Abstract
A fundamental impediment to understanding the brain is the availability of inexpensive and robust methods for targeting and manipulating specific neuronal populations. The need to overcome this barrier is pressing because there are considerable anatomical, physiological, cognitive, and behavioral differences between mice and higher mammalian species in which it is difficult to specifically target and manipulate genetically defined functional cell-types. In particular, it is unclear the degree to which insights from mouse models can shed light on the neural mechanisms that mediate cognitive functions in higher species including humans. Here we describe a novel recombinant adeno-associated virus (rAAV) that restricts gene expression to GABAergic interneurons within the telencephalon. We demonstrate that the viral expression is specific and robust, allowing for morphological visualization, activity monitoring and functional manipulation of interneurons in both mice and non-genetically tractable species, thus opening the possibility to study GABA-ergic function in virtually any vertebrate species.
INTRODUCTION
Inhibitory GABAergic interneurons, although sparser in number than glutamatergic neurons, play critical roles in all central processing and their dysfunction contributes to numerous neuropsychiatric disorders 1–3. To date, the ability to target interneurons has been essentially limited to the use of transgenic mice4. While certain cell types such as principal excitatory cells and glia have been successfully targeted5–7, an effective approach to target inhibitory interneurons is not currently available. Gene delivery using non-pathogenic rAAV is showing increasing promises for the treatment of both mono-allelic and complex diseases. Many phase I–III clinical trials using rAAV vectors have yielded encouraging results, and in 2012, the first rAAV based therapy received marketing approval by the European Union8. Despite its promise, the use of AAV as a gene-delivery method is hampered by its limited selectivity in expression. While it has been long recognized that the inclusion within rAAVs of cis-acting DNA-control elements, such as specific promoters or enhancers might provide the desired specificity for expression within particular target cells in the brain, the success of such strategies has been limited. With regard to rAAVs, the chief impediment to this approach is the fundamental incompatibility of the size of such control elements relative to the restricted packaging limit of AAVs9,10. Given the obligate minimal size of reporters (e.g. EGFP, dTomato) alone or in combination with effector elements (e.g. ChR2, DREADD), which average about 700bp to 2kb respectively, a maximum of ~2kb in packaging capacity remains for the insertion of a cis-acting DNA control element into a vector. In order to identify sufficiently small elements capable of restricting expression to a defined population of cells, we examined regulatory elements known in other contexts to be specific to interneurons11–13. The distal-less homeobox 5 and 6 (Dlx5/6) genes are specifically expressed by all forebrain GABAergic interneurons during embryonic development. These genes have an inverted orientation relative to one another and share a 400bp (mI56i or mDlx) and a 300bp (mI56ii) enhancer sequence in the 10kb non-coding intergenic region 3′ to each of them11,14. The high degree of conservation of these sequences across vertebrate species is suggestive of an important role in gene regulation. Indeed, the mDlx enhancer has been used in numerous contexts to reliably target reporter genes in a pattern very similar to the normal patterns of Dlx5/6 expression during embryonic development11,15–18.
RESULTS
The mDlx enhancer restricts reporter expression to GABAergic interneurons in vitro
We first aimed to determine whether the mDlx enhancer can restrict the expression of reporter genes to GABAergic interneuron in vitro using rAAV. We inserted the mDlx enhancer in front of a minimal promoter in an AAV backbone, utilizing a GCaMP6f construct as a reporter to monitor expression (rAAV-mDlx-GCaMP6f). Primary cortical neuronal cultures from mice were infected with rAAV-mDlx-GCaMP6f and cultured for 11 days post infection and examined for GCaMP6f expression. We observed that the majority of cells that expressed GCaMP6f also expressed the pan-interneuron marker GAD67 (~95%) (Fig. 1a,i). By contrast, rAAVs containing a promoter used to target excitatory neurons that is excluded from GABAergic interneurons (CamKII) had variable levels of expression and the expression of GCaMP6f was not restricted to GABAergic interneurons (Supplementary Fig. 1a). To test whether the level of GCaMP6f expression in interneurons was sufficient to allow their activity to be monitored, we performed in vitro calcium imaging. Consistent with their high level of expression, robust spontaneous calcium transients could be detected in primary culture of cortical neurons in vitro (Supplementary Fig. 1b). Taken together, these data demonstrate that the mDlx enhancer alone is sufficient to restrict the expression of a reporter to interneurons and is sufficient to allow for the monitoring of neuronal activity of interneurons in vitro when used to direct GCaMP6f expression.
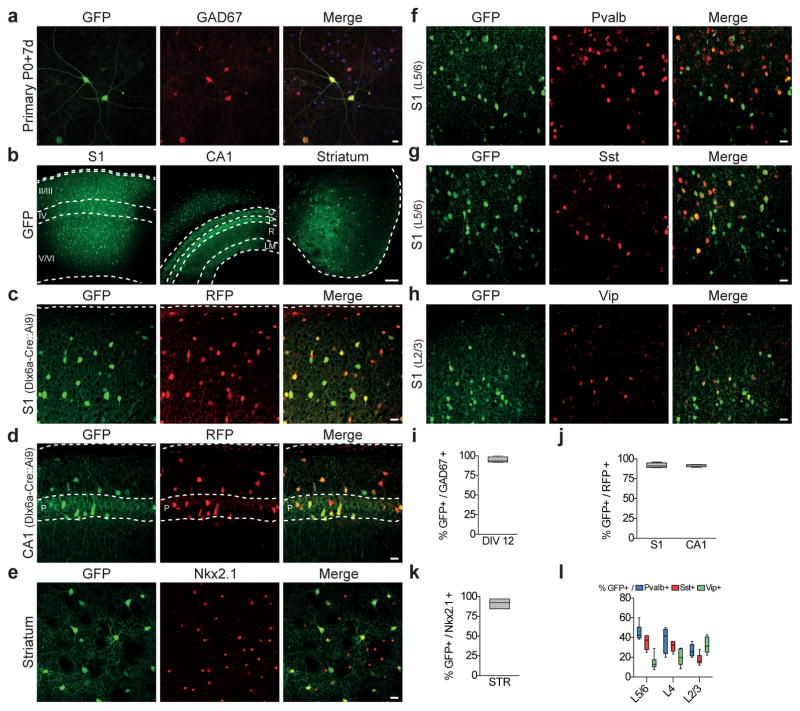
Figure 1. rAAV with mDlx enhancer restricts reporter expression to GABAergic interneurons.
(a) P0 Primary cortical cultures were infected with rAAV-mDlx-GCaMP6f at DIV8 and analyzed at DIV 19 by immunostaining using GFP and GAD67 antibodies. Representative example of co-localization between GFP and GAD67 (quantitation of this is shown in i). (b–h) Adult C57Bl6 (n=16) or Dlx6aCre::Ai9 (n=4) mice were stereotactically injected with 50–100nl of rAAV-mDlx-GFP in (c) somatosensory cortex (S1), (d) hippocampus (CA1) and (e) striatum, and were analyzed by immuno-staining for the indicated markers after 7 days. Representative example of GFP expression and co-localization between GFP and the indicated marker in the indicated brain region. (i) Quantitation of co-localization of rAAV-mediated viral expression of GFP and GAD67 in DIV19 cultured cortical neurons (94.7 ± 1.8%, n=875 cells from 5 coverslips). (j–l) Quantification of the proportion of cells co-expressing GFP and the indicated marker in the indicated anatomical regions. GFP/RFP, Dlx6a-Cre::Ai9, S1: 92.8 ± 1.2%, n= 657 cells from 4 animals; GFP/RFP, Dlx6a-Cre::Ai9, CA1: 91.8 ± 0.9%, n= 210 cells from 4 animals; GFP/NKX2.1, C57Bl6, Str: 36.4 ± 2.6%, n= 284 cells from 3 animals; GFP/PV, C57Bl6, S1: L2/3: 26.9 ± 2.5%; L4: 37.7 ± 5.1%; L5/6: 45.1 ± 3.2%; total n=577 from 3 animals; GFP/SST, C57Bl6, S1: L2/3: 17.8 ± 2.3%; L4: 32.3 ± 1.6%; L5/6: 35.4 ± 3.0%; total n=577 from 3 animals; GFP/VIP, C57Bl6, S1: L2/3: 32.6 ± 3.3%; L4: 20.0 ± 3.4%; L5/6: 14.6 ± 3.0%; total n=701 from 4 animals. Quantification are indicated in the text as mean ± s.e.m and are represented as box-and-whisker plot with upper and lower whiskers represent the maximum and minimum value respectively and the box represent upper, median and lower quartile. Dashed lines represent limits of the indicated anatomical structures. Roman numbers represent cortical layers, O - oriens, P - pyramidal, R - radiatum, LM - laconosum moleculare. Scale bars represent 10μm (a), 15μm (c–h) or 100μm (b).
