Abstract
Human exonuclease 1 (hEXO1) is an important nuclease involved in mismatch repair system that contributes to maintain genomic stability and modulate DNA recombination. This study is aimed to explore the associations between single-nucleotide polymorphisms (SNPs) of hEXO1 and the hereditary susceptibility of hepatocellular carcinoma (HCC). SNPs rs1047840, rs1776148, rs3754093, rs4149867, rs4149963, and rs1776181 of hEXO1 were examined from a hospital-based case-control study including 1,196 cases (HCC patients) and 1,199 controls (non-HCC patients) in Guangxi, China. We found the rs3754093 AG genotype decreased the risk of HCC (OR=0.714, 95% CI: 0.539∼0.946). According to the results of stratification analysis, rs3754093 mutant genotype AG/GG decreased the risk of HCC with some HCC protective factors such as non-smoking, non-alcohol consumption and non-HCC family history, but also decreased the risk of HCC with HBV infection. Moreover, it was correlated to non-tumor metastasis and increased the survival of HCC patients. The results from gene-environment interaction assay indicated all hEXO1 SNPs interacted with smoking, alcohol consumption, HBV infection in pathogenesis of HCC. However, gene-gene interaction assay suggested the interaction between rs3754093 and other 5 SNPs were associated with reducing the HCC risk. These results suggest rs3754093 exhibits a protective activity to decrease the incidence risk of HCC in Guangxi, China. In addition, all SNPs in this study interacted with environment risk factors in pathogenesis of HCC.
Keywords: hepatocellular carcinoma (HCC), human exonuclease 1 (hEXO1), single-nucleotide polymorphisms (SNPs), interaction
INTRODUCTION
Hepatocellular carcinoma (HCC) is a very common digestive system carcinoma which is the fifth most prevalent cancer and the third most frequent cause of cancer mortality globally [1]. Each year there are approximately 630,000 new cases of HCC in the world and more than half of the new cases occur in China [2]. It is noticed that the Southern Guangxi has one of the highest occurrence of liver cancer in China [3]. Epidemiological studies suggest that the etiology of HCC is a complicated disease caused by a multi-stage process involving multiple genetic or environmental factors including alcohol consumption, tobacco use, hepatitis B virus (HBV) and hepatitis C virus (HCV) infection, as well as HCC-family history [4–7]. However, only a minority of people with established risk factors eventually develop HCC, suggesting that other environmental and/or genetic factors may play a role in HCC development. In genomic levels, dysfunctions of some oncogenes and tumor suppressor genes induced the progressive growth of malignant cells, and caused HCC.
DNA mismatch repair (MMR) is a component of the DNA repair mechanism, which plays a key role in maintaining genomic stability, preventing gene mutation, the process of DNA replication, and modulates DNA recombination and mediates cell cycle arrest [8]. Human exonuclease 1 (hEXO1) which located at chromosome 1q42-q43, contains one untranslated exon followed by 13 coding exons and encodes an 846 amino acid protein [9–11]. EXO1 is an important member of DNA MMR system which has been well documented to implicate in DNA replication, DNA repair, DNA restructuring, and maintain the stability of telomeres [12]. EXO1 can interact physically with the MMR proteins MSH2 and MLH1 in yeast and human cells, and with MSH3 in human cells [9, 13–17]. Wei indicated that mammalian EXO1 is responsible for mutation prevention and mice EXO1 inactivation reduced survival time and increased risk of lymphoma [18]. In addition, some studies have indicated SNPs of hEXO1 were associated with various cancers including lung cancer, colorectal cancer, gastric cancer, breast cancer, oral cancer, and other tumors [10, 19–25]. A number of studies have reported a SNP of the hEXO1 gene, K589E (rs1047840), is associated with human lung cancer, breast cancer, oral cancer and gastric cancer risk in Chinese Taiwan population [24, 26–28]. Jin [29] and Luo [30] reported the hEXO1 K589E was associated with human lung cancer and cervical cancer susceptibility in Chinese Mainland population. The hEXO1 K589E polymorphism may be a genetic susceptibility factor of HCC in the Turkish population [31], and increase the risk of colorectal cancer in the Polish population [32].
