Abstract
HER2/ERBB2 amplification/overexpression determines the eligibility of breast cancer patients to HER2-targeted therapy. This study evaluates the agreement between ERBB2 copy number assessment by fluorescence in situ hybridization, a standard method recommended by the American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP), and newly available DNA extraction-based methods. A series of n=29 formalin-fixed paraffin-embedded breast cancers were subjected to ERBB2 copy number assessment by fluorescence in situ hybridization (FISH, Vysis, Abbott). Following macrodissection of invasive breast cancer tissue and DNA extraction, ERBB2 copy number was also determined by molecular inversion probe array analysis (MIP, OncoScan, Affymetrix) and next generation sequencing combined with normalized amplicon coverage analysis (NGS/NAC, AmpliSeq, Ion Torrent). ERBB2 copy number values obtained by MIP or NGS/NAC were tightly correlated with ERBB2 copy number values obtained by conventional FISH (rs = 0.940 and rs = 0.894, P < 0.001). Using ASCO/CAP guideline-conform thresholds for categorization of breast cancers as HER2-negative, equivocal or positive, nearly perfect concordance was observed for HER2 classification by FISH and MIP (93% concordant classifications, κ = 0.87). Substantial concordance was observed for FISH and NGS/NAC (83% concordant classifications, κ = 0.62). In conclusion, MIP facilitates precise ERBB2 copy number detection and should be considered as an ancillary method for clinical HER2 testing.
Keywords: HER2/ERBB2, OncoScan, breast cancer, copy number profiling, next generation sequencing
INTRODUCTION
The HER2/ERBB2 (erb-b2 receptor tyrosine kinase 2) proto-oncogene is activated by overexpression/amplification in 15% of breast cancers (BCs) [1]. The development of therapeutics targeting HER2 (trastuzumab, T-DM1, pertuzumab, lapatinib) has been a major breakthrough. According to current standard therapeutic regimes, ERBB2 amplification is the only actionable genetic alteration in BC. Eligibility of BC patients to HER2-targeted therapy depends on detection of HER2/ERBB2 overexpression/amplification in the tumor tissue by immunohistochemistry (IHC) and/or in situ hybridization (ISH). This methodology was introduced in now historical clinical trials, such as HERA and NSABP B-31 [2, 3]. Detailed guidelines for clinical HER2 testing are provided by the American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) [4]. From a technical point of view, ASCO/CAP guidelines admit a number of methodological variations for clinical HER2 testing, including dark field fluorescent ISH (FISH), bright field chromogenic ISH (CISH), ISH with just one probe for ERBB2 (chromosome 17q12) or ISH with two probes for ERBB2 and the centromeric region of chromosome 17 (CEP17). ASCO/CAP guidelines also define thresholds for a ERBB2 positive status, which take into consideration either the ERBB2/CEP17 ratio and/or the ERBB2 copy number (CN) per se [4].
HER2/ERBB2 assessment by IHC and FISH is subject to at least some inter-observer and inter-laboratory variability [5]. Moreover, BCs with equivocal results by IHC and ISH are notoriously difficult to classify with conventional methods [6–12]. Consequently, there is a vivid debate about ancillary or third line test methods [13]. So far, ASCO/CAP guidelines preclude mRNA-based ERBB2 expression assays, for which partly encouraging results, but also occasional misclassifications have been reported [14–19]. Meanwhile, innovative and rapidly developing DNA extraction-based methods have begun to play a substantial role in the classification of tumors [20]. Clinical HER2 testing might take advantage of these developments, which have made available new DNA-based methods for gene CN assessment. Here, we adopted two innovative DNA-based technologies, namely molecular inversion probe array analysis (MIP) [21, 22] and next generation sequencing with normalized amplicon coverage analysis (NGS/NAC) [23] for ERBB2 CN assessment in BC.
RESULTS
A series of n=29 BCs was subjected to ERBB2 CN assessment by FISH, MIP and NGS/NAC. FISH served as an ASCO/CAP guideline-conform standard method and was performed on whole-slide sections. No significant intratumoral heterogeneity for ERBB2 was observed by FISH in any of the BCs included. MIP and NGS/NAC were performed with DNA from macro-dissected tumor tissue. An average ERBB2 CN value was obtained by FISH and MIP. A ERBB2 NAC value, reflecting the average ERBB2 gene dosage, was obtained by NGS/NAC (Figure 1 and Table 1).
Figure 1.