The mDlx enhancer restricts reporter expression to adult GABAergic interneurons in the forebrain
A number of aspects of Dlx5/6 gene expression compromise their utility as markers for interneurons. Dlx5/6 genes are expressed at a high level in progenitors of GABAergic and post-mitotic interneurons during embryonic development but their expression strongly diminishes after birth. Moreover, in the striatum these genes are not specific to interneurons, as both interneurons and principal projection neurons, the medium spiny neurons (MSNs - 4% and 96% of the population respectively) express high levels of Dlx5/6 during embryonic development 11,12,19. In transgenic mice, groups which have utilized the mDlx enhancer have found that reporters under the control of this element are faithfully expressed in a pattern that closely matches endogenous patterns of Dlx5/6 expression, demonstrating the unsuitableness of using this approach to either label interneurons postnatally or differentiate between interneurons and MSNs within the striatum11,15–18. This prompted us to test whether rAAV-mDlx-GFP virus has similar limitations in its ability to target interneuronal populations. Surprisingly, immunofluorescence analyses of adult mice one week after injection with the rAAV-mDlx-GFP revealed sparse but strong GFP labeling in somatosensory cortex (S1), hippocampus and striatum (Fig. 1b) and the majority of infected cells in all these regions co-expressed GABA (data not shown). To confirm these findings, we injected rAAV-mDlx-GFP into the cortex and hippocampus of Dlx6aCre:Ai9 mice, where the persistence of the RFP reporters subsequent to the cessation of Dlx6 expression results in all interneurons within these areas being RFP-positive. Again, the majority of GFP-expressing cells co-expressed RFP (Fig. 1c,d,j). Moreover rAAV-mDlx-GFP expression within the striatum co-localized with NKX2.1, an interneuron marker within this structure (Fig. 1e,k). Further supporting that the expression of GFP directed from rAAV-mDlx-GFP is broadly and uniformly observed within all interneuron populations, within S1, the infected cells express parvalbumin (PVALB), somatostatin (SST) and vasoactive intestinal peptide (VIP) in proportions that correspond to the expected distribution of these 3 non-overlapping interneuron markers in this region1,20 (Fig. 1f–h,i). To investigate whether the Dlx enhancer could also drive expression in interneurons that are not derived from the Dlx5/6 lineage, we performed viral injections in the lumbar region of the spinal cord of P0 pups and analyzed the expression of the viral reporter after 7 days. In mice injected with rAAV-mDlx-GFP, the GFP expression appeared to be restricted to the dorsal horns of the lumbar spinal cord and virtually no cells expressing the viral reporter co-localize with PAX2, a marker exclusively expressed in virtually all GABAergic and Glycinergic interneurons of the dorsal horns of the spinal cord in both embryonic and adult animals21. By contrast, mice injected with an AAV expressing nuclear GFP under the control of the pan-neuronal promoter human synapsin1 (rAAV-hSyn1-nGFP) showed widespread expression of GFP within both ventral and dorsal spinal cord, with ~12% of cells co-localizing with PAX2 (Supplementary Fig. 2). These data show that the expression driven by the mDlx enhancer is excluded from the inhibitory neurons in the spinal cord, but restricted to a population of cells in the dorsal spinal cord that remains to be characterized. Altogether, these results demonstrate that infection of adult mice with rAAV-mDlx-GFP results in a highly selective expression within GABA-ergic interneurons from the Dlx5/6 lineage within the telencephalon.
rAAV-mDlx-Flex-GFP allows for the intersectional targeting of interneuron subtypes
Despite significant progress in recent years, the precise targeting and manipulation of specific interneuron subtypes is still essentially limited to the combinatorial use of transgenic mice expressing Cre and Flp recombinases in defined populations4,22. The specificity of targeting is thus hampered by the availability of driver lines and the time-consuming breeding associated with generating productive crosses on suitable genetic backgrounds. Therefore, we asked whether the specificity of the rAAV-Dlx for interneurons could be leveraged to develop an intersectional strategy that would require only one Cre-expressing mouse reporter. To test this idea, we injected an rAAV expressing a Cre-dependent GFP reporter under the control of the mDlx enhancer (rAAV-mDlx-Flex-GFP) into CCKCre mice. While Cck is expressed in specific interneurons subsets, its broader expression within glutamatergic neurons limits the usefulness of CCKCre mice for targeting interneurons4. In both somatosensory cortex S1 and hippocampal CA1, the majority of cells expressing GFP co-expressed the pan-interneuron marker GAD67 (Fig. 2). Importantly, CCKCre mice injected with an rAAV expressing a Cre-dependent GFP reporter under the control of an ubiquitous promoter (rAAV-CAG-Flex-GFP) showed that the GFP expression was not selective for interneurons and animals that do not express Cre-recombinase injected with rAAV-mDlx-Flex-GFP did not show GFP expression at the injection site (Supplementary Fig. 3). These data demonstrate that the mDlx enhancer restricts the expression of a recombinase-dependent reporter to interneurons and can be used as a simplified intersectional strategy.
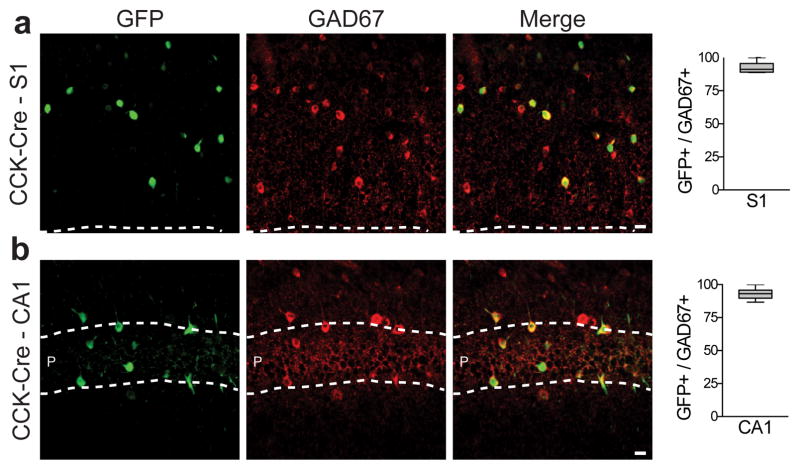
Figure 2. rAAV-mDlx-Flex-GFP allows intersectional targeting of CCK-expressing interneurons.
Adult CCK-Cre mice (n=4) were injected with rAAV-mDlx-Flex-GFP in somatosensory cortex (S1) or hippocampus (CA1) and were analyzed after 2 weeks by immunostaining for GAD67 immunoreactivity after 2 weeks. (a,b) Representative example of co-localization between GFP and GAD67 and corresponding quantifications (S1: 92.4 ± 1.4%, n= 319 cells from 4 animals; CA1: 93.2 ± 1.1%, n= 219 cells from 4 animals). Quantification are indicated in the text as mean ± s.e.m and are represented as box-and-whisker plot with upper and lower whiskers represent the maximum and minimum value respectively and the box represent upper, median and lower quartile. Dashed lines represent limits of the indicated anatomical structures. Scale bars represent 10μm.
