Abstract
Background
Although evidence exists for a selective component at transmission, it is clear that HIV-1 transmission is also to a large extent driven by drift. The variation in inoculum size among different risk groups therefore implies that the adaptation rate of HIV may vary between epidemics with different risk group compositions. Furthermore, factors that govern the rate of within-host evolution may also vary by risk group and therefore contribute to evolutionary differences at the epidemic level.
Methods
We adopted a population genetic approach to test whether the different proportions of multi-variant transmissions are reflected by varying proportions of transmitted diversity between men-having-sex-with-men (MSM), heterosexual (HET) and direct blood contact (BC) sub-populations. To this purpose, we collected all available transmission chain clonal sequence data sets (n = 70) available at the Los Alamos HIV website and through an extensive literature search. To assess evolutionary rate differences among different risk groups, we compiled risk group datasets for several subtypes and directly compared the absolute substitution rate and its synonymous and non-synonymous components.
Results
There was sufficient demographic signal to inform the transmission model in BEAST using env data to compare the transmission bottleneck size between the MSM and HET risk groups, i.e. the largest contributors to HIV spread. We find no indications for a different proportion of transmitted genetic diversity at the population level between these groups. In the direct rate comparisons between risk groups, however, we consistently recover a higher evolutionary rate in the male dominated risk group compared to the HET datasets.
Conclusions
We find that the risk group composition impacts the viral evolutionary rate and therefore potentially also the adaptation rate. In particular, risk group-specific sex ratios, and the variation in within-host evolutionary rates between males and females, imposes evolutionary rate differences at the epidemic level, but we cannot exclude a role of varying transmission rates.
Keywords: HIV, risk group, transmission bottleneck, evolutionary rate
Introduction
The determinants of the human immunodeficiency virus type 1 (HIV-1) evolutionary rate and its variability has been subject to extensive investigation. Because the process that controls which genetic variants survive the genetic bottleneck at transmission determines how the enormous within-host evolutionary potential of HIV-1 is translated into long-term evolution at a population scale level, the impact of transmission dynamics is central to our understanding of HIV evolution. The development of the single genome amplification (SGA) method made it possible to quantify the number of infecting variants and uncover their precise genetic characteristics early upon infection [1]. Studies using this technology have reported results that are in line with earlier findings of risk groups differing in the multiplicity of infection (see [2, 3] and references in [4]), and indicate that no single specific phenotypic trait seems to be consistently associated with transmission [5]. Even the transmission-associated co-receptor tropism may result from the overrepresentation of such variants in the genital tract [6]. It is therefore unsurprising that many of several distinct variants can efficiently start a productive infection, as was shown in a case study by English and colleagues [7]. In line with these findings, comparative analyses of the phenotypic properties have shown that there is no distinction between the efficiency by which the envelope portion of viruses from transmitting and non-transmitting breastfeeding mothers interacts with epithelial cells as well as between their sensitivity to neutralization [8]. The loose association between viral phenotypes and improved transmission was also demonstrated by a study of molecular infectious clone [9]. In the absence of clear differences in infectivity, there is little hope for predicting which variant(s) in the donor will become the founder strain(s) in a new infection [10].
Several lines of evidence however, indicate that transmission is not an entirely stochastic process. The first indications for this came from a number of seminal studies in the early ’90s [11–13] showing that the infecting virus(es) often do not simply mirror the circulating diversity in the donor at the time of transmission. Further evidence that evolution within a host is not always beneficial for transmission stems from studies indicating that, whereas the number and length of glycosylation sites increases over the course of infection, transmitted variants typically have less and/or shorter glycosylated envelope proteins than those found in the donor or during chronic infection [14]. While these patterns have not been consistently observed, recent in-depth examinations of the evolutionary rate difference of HIV-1 at various biological scales have further contributed to the evidence for a selective component at transmission. The slower evolutionary rate at the population level suggests that contemporary strains have a lower transmission efficiency than ancestral ones [15–17], supporting the notion that opposing selection pressures exist at the within and among host level [18, 19]. By examining many transmission pairs, Carlson and colleagues [20] recently corroborated the existence of a selection bias in heterosexual transmissions. Taken together, these observations indicate that selection at transmission can weed out mutations that reduce the efficiency of establishing new infections, but is not strong enough to eliminate stochastic effects on HIV-1 transmission.
Since transmission is not fully deterministic, differences in the magnitude of the virus population bottleneck at transmission - with multi-variant transmission estimated to be twice as common in MSM (~40%) than in HET contacts (~20%) [4] - may affect the long-term evolution of HIV at the epidemic level. As a case in point, when investigating the within versus between host rate difference for HIV-1, we found a markedly lower rate difference in our subtype C transmission chain (a ~2-fold rate difference) [17] as compared to earlier results based on subtype B data (a ~4 to 5-fold rate difference) [16]. Following the argumentation of Lythgoe and Fraser [15], we hypothesized that the dissimilarity in the magnitude of the rate difference follows from the differences in the underlying biological characteristics associated with transmission between the largely MSM driven subtype B epidemic and predominantly HET driven subtype C epidemic. That is, the smaller the number of viruses being transmitted (in HET), the higher the chance that new infections will be established by variants that avoided the accumulation of mutations in the donors. Consequently, the association between the number of transmitted variants and risk group can bring about risk group composition related differences in the tempo of HIV evolution. This is not limited to the host HLA background but can also involve resistance to antiretroviral therapy combinations (cART) and vaccines.
In addition to transmission dynamics, the factors that influence the overall amount of divergence accumulating between the founder strain and transmitted virus are also likely to impact the among-host evolutionary rate. Within-host evolutionary rates have for example been shown to vary with disease progression [21]. In this respect, it is interesting to note that men tend to have higher set point viral loads (spVL), a predictor of disease progression [22], than women [23–25], a difference that can persist for several years [26]. This suggests a potentially complex interplay between transmission and within-host evolutionary dynamics in determining the tempo of HIV-1 evolution.
