Abstract
The structure-specific nuclease human flap endonuclease-1 (hFEN1) plays a key role in DNA replication and repair and may be of interest as an oncology target. We present the first crystal structure of inhibitor-bound hFEN1 and show a cyclic N-hydroxyurea bound in the active site coordinated to two magnesium ions. Three such compounds had similar IC50 values but differed subtly in mode of action. One had comparable affinity for protein and protein–substrate complex and prevented reaction by binding to active site catalytic metal ions, blocking the unpairing of substrate DNA necessary for reaction. Other compounds were more competitive with substrate. Cellular thermal shift data showed engagement of both inhibitor types with hFEN1 in cells with activation of the DNA damage response evident upon treatment. However, cellular EC50s were significantly higher than in vitro inhibition constants and the implications of this for exploitation of hFEN1 as a drug target are discussed.
Introduction
Flap endonuclease 1 (FEN1) is the prototypical member of the 5′ nuclease superfamily,1,2 whose activities span a range of cellular pathways involved in DNA replication and genome maintenance.3,4 FEN1 is a structure-selective metallonuclease essential for Okazaki fragment maturation through efficient removal of 5′-flaps resulting from strand displacement during lagging-strand synthesis.5,6 This reaction produces nicked DNA suitable for ligation, thereby ensuring maintenance of genomic fidelity. FEN1 is also involved in long-patch base excision repair7–9 (LP-BER), amongst other pathways.
Given its critical replicative function, it is not surprising that FEN1 overexpression is characterized in multiple cancer types10–13 such that it has been suggested as both a biomarker relating to prognosis and disease progression, and a potential therapeutic target. Target validation studies have focused either on chemosensitization14,15 or synthetic lethal interactions16–19 with established oncogenes. Synthetic lethality arises when loss of function of either gene of an interacting pair is not cytotoxic, but mutation or inhibition of both does cause cell death; hence, targeting interacting partners of mutated genes in cancer offers potential for selective killing of cancer cells.
Therapeutic interest in FEN1 arises from its known synthetic lethal interactions with several genes frequently mutated in cancers.16,17,20 FEN1 inhibition selectively impairs proliferation of colon cancer cells deficient in Cdc4 and Mre11a,16,18 both frequently mutated in colorectal cancers. FEN1 has also emerged as a potential chemosensitizing target due to its role in LP-BER17 since it is critical for repair of MMS (methyl methanesulfonate)-induced alkylation damage,21 and its knockdown or inhibition increases sensitivity to TMZ (temozolomide) in glioblastoma13 and colorectal cancer14,16,18 cell lines.
This considerable interest in human FEN1 (hFEN1) as a drug target has prompted development of high-throughput screening procedures22,23 and the discovery of an N-hydroxyurea based series of hFEN1 inhibitors.24 We investigated the specificity and mode of action of these compounds and found they prevented access of the scissile phosphate diester of substrate DNA to catalytic metal ions. We also demonstrated cellular activity and target engagement in live cells, leading to activation of the DNA damage response and apoptosis.
Results
N-Hydroxyurea hFEN1 inhibitors bind catalytic site metals
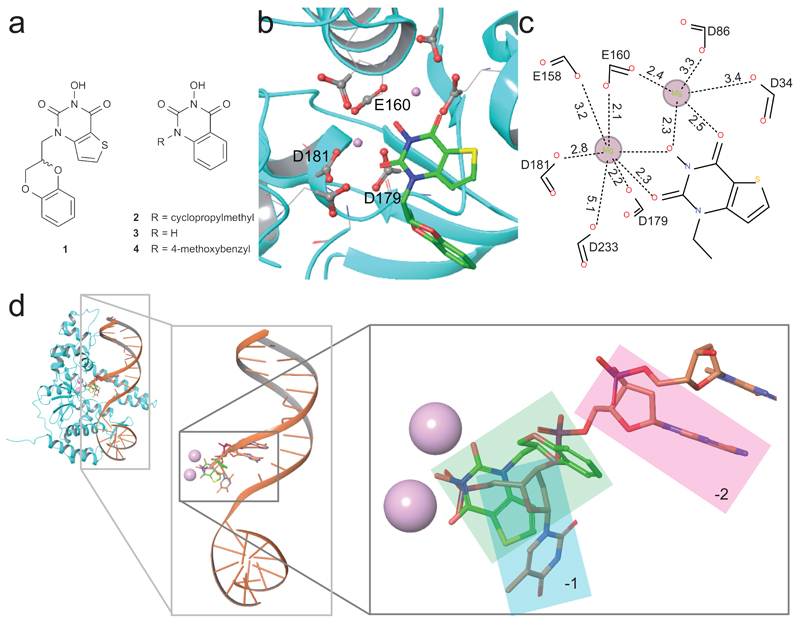
Inhibitor 124 (Figure 1a) was co-crystallized with hFEN1–Mg2+ truncated after residue 336 (hFEN1-336Δ), which retains all catalytic features but lacks the flexible 44 amino acid C-terminus.25,26 The crystal structure of the hFEN1-336Δ–inhibitor complex (Figure 1b) was solved at 2.84 Å resolution (Supplementary Results, Supplementary Table 1 and Supplementary Figure 1; PDB ID 5FV7) and resembled a kidney bean with the active site and requisite divalent metal ions residing at the indentation. The structure in the presence of the active site-bound inhibitor closely resembled that of hFEN1 in complex with proliferating cell nuclear antigen (PCNA).27 As with the PCNA-bound structure, no density was observed for the helical arch (α4 and α5) and α2-α3 loop regions, which are visible when co-crystallized with substrate or product DNA.2
Figure 1. Compounds used in this study and crystal structure of hFEN1-336Δ in complex with compound 1.
(a) Schematic illustration of compounds 1–4 that are inhibitors of hFEN1 phosphate diester hydrolysis. (b) Structure of hFEN1-336Δ nuclease active site (PDB ID 5FV7) showing the seven highly-conserved acidic residues (grey and red spheres represent carbonyl carbon and oxygen atoms, respectively), the two bound magnesium ions (pink spheres), and compound 1. (c) Schematic representation of the metal-coordination spheres of the two active site magnesium ions with distances reported in Ångstrom. (d) Structure of hFEN1-336Δ in complex with product DNA (PDB ID 3Q8K) superimposed with the hFEN1-336Δ in complex with compound 1 (protein not shown) to show that the inhibitor and terminal nucleotide of the product DNA interact with the divalent magnesium ions and share same pocket created by the protein. Metals are shown as pink spheres, terminal 5′ nucleotide (–1) highlighted in cyan box, penultimate nucleotide of the product DNA (–2) highlighted in the pink box, and compound 1 highlighted in the green box.
The inhibitor was situated in the protein’s nuclease active site with the N-hydroxyurea moiety directly coordinating two Mg2+ ions positioned 4.5 Å apart (Figure 1b), anchored by inner-sphere metal-coordinating contacts from carboxylates of E160, D179 and D181 and outer-sphere or water-mediated contacts from D34, D86, E158 and D233 (Figure 1c). The thiophene ring of the inhibitor filled a small hydrophobic pocket formed by M37, Y40 and V133, and the sulfur of M37 exhibited a short-distance (4Å) favorable contact to the electron deficient pyrimidine-2,4-dione ring of the ligand. The 2,3-dihydrobenzo[b][1,4]dioxine sidechain contacted M37 and Y40, though these contacts were less directional and mostly hydrophobic in nature. It was evident that different binding poses in the active site are possible for the N-hydroxyurea series of inhibitors, which goes some way to rationalizing the reported SAR.24 The relatively weak nature of protein contacts with the sidechain (N1-substituent) explained the modest improvement in IC50 values seen for compounds modified at this position.24 It is also understandable how substitutions restricting the conformational freedom of the sidechain—for example, introduction of a methyl group at the 7-position of the thieno[3,2-d]pyrimidine-2,4-dione system of 1—would significantly reduce binding affinity and therefore increase IC50, as is reported.24
Inhibitor binding pose suggests a possible mode-of-action
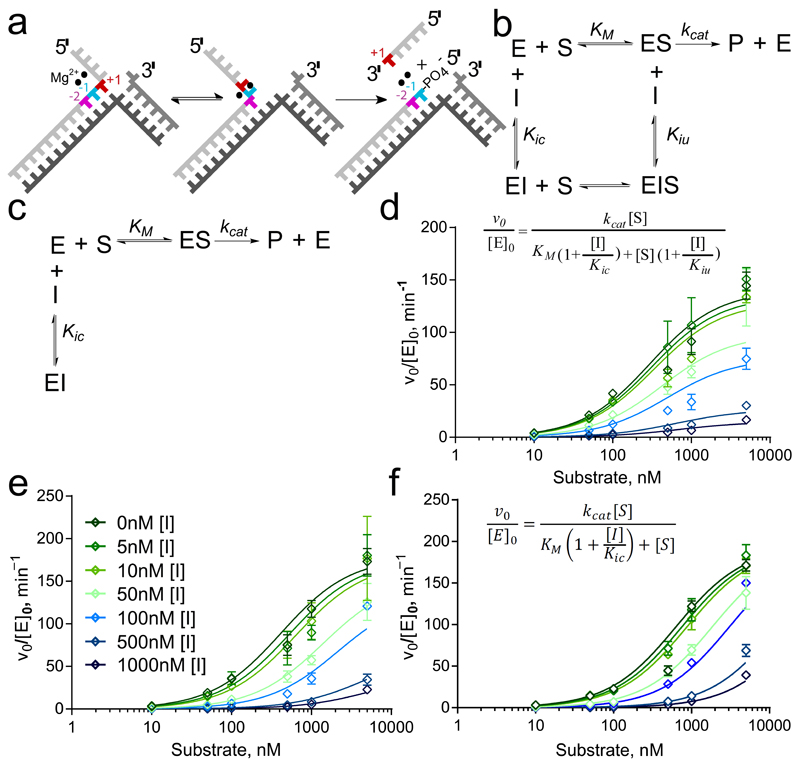
Coordination of 1 to the metal ions that catalyze specific phosphodiester hydrolysis of the substrate suggested a mode of action for this inhibitor. We modelled ternary protein–inhibitor–DNA complexes using the present hFEN1-336Δ–inhibitor structure together with the published hFEN1-336Δ–product DNA complex2 (Figure 1d). Alignment of product-bound and ligand-bound structures indicated that the inhibitor and the phosphate monoester of the product DNA strand both co-locate to bind the metal ions. Conversely, in the hFEN1-336Δ–substrate DNA complex2, the scissile bond is not in contact with active site metal ions because the DNA is base-paired. It is assumed a pre-reactive complex forms initially that requires the end of the DNA duplex to unpair and bind to metal ions as a prerequisite for cleavage.1,2,28 Hence, it was considered plausible that substrate could bind in the presence of inhibitor, but that this prevents DNA from accessing the catalytic metals as required for hydrolysis to occur (Figure 2a). An alternative hypothesis was that the inhibitor precludes DNA binding, although the compound was bound far from the other two main areas of protein–DNA interaction (K+/H2TH motif and 3′-flap binding pocket). We undertook further work to characterize the hFEN1-inhibitor interaction and establish whether the N-hydroxyurea inhibitors compete with substrate DNA binding.