rAAV-hDLX-Gq-DREADD allows for chemogenetic modulation of interneuronal activity
Having established that our strategy enables high expression of fluorescent reporters (GFP) and activity indicators (GCaMP6f) restricted to interneurons, we next sought to examine whether chemogenetic approaches could be used to manipulate interneuron activity. Designer receptors exclusively activated by designer drugs (DREADDs) are modified human muscarinic receptors that are activated by clozapine-N4-oxide (CNO), a pharmacologically inert and orally bioavailable drug23. To date their utilization in interneurons has been restricted to contexts involving the use of transgenic mice24–27. To test whether we could use the rAAV-hDlx to gain chemogenetic control of interneurons, we generated an rAAV-hDlx-Gq expressing a HA-tagged version of the Gq-DREADD followed by the nuclear red-fluorescent reporter NLS-dTomato, under the control of the humanized form of the mDlx enhancer element (Supplementary Fig. 4a). We first tested whether the use of the human form of the Dlx5/6 enhancer (hDlx) negatively impacts the targeting specificity of the virus. Immunofluorescence analyses of adult Dlx6aCre::RCE mice injected with the rAAV-hDlx-Gq in S1 and CA1 revealed that the majority of cells expressing dTomato also expressed GFP (where GFP expression within the cortex and hippocampus is restricted to interneurons; Supplementary Fig. 4b–d). In addition, consistent with the expected expression of a functional receptor, the Gq-DREADD was located at the membrane of the infected cells (Fig. 3a and Supplementary Fig. 4e). These data confirm that the rAAV-hDlx-Gq drives the expression of the Gq-DREADD exclusively within interneurons. We then formally tested the functionality of the Gq-DREADD within these cells. Upon bath application of CNO, all interneurons expressing Gq-DREADD, showed membrane potential depolarization within less than a minute, consistent with the expression of a functional receptor (Fig. 3b,c). Finally, voltage clamp recordings of a pyramidal cell in the vicinity of the Gq-DREADD expressing interneurons showed an increase in inhibitory post-synaptic currents (IPSCs) upon CNO application (Fig. 3d,e). These experiments demonstrate that rAAV-mDlx-Gq allows specific, functional and restricted expression of Gq-DREADD and that CNO-treatment effectively and selectively increases the activity of interneurons, allowing localized and pronounced increase in inhibitory tone within neighboring excitatory neurons.
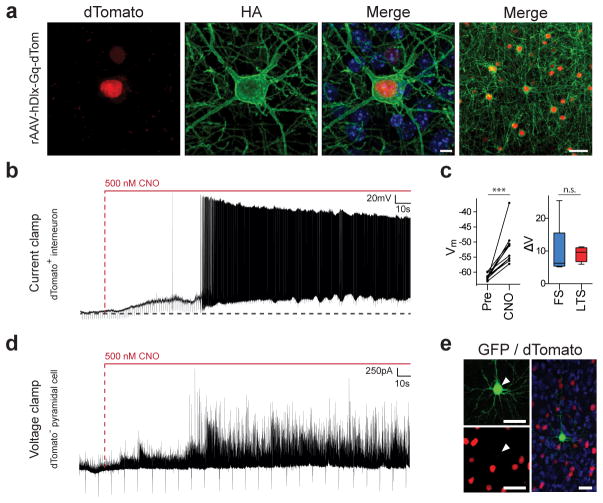
Figure 3. rAAV-hDLX-Gq-DREADD allows chemogenetic modulation of interneuronal activity in mice.
Adult C57Bl6 mice (n=18) were stereotactically injected with rAAV-hDlx-HA-Gq-DREADD-P2A-NLS-dTomato in somatosensory cortex and were either analyzed by immunostaining for the indicated markers after 7 days or sectioned for electrophysiological recording after 4 weeks. (a) Representative example of co-localization between dTomato and HA-tagged Gq-DREADD in somatosensory cortex layer VI. Note the expected membrane localization of the Gq-DREADD. (b) Effect of CNO on membrane potential and firing measured in current clamp configuration of a LTS interneuron. Horizontal dash line indicates baseline membrane potential (−60 mV). (c) Left panel: population data of effect of CNO on membrane potential. Average membrane potential (for a period of 25sec) 30min pre- and 2min post-CNO for individual cells (n=10 cells). Right panel: change in membrane potential for FS (10.2 ± 3.2mV, n=6 cells) and LTS cells (9.6 ± 0.7mV, n=4 cells). [AU QUERY: Please include test statistic and exact –values for these test] (d) Effect of CNO on inhibitory drive measured in voltage clamp configuration from a whole-cell recording of a pyramidal neuron within the area of viral infection. (e) Post-hoc immunostaining of a biocytin filled pyramidal neuron within the site of viral injection surrounded by interneurons infected by the rAAV-hDlx-HA-Gq-DREADD-P2A-NLS-dTomato. Nuclei were counterstained with Dapi (blue). Arrowheads point to the recorded cell. Unpaired T-test: *** = p-value <0.001, n.s. = non-significant. Quantification are indicated in the text as mean ± s.e.m and are represented as box-and-whisker plot with upper and lower whiskers represent the maximum and minimum value respectively and the box represent upper, median and lower quartile. Vertical dash lines in indicate CNO entry in the bath. Scale bars represent 5μm (a, left panels), or 20μm (a, right panel and e).
rAAV-mDlx is selectively expressed within GABAergic interneurons in a wide variety of vertebrate species
The high degree of conservation of the mDlx enhancer sequence across vertebrates suggests a conserved role in the transcriptional regulation of the Dlx5/6 genes14. We thus sought to test whether the use of rAAV-Dlx could be extended to direct the expression of a variety of different reporters and effectors specifically in interneurons in non-genetically tractable species. This proved successful, as we were able to achieve both comparable levels and the fidelity of expression of rAAV-Dlx in neurons from five different species. Similar to observations in mice, the expression of GFP or RFP in zebra finches, ferrets, gerbils and marmosets infected with the rAAV-Dlx co-localized with cells expressing the pan interneuron markers GABA or GAD67 (Fig. 4a–d). Most relevant to humans, infection of neurons obtained from patient-derived induced pluripotent stem-cells with the rAAV-mDlx-GFP showed that the majority of the cells expressing GFP co-expressed GABA (Fig. 5a). To confirm these findings, we introduced rAAV-Gq-dTomato into a co-culture containing both dorsalized human embryonic stem-cells (hESCs - mainly glutamatergic neurons), as well as a transgenic hESC reporter line expressing Citrine specifically in MGE-derived interneurons (hESC-Lhx6-Citrine). Immunostaining analysis showed that the majority of cells expressing the nuclear dTomato also expressed Citrine, while none of the dorsalized hESCs expressed dTomato (Fig. 5b). These results show that the Dlx enhancer restricts rAAV-mediated gene expression to interneurons in all vertebrate species tested including human. In addition, slice and in vivo recordings from zebra finches injected with the rAAV-mDlx-GFP show that the GFP expressing cells exhibited electrophysiological and morphological properties characteristic of interneurons (Supplementary Fig. 5). The success of this approach further highlights the potential of the rAAV-Dlx for the identification and characterization of interneuron subtypes in non-genetically tractable animal models. Using in vitro slice recordings from gerbils injected with rAAV-mDlx-ChR2-mCherry in the auditory cortex (A1), we could show that 1ms pulses of blue light elicited action potentials in cells expressing mCherry (Supplementary Fig. 6a,b). As expected, voltage-clamp recording of non-infected cells in the vicinity of the injection site show an abrupt interruption of action potentials upon light activation (Supplementary Fig. 6c,d). The rAAV-mDlx-ChR2-mCherry thus allows local optogenetic control of interneurons. In vivo calcium imaging of ferret visual cortex injected with rAAV-mDlx-GCaMP6f showed an activation of interneurons upon presentation of visual stimuli (Supplementary Fig. 7). This demonstrated that it is possible to use this tool to monitor the functional activity of neurons in the living brain while presenting behaviorally relevant stimuli to the animal.
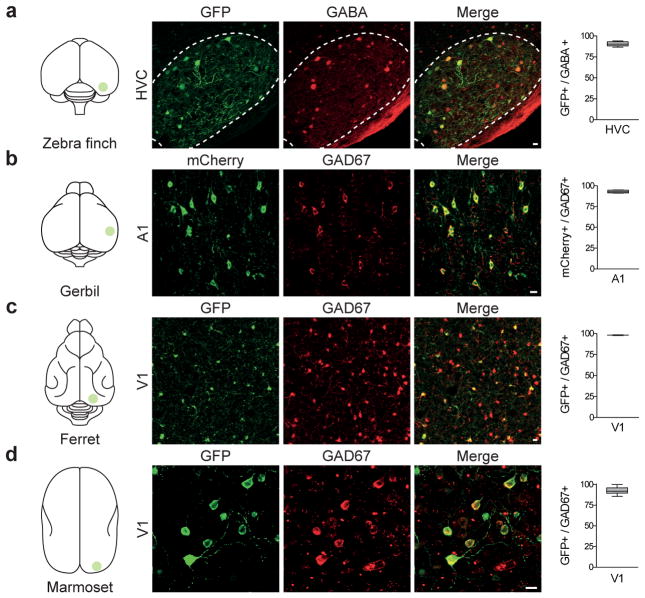
Figure 4. rAAV-mDlx is selectively expressed within GABAergic interneurons in various non-genetic model organisms.
