Abstract
During host infection, the foodborne pathogen Listeria monocytogenes must sense and respond to rapidly changing environmental conditions. Two transcriptional regulators, the alternative sigma factor B (σB) and the Positive Regulatory Factor A (PrfA), are key contributors to the transcriptomic responses that enable bacterial survival in the host gastrointestinal tract and invasion of host duodenal cells. Increases in temperature and osmolarity induce activity of these proteins; such conditions may be encountered in food matrices as well as within the host gastrointestinal tract. Differences in PrfA and σB activity between individual cells might affect the fate of a cell during host invasion, therefore, we hypothesized that PrfA and σB activities differ among individual cells under heat and salt stress. We used fluorescent reporter fusions to determine the relative proportions of cells with active σB or PrfA following exposure to 45°C heat or 4% NaCl. Activities of both PrfA and σB were induced stochastically, with fluorescence levels ranging from below detection to high among individual cells. The proportion of cells with active PrfA was significantly higher than the proportion with active σB under all tested conditions; under some conditions, nearly all cells had active PrfA. Our findings further support the growing body of evidence illustrating the stochastic nature of bacterial gene expression under conditions that are relevant for host invasion via food-borne, oral infection.
Keywords: Listeria monocytogenes, PrfA, sigB, single cell, stochastic gene expression, heat stress, salt stress
Introduction
Listeria monocytogenes is a foodborne pathogen responsible for bacterial infections in many species including ruminants and humans. While the incidence of listeriosis is relatively low [0.26 cases/1,000,000 in the USA in 2013 (Crim et al., 2015)], a high fatality rate, estimated to be ~15–30 deaths per 100 cases (de Valk et al., 2005; Popovic et al., 2014; Crim et al., 2015), makes L. monocytogenes a major food safety concern in many industrialized countries, particularly because it affects vulnerable populations including pregnant women, infants, and individuals with suppressed immune systems. Infection is typically acquired by ingestion of contaminated food with subsequent invasion of cells in the host duodenum. L. monocytogenes can enter the bloodstream and spread systemically, including to the placenta and the central nervous system. Consequently, listeriosis may present as gastroenteritis, septicemia, neonatal infection, or meningoencephalitis and can cause abortion in pregnant individuals (reviewed in Allerberger and Wagner, 2010).
L. monocytogenes is able to transition from a saprophytic lifestyle in the environment to an intracellular niche within host cells, which requires adaptation to the rapidly changing conditions encountered along the oral infection route, including shifts in temperature, pH, and osmolarity. These adaptation processes require substantial changes in gene expression. Two transcription factors, the alternative sigma factor B (σB) and the Positive Regulatory Factor A (PrfA), are central nodes in a complex regulatory network that governs the remodeling of the transcriptome during host infection (Chaturongakul et al., 2008; De las Heras et al., 2011). σB activates transcription of a large regulon that includes stress response as well as some virulence genes (Wiedmann et al., 1998); PrfA is the primary transcriptional regulator of virulence genes (Mengaud et al., 1991). A growing body of research shows that even within a clonal population of bacterial cells exposed to a superimposed environmental condition, bacterial gene expression in a population is inherently noisy, often showing stochastic patterns (reviewed in Kaern et al., 2005; Raj and van Oudenaarden, 2008). In L. monocytogenes, stochastic activity of σB in the presence of 0.5 M salt and in stationary phase has been previously demonstrated (Utratna et al., 2012). Such stochastic differences in gene expression can create specialized subpopulations that may serve the purpose of “labor-sharing,” where only a few cells carry the burden of producing a molecule that will profit the entire population, or “bet-hedging,” where the individual cells bearing the expressed molecules will have a relatively enhanced chance of surviving future abrupt changes in environmental parameters (Eldar and Elowitz, 2010).
A previous study in a pregnant guinea pig model has shown that a very small number of L. monocytogenes cells traffic from the maternal organs to the placenta; further, placental infection can originate from a single invading cell (Bakardjiev et al., 2006). Why would one individual cell succeed in invading the host (e.g., during the gastrointestinal stage of infection) while others do not? Previous high-resolution transcriptomics studies documenting the transcriptional changes in L. monocytogenes upon host infection (Camejo et al., 2009; Toledo-Arana et al., 2009) do not account for stochastic differences at the single cell level. Information on gene expression patterns in individual bacteria that succeed to establish infection could therefore provide novel insights into host-pathogen interactions at very high resolution. However, for L. monocytogenes there is no data on how differential gene expression affects the ability of individual cells to ultimately succeed in host invasion. The aim of this study was to take a first step toward addressing this question. Specifically, we hypothesized that σB and PrfA activities differ in individual bacteria in response to several environmental conditions. We chose two conditions shown previously to induce σB activity: heat or salt stress (i.e., exposure to 45°C or 4.4% NaCl) (Sue et al., 2003; Hu et al., 2007; Abram et al., 2008; Somolinos et al., 2010; Ait-Ouazzou et al., 2012; Ringus et al., 2012; Ribeiro et al., 2014). Water phase salt concentrations in the range of 4% are typical in food preservation, and salt is used in the preservation of foods linked to human listeriosis cases and outbreaks (e.g., cheese or deli meat; Gottlieb et al., 2006; Currie et al., 2015; Heiman et al., 2016; McIntyre et al., 2015). Heat stress at 45°C was used as a model for another, non-lethal stress condition. To probe the single cell response to the selected environmental conditions, we used fluorescent reporter fusions to visualize the activities of σB and PrfA in response to heat and salt stress.
Materials and methods
Bacterial strains and plasmids
E. coli was grown in Luria Bertani (LB) broth or on LB agar (at 37°C), while L. monocytogenes was grown in brain heart infusion (BHI) broth or on BHI agar at 37°C unless otherwise stated. Stock cultures of all strains (see Table 1 for a list of strains) were stored at −80°C in LB or BHI broth with 15% glycerol. From those stocks, cultures were streaked on LB or BHI agar plates to obtain single colonies to inoculate overnight cultures. For stress exposure experiments, cultures were started from frozen stock and grown on BHI agar. After overnight incubation, a single colony was inoculated into 5 ml BHI and grown for 6–8 h. This culture was then diluted 1:1000 into fresh BHI and grown for 15–17 h. This stationary phase culture was then diluted 1:100 into 100 ml BHI and the resulting culture was grown to either OD600 = 0.4 (representing mid-log phase) or OD600 = 1.0 + 1 h (early stationary phase). All cultures were grown with aeration (shaking at 200 rpm). Where appropriate, chloramphenicol (at 25 μg/ml) or kanamycin (at 30 μg/ml) was added to the growth media during the construction of the reporter strains.
Table 1.
Bacterial strains.
| Parental/host strain | Species | strain | Relevant Genotype |
|---|---|---|---|
| Listeria monocytogenes | 10403S | wt | Bishop and Hinrichs, 1987 |
| Listeria monocytogenes | 10403S | ΔsigB | Wiedmann et al., 1998 |
| Listeria monocytogenes | 10403S | ΔprfA | Wong and Freitag, 2004 |
| Listeria monocytogenes | EGDe | Plmo2230::eGFP | Utratna et al., 2012 |
| Listeria monocytogenes | 10403S | Phly::eGFP | this study |
| Listeria monocytogenes | 10403S | Plmo2230::eGFP | this study |
| Listeria monocytogenes | 10403S | constitutive GFP | this study |
| Listeria monocytogenes | 10403S | ΔsigB, Phly::eGFP | this study |
| Listeria monocytogenes | 10403S | ΔprfA, Phly::eGFP | this study |
| Listeria monocytogenes | 10403S | ΔsigB, Plmo2230::eGFP | this study |
| E.coli COB631 | Top ten | pKSV-7-lmo2230::eGFP | Utratna et al., 2012 |
| E.coli | DH5α | pPL2-hly::eGFP (pCG8) | this study |
| E.coli | SM10 | pPL2-hly::eGFP (pCG8) | this study |
| E.coli | DH5α | pPL2-lmo2230::eGFP (pCG1) | this study |
| E.coli | SM10 | pPL2-lmo2230::eGFP (pCG1) | this study |
Cloning techniques and construction of plasmids
Primers and plasmids used in this study are listed in Table 2, Supplementary Table 1. PCRs were carried out using Q5 polymerase (NEB, Ipswich, MA) according to the manufacturer's protocol; when necessary PCR products were purified using Qiagen (Valencia, CA) PCR Purification Kit No. 28106. For gel extractions, Invitrogen (Carlsbad, CA) Kit No. K210012 was used. Restriction digests were carried out using enzymes purchased from NEB. Plasmids were isolated using Qiagen Kit No. 27104.
Table 2.
Plasmids used in this study.
| Name | Relevant description | References |
|---|---|---|
| pPl2 | base for the construction of reporter strains | Lauer et al., 2002 |
| pKSV-7-Plmo2230::eGFP | source of the SigB reporter construct | Utratna et al., 2012 |
| pCG1 | pPL2-Plmo2230::eGFP | this study |
| pCG8 | pPL2-Phly::eGFP | this study |
| pAD-cGFP | used to make constitutively eGFP-expressing strain | Balestrino et al., 2010 |
Reporter fusions were constructed in pPL2, a site specific integration vector for L. monocytogenes. Specifically, an eGFP coding sequence codon-optimized for L. monocytogenes (Utratna et al., 2012) was used to construct two reporter fusions, including (i) a reporter fusion with a σB-dependent promoter (Plmo2230) to assess σB activity and (ii) a reporter fusion with a PrfA-dependent promoter (Phly) to assess PrfA activity. The promoters used for the reporter constructs were selected to represent genes regulated as exclusively as possible by the respective transcription factors. lmo2230, which encodes a putative arsenate reductase, is σB-regulated (Kazmierczak et al., 2003; Hain et al., 2008; Raengpradub et al., 2008; Oliver et al., 2009) and has been used previously in reporter systems for σB activity (Ondrusch and Kreft, 2011; Utratna et al., 2012). Although an early study suggested possible PrfA contributions to lmo2230 regulation (Milohanic et al., 2003), subsequent studies (e.g., Ollinger et al., 2009) determined lmo2230 to be solely σB-regulated. Specifically, lmo2230 is exclusively σB-regulated under NaCl stress (Utratna et al., 2011). The virulence gene hly encodes the PrfA-regulated hemolysin listeriolysin O (LLO) (Mengaud et al., 1991; Milohanic et al., 2003; Ollinger et al., 2009).