Figure 2. Differences in inhibition characteristics of the compounds.
(a) hFEN1-catalyzed reaction schematic showing double nucleotide unpairing at positions +1 and −1 (numbering relative to scissile phosphate). (b,c) Reaction schemes of mixed inhibition (b) and competitive inhibition (c) models. In each case, E, S, I and P represent enzyme, substrate, inhibitor and product, respectively. Kic is the dissociation constant of I from free enzyme (competitive with substrate) and Kiu is the dissociation constant of I from ES complex (uncompetitive). (d–f) Nonlinear regression plots of normalized initial rates of reaction vs. substrate concentration (open diamonds) for substrate DF1 at varying concentrations of compounds 1 (d; inset shows equation for mixed inhibition model), 2 (e; inset shows legend correlating color/symbol to inhibitor concentration) and 4 (f; inset shows equation for competitive inhibition model). Error bars represent standard errors from global fitting of combined data from two triplicate experiments (fits to alternative models are shown in Supplementary Figures S7–S9).
Inhibitor binding to hFEN1 requires magnesium ions
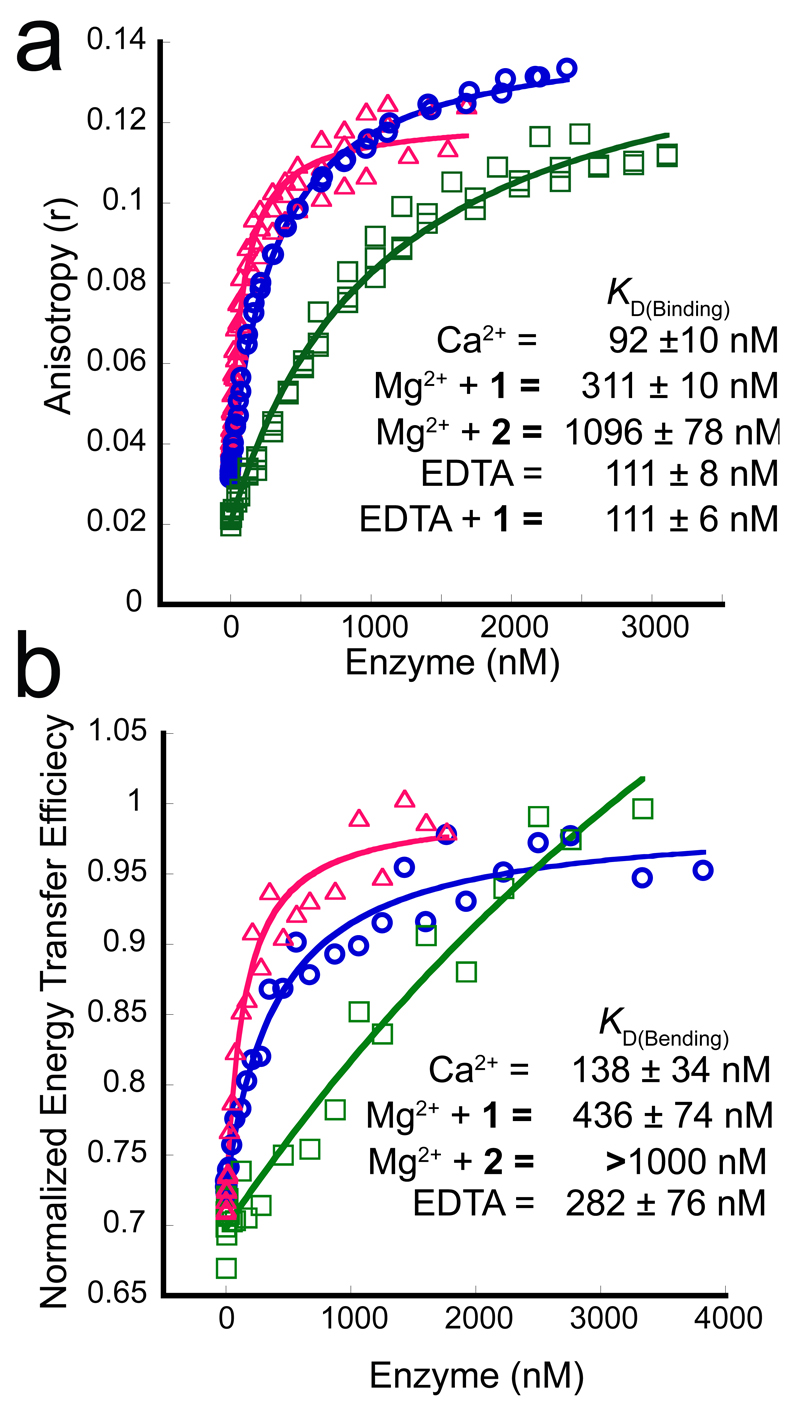
We quantified the interaction of 1, and related analogs 2 and 322 bearing a smaller or no sidechain (Figure 1a), with the substrate-free protein using isothermal titration calorimetry (ITC; Supplementary Table 2). Similar dissociation constants (KD) were obtained for 1 and 2 in the presence of Mg2+ with either hFEN1-336Δ (Supplementary Figure 2) or full-length hFEN1 (Supplementary Figure 3a,b) but the KD of 3 was approximately 10-fold higher, suggesting interactions between the sidechains of 1 and 2 and the protein contribute to binding.
Ca2+ ions are often employed as a nonviable cofactor in biophysical measurements with hFEN1 because they facilitate accommodation of the substrate DNA and its required conformational changes,28,29 but do not support catalysis. In fact, Ca2+ ions are a competitive inhibitor of 5′-nuclease reactions with respect to Mg2+,30,31 implying both ions occupy similar sites on the protein. However, KD values were drastically increased on replacement of Mg2+ with Ca2+ (Supplementary Figure 4), showing the latter did not support inhibitor binding. Thus, in accord with the crystal structure, interaction of 1 and 2 with hFEN1 was specific to the nuclease core domain and required Mg2+. To provide an estimate of residence time, we probed the interaction of 1 with hFEN1-336Δ using surface plasmon resonance (Supplementary Figure 2d) and obtained a dissociation constant similar to ITC with a residence time of 3 min.
Inhibitors bind to both protein and protein–DNA complex
Kinetic experiments were used to characterize hFEN1 inhibition by 1, 2 and 4. We measured rates of hFEN1-336Δ-catalyzed reaction with an optimal endonucleolytic double-flap substrate bearing a 5′-fluorescein label32 (DF1; Figure 2a, and Supplementary Figure 5a). At substrate concentration close to KM (100 nM), IC50 values for all three compounds were similar (Table 1), and a related exonucleolytic substrate gave similar IC50 results (Supplementary Figure 6a,b). Mode of inhibition was determined by globally fitting rates of reaction at varying inhibitor and double-flap substrate concentrations to four inhibition models: competitive, uncompetitive, non-competitive and mixed inhibition (Equations 2–5, online methods).
Table 1.
Kinetic parameters in absence and presence of inhibitors.
| Enzyme | Inhibitor | IC50, nM§ | kcat, min-1 | KM, nM | Kic, nM | Kiu, nM | kSTmax, min–1 | t1/2, min | ΔAICc |
|---|---|---|---|---|---|---|---|---|---|
| hFEN1 | None | n.a. | 165±9 | 20±3 | n.a. | n.a. | 916±49 | 7.57×10−4 | n.a. |
| hFEN1 | 1 | n.d. | n.d. | n.d. | n.d. | n.d. | 0.48±0.04 | 1.43 | n.a. |
| hFEN1 | 2 | n.d. | n.d. | n.d. | n.d. | n.d. | 1.52±0.09 | 0.46 | n.a. |
| hFEN1-336Δ | None | n.a. | 160±10 | 151±16 | n.a. | n.a. | 755±35 | 8.94±10−4 | n.a. |
| hFEN1-336Δ | 1 | 46.4±4.8 | 140±9 | 297±31 | 48±5 | 117±27 | n.d. | n.d. | 24.76* |
| hFEN1-336Δ | 2 | 30.0±6.0 | 182±13 | 422±50 | 17±2 | 306±125 | n.d. | n.d. | 10.21¶ |
| hFEN1-R100A | None | n.a. | n.d. | n.d. | n.d. | n.d. | 0.087±0.003 | 7.94 | n.a. |
| hFEN1-R100A | 1 | n.d. | n.d. | n.d. | n.d. | n.d. | ~4×10−4 | ≤1750 | n.a. |
| hFEN1-R100A | 2 | n.d. | n.d. | n.d. | n.d. | n.d. | ~2×10−3 | ≤360 | n.a. |
| hFEN1-336Δ | 4 | 16.9±1.2 | 194.5±11 | 630.8±53 | 26±2 | n.a. | n.d. | n.d. | Amb. |
IC50 values derived from rates at substrate concentration close to KM (100 nM). kSTmax is maximal reaction rate under single turnover conditions, used to calculate the substrate half-life (t1/2). ΔAICc is the difference between second order (corrected) Akaike Information Criteria values between models; if ≥6, the likelihood the incorrect model was selected is P < 0.0001. ΔAICc for 1* and 2¶ compares non-competitive with mixed-inhibition models and competitive with mixed-inhibition models, respectively. Mixed-inhibition is preferred for both. For 4, competitive inhibition was the only model whose fit was not ambiguous (Amb.).
The uncompetitive model—where the inhibitor can only bind to enzyme–substrate complex—afforded a poor fit for 1, which was unsurprising given the compound’s high affinity for free protein. The competitive model, where binding of inhibitor and substrate are mutually exclusive, also proved unsuitable but the mixed and non-competitive models produced acceptable fits (Figure 2b-d and Supplementary Figure 7). These models both assume the inhibitor can bind to DNA-free and DNA-bound forms of the enzyme, but the non-competitive model (Equation 4) assumes both complexes have equivalent ligand dissociation constants. Allowing dissociation constants to vary (mixed inhibition; Figure 2b and Equation 5) produced a marginally better data fit, yielding near-equivalent dissociation constants for 1(Table 1). Statistical model selection using Aikake’s Information Criteria (AIC) overwhelmingly preferred the mixed inhibition model.