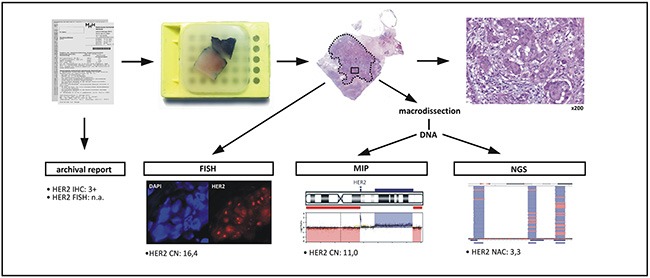
Work flow: An FFPE BC specimen was selected based on its HER2 status retrieved from archival reports. A representative FFPE tissue block was selected and was subjected to HER2/ERBB2 copy number (CN) assessment by fluorescence in situ hybridization (FISH). Following macrodissection of invasive tumor tissue (dotted line) and DNA extraction, the HER2/ERBB2 CN was also assessed by molecular inversion probe array analysis (MIP) and next generation sequencing with normalized amplicon coverage analysis (NGS).
Table 1. BC characteristics in detail.
| data from archival reports | ERBB2 re-evaluation in the present study, same FFPE block | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| case | ID | age | side | histology | pT | pN | grade | Ki67 | ER | PR | HER2 | ERBB2 FISH | FISH [CN] | MIP [CN] | NGS [NAC] | |||
| 1 | 2 | 74 | L | 8500/3 | 1c | x | 3 | 20 | 1 | 1 | 2 | equivocal | 4,1 | equivocal | 4,00 | equivocal | 0,55 | negative |
| 2 | 3 | 40 | L | 8500/3 | 2 | x | 3 | 25 | 1 | 0 | 3 | n.a. | 22,9 | positive | 11,67 | positive | 3,13 | positive |
| 3 | 4 | 35 | L | 8500/3 | 3 | 3a | 3 | 65 | 1 | 1 | 3 | n.a. | 16,4 | positive | 11,00 | positive | 3,28 | positive |
| 4 | 6 | 77 | L | 8500/3 | x | x | 3 | 10 | 1 | 0 | 3 | n.a. | 18,5 | positive | 11,67 | positive | 4,22 | positive |
| 5 | 7 | 83 | L | 8500/3 | 2 | 1a | 3 | 40 | 1 | 1 | 0 | n.a. | 2,0 | negative | 2,00 | negative | 0,48 | negative |
| 6 | 8 | 51 | R | 8500/3 | 1b | 0 | 3 | 30 | 1 | 1 | 0 | n.a. | 1,9 | negative | 2,00 | negative | 0,50 | negative |
| 7 | 9 | 53 | R | 8500/3 | 2 | 0 | 3 | 30 | 1 | 1 | 0 | n.a. | 2,1 | negative | 2,00 | negative | 0,49 | negative |
| 8 | 10 | 69 | L | 8520/3 | 2 | 0 | 3 | 30 | 1 | 0 | 2 | negative | 2,0 | negative | 2,00 | negative | 0,37 | negative |
| 9 | 11 | 47 | L | 8500/3 | 1c | 0 | 3 | 40 | 1 | 1 | 2 | negative | 3,7 | negative | 3,00 | negative | 0,97 | negative |
| 10 | 12 | 48 | L | 8500/3 | 3 | 3 | 3 | 30 | 1 | 1 | 0 | n.a. | 1,9 | negative | 1,67 | negative | 0,36 | negative |
| 11 | 13 | 38 | L | 8510/3 | 2 | 0 | 3 | 80 | 1 | 1 | 2 | equivocal | 2,9 | negative | 2,00 | negative | 0,59 | negative |
| 12 | 15 | 53 | L | 8500/3 | 1c | 0 | 3 | 25 | 1 | 1 | 3 | n.a. | 7,6 | positive | 6,00 | positive | 0,88 | negative |
| 13 | 16 | 68 | L | 8500/3 | 1c | 0 | 3 | 40 | 1 | 1 | 3 | n.a. | 11,2 | positive | 8,00 | positive | 1,12 | negative |
| 14 | 17 | 85 | R | 8500/3 | 1c | 1 | 3 | 35 | 1 | 1 | 3 | n.a. | 19,9 | positive | 7,67 | positive | 2,44 | positive |
| 15 | 18 | 70 | R | 8500/3 | 4b | 1 | 3 | 70 | 1 | 0 | 3 | n.a. | 11,5 | positive | 14,00 | positive | 2,29 | positive |
| 16 | 19 | 55 | R | 8500/3 | 2 | 1a | 3 | 60 | 1 | 1 | 3 | n.a. | 24,6 | positive | 10,33 | positive | 3,42 | positive |
| 17 | 22 | 57 | R | 8500/3 | 2 | 0 | 3 | 25 | 1 | 1 | 2 | negative | 2,5 | negative | 3,00 | negative | 0,52 | negative |
| 18 | 23 | 65 | L | 8500/3 | 1c | x | 2 | 50 | 1 | 1 | 2 | equivocal | 4,1 | equivocal | 5,00 | equivocal | 0,81 | negative |