(a) Adult Zebra Finches (n=6) were injected with rAAV-mDlx-GFP in HVC, and analyzed by immunostaining for GABA immunoreactivity after 2–4 weeks. Representative example of co-localization between GFP and GABA and corresponding quantification (90.6 ± 1.1%, n=104 cells, 4 animals). (b) Adult Gerbils (n=4) were injected with rAAV-mDlx-ChR2-mCherry in V1, and analyzed by immunostaining for GAD67 immunoreactivity after 2–4 weeks. Representative example of co-localization between mCherry and GAD67 and corresponding quantification (94.0 ± 0.3%, n=318 cells, 4 sections from 1 animal). (c) Juvenile ferrets (n=4) were injected with rAAV-mDlx-GCaMP6f in V1, and analyzed by immunostaining for GAD67 immunoreactivity after 2 weeks. Representative example of co-localization between GFP and GAD67 and corresponding quantification (98.2 ± 0.5%, n=1647 cells, 2 animals). (d) A marmoset (n=1) was injected with rAAV-mDlx-GFP in the visual cortex (V1) and analyzed by immunostaining for GAD67 immunoreactivity after 3 months. Representative example of co-localization between GFP and GAD67 and corresponding quantification (92.6 ± 1.2%, n=215 cells, 3 sections from 1 animal). Quantification are indicated in the text as mean ± s.e.m and are represented as box-and-whisker plot with upper and lower whiskers represent the maximum and minimum value respectively and the box represent upper, median and lower quartile. Dashed lines represent limits of the indicated anatomical structures. Scale bars represent 10μm.
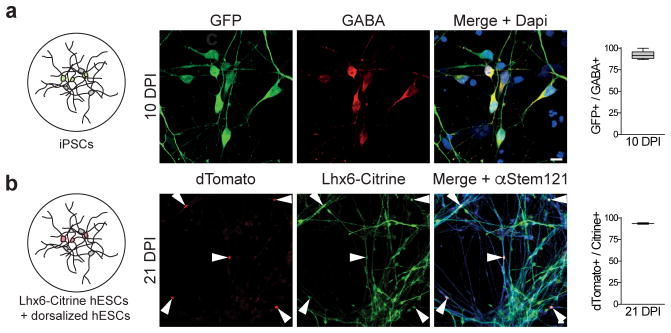
Figure 5. rAAV-Dlx restricts expression to interneurons derived from iPSCs and human embryonic stem cells.
(a) Neuronal cultures derived from human iPSCs were inoculated with rAAV-mDlx-GFP at DIV50 and analyzed by immunostaining for GABA immunoreactivity 10 days after inoculation (10 DPI). Representative example of co-localization between GFP and GABA and corresponding quantification (92.3 ± 1.3%, n=307 cells, 6 coverslips from 2 independent experiments). (b) Co-culture of excitatory neurons derived from hESCs and GABAergic interneurons derived from a transgenic hESC line expressing Citrine under the control of Lhx6 promoter were inoculated with rAAV-hDlx-Gq-DREADD at day1 of co-culture. Cells expressing dTomato were analyzed by immunostaining for Citrine and α-Stem-121 immunoreactivity 21 days after inoculation (21 DPI). Representative example of co-localization between dTomato and Citrine and corresponding quantification (93.6 ± 0.3%, n=128 cells, 2 coverslips from 2 independent experiments). Quantification are indicated in the text as mean ± s.e.m and are represented as box-and-whisker plot with upper and lower whiskers represent the maximum and minimum value respectively and the box represent upper, median and lower quartile. White arrowheads represent cells expressing the reporter dTomato. Scale bar represents 10μm (a) and 20μm (b).
DISCUSSION
Transgenic approaches in mice have revolutionized our understanding of circuits. However, this success highlights the limitations posed by our present inability to efficiently and reliably identify and manipulate specific neuronal subtypes in non-genetically tractable species. Here, we report a new approach, using recombinant adeno-associated viruses (rAAVs) that directly address this challenge. rAAVs have been previously used as a safe and effective method for gene delivery by researchers and clinicians in numerous contexts, but their use has been hampered by the limited cell-type selectivity that can be achieved by natural or engineered tropism8. Another major limitation of rAAVs is their naturally limited DNA payload (~4.7kb). This has reduced the range of regulatory sequences that can be used to achieve cellular specific expression9. One way to circumvent this limitation is to use short cis-regulatory sequences that can regulate gene expression. Enhancers are CIS-acting element composed of transcription-factor-binding-sites that are responsible for tissue-specific transcriptional regulation of gene expression. Several laboratories have taken advantage of the high degree of conservation of non-coding genomic sequences to identify putative enhancers with conserved activity between mice and humans 13,28. The mouse Dlx5/6 enhancer is composed of a core sequence of 297 nucleotides showing 98% identity with human and 75% with zebrafish and has been extensively used to drive gene expression exclusively in interneurons in various contexts11,15–18. A surprising finding from this work is the differential activity of the mDlx or hDlx enhancers when used in transgenic versus rAAV contexts. While the enhancer used in the context of rAAVs continues to direct interneuron expression in adult mice, it fails to do so endogenously or in transgenics. It seems likely that a combination of transcription factors cooperates to direct the expression of Dlx5/6 during embryonic development. The most likely cause of the loss of Dlx5/6 expression in adults is either that the transcription factors that bind and activate the mDlx enhancer decrease in expression level or that the enhancer itself is epigenetically silenced postnatally. Either possibility is tenable, as the virus by being episomal may escape epigenetic silencing or the large number of rAAV virions might be able to overcome reductions in transcription factor expression, such that rAAV expression can persist reliably in adults. Finally, in areas other than the telencephalon this virus showed reliable expression albeit in populations other than those expressing GABA. This likely reflects the presence within these cells of transcription factors able to specifically bind and activate the Dlx5/6 enhancer and opens the possibility to explore these cellular populations in the future.
Questions of how the mDlx/hDlx enhancer functions aside, our approach provides a toolbox for studying inhibitory GABAergic function across species in hippocampus, cortex and striatum and has the strong potential for utility for clinical intervention. In particular, we showed that administration of CNO, the ligand for Gq-DREADD, selectively increases the activity of interneurons, resulting in an increase of local inhibition on neighboring excitatory pyramidal neurons. In mice the use of optogenetic closed-loop recruitment of interneurons has proven effective for ameliorating seizures29. It has also been recently shown that reducing excitatory drive in pyramidal cells using the Gi-DREADD can reduce the severity of seizures in mouse models of focal epilepsy30. The combination of the selectivity of the rAAV-mDlx with the time control of the Gq-DREADD thus opens the possibility to locally increase the inhibitory drive of interneurons and could thus be beneficial to restore the E/I balance in patients with intractable focal epilepsy.
Our results demonstrate that the mDlx enhancer can be used in the context of rAAVs to broadly and specifically target and manipulate interneurons from the Dlx5/6 lineage. Our success in targeting these interneurons in mice, zebra finches, ferrets, gerbils, marmosets, as well as in both human iPSC-derived and ESCs-derived interneurons illustrates the potential of this approach for extending our understanding of interneuron function across a broad range of species. This raises the question of whether this approach can be generalized for the targeting of additional neuronal populations. Other enhancers with a similar degree of conservation have been already been identified28, suggesting this might be possible. In addition, the emergence of high throughput screening methods allowing genome-wide mapping of enhancers based on chromatin configuration and epigenetic marks has led to the discovery of many others13,31. It is tempting to speculate that the systematic screening of the sequences for their ability to direct expression in specific cell-types will provide an approach for the development of novel viral tools to target and manipulate individual functional cell types with unprecedented specificity.