However, the association between SNPs of hEXO1 and hereditary susceptibility of HCC has not been investigated in China. In this study, we conducted a screening on hEXO1 from NIEHS database to seek candidate SNPs in Chinese population. Minor allele frequency (MAF) of six SNPs (rs1047840, rs1776148, rs3754093, rs4149867, rs4149963, and rs1776181) was greater than 0.05 in Chinese population and had potential functions. Therefore, these 6 SNPs were selected to investigate their frequency distributions and associations with HCC in Guangxi, China. We expect the study will provide scientific basis for prevention and treatment of HCC.
RESULTS
Demographic information of HCC patients and controls
The demographic information of this study is presented in Table 1. There were no statistical differences in the age, sex and nation distribution between HCC patients and controls (P>0.05). However, 4 risk factors including HBV infection, alcohol consumption, smoking and HCC family history showed a significantly statistical differences between HCC patients and controls (P<0.05).
Table 1. Demographic information of the study objects.
| Characteristics | Controls (n=1199) | Cases (n=1196) | t/χ2 | P |
|---|---|---|---|---|
| Age(years, ± s) | 48.28±11.69 | 48.58±10.84 | -0.648 | 0.517 |
| Gender | 0.042 | 0.837 | ||
| Male | 1045 | 1039 | ||
| Female | 154 | 157 | ||
| Nation | 3.005 | 0.223 | ||
| Han | 753 | 791 | ||
| Zhuang | 423 | 386 | ||
| Others | 23 | 19 | ||
| Smoking | 140.90 | 0.000 | ||
| Yes | 1008 | 749 | ||
| No | 191 | 447 | ||
| Alcohol Consumption | 141.74 | 0.000 | ||
| Yes | 1036 | 785 | ||
| No | 163 | 411 | ||
| HBV infection | 1375.31 | 0.000 | ||
| Yes | 98 | 195 | ||
| No | 1101 | 1001 | ||
| HCC family history | 42.76 | 0.000 | ||
| Yes | 1189 | 1130 | ||
| No | 10 | 66 |
Distribution of genotypes and risk of HCC
The allele and genotype distributions of the six SNPs in this study and their associations with HCC risk are presented at Table 2. The distribution of all the SNPs’ genotypes in the controls obeyed Hardy-Weinberg equilibrium (HWE) (P> 0.05). The chi-square test also showed distribution of all the SNPs’ genotypes in the cases and the controls had no statistic difference (P>0.05). Following adjust the nation, gender, age, HBV infection, alcohol consumption, smoking, and HCC family history in Logistic regression models, the rs3754093AG (adjusted OR=0.714; 95% CI=0.539-0.946) genotypes exhibited an activity to reduce the risk of HCC (P<0.05).
Table 2. Distribution of genotypes and risk of HCC.
| Genotypes | Controls, n (%) | Cases, n (%) | OR (95%CI)a | OR (95%CI)b |
|---|---|---|---|---|
| rs1047840 | ||||
| AA | 54(4.50) | 64(5.35) | 1.000 | 1.000 |
| AG | 378(31.53) | 388(32.44) | 0.866(0.587∼1.278) | 0.769(0.419∼1.410) |
| GG | 767(63.97) | 744(62.21) | 0.818(0.562∼1.192) | 0.789(0.440∼1.417) |
| AG/GG | 1145(95.50) | 1132(94.65) | 0.834(0.575∼1.209) | 0.782(0.439∼1.394) |
| rs1776148 | ||||
| AA | 38(3.17) | 43(3.60) | 1.000 | 1.000 |
| AG | 362(30.19) | 355(29.68) | 0.867(0.547∼1.373) | 0.614(0.312∼1.207) |
| GG | 799(66.64) | 798(66.72) | 0.883(0.564∼1.380) | 0.547(0.284∼1.054) |
| AG/GG | 1161(96.83) | 1153(96.40) | 0.878(0.563∼1.368) | 0.568(0.297∼1.086) |
| rs3754093 | ||||
| AA | 401(33.44) | 441(36.87) | 1.000 | 1.000 |
| AG | 580(48.37) | 557 (46.57) | 0.873(0.731∼1.044) | 0.714(0.539∼0.946) |