The Phly::eGFP construct was created by amplifying the hly promoter (the first 204 bp upstream of the start codon, including a 123 bp 5′ untranslated region) from L. monocytogenes 10403S and the eGFP coding sequence from pKSV7-Plmo2230::eGFP, followed by a splicing by overlap-extension (SOE) PCR to join the two PCR products. The resulting PCR product was cloned into pPL2 creating pCG8 (Table 2). The Plmo2230::eGFP reporter was constructed using E. coli COB631 (obtained from Conor O'Bryne), which contains the plasmid pKSV7 with the lmo2230 promoter (the first 443 bp upstream of the start codon, including a 110 bp 5′ untranslated region) fused to a codon-optimized version of eGFP (pKSV7-Plmo2230::eGFP) (Utratna et al., 2012); this reporter construct was subcloned into pPL2, creating pCG1 (Table 2). The inserts in the resulting plasmids were verified by sequencing (Sanger Sequencing Core facilities, Cornell University). Plasmid construction was performed in E. coli DH5α and final plasmid constructs were introduced into E. coli SM10 as a source strain for conjugation into L. monocytogenes 10403S. Presence of the construct in L. monocytogenes 10403S was confirmed by PCR with primers CG17 and CG18 (Supplementary Table 1) and by verifying fluorescence using a Zeiss (Jena, Germany) LSM 510 confocal microscope. As a negative control, the reporter constructs were cloned into the appropriate null mutant backgrounds, creating strains 10403S ΔprfA, Phly::eGFP and 10403S ΔsigB, Plmo2230::eGFP (Table 1). As prfA also has a σB-dependent promoter (Freitag et al., 1993; Nadon et al., 2002; Ollinger et al., 2009), we cloned the PrfA reporter construct into a sigB null mutant background, creating strain 10403S ΔsigB, Phly::eGFP (Table 1), to investigate the influence of σB on PrfA activity.
To create constitutively GFP-expressing L. monocytogenes, pAD-cGFP was obtained from Balestrino et al. (2010), introduced into E. coli SM10 and conjugated into L. monocytogenes 10403S.
Salt and heat stress experiments
Stress exposure experiments were performed with bacteria grown, as described above, to either log or stationary phase. The σB and PrfA reporter strains reached the desired OD600 at the same rate as the parent and the constitutively GFP expressing control strains during growth of log and stationary phase cultures, indicating no growth defect associated with the reporter construct or the constitutive expression of GFP during growth in BHI at 37°C (Supplementary Figure 1). To compensate for potential differences between the strains during stress exposure, all analyses were performed based on the proportion of cells that were GFP positive within one strain.
All stress exposure experiments were performed either in phosphate buffered saline (PBS) to simulate nutrient-depleted conditions or in a nutrient-rich medium (BHI). L. monocytogenes survives well in PBS, showing a limited increase in bacterial numbers over 200 min (<1 log). All stress exposure experiments were conducted in 4 ml volumes using 16 by 125 mm disposable borosilicate glass tubes incubated with aeration (shaking at 200 rpm). Log or stationary phase cultures were used directly for the heat stress experiments in BHI. For the heat stress experiments in PBS, 200 μl of log or stationary phase culture was added to 4 ml PBS, which had been pre-warmed to 37°C. For heat stress experiments, cultures were incubated at 45°C while controls were incubated at 37°C. For the salt stress experiments in BHI, 3 ml log or stationary phase culture was added to 1 ml BHI with 17.6% w/v NaCl for a final concentration of 4.4% w/v, pre-warmed to 37°C; for the negative control, 3 ml culture was added to 1 ml BHI without added salt, pre-warmed to 37°C. For the salt stress experiments in PBS, 200 μl log or stationary phase culture was added to 4 ml pre-warmed (37°C) PBS with 4.6% w/v NaCl (final concentration 4.4% w/v); the negative control was 200 μl of culture added to 4 ml PBS without the added salt. Standard PBS contains 0.8% NaCl, while standard BHI contains 0.5% added NaCl. While we recognize that additional NaCl may originate from the tissue components inherently present in BHI, we opted to add the same final exogenous concentration of NaCl to both PBS and BHI.
Flow cytometry analysis
Flow cytometry (FCM) analysis of the reporter fusion strains exposed to different stress conditions was performed on cells collected right before (t = 0) or at 10, 20, 30, 60, 100, 150, and 200 min after initiation of salt or heat stress exposure. Cells collected at these time points were fixed in 1% paraformaldehyde and counter-stained with wheat-germ agglutinin conjugated to Alexa Fluor 633 (Invitrogen). For each experimental run, a sample of each culture was microscopically confirmed to be in single cell suspension; samples that did not meet this criterion were vortexed vigorously and reassessed. A confirmed sample was then diluted 1:400 in PBS and 50,000 individual cells were analyzed on a BD (San Jose, CA) FACS Aria Fusion flow cytometer to determine their eGFP status (positive/negative). Bacteria were gated on Alexa Fluor 633 so that no more than 0.1% of negative control bacteria (L. monocytogenes 10403S parent strain) and none of the PBS background registered as positive. Finally, the level of green spectrum autofluorescence in L. monocytogenes was determined by running an eGFP-negative parent strain control sample. The cutoff for eGFP was set so that no more than 1.5% of this negative control registered as eGFP positive. A strain constitutively expressing GFP was used as a positive control. All experiments were performed in at least three independent biological replicates (i.e., bacterial cells were grown and collected on different days).
Confocal microscopy
Confocal microscopy was performed to visualize the eGFP fluorescence of individual cells of the PrfA and σB reporter strains. Bacteria were collected after 200 min of heat stress in PBS and fixed in 1% PFA as described above. A 5 μl aliquot of the fixed cell suspension was streaked on a glass slide and a coverslip was mounted with glycergel (Invitrogen). Images were taken on a Zeiss LSM510 confocal microscope with a 63X objective.
RT-qPCRs
Reverse transcriptase (RT)-qPCR was used to: (i) confirm results from the reporter fusion experiments at the RNA level; and to (ii) assess the stability of the Phly::eGFP and Plmo2230::eGFP reporter mRNAs at 37 and 45°C. For all RT-qPCR experiments, cDNA was obtained from bacterial cells using a protocol previously detailed (Tang et al., 2015) with minor modifications. Briefly, 100 μl of a 1:10 mixture of acid-phenol chloroform in 100% EtOH was added to the bacterial cultures for each ml of culture. The cells were pelleted and lysed in lysozyme (20 mg/ml) and proteinase K (4 mg/ml) for 30 min, followed by bead-beating in Tri-Reagent (Life Technologies, Carlsbad CA), phase-separation with 1-bromo-3-chloropropane, and precipitation in isopropanol overnight at −80°C. The RNA pellet was then washed in 75% EtOH followed by treatment with Ambion Turbo DNAse and a second phenol-chloroform extraction, followed by overnight precipitation in 100% EtOH. The RNA pellet was washed in 70% EtOH and 500 ng of RNA was transcribed to cDNA using TaqMan® Reverse Transcription Reagents (Invitrogen Catalog Number N8080234) as per the manufacturer's manual. qPCRs were performed with SybrGreen (Invitrogen), with two technical replicates per sample, in optical 96-well plates on an ABI Prism 7,300 system (Applied Biosystems, Foster City, CA) using: (i) one cycle at 95.0°C for 10 min; (ii) 40 cycles at 95.0°C for 15 s and 55.0°C (except for eGFP PCRs, which used 60°C) for 1 min; and (iii) a dissociation curve. Genomic DNA isolated from L. monocytogenes 10403S was used to generate standard curves to determine optimal annealing temperatures of 55° or 60°C for eGFP and the efficiencies of the primer pairs used (i.e., primers targeting eGFP, prfA, sigB, hly, lmo2230, and rpoB). Serial dilutions were made to yield 10, 105 and 107 template copy numbers per reaction. The slope of the standard curve was determined and primer efficiency e was calculated as e = 2−1/slope (Supplementary Table 1).
RT-qPCR was used to confirm the results from the reporter fusion experiments performed on log phase cells exposed to heat stress in PBS. For these experiments, 3 ml of log phase culture was added to 7 ml PBS pre-warmed to 45°C (heat exposed) or 37°C (control), followed by incubation for 5, 20, or 40 min at either 45°C or 37°C. cDNA was isolated from cells collected at these time points and RT-qPCR was performed (as described above) to determine the transcript levels for prfA, sigB, hly and lmo2230. The fold change (fc) in transcript levels between heat exposed and control cells was calculated using the following formula where e is the efficiency of the respective target primers (see Supplementary Table 1): FC = . The reported values represent the means of three independent biological replicates.
To assess the stabilities of the Phly::eGFP and Plmo2230::eGFP reporter mRNAs at 37 and 45°C, 5 ml of log phase cells in BHI were incubated at either: (i) 45°C for 30 min to ensure strong induction of the eGFP mRNA; or (ii) 37°C for 30 min (as control). At this time, an initial sample of cells was collected (t = 0) and transcription was stopped by adding rifampicin (Invitrogen) to a final concentration of 50 μg/ml. Additional samples were collected at t = 5 and t = 20 min after the addition of rifampicin. mRNA isolation, cDNA preparation, and qPCR were performed as detailed above. The fraction of the mRNA remaining (f) at each time point was calculated using f = 2(ct eGFP(t = 0) − ct eGFP(t = x)), where ct eGFP(t = 0) is the ct-value at t = 0 and ct eGFP(t = x) the ct-value at t = 5 or 20 (Archambault et al., 2013). As our analyses were performed to define the decay rate, the ct-values for eGFP were not normalized to rpoB. The reported values represent the means of two independent biological replicates, each performed at 37° and 45°C, resulting in four data points per reporter.