ONLINE METHODS
Animals
Mice - Female C57BL/6J mice (Mus musculus; 10 weeks old) were obtained from Taconic (New York, NY). Zebra Finches - Adult male zebra finches (Taeniopygia guttata; 90d after hatching) were obtained from an outside breeder and maintained in a temperature- and humidity-controlled environment with a 12/12 h light/dark schedule. Gerbil - Adult gerbils (Meriones unguiculatus; 85 to 95 days old) were obtain from breeding pairs from Charles River Laboratories (Cambridge, MA). For mice, zebra finches and ferret, all animal maintenance and experimental procedures were performed according to the guidelines established by the Institutional Animal Care and Use Committee at the New York University Langone Medical Center. Ferret - Juvenile female ferrets (Mustela putorius furo; Postnatal day 27–30) were obtained from Marshall Farms. All procedures were approved by the Max Planck Florida Institute for Neuroscience Institutional Animal Care and Use Committee. Marmoset - A female marmoset (Callithrix jaccus; 4.3 years old) was obtained from the University of Utah. All procedures were approved by the Salk Institute for Biological Studies Institutional Animal Care and Use Committee. All procedures adhered to the standards of the National Institutes of Health. All the animals were maintained in a 12 light/12 dark cycle with a maximum of 5 animals per cage for mice and one animal per cage for all other species.
Surgery
Animals were anesthetized under isofluorane (1–3% in oxygen) and placed in a stereotactic head frame on a temperature-controlled heating pad. A craniotomy and a durotomy were performed above region of interest. The animals were injected with 50–500nl of the indicated virus at a rate of 10–25nl/min using a sharp glass pipette (25–35 mm diameter) that was left in place for 5–15 min after the injection to minimize backflow. The craniotomy site was covered with sterile bone wax, the surgical opening was closed with Vetbond and the animals were returned to their home cage for at least one week. The injection sites were defined by the following coordinates : Mice somatosensory cortex S1: 1.0mm posterior, 3.0mm lateral, 0.7 / 0.4mm ventral relative to bregma; Mice hippocampus CA1: 1.6mm posterior, 1.8mm lateral, 1.2mm ventral relative to bregma; Mice striatum: 0.5mm posterior, 2.0mm lateral, 3.2mm ventral relative to bregma; Zebra Finches HVC: 0,2mm anterior, 2.1 / 2.3 / 2.5mm lateral, 0.4mm ventral relative to the bifurcation of the sagittal sinus; Gerbils auditory cortex A1: 3.0mm anterior, 6.5mm lateral, 0.3mm ventral relative to lambda; Ferrets visual cortex V1: 2.0mm anterior, 7.5mm lateral, 0.25 / 0.4mm ventral relative to lambda. Marmosets visual cortex V1: 15mm anterior, 5mm lateral, 1mm ventral relative to bregma. For zebra finches, the craniotomy was covered with a silicone elastomer (Kwik-Cast; WPI). For the subset of animals used for in vivo recording, a thin cover glass (3 mm, #1 thickness, Warner Instruments) was affixed to the skull using cyanoacrylate to create a chronic optical window over HVC. For ferrets, the scalp was retracted and a custom titanium headplate adhered to the skull using C&B Metabond (Parkell). One coverglass (5mm diameter, #1.5 thickness, Electron Microscopy Sciences) was adhered to a custom titanium cannula using optical adhesive (Norland Products) and placed onto the brain to gently compress the underlying cortex and dampen biological motion during imaging. The cranial window was hermetically sealed using a stainless steel retaining ring (5/16″ internal retaining ring, McMaster-Carr), Kwik-Cast (World Precision Instruments), and Vetbond (3M). For marmosets, the craniotomy was covered with an artificial dura made from bovine cells and the bone flap was fixed in place using Vetbond (3M). Following placement of the bone flap, the skin was sutured closed.
rAAV cloning and production
Detailed maps, sequences and plasmids corresponding to the viral constructs were deposited at Addgene. To clone these constructs, the plasmid AAV-CaMKIIa-GCaMP6f-P2A-nls-dTomato (a gift from Jonathan Ting - Addgene plasmid # 51087) was used to create a backbone containing either the mDlx or the hDlx enhancer sequences. The mDlx enhancer sequence (530bp) was amplified by PCR from mouse genomic DNA using the following primers: 5′-tatacactcacagtggtttggc - 3′ and 5′-cttcctactgtgaaactttggg - 3′. The hDlx enhancer sequence (541bp) was amplified by PCR from human genomic DNA using the following primers: 5′-ttcagaatgttatgcactcaca - 3′ and 5′-cccaaagtttcacagtaggaag - 3′. The enhancers, reporters and effectors detailed below were subcloned using the Gibson Cloning Assembly Kit (NEB- E5510S) following standard protocol. For rAAV-mDlx-GFP, we amplified the GFP coding sequence from the plasmid pNeuroD-ires-GFP32 (a gift from Franck Polleux - Addgene plasmid # 61403). For the rAAV-mDlx-Flex-GFP, we amplified the GFP coding sequence and synthetized the fragments containing the non-compatible lox sites by primer annealing33. For rAAV-mDlx-GCaMP6f, we amplified the GCaMP6f coding sequence from the plasmid pGP-CMV-GCaMP6f34 (a gift from Douglas Kim - Addgene plasmid # 40755). For rAAV-mDlx-ChR2, we amplified the ChR2 coding sequence from the plasmid pAAV-Ef1a-DIO hChR2(E123T/T159C)-EYFP35 (a gift from Karl Deisseroth - Addgene plasmid # 35509) fused with the coding sequence for mCherry. For rAAV-hDlx-Gq-DREADD, we amplified the Gq-DREADD coding sequence from the plasmid pAAV-hSyn-DIO-hM3D(Gq)-mCherry36 (a gift from Bryan Roth - Addgene plasmid # 44361) and a P2A-NLS-dTomato from the plasmid AAV-hSyn1-GCaMP6s-P2A-nls-dTomato (a gift from Jonathan Ting - Addgene plasmid # 51084). The rAAVs were produced using standard production method. PEI was used for transfection and OptiPrep gradient (Sigma, USA) was used for viral particle purification. Titer was estimated by qPCR with primers annealing of the WPRE sequence that is common to all constructs. All batches produced were in the range of 10E+10 to 10E+12 viral genomes/ml.
Primary culture
Primary dissociated cortical neurons were prepared from C57BL/6J mice at postnatal day 0 (P0) using a previously described protocol 37. Dissociated neurons were plated onto coverslips pre-coated with polyethyleneimine (0.1%; Sigma, USA) plus laminin (20 μg/ml; Life Technologies, USA) at a density of 3000 cells/mm2. The cultures were treated with AraC (1 μg/ml; Sigma, USA) on day 5 in vitro and maintained for up to 21 days after plating. All primary cultures were tested for mycoplasma contamination using standard protocol.
iPSCs derived neuronal culture
The iPSC line iPSC49026 was generated at the NYSCF Research Institute by mRNA/miRNA reprogramming technology from skin fibroblasts of a de-identified male healthy donor38. Skin biopsy was obtained upon institutional review board approval and receipt of informed consent. iPSCs were maintained in feeder free conditions, seeded onto matrigel--coated plates (BD Biosciences) in the presence of mTeSR1 medium (StemCell Technologies). The IPSC derived neurons were generated using the Livesey protocol39 with minor modifications: the protocol was followed exactly up to the differentiation step where BrainPhys medium was used with 1uM dbcAMP, 40ng/ml BDNF, 40ng/ml GDNF and 1ug/ml Laminin. Cells were fixed with 4% PFA at 60 DIV. At day 50, the cells were inoculated with rAAV-mDlx-GFP at a MOI of ~150K vg/cell and fixed at day 60. All iPSC cultures were tested for mycoplasma contamination using standard protocol.
hESCs derived neuronal culture
Lhx6-Citrine GABAergic interneurons were differentiated from an Lhx6-citrine H9 reporter hESC line using a protocol as previously described40. Human excitatory neurons were obtained by forced expression of Ngn2 in a human iPSC line. At day 0, 200k hESC-Lhx6-Citirne interneurons and 200K Ngn2-induced excitatory neurons were plated on rat cortical feeders in a single 24-well. At day 1, the co-culture was inoculated with rAAV-hDlx-Gq-DREADD at a MOI of 100K vg/cell and fixed at day 21. All cells were manipulated and cultured in a sterile environment using ood cell culture practice and tested for mycoplasma using standard methods. All hESCs cultures were tested for mycoplasma contamination using standard protocol.
