Significance
HIV infection can be restricted by different host cell proteins. One such restriction factor is SAM domain and HD domain-containing protein 1 (SAMHD1), a triphosphohydrolase that cleaves dNTPs, which are required for HIV reverse transcription. The accessory lentiviral protein X (Vpx) from simian immunodeficiency viruses (SIV) of sooty mangabeys and rhesus macaques or from HIV-2 degrade SAMHD1. Here we show that Vpx proteins from SIVs of red-capped mangabeys and mandrills enhance HIV infection of resting CD4 T cells, but not macrophages, without affecting levels of either SAMHD1 or dNTPs. These Vpx proteins overcome a previously unrecognized SAMHD1-independent, cell-type–specific restriction at the level of reverse transcription and highlight the plasticity of lentiviruses to counteract the innate immune system.
Keywords: HIV, restriction factors, resting CD4 T cells, SAMHD1, Vpx
Abstract
Early after entry into monocytes, macrophages, dendritic cells, and resting CD4 T cells, HIV encounters a block, limiting reverse transcription (RT) of the incoming viral RNA genome. In this context, dNTP triphosphohydrolase SAM domain and HD domain-containing protein 1 (SAMHD1) has been identified as a restriction factor, lowering the concentration of dNTP substrates to limit RT. The accessory lentiviral protein X (Vpx) proteins from the major simian immunodeficiency virus of rhesus macaque, sooty mangabey, and HIV-2 (SIVsmm/SIVmac/HIV-2) lineage packaged into virions target SAMHD1 for proteasomal degradation, increase intracellular dNTP pools, and facilitate HIV cDNA synthesis. We find that virion-packaged Vpx proteins from a second SIV lineage, SIV of red-capped mangabeys or mandrills (SIVrcm/mnd-2), increased HIV infection in resting CD4 T cells, but not in macrophages, and, unexpectedly, acted in the absence of SAMHD1 degradation, dNTP pool elevation, or changes in SAMHD1 phosphorylation. Vpx rcm/mnd-2 virion incorporation resulted in a dramatic increase of HIV-1 RT intermediates and viral cDNA in infected resting CD4 T cells. These analyses also revealed a barrier limiting HIV-1 infection of resting CD4 T cells at the level of nuclear import. Single amino acid changes in the SAMHD1-degrading Vpx mac239 allowed it to enhance early postentry steps in a Vpx rcm/mnd-2–like fashion. Moreover, Vpx enhanced HIV-1 infection of SAMHD1-deficient resting CD4 T cells of a patient with Aicardi-Goutières syndrome. These results indicate that Vpx, in addition to SAMHD1, overcomes a previously unappreciated restriction for lentiviruses at the level of RT that acts independently of dNTP concentrations and is specific to resting CD4 T cells.
HIV-1 replication is restricted at different postentry steps in myeloid cells and resting CD4 T cells (1–7). A major barrier was consistently mapped at the level of reverse transcription (RT), where early, but not late RT products could be detected in infected cells (6, 8–10). Supplying the accessory lentiviral protein X (Vpx) in trans or in cis to HIV-infected monocytes, monocyte-derived dendritic cells, monocyte-derived macrophages (MDMs), or resting CD4 T cells boosted reverse transcription and allowed progression of the replication cycle in these primary target cells (2, 3, 11–15). SAM domain and HD domain-containing protein 1 (SAMHD1), a deoxynucleotide triphosphohydrolase (16), was identified as a target of Vpx (17, 18). SAMHD1 depletes dNTP pools, which prevents reverse transcription, leading to a block to early steps of HIV-1 infection in myeloid cells (19). Vpx proteins of the simian immunodeficiency virus of rhesus macaque, sooty mangabey, and HIV-2 (SIVmac/SIVsmm/HIV-2, respectively) lineage of primate lentiviruses antagonize SAMHD1 by targeting it for proteasomal degradation. Vpx directly binds to SAMHD1 and simultaneously to the DNA damage-binding protein 1 and Cullin-4A-associated factor 1 (DCAF1) substrate receptor of the cullin 4-RING ubiquitin ligase (CRL4) E3 ubiquitin ligase (CRL4DCAF1), thereby loading SAMHD1 onto this E3 complex for polyubiquitylation and subsequent degradation (17, 18).
Vpx also enhances HIV infection of resting CD4 T cells and infection enhancement by Vpx correlates with SAMHD1 depletion and a concomitant increase in cellular dNTP levels (11, 12). Notably, these studies had exclusively analyzed Vpx proteins of the SIVsmm/SIVmac/HIV-2 lineage. In the current work we find that Vpx proteins from the second Vpx+ lentiviral lineage, represented by SIVrcm and SIVmnd-2, are also able to enhance HIV-1 infection of resting CD4 T cells, but in a SAMHD1-independent manner that is uncoupled from alterations in cellular dNTP levels. Our results indicate that virion-incorporated Vpx can enhance infection of resting CD4 T cells by overcoming a previously unrecognized block to early RT.
Results
Vpx Proteins of the SIVrcm/mnd-2 Lineage Enhance HIV-1 Infection of Resting CD4 T Cells Without Degrading SAMHD1, Changing Its Phosphorylation or Elevating dNTP Pools.
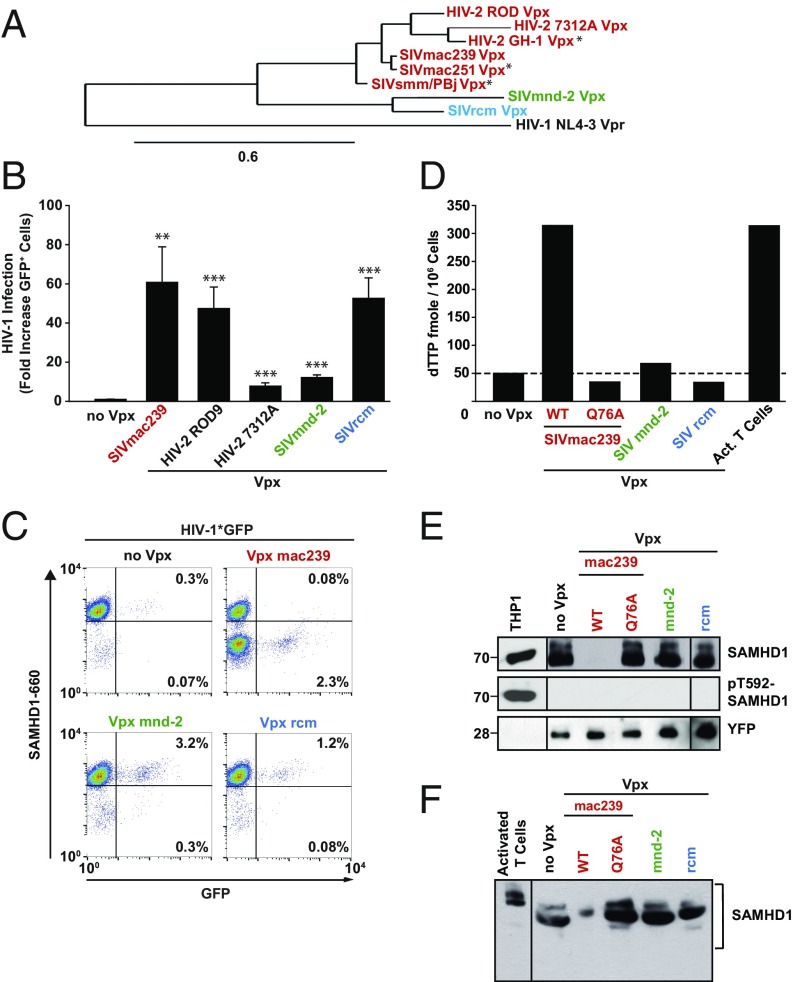
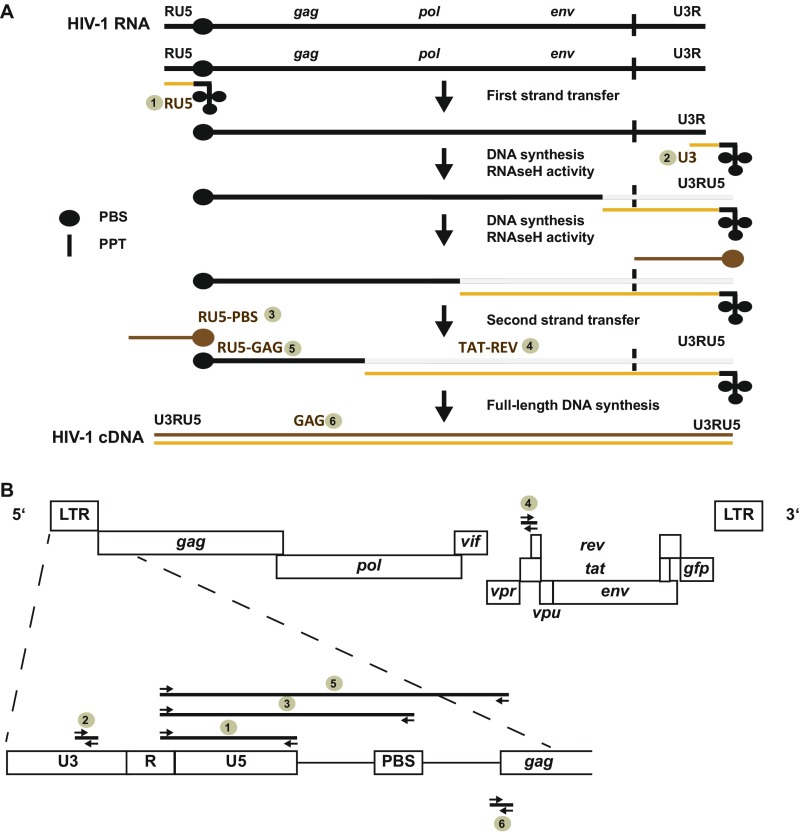
To assess the evolutionary breadth of the ability of Vpx to increase HIV-1 infection in resting CD4 T cells, we screened Vpx variants derived from HIV-2, SIVsmm, SIVmac, SIVmnd-2, and SIVrcm (Fig. 1A) (11, 20). Replication-competent, NL4-3–based HIV-1 reporter viruses encoding for GFP as a marker for early gene expression from integrated HIV and capable of trans-packaging tagged Vpx proteins via a Vpx-interaction motif in Gag p6 [referred to as CXCR-4 tropic (X4) HIV-1*GFP] (11) were produced (Fig. S1).