With compound 2, only the competitive (Equation 3) and mixed inhibition models produced acceptable fits (Figure 2e, Supplementary Figure 8). The same statistical criteria (AIC) again favored the mixed model, but in this case the derived dissociation constants (Kic and Kiu) varied by an order of magnitude (Table 1). For compound 4, only the competitive model produced an acceptable fit (Figure 2f, Table 1 and Supplementary Figure 9). Thus, whereas 1, 2 and 4 all bound to hFEN1–Mg2+ with similar efficiency, only 1 showed notable affinity for the enzyme-substrate complex (hFEN1–Mg2+–DNA), binding both DNA-free and DNA-bound forms of the enzyme with comparable dissociation constants.
Evidence for an hFEN1–Mg2+–Inhibitor–DNA complex
To verify formation of a quaternary complex of enzyme–Mg2+–inhibitor–DNA (E–Mg2+–I–DNA), we tested the ability of E–Mg2+–I to form complexes with DNA without significant hydrolysis of the substrate occurring. High concentrations of 1 or 2 (100 μM) slowed the rate of Mg2+-catalyzed reaction 10,000-fold under single-turnover conditions (Table 1 and Supplementary Figure 10), but appreciable substrate cleavage was still seen over the timescale required for biophysical measurements. Because Ca2+ did not support inhibitor binding (Supplementary Figure 4), substituting it in place of Mg2+ as a nonviable cofactor was not applicable. Instead, we employed a previously characterized hFEN1 mutant, R100A. Arg100 is strictly conserved in FEN1 proteins and its mutation to alanine slows reaction 7,000-fold.33 The half-life of substrate with R100A–Mg2+ and inhibitors was sufficiently long to permit measurements without significant product formation (Supplementary Figure 11), and ITC confirmed the mutation did not affect inhibitor binding (Supplementary Table 2).
Both 1 and 2 formed R100A–Mg2+–I–DNA complexes as demonstrated by increases in anisotropy (r) of DF1 substrate upon titration with R100A–Mg2+–I, with r reaching a common limiting value at high enzyme concentration (Figure 3a). Data fitting to a simple binding isotherm revealed similar trends in KD between R100A and its wt equivalent, with which the use of non-catalytic Ca2+ ions was necessary to prevent reaction (Supplementary Figures 11a, 12a–d). Competing away bound, FAM-labeled substrate with its unlabeled equivalent demonstrated specific interaction between R100A and this substrate (Supplementary Figures 5a,b, 12h). Substrate dissociation constants differed between quaternary complexes containing 1 or 2 (Figure 3a): with compound 1, R100A–Mg2+–1 displayed a KD only threefold greater than that for R100A–Ca2+. In contrast, the substrate bound 10-fold more weakly to R100A–Mg2+–2. These results were consistent with 1 having a closer Kiu value relative to Kic than 2, again suggesting 2 was more competitive than mixed in character.
Figure 3. Effect of inhibitors on substrate binding assessed by fluorescence anisotropy (FA) and FRET.
(a) Typical FA titration data for hFEN1-R100A binding DF1 in the presence of 10 mM Ca2+ (magenta, open triangles), 8 mM Mg2+ plus 100 μM compound 1 (blue, open circles) or 8 mM Mg2+ with 100 μM compound 2 (green, open squares); three independent titrations were carried out for all FA binding experiments. (b) Representative curves of typical normalized FRET binding data for DF1 and hFEN1-R100A. Experiments were conducted in triplicate, but only one data set and curve is shown here for each titration. Colours and symbols for each of the three plots are the same as in panel (a).
DNA is bent in complexes with or without inhibitors
hFEN1 possesses two juxtaposed double-stranded DNA binding sites that accommodate double-flap substrate DNA in a conformation with a 100° bend at the junction. To ascertain whether DNA bound similarly in the presence of inhibitor, we examined substrate bending using FRET. We labelled double-flap substrate with a rhodamine-fluorescein dye pair on its respective duplexes, and verified binding to hFEN1 produces an increase in FRET signal34 (Figure 3b and Supplementary Figures 5c-f, 13, 14). Titration of R100A–Ca2+ or R100A–Mg2+–1 into the labeled substrate produced comparable FRET efficiency start and end values (Figure 3b) confirming the enzyme had engaged both DNA binding sites with or without inhibitor. The substrate KD was raised by a factor of three in the presence of 1, whereas substrate binding was much weaker with 2 present (Figure 3b and Supplementary Table 3); hence, these results mirrored those obtained earlier by fluorescence anisotropy.
Inhibitors bound to catalytic metals block DNA unpairing
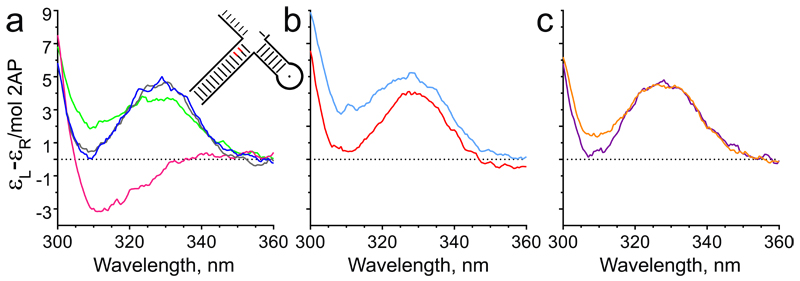
Unpairing of the reacting substrate duplex, which places the target phosphodiester onto active site metal ions, is a prerequisite for hFEN1-catalysed reaction one nucleotide into the double-stranded DNA (Figure 2a).28 This metal ion-dependent conformational change may be monitored using substrates containing a tandem 2-aminopurine (2AP) exciton pair at the –1 and –2 positions of the 5′-flap strand (DF3, Supplementary Figure 5g) by measuring changes in the low energy exciton-coupled CD spectrum resulting from the 2APs, usually in the presence of Ca2+ to prevent reaction.28
In adopting the reactive conformation, the +1 and −1 nucleotides are assumed to become extrahelical whereas the −2 nucleotide remains base-paired. In the absence of active site divalent ions (EDTA added), a strong maximum at 330 nm is observed from the R100A–DNA complex, due to the exciton pair and consistent with substrate remaining base-paired.28 With R100A–Ca2+–DNA, the DNA conformational change reverses the sign of the CD signal producing a deep minimum at 310 nm (Figure 4a). In the presence of 1 or 2, the measured CD signal of R100A–Mg2+–I–DNA did not differ significantly from that observed for R100A–DNA without divalent ions (Figure 4b,c), even though the DNA was assumed to be fully bound under these conditions (10 μM DNA, 12.5 μM R100A). This demonstrated that the inhibitors prevented substrate conformational rearrangements necessary for hydrolysis (Supplementary Figure 15).
Figure 4. N-Hydroxyurea inhibitors prevent FEN1 reaction by blocking substrate unpairing.
CD spectra recorded at pH 7.5 and 20 °C of (a) tandem 2-aminopurine containing substrate DF3 (illustrated schematically as inset, and Supplementary Figure 5g) alone in the presence of 10 mM Ca2+ (blue) or 25 mM EDTA (grey) and the same substrate bound to hFEN1-R100A in the presence of 10 mM Ca2+ (magenta) or 25 mM EDTA (green); (b) DF3 bound to hFEN1-R100A in the presence of Mg2+ plus excess compound 1 (cyan) or EDTA plus excess compound 1 (red); (c) DF3 bound to hFEN1-R100A with excess compound 2 in the presence of Mg2+ (orange) or EDTA (purple). Full DNA sequences are shown in Supplementary Tables 5,6 and Supplementary Figure 5g. Plots in panels a–c are representative of experiments repeated independently three times.
N-Hydroxyurea FEN1 inhibitors also target EXO1
FEN1 is the prototypical member of the structure-specific 5′-nuclease superfamily, also comprising exonuclease 1 (EXO1), gap endonuclease 1 (GEN1) and Xeroderma Pigmentosum complementation group G protein (XPG).1 Exoribonucleases XRN1 and 2 are also suggested members of the superfamily.1 These nucleases all share a similarly-folded nuclease domain with similar active site geometry and full conservation of essential catalytic residues.1,2 Consequently, it has been hypothesized that the substrate selectivity of these proteins stems from strict recognition of their respective DNA substrate structures, followed by double nucleotide unpairing to initiate scissile phosphate diester hydrolysis.1
It is known that hFEN1 inhibitors can exhibit limited but manageable promiscuity towards XPG.24 However, testing against human EXO1-352Δ (nuclease domain of EXO1)35 revealed that compounds 1 and 2 both inhibited this target with IC50 values similar to those against hFEN1 (Supplementary Figures 5k, 6a,e). Differential scanning fluorimetry experiments36 further confirmed binding of both compounds to both proteins in a divalent metal ion-dependent manner (Supplementary Figure 6g,h). In contrast, inhibitor 1 was found ineffective against bacteriophage T5 FEN (Supplementary Figures 5l, 6c) and Kluyveromyces lactis XRN1 (Supplementary Figure 16), both of which show a high level of active site conservation with the mammalian 5′-nuclease superfamily.1 Similarily, 1 did not inhibit the structurally unrelated DNA repair metallonuclease APE1 (Supplementary Figure 6f).
When hFEN1 acts in vivo it is usually associated with the toroidal clamp PCNA. PCNA increases the stability of FEN1–DNA complexes,34 suggesting that association with PCNA might allow FEN1 to overcome inhibition. However, when we added hPCNA to hFEN1 reactions inhibited by 1 or 4, the slow rates of reaction observed did not increase implying the FEN1 interaction partner does not dramatically influence the IC50 of either compound (Supplementary Figure 6d).