Data analysis
All statistical analyses were performed in R version 3.2.1 (R Core Team, 2015), using the packages lme4 version 1.1-12 (Bates et al., 2014), lmerTest version 2.0-32 (Kuznetsova et al., 2016), ggplot2 version 2.1.0 (Wickham, 2009), and dplyr version 0.5.0 (Wickham, 2011). For FCM-based data, analyses were performed using the proportion of 50,000 cells per sample that were eGFP positive as the dependent variable. The data were logit transformed (Warton and Hui, 2011) and separate linear models were fitted, using lme4, for each heat and salt stress data immediately before exposure to heat or salt stress (t = 0) and after 10, 20, 30, 60, 100, 150, and 200 min of stress exposure. Model selection was by AIC-based backwards stepwise removal process, beginning with a full model up to three-way interactions. Comparisons between (i) stress exposed cells and their controls and (ii) the wildtype strain and its isogenic ΔsigB mutant were made at t = 0, 60, 100, 150, 200 using lsmeans. Holm-Bonferroni-corrected p-values and an α = 0.05 were used as cutoff for statistical significance. The modeled logit predictions along with their standard errors and confidence intervals were back-transformed into probability values for plotting and discussion.
For the mRNA stability data, analyses were performed on the absolute ct-values. With exponential decay, these vary linearly over time. A linear model was fitted to the slopes of the mRNA decay using the lm function in R (R Core Team, 2015). The slopes were compared using lsmeans to obtain a Tukey-corrected p-value (averaged over temperature) with a cutoff of α = 0.05 for significance.
For mRNA induction data, a mixed effect linear model was fitted on the fold change, with a full three-way interaction between reporter, strain, and time effects. Fold change was log-transformed. To determine whether individual mRNA levels were increased (indicated by a fold change significantly larger than 1) after exposure to 45°C compared to 37°C at each time point, lsmeans was used to perform a one-sided effect test, with Holm-Bonferroni-corrected p-values and an α = 0.05 as cutoff for significance. To determine whether σB influenced prfA and hly transcript levels, comparisons between fold changes in transcript levels in the wt and ΔsigB mutant strain background at individual time-points (t = 5, 20, 40 min) were performed using lsmeans with Holm-Bonferroni-corrected p-values and α = 0.05 as cutoff for significance.
Results
Sigma B and PrfA activity are induced stochastically at the single cell level
To visualize the activities of σB and PrfA in individual L. monocytogenes cells, we constructed reporter fusions between promoters that are activated by either σB or PrfA and a gene encoding eGFP that had been codon-optimized for L. monocytogenes. Initial evaluation of the reporter fusion constructs by confocal microcopy showed that (i) the σB reporter strain (10403S::Plmo2230-eGFP) emitted fluorescence under conditions known to induce σB activity (i.e., stationary phase) and (ii) the PrfA reporter strain (10403S Phly::eGFP) emitted strong fluorescence in the presence of active PrfA (e.g., after 45°C heat exposure). Negative controls verified that fluorescence was induced exclusively by σB or PrfA, respectively, for lmo2230 and hly; the ΔsigB and ΔprfA strains containing reporter fusions (10403S ΔprfA, Phly::eGFP and 10403S ΔsigB, Plmo2230::eGFP) did not show any fluorescence. For the PrfA eGFP reporter, a positive control was possible; transformation of the Phly::eGFP construct into a strain carrying a mutation that constitutively activates PrfA (PrfA* G155S) (Shetron-Rama et al., 2003; Ollinger et al., 2009) resulted in eGFP fluorescence levels comparable to those of a control strain constitutively expressing GFP (Supplementary Figure 2). Creation of a positive control for the σB reporter strain is not possible, as strains that constitutively express σB grow poorly or die (Conor O'Byrne, personal communication).
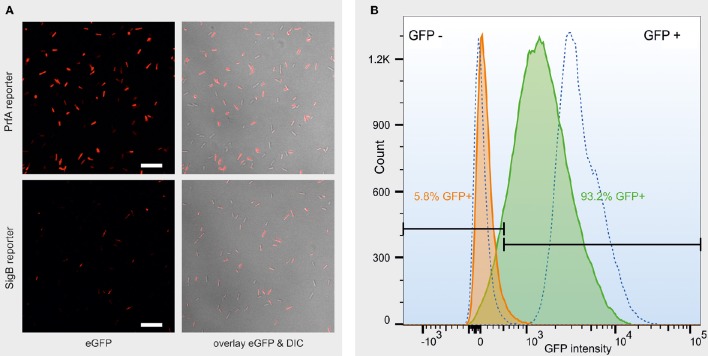
Confocal microscopy on L. monocytogenes log phase cells exposed to heat stress in PBS showed that both cells containing the σB or the PrfA reporter construct were heterogeneous with regard to eGFP expression, with some cells fluorescing strongly while others do not fluoresce at all (Figure 1A). Analysis of the reporter fusion strains by FCM showed a relatively broad peak, indicating various levels of eGFP fluorescence that fluctuated between “many cells off” to “many cells on” in response to heat and salt stress for both σB and PrfA, never fully reaching either 0 or 100% cells in either state. Representative FCM data are shown in Figure 1B, illustrating that log phase PrfA reporter cells were predominantly eGFP negative (5.8% eGFP positive) when incubated at 37°C in PBS for 100 min, while a majority of cells was eGFP positive after incubation at 45°C in PBS for 100 min (93.2% eGFP positive). Hence, both confocal microscopy and flow cytometry show that even under a single condition, L. monocytogenes cells are heterogeneous with regard to PrfA or σB activity, illustrating an inherent level of noise in the activities of PrfA and σB resulting from stochastic responses to changes in the environment. Interestingly, across all tested conditions, a significantly larger proportion of cells showed active PrfA than they did active σB. For example, when averaging across both growth phases (log and stationary phase) and media (BHI and PBS), after 200 min of heat stress, 55% of PrfA reporter cells, but only 5.5% of σB reporter cells, were eGFP positive (p < 0.0001).
Figure 1.
(A) Representative confocal images of a log phase PrfA reporter strain culture (top row) and a log phase σB reporter strain culture (bottom row) after 200 min exposure to heat stress in PBS. Left column: eGFP fluorescence, false-colored red for better contrast. Right column: overlay of the eGFP fluorescence with the differential interference contrast (DIC) image. Scale bar = 10 μm. (B) Representative fluorescence histogram from FCM data measuring the eGFP status of individual cells of a log phase PrfA reporter strain L. monocytogenes 10403S culture, after 100 min incubation in PBS at either 37°C (orange curve) or 45°C (green curve). 10403S wt (left dotted curve) and constitutive GFP (right dotted curve) are plotted as non-fluorescent and strongly fluorescent controls, respectively. GFP ± gates (black, horizontal bars) were set so that no more than 1.5% of the non-fluorescent controls is included in the GFP+ gate. The proportion of GFP positive cells is presented in the color that matches the curve that the data represent. The x-axis represents GFP intensity in arbitrary units, measured as GFP-area, in log scale.
The proportion of cells with active PrfA increased in response to heat stress in a growth-phase dependent manner
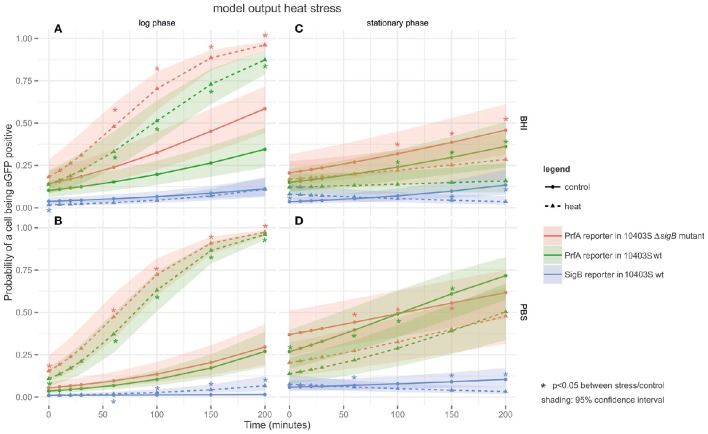
PrfA activity in response to heat stress was monitored at the single cell level under four different conditions: log phase cells in a rich medium (BHI) (Figure 2A) or in PBS (Figure 2B), and stationary phase cells in a rich medium (BHI) (Figure 2C) or in PBS (Figure 2D). A cell was defined as having active PrfA if it expressed eGFP fluorescence above the autofluorescence of the parent L. monocytogenes strain that does not contain a reporter fusion construct. Log phase cells exposed to heat in BHI showed significantly higher proportions of cells with active PrfA (at t = 60, 100, 150, and 200; Figure 2A) compared to the controls incubated at 37°C for both the PrfA reporter strain and its isogenic ΔsigB mutant. The same pattern of heat induction was observed for log-phase cells in PBS (Figure 2B). For example, PrfA activity was observed in 95.4% of log phase cells exposed to heat (at t = 200 min in PBS) compared to 24.7% in the respective control (t = 200, PBS, 37°C), p < 0.001.
Figure 2.
Output of the linear model fitted to the heat FCM data. The panels are tiled to represent cultures grown to different growth phases in either BHI or PBS: (A) log phase cultures in BHI; (B) log phase cultures in PBS; (C) stationary phase cultures in BHI; and (D) stationary phase cultures in PBS. The y-axis represents the probability of a cell being eGFP-positive (as a result of active PrfA or σB), while the x-axis represents time in min. Three different strains are plotted on the graphs: the PrfA-reporter construct in a L. monocytogenes 10403S background, the PrfA-reporter construct in a L. monocytogenes 10403S ΔsigB mutant background, and the σB-reporter construct in a L. monocytogenes 10403S background. Dotted lines represent data collected under the stress condition (45°C heat), the solid line represents the control condition (37°C), the shading around each line represents the 95% confidence interval. *Are color coded to match the strain they pertain to and indicate a significant difference in the probability that a cell is eGFP positive in the stress condition vs. the control condition within one strain at one time point.