| 19 | 24 | 51 | R | 8500/3 | 4b | 0 | 3 | 35 | 1 | 1 | 2 | equivocal | 2,0 | negative | 2,00 | negative | 0,37 | negative |
| 20 | 25 | 64 | R | 8500/3 | 1b | 0 | 2 | 10 | 1 | 1 | 2 | negative | 2,4 | negative | 2,33 | negative | 0,57 | negative |
| 21 | 26 | 73 | L | 8500/3 | 1b | 0 | 3 | 20 | 1 | 1 | 0 | n.a. | 3,7 | negative | 2,70 | negative | 0,63 | negative |
| 22 | 27 | 40 | R | 8500/3 | 1c | 0 | 3 | 10 | 1 | 1 | 2 | negative | 3,7 | negative | 4,00 | equivocal | 0,85 | negative |
| 23 | 28 | 56 | R | 8500/3 | 2 | 0 | 2 | 20 | 1 | 1 | 2 | negative | 2,1 | negative | 2,00 | negative | 0,33 | negative |
| 24 | 29 | 45 | L | 8500/3 | 1c | 0 | 3 | 30 | 1 | 1 | 2 | equivocal | 2,8 | negative | 2,50 | negative | 0,52 | negative |
| 25 | 30 | 53 | R | 8520/3 | 3 | 0 | 2 | 15 | 1 | 1 | 0 | n.a. | 1,6 | negative | 1,00 | negative | 0,34 | negative |
| 26 | 31 | 65 | R | 8520/3 | 2 | 0 | 2 | 15 | 1 | 1 | 0 | n.a. | 2,1 | negative | 2,00 | negative | 0,36 | negative |
| 27 | 32 | 57 | L | 8520/3 | 3 | 0 | 2 | 5 | 1 | 1 | 2 | equivocal | 4,9 | equivocal | 7,00 | positive | 0,73 | negative |
| 28 | 33 | 64 | R | 8500/3 | 2 | 0 | 2 | 15 | 1 | 1 | 0 | n.a. | 2,2 | negative | 2,00 | negative | 0,33 | negative |
| 29 | 34 | 54 | R | 8500/3 | 2 | 0 | 3 | 30 | 1 | 1 | 0 | n.a. | 2,8 | negative | 2,00 | negative | 0,36 | negative |
ERBB2 CN or NAC values obtained by MIP or NGS/NAC were tightly correlated with ERBB2 CN values obtained by FISH (Spearman correlation coefficients rs > 0.890, P < 0.001) (Figure 2). Counting of ERBB2 FISH signals in the tumor cells and ERBB2 CN values obtained by MIP showed a particularly tight correlation (rs = 0.940, P < 0.001) and an excellent agreement on a case-by-case basis (Figure 2A). FISH obtained slightly higher CN values in those cases with high level amplification (Figure 2A).
Figure 2. HER2/ERBB2 CN detection by three methods (FISH, MIP, NGS/NAC).
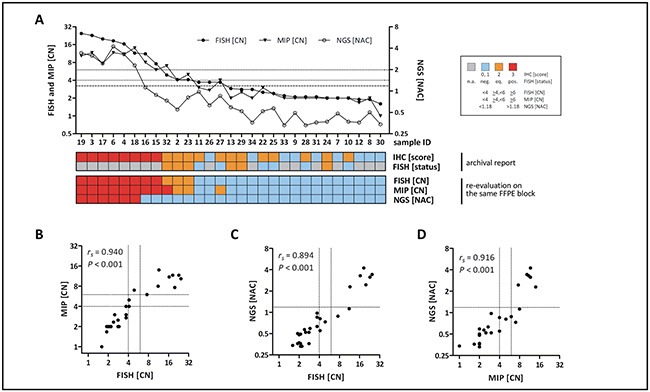
A. Diagrammatic representation of all cases ordered by FISH-based CN. Dotted lines indicate thresholds implemented for FISH and MIP. The dashed line indicates the threshold implemented fro NGS/NAC. B. Correlation of HER2/ERBB2 CN values obtained by FISH and MIP C. Correlation of HER2/ERBB2 CN values obtained by FISH and NGS/NAC D. Correlation of HER2/ERBB2 CN values obtained by MIP and NGS/NAC. Dotted lines indicate thresholds used to classify HER2 as negative, positive or equivocal.
Next, the HER2 status of each BC was categorized as negative, positive or equivocal, using ASCO/CAP 2013 CN thresholds for FISH and MIP data [4]. For NGS/NAC values, a provisional cutoff was implemented as detailed in the material and methods section. Nearly perfect concordance was observed for HER2 classification by FISH versus MIP (93% concordant classifications, κ = 0.87) (Table 2). Substantial concordance was observed for HER2 classification by FISH versus NGS/NAC (83% concordant classifications, κ= 0.62) (Table 2).