N-Hydroxyurea inhibitors engage with hFEN1 in live cells
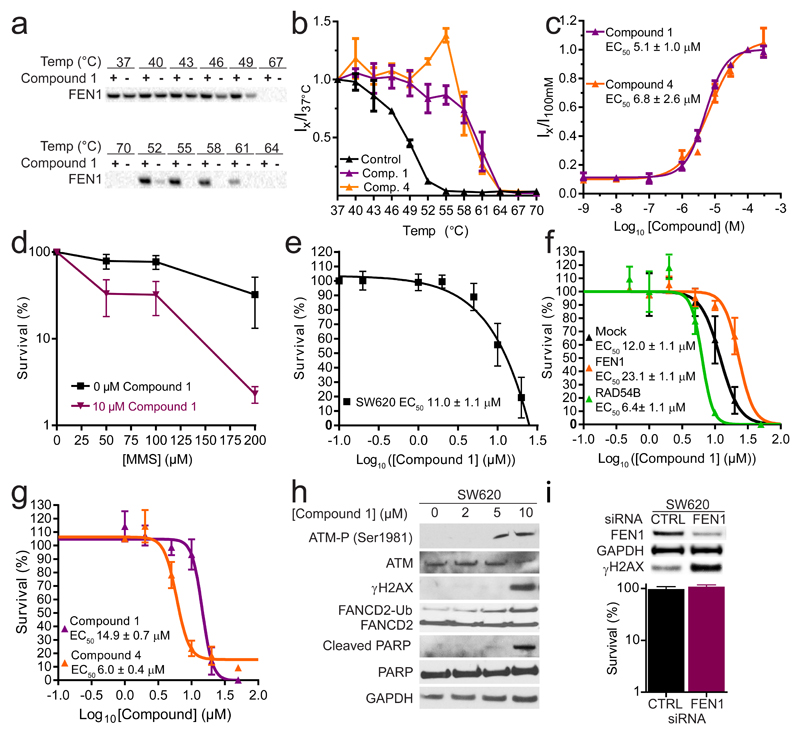
On the basis of contrasting inhibition modes, compounds 1 and 4 were selected for additional cellular studies. We employed the cellular thermal shift assay technique (CETSA)37 to establish whether they interacted with hFEN1 in SW620 colon cancer cells. CETSA detects changes in stability of a protein upon engagement with a ligand, like a biochemical thermal shift assay, but is performed with whole cells and a target-specific, label-free readout of engagement is obtained using a relevant antibody. Compounds 1 and 4 stabilized hFEN1 (Figure 5a-c and Supplementary Figure 17) with EC50 = 5.1 μM and 6.8 μM, respectively, in an isothermal concentration–response experiment, representing similar EC50s regardless of their differing modes of inhibition. Interestingly, these micromolar-range values represented a substantial drop-off in observed binding affinity compared with observations in prior biochemical assays (IC50 = 46 nM and 17 nM, respectively; Table 1) so we undertook a number of experiments to attempt to explain this. Cell permeability in MDCK and Caco-2 assays was not an issue (Supplementary Table 4); neither were other properties including solubility and chemical stability. The compounds’ affinity for free divalent metal ions in solution was insignificant, ruling out metal chelation as an explanation. Nonspecific protein binding may have contributed to the discrepancy between biochemical and phenotypic potency, although binding to other 5′-nuclease superfamily members represented the most obvious potential for off-target effects. Hence, we attempted further CETSA studies with 1 and 4 against hEXO1 but this was concluded to be a non-viable CETSA target (with only fragments of the protein detected on the blots), perhaps reflecting instability of the protein under the assay conditions, or its cellular context as a component of multi-protein complexes (which regulate its activity).
Figure 5. Cellular engagement and activity of hFEN1 inhibitors 1 and 4.
(a) Representative data of Western blot intensities from a melt curve for compound 1 ((+) indicates treated sample, (−) indicates control sample). (b) Melt and shift curve of FEN1 in intact SW620 cells with 100 μM 1 (purple), 4 (orange) and DMSO (control, black). (c) Ratio of hFEN1 protein isothermal shifts in cells with respect to concentration of compounds 1 (purple) or 4 (orange) after exposure of cells to 50 °C to indicate magnitude of target engagement of FEN1 in intact treated SW620 cells. (d) Dose-dependent sensitivity of SW620 cells to compound 1. (e) Sensitivity of HeLa cells stably expressing Fen1 (orange), Rad54b (green) or non-targeting (black) shRNA to compound 1. (f) MMS sensitivity of SW620 cells treated with continuous dose of 10 μM compound 1 (purple) or DMSO (control, black). (g) Dose-dependent sensitivity of HeLa cells to compounds 1 and 4. (h) Typical Western blots showing 1 induces a DNA damage response in a dose-dependent manner. (i) SW620 cells are insensitive to deletion of FEN1 by siRNA, but accumulate DNA damage. Panels (b) and (c) show data from three independent triplicate experiments, fitted globally (i.e. N = 3, n = 9) with standard error. Panels (d)–(g) and (i) show the mean of three independent experiments ± standard error. Full images of cut gels used to prepare panels (h) and (i) are included in Supplementary Figures 18 and 19, respectively.
hFEN1 inhibition activates the DNA damage checkpoint
High concentrations of compound 1 proved cytotoxic towards SW620 cells with an EC50 of 11 μM (Figure 5d), but HeLa cells stably expressing hFEN1-shRNA were 70% viable at 20 μM 1 (Figure 5e; purple curve). Mock-shRNA expressing HeLa cells were only 15% viable under the same conditions (Figure 5e; black curve), showing similar susceptibility to 1 as untransformed cells. Hence, a lack of hFEN1 conferred resistance to 1, suggesting on-target activity as the primary cause of cytotoxicity. SW620 cells also showed increased sensitivity to MMS when co-treated with 1, in a dose-dependent manner (Figure 5f), suggesting the compound inhibits the LP-BER function of FEN1 in a cellular context. Enhanced toxicity of 1 towards HeLa cells expressing Rad54b-shRNA (Figure 5e; green curve) was also observed with an EC50 of 6.4 μM compared to 14.9 μM against untransformed cells (Figure 5e,g), confirming the synthetic lethal interaction between Fen1 and Rad54b previously demonstrated by silencing of the former.18 Inhibitor 4 also proved cytotoxic to HeLa cells (EC50 6 μM; Figure 5g), appearing more potent than 1, whose EC50 of approximately 15 μM was in line with its toxicity against SW620 cells.
When treated with sub-lethal doses of 1, SW620 cells showed evidence of an induced DNA damage response (Figure 5h and Supplementary Figure 18) at concentrations consistent with the EC50 for target engagement observed by CETSA. The same compound effected a dose-dependent increase in ubiquitination of FANCD2, a marker for activation of the Fanconi anemia pathway recruited to stabilize stalled replication forks.38–40 At higher doses, accumulation of phosphorylated ATM and γH2AX was evident, indicating accumulation of unrepaired DNA double-strand breaks (DSBs). Cells treated with high concentrations of 1 also showed evidence of apoptosis, shown by the presence of cleaved PARP (Figure 5h). Knockdown of hFEN1 by siRNA activated a similar DNA damage response to treatment with 1; these cells accumulated γH2AX but otherwise remained viable (Figure 5i and Supplementary Figure 19). DNA damage response activation and apoptosis were consistent with loss of hFEN1 function, because the consequences of unprocessed Okazaki fragments would include stalled or collapsed replication forks, replication errors and double strand breaks.
Discussion
N-Hydroxyurea compounds 1, 2 and 4 prevented DNA cleavage with similar efficiency (Table 1), reflecting the SAR observed previously for similar-sized compounds24 inasmuch as comparable IC50 values were obtained despite notable differences in sidechain size and structure. These results were consistent with protein–inhibitor binding mediated primarily through interaction with active site Mg2+ ions, and a lack of strong contacts between the protein and inhibitor sidechain, as seen in the structure of 1 bound to hFEN1 (Figure 1 and Supplementary Table 2). Although the metal-coordinating headgroup clearly provided the predominant binding contribution, the elevated KD of 3 suggested interaction of the inhibitor sidechain with the protein was nonetheless important for optimal affinity. Further studies revealed subtle differences in mode of action on variation of the sidechain structure.
Although the DNA substrate bound in its usual conformation in the presence of compound 1, hydrolysis was impaired by prevention of double nucleotide unpairing through steric blocking of the catalytic metals (Figures 1b-d, 3, 4). These observations were reminiscent of the action of the HIV integrase inhibitor raltegravir.41 Raltegravir and functionally related compounds bind to active site metal ions of the integrase–DNA complex, similarly obstructing access of the reacting phosphodiester bond to the metals. In contrast, compounds 2 and 4, with altered sidechains, proved mostly competitive in character and primarily acted to reduce affinity of the enzyme for its DNA substrate.
The micromolar EC50s seen in CETSA experiments with 1 and 4 differed markedly from the compounds’ nanomolar potency against purified protein – though they were consistent with phenotypic potency in DNA damage induction and cytotoxicity assays. A clear explanation for this was not found, but the raised cellular EC50s might reflect a high local concentration of hFEN1 in the nucleus during S-phase, which could conceivably reach the micromolar range. The residence time of compound 1 on hFEN1 proved similar to that of raltegravir on its target (4.8 min),42 although this is short compared to the median of 51 min for a representative set of marketed drugs,42 so the short residence time of 1 may necessitate a high local drug concentration in the vicinity of the target for effective inhibition in cells.
Although hEXO1 is likely inhibited alongside hFEN1, the cellular concentration of hEXO1 is not expected to be significantly higher, so this seems an unlikely explanation for the raised EC50 values. The results with hFEN1-deficient cells (Figure 5e) did suggest some degree of target specificity, but previous cellular studies assuming selective inhibition of hFEN1 by the N-hydroxyurea series must nonetheless be interpreted with caution based on the likelihood of parallel hEXO1 inhibition, since it will not be possible to distinguish between phenotypes of hFEN1 and hEXO1 inhibition with this class of compounds. One such published24 inhibitor, related to 1-4, was employed to help validate a role for hFEN1 in homologous recombination (HR),43 demonstrating deficient HR upon treatment. However, hEXO1 is essential for competent HR,44–47 and the observed phenotype is explicable by inhibition of this enzyme alone. Although a role for hFEN1 in HR is otherwise supported in that study, we concluded that the N-hydroxyurea series should not be regarded as exclusive hFEN1 inhibitors.
The mixed inhibition mode of 1, which in theory permits ‘dead-end’ complexes of DNA and protein to form, did not confer any advantageous inhibition characteristics in cells. Unprocessed Okazaki fragments resulting from hFEN1 inhibition might be successfully repaired by the cell with apoptosis only resulting when the DNA damage response is overwhelmed. Some support for this notion was seen in SW620 cells treated with 1, where we observed dose-dependent activation of the Fanconi anemia pathway (Figure 5h). Because FANCD2 is recruited to stabilize stalled replication forks and initiate repair,38 treatment with 1 evidently did interrupt replication, prompting cells to activate other pathways to repair unprocessed Okazaki fragments directly. Failure to achieve this may cause collapse of replication forks into DSBs, and at higher doses of compound 1, we did see evidence for DSB repair pathway activation. These markers did not accumulate at lower doses, so the damage signal may only be obvious when the frequency of DSBs overwhelms the cell’s DNA damage response. Accumulation of cleaved PARP, indicating early apoptosis, also suggested cells exposed to 1 were accumulating DNA damage associated with hFEN1 and/or hEXO1 inhibition and signaling for apoptosis.