Fig. 1.
Vpx of SIVmnd-2 and SIVrcm enhance HIV-1 infection in resting CD4 T cells in the absence of degradation or phosphorylation of SAMHD1. (A) Phylogenetic tree analysis of Vpx proteins from HIV-2 and the two Vpx-carrying SIV lineages used in the current and previous studies. The figure was generated using the online tool on www.phylogeny.fr with MUSCLE for alignment and PhyML for the generation of the phylogenetic tree. Depicted are SAMHD1-degrading (red) and nondegrading (SIVmnd-2 in green; SIVrcm in blue) Vpx proteins of HIV-2 and SIV. HIV-1 NL4-3 Vpr was used as a reference. Stars denote previously analyzed Vpx alleles (11). (B and C) Resting CD4 T cells were challenged with equivalent infectious units of X4 HIV-1*GFP virions without (no Vpx; n = 15) or with incorporated Vpx from SIVmac239 (n = 15), HIV-2 ROD9 (n = 10), HIV-2 7312A (n = 11), SIVmnd-2 (n = 10), or SIVrcm (n = 12) and analyzed 3 d later for expression of GFP and SAMHD1, in principle as reported (11). (B) Factor of increase of Vpx-mediated HIV-1 infection enhancement. Shown are arithmetic means + SEM. (C) Dot plots of flow cytometric analysis of intracellular SAMHD1 and GFP expression for one representative donor. (D–F) Resting CD4 T cells were cotransfected with pDisplay-YFP and expression constructs for the indicated Vpx proteins, sorted for cells expressing YFP at the surface and subjected to (D) dTTP quantification, (E) Western blotting for SAMHD1 and pT592 SAMHD1, or (F) overall SAMHD1 phosphorylation using a phostag gel. (E and F) Images are from single gels, in which lanes with irrelevant samples were cut out. THP-1 cells and activated CD4 T cells served as controls for SAMHD1 phosphorylation, and YFP as loading control.
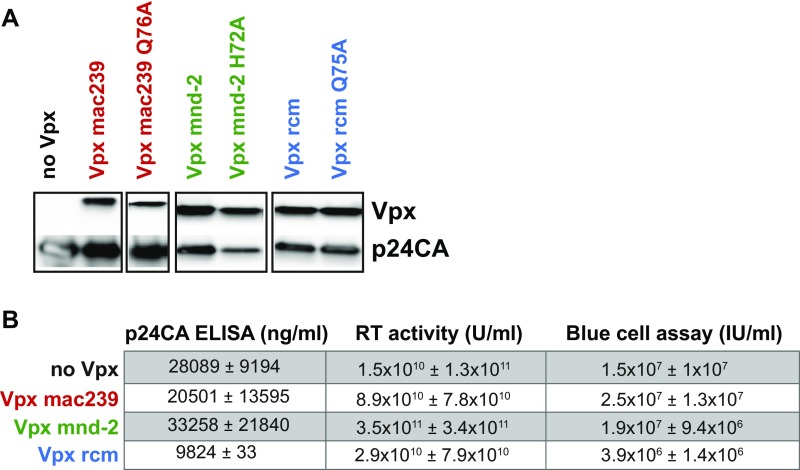
Fig. S1.
(A) Immunoblotting of X4 HIV-1*GFP virions for incorporated epitope-tagged Vpx proteins from SIVmac239 (WT and Q76A mutant; myc), SIVmnd-2 or SIVrcm (WT and corresponding H72A and Q75A mutant, respectively; both flag). HIV-1 p24CA served as a loading control. The use of different tags precluded quantitative comparisons of virion incorporation of these Vpx variants. myc-Vpx–tagged and flag-Vpx–tagged virions were run on the same membrane, respectively, and unrelated samples are not shown. (B) Quantification of physical particles via p24CA ELISA, RT activity via SG-PERT, and determination of the infectious titer on TZM-bl reporter cells via blue cell assay of sucrose-pelleted virions. Depicted is the mean ± SEM of two (p24CA ELISA) or three (RT activity and blue cell assay) independently produced virus stocks.
Resting CD4 T cells purified from peripheral blood of healthy donors were challenged with these X4 HIV-1* reporter viruses and, 3 d later, analyzed for GFP expression and intracellular SAMHD1 levels by flow cytometry. Whereas these CD25/CD69− CD4 T cells were largely refractory to infection with X4 HIV-1*GFP (<0.5% GFP+ cells), infection levels with all Vpx-carrying reporter viruses were increased (Fig. 1B). The magnitude of infection enhancement varied from 60-fold for the SIVmac239 Vpx, as previously reported (11), to 8- to 54-fold for SIVmnd-2 Vpx and SIVrcm Vpx, respectively (Fig. 1B). In contrast, all these Vpx proteins only mildly enhanced HIV-1 infection of activated CD4 T cells (Fig. S2A).
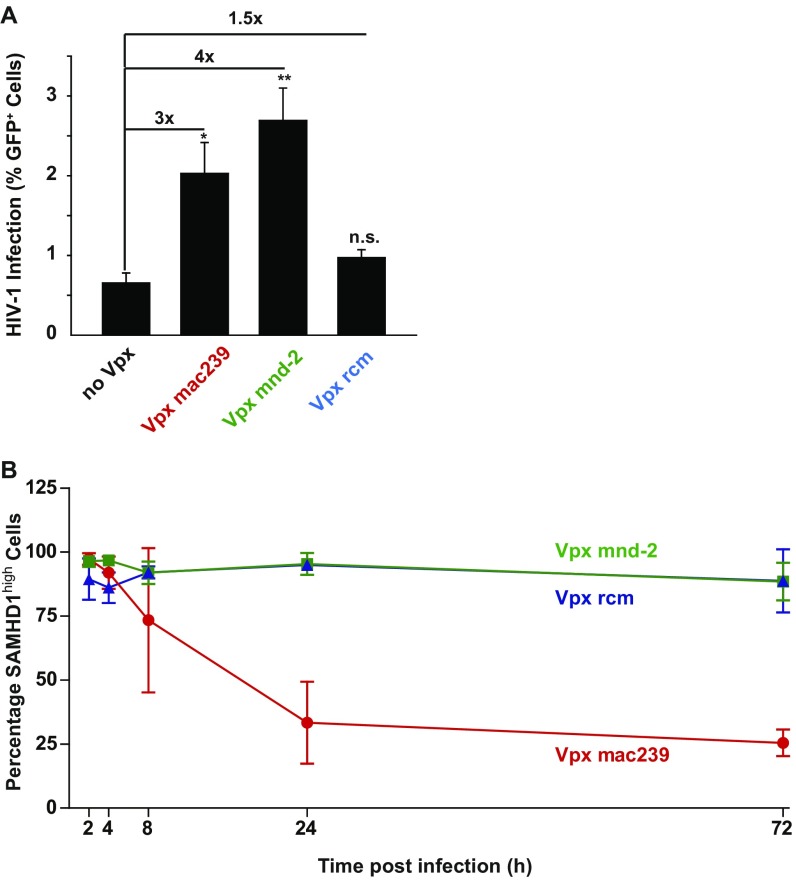
Fig. S2.
(A) Quantification of GFP expression levels 3 d postinfection of activated CD4 T cells challenged with equal TZM-bl infectious units of X4 HIV-1*GFP reporter virus carrying no Vpx or the indicated Vpx variants. Depicted is the mean ± SE of mean of seven donors, each measured in technical triplicates. (B) Time course of SAMHD1 expression in resting CD4 T cells after challenge with HIV-1*GFP with incorporated Vpx mac239 (n = 4 donors), Vpx mnd-2 (n = 4 donors), or Vpx rcm (n = 2 donors). Data points mark the mean percentages (±SD) of cells with high SAMHD1 expression levels.