Table 2. Concordance of ERBB2 classification by CN.
| FISH | MIP | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| neg | eq | pos | concordance | kappa | neg | eq | pos | concordance | kappa | |
| MIP | 27/29 (93%) | 0.87 | ||||||||
| neg | 17 | 0 | 0 | |||||||
| eq | 1 | 2 | 0 | |||||||
| pos | 0 | 1 | 8 | |||||||
| NGS | 24/29 (83%) | 0.62 | 23/29 (79%) | 0.56 | ||||||
| neg | 18 | 3 | 2 | 17 | 3 | 3 | ||||
| eq | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| pos | 0 | 0 | 6 | 0 | 0 | 6 | ||||
DISCUSSION
This is the first study, which determined agreement between MIP- or NGS/NAC-based ERBB2 CN assessment and FISH-based ERBB2 CN assessment in BC. Notably, MIP achieved nearly perfect agreement with FISH. Although this study was limited to comparatively few cases, our finding renders MIP a strong candidate for routine diagnostic application. A practical advantage of MIP is that ASCO/CAP CN thresholds are directly applicable to the data format generated by MIP.
NGS/NAC showed a slightly poorer performance. Two cases (IDs #15 and #16), were HER2/ERBB2-positive by IHC (3+), FISH, and MIP, but were classified as HER2-negative by NGS/NAC. Notably, these cases were closely above the CN threshold implemented for FISH and MIP and just below the CN threshold used for NGS/NAC. Of six cases showing an equivocal HER2 status by routine diagnostic assessment (IHC 2+/FISH-equivocal, namely IDs #32, #2, #23, #13, #29, and #24), three were confirmed as HER2 equivocal by repeated FISH in the context of this study. Two cases were confirmed as HER2 equivocal by MIP as well (IDs #2 and #23), one was assessed as HER2-positive by MIP (ID #32) showing a CN of 7 according to the TuScan algorithm, whereas positivity is defined as a CN ≥6. However, all these six cases were HER2-negative by NGS/NAC. Thus, NGS/NAC, as performed in this study, had a limited sensitivity for detection of borderline CN alterations and low level ERBB2 amplification. The performance of HER2 classification by NGS/NAC may be improved by fine-tuning of the NAC threshold defining ERBB2 positivity. This, however, requires additional studies in larger cohorts of HER2-positive BCs and HER2-negative controls. Clinical patient selection for HER2-targeted therapy should not be based on NGS/NAC analysis to this point.
An advantage of MIP and NGS/NAC is the possibility to determine ERBB2 CN and hot spot mutations in cancer related genes, such as PIK3CA or TP53, simultaneously (data no shown). Moreover, MIP assays generate genome-wide CN information. This may become clinically relevant, because distinct somatic mutations and CN variations may predict resistance to HER2-targeting therapy [25, 26]. The ability to gain genome wide CN variation information makes MIP a very strong method for ERBB2 CN classification and probably for patient stratification too. A potential disadvantage of MIP and NGS/NAC is that intratumoral heterogeneity for ERBB2 amplification could result in false-negative findings. According to ASCO/CAP guidelines, ERBB2 amplification in just 20 adjacent BC cells suffices to establish a HER2-positive status, even if all other parts of the tumor are negative for HER2 protein expression and ERBB2 amplification [4]. No such intratumoral heterogeneity was observed in the BC cases included in this study. However, DNA extraction-based methods certainly fail to classify such cases as ERBB2-positive. Intratumoral heterogeneity is often stressed in debates concerning in situ and extraction methods. Importantly, HER2-positive subclones do not dictate the course of the disease [15, 27].
BCs with an equivocal test result by IHC and ISH account for 5% to 15% of all BC cases [6–12]. ASCO/CAP guidelines warrant repeated testing by IHC or ISH in these instances and preclude alternative test types, such as mRNA-based ERBB2 expression assays [4]. Variable results and occasional misclassifications have been reported for Oncotype DX, a 21-gene-qRT-PCR expression test including ERBB2 [14, 16]. ERBB2 mRNA analysis has remained at least partly controversial, despite individual studies have documented substantial to almost perfect overall agreement between HER2 assessment by mRNA expression assays and conventional methods (κ= 0.73 – 0.84) [15, 17–19]. Detection of ERBB2 gene amplification is also possible by RNA in situ quantification using RNAscope and was shown to be superior to qPCR in cases with equivocal FISH results or intratumoral heterogeneity [28, 29]. Given the problematic acceptance of HER2 classification by mRNA expression analysis, MIP would represent an ideal ancillary test method, especially for cases with a HER2-equivocal status. First, MIP and ISH assess the same substrate (DNA). Second, ERBB2 CN assessment by ISH is limited to what is feasible to count, but MIP pools ten thousands of tumor cells and has the power to objectify or rectify prior IHC/ISH test result. In summary, this is the first study showing that MIP facilitates precise ERBB2 CN assessment in BC. MIP should be considered as an ancillary test type for clinical HER2 diagnostics.