Electrophysiological recordings - mice
Slice preparation. Virally injected mice were anesthetized with intraperitoneal injection of pentobarbital (100mg/kg body weigth). Upon loss of reflexes, mice were transcardially perfused with ice-cold oxygenated ACSF containing the following (in mM): 87 NaCl, 75 sucrose, 2.5 KCl, 1.25 NaH2PO4, 26 NaHCO3 10 glucose, 1 CaCl2 and 2 MgCl2. Mice were then decapitated and 300 μm thick coronal slices were sectioned using a Leica VT-1200-S vibratome and incubated in a holding chamber at 32–35°C for 15–30 min followed by continued incubation at room temperature for at least 45–60 min before physiological recordings. Slice containing the injection site were transferred in a recording chamber submerged with oxygenated ACSF containing the following (in mM): 125 NaCl, 2.5 KCl, 1.25 NaH2PO4, 26 NaHCO3, 10 glucose, 2 CaCl2 and 1 MgCl2 (pH = 7.4, bubbled with 95% O2 and 5% CO2). Current clamp. For interneuron recording, 10 μM CNQX, 25 μM AP-5 and 10 μM SR-95531 were also added to block AMPA, NMDA and GABAA receptors, respectively, in order to measure cell-intrinsic effect of CNO application. Whole-cell current clamp recordings were obtained from visually identified dTomato expressing cells using borosilicate pipettes (3–5 MΩ) containing (in mM): 130 K-gluconate, 6.3 KCl, 0.5 EGTA, 10 HEPES, 4 Mg-ATP, 0.3 Na-GTP and 0.3% biocytin (pH adjusted to 7.3 with KOH). Upon break-in, series resistance (typically 15–25 MΩ) was compensated and only stable recordings (<20% change) were included. Data were acquired using an MultiClamp 700B amplifier (Molecular Devices), sampled at 20 kHz and filtered at 10 kHz. All cells were held at −60mV with a DC current and current steps protocols were applied to obtain firing pattern and extract basic sub- and suprathreshold electrophysiological properties. Voltage clamp. dTomato negative cells were selected according to their pyramidal cell shaped soma under IR-DIC visualization and recorded with pipettes containing (in mM): 130 Cs-gluconate, 0.5 EGTA, 7 KCl, 10 HEPES, 4 Mg-ATP, 0.3 Na-GTP, 5 phosphocreatine, 5 QX-314 and 0.3% biocytin (pH adjusted to 7.3 with CsOH). Cells were held continuously at 0 mV for baseline and CNO application. For both current and voltage clamp recording, a baseline of at least 2 min was recorded before CNO (500 nM) was bath applied for at least 5 min. Small pulses (−20 pA or −5 mV, 100ms at 0.2 Hz or 0.5 Hz) were applied all throughout the baseline and CNO application to monitor series resistance changes. Data were analyzed off-line using Clampfit 10.2 software (Molecular Devises). Morphological identification. To confirm the morphology of recorded cells, slices were fixed for 1hour in 4% PFA following the recording, and then in 30% sucrose for storage. For biocytin staining, slices were washed with PBS and incubated with streptavidin conjugated Alexa (1:500) and 0.3% Triton X-100 in PBS overnight. Slices were then washed with PBS and mounted on microscope slides. Confocal image stacks were acquired using a Zeiss LSM510 microscope with a 20X objective.
Electrophysiological recordings - Zebra Finches
Slice preparation. All extracellular solutions were adjusted to 310 mOsm, pH 7.3–7.4, and aerated with a 95%/5% O2/CO2 mix. Zebra finches were first deeply anesthetized with an intramuscular injection of pentobarbital (40 mg/kg in saline). Once the animal was no longer responsive to a toe pinch, it was quickly decapitated and the brain was removed from the skull and submerged in cold (1–4°C) oxygenated zero-sodium ACSF containing the following (in mM): 225 sucrose, 3 KCl, 1.25 NaH2PO4, 26 NaHCO3, 10 D-(+)-glucose, 2 MgSO4, 2 CaCl2). The brain was then cut along the sagittal plane and the medial side of the right hemisphere was glued onto a custom-made slice block. The block was constructed such that the vibratome blade entered the brain posteriorly at a 12° angle of elevation with respect to the glued medial side. Slices (300 m) containing HVC were produced using a vibratome (VT1200S; Leica) outfitted with a ceramic blade (Cadence) and an advancing speed of 0.12 mm/s. After cutting, slices were transferred to a recovery chamber with reduced sodium ACSF containing the following (in mM): 60 NaCl, 75 sucrose, 2.5 KCl, 1.2 NaH2PO4, 30 NaHCO3, 25 D-(+)-glucose, 20 HEPES, 2 MgSO4, 2 CaCl2) at a temperature of 32°C for 25 min. The slices remained in the recovery chamber for at least another 40 min at room temperature (23°C) before electrophysiological recording. Current clamp. Whole-cell current clamp recordings were obtained from visually identified GFP expressing cells using borosilicate pipettes (3–5 MΩ) containing (in mM): 0.2 EGTA, 130 K-gluconate, 4 KCl, 2 NaCl, 10 HEPES, 4 ATP-Mg, 0.3 GTP-Tris, 14 phosphocreatine-Tris, 0.3% biocytin and brought to pH 7.25 and 292 mOsm. Upon break-in, series resistance (typically 15–25 MΩ) was compensated and only stable recordings (<20% change) were included. Data were acquired using an MultiClamp 700B amplifier, sampled at 20 kHz and filtered at 10 kHz. Current steps protocols were applied to obtain firing pattern and extract basic sub- and suprathreshold electrophysiological properties. Neuronal cell bodies were visualized for recording using an Axio Examiner A1 (Zeiss) fixed-stage upright microscope using IR-Dodt illumination through a 40 water-immersion objective. GFP positive neurons were targeted under visual guidance fluorescing in the free wavelength using an Axiocam MRm fluorescence camera (Zeiss). Morphological identification. To confirm the morphology of recorded cells, slices were fixed for 1hour in 4% PFA following the recording, and then in 30% sucrose for storage. For biocytin staining, slices were washed with PBS and incubated with streptavidin conjugated Alexa (1:500) and 0.3% Triton X-100 in PBS overnight. Slices were then washed with PBS and mounted on microscope slides. Confocal image stacks were acquired using a Zeiss LSM510 microscope with a 20x objective. Two-photon targeted whole-cell recordings. On the day of recording, a small hole (~100μm diameter) was carefully drilled into the implanted cover slip using a carbide bur (0.3 mm diameter) just lateral to the visually determined center of HVC and the recording was performed as previously described41.
Optogenetics and electrophysiological recordings - Gerbil
Slice preparation. Thalamo-cortical brain slice preparation (500 μm) have been previously described42. Current Clamp. The ACSF contained (in mM): 125 NaCl, 4 KCl, 1.2 KH2PO4, 1.3 MgSO4, 24 NaHCO3, 15 glucose, 2.4 CaCl2, and 0.4 L-ascorbic acid and bubbled with 95%O2-5%CO2 (pH 7.3). Before each whole-cell recording, the auditory cortex was identified by extracellular field responses evoked by stimulation of the auditory thalamus (Medial Geniculate Nucleus). Whole-cell current-clamp optogenetic. Current-clamp recordings were obtained from pyramidal, fast spiking, and low threshold spiking neurons in L2/3 of the ACx. Each neuron was verified as thalamo-recipient by recording an MG-evoked response, and all data were collected within layer 2/3. The internal recording solution contained (in mM): 5 KCl, 127.5 K-gluconate, 10 HEPES, 2 MgCl2, 0.6 EGTA, 2 ATP, 0.3 GTP, and 5 phosphocreatine (pH 7.2 with KOH). The tip resistance of the patch electrode filled with internal solution was 5–10 MΩ. Access resistance was 15–30 MΩ, and was compensated by ~70%. All data were acquired at a sampling rate of 10 kHz using a custom-designed IGOR (version 4.08; WaveMetrics, Lake Oswego, OR) macro on an iMac (Apple, Cupertino, CA). A second IGOR macro was used for offline analysis. Firing profiles were evaluated on the basis of responses to positive current pulses (1500 ms) of 400 to 600 pA. For pyramidal neurons inhibitory postsynaptic potentials (IPSPs) were evoked by passing positive current (400 to 600 pA) for 1.5 seconds coupled with 1 ms of blue light (20% intensity, 470 nm – lumen 1600 led: Olympus) at 500 ms. Putative inhibitory cells (fluorescing) were patched using IR-IDC and mCherry filters. Threshold to action potential was achieved by gradually increasing light (470 nm) exposure (1 ms) from 0 to 20% intensity. Input output functions were gathered by exposing the cell to increasing intensity of light (470 nm) exposure (1ms) from 0 to 100% in 10% steps. All neuronal images were collected in vitro with a fluorescent camera (Q-imaging) equipped with an mCherry filter cube (Olympus) using q-capture pro (Q-imaging).