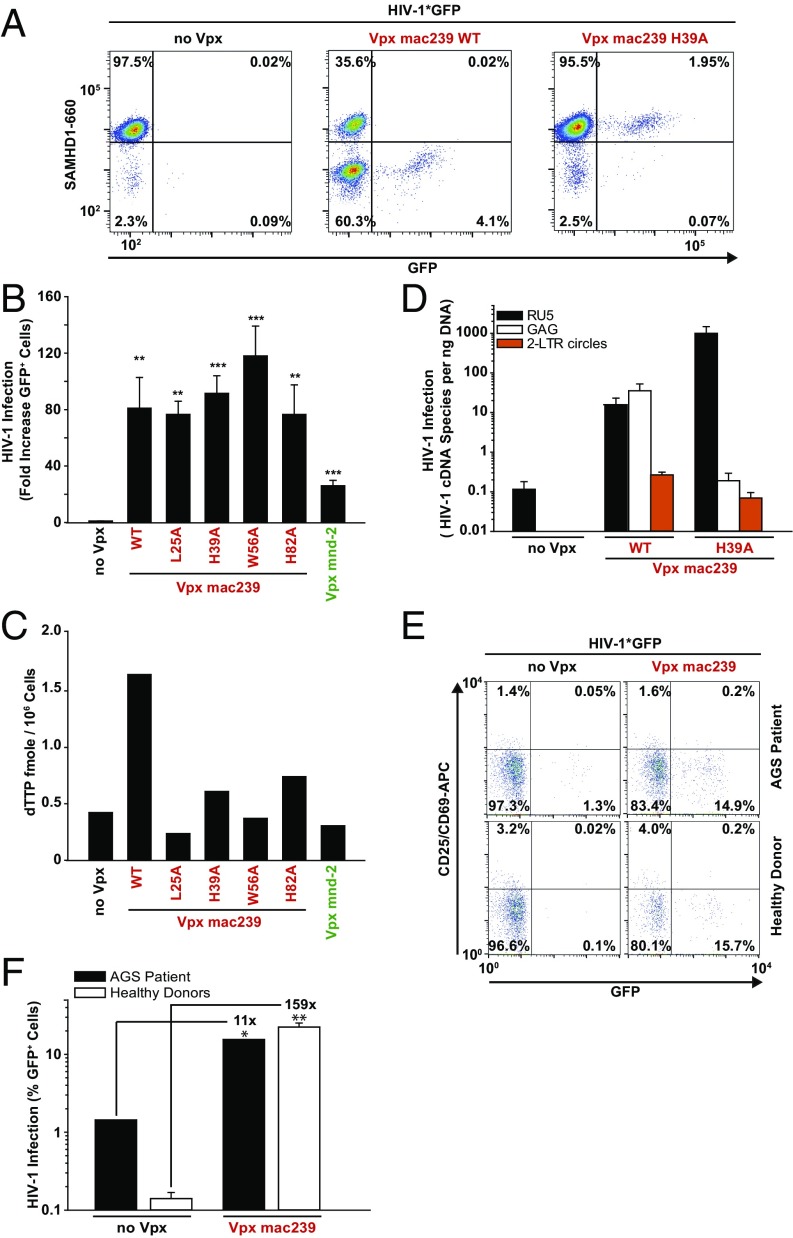
As reported previously (11), exposure to X4 HIV-1*GFP with packaged Vpx from SIVmac239 resulted in a massive depletion of SAMHD1 in a considerable fraction of resting CD4 T cells, and GFP expression was detected almost exclusively within this SAMHD1low population (Fig. 1C, Lower Right quadrant). Whereas incorporated Vpx proteins from the other lentiviral lineage, SIVmnd-2 and SIVrcm, also increased infection efficiencies of X4 HIV-1*GFP (Fig. 1 B and C), surprisingly, SAMHD1 levels were not reduced by these two Vpx alleles relative to control infections lacking Vpx (no Vpx), and GFP+ cells were observed in the SAMHD1high population (Fig. 1C). Kinetic analyses in resting CD4 T cells challenged with X4 HIV-1*GFP carrying SIVmac239 Vpx confirmed that SAMHD1 levels declined within hours after viral challenge and remained at a reduced level for at least 3 d (Fig. S2B and ref. 11). In contrast, virion-incorporated Vpx mnd-2 or Vpx rcm did not affect SAMHD1 levels throughout the experiment (Fig. S2B), excluding transient depletion of the enzyme by these alleles as a potential mode of action. Notably, whereas ectopic expression of Vpx mac239 WT, but not the CRL4DCAF1 interaction-deficient mutant Q76A, increased cellular dTTP concentrations to levels comparable to those in activated CD4 T cells, expression of Vpx mnd-2 or Vpx rcm did not affect the dTTP pool (Fig. 1D). These results suggest that the Vpx-mediated enhancement of HIV infection in this cell type can be mechanistically uncoupled from both SAMHD1 degradation and dNTPase activity.
Immunoblots of Vpx-expressing resting CD4 T cells confirmed that Vpx mnd-2 and Vpx rcm, in contrast to Vpx mac239, do not deplete SAMHD1 (Fig. 1E), consistent with our flow cytometry-based infection assays. Phosphorylation of SAMHD1 at threonine 592 (pSAMHD1-T592) correlates with a loss of its HIV-restrictive capacity (21, 22). Reprobing the immunoblot with an affinity-purified, T592 phosphosite-specific polyclonal rabbit antiserum (23) provided no evidence for an induction of this inactivating phosphorylation by any of the Vpx alleles, whereas cycling THP-1 cells displayed strong antibody reactivity (Fig. 1E). Moreover, no changes in overall SAMHD1 phosphorylation were induced by either Vpx mnd-2, Vpx rcm, or Vpx mac239 (Fig. 1F) and the CD4 T cells challenged with HIV-1 particles containing Vpx rcm or Vpx mnd-2 did not display increased levels of activation (Fig. S3 A–C). Together, these results show that Vpx proteins from the second Vpx+ lentiviral lineage, SIVrcm/mnd-2, overcome a postentry restriction for HIV-1 in resting CD4 T cells in the absence of SAMHD1 degradation, dNTP pool elevation, changes in SAMHD1 phosphorylation, or T-cell activation.
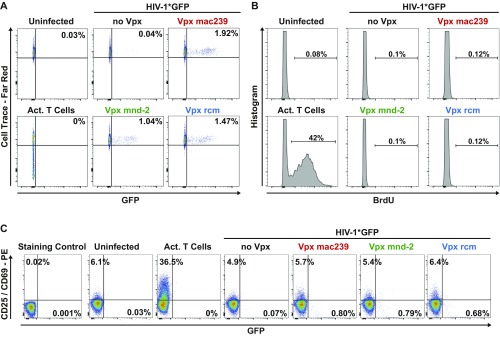
Fig. S3.
(A–C) Resting CD4 T cells were loaded with Cell Trace Dye (A) or BrdU (B) 1 d before infection and analyzed 3 d postinfection for Cell Trace-FarRed and BrdU expression levels, respectively. Alternatively, cells were stained for CD25/CD69 at the day of analysis (C). Depicted is one representative donor out of eight. Activated T cells served as positive control.
SIVrcm/mnd-2 Vpx Proteins Drastically Enhance Levels of Early Reverse Transcription Products.
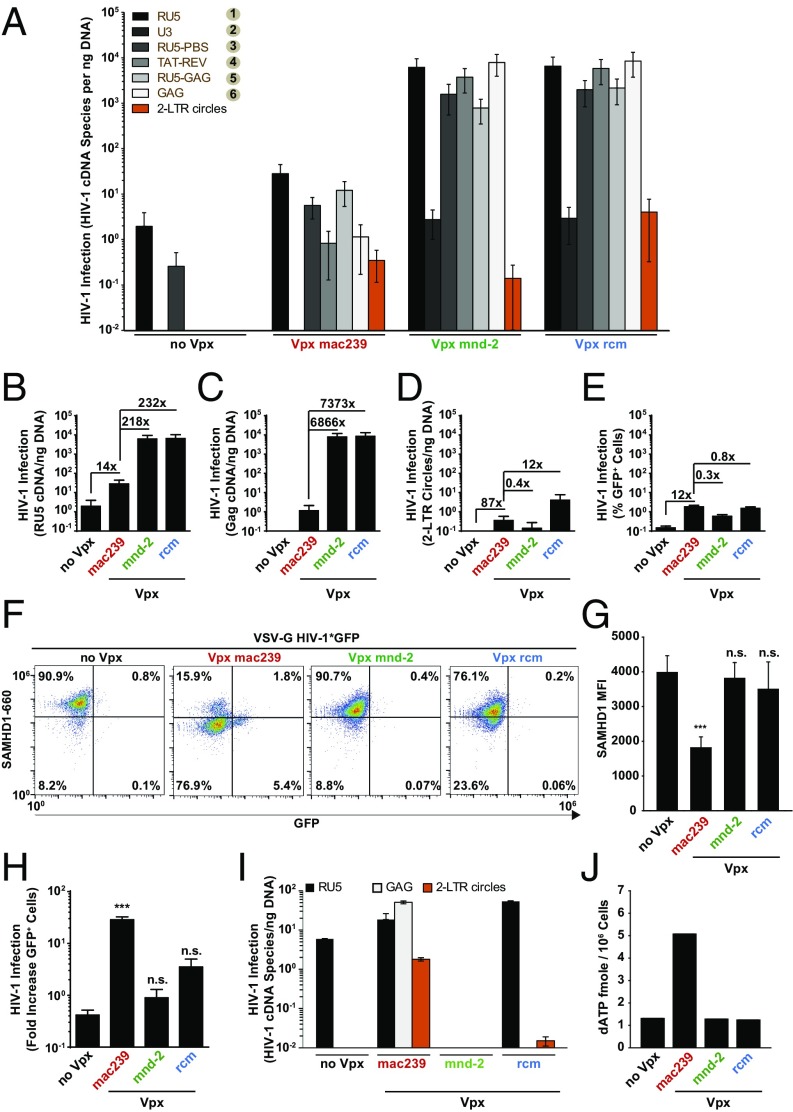
To gain insight into the mechanism by which Vpx mnd-2 and Vpx rcm facilitate X4 HIV-1*GFP infection, we determined the abundance of six different RT products, from first-strand transfer to full-length HIV-1 cDNA, using diagnostic qPCRs (see schematic representation in Fig. S4). In addition, GFP expression and 2 long-terminal repeat (2-LTR) circles, the latter a surrogate of completion of RT and successful nuclear entry, were quantified. Consistent with previous reports (6, 11, 12), challenge of resting CD4 T cells with X4 HIV-1 particles lacking Vpx gave rise to only low levels of early RT products, but late RT products, 2-LTR circles, and early gene expression above background could not be detected (Fig. 2 A–E, no Vpx, n = 10 donors). Virion-incorporated Vpx mac239 increased levels of early HIV-1 RT products ∼14-fold, relative to control cells and late RT products accumulated. Surprisingly, virion incorporation of Vpx mnd-2 or Vpx rcm elevated the abundance of early and late HIV-1 RT products in infected resting CD4 T cells to an even greater extent than Vpx mac239, ranging between 218- and 7,373-fold (Fig. 2 A–C). Of note, both levels of 2-LTR circles and percentage of GFP+ cells were similar for all three X4 HIV-1*GFP infections (Fig. 2 D and E) irrespective of the incorporated Vpx allele and apparent differences in levels of total HIV-1 cDNA (Fig. 2C). Thus, virion-incorporated Vpx rcm/mnd-2 that do not degrade SAMHD1 are highly effective in enhancing HIV-1 RT and the synthesis of full-length HIV-1 cDNA in infected quiescent CD4 T cells. A subsequent leveling of 2-LTR circle concentrations (orange bars in Fig. 2A) is suggestive of an additional barrier to nuclear import that likely limits the extent of downstream infection enhancement.