MATERIALS AND METHODS
BC specimens and DNA extraction
Formalin-fixed paraffin-embedded (FFPE) BC resection specimens were selected based on the HER2 status established by ASCO 2013 guideline-conform routine diagnostic assessment (RDA). To obtain a homogenous sample collection, cases were restricted to estrogen receptor (ER)-positive BCs. To support subsequent correlation analyses for ERBB2 CN, the series was evenly composed of HER2-positive, HER2-negative and HER2-equivocal cases (Table 3). All specimens were retrieved from the archive of the Institute of Pathology of the Hannover Medical School according to the guidelines of the local ethics committee and were made anonymous for scientific purposes. Subsequently, ERBB2 CN was assessed by FISH, MIP and NGS on the very same FFPE tissue block for all three assays. The HER2 IHC status was adopted from archival reports (Figure 1). For MIP and NGS, total DNA was extracted from macrodissected invasive tumor tissue (n=16 sections per case, 8 μm each). To guide macrodissection, an experienced pathologist (m.c.) encircled the entire invasive tumor with a pen on an extra HE-stained section taken from the middle of the section series cut for DNA extraction. Areas with carcinoma in situ were spared. DNA extraction was carried out with the DNeasy blood and tissue kit (Qiagen, Hilden, Germany) according to the manufacturers recommendations, with slight modifications. In brief, the xylol/ethanol proportion was slightly modified to1200 μl (instead of1000 μl) and, following resuspension of the pellet in ALT buffer, an additional incubation for 15 min at 98°C was included. Moreover, following proteinase K digest, an additional incubation in RNase A (250 μg/ml, AppliChem, Darmstadt, Germany) for 30 min at 37°C was included. DNA amount was quantified with a Qubit fluorometer (Thermo Fisher Scientific, Waltham, MA., U.S.A). A total of 29 out of 34 initially selected cases yielded sufficient DNA amount and quality for all assays (Table 3). An additional set of n=12 FFPE normal tonsils were included as controls for NGS/NAC analysis (see below).
Table 3. BC characteristics.
| number | percent | |
|---|---|---|
| all cases | 29 | 100 |
| age | ||
| <60 | 17 | 57 |
| >60 | 12 | 43 |
| pT stage | ||
| pT1 | 11 | 38 |
| pT2 | 11 | 38 |
| pT3/4 | 6 | 21 |
| pTx | 1 | 3 |
| pN stage | ||
| pN0 | 19 | 65 |
| pN1+ | 6 | 21 |
| pNx | 4 | 14 |
| grade | ||
| G1/G2 | 7 | 24 |
| G3 | 22 | 76 |
| ER | ||
| negative | 0 | 0 |
| positive | 29 | 100 |
| PR | ||
| negative | 4 | 14 |
| positive | 25 | 86 |
| HER2* | ||
| negative (IHC 0, 1+, FISH n.a.) | 9 | 31 |
| negative (IHC 2+, FISH -) | 6 | 21 |
| equivocal (IHC 2+, FISH equivoval) | 6 | 21 |
| positive (IHC 3+, FISH n.a.) | 8 | 27 |
HER2 status from archival reports
Fluorescence in situ hybridization (FISH)
FISH was performed on an extra whole slide FFPE sections (5 μm) using the Vysis PathVysion HER2 probe (Abbott, Wiesbaden, Germany). Two observer independently quantified ERBB2 signals in 50 tumor cells each. Clusters of ERBB2 signals were scored with an estimated ERBB2 CN per cluster. The average ERBB2 CN per cell was calculated as the sum of the counts of the two observers divided by 100. CN thresholds used for the definition of a negative, equivocal or positive HER2 status were <4, ≥4 to <6 and ≥6, as recommended by ASCO/CAP 2013 guidelines [4].
Molecular inversion probe (MIP) DNA array analysis
MIP array analysis was performed with 80 ng DNA and OncoScan® arrays following protocols provided by manufacturer (Affymetrix, Santa Clara, (CA), USA). Briefly, samples were split to separate (A/T) and (G/C) channels. After circularization, MIPs were linearized, cleaved and were then amplified by PCR. Amplicons were cleaved into two fragments (44 bp) with HaeIII. DNA fragments were subsequently hybridized to OncoScan® arrays at 58°C for 18 h. Next, arrays were stained and washed using the GeneChip® Fluidics station 450, and were scanned using the GeneChip® scanner 3000 7G (Affymetrix). Array fluorescence intensity (CEL) files were generated with Affymetrix® GeneChip® Command Console® (AGCC, Affymetrix). CEL files were processed with OncoScan Console software version 1.3.0.39 to produce OSCHP files and QC metrics. Samples passing QC criteria (MAPD ≤ 0.3, ndSNPQC ≥ 26) were further analyzed using OncoScan® assay SM calls and Chromosome Analyses Suite (ChAS) version 3.1.0.15 (r9069) for CN variation. The ERBB2 CN was determined with the TuScan algorithm. Based on B-allele frequencies (BAFs) and log2-ratios, the TuScan algorithm provides CN values adjusted for the estimated ploidy and percentage of aberrant cells included in the sample. However, if the sample is highly heterogeneous, TuScan provides average CN values. CN thresholds used for the definition of a negative, equivocal or positive HER2 status were adopted from ASCO/CAP 2013 ISH guidelines. A ERBB2 CN of <4 was considered negative, a CN of ≥4 to <6 was considered equivocal and a CN of ≥6 was considered positive [4]. The complete data series is deposited at GEO under the accession number GSE83916 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE83916).
