymphal blood feeding and spirochete transmission. Though we were able to make parallels to metabolites in other arthropod vector-pathogen interactions, additional research is necessary to investigate the impact of specific metabolites on the ability of Borrelia spirochetes to proliferate within and be transmitted by tick vectors.
Methods
Blood feeding by nymphal ticks
The ticks used in this study came from a colony maintained at The Centers for Disease Control and Prevention-Fort Collins, and originated from adults collected in multiple locations in Fairfield County, Connecticut in the fall of 2013. Nymphal ticks used in the experiments included uninfected I. scapularis nymphs and nymphs infected, by feeding on infectious mice in the preceding larval stage, with either Borrelia burgdorferi strain B3135 or the Borrelia mayonii type strain (MN14–1420)36. Nymphal blood meals were taken from 8–12 week old female Mus musculus CD-1 outbred mice (Charles River Laboratories, Wilmington, Massachusetts, USA). At the time of feeding, the nymphs were approximately 10 weeks post-molt.
Nymphs (n = 50) from each of the three experimental groups (B. burgdorferi-infected, B. mayonii-infected, and uninfected control) were collected before commencement of feeding to represent pre-feed sample specimens. Individual mice were exposed to feeding by 20 nymphs, allowed to select their feeding sites after being brushed onto anesthetized mice, from one of the three experimental groups, for a total of 9 mice infested by B. burgdorferi-infected nymphs, 12 mice by B. mayonii-infected nymphs and 8 mice by uninfected nymphs. Mouse numbers were determined based upon the infection rate of ticks, in order to generate the required numbers of infected nymphs per time point. Subsets of these mice were exposed to nymphal feeding for 24 hours, 48 hours, 72 hours and 96 hours, with the last time point representing complete feeding. At the assigned time point, all nymphs were removed from the mice or recovered from a water surface over which the mice were held to allow for collection for fed and detached ticks. No difference in the total blood meal volume (via tick weight) of replete ticks was detected (data not shown).
Processing of nymphs for analysis of metabolites
After being collected, ticks were quenched in ice-cold 100% HPLC-grade methanol (Sigma Aldrich, St. Louis, MO) and stored at −80 C. For processing, individual ticks were separated and added to tubes containing 100 μl of cold 100% methanol. Approximately 10–2.3 mm chrome steel beads (Biospec Products Inc., Bartlesville, OK) were added and the sample was homogenized using a Mini-beadbeater (Biospec Products Inc.) for 1 minute.
After bead beating, the samples were centrifuged at 10,000 xg for 1 minute. The supernatant was removed into a clean tube. To wash the beads, 100 μl of fresh 100% methanol was added to the tube containing the beads, briefly vortexed, and centrifuged at similar conditions. The supernatant was removed and added to the previously collected sample to produce a final volume of 150 μl of methanol. All samples were placed in a rotary evaporator and evaporated with vacuum until a sample volume of 50 μl was reached.
After homogenization, the remaining pellet was subjected to PCR to determine the infection status of individual ticks using combined detection of I. scapularis actin, as a control for both the DNA purification and the PCR testing, and the spirochete flagellar filament cap (fliD) target, which is present in both B. burgdorferi and B. mayonii6,37,38,39. A modified multiplex TaqMan PCR assay was used as described37,38,39. Any remaining methanol was evaporated from the pellet and the pellet resuspended in 50 μl DNase-free water. The pellet suspension was boiled at 95 °C for 15 minutes and briefly centrifuged to remove large debris. 4.8 μl of the pellet supernatant was removed to an Axygen 96 well plate (BioSpec Products, Inc.) to which 5.2 μl of master mix containing iQ Multiplex Powermix (Bio-Rad, Hercules, CA, USA), and primers in a final concentration of 300 nM and probes in a final concentration of 200 nM. The PCR cycling conditions were 95 °C for 3 minutes to denature DNA followed by 40 cycles of 95 °C for 10 seconds and 60 °C for 1 minute on a C1000 Touch thermal cycler with a CFX96 real time system (Bio-Rad). After determining the infection status of individual ticks, the metabolite-containing methanol extraction collected previously was combined to generate a pool of metabolites from 10 infected ticks per time point (pre-feed and after 24 hours, 48 hours, 72 hours or 96 hours of feeding) and infected experimental group (B. burgdorferi-infected or B. mayonii-infected). Uninfected control ticks fed concurrently alongside the infected-tick feed, generating 3 pools of 10 uninfected control ticks per time point. Uninfected control ticks were processed and tested individually via PCR as described above to confirm their uninfected status.
GC-MS Analysis
Metabolite extracts were dried under nitrogen, re-suspended in 50 μL of pyridine containing 15 mg/mL of methoxyamine hydrochloride, incubated at 60 °C for 45 minutes, sonicated for 10 minutes, and incubated for an additional 45 minutes at 60 °C. Next, 25 μL of N-methyl-N-trimethylsilyltrifluoroacetamide with 1% trimethylchlorosilane (MSTFA + 1% TMCS, Thermo Scientific, Waltham, MA) was added and samples were incubated at 60 °C for 30 minutes, centrifuged at 3000 xg for 5 minutes, cooled to room temperature, and 80 μL of the supernatant was transferred to a 150 μL glass insert in a GC-MS autosampler vial. Metabolites were detected using a Trace GC Ultra coupled to a Thermo ISQ mass spectrometer (Thermo Scientific). Samples were injected in a 1:10 split ratio twice in discrete randomized blocks. Separation was achieved using a 30 m TG-5MS column (Thermo Scientific, 0.25 mm i.d., 0.25 μm film thickness) with a 1.2 mL/minute helium gas flow rate, and the program consisted of 80 °C for 30 seconds, a ramp of 15 °C per minute to 330 °C, and an 8 minute hold. Masses between 50–650 m/z were scanned at 5 scans/second using electron impact ionization. The ionization source was cleaned and retuned and the injection liner replaced between injection replicates.
Data Analysis and Statistics
For each sample, raw data files were converted to.cdf format, and a matrix of molecular features as defined by retention time and mass (m/z) was generated using XCMS software in R40 for feature detection and alignment. Raw peak areas were normalized to total ion signal in R, outlier injections were detected based on total signal and PC1 of principle component analysis, and the mean area of the chromatographic peak was calculated among replicate injections (n = 2). Due to normalization of the dataset, the quantitation of each molecular feature is described using Relative Abundance (RA). Features were grouped using RAMClustR41, which groups features into spectra based coelution and covariance across the full dataset, whereby spectra are used to determine the identity of observed compounds in the experiment. Compounds were annotated based on spectral matching to NISTv14 and GOLM spectral databases, using the RAMSearch program42. GOLM retention index was plotted against retention time to provide additional retention based confidence to annotations. Annotation confidence is reported as defined by the Metabolomics Standards Initiative43, with the majority of annotations considered level 2, and a few level 3 (chemical class) based annotations. The peak areas for each feature in a spectrum were condensed via the weighted mean of all features in a spectrum into a single value for each compound. Analysis of variance was conducted on each compound using the aov function in R, and p-values were adjusted for false positives using the Bonferroni-Hochberg method in the p.adjust function in R44. PCA was conducted on mean-centered and pareto variance-scaled data using the pcaMethods package in R. The precision of the analytical method is described by the closeness of repeated individual measures of quality control (QC) samples. Using the coefficient of variation (CV) as a measure of precision in our dataset, 80% of the compounds in the dataset demonstrated coefficient of variation (CV) values for QC samples of less than 15.3%, indicating a high degree of precision (Supplementary Figure 6)45.