Fig. S4.
(A) Schematic of HIV-1 reverse transcription to illustrate the six different early to late HIV-1 RT products that were quantitatively detected by real-time PCR. (B) Positions of the different primer pairs are indicated and numbers refer to the order of detection shown in Fig. 2.
Fig. 2.
Virion-incorporated Vpx from SIVmnd-2 and SIVrcm boost levels of RT products in HIV-1–infected resting CD4 T cells, but not in primary macrophages. (A) Resting CD4 T cells were infected with equivalent infectious units of DNase-treated X4 HIV-1*GFP stocks and harvested on days 1 and 3 postchallenge for qPCR analyses. Levels of the different HIV-1 RT DNA products and episomal 2-LTR circles are presented as arithmetic means + SEM of 10 donors. Each measurement was normalized to an efavirenz control to account for coquantification of residual plasmid DNA contaminants. (B–D) Same data as in A but plotted separately for individual RT products: (B) RU5, (C) Gag, and (D) 2-LTR circles. (E) Percentage of GFP+ cells. (F–H) MDMs were challenged with equivalent infectious units of VSV-G pseudotyped HIV-1*GFP virions without (no Vpx) or with incorporated Vpx from SIVmac239, SIVmnd-2, or SIVrcm and analyzed 3 d later for expression of GFP and SAMHD1. (F) Dot plots of flow cytometric analysis for one representative donor of 7 donors tested 24 h postchallenge. (G) SAMHD1 levels represent the median fluorescence intensity (arithmetic means + SEM) of flow cytometric analyses for 7 donors. (H) Factor of increase of Vpx-mediated HIV-1 infection enhancement 3 d postchallenge. Shown are arithmetic means + SEM of 7 donors. (I) MDMs were infected with equivalent infectious units of the indicated DNase-treated virus stocks and harvested on days 1 and 3 postchallenge for qPCR analyses. Levels of the indicated HIV-1 RT DNA products and 2-LTR circles are shown and expressed as arithmetic means + SEM of triplicate infections for cells of 1 of 2 donors tested. (J) dATP levels of MDMs challenged 2 d earlier with VSV-G pseudotyped HIV-1*GFP virions without (no Vpx) or with incorporated Vpx from SIVmac239, SIVmnd-2, or SIVrcm. Depicted are arithmetic means of pooled triplicates from 2 donors.
Vpx Allele- and dNTP-Dependent Differences of HIV-1 Infection Enhancement in Resting CD4 T Cells and Macrophages.
Because a Vpx-sensitive, early-postentry barrier to HIV infection was originally identified in myeloid cells (2, 3) that was later causally linked to the SAMHD1 restriction (17, 18), we wondered whether virion-incorporated Vpx mnd-2 or Vpx rcm were capable of enhancing HIV-1 susceptibility also in terminally differentiated MDMs. Expectedly, infection of MDMs with vesicular stomatitis virus G protein (VSV-G) HIV-1*GFP lacking Vpx (no Vpx) resulted only in a low-level infection as determined by GFP expression analysis at 24 h (Fig. 2F) and 72 h (Fig. 2H) postchallenge. This replication block occurred subsequent to the initiation of RT (Fig. 2I and Fig. S4), in line with previous studies (2, 3, 8, 13, 17, 18). Virion-incorporated Vpx mac239 efficiently depleted cellular SAMHD1 (Fig. 2 F and G), elevated cellular dNTP pools (Fig. 2J, dATP concentration shown), and resulted in the expression of the GFP reporter from integrated provirus in a subfraction of MDMs at early (Fig. 2F) and in 29% of cells at later time points (Fig. 2H; n = 13). This infection enhancement was mirrored in the accumulation of high levels of total HIV-1 cDNA and episomal 2-LTR circles (Fig. 2I). In contrast, challenge of MDMs with VSV-G HIV-1*GFP virions carrying Vpx mnd-2 or Vpx rcm did not deplete SAMHD1 (Fig. 2F, variation of staining efficiency in individual samples (Vpx rcm in Fig. 2F) were not statistically significant, Fig. 2G), failed to enhance levels of RT products (Fig. 2I), or dNTP pools (Fig. 2J), and, importantly, did not render these myeloid target cells permissive to HIV-1 infection (Fig. 2H). These results confirm that Vpx proteins from the Vpx rcm/mnd-2 lineage are unable to overcome the likely SAMHD1-imposed restriction in myeloid cells (2, 24) and highlight a pronounced cell-type–dependent effect of Vpx alleles on HIV-1*GFP susceptibility.
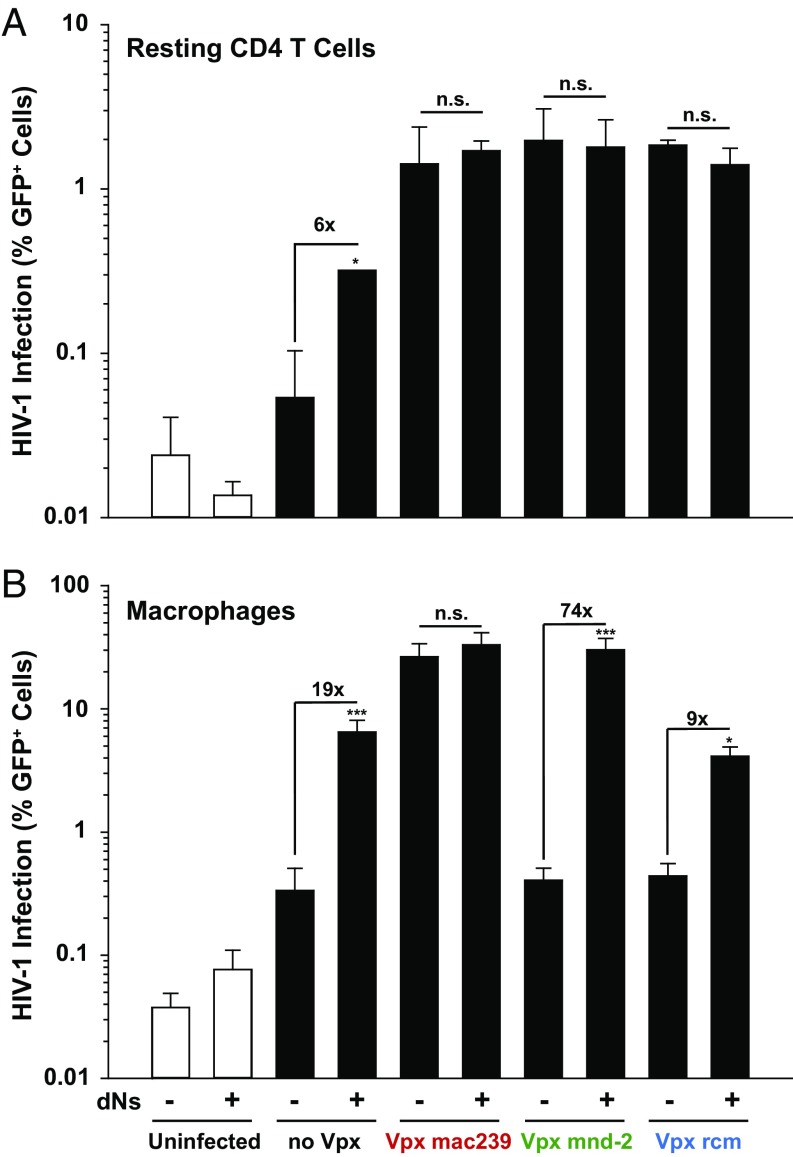
To further probe the importance of the dNTP pool as a rate-limiting factor for infection in primary HIV target cells, we added pyrimidine and purine deoxynucleosides (dNs) as metabolic precursors to the culture medium to artificially increase intracellular dNTP concentrations (11, 19, 25). In both MDMs and resting CD4 T cells, dN treatment elevated infection levels of HIV-1*GFP, in the absence of Vpx (no Vpx), by 19- and 6-fold, respectively (Fig. 3 A and B), consistent with previous findings (11, 26). Although virion-incorporated Vpx alleles from the two major lentiviral lineages had either an enhancing (SIVmac) or no direct (SIVrcm, SIVmnd-2) effect on dNTP levels in resting CD4 T cells (Fig. 1E), none of their infection-enhancing activities was boosted by additional dN treatment (Fig. 3A). In MDMs, the inability of both Vpx mnd-2 and Vpx rcm to increase HIV-1*GFP infection (Fig. 2H) and dNTP levels (Fig. 2J) was effectively compensated by dN addition to the medium, but did not further elevate infection levels of virions carrying Vpx mac239 (Fig. 3B). Thus, Vpx from SIVrcm and SIVmnd-2 is only able to enhance infection of resting CD4 T cells, but not of MDMs, and intracellular dNTP concentrations play a far greater role in the early-postentry phase of HIV-1 in MDMs than in resting CD4 T cells.