Here, we investigate whether the risk group composition can affect the viral evolutionary rate at a population scale. We do this by examining what factor - the transmission dynamics, within-host evolution, or a combination of both - imposes HIV-1 evolutionary rate differences among different risk groups. To explore the impact of the transmission bottleneck, we use a population genetic approach to contrast the loss of genetic diversity at transmission between risk groups. To this end, we compiled a large collection of datasets for previously described HIV-1 transmission chains. We describe the viral evolutionary histories with a recently introduced transmission model in BEAST [17] and test for transmission bottleneck size differences with a Bayesian hierarchical phylogenetic model (HPM) approach [27] that incorporates fixed effects [28]. We find no support for a difference in the loss of genetic diversity between the HET and MSM groups. To assess the impact of within-host evolution, we compiled risk group-specific datasets of subtypes A1, B and CRF01 AE and tested for differences in substitution rate. For subtype B and CRF01 AE we find that HIV evolves slower in HET than in MSM epidemics, and that for subtype A1 the evolutionary rate is also lower in the HET than in the injecting drug users (IDU) sub-epidemics, and this may be associated with the varying proportions of males in the examined datasets. These estimates indicate that within-host evolutionary processes can impact differences in between-host evolutionary rates.
Results
Dataset compilation
We compiled a large collection of datasets for the most important risk groups involved in HIV spread: the MSM, HET and BC risk groups. The distribution of transmission chains by risk group and the genomic fragments sequenced are listed in Table 1. The time between infection and sampling of the recipient patients is an important variable for accurately quantifying the loss of diversity at transmission using an population genetic approach [17]. The available number of samples is also important as this determines the amount of information available to the phylogenetic reconstructions. A number of descriptive statistics of these two parameters are summarized in Additional Table 1. Briefly, most recipients were sampled at one time point and both MSM and HET are more frequently sampled earlier since time of infection as compared to BC. This suggests that the demographic events during the initial stages of infection are likely to be better captured in the MSM and HET risk group datasets.
Table 1.
Overview of the dataset composition for assessment of between risk-group bottleneck size differences.
| risk group | # transmission chains | gag | pol | env |
|---|---|---|---|---|
| BC | 7 (8) | 3 | - | 7 |
| HET | 39 (50) | - | 2 | 39 |
| MSM | 24 (26) | 7 | 5 | 23 |
| total | 70 (84) | 10 | 7 | 69 |
The number of transmission chains is listed, with the total number of transmission events between brackets. The majority of the chains (60/70, 85.7%) represent a single transmission event (range: 1 to 6). In some cases, data for multiple genomic regions was available. The number of chains is also given for each genomic region. Most data are available of the env region.
Support for a transmission bottleneck
We first set out to test whether the transmission chain datasets we collected support a bottleneck at transmission. To this purpose, we apply a genealogical transmission chain model that allows the effective viral population size to change upon transmission according to different coalescent models [17]: constant population size (CON, no bottleneck), exponential growth (EXP, the population size upon transmission is an estimable proportion of the donor population size and grows exponentially in the new recipient) and logistic growth (LOG, as in EXP but with logistic growth in the recipient). We independently fit the transmission model with the three different demographic functions to each data set and compare their model fit using marginal likelihood testing [29]. This indicates that a model accommodating a bottleneck (EXP or LOG) is strongly supported by 73% of the transmission chain data sets (Additional Figure 1). There is however a marked difference between the risk groups: whereas we find strong Bayes factor support in 80% of the HET and 77% of the MSM transmission events for a model with a bottleneck, this drops to 30% for the BC risk group. This likely reflects less informative datasets due to longer times between time of infection and time of sampling for the recipients in the BC transmission pairs (see Methods and Additional Table 1).
Both PS and SS sampling estimators converged on the same model for all but one dataset that was retained in the fixed-effects analysis (see below). The small difference in log likelihood between the exponential and logistic growth model (<0.5) cannot be considered as strong support and likely results from the variance of the marginal likelihood estimators. We chose the exponential model for this dataset because the first sample was taken during Fiebig stage II, which is before or at the usual time of peak viral load.
Similar amounts of drift and selection in all risk groups
Despite the evidence for a bottleneck in the overwhelming majority of the datasets, unambiguously estimating its size using the transmission model proved difficult in many cases (see Methods). We therefore restricted our approach to test differences between risk groups to the most informative subset of transmission chains for the env region, which were only available for the HET and MSM risk groups. The demographic function was parametrized according to the best fitting demographic model in the ‘best fit’ analysis (with the exception described above). To test the robustness of our bottleneck size estimates to demographic model specification, we also performed an analysis consistently applying either a EXP or a LOG function to all datasets in the ‘exponential’ and ‘logistic’ analysis respectively. Using our population genetic approach, the magnitude of the bottleneck is estimated as the proportion of the donor effective population size that is transmitted to recipient, but we report the complement of this proportion as the percentage of loss in diversity at transmission.
In our test approach, we allow for sharing of information on the demographic parameters across individuals, but model potential differences in bottleneck sizes among risk groups using a fixed effect. We do not find Bayes factor support for the risk group fixed effect indicating no difference in the loss of genetic diversity in env for both risk groups. The severity of the bottleneck is estimated >99% for both the ‘best fit’ , ‘exponential’ and ‘logistic’ analyses, with individual patient estimates ranging from 34% to 99.9%.
The average difference in ancestral proportion size estimates between the ‘exponential’ and ‘logistic’ analyses was 0.17%, and there was no trend for either model to consistently estimate higher or lower values for this parameter, indicating that our estimates are robust to the demographic parametrization.