For stationary phase cells in BHI, no significant activation of PrfA by heat was observed. Stationary phase cells exposed to 45°C showed significantly lower proportions of cells with active PrfA at t = 100, 150, and 200 (Figure 2C) compared to the 37°C control condition for both the PrfA reporter and its isogenic ΔsigB mutant. For example, PrfA activity was observed in 15.6% of heat exposed stationary phase cells (at t = 200 in BHI) compared to 37.9% in the respective control (t = 200 in BHI at 37°C), p < 0.001. For stationary phase cells in PBS, the same pattern was observed (Figure 2D).
Across all heat stress conditions (Figure 2), there was no significant difference in the proportion of cells with active PrfA when comparing the 10403S PrfA reporter strain and its isogenic ΔsigB mutant, suggesting no significant additional contributions of σB to PrfA activity under the conditions studied.
The proportion of cells with active PrfA increased in log phase cells exposed to salt stress under nutrient limiting conditions
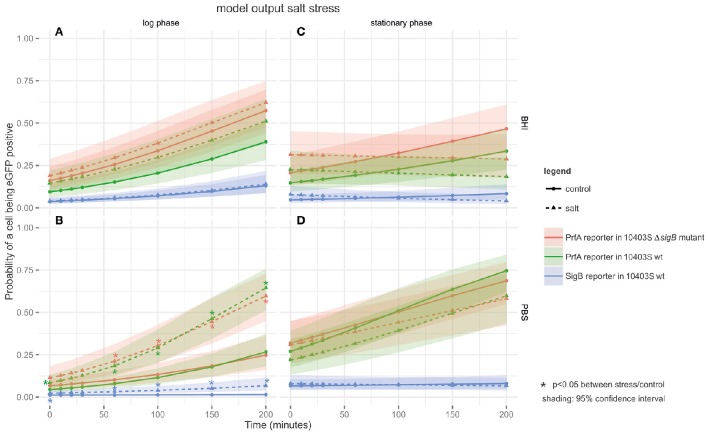
For log phase cells grown in BHI, exposure to salt (4% NaCl) only had a mild effect on PrfA activity; log phase cells exposed to salt in BHI (Figure 3A) showed no significant difference in the proportions of cells with active PrfA compared to the controls without the added salt. In contrast, log phase cells exposed to salt in PBS (Figure 3B), showed significantly higher proportions of cells with active PrfA at all time points in both the PrfA reporter and its isogenic ΔsigB mutant. For example, PrfA activity was observed in 69.5% of salt exposed log phase cells (at t = 200 in PBS) compared to 30.8% in the respective control conditions (t = 200, PBS, no salt, p < 0.001).
Figure 3.
Output of the linear model fitted to the salt stress FCM data. The panels are tiled to represent cultures grown to different growth phases in either BHI or PBS: (A) log phase cultures in BHI; (B) log phase cultures in PBS; (C) stationary phase cultures in BHI; and (D) stationary phase cultures in PBS. The y-axis represents the probability of a cell being eGFP-positive (as a result of active PrfA or σB), while the x-axis represents time in min. Three different strains are plotted on the graphs: the PrfA-reporter construct in a L. monocytogenes 10403S background, the PrfA-reporter construct in a L. monocytogenes 10403S ΔsigB mutant background, and the σB-reporter construct in a L. monocytogenes 10403S background. Dotted lines represent data collected under the stress condition (4.4% salt stress), the solid line represents the control condition (no salt), the shading around each line represents the 95% confidence interval. *Are color coded to match the strain they pertain to and indicate a significant difference in the probability that a cell is eGFP positive in the stress condition vs. the control condition within one strain at one time point.
Stationary phase cells exposed to salt stress in BHI (Figure 3C) showed no significant difference in the proportions of cells with active PrfA compared to the controls without the added salt for both the PrfA reporter and its isogenic ΔsigB mutant. Therefore, exposure to salt stress did not induce PrfA activity in this condition. The same pattern was observed in stationary phase cells exposed to salt in PBS (Figure 3D). For example, PrfA activity was observed in 54.2% of salt exposed stationary phase cells (at t = 200 in PBS) compared to 54.5% in the respective control (t = 200, PBS, no salt, p = 0.71).
Across all salt stress conditions (Figure 3), there was no significant difference in the proportion of cells with active PrfA when comparing the PrfA reporter to its isogenic ΔsigB mutant.
Heat stress lead to a significant increase in the proportion of cells with active σB in log phase cells in PBS, but not in log phase cells grown in BHI or in stationary phase cells
σB activity after heat stress was measured under the same conditions used to assess PrfA activity: log phase cells in a rich medium (BHI) (Figure 2A) or in PBS (Figure 2B), and stationary phase cells in a rich medium (BHI) (Figure 2C) or in PBS (Figure 2D). A cell was defined as having active σB if it expressed eGFP fluorescence above the autofluorescence of the parent L. monocytogenes strain. Across conditions, the proportion of cells with active σB was markedly lower than those with active PrfA (Figure 2).
Log phase cells exposed to 45°C in BHI (Figure 2A) showed no significant difference in the proportion of cells with active σB compared to the controls incubated at 37°C. However, log phase cells exposed to 45°C heat stress in PBS (Figure 2B) showed significantly higher proportions of cells with active σB at all time-points compared to the 37°C controls. For example, σB activity was observed in 3.7% of heat exposed log phase cells (at t = 200 in PBS) compared to 0.7% in the respective control (t = 200, PBS, 37°C, p < 0.001).
In stationary phase cells, heat stress did not induce σB activity in BHI (Figure 2C) or in PBS (Figure 2D). In fact, stationary phase cells exposed to 45°C heat in both media (Figures 2C,D) showed significantly lower proportions of cells with active σB (at t = 60, 150, and 200) compared to the 37°C controls. For example, σB activity was observed in 2.7% of heat exposed stationary phase cells (at t = 200 in BHI) compared to 13.5% in the respective control (t = 200, BHI, 37°C, p < 0.001).
Salt stress increased the proportion of cells with active σB in log phase cells in PBS, but not under any of the other tested conditions
σB activity in response to salt stress was monitored under the same conditions as for heat stress (Figure 3). In log phase cells exposed to salt (4% NaCl) in BHI, the salt exposed cells did not show significantly higher proportions of cells with active σB compared to the controls without the added salt (Figure 3A). However, log phase cells exposed to salt in PBS (Figure 3B) showed significantly higher proportions of cells with active σB at all-time points (t = 60, 100, 150, 200) compared to the controls without the added salt. For example, σB activity was observed in 8.4% of salt exposed log phase cells (at t = 200 in PBS) compared to 2.6% in the respective control (t = 200, PBS, no salt, p < 0.001).
In stationary phase cells exposed to salt stress in BHI (Figure 3C), the proportion of cells with active σB did not increase compared to the controls without the added salt. For example, σB activity was observed in 7.5% of salt exposed stationary phase cells (at t = 200 in BHI) compared to 14.4% in the respective control (t = 200, BHI, no salt, p = 0.091). Stationary phase cells exposed to salt in PBS (Figure 3D) showed no significant changes in the proportions of cells with active σB.
RT-qPCR shows no difference in the stability of PrfA reporter fusion mRNA compared to σB reporter fusion mRNA
As the FCM data showed that generally a much larger proportion of cells had active PrfA compared to the proportion of cells that had active σB (e.g., in log phase cells exposed to 45°C heat for 200 min in PBS, on average, 95.5% showed active PrfA while 3.7% of cells showed active σB), we performed RT-qPCR experiments to assess the relative mRNA stability of the PrfA and σB reporter fusions, using qPCR primers targeting eGFP. Specifically, we hypothesized that the PrfA reporter construct (Phly::eGFP) may be more stable than the σB reporter construct due to a longer native 5′ untranslated region (UTR) (123 nt) in this construct as compared to a shorter (51 nt) 5′ UTR in the σB reporter construct (Plmo2230::eGFP). qPCR data showed a clear reduction of mRNA levels for both reporter constructs after rifampicin treatment to arrest transcription. The data fit a general linear model that allowed us to test whether (i) temperature and (ii) the choice of reporter promoter including its 5′UTR (i.e., Plmo2230::eGFP as reporter for σB activity and Phly::eGFP as reporter for PrfA activity) had a significant effect on mRNA decay (which measures RNA stability); a p-value of 0.25 for the effect of reporter gene on RNA decay showed that there was no significant difference in mRNA stability between the two reporter genes (Supplementary Figure 3).
RT-qPCR supports differences in PrfA and σB activation by heat
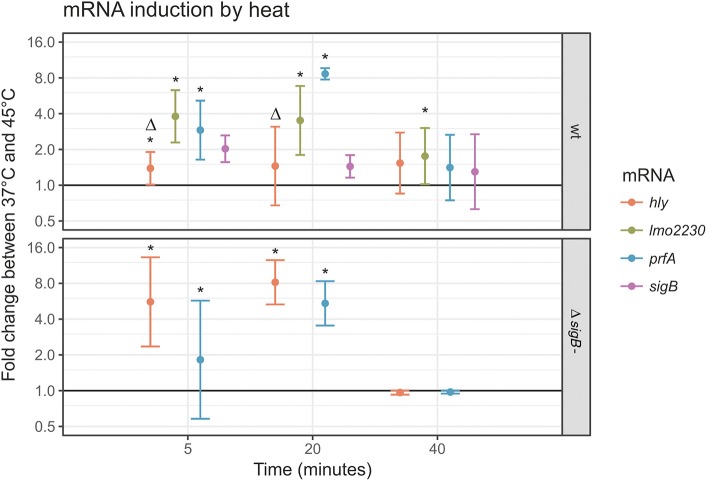
To confirm the results obtained with the reporter fusions, transcript levels for four genes (prfA and the PrfA-dependent hly; sigB and the σB-dependent lmo2230) were determined by RT-qPCR in log phase cells exposed to heat stress in PBS (Figure 3). This condition was selected as our reporter fusion experiments indicated that it induced both PrfA and σB activity (Figure 2B). RT-qPCR data showed clear evidence for increased transcript levels for prfA after 45°C heat stress (2.9-fold increase at t = 5 and 8.5-fold increase at t = 20 compared to the 37°C controls, p < 0.001); Figure 4, top panel. By comparison, the PrfA-dependent hly showed only a mild increase in transcript levels after 45°C heat exposure (1.3-fold increase at t = 5 and 1.4-fold increase at t = 20 as compared to the 37°C control, p > 0.05). Overall, these data suggest that for the PrfA-mediated response to heat stress, an increase in prfA transcript levels is at least a part of the process. RT-qPCR data also showed evidence for mildly increased transcript levels for sigB after 45°C heat exposure (2.2 (p = 0.009) fold increase at t = 5 min, and 1.6 (p = 0.08) fold increase at t = 20 min compared to the 37°C controls). In contrast, the transcript levels for the σB-dependent lmo2230 increased in response to 45°C heat exposure (4.2-fold increase at t = 5 and 3.8-fold increase at t = 20 compared to the 37°C control, p < 0.001). Our finding that the fold change in transcript levels for the σB-dependent lmo2230 exceeds the fold change for sigB transcript levels is consistent with the fact that σB is post-translationally activated by a phosphorylation cascade involving anti-sigma factors (reviewed in O'Byrne and Karatzas, 2008). As a consequence, increased expression of the σB regulon does not necessarily require a parallel increase in sigB transcription levels.