Calcium imaging in mice primary culture and in ferrets
GCaMP6f imaging were performed as previously described37,43. For primary culture, rAAV-mDlx-GCaMP6f (titer was 1×10E+12 vg/ml, 1μl per well in 24-well plate) was added to neuronal cultures at DIV8, and GCaMP imaging was performed 11 days after infection.
Immunohistochemistry
Citations with validation data for each antibod is reported on the provider’s website. For primary culture of mice neurons, iPSC-derived neuronal cultures and hESC-derived neuronal culture, neurons were fixed by incubation with 4% paraformaldehyde and 4% sucrose in phosphate-buffered saline (PBS) at room temperature for 5min and then washed three times with PBS. Fixed neurons were permeabilized with 0.1% Triton X-100/PBS for 5 min, washed three times with PBS, and incubated in Blocking Buffer (3% bovine serum albumin plus 10% normal goat serum in PBS) for 1 h. The cells were then incubated overnight in Blocking Buffer with the indicated combination of the following primary antibodies at 4 °C: rabbit anti-GFP at 1:1000 (Invitrogen - A6455, USA); mouse anti-GAD67 at 1:1000 (Millipore - 1G10.2, USA); mouse anti-Stem-121 (Clontech - Y40410, USA); mouse anti-GABA at 1:1000 (Sigma - A0310, USA). The cells were then washed three times with PBS, they were incubated with Alexa conjugated secondary antibodies (Invitrogen, USA). For the animals injected with the virus, animals were euthanized with Euthasol (source) and transcardially perfused with 4% paraformaldehyde (PFA). The brains were placed in 4% PFA overnight then sectioned at 50–60μm using a Leica VTS1000 vibrosector. Floating sections were permeabilized with 0.1% Triton X-100 and PBS for 30min, washed three times with PBS, and incubated in Blocking Buffer (3% bovine serum albumin plus 5% normal goat serum plus 5% normal donkey serum in PBS) for 1 h. The sections were then incubated overnight in Blocking Buffer with the indicated combination of the following primary antibodies at 4 °C: chicken anti-GFP at 1:1000 (Abcam - ab13970, USA); rabbit anti-GABA at 1:500 (Sigma - A2052, USA); rabbit anti-dsRED at 1:1000 (Clontech - 632496, USA); rabbit anti-Nkx2.1 at 1:2000 (Abcam - ab76013, USA); goat anti-PV at 1:1000 (Swant - PVG-213, USA); rat anti-SST at 1:400 (Millipore – MAB354, USA); rabbit anti-VIP at 1:500 (Immunostar - 20077, USA); goat anti-HA at 1:1000 (Abcam - ab9134, USA) and rabbit anti-GAD65/67 at 1:1000 (Abcam - ab11070, USA) - for ferret. P7 spinal cord preparation were fixed by incubation with 4% paraformaldehyde and incubated overnight in 4% sucrose in phosphate-buffered saline (PBS), then washed three times with PBS and cryosectioned at 30um. Cryosections were incubated in Blocking Buffer (3% bovine serum albumin plus 10% normal goat serum in PBS) for 1h then incubated overnight in Blocking Buffer with the indicated combination of the following primary antibodies at 4 °C: chicken anti-GFP at 1:1000 (Invitrogen - A10262, USA) and rabbit anti-Pax2 1:500 (Invitrogen - 71-6000, USA). The sections were then washed three times with PBS, incubated with Alexa conjugated secondary antibodies at 1:500 (Invitrogen, USA), counterstained with Dapi (Sigma, USA) and mounted to a glass slide using Fluoromount-G (Sigma, USA). Note that for mouse and marmoset immunostaining for GAD67, mouse anti-GAD67 at 1:200 (Millipore - MAB506, USA) was incubated during 48h at room temperature. Incubation of primary, secondary and intermediate washes were performed in 0,1M PB without detergent. Confocal images were acquired using a Zeiss LSM510 and LSM800 confocal microscopes.
Quantification and statistics
For quantification of co-localization, cells expressing the indicated reporter were counted using only the corresponding color channel, then among these cells, the number of cells co-expressing the marker of interest were counted. A cell was considered to be positive for a given marker if the corresponding signal was significantly above background fluorescence. For animals, the brain sections containing the highest number of cells expressing the viral reporter were used for quantification. Several sections from the same animal were used when indicated. For in vitro studies, all the cells from randomly selected fields were counted. Data collection and analysis were not performed blind to the conditions of the experiments, but experimenters from different research groups performed the quantification. The ratio of cells co-expressing both markers over the total number of cells expressing only the reporter was then calculated, reported in the text as mean ± s.e.m and represented in the figures as box-and-whisker plot with upper and lower whiskers represent the maximum and minimum value respectively and the box represent upper, median and lower quartile. Quantifications were performed using a minimum of 2 independent biological replicates (the specific number of cells, animals and conditions are indicated for each individual quantification in the figure legends). An unpaired t-test was performed to estimate the statistical difference in Fig. 3c right and left panels (equal variance between the two populations was not tested; df=8 and 18 respectively). No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications.
Supplementary Material
Acknowledgments
We wish to thank Stephanie Gerard, Luke Sjulson and Timothy Petros for helpful discussions and comments on the manuscript. This work was supported by grants from the National Institutes of Health (NIH): MH071679, NS08297, NS074972, MH095147, as well as support from the Simons Foundation (SFARI) (GF); MH066912 (SSA); EY022577 and MH063912 (EMC).
Footnotes
ACCESSION CODES
The plasmids for the production of rAAV are available at Addgene with the following accession numbers: pAAV-mDlx-GFP #83900; pAAV-mDlx-GCaMP6f #83899; pAAV-mDlx-ChR2-mCherry #83898; pAAV-hDlx-Gq-DREADD-dTomato #83897; pAAV-hDlx-Flex-GFP #83895.
AUTHOR CONTRIBUTIONS
J.D. conceived and designed the viral constructs, designed and performed experiments, analyzed data, prepared figures and wrote the manuscript. Q.C., C.L. and R.L. performed experiments, analyzed data, prepared figures related to mouse primary cultures. R.T performed the slice recording experiments, analyzed the data, and prepared the associated figures and text. GA.S. and S.R. performed viral injections and IHC on mice. Q.X. and L.G. produced the viruses at NYUAD using plasmids conceived and generated at NYU. G.K. performed experiments related to zebra finch. A.L.J, G.B.S and D.W. performed experiments related to ferrets. V.S.K and T.M.M performed experiments related to gerbils. J.H.R., M.C.A and M.S.R. performed experiments related to marmosets. I.K. and T.R. performed experiments related to iPSCs. J.C. and S.A. performed experiments related to hESCs, B.M and D.J.S performed experiments related to spinal cord. S.S.A, E.C., D.J.S, D.F., V.F, M.A.L, S.N., J.H.R., D.H.S., G.F., B.R. and G.Fg helped with the study design, G.Fi. helped with study design, manuscript and figure preparation and supervised the project. All authors edited and approved the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: The New York University Langone Medical Center has filed patent applications related to this work with J.D and G.F listed as inventors.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon request.