Gender ratio drives risk group evolutionary rate differences
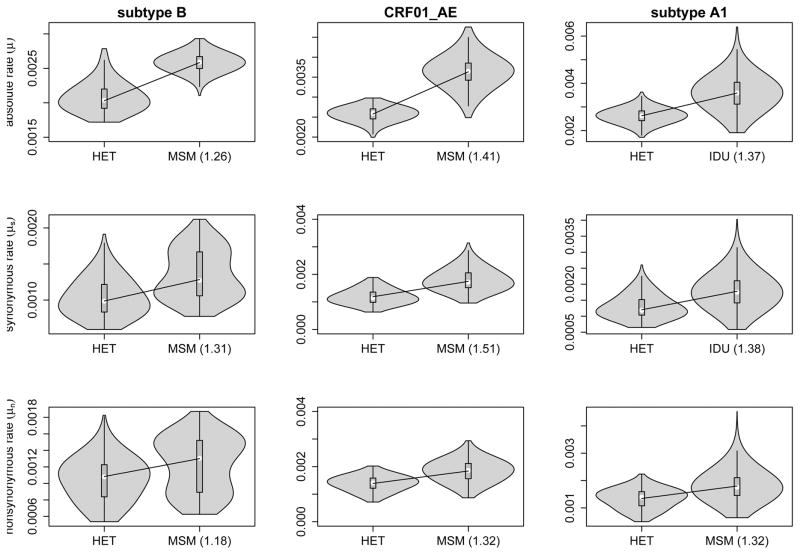
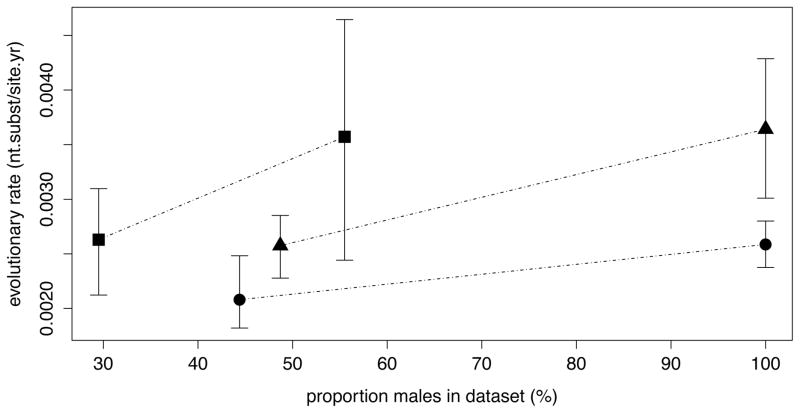
To test for HIV-1 evolutionary rate differences among different risk groups, we collected near full genome data from HET risk groups for subtype A1, B and CRF01 AE, from MSM risk groups for subtype B and CRF01 AE and from IDUs for subtype A1 (Table 2). We consistently find slower HIV evolutionary rates in the HET data sets as compared to MSM. For subtype A1, the HIV evolutionary rate is also slower in the HET risk group compared to the IDU risk group (Figure 1). In order to asses whether the rate differences reflect variation in selective pressure and/or replication rate (generation time), we follow the approach from [21] to obtain posterior estimates of the absolute synonymous (μS) and non-synonymous (μN) rates for all risk group datasets (see Methods). This reveals both elevated μS and μN rates in the MSM and IDU datasets when compared to the HET groups for all subtypes, suggesting that the underlying replication rate is lower in HET groups. By comparing the rates with the proportion of males in the datasets, we see a consistently higher rate for a higher proportion of males within each subtype (Figure 2), and find strong Bayes factor support for this (Table 3). There is however no clear linear relationship between rate and proportion of males independent of the subtype (Figure 2 and Table 3), suggesting that other factors confound evolutionary rate differences associated with gender composition at the epidemiological level.
Table 2.
Risk group evolutionary rate dataset characteristics.
| subtype | risk group (# taxa) | date range | % male |
|---|---|---|---|
| A1 | HET (44) | 1986–2008 | 29.5 |
| IDU (23) | 1997–2007 | 55.5 | |
| B | HET (54) | 1996–2009 | 44.4 |
| MSM (73) | 1983–2012 | 100 | |
| CRF01 AE | HET (77) | 1990–2009 | 48.71 |
| MSM (57) | 2007–2011 | 100 |
In these datasets there were a number of taxa with unknown gender origins. The reported male proportion does not take these into account.
Figure 1. Violin plot representation of the risk group evolutionary rates.
HET, MSM and IDU label the estimate of the evolutionary rate for the respective risk group. The means of each rate estimate are indicate by a white circle. Numbers between brackets indicate the fold increase of the mean relative to the HET mean rate estimate. All rates are in units of nucleotide substitutions per site per year * 10−3.
Figure 2. Illustration of the effect of the proportion of males on the rate estimate.
The means of the rate estimates are indicated by symbols, and the whiskers delimitate the credible interval. Squares: subtA1; Triangles: CRF01 AE; Circles: subtB. The impact of the male proportion is visualized as the slope between the mean of the rate estimates of the risk groups per subtype.
Table 3.
Overview of the rate effect of male proportion on the evolutionary rate.
| risk group | Bayes factor1 | slope2 |
|---|---|---|
| subtA1 | 8.2 | 3.57 × 10−5 ( −1.07 × 10−5 – 9.45 × 10−5) |
| subtB | 33.5 | 8.94 × 10−6 ( 1.26 × 10−6 – 1.54 × 10−5) |
| CRF01_AE | 203.2 | 2.09 × 10−5 ( 7.31 × 10−6 – 3.42 × 10−5) |
The Bayes factor expresses the posterior odds over the prior odds that the evolutionary rate in the male-dominated dataset is higher [57].
Uncertainty in the estimate of the slope is reflected in the 95% HPD interval range between brackets.
Discussion
In this study, we set out to investigate whether the population genetic dynamics associated with transmission, within-host viral replication, or a combination of both, are responsible for evolutionary rate differences between risk groups. For the transmission dynamics to impose such differences, we would expect a difference in bottleneck size among the risk groups and we evaluated this based on a large collection of HIV-1 transmission chains for the three most important risk groups (HET, MSM and BC) involved in HIV spread. Although many transmission chains provide evidence for a transmission-associated population bottleneck, we had to restrict the estimation of its size to the most informative env data sets for HET and MSM risk groups. Formal testing between the two risk groups does not provide any evidence for differences in loss of diversity at transmission, which is very high in both HET and MSM. Although we cannot see any reason for a bias against including datasets with high multiplicity of infection in either risk groups, we acknowledge that our conclusion is restricted to the relatively limited subset of informative data sets.