Regulatory compliance
Animal use and experimental procedures were in accordance with an approved protocol on file with the Centers for Disease Control and Prevention Division of Vector-Borne Diseases Animal Care and Use Committee.
Results and Discussion
In this study, we describe the metabolic phenotypes associated with infection by the Lyme disease spirochetes B. burgdorferi and B. mayonii in I. scapularis nymphal ticks. All metabolites discussed herein are significant by ANOVA by both treatment and time. In our dataset, 480 of 567 (85%) compounds in the measured or analyzed metabolome were significantly different, typical of many untargeted metabolomics studies. We were able to annotate 114 of 567 (20%) metabolites. The statistically significant metabolites detected are reported in Table 1.
Table 1. Table of significant features for uninfected (Control) Ixodes scapularis nymphs versus nymphs infected with the Lyme disease spirochetes Borrelia burgdorferi (Bb) or Borrelia mayonii (Bm).
| Annotation |
p-value |
Prefeed-Fold Change |
Day 1-Fold Change |
Day 2-Fold Change |
Day 3-Fold Change |
Day 4-Fold Change |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | Functional Class | KEGG ID | MSI Conf. | trt | day | trt:day | Control /Bb | Control /Bm | Control /Bb | Control /Bm | Control /Bb | Control /Bm | Control/Bb | Control/Bm | Control/Bb | Control/Bm |
| Adenosine, alpha- (4TMS) MP | Nucleic Acid | C00212 | 2 | 0.0071 | 0.0014 | 0.0656 | 2.5 | 3.6 | 1.2 | 0.9 | 5.8 | 7.7 | 1.5 | 1.6 | 0.7 | 2.6 |
| Adenosine-5-monophosphate (5TMS) | Nucleic Acid | C00020 | 2 | 0.0219 | <0.0001 | 0.1554 | 0.8 | 0.8 | 1.3 | 0.9 | 0.8 | 0.8 | 1.0 | 1.7 | 1.0 | 3.2 |
| Alanine (2TMS) | Amino Acids | C00041 | 2 | 0.0048 | <0.0001 | 0.0193 | 1.1 | 1.0 | 1.4 | 0.5 | 2.3 | 1.9 | 1.3 | 1.1 | 1.1 | 2.3 |
| Allantoin (4TMS) | Ureide | C02350 | 2 | 0.1195 | 0.0001 | 0.5881 | 1.0 | 1.0 | 1.4 | 0.7 | 2.2 | 1.8 | 1.3 | 1.5 | 0.9 | 2.0 |
| Benzoic acid, (1TMS) | Co-factor | C00180 | 1 | 0.0027 | 0.0003 | 0.0041 | 1.6 | 0.5 | 3.6 | 1.0 | 1.2 | 2.0 | 0.9 | 0.7 | 1.5 | 3.7 |
| Campesterol (1TMS) | Lipid | C01789 | 2 | 0.0281 | <0.0001 | 0.0017 | 0.8 | 0.8 | 1.2 | 1.3 | 0.9 | 1.5 | 0.7 | 0.6 | 0.8 | 2.8 |
| Carbodiimide (2TMS) | Xenobiotic | C01566 | 2 | 0.0005 | 0.0002 | 0.0106 | 2.1 | 1.8 | 0.3 | 1.7 | 0.7 | 3.7 | 1.5 | 1.5 | 1.2 | 2.2 |
| Cholesterol (1TMS) | Lipid | C00187 | 2 | 0.0004 | <0.0001 | 0.0022 | 2.2 | 1.4 | 2.1 | 1.2 | 1.7 | 2.3 | 0.8 | 1.0 | 1.0 | 2.6 |
| Cystathionine (4TMS) | Amino Acids | C02291 | 2 | 0.0010 | <0.0001 | 0.0013 | 0.9 | 0.8 | 0.7 | 0.9 | 0.9 | 1.7 | 1.4 | 1.2 | 1.2 | 7.2 |
| Dodecanoic acid (1TMS) | Lipid | C02679 | 2 | <0.0001 | 0.0003 | 0.0001 | 1.8 | 1.0 | 2.3 | 1.3 | 1.3 | 1.9 | 1.2 | 1.2 | 1.1 | 3.2 |
| Ethanolamine (3TMS) | Amine | C00189 | 2 | 0.0023 | <0.0001 | 0.0146 | 1.6 | 1.3 | 1.4 | 1.8 | 1.5 | 1.6 | 1.6 | 2.5 | 1.0 | 5.7 |
| Ethanolaminephosphate (4TMS) | Lipid | C00346 | 2 | 0.0083 | <0.0001 | 0.0476 | 1.7 | 0.8 | 1.5 | 0.8 | 2.6 | 2.1 | 1.2 | 1.2 | 1.0 | 4.2 |
| Galactose (1MEOX) (5TMS) BP.1 | Carbohydrates | C00124 | 2 | 0.0265 | 0.0001 | 0.0305 | 1.2 | 1.6 | 1.6 | 1.0 | 1.6 | 1.5 | 1.6 | 1.0 | 1.7 | 11.2 |
| Gentiobiose (1MEOX) (8TMS) BP | Carbohydrates (Xeno) | C08240 | 2 | 0.0080 | 0.0001 | 0.0155 | 1.1 | 0.9 | 1.2 | 1.5 | 4.7 | 5.1 | 6.5 | 3.1 | 0.9 | 2.9 |
| Glutamic acid (3TMS) | Amino Acids | C00025 | 2 | <0.0001 | <0.0001 | 0.0001 | 1.5 | 0.7 | 1.1 | 0.3 | 2.1 | 1.7 | 1.2 | 1.3 | 1.0 | 2.4 |
| Glutamine, DL- (3TMS) | Amino Acids | C00064 | 2 | 0.1856 | <0.0001 | 0.0249 | 4.7 | 0.9 | 1.8 | 0.7 | 2.4 | 1.8 | 3.0 | 1.0 | 0.6 | 1.7 |
| Glycerol (3TMS) | Carbohydrates | C00116 | 2 | 0.0004 | <0.0001 | 0.0066 | 1.5 | 1.5 | 1.2 | 1.1 | 1.7 | 2.1 | 1.4 | 1.3 | 1.4 | 3.0 |
| Glycerol-3-phosphate (4TMS) | Lipid | C00093 | 2 | 0.0004 | <0.0001 | 0.0025 | 1.7 | 0.9 | 4.2 | 1.9 | 3.0 | 2.6 | 1.2 | 1.3 | 0.8 | 2.8 |
| Glycine (3TMS) | Amino Acids | C00037 | 2 | <0.0001 | <0.0001 | 0.0009 | 1.6 | 1.4 | 1.4 | 0.5 | 2.3 | 2.0 | 1.4 | 1.1 | 1.2 | 2.3 |
| Guanine (3TMS) | Nucleic Acid | C00242 | 2 | 0.0214 | 0.0001 | 0.0797 | 2.1 | 1.4 | 2.1 | 0.8 | 2.9 | 5.0 | 1.4 | 2.3 | 0.6 | 3.0 |
| Guanosine (5TMS) | Nucleic Acid | C00387 | 2 | 0.0025 | 0.0003 | 0.0856 | 4.2 | 4.2 | 1.8 | 0.7 | 5.2 | 7.3 | 1.6 | 2.0 | 0.7 | 3.2 |