Without exposure to inhibitor, both SW620 cells treated with hFEN1-siRNA and HeLa cells stably expressing hFEN1-shRNA showed viability indistinguishable from untransformed controls yet constitutively initiated a DNA damage response (Figure 5i). The hFEN1-shRNA cells showed reduced sensitivity to 1, suggesting a degree of selectivity and on-target activity for the compound since the DNA damage reponse remained competent. Our data suggests removal of functional hFEN1 alone did not induce toxicity and that damage associated with its loss is successfully repaired until such mechanisms become overwhelmed. This result, alongside our other observations in human cells, suggests targeting of hFEN1 in cancer will not prove effective as a monotherapy, but could be useful in exploiting synthetic lethal vulnerabilities. Synthetic lethal interactions between hFEN1 and Rad54b,18 Cdc416 and Mre11a16 are established, and other such interactions with potential clinical relevance are proposed.16,38 We confirmed synthetic lethal interaction with Rad54b, previously established using hFEN1 knockdown,18 through inhibition of the latter by 1. Thus, hFEN1 inhibitors might prove beneficial as a component of targeted or personalized therapies, provided selectivity over hEXO1 and the other 5′-endonuclease superfamily members can be realized.
Online Methods
Protein Expression and Purification
hFEN1-Wild-type hFEN1 and the mutant hFEN1 protein, R100A, were expressed from previously-prepared pET28b vectors containing the appropriate sequences for WT or R100A and subsequently purified and stored as described previously.2 The C-terminally truncated counterparts of wt-hFEN1 and R100A (i.e. hFEN1-Δ336 and R100A-Δ336 respectively) were expressed from previously-prepared pET29b vectors containing the respective hFEN1-336 sequence in-frame with a PreScission protease site and (His)6-tag after residue 336 (removing 44 residues). The proteins were then purified and stored as previously described.2 T5FEN protein was expressed and purified as previously described.29
hEXO1-To create a vector for the expression of truncated, wild-type hEXO1-352 bearing an in-frame TEV protease site and C-terminal (His)6-tag, primers (5'-gtctctcccatggggatacagggattgctac-3' and 5'-ggttctccccagctcttgaatgggcaggcatagc-3')—to amplify hEXO1-352 DNA bearing leader sequences necessary for ligation independent cloning (LIC) with SmaI-digested pMCSG28 vector (DNASU plasmid repository)—were utilized according to protocol.48 The DNA sequence encoding hEXO1-352-TEV-(His)6-Stop was then subcloned from the pMCSG28-hEXO1-352 vector into a pET21a vector using the NdeI and NotI restriction sites with appropriate primers (5'-ggaattccatatggggatacagggattgctac-3' and 5'-ggataagaatgcggccgcttaatgatgatgatggtggtgcc-3'). The hEXO1-352-TEV-(His)6 protein was expressed in BL21(DE3)-RIPL E. coli using autoinduction media as described. The protein was purified by Co2+-immobilized affinity and anion exchange chromatography in a manner analogous to that described previously for hFEN1.2 Fractions containing hEXO1-352-TEV-(His)6 were pooled, concentrated using an Amicon ultrafiltration device with a 5,000 MWCO membrane and then dialyzed into 2X 2L 50 mM Tris pH 7.0, 50 mM NaCl, 5 mM DTT, 1 mM EDTA, 5% glycerol containing 1000U of TurboTEV (BioVision) to remove the (His)6-tag. The dialysate was treated with MagneGST™ glutathione particles to remove the TurboTEV, and then the protein was further purified using a Heparin affinity column and a salt gradient from 0 to 1 M NaCl as described previously.35 hEXO1-352-containing fractions were pooled, concentrated by ultrafiltration as before and then applied to a 16/60 Sephacryl™ S-100 HR (GE Lifesciences) column. Fractions containing the protein were concentrated and finally stored at 100 µM at –20 °C in 50 mM Tris pH 8.0, 50 mM NaCl, 1 mM DTT, 50 μM EDTA, 50% v/v glycerol.
PCNA-The vector for human PCNA sub-cloned in-frame with a C-terminal-(His)8-tag into pET41b using the NdeI and XhoI restricition sites was a kind gift of Professor Binghui Shen (Beckman Research Institute, City of Hope). The hPCNA-(His)8 protein was expressed overnight at 37 °C in BL21(DE3)-RIPL E. coli using autoinduction media as described.49 The cells were collected by centrifugation at 6000 g and resuspended in ice-cold PBS buffer. The cells were pelleted again and the supernatant was removed. The cell pellet was resuspended in Buffer A (25 mM Tris pH=7.4, 0.02% NaN3, 5 mM imidazole, 2 mM β-mercaptoethanol) containing 1M NaCl, 1X EDTA-free protease inhibitor cocktail, and 0.1 mg/mL lysozyme. After cell lysis by freeze thaw and sonication, Buffer A containing 1% Tween-20 (10% of the total volume of the lysate) was added. The lysate was clarified by centrifugation at 30,000 g for 30 minutes at 4 °C. The supernatant was then applied to Co2+-TALON immobilized affinity column and washed with 5 column volumes of Buffer A. The column was then washed with 5 CV of Buffer A containing, 200 mM NaCl, and 0.01% NP-40. The protein was then eluted in buffer B (25 mM Tris pH=7.4, 0.02% NaN3, 200 mM NaC,l 250 mM imidazole, 2 mM β-mercaptoethanol, 0.01% NP-40%). The eluate was directly applied to two tandem 5 mL Hi-Trap Q columns and further purified as described.50 Briefly, the fractions containing hPCNA were pooled and dialysed 2 X 2L into Buffer C (25 mM KPO4 pH=7.0, 0.01% NP-40%, 10% glycerol, 10 mM NaHSO3, 5 mM DTT, 0.02% NaN3). The dialysate was passed through a 5 mL Hi-Trap S HP column that was pre-equilibrated with Buffer C to remove impurities, but hPCNA was found exclusively in the flow-through. The flow-through was loaded onto a hydroxylapatite column (BioSepra HA Ultrogel, 11 cm by 2.6 cm) and then eluted using a 20-column volume gradient from 0.025 and 0.5 M KPO4 in Buffer C. The eluate was dialysed 2 X 2Lt into Buffer D (25 mM potassium phosphate pH 7.0, 1.5 M (NH4)2SO4, 0.02% NaN3). The dialysate was centrifuged at 3,300 g for 10 minutes at 4 °C to remove any precipitate and then loaded onto a HiPrep Phenyl-Sepharose FF (high sub) column and eluted using a 20 column volume inverse gradient using Buffer D and Buffer E (25 mM potassium phosphate pH 7.0, 10% glycverol, 0.02% NaN3). The isolated PCNA was then dialysed into Buffer F (100 mM HEPES pH=7.5, 200 mM KCl, 10 mM DTT, 0.1 mM EDTA, 0.04% NaN3), and concentrated to provide 200 μM PCNA trimer (i.e. 600 μM monomer) before the addition of glycerol to 50% v/v and storage at –20 °C.
KlXRN1-The vector corresponding to residues 1–1245 of Kluyveromyces lactis Xrn1 that was subcloned in-frame with a C-terminal hexahistidine tag into pET-26b was a kind gift of Professor Liang Tong laboratory (Columbia University). The protein was expressed in Rosetta E.coli according to protocol51 and purified as described for hFEN1. Once purified, the protein was stored in 20 mM Tris pH = 7.5, 200 mM NaCl, 2 mM DTT and 50% glycerol. The purity of all proteins used was assessed by SDS-PAGE (Supplementary Figure 20).
Crystallisation and Structure Determination
The C-terminally truncated protein was crystallized using the hanging drop vapor diffusion method. Briefly, the protein was concentrated to approximately 8 mg/mL in a buffer containing 50 mM Tris pH 7.5, 200 mM NaCl, 10 mM MgCl2, 1 mM TCEP with 5 mM inhibitor 1 added. The crystallization well contained 25% PEG 3350, 0.1 M MOPS pH 7.0, 5% 2-propanol and 2% glycerol. Crystals appeared after 3 days at room temperature. Data were collected at the ERSF synchrotron on station ID23 (T = 100 K). Data were processed and scaled using the XDS and SCALA software packages.52 The crystals diffracted to 2.8 Å resolution, belong to Space Group P1 and having unit cell dimensions of a = 43.3Å, b = 50.2 Å, c = 66.9 Å, α = 102.1°, β = 94.0°, γ = 90.7°. The structure was solved by molecular replacement, model rebuilding was conducted using COOT53 and the structure was refined using the BUSTER software.54 The final model has good geometry with 92% of residues in the favored region of the Ramachandran plot, 7% in the allowed regions and 1% in the disallowed regions as defined by PROCHECK.52 At convergence a final crystallographic R-factor of 23.3% was achieved. Full data and refinement statistics are shown in Supplementary Table 1 and ligand electron density in Supplementary Figure 1.
Isothermal Titration Calorimetry (ITC)
Binding affinities of wt hFEN1, hFEN1-336Δ and hFEN1-R100A for compounds 1 and 2 were measured using either a VP-ITC microcalorimeter (GE Healthcare) or NANO-ITC (TA Instruments). The appropriate protein was exchanged from storage buffer into 100 mM KCl, 1 mM DTT, 50 mM HEPES pH 7.5 containing 8 mM MgCl2 or 10 mM CaCl2 using a HiPrep 26/10 desalting column at 4 °C. Subsequently, the protein was dialyzed overnight at 4 °C against the same buffer composition. In all cases, the dialysate was used prepare a solution with final protein concentration 18 μM (based on A280 using extinction coefficients calculated using the ExPASy ProtParam tool, http://web.expasy.org/protparam/) and final inhibitor concentration 200 μM, diluted from DMSO stock solution to a final DMSO concentration of 1%. Twenty-five injections were performed with 180 s spacing time at 25 °C. Titration traces were integrated by NITPIC55 and the resultant curves were globally fit by SEDFIT.56 The figures were prepared using GUSSI (http://biophysics.swmed.edu/MBR/software.html).