The absence of a bottleneck size difference is in agreement with many founder effects in HET (~80%) and MSM (~60%) transmission [5], a scenario under which many transmissions will generally be represented by a single phylogenetic branch connecting the donor and recipient viral populations. In such circumstances, the measured loss in genetic diversity at transmission will critically rely on the sampled diversity in the donor and recipient. In this respect, it is important to point out that our comparative analysis included both plasma and PBMC samples. Plasma samples reflect the freely circulating virus, which is usually interpreted as recently generated diversity. PBMCs however may represent an archive of both past and current HIV-1 diversity. These cells can also be co-cultured before DNA-extraction and the resulting in vitro picture may not accurately mirror the in vivo diversity. Reassuringly however, it has been shown that the different experimental sampling approaches lead to a similar viral genetic composition [30], and there were no specific biases of either plasma or PBMC between our risk group samples. Other biases may result from differences in the length of infection [31] as well as the therapy history differences in the donor. Unfortunately, we have very limited information on these variables for the datasets we investigate here, which makes it impossible to currently address these issues.
We estimate the size of the bottleneck in env to >99%. This is in agreement with the findings by Edwards et al. (2006) who, also relying on a coalescent approach to infer the decrease in genetic diversity at transmission, estimated a loss of ≥99% of env and gag diversity in 1 MSM couple [32]. By comparing the estimated diversity at transmission in 9 MSM and 27 mother-to-child (MTC) transmissions they also found that the mode of transmission does not seem to impact the severeness of the transmission associated bottleneck [32]. Taken together with our findings, this indicates that the difference in frequency of multi-variant transmission (~20% between MSM and HET, ~10% between MSM and MTCT [5]) is too limited to set apart the overall transmitted diversity between the risk groups. The comparable amounts of transmitted diversity imply a similar interplay of drift and selection at transmission in the different risk groups and are therefore not expected to lead to evolutionary rate differences at the epidemic level.
The evolutionary rate comparison among risk groups indicates differences among HET and MSM and among HET and IDU, but the direction of these differences may seem counterintuitive in the light of previous findings. There is increasing evidence that the transmission/establishment advantage of ancestral variants leaves its footprint by slowing down the divergence rate among hosts [15–17, 33]. Given that this effect is larger for subtype B than for subtype C [17], we hypothesized that differences in proportion of multi-variant transmission may play an important role. Following this reasoning, we would expect the following overall ranking in the evolutionary rate between risk groups: HET > MSM > IDU. We find however that the evolutionary rate is lower in HET compared to MSM and also lower in HET compared to IDU, and this is the case for both absolute synonymous and non-synonymous rates (Figure 1). Because synonymous substitution rates - as a marker of viral replication rates - are are associated with HIV-1 disease progression and males are predicted to have higher disease progression rates by their viral set-point, we hypothesize that gender ratios may be key to explaining the risk group associated evolutionary rate differences. This implies that within-host divergence rates, together with a general transmission-associated rate slowdown, impact the tempo of evolution at the population level.
Despite higher rates for in risk groups with a larger proportion of males within each subtype, this relationship is less clear independent of the subtypes. On the one hand, the individuals sampled in our data sets may not accurately reflect the composition of the risk group population from which the samples were taken. On the other hand, the impact of differences in sex-specific within-host evolution may also be confounded by varying transmission rates by stage of infection. As suggested by Maljkovic Berry et al. (2007), in risk groups that generally transmit at an earlier stage of infection, before selective pressures of the immune system have much impact, the rate of HIV-1 evolution on the population level is expected to be lower compared to the rate when transmission usually occurs throughout asymptomatic infection. This provided an explanation for lower HIV-1 subtype A1 rate estimates among IDUs in the former Soviet Union (FSU) as compared to heterosexual transmission in Africa. Although we find opposite differences here between our HET and IDU data set, this does not necessarily contradict their findings because our IDU sampling is fundamentally different. Whereas Maljkovic Berry et al. (2007) have focused on sequence data from specific IDU transmission in the FSU, which should capture the IDU transmission dynamics very well, we analyzed all available sequences encompassing a far broader geographic area. Therefore, much of the ancestral history in our genealogical reconstruction of this sample will not specifically reflect IDU transmission. As a consequence, the differences we pick up are likely to be caused by the more recent, within-host evolutionary dynamics, explaining why we find a higher evolutionary rate in our male-dominated IDU sample. Knowledge of the transmission dynamics may further assist in explaining evolutionary rate differences at the epidemic level, but consistent dynamics appear to vary or difficult to establish. For example, whereas transmission rates early after infection were found to be similar among risk groups in the Quebec population [34], these were twice a high in the MSM population than in the HET group in a UK based survey [35].
In summary, we show that the effect of within-host evolution on the between-host rate dominates over transmission-related events. Because of this, we propose varying gender ratios as driving risk group related evolutionary rate differences.
Methods
Data compilation - transmission chains
To obtain a comprehensive collection of genetic data sets from HIV-1 transmission chains, we employed the search feature of the HIV database (http://www.hiv.lanl.gov/) that allows to retrieve intra-host data sets. At the time of the query (November 2012), 953 sets were reported without specific selection criteria. The title and abstract of the published studies were screened and each study that potentially involved clonal sequence data together with known transmission route and/or known infection time frame was subjected to a more detailed screening. We also undertook a literature search based on the references with high potential from the selected studies.
All transmission pairs for which time-stamped clonal sequences of both donor and recipient was available together with at least an upper bound on the transmission interval were grouped under the following risk groups: heterosexual (HET), men having sex with men (MSM) and blood contact (BC). The denominator ‘BC’ groups together not only those pairs infected through blood transfusion, but also contains transmission pairs involving a bite [36], a knife-fight [37], surgical procedures [38] and malignant injection [39].
Data compilation - risk group data sets for evolutionary rate estimation
Subtypes have been shown to evolve at different rates [40]. To compare the evolutionary rate between risk groups within subtypes, we downloaded near complete genome datasets for subtype A1, B and CRF01 AE from the Los Alamos HIV database (Table 3). The other subtypes lacked sufficient data of at least 2 risk groups to allow for a meaningful comparative analysis among risk groups.