| Hexadecanoic acid (1TMS) | Lipid | C00249 | 2 | <0.0001 | <0.0001 | <0.0001 | 1.5 | 0.3 | 1.0 | 0.3 | 1.1 | 0.3 | 0.9 | 0.4 | 1.0 | 1.1 |
| Hypoxanthine (2TMS) | Nucleic Acid | C00262 | 2 | 0.1108 | 0.0002 | 0.5390 | 1.0 | 1.0 | 1.5 | 0.8 | 8.4 | 8.4 | 1.1 | 1.2 | 1.0 | 2.4 |
| Inosine (4TMS) | Nucleic Acid | C00294 | 2 | 0.0595 | <0.0001 | 0.3658 | 74.9 | 4.5 | 39.1 | 29.3 | 0.0 | 0.0 | 0.5 | 1.6 | 1.8 | 17.2 |
| Isoleucine (2TMS) | Amino Acids | C00407 | 2 | 0.0045 | <0.0001 | 0.0211 | 1.5 | 0.4 | 2.1 | 0.2 | 3.4 | 2.6 | 2.1 | 1.2 | 1.2 | 3.9 |
| Leucine (1TMS) | Amino Acids | C00123 | 2 | 0.0017 | <0.0001 | 0.0134 | 1.5 | 1.1 | 1.9 | 0.5 | 3.7 | 4.2 | 1.8 | 1.1 | 1.3 | 4.1 |
| Lysine (4TMS) | Amino Acids | C00047 | 2 | 0.0009 | <0.0001 | 0.0011 | 1.6 | 1.5 | 0.9 | 0.4 | 2.4 | 1.5 | 1.5 | 1.5 | 1.0 | 3.2 |
| Maltose (1MEOX) (8TMS) MP | Carbohydrates | C00208 | 2 | 0.0036 | <0.0001 | 0.0071 | 1.5 | 1.9 | 1.3 | 1.6 | 4.8 | 6.7 | 5.5 | 3.3 | 0.8 | 2.9 |
| myo-Inositol-1-phosphate (7TMS) | Co-factor | C01177 | 2 | 0.0016 | <0.0001 | 0.0184 | 1.5 | 1.5 | 1.1 | 0.8 | 2.6 | 2.1 | 1.0 | 1.1 | 1.0 | 2.3 |
| NA114002 (classified unknown) | unknown | unknown | 2 | 0.0005 | <0.0001 | 0.0012 | 1.3 | 0.9 | 1.1 | 1.0 | 1.2 | 1.2 | 1.1 | 1.0 | 1.0 | 1.1 |
| Octadecenoic acid, 9-(Z)- (1TMS) | Lipid | C00712 | 2 | <0.0001 | <0.0001 | 0.0017 | 1.4 | 0.7 | 1.9 | 1.4 | 1.7 | 2.7 | 1.3 | 1.2 | 1.2 | 3.6 |
| Ornithine (4TMS) | Amino Acids | C00077 | 2 | 0.0045 | <0.0001 | 0.0072 | 2.5 | 3.2 | 1.5 | 0.7 | 1.7 | 1.5 | 1.5 | 1.1 | 1.0 | 3.8 |
| Pantothenic acid, D- (3TMS) | Vitamins | C00864 | 2 | <0.0001 | <0.0001 | 0.0002 | 1.1 | 0.8 | 1.6 | 0.8 | 3.6 | 2.9 | 2.1 | 1.8 | 1.1 | 3.0 |
| Phenylalanine (2TMS) | Amino Acids | C00079 | 2 | 0.0003 | <0.0001 | 0.0017 | 1.8 | 0.9 | 1.9 | 0.4 | 1.9 | 2.5 | 2.2 | 1.4 | 1.4 | 4.7 |
| Phosphoric acid (3TMS) | Co-factor | C00009 | 2 | 0.0004 | <0.0001 | 0.0015 | 1.0 | 1.1 | 0.9 | 1.3 | 1.5 | 1.7 | 1.0 | 1.0 | 1.0 | 2.0 |
| Phosphoric acid monomethyl ester (2TMS) | Xenobiotic | C19998 | 2 | 0.0042 | <0.0001 | 0.0095 | 1.0 | 1.2 | 1.0 | 0.7 | 4.0 | 2.4 | 0.6 | 1.2 | 0.9 | 3.3 |
| Proline (1TMS) | Amino Acids | C00148 | 2 | <0.0001 | <0.0001 | <0.0001 | 1.1 | 1.0 | 1.0 | 1.0 | 2.3 | 1.6 | 1.1 | 0.8 | 1.1 | 4.7 |
| Pyroglutamic acid (2TMS) | Amino Acids | C01879 | 2 | <0.0001 | <0.0001 | 0.0006 | 2.6 | 1.2 | 1.4 | 0.8 | 2.0 | 1.7 | 1.3 | 1.6 | 0.9 | 2.8 |
| Serine (3TMS) | Amino Acids | C00065 | 2 | 0.0012 | <0.0001 | 0.0052 | 1.8 | 1.0 | 1.1 | 0.5 | 1.9 | 1.9 | 1.7 | 1.4 | 1.0 | 3.3 |
| Sitosterol, beta- (1TMS) | Lipid | C01753 | 2 | 0.0874 | <0.0001 | 0.0078 | 0.9 | 1.4 | 1.0 | 1.5 | 0.5 | 1.5 | 0.5 | 0.4 | 0.8 | 3.3 |
| Sucrose (8TMS) | Carbohydrates | C00089 | 2 | 0.0010 | 0.2407 | 0.0043 | 2.2 | 1.0 | 1.1 | 0.1 | 1.2 | 0.1 | 1.7 | 1.2 | 1.0 | 1.4 |
| Threonine (3TMS) | Amino Acids | C00188 | 2 | 0.0024 | <0.0001 | 0.0236 | 1.5 | 1.1 | 1.2 | 0.5 | 2.0 | 2.4 | 1.9 | 1.1 | 1.3 | 3.2 |
| Tryptophan (3TMS) | Amino Acids | C00078 | 2 | 0.1798 | <0.0001 | 0.0103 | 1.3 | 0.5 | 1.0 | 0.6 | 1.5 | 0.8 | 1.0 | 0.9 | 0.9 | 1.6 |
| Tyrosine (3TMS) | Amino Acids | C00082 | 2 | 0.0028 | <0.0001 | 0.0136 | 1.2 | 1.2 | 1.6 | 0.5 | 1.6 | 1.6 | 1.5 | 1.4 | 1.0 | 3.3 |
| Uracil (2TMS) | Nucleic Acid | C00106 | 2 | 0.0021 | <0.0001 | 0.0471 | 1.2 | 0.8 | 0.5 | 0.5 | 2.4 | 2.0 | 1.1 | 1.1 | 1.2 | 2.1 |
| Urea (2TMS) | Amino Acids | C00086 | 2 | 0.0045 | <0.0001 | 0.0366 | 1.1 | 0.5 | 2.3 | 0.6 | 2.9 | 2.9 | 1.0 | 1.1 | 1.3 | 2.8 |
| Valine (2TMS) | Amino Acids | C00183 | 2 | <0.0001 | <0.0001 | 0.0002 | 1.4 | 0.7 | 1.2 | 0.4 | 3.1 | 2.3 | 2.1 | 1.4 | 1.1 | 3.8 |
| Xanthine (3TMS) | Nucleic Acid | C00385 | 2 | 0.0001 | <0.0001 | 0.0001 | 1.1 | 0.8 | 1.0 | 0.6 | 6.5 | 4.4 | 0.8 | 1.2 | 0.7 | 0.7 |
| Xanthosine (5TMS) | Nucleic Acid | C01762 | 2 | 0.2085 | <0.0001 | 0.0340 | 2.1 | 0.9 | 1.3 | 0.2 | 7.0 | 1.3 | 1.4 | 1.4 | 0.4 | 1.6 |
A. KEGG: Kyoto Encyclopedia of Genes and Genomes; B. MSI Conf. Metabolomics Standards Initiative confidence of annotation; C. p-value significance by ANOVA analysis of treatment (trt: Borrelia infection status), time, and treatment:time (trt:time). D. Fold change determined by dividing relative abundance of selected metabolite in control ticks by relative abundance of metabolite in the Borrelia infected groups.