Next generation sequencing (NGS) and normalized amplicon coverage (NAC) analysis
Library preparation was performed with Ion AmpliSeq library kit 2.0. Quantification of prepared libraries was conducted by qPCR using the Ion Library Quantification Kit. For template preparation using the Ion OneTouch 2 instrument, 12 BC samples were pooled (100 pM each). Sequencing was performed with Ion PGM Hi-Q Kit v2 and using 318 v2 Chips. Analyses of sequencing raw data were performed with Torrent server software (version 4.2.1). The mean sequencing depths for the n=29 BCs was 3869 reads. The mean sequencing depths for the n=12 tonsil specimens, which served to define a NAC cutoff for ERBB2 positivity, was 2484 reads. For the calculation of the normalized amplicon coverage, the read count for a given amplicon was divided by the mean read count for this sample. The mean value for the three amplicons covering ERBB2 was calculated. The same value was calculated for the 8 amplicons covering TP53 and the ratio of these two values was computed. The mean value plus two times the standard deviation obtained from the n=12 tonsil samples (=1.18) was implemented as a threshold defining an ERBB2 CN gain.
Statistics
Correlation analyses were performed with GraphPad Prims software (version 5, Graph Pad Inc. San Diego, U.S.A). Concordance of ERBB2 classification was assessed with JMP Pro10 software (SAS, Marlow, UK) and Cohens unweighted κ [24].
Acknowledgments
The authors gratefully acknowledge Hildegard Frye-Boukhriss and Henriette Christgen for excellent technical assistance.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicts of interest to disclose.
REFERENCES
- 1.Bilous M, Morey AL, Armes JE, Bell R, Button PH, Cummings MC, Fox SB, Francis GD, Waite B, McCue G, Raymond WA, Robbins PD, Farshid G. Assessing HER2 amplification in breast cancer: findings from the Australian In Situ Hybridization Program. Breast Cancer Res Treat. 2012;134:617–624. doi: 10.1007/s10549-012-2093-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Piccart-Gebhart MJ, Procter M, Leyland-Jones B, Goldhirsch A, Untch M, Smith I, Gianni L, Baselga J, Bell R, Jackisch C, Cameron D, Dowsett M, Barrios CH, Steger G, Huang CS, Andersson M, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353:1659–1672. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 3.Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE, Jr, Davidson NE, Tan-Chiu E, Martino S, Paik S, Kaufman PA, Swain SM, Pisansky TM, Fehrenbacher L, Kutteh LA, Vogel VG, Visscher DW, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 4.Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M, Fitzgibbons P, Hanna W, Jenkins RB, Mangu PB, Paik S, Perez EA, Press MF, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31:3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 5.McCullough AE, Dell'orto P, Reinholz MM, Gelber RD, Dueck AC, Russo L, Jenkins RB, Andrighetto S, Chen B, Jackisch C, Untch M, Perez EA, Piccart-Gebhart MJ, Viale G. Central pathology laboratory review of HER2 and ER in early breast cancer: an ALTTO trial [BIG 2-06/NCCTG N063D (Alliance)] ring study. Breast Cancer Res Treat. 2014;143:485–492. doi: 10.1007/s10549-013-2827-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varga Z, Noske A. Impact of Modified 2013 ASCO/CAP Guidelines on HER2 Testing in Breast Cancer. One Year Experience. PLoS One. 2015;10:e0140652. doi: 10.1371/journal.pone.0140652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim TH, Lim AS, Thike AA, Tien SL, Tan PH. Implications of the Updated 2013 American Society of Clinical Oncology/College of American Pathologists Guideline Recommendations on Human Epidermal Growth Factor Receptor 2 Gene Testing Using Immunohistochemistry and Fluorescence In Situ Hybridization for Breast Cancer. Arch Pathol Lab Med. 2016;140:140–147. doi: 10.5858/arpa.2015-0108-OA. [DOI] [PubMed] [Google Scholar]
- 8.Fan YS, Casas CE, Peng J, Watkins M, Fan L, Chapman J, Ikpatt OF, Gomez C, Zhao W, Reis IM. HER2 FISH classification of equivocal HER2 IHC breast cancers with use of the 2013 ASCO/CAP practice guideline. Breast Cancer Res Treat. 2016;155:457–462. doi: 10.1007/s10549-016-3717-z. [DOI] [PubMed] [Google Scholar]