Figure 4.
prfA, hly, sigB and lmo2230 transcript levels in response to 45°C heat stress in log phase cells. RT-qPCR was performed with primers for prfA, hly, sigB and lmo2230 on log phase L. monocytogenes 10403S parent strain cultures (top panel) or its otherwise isogenic ΔsigB strain (bottom panel). The y-axis represents the log fold change between cells incubated in PBS at 37° or 45°C, determined by the delta-delta ct method. A value of 1 represents no change (horizontal line). For each time point, the average ± SD were plotted. *Indicates a statistically significant fold change between 37° and 45°C for this mRNA at this time point (p < 0.05), Δ indicates a significant difference between the wt and the ΔsigB mutant strain at this time point (p < 0.05).
To assess the effect of active σB on expression of the PrfA regulon, prfA and hly mRNA levels were also determined in the 10403S ΔsigB mutant strain (Figure 4, bottom panel). As expected, no sigB mRNA was detected in the 10403S ΔsigB mutant, and only traces of lmo2230 RNA were detectable (absolute ct > 29 in all samples). In the parent strain, the fold change in prfA transcript levels in response to 45°C heat stress was numerically, but not statistically significantly higher than in the 10403S ΔsigB mutant strain, with the highest measurement at 8.5 vs. 5.2-fold change compared to the 37°C controls at t = 20 min; not significant at p = 0.34. In contrast, the hly transcript levels that depend on active PrfA showed a smaller increase in the parent strain after heat treatment than in the ΔsigB mutant strain (with the highest measurement at 1.4 vs. 8.2-fold change after 20 min, p < 0.001). These results reflect the known dual role for σB in PrfA regulation, including (i) positive regulation of prfA transcription (Nadon et al., 2002; Rauch et al., 2005; Schwab et al., 2005) and (ii) indirect negative regulation of PrfA activity (Ollinger et al., 2009).
Discussion
Using reporter fusions that allow monitoring either PrfA or σB activity at the single cell level, we show the dynamic response of individual cells of L. monocytogenes to heat and salt stress. Our data provide a number of novel insights into the regulation of the activity of these two important transcriptional regulators, including evidence for their stochastic activation.
PrfA and σB activity are induced stochastically
Differential and stochastic gene expression in clonal microbial populations allow for bet-hedging and labor-sharing within the population (Elowitz et al., 2002). Our data support that neither PrfA nor σB activity is induced by deterministic circuits that would turn these transcriptional regulators either “on” or “off” in all cells. Rather, confocal microscopy as well as FCM results presented here showed stochastic induction of PrfA and σB activity, which produced an inherent level of noise. As PrfA is essential for virulence and as σB contributes to resilience in L. monocytogenes (O'Byrne and Karatzas, 2008; De las Heras et al., 2011), it is conceivable that stochasticity in PrfA and σB activity is a mechanism of bet-hedging, creating sub-populations that are pre-adapted to enable survival under changing environmental conditions. While stochastic activation following salt stress has been demonstrated previously for σB (Utratna et al., 2012), our data provide novel insights into PrfA activation. Stochastic regulation of virulence gene expression has been demonstrated in other microbial species, including the cap genes in S. aureus (George et al., 2015), the type three secretion system 1 in Salmonella Typhi (Arnoldini et al., 2014), and flagella synthesis in S. enterica (Stewart and Cookson, 2014). The PrfA operon includes genes that encode secreted factors, e.g., the pore-forming toxin, LLO, which facilitates L. monocytogenes escape from the phagocytic vacuole in host cells, a mechanism that potentially lends itself to labor-sharing. However, the PrfA regulon also includes a number of genes that encode surface proteins (e.g., ActA, internalin A and B) for which advantages from labor-sharing are less obvious, as presumably each individual cell would need to express these proteins to successfully proliferate in the host (Gaillard et al., 1991). The necessity of PrfA-dependent surface proteins for successful host cell invasion thus may be a reason why a large proportion of individual L. monocytogenes cells were found to show active PrfA under some of the conditions in this study. A possible model is that stochastic expression of PrfA in a relatively small subpopulation of cells in a certain environment (e.g., outside a host or before initial invasion into host cells) provides for bet-hedging, while expression of PrfA in a majority of cells under other conditions (e.g., inside a host cell) reflects the importance of virulence gene expression during specific phases of infection. Future studies will be necessary to determine correlations among PrfA activity, σB activity and bacterial phenotype in terms of stress resistance, cell-cell interaction, growth, and host cell invasiveness.
Heat and salt stress represent newly identified activator signals for PrfA
The reporter fusion data reported here show that the proportion of cells with active PrfA strongly increased after heat stress exposure of log-phase cells, but not in stationary phase cells. The same was true for the proportion of cells with active σB, indicating the complexity of the underlying regulatory networks and the probable involvement of other, yet unknown regulatory factors that render the response to heat stress growth-phase dependent. We initially included a mild heat stress of 45°C to determine its effect on σB activity, since a ΔsigB mutant is less heat resistant than the wildtype and σB is involved in the heat shock response (Hu et al., 2007). Somewhat surprisingly, we found that in log phase cells after heat stress, the proportion of individual cells with active PrfA reached nearly 100%, which was higher than the proportion of cells with active σB under any conditions (the maximum observed was 14.8% of log phase cells with active σB after exposure to salt stress in BHI for 200 min). RT-qPCR confirmed heat induction of PrfA and showed an increase of prfA transcripts after 45°C heat exposure. While these data suggest that increased transcription of prfA may at least partially contribute to the increased levels of active PrfA observed after heat stress, the specific mechanism of 45°C heat stress induction of PrfA activity remains to be determined. PrfA activity is regulated at the transcriptional level through an autoregulatory feedback loop (Mengaud et al., 1991; Chakraborty et al., 1992) as well as at the post-transcriptional and post-translational levels, allowing for potential induction at various levels. A temperature-dependent post-transcriptional mechanism regulates prfA translation. The monocistronic prfA mRNA acts as a thermosensor by forming a temperature-dependent secondary structure in the 5′UTR that masks the ribosomal binding site at 30°C (Johansson et al., 2002), but not at 37°C, ensuring that PrfA-controlled virulence genes are expressed at typical mammalian host body temperatures. It is thus tempting to speculate that increases in temperatures above 37°C may further enhance prfA translation with subsequent enhanced prfA transcription through the auto-regulatory feedback loop. Alternatively, 45°C heat stress may also contribute to post-translational activation of PrfA. A previous study (Herbert and Foster, 2001) found no effect of a prfA null mutation on heat resistance. While 45°C may not be encountered frequently by L. monocytogenes during infection, heat stress, in general, could be encountered by L. monocytogenes during transmission, including in food processing-associated environments (e.g., hot water cleaning), foods (e.g., sublethal heat treatment), and, potentially, in certain hosts with higher body temperatures, e.g., chickens (Speer, 2006).
Alternatively, increased activity at 45°C may simply result from the complex regulation of PrfA activity, which is not fully understood, but appears to involve transcriptional upregulation mediated by the presence of branched chain amino acids (sensed via CodY; Lobel et al., 2015) and glucose-1-phosphate carbohydrate (via the mannose-type phosphotransferase systems; Ake et al., 2011). Furthermore, the presence of glutathione (produced by a Listeria glutathione synthase) also appears to activate PrfA (Reniere et al., 2015).
Our data also indicated that exposure of log phase cells to salt stress increased the proportion of cells with active PrfA, albeit this induction was considerably less pronounced than that observed in response to heat stress. While previous studies have shown that a prfA null deletion had no effect on L. monocytogenes survival of 11% salt stress (Ribeiro et al., 2014), an increase in osmolarity may serve as a signal for the imminent encounter of the pathogen with host cells in the duodenum, where the osmolarity is higher than in previously encountered gastrointestinal compartments due to the breakdown of gastric contents into smaller molecules. Therefore, increased osmolarity may represent an environmental signal that is used by L. monocytogenes to sense its presence in a host environment where induction of PrfA activity is needed to facilitate invasion of host cells. Whether results obtained in liquid cell culture media, such as in our experiments, translate to L. monocytogenes growing on food matrices, and particularly on solid ones, remain to be determined.
PrfA activity was not significantly altered in a ΔsigB mutant compared to the parent strain, although an intact σB contributed to higher prfA mRNA levels under heat stress. Thus, our reporter fusion data reported here further support the previously reported (Nadon et al., 2002; Ollinger et al., 2009) dual role of σB in regulating prfA transcription and expression of the PrfA regulon, including (i) σB-dependent upregulation of prfA transcription through the prfAP1 promotor and (ii) σB-dependent downregulation of PrfA-dependent genes (including hly) in the presence of active PrfA through a yet-to-be determined mechanism. The positive influence of σB on prfA transcription is supported by our RT-qPCR data showing higher prfA transcript levels in the parent strain compared to the ΔsigB mutant. The indirect negative influence of σB on PrfA activity is visible in the reporter fusion data that consistently showed a trend of higher PrfA activity in a sigB null background (Figure 2). This conclusion is further supported by RT-qPCR data, which showed lower transcript levels of the PrfA dependent hly in the parent strain compared to the ΔsigB mutant under heat stress (Figure 4). Future experiments with a double reporter strain for PrfA and σB activity may provide an opportunity to gain higher-resolution insights into the relation between PrfA and σB activity and to investigate the overlap between cells with high PrfA and σB activity.