Purine metabolism
Purine-based nucleotides, present in all living organisms, are fundamental components of many crucial biomolecules such as DNA, RNA, ATP, and coenzymes46. For many bacterial pathogens, purine metabolism is required for growth and virulence47,48,49. While mammals can synthesize purines de novo, many bacterial pathogens lack the necessary molecular machinery and must salvage purines from their host. The role of purine salvage has been well recognized in Salmonella typhimurium during human infections50 and for a variety of vector-borne protozoa and helminths46.
The genome of B. burgdorferi does not contain the genes encoding the enzymes required for de novo purine synthesis nor for the classical salvage pathway47. B. burgdorferi obtains both purine bases and deoxynucleotides via novel purine salvage pathways, directly from the host while residing in a vertebrate reservoir, or from tick-ingested vertebrate blood while residing within the tick vector34,47,48. The most abundant purine in mammalian blood is hypoxanthine51. The specific purine salvage abilities of B. mayonii are yet-undetermined.
Many compounds involved in purine metabolism were statistically significant by ANOVA by both treatment and time in our dataset (Table 1). These data provide evidence that, during colonization and replication of B. burgdorferi and B. mayonii in feeding I. scapularis ticks, these spirochetes exert a significant demand on purines from the incoming vertebrate blood meal compared to the uninfected control ticks (Fig. 1, Supplementary Figure 1). Additionally, the data indicate possible differences in purine metabolism between the two species (Fig. 1, Supplementary Figure 1). Because Borrelia are able to sequester metabolites from the vector and the vertebrate host blood meal in lieu of de novo synthesis, the spirochetes have lost many genes encoding biosynthetic machinery. While incidences of gene loss from more metabolically competent ancestors have been previously identified in B. burgdorferi and other pathogenic host-associated bacteria22,52, additional research is required to evaluate the full metabolic capacity of B. mayonii.
Figure 1. Purine Pathways.
Accumulation over time of statistically significant (ANOVA p ≤ 0.05) metabolites involved in purine metabolic pathway in uninfected (control) Ixodes scapularis nymphs versus nymphs infected with the Lyme disease spirochetes Borrelia burgdorferi or Borrelia mayonii.
Amino Acids
A wide variety of arthropod-borne pathogens place significant metabolic demands on their vectors. The dependence on the vector, and especially the vertebrate blood meal it ingests, is evidenced by the lack of de novo synthesis capabilities of amino acids. In the vertebrate host, Plasmodium falciparum malaria, as well as Leptospira and Treponema spirochetes, harvest essential amino acids from plasma and cell hemoglobin53,54. In the sand fly vector, Leishmania major utilizes amino acids from the ingested blood meal as an energy source for glycolysis and related metabolic cycles55, while the plague bacterium Yersinia pestis catabolizes amino acids from the flea vector blood meal as a primary carbon source during colonization of the flea gut56.
B. burgdorferi also lacks the capacity for the de novo synthesis of amino acids and must sequester them from the vertebrate host or from the blood meal ingested by the tick vector22. Isoleucine, leucine, lysine, serine, and asparagine are the most common amino acids within the B. burgdorferi genome sequence22, but very little is known about how they are acquired within I. scapularis. In our dataset, the aliphatic and hydrophobic amino acid classes demonstrate the most significant differential abundance among treatment groups (Table 1, Supplementary Figure 2).
Interestingly, all statistically significant amino acids for both B. burgdorferi and B. mayonii display the same general trend (Supplementary Figure 2). In most cases, amino acid abundance in tick groups infected with B. burgdorferi remains at a lower level than uninfected controls for the duration of the blood meal (Supplementary Figures 2 and 3). Amino acid abundance in tick groups infected with B. mayonii follows the same trend, but sharply declines for each amino acid around day 3 (Supplementary Figure 2). Amino acid abundance in both uninfected tick groups and B. burgdorferi-infected tick groups follow the same general trend for most amino acids, with the exception of differences in the utilization of glutamine and urea (Supplementary Figure 2H,Q). Since the spirochetes must harvest amino acids from the tick’s blood meal, it seems appropriate that the abundance of amino acids would continue to decrease as feeding continues. Overall, our data show a difference in the impacts of B. mayonii and B. burgdorferi on the tick amino acid metabolism (Supplementary Figures 2 and 3).
Carbohydrates
B. burgdorferi can utilize a finite number of carbohydrates as energy sources, including glucose, maltose, glycerol, mannose, trehalose, chitobiose, and N-acetylglucosamine (GlcNAc)30,57,58. The presence of three putative phosphotransferase-type glucose transporter genes in the genome of B. burgdorferi suggests that glucose is likely a primary energy source30,59. This is especially true for spirochetes within vertebrate reservoir hosts and when colonizing I. scapularis after a blood meal, as glucose is the most prevalent carbohydrate and most efficient energy source in mammalian blood33,60,61. During the period prior to nymphal feeding, when glucose is scarce, B. burgdorferi relies on glycerol and a finite supply of chitobiose to fuel glycolysis33. Furthermore, glycerol is required by Borrelia for maximum fitness during the tick phase of the enzootic cycle and subsequent transmission33. Maltose is not required by B. burgdorferi during mammalian infection57. However, the B. burgdorferi genome encodes several phosphoenolpyruvate:phosphotransferase (PTS) systems used to acquire maltose, suggesting that maltose may also be used in glycolysis22,59. Although previous studies hypothesized that Borrelia could utilize galactose as an energy source22, no evidence has been found to support this hypothesis30.