Synthesis and Purification of DNA constructs
The DNA oligonucleotides detailed in Supplementary Table 5, including those synthesised with 5′-fluorescein-CE-phosphoramidite (6-FAM), internal dSpacer-CE-phosphoramidite (dS) or containing site-specific 2-aminopurine (2AP) substitutions, were purchased with HPLC purification from DNA Technology A/S (Risskov, Denmark). MALDI–TOF spectrometry confirmed experimental molecular weights were all within 3 Da of calculated values (data not shown). The concentration of individual oligonucleotides was determined by measuring the absorbance at 260 nm (20 °C), using an extinction coefficient (ε260) calculated with OligoAnalyzer 3.1 (https://eu.idtdna.com/calc/analyzer). Heteroduplex substrates were prepared by heating the appropriate flap (or exo) strand with the complementary template in a 10:11 ratio at 95 °C for 5 min in 100 mM KCl, 50 mM HEPES pH 7.5 with subsequent cooling to room temperature (Supplementary Table 6 and Supplementary Figure 5).
Steady-state kinetic experiments
Reaction mixtures containing twelve different concentrations of FAM-labeled DF1 (Supplementary Figure 5a) substrate were prepared in reaction buffer (RB; 55 mM HEPES pH 7.5, 110 mM KCl, 80 mM MgCl2, 0.1 mg/mL bovine serum albumin, 1 mM DTT) and incubated at 37 °C for 10 min. Reactions were initiated by the addition of hFEN1-336Δ in RB. Reactions were sampled at seven time intervals between 2–20 min and quenched with excess EDTA (250 mM) with reaction progress being monitored by dHPLC equipped with a fluorescence detector (Wave® fragment analysis system, Transgenomic UK) as described.32 All reactions were independently repeated four times. Initial rates (v0, nM min−1) were determined by linear regression of plots of the amount of product concentration versus time up to 10% product formation. Kinetic parameters kcat and KM were determined by generalized nonlinear least squares using a Michaelis–Menten model (Equation 1), from plots of normalized initial rates (v0/[E]0, min−1) as a function of substrate concentration. The error distribution was assumed to be Gaussian, but to account for the unequal variance with increasing substrate concentration the variance was weighted to 1/Y2. All graph fitting and statistical analyses were done using GraphPad Prism 6.04 (GraphPad Software, Inc.).
| Equation 1 |
Inhibition Studies
The steady-state kinetic parameters of hFEN1-336Δ with DF1 were determined as above at various concentrations of 1, 2 and 4 (0, 5, 10, 50, 100, 500, 1000 nM) diluted from DMSO stock solutions as required. For each inhibitor concentration, reactions were followed in triplicate (each replicate using an independent serial dilution of enzyme) at six different concentrations of DF1 (10, 50, 100, 500, 1000, 5000 nM). Each experiment was independently conducted twice in triplicate. RB was used with a final DMSO concentration of 1% (this DMSO concentration did not affect reaction rates in the absence of inhibitor). Reactions were assayed, and normalized initial rates were determined, as described for steady-state analyses. Kinetic parameters kcat and KM were determined globally for the four simplest types of reversible linear inhibition: uncompetitive (Equation 2), competitive (Equation 3), non-competitive (Equation 4) and mixed (Equation 5) by non-linear regression plots of normalized initial rates (νo/[E]o, min−1) versus the substrate concentration for each concentration of inhibitor. The same weighting as above (1/Y2) was applied in each case. In addition to the goodness of fit of these calculated slopes to the raw normalized initial rates, statistical analyses were done using GraphPad Prism. Akaike information criteria (AIC) was employed as a statistical test to aid model selection (e.g. non-competitive versus competitive). Unless the more complex model gave a difference in AIC of more than –6 (95% probability), the less complex model was preferred as the appropriate one. This type of analysis penalizes the more parameterized model unless the sum-of-squares is significantly reduced. As an additional check, the residuals from both the non-competitive and mixed inhibition models were inspected. IC50 values for inhibition of hFEN1-336Δ by compounds 1, 2 and 4 (reported in Table 1) were derived from data obtained at 100 nM substrate DF1 and the same concentrations of inhibitor as above, using nonlinear regression in GraphPad Prism.
| Equation 2 |
| Equation 3 |
| Equation 4 |
| Equation 5 |
The rates of reaction of hFEN1-, hFEN1–PCNA-, hEXO1- and T5FEN-catalysed reactions of SF, DF4, EO and pY7 (Supplementary Tables 5,6 and Supplementary Figure 5h,i), respectively, were also determined at varying concentrations of compounds 1 and 4 (hFEN1–PCNA), 1 and 2 (hEXO1) or 1 only (T5FEN) in an analogous fashion at fixed concentrations of substrate as detailed in Supplementary Figure 6b–e.
RNA and DNA oligonucleotides used in XRN1 assays were ordered purified using reverse-phase HPLC and synthesised by DNA Technology (Risskov, Denmark), using standard phosphoramidites. Reactions were performed as described,51 but were monitored by denaturing PAGE using a Chemidoc system (Bio-Rad) to visualize the FAM and TAMRA labelled oligos (Supplementary Figure 16).
Human APE1 was purchased from Sino Biologicals via Life Technologies. APE1 was assayed with the AP1 substrate57 in 50 mM HEPES-KOH pH 7.5, 25 mM KCl, 5 mM MgCl2 and 0.1 mg/mL BSA. The reaction was monitored by dHPLC in a manner analogus to FEN1.
Determination of kSTmax of hFEN1, hFEN1-336Δ and hFEN1-R100A in the presence and absence of inhibitors
Maximal single turnover rates of reaction were determined using rapid quench apparatus, or manual sampling where appropriate, in triplicate (technical replicates) at 37 °C as described.32 To initiate reaction, enzyme at a final concentration of at least 10 × Kd of the substrate (DF1; Supplementary Tables 56 and Supplementary Figure 5a) in RB was added to an equal volume of substrate in the same buffer. To determine kSTmax in the presence of the inhibitor 1 or 2, reaction mixtures were prepared as above but containing 100 μM (1% DMSO) of either inhibitor. Samples were quenched (1.5 M NaOH, 80 mM EDTA) over a range of different time intervals and reaction progress monitored as above.32 The first-order rate constant (kSTmax) of reaction was determined by plotting the appearance of product against time (Pt) and applying nonlinear regression to Equation 6, where P∞ is the amount of product at endpoint.
| Equation 6 |
Fluorescence Anisotropy
Dissociation constants for free enzyme and the enzyme–inhibitor complex with the DNA substrate (DF1; Supplementary Tables 5,6 and Supplementary Figure 5a) were measured under equilibrium conditions by fluorescence anisotropy using a Horiba Jobin Yvon FluoroMax-3® spectrofluorometer with automatic polarizers. The excitation wavelength was 490 nm (slit width 5 nm) with emission detected at 510 nm (slit width 5 nm). Samples contained 10 mM CaCl2 or 2 mM EDTA (or when inhibitors were present 8 mM MgCl2) and 10 nM DF1, 110 mM KCl, 55 mM HEPES pH 7.5, 0.1 mg/mL bovine serum albumin, 1 mM DTT and 1% DMSO. Inhibitors 1 and 2 were added at 100 µM as appropriate. This solution containing substrate was incubated at 37 °C for a minimum of 10 min before the first measurement at 0 nM protein with subsequent readings taken on the cumulative addition of enzyme in a matched buffer, with corrections made for dilution. Data were modeled by nonlinear least squares regression in KaleidaGraph 4.0 using Equation 7, where r is the measured anisotropy at a particular total concentration of enzyme ([E]) and fluorescent substrate ([S]), with rmin giving the minimum anisotropy, of free DNA, and rmax the maximum anisotropy, the anisotropy of the saturated substrate.
| Equation 7 |
The equilibrium dissociation constant Kd(binding) is extracted from this analysis. Each measurement was independently repeated in triplicate (Supplementary Figure 10), and samples were taken after completion of the titration and analyzed by dHPLC to determine the amount of product produced (Supplementary Figure 11a).
Fluorescence Anisotropy Competition Experiments
Samples were prepared and anisotropy readings taken as described for the protein–DNA equilibrium binding measurements above. Enzyme was added cumulatively up to ~ 80% saturation of the substrate (DF1; Supplementary Tables 5,6 and Supplementary Figure 5a). At this point unlabeled DNA in the same buffer (DF2; Supplementary Table 5,6 and Supplementary Figure 5b) was added in a stepwise manner with readings taken after each addition of the competitor until the anisotropy value reached that of oligonucleotide in the absence of any protein (Supplementary Figure 11h).
Fluorescence Resonance Energy Transfer (FRET)
FRET energy transfer efficiencies (E) were determined using the (ratio)A method58 by measuring the enhanced acceptor fluorescence. The steady state fluorescent spectra of 10 nM non-labeled trimolecular, donor-only labeled and doubly-labeled DNA substrates (Supplementary Figure 5c,d,f) were recorded using a Horiba Jobin Yvon FluoroMax-3® fluorometer and normalized for lamp and wavelength variations. For direct excitation of the donor (fluorescein, DOL) or acceptor (rhodamine, AOL; Supplementary Figure 5e), the sample was excited at 490 nm or 560 nm (2 nm slit width) and the emission signal collected form 515–650 nm or 575–650 nm (5 nm slit width). Emission spectra were corrected for buffer and enzyme background signal by subtracting the signal form the non-labeled (NL) DNA sample. In addition to 10 nM of the appropriate DNA construct samples contained 10 mM CaCl2 or when inhibitor was present 8 mM MgCl2 or 2 mM EDTA and 110 mM KCl, 55 mM HEPES pH 7.5, 0.1 mg/mL bovine serum albumin, 1 mM DTT, 1% DMSO and 100 μM inhibitor 1or 2 as appropriate. The first measurement was taken prior to the addition of protein (either hFEN1-WT or hFEN1-R100A) with subsequent readings taken on the cumulative addition of enzyme, with corrections made for dilution. Transfer efficiencies (E) were determined according to Equation 8, where FDA and FD represent the fluorescent signal of the doubly-labeled DNA (DAL) and donor-only-labeled DNA (DOL) at the given wavelengths, respectively; εD and εA are the molar absorption coefficients of donor and acceptor at the given wavelengths; and εD(490)/εA(560) and εA(490)/εA(560) are determined from the absorbance spectra of doubly-labeled molecules (DAL) and the excitation spectra of singly rhodamine-only-labeled molecules (AOL). Energy transfer efficiency (E) was fit by non-linear regression to Equation 9, where Emin and Emax are the minima and maxima of energy transfers, [S] is the substrate concentration, [P] is the protein concentration and KD is the bending equilibrium dissociation constant of the protein substrate [PS] complex.
| Equation 8 |
| Equation 9 |
2-Aminopurine Exciton-Coupled Circular Dichroism (ECCD) Spectroscopy
Spectra were recorded of samples containing 10 μM DF3 (Supplementary Figure 5g), 110 mM KCl, 55 mM HEPES pH 7.5, 1 mM DTT and either 10 mM CaCl2; 10 mM CaCl2 + 25 mM EDTA; 8 mM MgCl2 + 100 µM compound 1 or 2; or 8 mM MgCl2 + 100 µM compound 1 or 2 + 25 mM EDTA; and, where appropriate, 12.5 μM protein, using a JASCO J-810 CD spectrophotometer (300–480 nm) at 20 °C as described.28 In samples containing either inhibitor 1 or 2, the enzyme was pre-incubated with the inhibitor before addition of the substrate. The CD spectra were plotted as Δε per mol of 2AP residue versus wavelength. Each measurement was independently repeated (typically in triplicate) and gave similar results. After measurements were recorded aliquots were taken and the amount of product produced was checked by dHPLC (Supplementary Figure 12b).