References
- 1.Rudy B, Fishell G, Lee S, Hjerling-Leffler J. Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Dev Neurobiol. 2011;71:45–61. doi: 10.1002/dneu.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marín O. Interneuron dysfunction in psychiatric disorders. Nat Rev Neurosci. 2012:1–14. doi: 10.1038/nrn3155. [DOI] [PubMed] [Google Scholar]
- 3.Kepecs A, Fishell G. Interneuron cell types are fit to function. Nature. 2014;505:318–326. doi: 10.1038/nature12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taniguchi H, et al. A Resource of Cre Driver Lines for Genetic Targeting of GABAergic Neurons in Cerebral Cortex. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nathanson JL, Yanagawa Y, Obata K, Callaway EM. Preferential labeling of inhibitory and excitatory cortical neurons by endogenous tropism of adeno-associated virus and lentivirus vectors. Neuroscience. 2009;161:441–450. doi: 10.1016/j.neuroscience.2009.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han X, et al. Millisecond-timescale optical control of neural dynamics in the nonhuman primate brain. Neuron. 2009;62:191–198. doi: 10.1016/j.neuron.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nassi JJ, Avery MC, Cetin AH, Roe AW, Reynolds JH. Optogenetic Activation of Normalization in Alert Macaque Visual Cortex. Neuron. 2015;86:1504–1517. doi: 10.1016/j.neuron.2015.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kotterman MA, Schaffer DV. Engineering adeno-associated viruses for clinical gene therapy. Nat Rev Genet. 2014;15:445–451. doi: 10.1038/nrg3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Z, Yang H, Colosi P. Effect of Genome Size on AAV Vector Packaging. Molecular Therapy. 2009;18:80–86. doi: 10.1038/mt.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi JH, et al. Optimization of AAV expression cassettes to improve packaging capacity and transgene expression in neurons. Mol Brain. 2014;7:17. doi: 10.1186/1756-6606-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zerucha T, et al. A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. Journal of Neuroscience. 2000;20:709–721. doi: 10.1523/JNEUROSCI.20-02-00709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghanem N, Yu M, Poitras L, Rubenstein JLR, Ekker M. Characterization of a distinct subpopulation of striatal projection neurons expressing the Dlx genes in the basal ganglia through the activity of the I56ii enhancer. Developmental Biology. 2008;322:415–424. doi: 10.1016/j.ydbio.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 13.Visel A, et al. A High-Resolution Enhancer Atlas of the Developing Telencephalon. Cell. 2013;152:895–908. doi: 10.1016/j.cell.2012.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghanem N, et al. Regulatory roles of conserved intergenic domains in vertebrate Dlx bigene clusters. Genome Research. 2003;13:533–543. doi: 10.1101/gr.716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stühmer T, Puelles L, Ekker M, Rubenstein JLR. Expression from a Dlx gene enhancer marks adult mouse cortical GABAergic neurons. Cerebral Cortex. 2002;12:75–85. doi: 10.1093/cercor/12.1.75. [DOI] [PubMed] [Google Scholar]
- 16.Stenman J, Toresson H, Campbell K. Identification of two distinct progenitor populations in the lateral ganglionic eminence: implications for striatal and olfactory bulb neurogenesis. Journal of Neuroscience. 2003;23:167–174. doi: 10.1523/JNEUROSCI.23-01-00167.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monory K, et al. The endocannabinoid system controls key epileptogenic circuits in the hippocampus. 2006;51:455–466. doi: 10.1016/j.neuron.2006.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyoshi G, et al. Genetic Fate Mapping Reveals That the Caudal Ganglionic Eminence Produces a Large and Diverse Population of Superficial Cortical Interneurons. Journal of Neuroscience. 2010;30:1582–1594. doi: 10.1523/JNEUROSCI.4515-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cobos I, Long JE, Thwin MT, Rubenstein JL. Cellular patterns of transcription factor expression in developing cortical interneurons. Cerebral Cortex. 2006;16(Suppl 1):i82–8. doi: 10.1093/cercor/bhk003. [DOI] [PubMed] [Google Scholar]
- 20.Xu X, Roby KD, Callaway EM. Mouse cortical inhibitory neuron type that coexpresses somatostatin and calretinin. J Comp Neurol. 2006;499:144–160. doi: 10.1002/cne.21101. [DOI] [PubMed] [Google Scholar]
- 21.Punnakkal P, von Schoultz C, Haenraets K, Wildner H, Zeilhofer HU. Morphological, biophysical and synaptic properties of glutamatergic neurons of the mouse spinal dorsal horn. J Physiol (Lond) 2014;592:759–776. doi: 10.1113/jphysiol.2013.264937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fenno LE, et al. Targeting cells with single vectors using multiple-feature Boolean logic. Nature Methods. 2014;11:763–772. doi: 10.1038/nmeth.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roth BL. DREADDs for Neuroscientists. Neuron. 2016;89:683–694. doi: 10.1016/j.neuron.2016.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc Natl Acad Sci USA. 2007;104:5163–5168. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexander GM, et al. Remote control of neuronal activity in transgenic mice expressing evolved G protein-coupled receptors. 2009;63:27–39. doi: 10.1016/j.neuron.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou D, et al. DREADD in parvalbumin interneurons of the dentate gyrus modulates anxiety, social interaction and memory extinction. Curr Mol Med. 2016;16:91–102. doi: 10.2174/1566524016666151222150024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Urban DJ, Roth BL. DREADDs (Designer Receptors Exclusively Activated by Designer Drugs): Chemogenetic Tools with Therapeutic Utility. Annu Rev Pharmacol Toxicol. 2015;55:399–417. doi: 10.1146/annurev-pharmtox-010814-124803. [DOI] [PubMed] [Google Scholar]
- 28.Nord AS, Pattabiraman K, Visel A, Rubenstein JLR. Genomic perspectives of transcriptional regulation in forebrain development. Neuron. 2015;85:27–47. doi: 10.1016/j.neuron.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krook-Magnuson E, Armstrong C, Oijala M, Soltesz I. On-demand optogenetic control of spontaneous seizures in temporal lobe epilepsy. Nat Commun. 2013;4:1376. doi: 10.1038/ncomms2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kätzel D, Nicholson E, Schorge S, Walker MC, Kullmann DM. Chemical-genetic attenuation of focal neocortical seizures. Nat Commun. 2014;5:3847. doi: 10.1038/ncomms4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whalen S, Truty RM, Pollard KS. Enhancer-promoter interactions are encoded by complex genomic signatures on looping chromatin. Nat Genet. 2016:1–10. doi: 10.1038/ng.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guerrier S, et al. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell. 2009;138:990–1004. doi: 10.1016/j.cell.2009.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saunders A, Johnson CA, Sabatini BL. Novel recombinant adeno-associated viruses for Cre activated and inactivated transgene expression in neurons. Front Neural Circuits. 2012;6 doi: 10.3389/fncir.2012.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen TW, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mattis J, et al. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nature Methods. 2012;9:159–172. doi: 10.1038/nmeth.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krashes MJ, et al. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J Clin Invest. 2011;121:1424–1428. doi: 10.1172/JCI46229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu C, et al. Micro-electrode array recordings reveal reductions in both excitation and inhibition in cultured cortical neuron networks lacking Shank3. Mol Psychiatry. 2016;21:159–168. doi: 10.1038/mp.2015.173. [DOI] [PubMed] [Google Scholar]
- 38.Paull D, et al. Automated, high-throughput derivation, characterization and differentiation of induced pluripotent stem cells. Nature Methods. 2015;12:885–892. doi: 10.1038/nmeth.3507. [DOI] [PubMed] [Google Scholar]
- 39.Shi Y, Kirwan P, Livesey FJ. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat Protoc. 2012;7:1836–1846. doi: 10.1038/nprot.2012.116. [DOI] [PubMed] [Google Scholar]
- 40.Maroof AM, et al. Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell. 2013;12:559–572. doi: 10.1016/j.stem.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vallentin D, Kosche G, Lipkind D, Long MA. Neural circuits. Inhibition protects acquired song segments during vocal learning in zebra finches. Science. 2016;351:267–271. doi: 10.1126/science.aad3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kotak VC, et al. Hearing loss raises excitability in the auditory cortex. Journal of Neuroscience. 2005;25:3908–3918. doi: 10.1523/JNEUROSCI.5169-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Q, et al. Imaging neural activity using Thy1-GCaMP transgenic mice. Neuron. 2012;76:297–308. doi: 10.1016/j.neuron.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.