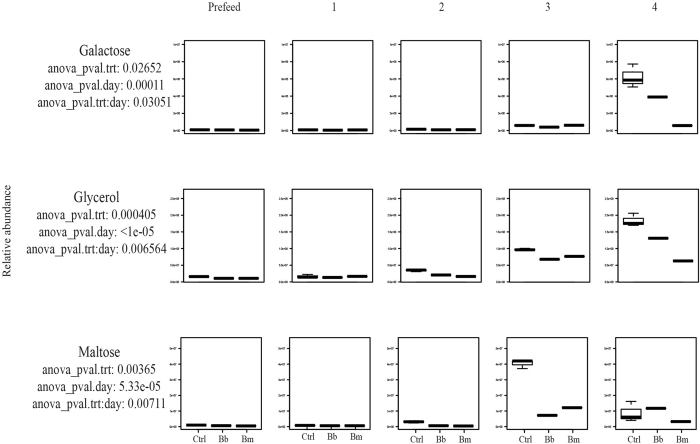
In our study, galactose, glycerol, and maltose are statistically significant by both treatment and time (Table 1). Our data show that all three I. scapularis groups utilize all detectable galactose until day 3, when the relative abundance begins to increase sharply in B. burgdorferi-infected tick groups (Fig. 2, Supplementary Figure 4). By day 4 of feeding, in tick groups infected with B. burgdorferi, the abundance of galactose decreases more so than within uninfected control tick groups, but less than B. mayonii-infected tick groups, which use significantly more galactose compared to the other groups (Fig. 2, Supplementary Figure 4). These data suggest that galactose may be an important carbohydrate involved in the development of spirochetes within the tick, represented by the lower relative abundance of galactose in tick groups infected with B. burgdorferi or B. mayonii compared to uninfected control ticks (Fig. 2, Supplementary Figure 4). Our data also suggest that B. mayonii and B. burgdorferi may differ in their metabolic capacities within vector ticks during the later stages of infection, especially with regard to specific carbohydrate utilization and availability (Fig. 2, Supplementary Figure 4).
Figure 2. Carbohydrates.
Accumulation over time of statistically significant (ANOVA p ≤ 0.05) carbohydrates in uninfected (control) Ixodes scapularis nymphs versus nymphs infected with the Lyme disease spirochetes Borrelia burgdorferi or Borrelia mayonii.
Within I. scapularis, glycerol is involved in glycerophospholipid metabolism (KEGG map00564), glycerolipid metabolism (KEGG map00561), and ether lipid metabolism (KEGG map00565). While there is a low abundance of glycerol present in the early stages of feeding glycerol availability begins to spike upward at day 2 of feeding (Fig. 2, Supplementary Figure 4). As the blood meal arrives and is processed within the tick, ATP input is required to convert glycerol into a more usable form: glycerol-3-phosphate (also significant by treatment, time, and treatment:time) (Table 1). Glycerol-3-phosphate can then be converted into dihydroxyacetone phosphate by glycerol-3-phosphate dehydrogenase, using energy cofactors NADH and NAD+.
In both I. scapularis and in B. burgdorferi, maltose is involved with starch and sucrose metabolism, phosphotransferase systems, and ABC transporters (KEGG C00208). Our data show that the abundance of maltose in the uninfected control tick groups and the Borrelia-infected tick groups is similar early in the feeding process, but differs vastly 3 days after attachment (Fig. 2, Supplementary Figure 4). Post attachment (day 3), a significantly higher abundance of maltose is present in uninfected tick groups than Borrelia-infected tick groups, falling to a comparable abundance in all groups by day 4 (Fig. 2, Supplementary Figure 4). Because maltose accumulates more rapidly in the uninfected tick groups than in the infected tick groups, B. burgdorferi-infected tick groups likely have a greater demand for maltose from the tick early on in feeding. B. mayonii follows the same general trend as the uninfected control albeit with a lower relative abundance, suggesting that B. mayonii-infected tick groups and B. burgdorferi-infected tick groups may differ slightly in their maltose requirements during feeding (Fig. 2, Supplementary Figure 4). Maltose may be used as a carbon source until a certain threshold, when it may become involved in metabolite shuttling, as has been demonstrated in Borrelia phosphoenolpyruvate phosphotransferase systems (PEP-PTS)59,61, and in other bacteria57.
Fatty acids
Fatty acids are a major component of phospholipid bilayers in cell membranes and play important roles as energy sources for many organisms including bacteria62. Some bacteria, such as Coxiellia burnetii, and Chlamydia spp. acquire lipids from their vertebrate host to establish and maintain infections63,64. Within bacterial and mammalian cell membranes and phospholipids, phosphatidylethanolamine is readily available65 to be broken down into ethanolamine and glycerol for use in glycerophospholipid metabolism, glycosylphosphatidylinositol-anchor biosynthesis, and pathogenesis66. Notably, ethanolamine is a critical metabolite used by enteric bacteria including S. typhimurium67, Escherichia coli68, EHEC (E. coli O157:H7)69, and Enterococcus faecalis70, in pathogenesis 69,71,72,73,74,75 and for energy71.
Because B. burgdorferi are not able to synthesize fatty acids or cholesterol, elongate fatty acid chains, nor beta-oxidize exogenous fatty acids de novo76,77, they rely heavily on their vertebrate hosts or the tick blood meal for these compounds22. Outer membranes of B. burgdorferi contain phosphatidylcholine, phosphatidylglycerol, and lipoproteins, but are mainly composed of cholesterol, which is processed to make cholesterol-glycolipids77,78,79,80,81. Free cholesterol and cholesterol-glycolipids within B. burgdorferi likely play important roles in the mediation of tick-cell interactions and create opportunities for lipid-lipid interactions with eukaryotic-like lipid rafts78,82. Lipid rafts may have implications during the enzootic cycle83.
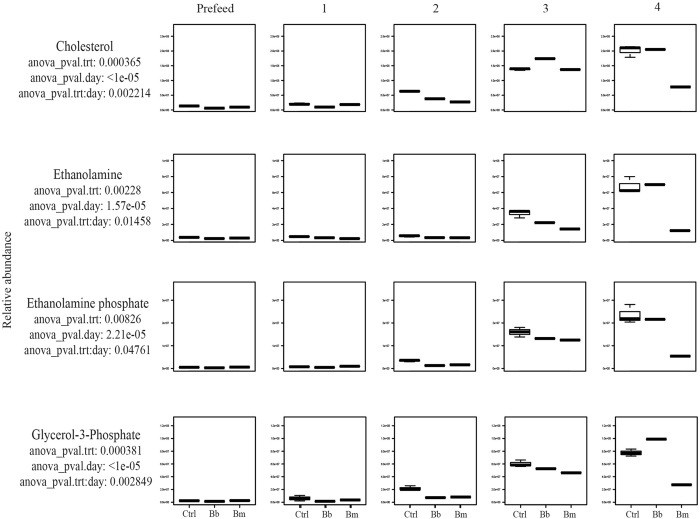
In our dataset, total cholesterol abundance is low in all 3 tick groups at days 0 and 1 (Fig. 3, Supplementary Figure 5). After day 1, cholesterol begins to accumulate in the uninfected tick groups while B. burgdorferi and B. mayonii-infected tick groups demonstrate a lower abundance, potentially indicating increased acquisition of cholesterol by the spirochetes (Fig. 3, Supplementary Figure 5). By day 3, the cholesterol abundance in B. burgdorferi-infected tick groups rises above both B. mayonii-infected tick groups and control tick groups (Fig. 3, Supplementary Figure 5). The trend over time in B. burgdorferi-infected tick groups may reflect incoming cholesterol being used for tick membrane remodeling and for metabolic energy in the beginning of the feeding cycle. Since free cholesterol has been shown to be required for B. burgdorferi-eukaryotic cell interactions via lipid rafts78, this could potentially explain why cholesterol increases in abundance towards the end of the feeding cycle.
Figure 3. Fatty Acids.
Accumulation over time of statistically significant (ANOVA p ≤ 0.05) lipids in uninfected (control) Ixodes scapularis nymphs versus nymphs infected with the Lyme disease spirochetes Borrelia burgdorferi or Borrelia mayonii.