Incorporating the uncertainty of sampling dates and transmission times
In order to apply the same time-scale for all analyses, all sampling and transmission dates were specified in units of days. Often however, sampling dates or transmission intervals are reported only approximately. Arbitrarily choosing for example the midpoint of the potential interval can introduce biases in the bottleneck size estimations because, especially in situations of recipient sampling close to the time of infection, there can be an interaction between the bottleneck size parameter and transmission/divergence times [17]. To avoid this, we made use of the flexibility offered by BEAST [41] to integrate out both the transmission and sampling dates constrained by a time interval [42]. However, an extension of the standard approach [42] was required to accommodate sampling of a single single date for a set of taxa representing a single clonal sample. We therefore extended BEAST [41] to handle such situations, and specified uniform priors over a transmission or sampling interval with known boundaries.
In some cases, it was possible to approximate the boundaries for the transmission interval using the Fiebig stage information [43]. Specifically, the earliest and latest possible infection dates were taken as the cumulative lower and upper boundaries of the 95% confidence intervals of the duration of the stages up to (lower boundary) or up to and including (upper boundary) the stage the recipient was in at the moment of sampling. As an example, suppose the first sample was taken while the recipient was in Fiebig stage II. Our approach assumes that stage II could have begun at the earliest 10 days after infection (5 days eclipse phase + 5 days phase I) and could have ended at the latest 18 days later (10 days eclipse phase + 10 days phase I + 8 days phase II). Therefore, infection could have taken place 10 to 28 days before the sampling date.
We took a similar approach when the time to seroconversion was known. This was cautiously interpreted as Fiebig stage III/IV because it is not always communicated which antibody detection test(s) was/were used and sensitivity of the tests has increased. In addition, because symptoms of primary or acute HIV infection represent terms that do not entail widely accepted definitions and reflect time windows after infection that are subject to the sensitivity of the tests [44], we calculated the transmission time interval using Fiebig stage I/II boundaries in such cases.
Phylogenetic inference
We used the BEAST software package [41] for all phylogenetic and population genetic inferences. The substitution process was described using a Hasegawa-Kishino-Yano (HKY, [45]) substitution model with discrete Γ-distribution to model among-site rate heterogeneity [46, 47]. We used a recently described transmission model [17] to accommodate the transmission history and to quantify the transmission bottleneck in terms of the loss of genetic diversity at transmission. We adopted a Bayesian hierarchical phylogenetic model (HPM) procedure to share information across transmission chains and specified hierarchical prior distributions on all parameters of the coalescent model. To formally test for differences in bottleneck size among the risk groups we specified a fixed effect on the bottleneck size parameter [28]. The sparse within-host sampling scheme of most transmission chains (Additional Table 1) resulted in poor temporal information to calibrate the molecular clock in many datasets. We therefore follow a slight modification of the approach by Keele et al. [1] to arrive at an informative prior distribution on the evolutionary rate on very short time scales. Specifically, we transformed the standard generation time curve [48] to a rate distribution using the experimentally derived reverse transcriptase error rate [49] and set the upper limit to the rate at a generation time of 18 hours [48].
Model selection
For all datasets, the log marginal likelihood was estimated for all demographic functions currently available under the transmission model (constant population size, exponential and logistic growth) [17]. To this end, we made use of the path sampling (PS) [50] and stepping-stone (SS) sampling estimators [51] of the (log) marginal likelihood as implemented in BEAST [29, 52, 53]. Both estimators have shown considerable increases in accuracy over so-called posterior-based (log) marginal likelihood estimators such as the harmonic mean estimators (HME and sHME), albeit at the expense of increased computational demands. All (log) marginal likelihood estimates were checked for convergence by performing multiple runs with different computational settings. We consider a model to be better supported by the data than the competing hypothesis when the difference in log Bayes factor (logBF) support exceeds 5 [54].
Selection of informative transmission chains for the fixed effects analysis
The parameterization of the population genetic dynamics throughout transmission in the transmission chain model may influence the bottleneck size estimations. Although we can select the best fitting parametrization using marginal likelihood estimation, the preference for a particular coalescent model may depend on the amount of available information. And whereas a simple model may be required for independent estimation, a more complex model may still be suitable when sharing information in a hierarchical modeling setting. We therefore assess the robustness of the results with respect to demographic model specification, by first performing the HPM + fixed effects analysis with the within-host population dynamics described by the function obtained by model selection (‘best fit’). Next, we also ran the fixed effects analyses on these datasets consistently applying either a logistic or an exponential model for the demographic process to each transmission chain (‘logistic’ and ‘exponential’). In order to avoid a higher weight for transmission chains for which multiple genomic regions are available, the genomic regions were analyzed separately.
Our previous results using the transmission chain model [17] indicate that the model is best informed by data that capture the early population dynamics. The poor mixing for the gag and env genomic regions for many datasets revealed that not all the transmission chain data sets can be used to properly inform the model. For the pol datasets, mixing was less of an issue but here the bottleneck size estimates were highly dependent on the specified model. Because of these issues we only focused on those datasets with first sampling close to the time of transmission (first recipient sampling ≤23 days, which is before or around the approximate time of peak viral load [43]) and with good mixing properties for further analyses. This filtering step retained 17 HET and 9 MSM chains with env data for the HPM + fixed effects analysis.
Among risk group evolutionary rate comparison
We visually inspected a regression of root-to-tip divergences as a function of sampling time using Path-O-Gen (http://tree.bio.ed.ac.uk/software/pathogen/). This revealed that sufficient temporal signal was present in all but the full genome subtype B datasets. To remove the noise from this dataset, we followed the same procedure as in [17] to arrive at a balanced dataset with clear temporal signal (R2 for both ≥ 0.48). The evolutionary rate for all datasets was estimated under a relaxed clock model using a lognormal distribution [55] by fitting the HKY model [45] while allowing for gamma-distributed among-site rate heterogeneity [46, 47]. The genome was split into gene-specific partitions to also allow for among-gene rate variation. A Skygrid model was specified as a flexible tree prior [56]. To estimate absolute synonymous and non-synonymous rates for each dataset we applied a renaissance counting procedure [21] following [17] to the non-overlapping parts of the open reading frames (except for nef for which data was missing for many taxa).