- 9.Shah MV, Wiktor AE, Meyer RG, Tenner KS, Ballman KV, Green SJ, Sukov WR, Ketterling RP, Perez EA, Jenkins RB. Change in Pattern of HER2 Fluorescent in Situ Hybridization (FISH) Results in Breast Cancers Submitted for FISH Testing: Experience of a Reference Laboratory Using US Food and Drug Administration Criteria and American Society of Clinical Oncology and College of American Pathologists Guidelines. J Clin Oncol. 2016 doi: 10.1200/JCO.2015.61.8983. in press. [DOI] [PubMed] [Google Scholar]
- 10.Press MF, Sauter G, Buyse M, Fourmanoir H, Quinaux E, Tsao-Wei DD, Eiermann W, Robert N, Pienkowski T, Crown J, Martin M, Valero V, Mackey JR, Bee V, Ma Y, Villalobos I, et al. HER2 Gene Amplification Testing by Fluorescent In Situ Hybridization (FISH): Comparison of the ASCO-College of American Pathologists Guidelines With FISH Scores Used for Enrollment in Breast Cancer International Research Group Clinical Trials. J Clin Oncol. 2016 doi: 10.1200/JCO.2016.66.6693. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Overcast WB, Zhang J, Zynger DL, Tozbikian GH. Impact of the 2013 ASCO/CAP HER2 revised guidelines on HER2 results in breast core biopsies with invasive breast carcinoma: a retrospective study. Virchows Arch. 2016;469:203–212. doi: 10.1007/s00428-016-1951-8. [DOI] [PubMed] [Google Scholar]
- 12.Singh K, Tantravahi U, Lomme MM, Pasquariello T, Steinhoff M, Sung CJ. Updated 2013 College of American Pathologists/American Society of Clinical Oncology (CAP/ASCO) guideline recommendations for human epidermal growth factor receptor 2 (HER2) fluorescent in situ hybridization (FISH) testing increase HER2 positive and HER2 equivocal breast cancer cases; retrospective study of HER2 FISH results of 836 invasive breast cancers. Breast Cancer Res Treat. 2016;157:405–411. doi: 10.1007/s10549-016-3824-x. [DOI] [PubMed] [Google Scholar]
- 13.Gunn S, Yeh IT, Lytvak I, Tirtorahardjo B, Dzidic N, Zadeh S, Kim J, McCaskill C, Lim L, Gorre M, Mohammed M. Clinical array-based karyotyping of breast cancer with equivocal HER2 status resolves gene copy number and reveals chromosome 17 complexity. BMC Cancer. 2010;10:396. doi: 10.1186/1471-2407-10-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dabbs DJ, Klein ME, Mohsin SK, Tubbs RR, Shuai Y, Bhargava R. High false-negative rate of HER2 quantitative reverse transcription polymerase chain reaction of the Oncotype DX test: an independent quality assurance study. J Clin Oncol. 2011;29:4279–4285. doi: 10.1200/JCO.2011.34.7963. [DOI] [PubMed] [Google Scholar]
- 15.Lehmann-Che J, Amira-Bouhidel F, Turpin E, Antoine M, Soliman H, Legres L, Bocquet C, Bernoud R, Flandre E, Varna M, de Roquancourt A, Plassa LF, Giacchetti S, Espie M, de Bazelaire C, Cahen-Doidy L, et al. Immunohistochemical and molecular analyses of HER2 status in breast cancers are highly concordant and complementary approaches. Br J Cancer. 2011;104:1739–1746. doi: 10.1038/bjc.2011.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christgen M, Harbeck N, Gluz O, Nitz U, Kreipe HH. Recognition and Handling of Discordant Negative Human Epidermal Growth Factor Receptor 2 Classification by Oncotype DX in Patients With Breast Cancer. J Clin Oncol. 2012;30:3313–3314. doi: 10.1200/JCO.2012.42.1990. [DOI] [PubMed] [Google Scholar]
- 17.Denkert C, Huober J, Loibl S, Prinzler J, Kronenwett R, Darb-Esfahani S, Brase JC, Solbach C, Mehta K, Fasching PA, Sinn BV, Engels K, Reinisch M, Hansmann ML, Tesch H, von Minckwitz G, et al. HER2 and ESR1 mRNA expression levels and response to neoadjuvant trastuzumab plus chemotherapy in patients with primary breast cancer. Breast Cancer Res. 2013;15:R11. doi: 10.1186/bcr3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viale G, Slaets L, Bogaerts J, Rutgers E, van't Veer L, Piccart-Gebhart MJ, de Snoo FA, Stork-Sloots L, Russo L, Dell'Orto P, van den Akker J, Glas A, Cardoso F. High concordance of protein (by IHC), gene (by FISH; HER2 only), and microarray readout (by TargetPrint) of ER, PgR, and HER2: results from the EORTC 10041/BIG 03-04 MINDACT trial. Ann Oncol. 