Also statistically significant (ANOVA p ≤ 0.05 by treatment, time, and treatment: time) in our dataset are ethanolamine phosphate, ethanolamine, and, as mentioned previously, glycerol-3-phosphate (Fig. 3, Supplementary Figure 5), all of which are involved in glycerophospholipid metabolism (KEGG map00564). These metabolites follow the same general trend in all 3 tick groups during days 0–3. The abundance of ethanolamine phosphate, ethanolamine, and glycerol-3-phosphate are low during days 0–2, but by day 3 post attachment, the abundance of each metabolite sharply increases in all three tick groups, followed by a rapid decline in only the B. mayonii-infected tick groups (Fig. 3, Supplementary Figure 5). Similar to the trend of cholesterol abundance over time, the abundance of ethanolamine, ethanolamine phosphate, and glycerol-3-phosphate during feeding may be described by the changing roles of these particular metabolites. At the start of the blood meal, exogenous carbon sources are necessary to synthesize new membrane components; as the tick reaches repletion, ethanolamine and glycerol-related metabolites likely become available for use.
Study limitations
One notable limitation of this study that needs to be addressed in future work is that we did not determine the mechanism(s) underlying the observed differences between ticks infected with B. mayonii as compared with B. burgdorferi. The more substantial decrease in the abundance of purine-related compounds, amino acids, carbohydrates, and fatty acids throughout the blood meal of B. mayonii-infected ticks compared to B. burgdorferi-infected ticks may suggest that B. mayonii places a greater demand for these molecules contained in the blood within the tick vector than B. burgdorferi. Alternative explanations could be that the number of spirochetes present within the infected ticks typically is higher for B. mayonii than for B. burgdorferi, or some as yet unknown aspect of the physiology of B. mayonii differing from B. burgdorferi. Graphical interpretation of results from the PCR conducted to confirm infection status of individual ticks are suggestive of higher spirochete numbers within ticks infected with B. mayonii, as compared with B. burgdorferi, but this is not conclusive as the assay was not specifically designed to quantify the amount of spirochete genetic material in the ticks. Another limitation of the study is that we used a single strain of each Borrelia species. Examination of additional spirochete strains is required to more confidently generalize our finding for the examined Borrelia species. Another important aspect to consider in future studies is whether the observed differences between ticks infected with B. mayonii as compared with B. burgdorferi actually result in reduced fitness of infected blood fed ticks, for example manifesting as reduced likelihood of B. mayonii-infected nymphs to molt to the adult stage or reduced size of female ticks.
Conclusions
We present the first account of a metabolomic profile of ticks infected with Lyme disease spirochetes, B. burgdorferi and B. mayonii, which provides insight into the strategies these important human pathogens may employ to adapt to and modify the environment within the tick vector in order to establish infection and continue the transmission cycle. Analogous pathways involved in infection in other pathogenic bacteria, especially enteropathogenic bacteria, may imply that these metabolites play similar roles during Borrelia growth and transmission of Borrelia by infected ticks. Although the specific roles and significance of these metabolic pathways may be established in other organisms, their functions are not yet fully understood within arthropod vectors. Therefore, the discussions and conclusions made herein remain putative, however, these data do suggest that there are differences in the effects of these two species of Lyme disease spirochetes on the tick metabolome. Future studies repeated over longer time periods could provide insight into aspects of post drop-off metabolism and their potential influence on differences related to transstadial survival and potential transovarial transmission of Borrelia, and differences related to subsequent transmission efficacy of the spirochetes.
Additional Information
How to cite this article: Hoxmeier, J. C. et al. Metabolomics of the tick-Borrelia interaction during the nymphal tick blood meal. Sci. Rep. 7, 44394; doi: 10.1038/srep44394 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The authors would like to thank Martin Williams for CDC tick colony maintenance. J.C.H. and A.C.F. are grateful for the support of the research participation program at the Centers for Disease Control and Prevention, U.S. Environmental Protection Agency, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and EPA.
Footnotes
The authors declare no competing financial interests.
Author Contributions J.C.H. performed the tick experiments; J.C.H., A.C.F., and C.B. performed the data analysis. J.C.H., A.C.F., C.B., J.P., M.C.D., K.L.G. and L.E. contributed to the preparation of the manuscript.
References
- Hinckley A. F. et al. Lyme disease testing by large commercial laboratories in the United States. Clinical Infectious Diseases, ciu397 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead P. S. Epidemiology of Lyme disease. Infect Dis Clin N Am 29, 187–210, doi: 10.1016/j.idc.2015.02.010 (2015). [DOI] [PubMed] [Google Scholar]
- Nelson C. A. et al. Incidence of clinician-diagnosed Lyme disease, United States, 2005-2010. Emerg Infect Dis 21, 1625–1631, doi: 10.3201/eid2109.150417 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piesman J. & Eisen L. Prevention of tick-borne diseases. Ann Rev Entomol 53, 323–343, doi: 10.1146/annurev.ento.53.103106.093429 (2008). [DOI] [PubMed] [Google Scholar]
- Eisen R. J., Eisen L. & Beard C. B. County-scale distribution of Ixodes scapularis and Ixodes pacificus (Acari: Ixodidae) in the continental United States. J Med Entomol 53, 349–386, doi: 10.1093/jme/tjv237 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan M. C. et al. Vector competence of the blacklegged tick, Ixodes scapularis, for the recently recognized Lyme borreliosis spirochete Candidatus Borrelia mayonii. Ticks Tick Borne Dis 7, 665–669, doi: 10.1016/j.ttbdis.2016.02.012 (2016). [DOI] [PubMed] [Google Scholar]
- Pritt B. S. et al. Identification of a novel pathogenic Borrelia species causing Lyme borreliosis with unusually high spirochaetaemia: a descriptive study. Lancet Infect Dis 16, 556–564, doi: 10.1016/s1473-3099(15)00464-8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritt B. S. et al. Borrelia mayonii sp. nov., a member of the Borrelia burgdorferi sensu lato complex, detected in patients and ticks in the upper midwestern United States. Int J Syst Evol Microbiol, doi: 10.1099/ijsem.0.001445 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmanan V., Rhee K. Y. & Daily J. P. Metabolomics and malaria biology. Mol Biochem Parasitol 175, 104–111, doi: 10.1016/j.molbiopara.2010.09.008 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafsack B. F. & Llinas M. Eating at the table of another: metabolomics of host-parasite interactions. Cell Host Microbe 7, 90–99, doi: 10.1016/j.chom.2010.01.008 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes L. C. et al. Metabolic signatures of triatomine vectors of Trypanosoma cruzi unveiled by metabolomics. PLoS One 8, e77283, doi: 10.1371/journal.pone.0077283 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Andrade J. B. et al. Perspectives for diagnosis and control of leishmaniasis based on volatile organic compounds. doi: 10.5772/57279 (2014). [DOI] [Google Scholar]