| GG | 218(18.18) | 198(16.55) | 0.826(0.653∼1.045) | 0.822(0.567∼1.193) |
| AG/GG | 798(66.56) | 755(63.12) | 0.906(0.766∼1.071) | 0.741 (0.569∼0.966) |
| rs4149867 | ||||
| CC | 858(71.56) | 718(68.39) | 1.000 | 1.000 |
| CT | 316(26.35) | 347 (29.01) | 1.312(1.094∼1.574) | 0.962 (0.723∼1.281) |
| TT | 25(2.09) | 31(2.59) | 1.482(0.867∼2.533) | 0.767(0.337∼1.749) |
| CT/TT | 341(28.44) | 378(31.60) | 1.325(1.110∼1.581) | 0.945 (0.715∼1.248) |
| rs4149963 | ||||
| CC | 1057(88.15) | 1047(87.54) | 1.000 | 1.000 |
| CT | 129(10.76) | 124(10.37) | 0.970(0.748∼1.260) | 0.805 (0.531∼1.222) |
| TT | 13(1.09) | 25(2.09) | 1.941(0.988∼3.815) | 1.270(0.410∼3.932) |
| CT/TT | 142(11.85) | 149(12.46) | 1.059(0.829∼1.354) | 0.846 (0.570∼1.257) |
| rs1776181 | ||||
| CC | 644(53.71) | 609(50.92) | 1.000 | 1.000 |
| CT | 445(37.11) | 447(37.37) | 1.062(0.895∼1.261) | 0.939 (0.716∼1.230) |
| TT | 110(9.17) | 140(11.71) | 1.346(1.024∼1.769) | 0.873(0.563∼1.353) |
| CT/TT | 555(46.28) | 587(49.08) | 1.118(0.953∼1.313) | 0.925 (0.717∼1.192) |
a:OR and 95%CI without adjusting. b:Adjusted by logistic regression for age, gender, nations, smoking, alcohol consumption, HBV infection, and HCC family history.
Stratification analysis
The distribution of HCC risk factors including smoking, alcohol consumption, HBV infection, and HCC family history in the cases and the controls had statistic difference. Therefore, we conducted a stratification analysis based on these factors (Table 3 and Supplementary Tables S1-S5). As shown in Table 3, the interaction of rs3754093 and HCC risk factors was assessed for the possible combined effect on HCC risk.
Table 3. Results of rs3754093 stratification analysis.
| Variables | Genotypes | Controls (n=1199) | Cases (n=1196) | OR (95%CI)a | P |
|---|---|---|---|---|---|
| Smoking | |||||
| No | AA | 336 | 278 | 1.000 | 0.029 |
| Yes | AA | 65 | 163 | 1.000 | 0.246 |
| Alcohol Consumption | |||||
| No | AA | 345 | 296 | 1.000 | 0.005 |
| Yes | AA | 56 | 145 | 1.000 | 0.716 |
| HBV Infection | |||||
| No | AA | 377 | 73 | 1.000 | 0.417 |
| Yes | AA | 24 | 368 | 1.000 | 0.010 |
| HCC family history | |||||
| No | AA | 399 | 413 | 1.000 | 0.039 |
| Yes | AA | 2 | 28 | 1.000 | 0.544 |
a: Adjusted by logistic regression for age, gender, nations, smoking, alcohol consumption, HBV infection, and HCC family history.
Associations between hEXO1 SNPs and the clinicopathological features of HCC
We evaluated the associations of hEXO1 SNPs with various clinicopathological features of HCC including: tumor size, tumor number, TNM staging (tumor-node-metastasis staging, a cancer staging notation system that describes the stage of a cancer which originates from a solid tumor with alphanumeric codes), combined with liver cirrhosis, AFP level, and tumor metastasis (Table 4∼9). The G allele (AG/GG genotype) of rs3754093 was associated with non-tumor metastasis (adjusted OR: 1.535, 95% CI: 1.023–2.538, P<0.05). However, other 5 SNPs of hEXO1 were not significantly associated with any of the clinicopathological features. Because tumor metastasis is directly related to the prognosis of HCC patients, we performed a univariate survival analysis shown in Figure 1. In accordance with above results, HCC patients with allele G (AG/GG genotype) had a significantly increased risk for death than patients without allele G (AA genotype) (HR= 1.493, 95% CI = 1.102–2.024, P=0.01) (Figure 1).