Differential Scanning Fluorimetry (DSF)
The stability of purified hFEN1 and hEXO1-352 with and without available Mg2+ was assessed as a function of inhibitor concentration by DSF36 using the fluorescent probe SYPRO® Orange (Sigma–Aldrich). Final volumes of 20 μL containing 2.5 μM hFEN1 or hEXO1-352 in 50 mM HEPES–KOH pH 7.5, 100 mM KCl, 8 mM MgCl2, 1× SYPRO® Orange with either 25 mM EDTA or 25 mM NaCl and various concentrations of compound 1 or 2 (0, 1, 2, 4, 6, 8, 10, 20, 40, 60, 80, 100 μM) were mixed in white 96-well PCR-plates (Starlab) and sealed with StarSeal Advanced Polyolefin Film (Starlab). The plates were inserted into an Agilent MX3005P QPCR instrument for thermal denaturation. The emission at 610 nm (excitation 492 nm) from each well was recorded from 25 to 95 °C at a scan rate of 1 °C/min with a filter set gain multiplier of ×4. Analysis of the resulting thermal denaturation curves was accomplished using the DSF Analysis Excel36 script as described (ftp://ftp.sgc.ox.ac.uk/pub/biophysics) in combination with GraphPad Prism 6.04, which provided the nonlinear regression function with the Boltzman equation (Equation 10).
| Equation 10 |
Cellular Thermal Shift Assay (CETSA)
CETSA was performed as described37 by first establishing melt curves and ligand-induced shifts followed by testing of the compounds with increasing concentrations of 1 or 4 at a single temperature to establish the CETSA EC50 of target engagement. Target engagement was determined by isothermal concentration–response (IsoT C–R) stabilization curves for compound 1 and 4 on hFEN1 in treated intact cells. Western blots were performed using an iBlot2 device (Life Technologies) on nitrocellulose membranes. Transfer was set to 8 minutes at 25 V. Blocking and dilution of antibodies were performed in 5% non-fat milk in Tris Buffered Saline–Tween (TBST). A commercially available primary antibody against hFEN1 (ab109132, Abcam) was diluted at 1:5000 and incubated at 4 °C overnight. Specific hFEN1 bands were then detected using the horseradish peroxidase (HRP) conjugated secondary antibody sc-2374 (Santa Cruz Biotechnology) together with Clarity Western ECL substrate (BioRad).
Melt and shift curves (Figure 5a,b) for FEN1 in intact SW-620 cells were determined by washing cells with HBSS followed by trypsinization using TrypLE (Gibco) and pelleting by centrifugation. The pellet was washed with HBSS, pelleted and re-suspended in HBSS to a cell density of 20 million cells/mL. Compound incubation was performed during 60 minutes at 37 °C at 100 μM final concentration, whereas 0.2% DMSO was used as negative control. The samples were gently mixed every 10 min. Cell viability was measured before and after compound incubation. The treated cells were divided into 50 μL aliquots and subjected to a 12-step heat challenge between 37 and 70 °C for 3 min, followed by immediate cell lysis by 3 rounds of freeze–thawing. Precipitated protein was pelleted by centrifugation at 20,000 g for 20 min, then 30 μL of the supernatant was mixed with 15 μL gel loading buffer (NuPAGE LDS sample buffer, Life Technologies) and 10 μL/lane of the mixture was loaded to a gel. Protein amounts were detected using Western blot techniques as described above.
Isothermal concentration response curves (Figure 5c) were determined with intact SW-620 cells treated as above, but at a final concentration of 40 million cells/mL. The cell suspension was divided into 30 μL aliquots and an equal volume of HBSS containing 2× the intended compound concentration was added, resulting in a final cell concentration of 20 million cells/mL at the correct concentration. A 7-step dilution concentration response series of the ligands in 0.2% DMSO was applied together with 0.2% DMSO as control. The log10 dilution series ranged from 100 pM to 100 μM. An additional 7-step series was applied, ranging from 100 nM to 300 μM. The cells were incubated with ligand at 37 °C for 60 min, with gentle mixing every 10 min. The aliquots were heated to a single specific temperature, 50 °C, as determined from the previously established FEN1 melt and shift curves, for 3 min, and lysed by 3 cycles of freeze–thawing. Precipitated protein and cellular debris were pelleted by centrifugation at 20.000 g for 20 min then 40 μL of the supernatant was mixed with 20 μL LDS sample buffer. Protein amounts were detected after loading 10 μL/lane of the supernatant/LDS mixture per on a gel using standard Western blot techniques.
The Western blot intensities were obtained by measuring the chemiluminescence counts per mm2 (I = count/mm2). The obtained intensities were plotted in GraphPad Prism for melt curves, with the luminescence count normalized to the control count at 37 °C. The IsoT C–R data was analyzed and normalized to the maximum compound concentration. The normalized intensities were plotted and analyzed using GraphPad Prism. Data points are shown as mean values with error bars indicating the standard error of the mean. Concentration–response curves were fitted using the modified logistic Hill equation algorithm included in the GraphPad Prism software. The obtained CETSA™ EC50 concentration response values represent the half maximal concentration of the ligands for stabilizing hFEN1 at 50 °C. The quoted EC50 with 95% confidence intervals is therefore a relative measure of target engagement of compound available for interaction with FEN1 in intact SW-620 cells.
Cytotoxicity Assay
SW620 cells were obtained from ATCC and HeLa SilenciX cell lines stably expressing shRNA against Fen1, Rad54b or a non-targeting control were obtained from Tebu Biosciences. Cell-line identity was confimed by short tandem repeat fingerprinting prior to banking and cells are routinely tested for mycoplasma contamination. SilenciX gene knockdown was confirmed by quantitative PCR. Exponentially growing cells were split into 6-well plates at an appropriate density in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 2 mM L-glutamine and 10% foetal calf serum (FCS) and incubated for 24 h to allow cells to adhere. Cells were treated with compound 1 or 4 (diluted from DMSO stock solution) at the concentration stated. For the MMS sensitivity assay, cells were pre-treated with 100 μM MMS in DMEM for 2 h before replacing the media with DMEM containing the stated concentration of 1 or 4. For siRNA survival assays, Fen1 knockdown was achieved by treating with targeting and non-targeting siRNA pools (Dharmacon) for 24 h using RNAiMAX lipofectamine transfection reagent (Life Technologies) before cells were allowed to recover in fresh media. In all cases, plates were incubated for 10–14 days to allow for colony formation. Colonies were stained with crystal violet and colony frequencies determined using the GelCount automated system (Oxford Optronix). Survival is expressed as a percentage of a mock-treated control. Knockdown of Fen1 by siRNA was confirmed by Western blot.
DNA Damage Induction Assay
Exponentially growing SW620 cells were seeded in 6-well plates and incubated for 4 days with compound 1 at the stated dose. Cells were subsequently washed, trypsinized and lysed in Cell Panel Lysis Buffer (5 mM Tris-HCl, 3 mM EDTA, 3 mM EGTA, 50 mM NaF, 2 mM sodium orthovanadate, 0.27 M sucrose, 10 mM β-glycerophosphate, 5 mM sodium pyrophosphate, and 0.5% Triton X-100) supplemented with complete protease and phosSTOP phosphotase inhibitors (both Roche). Proteins were separated by gel electrophoresis and transferred to nitrocellulose membrane by Western blot. Membranes were probed, at a concentration of 1:1000 unless stated otherwise, for cleaved PARP (#9541, Cell Signaling Technology), γH2AX (#2577, Cell Signaling Technology; 1:500), GAPDH (#3683, Cell Signaling Technology; 1:5000), FEN1 (ab109132, Abcam), phospho-ATM (Ser1981) (ab81292, Abcam), PARP (51-6639GR, BD Biosciences), ATM (sc-23921, Santa Cruz Biotechnology) and FANCD2 (sc-20022, Santa Cruz Biotechnology).
Accession Codes
The PDB accession code for the X-ray crystal structure of compound 1 bound to human FEN1, as detailed above, is 5FV7.
Supplementary Material
Acknowledgements
This work was supported by BBSRC grants BB/K009079/1 and BB/M00404X/1 (both to JAG) and AstraZeneca. JCE thanks the EPSRC and AstraZeneca for a studentship. The authors thank Dr Chris Phillips for assistance with submissions of the crystallographic data and Dr Thomas McGuire for synthetic support.
Footnotes
Accession Codes – The PDB accession code for the crystal structure presented in Figure 1 is 5FV7.
Author Contributions
CJ designed and synthesized inhibitors, JCE, MJT, LDF and SJS carried out kinetic and biophysical experiments. JCE, MJT, LDF, CM, JWMN, MA and JAG designed experiments and analyzed this data. JD and JWMN obtained and analyzed structures. CLBS and DMM performed the CETSA assays. TAW carried out other cellular experiments, and TAW and STD analyzed data. All authors contributed to the preparation of the manuscript.
Competing Financial Interest Statement
MA, CM, JD, JWMN, TAW and STD are employees of AstraZeneca. CJ was an employee of AstraZeneca at the time of writing. CLBS and DMM are employees of Pelago Bioscience AB.