- Hoxmeier J. C. et al. Analysis of the metabolome of Anopheles gambiae mosquito after exposure to Mycobacterium ulcerans. Sci Rep 5, 9242, doi: 10.1038/srep09242 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molins C. R. et al. Development of a metabolic biosignature for detection of early Lyme disease. Clin Infect Dis 60, 1767–1775, doi: 10.1093/cid/civ185 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano M. J., Drecktrah D., Kung F. & Samuels D. S. Interaction of the Lyme disease spirochete with its tick vector. Cell Microbiol 18, 919–927, doi: 10.1111/cmi.12609 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushtaq M. Y., Choi Y. H., Verpoorte R. & Wilson E. G. Extraction for metabolomics: access to the metabolome. Phytochem Anal 25, 291–306, doi: 10.1002/pca.2505 (2014). [DOI] [PubMed] [Google Scholar]
- Gao P. & Xu G. Mass-spectrometry-based microbial metabolomics: recent developments and applications. Anal Bioanal Chem 407, 669–680, doi: 10.1007/s00216-014-8127-7 (2015). [DOI] [PubMed] [Google Scholar]
- Patti G. J., Yanes O. & Siuzdak G. Innovation: metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol 13, 263–269, doi: 10.1038/nrm3314 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- German J. B., Hammock B. D. & Watkins S. M. Metabolomics: building on a century of biochemistry to guide human health. Metabolomics 1, 3–9, doi: 10.1007/s11306-005-1102-8 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay B. B. & Cossart P. Exploitation of mammalian host cell functions by bacterial pathogens. Science 276, 718–725, doi: 10.1126/science.276.5313.718 (1997). [DOI] [PubMed] [Google Scholar]
- Gherardini F., Boylan J., Lawrence K. & Skare J. In Borrelia: molecular biology, host interaction and pathogenesis (ed. Justin D. Radolf) 103–138 (2010). [Google Scholar]
- Fraser C. M. et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390, 580–586, doi: 10.1038/37551 (1997). [DOI] [PubMed] [Google Scholar]
- Tilly K., Rosa P. A. & Stewart P. E. Biology of infection with Borrelia burgdorferi. Infect Dis Clin North Am 22, 217–234, v, doi: 10.1016/j.idc.2007.12.013 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenshine D. Biology of ticks, vol. II. (Oxford University Press, New York, NY, 1993). [Google Scholar]
- Iyer R. et al. Stage-specific global alterations in the transcriptomes of Lyme disease spirochetes during tick feeding and following mammalian host adaptation. Molec Microbiol 95, 509–538, doi: 10.1111/mmi.12882 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal U. & Fikrig E. In Borrelia: molecular biology, host interaction and pathogenesis (ed. Samuels D. S., Radolf J. D.) 279–298 (Caister Academic, 2010). [Google Scholar]
- Piesman J., Schneider B. S. & Zeidner N. S. Use of quantitative PCR to measure density of Borrelia burgdorferi in the midgut and salivary glands of feeding tick vectors. J Clin Microbiol 39, 4145–4148 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham-Ems S. M. et al. Live imaging reveals a biphasic mode of dissemination of Borrelia burgdorferi within ticks. J Clin Invest 119, 3652–3665 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piesman J., Oliver J. R. & Sinsky R. J. Growth kinetics of the Lyme disease spirochete (Borrelia burgdorferi) in vector ticks (Ixodes dammini). Am J Trop Med Hyg 42, 352–357 (1990). [DOI] [PubMed] [Google Scholar]
- von Lackum K. & Stevenson B. Carbohydrate utilization by the Lyme borreliosis spirochete, Borrelia burgdorferi. FEMS Microbiol Lett 243, 173–179, doi: 10.1016/j.femsle.2004.12.002 (2005). [DOI] [PubMed] [Google Scholar]
- Van Laar T. A. et al. Effect of levels of acetate on the mevalonate pathway of Borrelia burgdorferi. PLoS One 7, e38171, doi: 10.1371/journal.pone.0038171 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H. et al. Role of acetyl-phosphate in activation of the Rrp2-RpoN-RpoS pathway in Borrelia burgdorferi. PLoS Pathog 6, e1001104, doi: 10.1371/journal.ppat.1001104 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas C. J. et al. Borrelia burgdorferi requires glycerol for maximum fitness during the tick phase of the enzootic cycle. PLoS Pathog 7, e1002102, doi: 10.1371/journal.ppat.1002102 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson J. et al. Purine salvage pathways among Borrelia species. Infect Immun 75, 3877–3884, doi: 10.1128/IAI.00199-07 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piesman J. Standard system for infecting ticks (Acari: Ixodidae) with the Lyme disease spirochete, Borrelia burgdorferi. J Med Entomol 30, 199–203 (1993). [DOI] [PubMed] [Google Scholar]
- Pritt B. S. et al. Identification of a novel pathogenic Borrelia species causing Lyme borreliosis with unusually high spirochaetaemia: a descriptive study. Lancet Infect Dis, doi: 10.1016/S1473-3099(15)00464-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan M. C. et al. Elimination of Borrelia burgdorferi and Anaplasma phagocytophilum in rodent reservoirs and Ixodes scapularis ticks using a doxycycline hyclate-laden bait. Am J Trop Med Hyg 85, 1114–1120, doi: 10.4269/ajtmh.2011.11-0292 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard J., Embers M., Hojgaard A. & Piesman J. Comparison of tick feeding success and vector competence for Borrelia burgdorferi among immature Ixodes scapularis (Ixodida: Ixodidae) of both southern and northern clades. J Med Entomol 52, 81–85, doi: 10.1093/jme/tju005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojgaard A., Lukacik G. & Piesman J. Detection of Borrelia burgdorferi, Anaplasma phagocytophilum and Babesia microti, with two different multiplex PCR assays. Ticks Tick Borne Dis 5, 349–351, doi: 10.1016/j.ttbdis.2013.12.001 (2014). [DOI] [PubMed] [Google Scholar]
- Smith C. A., Want E. J., O’Maille G., Abagyan R. & Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem 78, 779–787, doi: 10.1021/ac051437y (2006). [DOI] [PubMed] [Google Scholar]
- Broeckling C. D., Afsar F. A., Neumann S., Ben-Hur A. & Prenni J. E. RAMClust: A novel feature clustering method enables spectral-matching-based annotation for metabolomics data. Anal Chem 86, 6812–6817, doi: 10.1021/ac501530d (2014). [DOI] [PubMed] [Google Scholar]
- Broeckling C. D. et al. Annotation from the MS1-spectrum and time predictions approach to LC-MS metabolomics data. Anal Chem, doi: 10.1021/acs.analchem.6b02479 (2016). [DOI] [PubMed] [Google Scholar]
- Sumner L. W. et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3, 211–221, doi: 10.1007/s11306-007-0082-2 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y. & Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57, 289–300 (1995). [Google Scholar]
- Wani T. A. & Zargar S. New ultra-performance liquid chromatography-tandem mass spectrometry method for the determination of irbesartan in human plasma. J Food Drug Anal 23, 569–576, doi: 10.1016/j.jfda.2015.02.008 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berens R. L., Krug E. C. & Marr J. J. In Biochemistry and Molecular Biology of Parasites (ed. Marr J. & Muller M.) 89–117 (1995). [Google Scholar]