Fig. 3.
Exogenous addition of dNs has a different impact on HIV-1 infection in resting CD4 T cells and macrophages. (A) Resting CD4 T cells were challenged with equal infectious units of X4 HIV-1*GFP virions with or without (no Vpx) incorporated Vpx from SIVmac239, SIVmnd-2, or SIVrcm in the presence or absence of 2 mM dNTP precursors (dNs) and analyzed on day 3 postinfection for GFP expression and SAMHD1 levels. The percentages of GFP+ cells are shown for one representative donor out of three. (B) MDMs were challenged with equal infectious units of VSV-G pseudotyped HIV-1*GFP virions with or without (no Vpx) incorporated Vpx from SIVmac239, SIVmnd-2, or SIVrcm in the presence or absence of 4 mM dNs and analyzed on day 3 postinfection for GFP expression and SAMHD1 levels. The arithmetic means + SEM of four donors are depicted for the percentage of GFP+ cells.
Antagonism of the SAMHD1-Independent RT Block in Resting CD4 T Cells Is a Conserved Feature of Vpx Proteins.
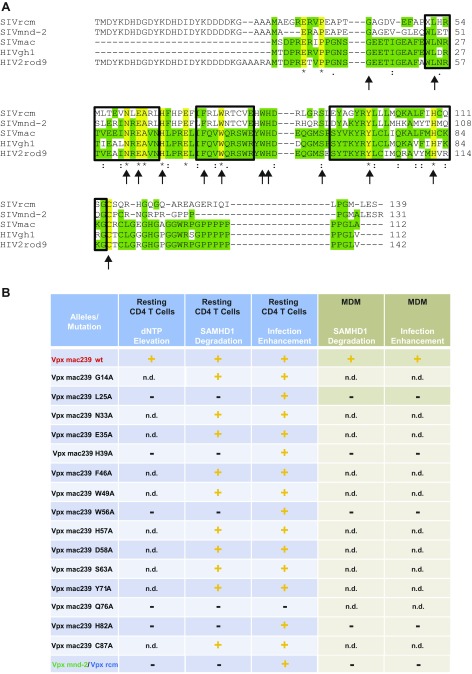
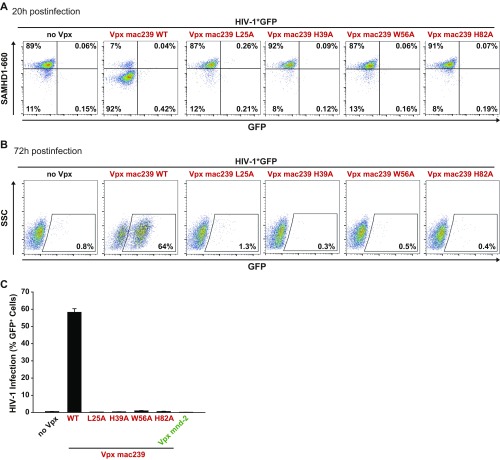
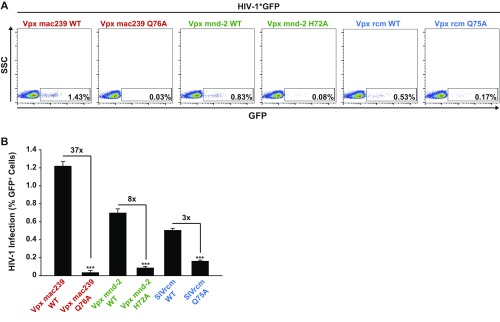
We next asked whether the Vpx protein from SIVmac239 exclusively harbors the activity for enhancing early steps of HIV-1 infection of resting CD4 T cells dependent on SAMHD1 degradation and dNTP elevation or, potentially, also carries a Vpx rcm/mnd-2–like activity of additionally enhancing infection of resting CD4 T cells in the absence of SAMHD1 degradation. Therefore, we generated a panel of Vpx mac239 point mutants with a focus on residues that are conserved among Vpx proteins of the two lentiviral lineages and are thought to be involved in binding to SAMHD1 (L25) or zinc (H39 and H82) or fail to target SAMHD1 for degradation for unknown reasons (W56) (27–30) (Fig. S5A). Following virion packaging, these mutants were characterized for their capacity to enhance X4 HIV-1*GFP infection, degrade SAMHD1, and elevate dNTPs in resting CD4 T cells. A number of mutants retained the ability of Vpx mac239 WT to deplete SAMHD1 and these proteins also enhanced infection (Fig. S5B). Remarkably, several Vpx mac239 mutants, including L25A, H39A, W56A, and H82A, boosted resting CD4 T-cell infection in the absence of SAMHD1 degradation (Fig. 4 A and B and Fig. S5B). Importantly, these non–SAMHD1-degrading Vpx mac239 mutants, in contrast to the WT protein, did not elevate cellular dTTP levels (Fig. 4C), but strongly boosted levels of RT products and 2-LTR circles (Fig. 4D). Consistent with the cell-type specificity of HIV-1’s postentry restriction and Vpx antagonism, all Vpx mac239 mutants that failed to degrade SAMHD1 were unable to facilitate MDM infection (Figs. S5B and S6). Single amino acid replacements in Vpx mac239 that prevented functional interactions with SAMHD1 thus resulted in accessory proteins that phenocopied the infection-enhancing ability of Vpx mnd-2 and Vpx rcm. Introducing analogous mutations to the DCAF interaction disrupting Q76A of Vpx mac239 into Vpx mnd-2 (H72A) or Vpx rcm (Q75A) abrogated their ability to enhance infection of resting CD4 T cells (Fig. S7), indicating that interactions with the proteasomal degradation machinery are critical for this activity. This finding reveals that enhancement of early reverse transcription in noncycling CD4 T cells by a mechanism that is independent of SAMHD1 depletion or elevation of cellular dNTP pools is, in principle, a conserved activity of Vpx proteins from both Vpx+ lentiviral lineages and likely involves degradation of Vpx targets distinct from SAMHD1.
Fig. S5.
(A) Sequence aligment generated with ClustalW of Vpx proteins from SIVrcm, SIVmnd-2, SIVmac239, HIV-2 GH-1, and HIV-2 ROD9. Boxes highlight helical structures. Arrows indicate positions for site-directed mutagenesis in SIVmac239 Vpx. Note that for Vpx rcm, mnd-2, and Rod9, the N-terminal flag tag and linker sequences are included in the alignment. (B) Summary of functional analyses of Vpx mutants and variants in resting CD4 T cells and monocyte-derived macrophages (MDMs). + or − delineate the ability or inability, respectively, of the depicted virion-incorporated Vpx mutants or variants to lead to dNTP elevation, SAMHD1 degradation, and HIV-1*GFP infection enhancement. n.d., not done.
Fig. 4.
Antagonism of a SAMHD1-independent early postentry restriction for HIV-1 in resting CD4 T cells is a conserved feature of Vpx proteins. (A and B) Resting CD4 T cells from healthy donors were challenged with equivalent infectious units of X4 HIV-1*GFP virions without (no Vpx) or with incorporation of the indicated Vpx alleles and point mutants and analyzed 3 d later for GFP expression and SAMHD1 levels. (A) Dot plots of flow cytometric analysis of intracellular SAMHD1 and GFP levels for one representative donor. (B) Factor of increase of Vpx-mediated HIV-1 infection enhancement 3 d postchallenge. Shown are arithmetic means + SEM of data from at least three donors. (C) Resting CD4 T cells were cotransfected with pDisplay-YFP and expression constructs for the indicated Vpx constructs, sorted for YFP surface expression, and analyzed for dTTP levels. Shown are the arithmetic means from two independent experiments. (D) Resting CD4 T cells were challenged with equivalent infectious units of the indicated DNase-treated virus stocks and harvested 3 d later for qPCR analyses. Shown are levels of early (RU5) and late (GAG) RT products as well as 2-LTR circles presented as arithmetic means + SEM of five donors. (E and F) Resting CD4 T cells from a patient with AGS with SAMHD1 deficiency and from two healthy donors were challenged with equivalent infectious units of X4 HIV-1*GFP virions without (no Vpx) or with incorporation of Vpx from SIVmac239 and analyzed 3 d later for expression of GFP and activation markers CD25/CD69. (E) Representative FACS dot plots and (F) arithmetic means of the percentages of GFP+ cells of duplicate infections.
Fig. S6.
MDMs were infected with VSV-G pseudotyped HIV-1*GFP with incorporated SIVmac239 WT or the depicted mutants using identical TZM-bl infectious units. (A and B) Primary FACS dot plots for SAMHD1 expression and GFP of MDMs from one donor 20 h postinfection (A) and 72 h postinfection (B). (C) Quantification of the percentage of GFP+ cells 72 h postinfection. Depicted is the arithmetic mean + SEM of four donors, each measured in technical duplicates.
Fig. S7.
(A) Resting CD4 T cells were challenged with X4 HIV-1*GFP carrying the indicated Vpx WT variants and mutants, the latter incapable of interaction with the proteasome complex. Cells were challenged with identical TZM-bl infectious units and analyzed for GFP expression 3 d postinfection. Primary dot plots for GFP expression are shown for one representative donor. (B) Quantification of the percentage of GFP+ cells shown in A. Depicted are arithmetic means + SEM of analyses of eight (SIVmnd-2WT and SIVmnd-2H72A) and four donors (SIVrcmWT and SIVrcmQ75A), respectively, each measured in technical triplicates.