Table 4. The associations between rs3754093 and clinical characteristics of hepatocellular carcinoma patients.
| Variables | AA | AG/GG | OR(95%CI)a | OR(95%CI)b |
|---|---|---|---|---|
| tumor size | ||||
| ≥5cm | 85(34.56) | 161(63.44) | 1.000 | 1.000 |
| <5cm | 217(29.17) | 527(70.83) | 1.282(0.944∼1.742) | 1.165 (0.876∼1.963) |
| tumor number | ||||
| solitary | 267(30.58) | 606(69.42) | 1.000 | 1.000 |
| multiple | 35(29.91) | 82(70.09) | 1.032(0.677∼1.573) | 0.972 (0.596∼2.248) |
| TNM staging | ||||
| T1+T2 | 262(31.30) | 575(68.70) | 1.000 | 1.000 |
| T3+T4 | 40(26.14) | 113(74.86) | 1.287(0.872∼1.899) | 1.196 (0.756∼1.972) |
| Combined with liver cirrhosis | ||||
| Yes | 100(23.98) | 317(76.02) | 1.000 | 1.000 |
| No | 202(35.25) | 371(64.75) | 0.579(0.437∼0.769) | 0.585 (0.446∼0.756) |
| AFP level (ng/ml) | ||||
| ≥400 | 205 (29.45) | 491(70.55) | 1.000 | 1.000 |
| <400 | 97 (37.16) | 164(62.84) | 0.706 (0.523∼0.952) | 0.681 (0.434∼1.051) |
| tumor metastasis | ||||
| Yes | 53(40.15) | 79(59.85) | 1.000 | 1.000 |
| No | 249(29.02) | 609(70.98) | 1.641(1.124∼2.394) | 1.535 (1.023∼2.538) |
a: OR (95% CI) not adjusted; b: OR (95% CI) adjusted for age, gender, nations, smoking, alcohol consumption, HBV infection, and HCC family history.
Table 9. The associations between rs1776181 and clinical characteristics of hepatocellular carcinoma patients.
| Variables | AA | AG/GG | OR(95%CI)a | OR(95%CI)b |
|---|---|---|---|---|
| tumor size | ||||
| ≥5cm | 390(52.42) | 354(41.58) | 1.000 | 1.000 |
| <5cm | 120(48.78) | 126(51.22) | 1.157(0.867∼1.544) | 1.183 (0.711∼1.966) |
| tumor number | ||||
| solitary | 450(51.55) | 423(48.45) | 1.000 | 1.000 |
| multiple | 60(51.28) | 57(48.72) | 1.101(0.687∼1.487) | 0.919 (0.464∼1.819) |
| TNM staging | ||||
| T1+T2 | 427(49.82) | 410(50.18) | 1.000 | 1.000 |
| T3+T4 | 80(60.00) | 70(40.0) | 0.911(0.643∼1.291) | 0.850 (0.545∼1.426) |
| Combined with liver cirrhosis | ||||
| Yes | 231(55.80) | 183(44.20) | 1.000 | 1.000 |
| No | 279(48.69) | 294(51.31) | 1.330(1.032∼1.715) | 1.295 (0.828∼2.028) |
| AFP level (ng/ml) | ||||
| ≥400 | 357(51.29) | 339(48.71) | 1.000 | 1.000 |
| <400 | 153(52.04) | 141(47.96) | 0.971(0.739∼1.275) | 0.882 (0.597∼1.558) |
| tumor metastasis | ||||
| Yes | 72(54.55) | 60(45.45) | 1.000 | 1.000 |
| No | 435(51.06) | 423(48.94) | 0.857(0.593∼1.239) | 0.857 (0.450∼1.632) |
a: OR (95% CI) not adjusted; b: OR (95% CI) adjusted for age, gender, nations, smoking, alcohol consumption, HBV infection, and HCC family history
Figure 1. Kaplan-Meier overall survival curve for HCC patients based on rs3754093 genotypes.