- Jain S., Sutchu S., Rosa P. A., Byram R. & Jewett M. W. Borrelia burgdorferi harbors a transport system essential for purine salvage and mammalian infection. Infect Immun 80, 3086–3093, doi: 10.1128/IAI.00514-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett M. W. et al. GuaA and GuaB are essential for Borrelia burgdorferi survival in the tick-mouse infection cycle. J Bacteriol 191, 6231–6241, doi: 10.1128/JB.00450-09 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis N., Hogan D., Tilly K. & Rosa P. A. Plasmid location of Borrelia purine biosynthesis gene homologs. J Bacteriol 176, 6427–6432 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields P. I., Swanson R. V., Haidaris C. G. & Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci USA 83, 5189–5193 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwick R. A., Krstulovic A. M. & Brown P. R. Identification and quantitation of nucleosides, bases and other UV-absorbing compounds in serum, using reversed-phase high-performance liquid chromatography. II. Evaluation of human sera. J Chromatogr 186, 659–676 (1979). [DOI] [PubMed] [Google Scholar]
- Moran N. A. Microbial minimalism: genome reduction in bacterial pathogens. Cell 108, 583–586, doi: 10.1016/S0092-8674(02)00665-7 (2002). [DOI] [PubMed] [Google Scholar]
- Sherman I. W. Amino acid metabolism and protein synthesis in malarial parasites. Bull World Health Organ 55, 265 (1977). [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C. Biology of parasitic spirochaetes. (Academic Press, 2013). [Google Scholar]
- Opperdoes F. R. & Coombs G. H. Metabolism of Leishmania: proven and predicted. Trends Parasitol 23, 149–158, doi: 10.1016/j.pt.2007.02.004 (2007). [DOI] [PubMed] [Google Scholar]
- Chouikha I. & Hinnebusch B. J. Yersinia–flea interactions and the evolution of the arthropod-borne transmission route of plague. Curr Opin Microbiol 15, 239–246, doi: 10.1016/j.mib.2012.02.003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoon-Hanks L. L. et al. Borrelia burgdorferi malQ mutants utilize disaccharides and traverse the enzootic cycle. FEMS Immunol Med Microbiol 66, 157–165, doi: 10.1111/j.1574-695X.2012.00996.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Schmid G. P., Hyde F. W., Steigerwalt A. & Brenner D. J. Borrelia burgdorferi sp. nov.: etiologic agent of Lyme disease. Int J Sys Bacteriol 34, 496–497 (1984). [Google Scholar]
- Troy E. B. et al. Global Tn-seq analysis of carbohydrate utilization and vertebrate infectivity of Borrelia burgdorferi. Mol Microbiol, doi: 10.1111/mmi.13437 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D. S., Harris E. K. & Cotlove E. Biological and analytic components of variation in long-term studies of serum constituents in normal subjects. IV. Results of a study designed to eliminate long-term analytic deviations. Clin Chem 17, 403–410 (1971). [PubMed] [Google Scholar]
- Khajanchi B. K. et al. Phosphoenolpyruvate phosphotransferase system components modulate gene transcription and virulence of Borrelia burgdorferi. Infect and Immun 84, 754–764, doi: 10.1128/iai.00917-15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Matsuoka H. & Hirooka K. Regulation of fatty acid metabolism in bacteria. Mol Microbiol 66, 829–839, doi: 10.1111/j.1365-2958.2007.05947.x (2007). [DOI] [PubMed] [Google Scholar]
- Elwell C. A. & Engel J. N. Lipid acquisition by intracellular Chlamydiae. Cell Microbiol 14, 1010–1018, doi: 10.1111/j.1462-5822.2012.01794.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe D. & Heinzen R. A. Coxiella burnetii inhabits a cholesterol-rich vacuole and influences cellular cholesterol metabolism. Cell Microbiol 8, 496–507, doi: 10.1111/j.1462-5822.2005.00641.x (2006). [DOI] [PubMed] [Google Scholar]
- Randle C. L., Albro P. W. & Dittmer J. C. The phosphoglyceride composition of Gram-negative bacteria and the changes in composition during growth. Biochim Biophys Acta 187, 214–220 (1969). [DOI] [PubMed] [Google Scholar]
- Larson T. J., Ehrmann M. & Boos W. Periplasmic glycerophosphodiester phosphodiesterase of Escherichia coli, a new enzyme of the glp regulon. J Biol Chem 258, 5428–5432 (1983). [PubMed] [Google Scholar]
- Chang G. W. & Chang J. T. Evidence for the B12-dependent enzyme ethanolamine deaminase in Salmonella. Nature 254, 150–151 (1975). [DOI] [PubMed] [Google Scholar]
- Blackwell C. M., Scarlett F. A. & Turner J. M. Ethanolamine catabolism by bacteria, including Escherichia coli. Biochem Soc Trans 4, 495–497 (1976). [DOI] [PubMed] [Google Scholar]
- Bertin Y. et al. Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ Microbiol 13, 365–377, doi: 10.1111/j.1462-2920.2010.02334.x (2011). [DOI] [PubMed] [Google Scholar]
- Del Papa M. F. & Perego M. Ethanolamine activates a sensor histidine kinase regulating its utilization in Enterococcus faecalis. J Bacteriol 190, 7147–7156, doi: 10.1128/jb.00952-08 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garsin D. A. Ethanolamine utilization in bacterial pathogens: roles and regulation. Nat Rev Microbiol 8, 290–295, doi: 10.1038/nrmicro2334 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C. J., Clark D. E., Adli M. & Kendall M. M. Ethanolamine signaling promotes Salmonella niche recognition and adaptation during infection. PLoS Pathog 11, e1005278, doi: 10.1371/journal.ppat.1005278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C. J., Clark D. E., Adli M. & Kendall M. M. Salmonella senses ethanolamine to gauge distinct host environments and coordinate gene expression. Microbial Cell 3, 89–91, doi: 10.15698/mic2016.02.479 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall M. M., Gruber C. C., Parker C. T. & Sperandio V. Ethanolamine controls expression of genes encoding components involved in interkingdom signaling and virulence in enterohemorrhagic Escherichia coli O157:H7. mBio 3, doi: 10.1128/mBio.00050-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikumar S. & Fuchs T. M. Ethanolamine utilization contributes to proliferation of Salmonella enterica serovar Typhimurium in food and in nematodes. Appl Environ Microbiol 77, 281–290, doi: 10.1128/AEM.01403-10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore B. P., Bey R. F. & Johnson R. C. Lipid metabolism of Borrelia hermsi. Infect Immun 20, 215–220 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C. The spirochetes. Annu Rev in Microbiol 31, 89–106 (1977). [DOI] [PubMed] [Google Scholar]
- Crowley J. T. et al. Lipid exchange between Borrelia burgdorferi and host cells. PLoS Pathog 9, e1003109, doi: 10.1371/journal.ppat.1003109 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belisle J. T., Brandt M. E., Radolf J. D. & Norgard M. V. Fatty acids of Treponema pallidum and Borrelia burgdorferi lipoproteins. J Bacteriol 176, 2151–2157 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radolf J. D. et al. Characterization of outer membranes isolated from Borrelia burgdorferi, the Lyme disease spirochete. Infect and Immun 63, 2154–2163 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain H., Wellensiek H.-J., Geyer R. & Lochnit G. Structural analysis of glycolipids from Borrelia burgdorferi. Biochimie 83, 683–692, doi: 10.1016/S0300-9084(01)01296-2 (2001). [DOI] [PubMed] [Google Scholar]
- LaRocca T. J. et al. Proving lipid rafts exist: membrane domains in the prokaryote Borrelia burgdorferi have the same properties as eukaryotic lipid rafts. PLoS Pathog 9, e1003353 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRocca T. J. et al. Cholesterol lipids of Borrelia burgdorferi form lipid rafts and are required for the bactericidal activity of a complement-independent antibody. Cell Host Microbe 8, 331–342, doi: 10.1016/j.chom.2010.09.001 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.