Finally, we wanted to determine whether HIV-1 susceptibility of resting CD4 T cells could be further increased by virion-packaging of Vpx in the complete absence of SAMHD1. To this end, we had the opportunity to analyze resting CD4 T cells with a nonsense mutation in SAMHD1 obtained from one patient with Aicardi-Goutières syndrome (AGS) (11, 28). Consistent with our previous report (11), CD25–CD69– CD3+CD4+ T cells from this donor were intrinsically more permissive for X4 HIV-1*GFP infection compared with resting CD4 T cells from healthy donors (Fig. 4 E and F), underscoring the relevance of SAMHD1 in this process. Incorporation of Vpx mac239 WT increased HIV-1 infection of noncycling CD4 T cells from healthy donors by 159-fold, but, importantly, Vpx also boosted infection of SAMHD1-deficient AGS cells by 11-fold (Fig. 4 E and F). Due to highly limited fresh cell samples available and the low survival rate of previously cryopreserved resting CD4 T cells from patients with AGS, Vpx rcm/mnd-2 could, unfortunately, not be analyzed in this experiment and cells from additional donors were not accessible. These results provide direct evidence that Vpx can target a SAMHD1-independent restriction in resting CD4 T cells.
Discussion
Our functional analysis of virion-packaged Vpx proteins from the two Vpx-carrying lineages of lentiviruses provided insight into the cell-type specificity and mode of action of postentry restrictions for HIV-1 in resting CD4 T cells and macrophages. Consistent with previous reports (2, 3, 8, 13) only Vpx alleles with SAMHD1-degrading activity were capable of enhancing HIV-1 infection in macrophages and this strictly correlated with an elevation of intracellular dNTP levels. In contrast to Vpx from SIVmac239, the accessory proteins from the Vpx rcm/mnd-2 lineage that cannot degrade human SAMHD1 (31, 32) and that do not affect dNTP pools, failed to increase HIV-1 infection in this cell type.
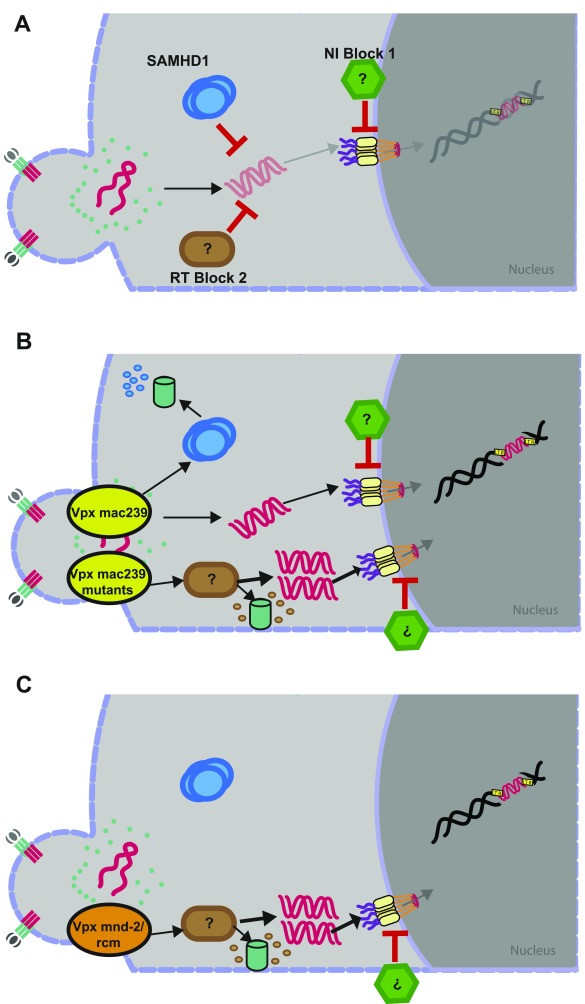
In resting CD4 T cells, surprisingly, Vpx mnd-2 and Vpx rcm overcame a postentry restriction for HIV-1 at the level of reverse transcription characterized by a drastic increase of RT products in the absence of SAMHD1 degradation, dNTP pool elevation, or changes in SAMHD1 phosphorylation. Importantly, virion-packaged Vpx also boosted HIV-1 infection of resting CD4 T cells from a patient with AGS who does not express SAMHD1 (11, 28). This finding strongly suggests that lentiviral Vpx proteins can antagonize a second, SAMHD1-independent cellular restriction, we refer to as “RT block 2,” that acts at the level of early reverse transcription in a dNTP-independent fashion in noncycling CD4 T cells (Fig. S8 A–C).
Fig. S8.
Proposed model for HIV restrictions in primary resting CD4 T cells and counteraction by SIV Vpx variants. (A) Both SAMHD1 (RT block 1) and an unknown factor (RT block 2) are able to restrict HIV at the level of reverse transcription. Downstream, an unknown factor limits nuclear import of the preintegration complex (NI block 1). (B) SIVmac239 Vpx WT targets SAMHD1 for degradation to overcome RT block 1. In the presence of SAMHD1 and RT block 2, SAMHD1 is the preferred target of SIVmac239 Vpx WT, but in the absence of SAMHD1, RT block 2 is targeted. SAMHD1 degradation-deficient mutants of SIVmac239 Vpx target RT block 2 similarly to SIVmnd-2 and SIVrcm Vpx through a mechanism that likely involves proteasomal degradation. (C) SIVmnd-2 and SIVrcm Vpx are unable to target SAMHD1 for degradation but apparently overcome the major restriction at the level of reverse transcription by targeting RT block 2. Despite highly efficient reverse transcription, levels of 2-LTR circles are similar to those observed with SIVmac239 Vpx, indicating that also under these conditions, nuclear import is restricted by NI block 1.
Surmounting either of these cellular postentry barriers in resting CD4 T cells, through either degrading Vpx variants by targeting SAMHD1- or non–SAMHD1-degrading Vpx variants by targeting RT block 2, resulted in a marked increase of early HIV-1 RT products. Notably, SAMHD1 degradation by Vpx from SIVmac239 was associated with a significantly milder increase in HIV-1 RT products (∼14-fold) over control infections than antagonism of RT block 2 by non–SAMHD1-degrading Vpx proteins (>1,000-fold increase). This quantitative difference together with the observation that single amino acid changes can transform Vpx mac239 into a non–SAMHD1-degrading, putative antagonist of RT block 2 indicates that individual Vpx proteins probably cannot, or only with low efficacy simultaneously or sequentially, antagonize both restrictions. SAMHD1-binding–competent, virion-packaged Vpx may, following HIV entry, first encounter and degrade SAMHD1 and thus typically not be available for interaction with RT block 2. In line with this scenario, the Vpx mac239 mutants that phenocopy Vpx rcm/mnd-2 are expected to lack functional interactions with SAMHD1, which may render them available for antagonism of RT block 2 (Fig. S8B).
Whereas the identity of RT block 2 remains unknown, our results allow us to speculate about its characteristics. Vpx resides within the incoming core and is expected to get access to the host cell cytoplasm only after at least partial uncoating has occurred (33). HIV-1 core stability is intrinsically coupled to the efficiency of RT and the timing and subcellular localization at which uncoating occurs likely determines the accessibility of inner core components for cellular factors. The tempospatial regulation of these events in reporter cell lines and myeloid cells is currently intensely debated (34, 35), and even less information is available about these processes in resting CD4 T cells. Our results are consistent with a model in which partial uncoating of HIV-1 cores allows, on the one hand, Vpx to interact with cytoplasmic cellular factors and, on the other hand, restriction factors to access RT complexes to directly inhibit early HIV-1 RT or degrade HIV RNA and early RT products. The apparent cell-type–specific difference in the capacity of Vpx variants for antagonism of RT block 2 may reflect differences in expression of this cellular factor or in the local milieu at which uncoating occurs in resting CD4 T cells and myeloid cells. Because SAMHD1-nondegrading Vpx proteins depend on residues predicted to couple them to the proteasome machinery, antagonism of RT block 2 likely also involves targeted degradation of this cellular factor. The apparent conservation of antagonism of RT block 2 among both Vpx+ lentiviral lineages argues for a high physiological relevance of this barrier.
The current findings challenge the dogma that restriction of HIV RT and Vpx antagonism thereof are exclusively accomplished through a modulation of cellular dNTP concentrations and allow a refined, cell-type–specific assessment: Both the infection enhancement of SAMHD1-degrading Vpx proteins and exogenous dN treatment point to a pivotal regulatory role of dNTP levels in macrophages (20–40 nM) (reviewed in ref. 36) for HIV-1 restriction. In noncycling CD4 T cells, in which dNTP concentrations are naturally ∼10- to 100-fold higher (300–5,000 nM) (36), Vpx can, however, boost infection in the absence of dNTP elevation to levels that are not further elevated by addition of exogenous dNs. The concentrations of the RT substrates are thus not restrictive but rather rate limiting; a further increase of available dNTPs by dN treatment, SAMHD1-degrading Vpx proteins, or T-cell activation enhances infection.