Supplementary Material
Additional Table 1 — Dataset characteristics
Additional Figure 1 — Illustration of the signal strength in the datasets for an exponential or logistic demographic history
Acknowledgments
BV was supported by a PhD grant from the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT Vlaanderen). MAS is partially supported by National Science Foundation grant DMS-1264153. This work was made possible by funding of the Onderzoeksfonds KU Leuven Research Fund KU Leuven. The research leading to these results has received funding from the European Union Seventh Framework Programme [FP7/2007–2013] under Grant Agreement nr. 278433-PREDEMICS and ERC Grant Agreement nr. 260864. This work was supported in part by grants from the Fonds voor Wetenschappelijk Onderzoek Vlaanderen (FWO G.0692.14), by a grant from the Interuniversity Attraction Poles Programme, Belgian State, Belgian Science Policy (IUAP-VI P6/41), by the European Community’s Seventh Framework Programme (FP7/2007–2013) under the project ‘Collaborative HIV and Anti-HIV Drug Resistance Network’ (CHAIN, grant 223131), by KU Leuven (Program Financing no. PF/10/018). Some of the computational resources (Stevin Supercomputer Infrastructure) and services used in this work were provided by Ghent University, the Hercules Foundation and the Flemish Government - department EWI.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Author’s contributions
BV conceived and performed the experiments, drafted and wrote the manuscript. MAS implemented the computational developments. PL participated in the design of the study and helped with the analysis. GB helped with the analyses. PL, GB, KVL and AVD contributed to writing the manuscript. All authors read and approved the final manuscript.
References
- 1.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. Identification and characterization of transmitted and early founder virus envelopes in primary hiv-1 infection. Proc Natl Acad Sci U S A. 2008;105(21):7552–7. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Long EM, Martin HL, Jr, Kreiss JK, Rainwater SM, Lavreys L, Jackson DJ, Rakwar J, Mandaliya K, Overbaugh J. Gender differences in hiv-1 diversity at time of infection. Nat Med. 2000;6(1):71–5. doi: 10.1038/71563. [DOI] [PubMed] [Google Scholar]
- 3.Sagar M, Kirkegaard E, Long EM, Celum C, Buchbinder S, Daar ES, Overbaugh J. Human immunodeficiency virus type 1 (hiv-1) diversity at time of infection is not restricted to certain risk groups or specific hiv-1 subtypes. J Virol. 2004;78(13):7279–83. doi: 10.1128/JVI.78.13.7279-7283.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H, Bar KJ, Wang S, Decker JM, Chen Y, Sun C, Salazar-Gonzalez JF, Salazar MG, Learn GH, Morgan CJ, Schumacher JE, Hraber P, Giorgi EE, Bhattacharya T, Korber BT, Perelson AS, Eron JJ, Cohen MS, Hicks CB, Haynes BF, Markowitz M, Keele BF, Hahn BH, Shaw GM. High multiplicity infection by hiv-1 in men who have sex with men. PLoS Pathog. 2010;6(5):1000890. doi: 10.1371/journal.ppat.1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaw GM, Hunter E. Hiv transmission. Cold Spring Harb Perspect Med. 2012;2(11) doi: 10.1101/cshperspect.a006965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soulie C, Calvez V, Marcelin AG. Coreceptor usage in different reservoirs. Curr Opin HIV AIDS. 2012;7(5):450–5. doi: 10.1097/COH.0b013e328356e9c2. [DOI] [PubMed] [Google Scholar]
- 7.English S, Katzourakis A, Bonsall D, Flanagan P, Duda A, Fidler S, Weber J, McClure M, Phillips R, Frater J SPARTAC Trial Investigators. Phylogenetic analysis consistent with a clinical history of sexual transmission of hiv-1 from a single donor reveals transmission of highly distinct variants. Retrovirology. 2011;8:54. doi: 10.1186/1742-4690-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fouda GG, Mahlokozera T, Salazar-Gonzalez JF, Salazar MG, Learn G, Kumar SB, Dennison SM, Russell E, Rizzolo K, Jaeger F, Cai F, Vandergrift NA, Gao F, Hahn B, Shaw GM, Ochsenbauer C, Swanstrom R, Meshnick S, Mwapasa V, Kalilani L, Fiscus S, Montefiori D, Haynes B, Kwiek J, Alam SM, Permar SR. Postnatally-transmitted hiv-1 envelope variants have similar neutralization-sensitivity and function to that of nontransmitted breast milk variants. Retrovirology. 2013;10:3. doi: 10.1186/1742-4690-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parrish NF, Gao F, Li H, Giorgi EE, Barbian HJ, Parrish EH, Zajic L, Iyer SS, Decker JM, Kumar A, Hora B, Berg A, Cai F, Hopper J, Denny TN, Ding H, Ochsenbauer C, Kappes JC, Galimidi RP, West AP, Jr, Bjorkman PJ, Wilen CB, Doms RW, O’Brien M, Bhardwaj N, Borrow P, Haynes BF, Muldoon M, Theiler JP, Korber B, Shaw GM, Hahn BH. Phenotypic properties of transmitted founder hiv-1. Proc Natl Acad Sci U S A. 2013;110(17):6626–33. doi: 10.1073/pnas.1304288110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frange P, Meyer L, Jung M, Goujard C, Zucman D, Abel S, Hochedez P, Gousset M, Gascuel O, Rouzioux C, Chaix ML ANRS PRIMO Cohort Study Group. Sexually-transmitted/founder hiv-1 cannot be directly predicted from plasma or pbmc-derived viral quasispecies in the transmitting partner. PLoS One. 2013;8(7):69144. doi: 10.1371/journal.pone.0069144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolinsky SM, Wike CM, Korber BT, Hutto C, Parks WP, Rosenblum LL, Kunstman KJ, Furtado MR, Muñoz JL. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science. 1992;255(5048):1134–7. doi: 10.1126/science.1546316. [DOI] [PubMed] [Google Scholar]
- 12.Wolfs TF, Zwart G, Bakker M, Goudsmit J. Hiv-1 genomic rna diversification following sexual and parenteral virus transmission. Virology. 1992;189(1):103–10. doi: 10.1016/0042-6822(92)90685-i. [DOI] [PubMed] [Google Scholar]