2014;25:816–823. doi: 10.1093/annonc/mdu026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Viale G, Slaets L, de Snoo FA, Bogaerts J, Russo L, Van't Veer L, Rutgers EJ, Piccart-Gebhart MJ, Stork-Sloots L, Dell'Orto P, Glas AM, Cardoso F. Discordant assessment of tumor biomarkers by histopathological and molecular assays in the EORTC randomized controlled 10041/BIG 03-04 MINDACT trial breast cancer : Intratumoral heterogeneity and DCIS or normal tissue components are unlikely to be the cause of discordance. Breast Cancer Res Treat. 2016;155:463–469. doi: 10.1007/s10549-016-3690-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahn S, Hong M, Van Vrancken M, Lyou YJ, Kim ST, Park SH, Kang WK, Park YS, Jung SH, Woo M, Lee J, Kim KM. An Counter CNV Assay to Detect HER2 Amplification: A Correlation Study with Immunohistochemistry and In Situ Hybridization in Advanced Gastric Cancer. Mol Diagn Ther. 2016;20:375–383. doi: 10.1007/s40291-016-0205-4. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Cottman M, Schiffman JD. Molecular inversion probes: a novel microarray technology and its application in cancer research. Cancer Genet. 2012;205:341–355. doi: 10.1016/j.cancergen.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 22.Foster JM, Oumie A, Togneri FS, Vasques FR, Hau D, Taylor M, Tinkler-Hundal E, Southward K, Medlow P, McGreeghan-Crosby K, Halfpenny I, McMullan DJ, Quirke P, Keating KE, Griffiths M, Spink KG, et al. Cross-laboratory validation of the OncoScan(R) FFPE Assay, a multiplex tool for whole genome tumour profiling. BMC Med Genomics. 2015;8:5. doi: 10.1186/s12920-015-0079-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh RR, Patel KP, Routbort MJ, Aldape K, Lu X, Manekia J, Abraham R, Reddy NG, Barkoh BA, Veliyathu J, Medeiros LJ, Luthra R. Clinical massively parallel next-generation sequencing analysis of 409 cancer-related genes for mutations and copy number variations in solid tumours. Br J Cancer. 2014;111:2014–2023. doi: 10.1038/bjc.2014.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- 25.Cescon DW, Bedard PL. PIK3CA genotype and treatment decision in human epidermal growth factor receptor 2-positive breast cancer. J Clin Oncol. 2015;33:1318–1321. doi: 10.1200/JCO.2014.59.3160. [DOI] [PubMed] [Google Scholar]
- 26.Gogas H, Kotoula V, Alexopoulou Z, Christodoulou C, Kostopoulos I, Bobos M, Raptou G, Charalambous E, Tsolaki E, Xanthakis I, Pentheroudakis G, Koutras A, Bafaloukos D, Papakostas P, Aravantinos G, Psyrri A, et al. MYC copy gain, chromosomal instability and PI3K activation as potential markers of unfavourable outcome in trastuzumab-treated patients with metastatic breast cancer. J Transl Med. 2016;14:136. doi: 10.1186/s12967-016-0883-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moeder CB, Giltnane JM, Harigopal M, Molinaro A, Robinson A, Gelmon K, Huntsman D, Camp RL, Rimm DL. Quantitative justification of the change from 10% to 30% for human epidermal growth factor receptor 2 scoring in the American Society of Clinical Oncology/College of American Pathologists guidelines: tumor heterogeneity in breast cancer and its implications for tissue microarray based assessment of outcome. J Clin Oncol. 2007;25:5418–5425. doi: 10.1200/JCO.2007.12.8033. [DOI] [PubMed] [Google Scholar]
- 28.Wang Z, Portier BP, Gruver AM, Bui S, Wang H, Su N, Vo HT, Ma XJ, Luo Y, Budd GT, Tubbs RR. Automated quantitative RNA in situ hybridization for resolution of equivocal and heterogeneous ERBB2 (HER2) status in invasive breast carcinoma. J Mol Diagn. 2013;15:210–219. doi: 10.1016/j.jmoldx.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 29.Zhu Y, Lu D, Lira ME, Xu Q, Du Y, Xiong J, Mao M, Chung HC, Zheng G. Droplet digital polymerase chain reaction detection of HER2 amplification in formalin fixed paraffin embedded breast and gastric carcinoma samples. Exp Mol Pathol. 2016;100:287–293. doi: 10.1016/j.yexmp.2015.11.027. [DOI] [PubMed] [Google Scholar]