The quantitative analysis of consecutive steps of the HIV replication cycle in resting CD4 T cells in the presence of Vpx variants that target either SAMHD1 or RT block 2 allowed us to describe another potent restriction. Both levels of episomal 2-LTR circles and early gene expression were comparable for the two mechanistically distinct types of Vpx proteins, despite impressive differences in absolute levels of early to late RT products (ratio 2-LTR circles per total HIV-1 cDNA: 0.304 for Vpx mac239, 0.000018 for Vpx mnd-2, and 0.00048 for Vpx rcm). This finding suggests a potent, rate-limiting barrier to nuclear import of the preintegration complex in resting CD4 T cells that is not easily titratable and that we tentatively refer to as “nuclear import block 1” (NI block 1) (Fig. S8). The relatively milder impact of this barrier on particles that degrade SAMHD1 lets us hypothesize that distinct transport pathways may exist downstream of RT blocks 1 and 2. In this scenario, events subsequent to overcoming the SAMHD1 barrier may be less prone to inhibition by NI block 1, which may represent one reason why degradation of SAMHD1 is selected in evolution. Our study reveals a thus far unrecognized complexity and target cell specificity of lentiviral Vpx proteins in their ability to overcome early-postentry restrictions of HIV infection. Assessing these Vpx activities in the natural viral context and in target cells of the natural host as well as identifying the cellular factors responsible for RT block 2 and NI block 1 in resting CD4 T cells represent important goals of future studies aiming at dissecting the molecular events that regulate the postentry susceptibility of this major target cell population.
Materials and Methods
The generation of the infectious HIV-1*GFP proviral clone was previously described (11). The following Vpx expression constructs were used: pcDNA3.1, Vpx SIVmac239-myc (WT and Q76A) (37), pcDNA3.1-flag Vpx SIVmnd-2 (20), and pcDNA3.1-flag Vpx SIVrcm (20). SIVmac239 alanine mutants and the Q76A-analogous mutants of SIVmnd-2 (H72A) and SIVrcm (Q75A) were generated by site-directed mutagenesis. pDisplay-YFP was a gift from Barbara Müller, Department of Infectious Diseases, University Hospital Heidelberg, Heidelberg, Germany.
SI Materials and Methods
Cells and Reagents.
Peripheral blood mononuclear cells (PBMCs) from healthy blood donors were purified by Ficoll-Hypaque gradient centrifugation (11). Resting CD4 T cells were isolated from PBMCs by negative selection with the RosetteSep Human CD4 T Cells Enrichment Mixture (StemCell Technologies). Resting CD4 T cells were cultured at a density of 2 × 106 cells/mL in RPMI-1640 medium (Gibco) supplemented with 10% (vol/vol) heat-inactivated FCS, glutamine (2 mM), and antibiotics (100 units/mL penicillin, 100 mg/mL streptomycin). For T-cell activation, phytohemagglutinin-P (PHA-P) (5 µg/mL) (Sigma-Aldrich) was added to the culture medium for 2–3 days together with 100 IU/mL IL2 (Biomol). Monocyte-derived macrophages were isolated from PBMCs by negative selection with Monocyte Isolation Kit II (Miltenyi Biotec) and separated by the autoMACS Pro Separator. Monocytes were differentiated in coated 24-well plates (Greiner) containing DMEM (Gibco) supplemented with 10% (vol/vol) heat-inactivated FCS, glutamine (2 mM), antibiotics (100 units/mL penicillin, 100 mg/mL streptomycin), and 10% (vol/vol) human AB-serum (Sigma-Aldrich). The human cell lines THP-1 and 293T were cultivated as reported (28). The following fluorochrome-conjugated antibodies were used: anti-human CD25 (clone M-A251) and anti-human CD69 (clone FN50) (both from BD Pharmingen). Additional antibodies were: sheep anti-p24 capsid (kindly provided by Barbara Müller), rabbit anti-SAMHD1 (Proteintech), mouse anti-YFP (Sigma-Aldrich), mouse anti-myc (Santa Cruz Biotechnology), mouse anti-Flag (Sigma-Aldrich), and phospho-SAMHD1 T592 (23). Efavirenz (Bristol-Myers Squibb) was used as described (38).
Virus Production.
Sucrose cushion purified HIV-1 virus stocks were produced as previously described (11). HIV-1*GFP virus stocks, which allow incorporation of Vpx, were produced by cotransfection of the proviral HIV-1*GFP DNA and the indicated Vpx expression constructs (10). The infectious titer was determined in a standardized TZM-bl infectivity assay (39), and the amount of physical particles determined by quantification of virion-associated RT activity (SG-PERT) (40) and/or by p24 ELISA (39).
HIV-1 Infection.
Resting primary CD4 T cells were infected with identical infectious units, determined on TZM-bl cells, of HIV-1*GFP ± Vpx using spin infection for 90 min at 20 × g at room temperature. The medium was exchanged 4–6 h p.i. Primary monocyte-derived macrophages were infected overnight with identical infectious units of VSV-G pseudotyped HIV-1*GFP ± Vpx.
Flow Cytometry.
For fixation and antibody staining, infected CD4 T cells and detached primary monocyte-derived macrophages were fixed for 90 min at room temperature with BD Cytofix Fixation Buffer and permeabilized in BD Phosflow Perm Buffer III for 2 min at 4 °C in the dark. After two washes in PBS/1% FCS/0.09% sodium azide, cells were stained with the rabbit anti-SAMHD1 antiserum followed by a goat anti-rabbit Alexa 660 antibody. A FACSCalibur flow cytometer and FACSVerse flow cytometer and FlowJo software (TreeStar) were used for analyses. For CD25/69 staining, anti-CD25 and CD69-PE antibodies were added 1:40 at the same incubation step as the primary SAMHD1 antibody. Staining with CellTrace Far Red DDAO-SE (Invitrogen) and BrdU (BrdU Cell Proliferation Assay Kit, BD Pharmingen) were performed according to the manufacturer’s protocol.
Analysis of SAMHD1 Levels by Western Blotting.
Resting CD4 T cells were nucleofected (Amaxa) with expression plasmids for Vpx as well as Display-YFP and YFP-expressing cells isolated and analyzed by Western blotting as described (11). For the analysis of the overall SAMHD1 phosphorylation pattern, lysates of YFP+ cells were subjected to SDS/PAGE on phos-tag acrylamide (Wako Chemicals) gels according to the manufacturer’s instructions.
PCR Analyses.
TaqMan-based quantification of HIV-1 2-LTR circles in DNA extracts from infected resting CD4 T cells and primary monocyte-derived macrophages was performed as described (38). The following quantitative PCRs were designed and validated based on the primer regions chosen by Zack et al. (6). Quantification of U3 region: forward primer M665 (5′-TGT TAC ACC CTA TGA GCC AGC AT-3′), reverse primer M666 (5′-AGG CCA CAC CTC CCT GGA-3′), and fluorescent-labeled probe M665 (FAM-AGA GCT GCA TCC GGA GTA CTA CAA AGA CTG CT-TAMRA). Cycling conditions for U3 were: 50 °C 2 min, 95 °C 10 min, 40 cycles with a denaturation step at 95 °C for 15 s, and annealing and elongation step at 62 °C for 1 min. Quantification of RU5 region is as follows: forward primer M667 (5′-CTA ACT AGG GAA CCC ACT GCT TAA-3′), reverse primer AA55 (5′-TGC TAG AGA TTT TCC ACA CTG ACT AA-3′), and fluorescent-labeled probe M667 (FAM-TTG AGT GCT CAA AGT AGT GTG TGC CCG TCT GTT-TAMRA). Cycling conditions were similar to RU5-gag (11). Quantification of RU5-PBS region is as follows: forward primer M667, reverse primer BB301 (5′-TTT CGC TTT CAA GTC CCT GTT-3′), and FAM-labeled probe M667. Cycling conditions for RU5-PBS were as follows: 50 °C 2 min, 95 °C 10 min, 40 cycles with a denaturation step at 95 °C for 15 s, annealing step at 50 °C for 30 s and elongation step at 72 °C for 1 min. Data acquisition was performed at the elongation step. Quantification of RU5-gag was as follows: forward primer M667, reverse primer M661 (5′-TCG AGA GAT CTC CTC TGG CTT-3′), and FAM-labeled probe M667. Cycling conditions were already described (11). Quantification of gag is as follows: forward primer LA8 (5′-CAG GAC TCG GCT TGC TGA A-3′), reverse primer LA9 (5′-CTC GCA CCC ATC TCT CTC CTT-3′), and fluorescent-labeled probe LA8 (FAM-TCC GCT AGT CAA AAT TTT TGG CGT ACT CAC-TAMRA). Cycling conditions were default settings with an annealing and elongation step at 60 °C. Quantification of tat/rev was as follows: forward primer LA45 (5′-GAC AAA AGC CTT AGG CAT CTC CTA T-3′ ), reverse primer LA64 (5′-ATA GGT TGC ATT ACA TGT ACT ACT TAC TGC T-3′), and fluorescent-labeled probe LA45 (FAM-TGA TGA GTC TGA CTG TTC TGA TGA GCT CTT CGT-TAMRA). Cycling conditions were default settings with an annealing and elongation step at 60 °C. RNAseP quantification served as endogenous control. To account for residual plasmid DNA contaminations in virus preparations and to quantify only de novo synthesized HIV-1 cDNA, cells were treated with the RT inhibitor efavirenz, infected and measured in parallel. Data sets presented in Figs. 2 and 4 show normalized values, where input DNA contamination accounted for at maximum 30% of the absolute signal, and these values were subtracted from the values of cells in the absence of inhibitor.
dNTP Assay.
dNTPs from primary cells were extracted and quantified using the protocol previously described by Diamond et al. (41). Briefly, this assay uses a 5′ P32 radiolabeled 23-mer primer annealed to one of four distinct 24-mer templates. The single nucleotide overhang on the 24-mer template (A, C, G, or T) determines the dNTP to be measured upon the dNTP incorporation. The template/primer was incubated with extracted cellular dNTPs and purified HIV-1 RT for 5 min at 37 °C and then quenched with 40 mM EDTA and 99% (vol/vol) formamide at 95 °C for 2 min. The reactions were resolved on a 20% (wt/vol) urea-PAGE gel (American Bio Sequel NE reagent) and imaged using Pharos FX molecular imager (Bio-Rad Laboratories). The increase in radiolabeled 24-mer product indicates that the dNTP specific for the template has been incorporated, and the primer extension amounts were used to measure the dNTP levels in the samples.