- 13.Zhu T, Mo H, Wang N, Nam DS, Cao Y, Koup RA, Ho DD. Genotypic and phenotypic characterization of hiv-1 patients with primary infection. Science. 1993;261(5125):1179–81. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]
- 14.Sagar M. Hiv-1 transmission biology: selection and characteristics of infecting viruses. J Infect Dis. 2010;202(Suppl 2):289–96. doi: 10.1086/655656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lythgoe KA, Fraser C. New insights into the evolutionary rate of hiv-1 at the within-host and epidemiological levels. Proc Biol Sci. 2012;279(1741):3367–75. doi: 10.1098/rspb.2012.0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alizon S, Fraser C. Within-host and between-host evolutionary rates across the hiv-1 genome. Retrovirology. 2013;10(1):49. doi: 10.1186/1742-4690-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vrancken B, Rambaut A, Suchard MA, Drummond A, Baele G, Van Wijngaerden E, Vandamme AM, Van Laethem K, Lemey P. The genealogical population dynamics of hiv-1 in a large transmission chain: bridging within and among host evolutionary rates. PLoS Computational Biology. 2014;10(4):1003505. doi: 10.1371/journal.pcbi.1003505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lythgoe KA, Pellis L, Fraser C. Is hiv short-sighted? insights from a multistrain nested model. Evolution. 2013;67(10):2769–82. doi: 10.1111/evo.12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fraser C, Lythgoe K, Leventhal GE, Shirreff G, Hollingsworth TD, Alizon S, Bonhoeffer S. Virulence and pathogenesis of hiv-1 infection: an evolutionary perspective. Science. 2014;343(6177):1243727. doi: 10.1126/science.1243727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carlson JM, Schaefer M, Monaco DC, Batorsky R, Claiborne DT, Prince J, Deymier MJ, Ende ZS, Klatt NR, DeZiel CE, Lin TH, Peng J, Seese AM, Shapiro R, Frater J, Ndung’u T, Tang J, Goepfert P, Gilmour J, Price MA, Kilembe W, Heckerman D, Goulder PJR, Allen TM, Allen S, Hunter E. Hiv transmission. selection bias at the heterosexual hiv-1 transmission bottleneck. Science. 2014;345(6193):1254031. doi: 10.1126/science.1254031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemey P, Kosakovsky Pond SL, Drummond AJ, Pybus OG, Shapiro B, Barroso H, Taveira N, Rambaut A. Synonymous substitution rates predict hiv disease progression as a result of underlying replication dynamics. PLoS Comput Biol. 2007;3(2):29. doi: 10.1371/journal.pcbi.0030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mellors JW, Rinaldo CR, Jr, Gupta P, White RM, Todd JA, Kingsley LA. Prognosis in hiv-1 infection predicted by the quantity of virus in plasma. Science. 1996;272(5265):1167–70. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 23.Farzadegan H, Hoover DR, Astemborski J, Lyles CM, Margolick JB, Markham RB, Quinn TC, Vlahov D. Sex differences in hiv-1 viral load and progression to aids. Lancet. 1998;352(9139):1510–4. doi: 10.1016/S0140-6736(98)02372-1. [DOI] [PubMed] [Google Scholar]
- 24.Langford SE, Ananworanich J, Cooper DA. Predictors of disease progression in hiv infection: a review. AIDS Res Ther. 2007;4:11. doi: 10.1186/1742-6405-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alizon S, von Wyl V, Stadler T, Kouyos RD, Yerly S, Hirschel B, Böni J, Shah C, Klimkait T, Furrer H, Rauch A, Vernazza PL, Bernasconi E, Battegay M, Bürgisser P, Telenti A, Günthard HF, Bonhoeffer S Swiss HIV Cohort Study. Phylogenetic approach reveals that virus genotype largely determines hiv set-point viral load. PLoS Pathog. 2010;6(9):1001123. doi: 10.1371/journal.ppat.1001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sterling TR, Vlahov D, Astemborski J, Hoover DR, Margolick JB, Quinn TC. Initial plasma hiv-1 rna levels and progression to aids in women and men. N Engl J Med. 2001;344(10):720–5. doi: 10.1056/NEJM200103083441003. [DOI] [PubMed] [Google Scholar]
- 27.Suchard MA, Kitchen CMR, Sinsheimer JS, Weiss RE. Hierarchical phylogenetic models for analyzing multipartite sequence data. Syst Biol. 2003;52(5):649–64. doi: 10.1080/10635150390238879. [DOI] [PubMed] [Google Scholar]
- 28.Edo-Matas D, Lemey P, Tom JA, Serna-Bolea C, van den Blink AE, van ’t Wout AB, Schuitemaker H, Suchard MA. Impact of ccr5delta32 host genetic background and disease progression on hiv-1 intrahost evolutionary processes: efficient hypothesis testing through hierarchical phylogenetic models. Mol Biol Evol. 2011;28(5):1605–16. doi: 10.1093/molbev/msq326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baele G, Lemey P, Bedford T, Rambaut A, Suchard MA, Alekseyenko AV. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol Biol Evol. 2012;29(9):2157–67. doi: 10.1093/molbev/mss084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edo-Matas D, van Gils MJ, Bowles EJ, Navis M, Rachinger A, Boeser-Nunnink B, Stewart-Jones GB, Kootstra NA, van ’t Wout AB, Schuitemaker H. Genetic composition of replication competent clonal hiv-1 variants isolated from peripheral blood mononuclear cells (pbmc), hiv-1 proviral dna from pbmc and hiv-1 rna in serum in the course of hiv-1 infection. Virology. 2010;405(2):492–504. doi: 10.1016/j.virol.2010.06.029. [DOI] [PubMed] [Google Scholar]
- 31.Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Farzadegan H, Gupta P, Rinaldo CR, Learn GH, He X, Huang XL, Mullins JI. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73(12):10489–502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edwards CTT, Holmes EC, Wilson DJ, Viscidi RP, Abrams EJ, Phillips RE, Drummond AJ. Population genetic estimation of the loss of genetic diversity during horizontal transmission of hiv-1. BMC Evol Biol. 2006;6:28. doi: 10.1186/1471-2148-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Redd AD, Collinson-Streng AN, Chatziandreou N, Mullis CE, Laeyendecker O, Martens C, Ricklefs S, Kiwanuka N, Nyein PH, Lutalo T, Grabowski MK, Kong X, Manucci J, Sewankambo N, Wawer MJ, Gray RH, Porcella SF, Fauci AS, Sagar M, Serwadda D, Quinn TC. Previously transmitted hiv-1 strains are preferentially selected during subsequent sexual transmissions. J Infect Dis. 2012;206(9):1433–42. doi: 10.1093/infdis/jis503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brenner BG, Roger M, Routy JP, Moisi D, Ntemgwa M, Matte C, Baril JG, Thomas R, Rouleau D, Bruneau J, Leblanc R, Legault M, Tremblay C, Charest H, Wainberg MA Quebec Primary HIV Infection Study Group. High rates of forward transmission events after acute/early hiv-1 infection. J Infect Dis. 2007;195(7):951–9. doi: 10.1086/512088. [DOI] [PubMed] [Google Scholar]