Heat and salt stress act as activators of σB under select conditions
Our data showed that heat stress exposure of log phase cells in PBS led to a significantly increased proportion of cells with active σB; this effect was not seen for log phase cells grown in BHI or for stationary phase cells. Previous studies suggest that σB is involved in the heat stress response; σB has been shown to directly or indirectly regulate expression of several heat shock proteins, such as ClpP, and regulators of heat survival, including CtsR and HrcA (Hu et al., 2007). For example, an hrcA mutant was significantly less heat resistant than its parent strain and a σB-dependent promoter was computationally identified upstream of hrcA (Hu et al., 2007). However, results from studies that investigated the impact of sigB null deletions on heat stress survival have varied from minor (Ferreira et al., 2001) to significant effects (Hu et al., 2007; Somolinos et al., 2010; Ait-Ouazzou et al., 2012). σB activity is governed by a complex regulatory network and is affected by multiple factors including nutrients and temperature (Guariglia-Oropeza et al., 2014), therefore different outcomes across studies likely reflect the different conditions, strains and media used. Overall, we provide further support that σB contributes to heat stress response in L. monocytogenes.
Our data showed an increased proportion of log phase cells with active σB in response to salt stress in PBS (but not in BHI), however, the proportion of cells with active σB did not exceed 15% in any of the tested conditions. By comparison, a study by Utratna et al. (2012) found about 42% of cells displaying σB-dependent fluorescence after growing L. monocytogenes EGDe to log phase in 3.5% NaCl in BHI, compared to 13% with σB-dependent fluorescence in our study. These differences could result from strain differences between 10403S and EGDe, but could also reflect different FCM instruments and protocols, or the different site in the genome where the reporter construct was integrated. Salt concentrations in the range of 0.5 M (2.9%) to 1.9 M (11%) have previously been shown to induce σB activity on a population level (Sue et al., 2003; Ringus et al., 2012) or to significantly inhibit growth of ΔsigB mutants (Abram et al., 2008; Ribeiro et al., 2014). Additionally, previous studies indicate that the type of osmotic stress used here strongly increases transcription of σB-dependent genes (Sue et al., 2003, 2004). If σB activity is crucial for survival under salt stress, but a measurable increase in σB activity only occurs in a small proportion of cells, it is possible that the effect observed at the population-level results from increased σB activity in relatively few cells. This surprising finding could indicate either (i) a bet-hedging mechanism (as discussed in more detail below) or (ii) the existence of a mechanism for the whole population to profit from active σB in a proportion of cells in a labor-sharing fashion. Unlike extracytoplasmatic sigma factors such as σC, σB is not secreted. However, one could speculate that a σB-dependent, secreted factor may serve the whole population. While there are no known σB-dependent proteins with an obvious potential for labor-sharing in L. monocytogenes, there are σB-dependent factors produced by L. monocytogenes that do get secreted, including the products of fri, sodA, lmo1068, and lmo0796 (Trost et al., 2005). Fri (LMRG_0241) is a homolog of the E. coli dps (DNA-binding protein from starving cells; Olsen et al., 2005) that prevents generation of hydroxyl radicals under anaerobic conditions by sequestering iron from Fenton reactions (Ilari et al., 2002). The σB-dependent promoter upstream of fri (Polidoro et al., 2002) has been experimentally confirmed (Olsen et al., 2005). The superoxide dismutase encoded by sodA has a σB-dependent promoter (Raengpradub et al., 2008) and is secreted by the auxiliary protein secretion system SecA (Archambaud et al., 2006). Much less is known about the functions of lmo0796 (a ycel-family like protein) and the hypothetical protein lmo1068, both of which have been shown to be secreted (Trost et al., 2005) and to have σB-dependent promoters (Hain et al., 2008; Oliver et al., 2010; Mujahid et al., 2013).
The observation that few individual cells induce σB activity to a detectable level, even in conditions known to strongly activate σB, also has implications on the interpretation of transcriptional data generated at the population level (e.g., through microarrays). In any method that detects the cumulative signal from a population, it is possible that a relatively small number of cells with strong σB activity produce the signal while the remainder of cells stay “silent,” do not activate σB and hence do not induce transcription of the σB regulon. In summary, the following model could explain our observations for σB activity: generally, very few cells show high σB activity. Under conditions known to activate σB, the number of cells with active σB increases, while a majority of cells still show undetectable levels of σB activity. It remains to be determined whether σB activity at levels below the detection limit of our assay can exert cellular functions, while keeping the “cost” (as discussed below) to the cell low.
Induction of PrfA activity appears to be population-wide, while σB activity appears restricted to a minority subpopulation, indicating stronger bet-hedging, and/or labor sharing for σB
Interestingly, we found that under all conditions evaluated, the proportion of individual cells that express detectable levels of active PrfA was significantly higher than for σB. Other authors previously reported much higher levels of prfA mRNA compared to sigB mRNA in stationary phase cells (Olesen et al., 2010). The differences in proportion of cells with active PrfA and σB could indicate that it is advantageous to have a large number of cells with active PrfA under certain conditions, while it is advantageous for L. monocytogenes to activate σB in only a smaller subpopulation. Alternatively, our system may not be sensitive enough to pick up subtle changes in σB activity; if the effect of a transcription factor is amplified by a positive feedback loop, as with σB, a relatively small number of active molecules can have a large effect. It is therefore possible that σB activity stays below the detection limit of our assay but still has an impact on the transcriptome. It is also possible that σB may be activated in large subpopulations, but under conditions other than those tested here.
The fact that the proportion of cells with active σB was found to be limited under stress conditions that have been shown previously to strongly induce σB activity is consistent with observations on the “fitness cost” of expressing active forms of σB and PrfA. Previous data clearly indicate that σB activity comes at a cost to the cell as supported by the fact that L. monocytogenes with either a sigB deletion or a deletion of regulators that activate σB show more rapid log phase growth as compared to wildtype strains (Chaturongakul and Boor, 2004). Also, attempts to create mutations with constitutively active σB have failed in the past or show very poor growth in L. monocytogenes (Conor O'Byrne, personal communication) as well as in B. subtilis (Redfield and Price, 1996). In contrast, constitutive activation of PrfA is possible and does not lead to growth deficiencies of comparable proportions. In fact, there are several known mutations that render PrfA constitutively active (PrfA*) (Ripio et al., 1997; Monk et al., 2008). Characterization of these mutants did not show any growth defects when grown in monoculture in rich media, however a fitness defect was detectable in competitive growth assays with the isogenic wildtype strains (Bruno and Freitag, 2010). This proposed model might help explain why σB activity is restricted to a relatively small number of individual cells compared to PrfA activity, resulting in stronger bet-hedging or labor-sharing in the case of σB. We thus conclude that our data convincingly suggest that the dynamics and stochasticity of regulation of PrfA and σB activity differ at the single cell level.
Author contributions
This study was conceived and designed by MW, KB, and CG. CG and SH conducted the labwork. Data analysis and interpretation was done by CG, DK, VG, MW. CG and MW wrote the manuscript with critical and crucial input by VG, KB, SH, and DK.