Acknowledgments
We are grateful to Ulrike Protzer for support and access to the biosafety laboratory level 3 (BSL3) at the Technische Universität Munich (TUM). We thank Nathaniel Landau for providing reagents and Elina Erikson for help with initial FACS analyses. This work was supported by grants from the Deutsche Forschungsgemeinschaft [projects in priority program SPP1923 1923 (KE 742-/7-1 to O.T.K., FA 378/17-1 to O.T.F.], Grants FA 378/11-1 (to O.T.F.), KE 742/4-1 (to O.T.K.), NIH Grant R01GM110570 (to O.I.F., E.S.L., and M.E.), Grants AI049781 and GM104198 to B.K.), and the German Center for Infection Research (DZIF) (R.K., O.T.F., O.T.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613635114/-/DCSupplemental.
References
- 1.Bakri Y, et al. The maturation of dendritic cells results in postintegration inhibition of HIV-1 replication. J Immunol. 2001;166(6):3780–3788. doi: 10.4049/jimmunol.166.6.3780. [DOI] [PubMed] [Google Scholar]
- 2.Goujon C, et al. Characterization of simian immunodeficiency virus SIVSM/human immunodeficiency virus type 2 Vpx function in human myeloid cells. J Virol. 2008;82(24):12335–12345. doi: 10.1128/JVI.01181-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goujon C, et al. SIVSM/HIV-2 Vpx proteins promote retroviral escape from a proteasome-dependent restriction pathway present in human dendritic cells. Retrovirology. 2007;4:2. doi: 10.1186/1742-4690-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaushik R, Zhu X, Stranska R, Wu Y, Stevenson M. A cellular restriction dictates the permissivity of nondividing monocytes/macrophages to lentivirus and gammaretrovirus infection. Cell Host Microbe. 2009;6(1):68–80. doi: 10.1016/j.chom.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stevenson M, Stanwick TL, Dempsey MP, Lamonica CA. HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J. 1990;9(5):1551–1560. doi: 10.1002/j.1460-2075.1990.tb08274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zack JA, et al. HIV-1 entry into quiescent primary lymphocytes: Molecular analysis reveals a labile, latent viral structure. Cell. 1990;61(2):213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 7.Pan X, Baldauf HM, Keppler OT, Fackler OT. Restrictions to HIV-1 replication in resting CD4+ T lymphocytes. Cell Res. 2013;23(7):876–885. doi: 10.1038/cr.2013.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergamaschi A, et al. The human immunodeficiency virus type 2 Vpx protein usurps the CUL4A-DDB1 DCAF1 ubiquitin ligase to overcome a postentry block in macrophage infection. J Virol. 2009;83(10):4854–4860. doi: 10.1128/JVI.00187-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujita M, et al. Vpx is critical for reverse transcription of the human immunodeficiency virus type 2 genome in macrophages. J Virol. 2008;82(15):7752–7756. doi: 10.1128/JVI.01003-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sunseri N, O’Brien M, Bhardwaj N, Landau NR. Human immunodeficiency virus type 1 modified to package Simian immunodeficiency virus Vpx efficiently infects macrophages and dendritic cells. J Virol. 2011;85(13):6263–6274. doi: 10.1128/JVI.00346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baldauf HM, et al. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat Med. 2012;18(11):1682–1687. doi: 10.1038/nm.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Descours B, et al. SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4(+) T-cells. Retrovirology. 2012;9:87. doi: 10.1186/1742-4690-9-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pertel T, Reinhard C, Luban J. Vpx rescues HIV-1 transduction of dendritic cells from the antiviral state established by type 1 interferon. Retrovirology. 2011;8:49. doi: 10.1186/1742-4690-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Srivastava S, et al. Lentiviral Vpx accessory factor targets VprBP/DCAF1 substrate adaptor for cullin 4 E3 ubiquitin ligase to enable macrophage infection. PLoS Pathog. 2008;4(5):e1000059. doi: 10.1371/journal.ppat.1000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu XF, Yu QC, Essex M, Lee TH. The vpx gene of simian immunodeficiency virus facilitates efficient viral replication in fresh lymphocytes and macrophage. J Virol. 1991;65(9):5088–5091. doi: 10.1128/jvi.65.9.5088-5091.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldstone DC, et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480(7377):379–382. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- 17.Hrecka K, et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474(7353):658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laguette N, et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474(7353):654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lahouassa H, et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat Immunol. 2012;13(3):223–228. doi: 10.1038/ni.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim ES, et al. The ability of primate lentiviruses to degrade the monocyte restriction factor SAMHD1 preceded the birth of the viral accessory protein Vpx. Cell Host Microbe. 2012;11(2):194–204. doi: 10.1016/j.chom.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cribier A, Descours B, Valadão AL, Laguette N, Benkirane M. Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Reports. 2013;3(4):1036–1043. doi: 10.1016/j.celrep.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 22.White TE, et al. The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe. 2013;13(4):441–451. doi: 10.1016/j.chom.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt S, et al. SAMHD1's protein expression profile in humans. J Leukoc Biol. 2015;98(1):5–14. doi: 10.1189/jlb.4HI0714-338RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reinhard C, Bottinelli D, Kim B, Luban J. Vpx rescue of HIV-1 from the antiviral state in mature dendritic cells is independent of the intracellular deoxynucleotide concentration. Retrovirology. 2014;11:12. doi: 10.1186/1742-4690-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plesa G, et al. Addition of deoxynucleosides enhances human immunodeficiency virus type 1 integration and 2LTR formation in resting CD4+ T cells. J Virol. 2007;81(24):13938–13942. doi: 10.1128/JVI.01745-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim B, Nguyen LA, Daddacha W, Hollenbaugh JA. Tight interplay among SAMHD1 protein level, cellular dNTP levels, and HIV-1 proviral DNA synthesis kinetics in human primary monocyte-derived macrophages. J Biol Chem. 2012;287(26):21570–21574. doi: 10.1074/jbc.C112.374843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaller T, Bauby H, Hué S, Malim MH, Goujon C. New insights into an X-traordinary viral protein. Front Microbiol. 2014;5:126. doi: 10.3389/fmicb.2014.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berger A, et al. SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutières syndrome are highly susceptible to HIV-1 infection. PLoS Pathog. 2011;7(12):e1002425. doi: 10.1371/journal.ppat.1002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwefel D, et al. Molecular determinants for recognition of divergent SAMHD1 proteins by the lentiviral accessory protein Vpx. Cell Host Microbe. 2015;17(4):489–499. doi: 10.1016/j.chom.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwefel D, et al. Structural basis of lentiviral subversion of a cellular protein degradation pathway. Nature. 2014;505(7482):234–238. doi: 10.1038/nature12815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fregoso OI, et al. Evolutionary toggling of Vpx/Vpr specificity results in divergent recognition of the restriction factor SAMHD1. PLoS Pathog. 2013;9(7):e1003496. doi: 10.1371/journal.ppat.1003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, et al. Characterization of the interactions between SIVrcm Vpx and red-capped mangabey SAMHD1. Biochem J. 2015;468(2):303–313. doi: 10.1042/BJ20141331. [DOI] [PubMed] [Google Scholar]
- 33.Kewalramani VN, Emerman M. Vpx association with mature core structures of HIV-2. Virology. 1996;218(1):159–168. doi: 10.1006/viro.1996.0176. [DOI] [PubMed] [Google Scholar]
- 34.Ambrose Z, Aiken C. HIV-1 uncoating: Connection to nuclear entry and regulation by host proteins. Virology. 2014;454-455:371–379. doi: 10.1016/j.virol.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campbell EM, Hope TJ. HIV-1 capsid: The multifaceted key player in HIV-1 infection. Nat Rev Microbiol. 2015;13(8):471–483. doi: 10.1038/nrmicro3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ayinde D, Casartelli N, Schwartz O. Restricting HIV the SAMHD1 way: Through nucleotide starvation. Nat Rev Microbiol. 2012;10(10):675–680. doi: 10.1038/nrmicro2862. [DOI] [PubMed] [Google Scholar]
- 37.Gramberg T, Sunseri N, Landau NR. Evidence for an activation domain at the amino terminus of simian immunodeficiency virus Vpx. J Virol. 2010;84(3):1387–1396. doi: 10.1128/JVI.01437-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goffinet C, Allespach I, Keppler OT. HIV-susceptible transgenic rats allow rapid preclinical testing of antiviral compounds targeting virus entry or reverse transcription. Proc Natl Acad Sci USA. 2007;104(3):1015–1020. doi: 10.1073/pnas.0607414104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keppler OT, et al. Rodent cells support key functions of the human immunodeficiency virus type 1 pathogenicity factor Nef. J Virol. 2005;79(3):1655–1665. doi: 10.1128/JVI.79.3.1655-1665.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pizzato M, et al. A one-step SYBR Green I-based product-enhanced reverse transcriptase assay for the quantitation of retroviruses in cell culture supernatants. J Virol Methods. 2009;156(1–2):1–7. doi: 10.1016/j.jviromet.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 41.Diamond TL, et al. Macrophage tropism of HIV-1 depends on efficient cellular dNTP utilization by reverse transcriptase. J Biol Chem. 2004;279(49):51545–51553. doi: 10.1074/jbc.M408573200. [DOI] [PMC free article] [PubMed] [Google Scholar]