References
- 1.Grasby JA, Finger LD, Tsutakawa SE, Atack JM, Tainer JA. Unpairing and gating: sequence-independent substrate recognition by FEN superfamily nucleases. Trends Biochem Sci. 2012;37:74–84. doi: 10.1016/j.tibs.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsutakawa SE, et al. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011;145:198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim CY, Shen BH, Park MS, Olah GA. Structural changes measured by X-ray scattering from human flap endonuclease-1 complexed with Mg2+ and flap DNA substrate. J Biol Chem. 1999;274:1233–1239. doi: 10.1074/jbc.274.3.1233. [DOI] [PubMed] [Google Scholar]
- 4.Zheng L, et al. Functional regulation of FEN1 nuclease and its link to cancer. Nucleic Acids Res. 2011;39:781–794. doi: 10.1093/nar/gkq884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waga S, Bauer G, Stillman B. Reconstitution of complete SV40 SNA replication with purified replication factors. J Biol Chem. 1994;269:10923–10934. [PubMed] [Google Scholar]
- 6.Bambara RA, Murante RS, Henricksen LA. Enzymes and reactions at the eukaryotic DNA replication fork. J Biol Chem. 1997;272:4647–4650. doi: 10.1074/jbc.272.8.4647. [DOI] [PubMed] [Google Scholar]
- 7.Kim K, Biade S, Matsumoto Y. Involvement of flap endonuclease 1 in base excision DNA repair. J Biol Chem. 1998;273:8842–8848. doi: 10.1074/jbc.273.15.8842. [DOI] [PubMed] [Google Scholar]
- 8.Parikh SS, Mol CD, Hosfield DJ, Tainer JA. Envisioning the molecular choreography of DNA base excision repair. Curr Opin Struct Biol. 1999;9:37–47. doi: 10.1016/s0959-440x(99)80006-2. [DOI] [PubMed] [Google Scholar]
- 9.Beard WA, Wilson SH. Structure and mechanism of DNA polymerase beta. Chem Rev. 2006;106:361–382. doi: 10.1021/cr0404904. [DOI] [PubMed] [Google Scholar]
- 10.Mohan V, Srinivasan M. New Research Directions in DNA Repair. In: Chen Clark., editor. InTech; 2013. [Google Scholar]
- 11.Singh P, et al. Overexpression and hypomethylation of flap endonuclease 1 gene in breast and other cancers. Mol Cancer Res. 2008;6:1710–1717. doi: 10.1158/1541-7786.mcr-08-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lam JS, et al. Flap endonuclease 1 is overexpressed in prostate cancer and is associated with a high Gleason score. BJU Int. 2006;98:445–451. doi: 10.1111/j.1464-410X.2006.06224.x. [DOI] [PubMed] [Google Scholar]
- 13.Nikolova T, Christmann M, Kaina B. FEN1 is overexpressed in testis, lung and brain tumors. Anticancer Res. 2009;29:2453–2459. [PubMed] [Google Scholar]
- 14.Panda H, et al. Amino acid Asp181 of 5'-flap endonuclease 1 is a useful target for chemotherapeutic development. Biochemistry. 2009;48:9952–9958. doi: 10.1021/bi9010754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshimoto K, et al. Complex DNA repair pathways as possible therapeutic targets to overcome temozolomide resistance in glioblastoma. Front Oncol. 2012;2:186. doi: 10.3389/fonc.2012.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Pel DM, et al. An evolutionarily conserved synthetic lethal interaction network identifies FEN1 as a broad-spectrum target for anticancer therapeutic development. PLoS Genet. 2013;9:11. doi: 10.1371/journal.pgen.1003254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Illuzzi JL, Wilson DM. Base excision repair: contribution to tumorigenesis and target in anticancer treatment paradigms. Curr Med Chem. 2012;19:3922–3936. doi: 10.2174/092986712802002581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McManus KJ, Barrett IJ, Nouhi Y, Hieter P. Specific synthetic lethal killing of RAD54B-deficient human colorectal cancer cells by FEN1 silencing. Proc Natl Acad Sci USA. 2009;106:3276–3281. doi: 10.1073/pnas.0813414106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durant ST. Telomerase-independent paths to immortality in predictable cancer subtypes. J Canc. 2012;3:67–82. doi: 10.7150/jca.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang J-C, et al. The overexpression of FEN1 and RAD54B may act as independent prognostic factors of lung adenocarcinoma. PLoS ONE. 2015;10:e0139435. doi: 10.1371/journal.pone.0139435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shibata Y, Nakamura T. Defective flap endonuclease 1 activity in mammalian cells is associated with impaired DNA repair and prolonged S phase delay. J Biol Chem. 2002;277:746–754. doi: 10.1074/jbc.M109461200. [DOI] [PubMed] [Google Scholar]
- 22.McWhirter C, et al. Development of a high-throughput fluorescence polarization DNA cleavage assay for the identification of FEN1 inhibitors. J Biomol Screen. 2013;18:567–575. doi: 10.1177/1087057113476551. [DOI] [PubMed] [Google Scholar]
- 23.Dorjsuren D, Kim D, Maloney DJ, Wilson DM, Simeonov A. Complementary non-radioactive assays for investigation of human flap endonuclease 1 activity. Nucleic Acids Res. 2011;39:e11. doi: 10.1093/nar/gkq1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tumey LN, et al. The identification and optimization of a N-hydroxy urea series of flap endonuclease 1 inhibitors. Bioorg Med Chem Lett. 2005;15:277–281. doi: 10.1016/j.bmcl.2004.10.086. [DOI] [PubMed] [Google Scholar]
- 25.Finger LD, et al. The wonders of flap endonucleases: structure, function, mechanism and regulation. Sub Cell Biochem. 2012;62:301–326. doi: 10.1007/978-94-007-4572-8_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo ZG, et al. Sequential posttranslational modifications program FEN1 degradation during cell-cycle progression. Mol Cell. 2012;47:444–456. doi: 10.1016/j.molcel.2012.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakurai S, et al. Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 2005;24:683–693. doi: 10.1038/sj.emboj.7600519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finger LD, et al. Observation of unpaired substrate DNA in the flap endonuclease-1 active site. Nucleic Acids Res. 2013;41:9839–9847. doi: 10.1093/nar/gkt737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel N, et al. Flap endonucleases pass 5'-flaps through a flexible arch using a disorder-thread-order mechanism to confer specificity for free 5'-ends. Nucleic Acids Res. 2012;40:4507–4519. doi: 10.1093/nar/gks051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Syson K, et al. Three metal ions participate in the reaction catalyzed by T5 flap endonuclease. J Biol Chem. 2008;283:28741–28746. doi: 10.1074/jbc.M801264200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomlinson CG, et al. Neutralizing mutations of carboxylates that bind metal 2 in T5 flap endonuclease result in an enzyme that still requires two metal ions. J Biol Chem. 2011;286:30878–30887. doi: 10.1074/jbc.M111.230391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finger LD, et al. The 3'-flap pocket of human flap endonuclease 1 is critical for substrate binding and catalysis. J Biol Chem. 2009;284:22184–22194. doi: 10.1074/jbc.M109.015065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel N, et al. Proline scanning mutagenesis reveals a role for the flap endonuclease-1 helical cap in substrate unpairing. J Biol Chem. 2013;288:34239–34248. doi: 10.1074/jbc.M113.509489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Craggs TD, Hutton RD, Brenlla A, White MF, Penedo JC. Single-molecule characterization of Fen1 and Fen1/PCNA complexes acting on flap substrates. Nucleic Acids Res. 2014;42:1857–1872. doi: 10.1093/nar/gkt1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orans J, et al. Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell. 2011;145:212–223. doi: 10.1016/j.cell.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protocol. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 37.Molina DM, et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341:84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- 38.Moldovan G-L, D'Andrea AD. How the fanconi anemia pathway guards the genome. Annu Rev Genet. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeo JE, Lee EH, Hendrickson EA, Sobeck A. CtIP mediates replication fork recovery in a FANCD2-regulated manner. Hum Mol Genet. 2014;23:3695–3705. doi: 10.1093/hmg/ddu078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Di Santo R. Inhibiting the HIV integration process: past, present, and the future. J Med Chem. 2014;57:539–566. doi: 10.1021/jm400674a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dahl G, Akerud T. Pharmacokinetics and the drug–target residence time concept. Drug Discov Today. 2013;18:697–707. doi: 10.1016/j.drudis.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 43.Fehrmann RSN, et al. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat Genet. 2015;47:115–125. doi: 10.1038/ng.3173. [DOI] [PubMed] [Google Scholar]
- 44.Bolderson E, et al. Phosphorylation of Exo1 modulates homologous recombination repair of DNA double-strand breaks. Nucleic Acids Res. 2010;38:1821–1831. doi: 10.1093/nar/gkp1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Desai A, Qing Y, Gerson SL. Exonuclease 1 is a critical mediator of survival during DNA double strand break repair in non-quiescent hematopoietic stem and progenitor cells. Stem Cells. 2014;32:582–593. doi: 10.1002/stem.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomimatsu N, et al. Phosphorylation of EXO1 by CDKs 1 and 2 regulates DNA end resection and repair pathway choice. Nat Comm. 2014;5 doi: 10.1038/ncomms4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomimatsu N, et al. Exo1 plays a major role in DNA end resection in humans and influences double-strand break repair and damage signaling decisions. DNA Repair. 2012;11:441–448. doi: 10.1016/j.dnarep.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eschenfeldt WH, et al. Cleavable C-terminal His-tag vectors for structure determination. J Struct Funct Genom. 2010;11:31–39. doi: 10.1007/s10969-010-9082-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Studier FW. Protein production by auto-induction in high-density shaking cultures. Protein Express Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 50.Fien K, Stillman B. Identification of replication factor C from Saccharomyces cerevisiae: a component of the leading-strand DNA replication complex. Mol Cell Biol. 1992;12:155–163. doi: 10.1128/mcb.12.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang JH, Xiang S, Xiang K, Manley JL, Tong L. Structural and biochemical studies of the 5′→ 3′ exoribonuclease Xrn1. Nat Struct Mol Biol. 2011;18:270–276. doi: 10.1038/nmsb.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 53.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buster version 2.11.2. Global Phasing Ltd; Cambridge, United Kingdom: 2011. [Google Scholar]
- 55.Keller S, et al. High-precision isothermal titration calorimetry with automated peak-shape analysis. Anal Chem. 2012;84:5066–5073. doi: 10.1021/ac3007522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Houtman JCD, et al. Studying multisite binary and ternary protein interactions by global analysis of isothermal titration calorimetry data in SEDPHAT: Application to adaptor protein complexes in cell signaling. Protein Sci. 2007;16:30–42. doi: 10.1110/ps.062558507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Izumi T, Mitra S. Deletion analysis of human AP-endonuclease: minimum sequence required for the endonuclease activity. Carcinogenesis. 1998;19:525–527. doi: 10.1093/carcin/19.3.525. [DOI] [PubMed] [Google Scholar]
- 58.Clegg RM, et al. Fluorescence resonance energy transfer analysis of the structure of the four-way DNA junction. Biochemistry. 1992;31:4846–4856. doi: 10.1021/bi00135a016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.