Funding
This work was funded by a grant of the Swiss National Science Foundation to Claudia Guldimann (Grant No. P2BEP3_148881)
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are thankful to Erika Mudrak from the Cornell Statistical Consulting Unit for her support on the statistical analysis and Barbara Bowen (Food Safety Laboratory, Cornell University, USA) for her expertise on cloning methods. The plasmid pAD::eGFP was a gift from Damien Balestrino (Laboratoire de Bactériologie, Université de l'Auvergne, France), and Conor O'Byrne (National University of Ireland Galway, Ireland) provided the Plmo2230::eGFP reporter construct.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00348/full#supplementary-material
References
- Abram F., Starr E., Karatzas K. A., Matlawska-Wasowska K., Boyd A., Wiedmann M., et al. (2008). Identification of components of the sigma B regulon in Listeria monocytogenes that contribute to acid and salt tolerance. Appl. Environ. Microbiol. 74, 6848–6858. 10.1128/AEM.00442-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ait-Ouazzou A., Manas P., Condón S., Págan R., García-Gonzalo D. (2012). Role of general stress-response alternative sigma factors sigma(S) (RpoS) and sigma(B) (SigB) in bacterial heat resistance as a function of treatment medium pH. Int. J. Food. Microbiol. 153, 358–364. 10.1016/j.ijfoodmicro.2011.11.027 [DOI] [PubMed] [Google Scholar]
- Ake F. M., Joyet P., Deutscher J., Milohanic E. (2011). Mutational analysis of glucose transport regulation and glucose-mediated virulence gene repression in Listeria monocytogenes. Mol. Microbiol. 81, 274–293. 10.1111/j.1365-2958.2011.07692.x [DOI] [PubMed] [Google Scholar]
- Allerberger F., Wagner M. (2010). Listeriosis: a resurgent foodborne infection. Clin. Microbiol Infect. 16, 16–23. 10.1111/j.1469-0691.2009.03109.x [DOI] [PubMed] [Google Scholar]
- Archambaud C., Nahori M. A., Pizarro-Cerda J., Cossart P., Dussurget O. (2006). Control of Listeria superoxide dismutase by phosphorylation. J. Biol. Chem. 281, 31812–31822. 10.1074/jbc.M606249200 [DOI] [PubMed] [Google Scholar]
- Archambault L., Borchert J. S., Bergeron J., Snow S., Schlax P. J. (2013). Measurements of mRNA degradation in Borrelia burgdorferi. J. Bacteriol. 195, 4879–4887. 10.1128/JB.00659-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoldini M., Vizcarra I. A., Peña-Miller R., Stocker N., Diard M., Vogel V., et al. (2014). Bistable expression of virulence genes in Salmonella leads to the formation of an antibiotic-tolerant subpopulation. PLoS. Biol. 12:e1001928. 10.1371/journal.pbio.1001928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakardjiev A. I., Theriot J. A., Portnoy D. A. (2006). Listeria monocytogenes traffics from maternal organs to the placenta and back. PLoS. Pathog. 2:e66. 10.1371/journal.ppat.0020066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrino D., Hamon M. A., Dortet L., Nahori M. A., Pizarro-Cerda J., Alignani D., et al. (2010). Single-cell techniques using chromosomally tagged fluorescent bacteria to study Listeria monocytogenes infection processes. Appl. Environ. Microbiol. 76, 3625–3636. 10.1128/AEM.02612-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D., Maechler M., Bolker B., Walker S. (2014). Lme4: Linear Mixed-Effects Models Using Eigen and S4. J. Stat. Softw. 67, 1–48. 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- Bishop D. K., Hinrichs D. J. (1987). Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 139, 2005–2009. [PubMed] [Google Scholar]
- Bruno J. C., Jr., Freitag N. E. (2010). Constitutive activation of PrfA tilts the balance of Listeria monocytogenes fitness towards life within the host versus environmental survival. PLoS ONE 5:e15138. 10.1371/journal.pone.0015138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camejo A., Buchrieser C., Couve E., Carvalho F., Reis O., Ferreira P., et al. (2009). In vivo transcriptional profiling of Listeria monocytogenes and mutagenesis identify new virulence factors involved in infection. PLoS. Pathog. 5:e1000449. 10.1371/journal.ppat.1000449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty T., Leimeister-Wächter M., Domann E., Hartl M., Goebel W., Nichterlein T., et al. (1992). Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J. Bacteriol. 174, 568–574. 10.1128/jb.174.2.568-574.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturongakul S., Boor K. J. (2004). RsbT and RsbV contribute to sigmaB-dependent survival under environmental, energy, and intracellular stress conditions in Listeria monocytogenes. Appl. Environ. Microbiol. 70, 5349–5356. 10.1128/AEM.70.9.5349-5356.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturongakul S., Raengpradub S., Wiedmann M., Boor K. J. (2008). Modulation of stress and virulence in Listeria monocytogenes. Trends Microbiol. 16, 388–396. 10.1016/j.tim.2008.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crim S. M., Griffin P. M., Tauxe R., Marder E. P., Gilliss D., Cronquist A. B., et al. (2015). Preliminary incidence and trends of infection with pathogens transmitted commonly through food - Foodborne Diseases Active Surveillance Network, 10 U.S. sites, 2006-2014. MMWR. Morb. Mortal. Wkly. Rep. 64, 495–499. Available online at: https://www.cdc.gov/mmwr/preview/mmwrhtml/mm6418a4.htm [PMC free article] [PubMed] [Google Scholar]
- Currie A., Farber J. M., Nadon C., Sharma D., Whitfield Y., Gaulin C., et al. (2015). Multi-province listeriosis outbreak linked to contaminated deli meat consumed primarily in institutional settings, Canada, 2008. Foodborne. Pathog. Dis. 12, 645–652. 10.1089/fpd.2015.1939 [DOI] [PubMed] [Google Scholar]
- De las Heras A., Cain R. J., Bielecka M. K., Vázquez-Boland J. A. (2011). Regulation of Listeria virulence: PrfA master and commander. Curr. Opin. Microbiol. 14, 118–127. 10.1016/j.mib.2011.01.005 [DOI] [PubMed] [Google Scholar]
- de Valk H., Jacquet C., Goulet V., Vaillant V., Perra A., Simon F., et al. (2005). Surveillance of Listeria infections in Europe. Euro. Surveill. 10, 251–255. Available online at: http://www.eurosurveillance.org/images/dynamic/EQ/v05n04/v05n04.pdf [PubMed] [Google Scholar]
- Eldar A., Elowitz M. B. (2010). Functional roles for noise in genetic circuits. Nature 467, 167–173. 10.1038/nature09326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz M. B., Levine A. J., Siggia E. D., Swain P. S. (2002). Stochastic gene expression in a single cell. Science 297, 1183–1186. 10.1126/science.1070919 [DOI] [PubMed] [Google Scholar]
- Ferreira A., O'Byrne C. P., Boor K. J. (2001). Role of sigma(B) in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl. Environ. Microbiol. 67, 4454–4457. 10.1128/AEM.67.10.4454-4457.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag N. E., Rong L., Portnoy D. A. (1993). Regulation of the prfA transcriptional activator of Listeria monocytogenes: multiple promoter elements contribute to intracellular growth and cell-to-cell spread. Infect. Immun. 61, 2537–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard J. L., Berche P., Frehel C., Gouin E., Cossart P. (1991). Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell 65, 1127–1141. 10.1016/0092-8674(91)90009-N [DOI] [PubMed] [Google Scholar]
- George S. E., Nguyen T., Geiger T., Weidenmaier C., Lee J. C., Liese J., et al. (2015). Phenotypic heterogeneity and temporal expression of the capsular polysaccharide in Staphylococcus aureus. Mol. Microbiol. 98, 1073–1088. 10.1111/mmi.13174 [DOI] [PubMed] [Google Scholar]
- Gottlieb S. L., Newbern E. C., Griffin P. M., Graves L. M., Hoekstra R. M., Baker N. L., et al. (2006). Multistate outbreak of Listeriosis linked to turkey deli meat and subsequent changes in US regulatory policy. Clin. Infect. Dis. 42, 29–36. 10.1086/498113 [DOI] [PubMed] [Google Scholar]
- Guariglia-Oropeza V., Orsi R. H., Yu H., Boor K. J., Wiedmann M., Guldimann C. (2014). Regulatory network features in Listeria monocytogenes -changing the way we talk. Front. Cell Infect. Microbiol. 4:14. 10.3389/fcimb.2014.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hain T., Hossain H., Chatterjee S. S., Machata S., Volk U., Wagner S., et al. (2008). Temporal transcriptomic analysis of the Listeria monocytogenes EGD-e SigmaB regulon. BMC Microbiol. 8:20. 10.1186/1471-2180-8-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman K. E., Garalde V. B., Gronostaj M., Jackson K. A., Beam S., Joseph L., et al. (2016). Multistate outbreak of listeriosis caused by imported cheese and evidence of cross-contamination of other cheeses, USA, 2012. Epidemiol. Infect. 144, 2698–2708. 10.1017/S095026881500117X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert K. C., Foster S. J. (2001). Starvation survival in Listeria monocytogenes: characterization of the response and the role of known and novel components. Microbiology 147, 2275–2284. 10.1099/00221287-147-8-2275 [DOI] [PubMed] [Google Scholar]
- Hu Y., Oliver H. F., Raengpradub S., Palmer M. E., Orsi R. H., Wiedmann M., et al. (2007). Transcriptomic and phenotypic analyses suggest a network between the transcriptional regulators HrcA and sigmaB in Listeria monocytogenes. Appl. Environ. Microbiol. 73, 7981–7991. 10.1128/AEM.01281-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilari A., Ceci P., Ferrari D., Rossi G. L., Chiancone E. (2002). Iron incorporation into Escherichia coli Dps gives rise to a ferritin-like microcrystalline core. J. Biol. Chem. 277, 37619–37623. 10.1074/jbc.M206186200 [DOI] [PubMed] [Google Scholar]
- Johansson J., Mandin P., Renzoni A., Chiaruttini C., Springer M., Cossart P. (2002). An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110, 551–561. 10.1016/S0092-8674(02)00905-4 [DOI] [PubMed] [Google Scholar]
- Kaern M., Elston T. C., Blake W. J., Collins J. J. (2005). Stochasticity in gene expression: from theories to phenotypes. Nat. Rev. Genet. 6, 451–464. 10.1038/nrg1615 [DOI] [PubMed] [Google Scholar]
- Kazmierczak M. J., Mithoe S. C., Boor K. J., Wiedmann M. (2003). Listeria monocytogenes sigma B regulates stress response and virulence functions. J. Bacteriol. 185, 5722–5734. 10.1128/JB.185.19.5722-5734.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsova A., Bruun Brockhoff P., Bojesen Christensen H. (2016). lmerTest: Tests in Linear Mixed Effects Models. Available online at: https://CRAN.R-project.org/package=lmerTest