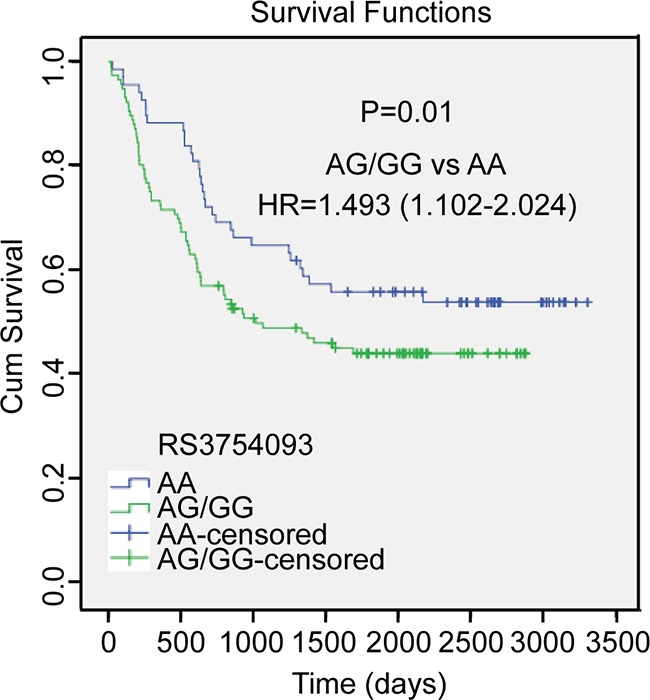
P value is from the log-rank test. HR with 95% CI was from univariate analysis of OS and DFS.
Table 5. The associations between rs1047840 and clinical characteristics of hepatocellular carcinoma patients.
| Variables | AA | AG/GG | OR(95%CI)a | OR(95%CI)b |
|---|---|---|---|---|
| tumor size | ||||
| ≥5cm | 18(7.32) | 228(92.68) | 1.000 | 1.000 |
| <5cm | 33(4.64) | 711(95.36) | 0.588(0.325∼1.064) | 0.582(0.202∼1.678) |
| tumor number | ||||
| solitary | 48(5.50) | 825(94.50) | 1.000 | 1.000 |
| multiple | 3(2.56) | 114(97.44) | 2.211(0.678∼7.215) | 2.131(0.268∼16.913) |
| TNM staging | ||||
| T1+T2 | 48(5.73) | 789(94.27) | 1.000 | 1.000 |
| T3+T4 | 3(1.96) | 150(98.04) | 3.042(0.935∼9.893) | 2.828(0.359∼22.287) |
| Combined with liver cirrhosis | ||||
| Yes | 21(5.04) | 396(94.36) | 1.000 | 1.000 |
| No | 30(5.24) | 543(94.86) | 0.960(0.541∼1.702) | 1.115(0.402∼3.089) |
| AFP level (ng/ml) | ||||
| ≥400 | 33(4.74) | 663(95.26) | 1.000 | 1.000 |
| <400 | 18(6.89) | 243(93.10) | 0.672(0.371∼1.216) | 0.749(0.264∼2.130) |
| tumor metastasis | ||||
| Yes | 6(4.55) | 126(95.45) | 1.000 | 1.000 |
| No | 45(5.24) | 813(94.76) | 1.162(0.486∼2.781) | 1.201(0.257∼5.681) |
a: OR (95% CI) not adjusted; b: OR (95% CI) adjusted for age, gender, nations, smoking, alcohol consumption, HBV infection, and HCC family history.
Table 6. The associations between rs1776148 and clinical characteristics of hepatocellular carcinoma patients.
| Variables | AA | AG/GG | OR(95%CI)a | OR(95%CI)b |
|---|---|---|---|---|
| tumor size | ||||
| ≥5cm | 6(2.44) | 240(97.56) | 1.000 | 1.000 |
| <5cm | 24(3.23) | 720(96.77) | 1.333(0.539∼3.301) | 1.587(0.317∼7.937) |
| tumor number | ||||
| solitary | 27(3.09) | 846(96.91) | 1.000 | 1.000 |
| multiple | 3(2.56) | 114(97.44) | 1.213(0.362∼4.062) | 1.028(0.121∼8.760) |
| TNM staging | ||||
| T1+T2 | 30(3.58) | 807(96.42) | 1.000 | 1.000 |
| T3+T4 | 6(3.92) | 147(96.08) | 0.911(0.373∼2.227) | 1.128(0.569∼2.233) |
| Combined with liver cirrhosis | ||||
| Yes | 3(0.72) | 414(99.28) | 1.000 | 1.000 |
| No | 27(4.71) | 546(95.29) | 0.147(0.044∼0.486) | 0.154(0.019∼1.260) |
| AFP level (ng/ml) | ||||
| ≥400 | 27(3.88) | 669(96.12) | 1.000 | 1.000 |
| <400 | 3(1.02) | 291(98.98) | 3.915(1.178∼13.007) | 4.391(0.536∼35.971) |
| tumor metastasis | ||||
| Yes | 6(4.55) | 126(95.45) | 1.000 | 1.000 |
| No | 30(3.50) | 828(96.50) | 0.761(0.310∼1.865) | 0.764(0.164∼3.606) |
a: OR (95% CI) not adjusted; b: OR (95% CI) adjusted for age, gender, nations, smoking, alcohol consumption, HBV infection, and HCC family history.