- 35.Hughes GJ, Fearnhill E, Dunn D, Lycett SJ, Rambaut A, Leigh Brown AJ UK HIV Drug Resistance Collaboration. Molecular phylodynamics of the heterosexual hiv epidemic in the united kingdom. PLoS Pathog. 2009;5(9):1000590. doi: 10.1371/journal.ppat.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andreo SMS, Barra LAC, Costa LJ, Sucupira MCA, Souza IEL, Diaz RS. Hiv type 1 transmission by human bite. AIDS Res Hum Retroviruses. 2004;20(4):349–50. doi: 10.1089/088922204323048087. [DOI] [PubMed] [Google Scholar]
- 37.Kao CF, Hsia KT, Chang SY, Chang FY, Nelson K, Yang CH, Huang YF, Fu TY, Yang JY. An uncommon case of hiv-1 transmission due to a knife fight. AIDS Res Hum Retroviruses. 2011;27(2):115–22. doi: 10.1089/aid.2010.0044. [DOI] [PubMed] [Google Scholar]
- 38.Blanchard A, Ferris S, Chamaret S, Guétard D, Montagnier L. Molecular evidence for nosocomial transmission of human immunodeficiency virus from a surgeon to one of his patients. J Virol. 1998;72(5):4537–40. doi: 10.1128/jvi.72.5.4537-4540.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Metzker ML, Mindell DP, Liu XM, Ptak RG, Gibbs RA, Hillis DM. Molecular evidence of hiv-1 transmission in a criminal case. Proc Natl Acad Sci U S A. 2002;99(22):14292–7. doi: 10.1073/pnas.222522599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abecasis AB, Vandamme AM, Lemey P. Quantifying differences in the tempo of human immunodeficiency virus type 1 subtype evolution. J Virol. 2009;83(24):12917–24. doi: 10.1128/JVI.01022-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with beauti and the beast 1.7. Mol Biol Evol. 2012;29(8):1969–73. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shapiro B, Ho SYW, Drummond AJ, Suchard MA, Pybus OG, Rambaut A. A bayesian phylogenetic method to estimate unknown sequence ages. Mol Biol Evol. 2011;28(2):879–87. doi: 10.1093/molbev/msq262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fiebig EW, Wright DJ, Rawal BD, Garrett PE, Schumacher RT, Peddada L, Heldebrant C, Smith R, Conrad A, Kleinman SH, Busch MP. Dynamics of hiv viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary hiv infection. AIDS. 2003;17(13):1871–9. doi: 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- 44.Cohen MS, Gay CL, Busch MP, Hecht FM. The detection of acute hiv infection. J Infect Dis. 2010;202(Suppl 2):270–7. doi: 10.1086/655651. [DOI] [PubMed] [Google Scholar]
- 45.Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial dna. J Mol Evol. 1985;22(2):160–74. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 46.Yang Z. Among-site rate variation and its impact on phylogenetic analyses. Trends Ecol Evol. 1996;11(9):367–72. doi: 10.1016/0169-5347(96)10041-0. [DOI] [PubMed] [Google Scholar]
- 47.Yang Maximum-likelihood models for combined analyses of multiple sequence data. J Mol Evol. 1996;42(5):587–96. doi: 10.1007/BF02352289. [DOI] [PubMed] [Google Scholar]
- 48.Althaus CL, De Vos AS, De Boer RJ. Reassessing the human immunodeficiency virus type 1 life cycle through age-structured modeling: life span of infected cells, viral generation time, and basic reproductive number, r0. J Virol. 2009;83(15):7659–67. doi: 10.1128/JVI.01799-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mansky LM, Temin HM. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69(8):5087–94. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lartillot N, Philippe H. Computing bayes factors using thermodynamic integration. Syst Biol. 2006;55(2):195–207. doi: 10.1080/10635150500433722. [DOI] [PubMed] [Google Scholar]
- 51.Xie W, Lewis PO, Fan Y, Kuo L, Chen MH. Improving marginal likelihood estimation for bayesian phylogenetic model selection. Syst Biol. 2011;60(2):150–60. doi: 10.1093/sysbio/syq085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baele G, Li WLS, Drummond AJ, Suchard MA, Lemey P. Accurate model selection of relaxed molecular clocks in bayesian phylogenetics. Mol Biol Evol. 2013;30(2):239–43. doi: 10.1093/molbev/mss243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baele G, Lemey P. Bayesian evolutionary model testing in the phylogenomics era: matching model complexity with computational efficiency. Bioinformatics. 2013;29(16):1970–1979. doi: 10.1093/bioinformatics/btt340. [DOI] [PubMed] [Google Scholar]
- 54.Kass RE, Raftery AE. Bayes factors. journal of the american Statistical Association. 1995;90(430):773–795. [Google Scholar]
- 55.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4(5):88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gill MS, Lemey P, Faria NR, Rambaut A, Shapiro B, Suchard MA. Improving bayesian population dynamics inference: a coalescent-based model for multiple loci. Mol Biol Evol. 2013;30(3):713–24. doi: 10.1093/molbev/mss265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suchard MA, Weiss RE, Sinsheimer JS. Models for estimating bayes factors with applications to phylogeny and tests of monophyly. Biometrics. 2005;61(3):665–73. doi: 10.1111/j.1541-0420.2005.00352.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Table 1 — Dataset characteristics
Additional Figure 1 — Illustration of the signal strength in the datasets for an exponential or logistic demographic history