- Lauer P., Chow M. Y., Loessner M. J., Portnoy D. A., Calendar R. (2002). Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184, 4177–4186. 10.1128/JB.184.15.4177-4186.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobel L., Sigal N., Borovok I., Belitsky B. R., Sonenshein A. L., Herskovits A. A. (2015). The metabolic regulator CodY links Listeria monocytogenes metabolism to virulence by directly activating the virulence regulatory gene prfA. Mol. Microbiol. 95, 624–644. 10.1111/mmi.12890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre L., Wilcott L., Naus M. (2015). Listeriosis outbreaks in British Columbia, Canada, caused by soft ripened cheese contaminated from environmental sources. Biomed. Res. Int. 2015:131623. 10.1155/2015/131623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengaud J., Dramsi S., Gouin E., Vazquez-Boland J. A., Milon G., Cossart P. (1991). Pleiotropic control of Listeria monocytogenes virulence factors by a gene that is autoregulated. Mol. Microbiol. 5, 2273–2283. 10.1111/j.1365-2958.1991.tb02158.x [DOI] [PubMed] [Google Scholar]
- Milohanic E., Glaser P., Coppée J. Y., Frangeul L., Vega Y., Vazquez-Boland J. A., et al. (2003). Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 47, 1613–1625. 10.1046/j.1365-2958.2003.03413.x [DOI] [PubMed] [Google Scholar]
- Monk I. R., Gahan C. G., Hill C. (2008). Tools for functional postgenomic analysis of Listeria monocytogenes. Appl. Environ. Microbiol. 74, 3921–3934. 10.1128/AEM.00314-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujahid S., Orsi R. H., Vangay P., Boor K. J., Wiedmann M. (2013). Refinement of the Listeria monocytogenes sigmaB regulon through quantitative proteomic analysis. Microbiology 159, 1109–1119. 10.1099/mic.0.066001-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadon C. A., Bowen B. M., Wiedmann M., Boor K. J. (2002). Sigma B contributes to PrfA-mediated virulence in Listeria monocytogenes. Infect. Immun. 70, 3948–3952. 10.1128/IAI.70.7.3948-3952.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Byrne C. P., Karatzas K. A. (2008). The role of sigma B (sigma B) in the stress adaptations of Listeria monocytogenes: overlaps between stress adaptation and virulence. Adv. Appl. Microbiol. 65, 115–140. 10.1016/S0065-2164(08)00605-9 [DOI] [PubMed] [Google Scholar]
- Olesen I., Thorsen L., Jespersen L. (2010). Relative transcription of Listeria monocytogenes virulence genes in liver pates with varying NaCl content. Int. J. Food. Microbiol. 141(Suppl. 1), S60–S68. 10.1016/j.ijfoodmicro.2010.01.042 [DOI] [PubMed] [Google Scholar]
- Oliver H. F., Orsi R. H., Ponnala L., Keich U., Wang W., Sun Q., et al. (2009). Deep RNA sequencing of L. monocytogenes reveals overlapping and extensive stationary phase and sigma B-dependent transcriptomes, including multiple highly transcribed noncoding RNAs. BMC Genomics 10:641. 10.1186/1471-2164-10-641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver H. F., Orsi R. H., Wiedmann M., Boor K. J. (2010). Listeria monocytogenes sigma B has a small core regulon and a conserved role in virulence but makes differential contributions to stress tolerance across a diverse collection of strains. Appl. Environ. Microbiol. 76, 4216–4232. 10.1128/AEM.00031-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollinger J., Bowen B., Wiedmann M., Boor K. J., Bergholz T. M. (2009). Listeria monocytogenes sigmaB modulates PrfA-mediated virulence factor expression. Infect. Immun. 77, 2113–2124. 10.1128/IAI.01205-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen K. N., Larsen M. H., Gahan C. G., Kallipolitis B., Wolf X. A., Rea R., et al. (2005). The Dps-like protein Fri of Listeria monocytogenes promotes stress tolerance and intracellular multiplication in macrophage-like cells. Microbiology 151, 925–933. 10.1099/mic.0.27552-0 [DOI] [PubMed] [Google Scholar]
- Ondrusch N., Kreft J. (2011). Blue and red light modulates SigB-dependent gene transcription, swimming motility and invasiveness in Listeria monocytogenes. PLoS ONE 6:e16151. 10.1371/journal.pone.0016151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polidoro M., De B. D., Montagnini B., Guarrera L., Cavallo S., Valenti P., et al. (2002). The expression of the dodecameric ferritin in Listeria spp. is induced by iron limitation and stationary growth phase. Gene 296, 121–128. 10.1016/S0378-1119(02)00839-9 [DOI] [PubMed] [Google Scholar]
- Popovic I., Heron B., Covacin C. (2014). Listeria: an Australian perspective (2001-2010). Foodborne. Pathog. Dis. 11, 425–432. 10.1089/fpd.2013.1697 [DOI] [PubMed] [Google Scholar]
- Raengpradub S., Wiedmann M., Boor K. J. (2008). Comparative analysis of the sigma B-dependent stress responses in Listeria monocytogenes and Listeria innocua strains exposed to selected stress conditions. Appl. Environ. Microbiol. 74, 158–171. 10.1128/AEM.00951-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A., van Oudenaarden A. (2008). Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135, 216–226. 10.1016/j.cell.2008.09.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch M., Luo Q., Müller-Altrock S., Goebel W. (2005). SigB-dependent in vitro transcription of prfA and some newly identified genes of Listeria monocytogenes whose expression is affected by PrfA in vivo. J. Bacteriol. 187, 800–804. 10.1128/JB.187.2.800-804.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2015). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Redfield A. R., Price C. W. (1996). General stress transcription factor sigmaB of Bacillus subtilis is a stable protein. J. Bacteriol. 178, 3668–3670. 10.1128/jb.178.12.3668-3670.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reniere M. L., Whiteley A. T., Hamilton K. L., John S. M., Lauer P., Brennan R. G., et al. (2015). Glutathione activates virulence gene expression of an intracellular pathogen. Nature 517, 170–173. 10.1038/nature14029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro V. B., Mujahid S., Orsi R. H., Bergholz T. M., Wiedmann M., Boor K. J., et al. (2014). Contributions of sigma(B) and PrfA to Listeria monocytogenes salt stress under food relevant conditions. Int. J. Food. Microbiol. 177, 98–108. 10.1016/j.ijfoodmicro.2014.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringus D. L., Ivy R. A., Wiedmann M., Boor K. J. (2012). Salt stress-induced transcription of sigmaB- and CtsR-regulated genes in persistent and non-persistent Listeria monocytogenes strains from food processing plants. Foodborne. Pathog. Dis. 9, 198–206. 10.1089/fpd.2011.1000 [DOI] [PubMed] [Google Scholar]
- Ripio M. T., Dominguez-Bernal G., Lara M., Suarez M., Vazquez-Boland J. A. (1997). A Gly145Ser substitution in the transcriptional activator PrfA causes constitutive overexpression of virulence factors in Listeria monocytogenes. J. Bacteriol. 179, 1533–1540. 10.1128/jb.179.5.1533-1540.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab U., Bowen B., Nadon C., Wiedmann M., Boor K. J. (2005). The Listeria monocytogenes prfAP2 promoter is regulated by sigma B in a growth phase dependent manner. FEMS Microbiol. Lett. 245, 329–336. 10.1016/j.femsle.2005.03.025 [DOI] [PubMed] [Google Scholar]
- Shetron-Rama L. M., Mueller K., Bravo J. M., Bouwer H. G., Way S. S., Freitag N. E. (2003). Isolation of Listeria monocytogenes mutants with high-level in vitro expression of host cytosol-induced gene products. Mol. Microbiol. 48, 1537–1551. 10.1046/j.1365-2958.2003.03534.x [DOI] [PubMed] [Google Scholar]
- Somolinos M., Espina L., Pagan R., Garcia D. (2010). sigB absence decreased Listeria monocytogenes EGD-e heat resistance but not its Pulsed Electric Fields resistance. Int. J. Food. Microbiol. 141, 32–38. 10.1016/j.ijfoodmicro.2010.04.023 [DOI] [PubMed] [Google Scholar]
- Speer B. (2006). Pet chicken medicine and surgery, in NAVC Proceedings. Gainsville, FL. [Google Scholar]
- Stewart M. K., Cookson B. T. (2014). Mutually repressing repressor functions and multi-layered cellular heterogeneity regulate the bistable Salmonella fliC census. Mol. Microbiol. 94, 1272–1284. 10.1111/mmi.12828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sue D., Boor K. J., Wiedmann M. (2003). Sigma(B)-dependent expression patterns of compatible solute transporter genes opuCA and lmo1421 and the conjugated bile salt hydrolase gene bsh in Listeria monocytogenes. Microbiology 149, 3247–3256. 10.1099/mic.0.26526-0 [DOI] [PubMed] [Google Scholar]
- Sue D., Fink D., Wiedmann M., Boor K. J. (2004). sigmaB-dependent gene induction and expression in Listeria monocytogenes during osmotic and acid stress conditions simulating the intestinal environment. Microbiology 150, 3843–3855. 10.1099/mic.0.27257-0 [DOI] [PubMed] [Google Scholar]
- Tang S., Orsi R. H., den Bakker H. C., Wiedmann M., Boor K. J., Bergholz T. M. (2015). Transcriptomic analysis of the adaptation of Listeria monocytogenes to growth on vacuum-packed cold smoked salmon. Appl. Environ. Microbiol. 81, 6812–6824. 10.1128/AEM.01752-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Arana A., Dussurget O., Nikitas G., Sesto N., Guet-Revillet H., Balestrino D., et al. (2009). The Listeria transcriptional landscape from saprophytism to virulence. Nature 459, 950–956. 10.1038/nature08080 [DOI] [PubMed] [Google Scholar]
- Trost M., Wehmhöner D., Kärst U., Dieterich G., Wehland J., Jansch L. (2005). Comparative proteome analysis of secretory proteins from pathogenic and nonpathogenic Listeria species. Proteomics 5, 1544–1557. 10.1002/pmic.200401024 [DOI] [PubMed] [Google Scholar]
- Utratna M., Cosgrave E., Baustian C., Ceredig R., O'Byrne C. (2012). Development and optimization of an EGFP-based reporter for measuring the general stress response in Listeria monocytogenes. Bioeng. Bugs 3, 93–103. 10.4161/bbug.19476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utratna M., Shaw I., Starr E., O'Byrne C. P. (2011). Rapid, transient, and proportional activation of sigma(B) in response to osmotic stress in Listeria monocytogenes. Appl. Environ. Microbiol. 77, 7841–7845. 10.1128/AEM.05732-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warton D. I., Hui F. K. (2011). The arcsine is asinine: the analysis of proportions in ecology. Ecology 92, 3–10. 10.1890/10-0340.1 [DOI] [PubMed] [Google Scholar]
- Wickham H. (2009). ggplot2: Elegant Graphics for Data Analysis. New York, NY: Springer. [Google Scholar]
- Wickham H. (2011). The split-apply-combine strategy for data analysis. J. Stat. Softw. 40, 1–29. 10.18637/jss.v040.i01 [DOI] [Google Scholar]
- Wiedmann M., Arvik T. J., Hurley R. J., Boor K. J. (1998). General stress transcription factor sigmaB and its role in acid tolerance and virulence of Listeria monocytogenes. J. Bacteriol. 180, 3650–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K. K., Freitag N. E. (2004). A novel mutation within the central Listeria monocytogenes regulator PrfA that results in constitutive expression of virulence gene products. J. Bacteriol. 186, 6265–6276. 10.1128/JB.186.18.6265-6276.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.