Table 7. The associations between rs4149867 and clinical characteristics of hepatocellular carcinoma patients.
| Variables | AA | AG/GG | OR(95%CI)a | OR(95%CI)b |
|---|---|---|---|---|
| tumor size | ||||
| ≥5cm | 165(67.07) | 81(32.93) | 1.000 | 1.000 |
| <5cm | 501(67.34) | 243(32.66) | 1.125(0.533∼2.373) | 0.638(0.357∼1.138) |
| tumor number | ||||
| solitary | 609(69.76) | 264(30.24) | 1.000 | 1.000 |
| multiple | 78(66.66) | 39(33.34) | 1.153(0.765∼1.739) | 1.080(0.521∼2.238) |
| TNM staging | ||||
| T1+T2 | 588(70.25) | 249(29.75) | 1.000 | 1.000 |
| T3+T4 | 96(64.00) | 54(36.00) | 1.328(0.922∼1.913) | 1.250(0.646∼2.419) |
| Combined with liver cirrhosis | ||||
| Yes | 273(65.94) | 141(34.06) | 1.000 | 1.000 |
| No | 414(72.25) | 159(27.05) | 0.744(0.566∼0.977) | 0.781(0.480∼1.270) |
| AFP level (ng/ml) | ||||
| ≥400 | 483(69.40) | 213(30.60) | 1.000 | 1.000 |
| <400 | 204(69.39) | 90(30.61) | 1.000(0.744∼1.345) | 0.988(0.587∼1.665) |
| tumor metastasis | ||||
| Yes | 99(75.00) | 33(25.00) | 1.000 | 1.000 |
| No | 582(68.31) | 270(31.69) | 0.719(0.472∼1.093) | 0.726(0.348∼1.515) |
a: OR (95% CI) not adjusted; b: OR (95% CI) adjusted for age, gender, nations, smoking, alcohol consumption, HBV infection, and HCC family history.
Table 8. The associations between rs4149963 and clinical characteristics of hepatocellular carcinoma patients.
| Variables | AA | AG/GG | OR(95%CI)a | OR(95%CI)b |
|---|---|---|---|---|
| tumor size | ||||
| ≥5cm | 218(88.62) | 28(11.38) | 1.000 | 1.000 |
| <5cm | 669(89.91) | 75(10.08) | 1.146(0.723∼1.815) | 1.648(0.775∼3.508) |
| tumor number | ||||
| solitary | 780(89.34) | 93(10.65) | 1.000 | 1.000 |
| multiple | 104(88.89) | 13(11.11) | 1.048(0.567∼1.940) | 1.610(0.614∼4.220) |
| TNM staging | ||||
| T1+T2 | 738(88.17) | 99(11.83) | 1.000 | 1.000 |
| T3+T4 | 131(87.33) | 19(2.67) | 1.081(0.640∼1.828) | 0.586(0.189∼1.814) |
| Combined with liver cirrhosis | ||||
| Yes | 360(86.96) | 51(13.04) | 1.000 | 1.000 |
| No | 516(90.05) | 57(9.95) | 0.780(0.522∼1.164) | 0.759(0.378∼1.527) |
| AFP level (ng/ml) | ||||
| ≥400 | 603(86.64) | 93(13.36) | 1.000 | 1.000 |
| <400 | 252(85.71) | 42(14.29) | 1.081(0.730∼1.601) | 0.420(0.168∼1.049) |
| tumor metastasis | ||||
| Yes | 117(88.64) | 15(11.36) | 1.000 | 1.000 |
| No | 762(88.81) | 96(11.19) | 1.018(0.571∼1.814) | 1.308(0.506∼3.382) |
a: OR (95% CI) not adjusted; b: OR (95% CI) adjusted for age, gender, nations, smoking, alcohol consumption, HBV infection, and HCC family history.
Gene-environment interaction and SNP-SNP interaction
As shown in Table 10, all the SNPs were found to interact with smoking, alcohol consumption, HBV infection in the pathogenesis of HCC. The results of SNP-SNP interaction suggested the interaction between rs3754093 and other 5 SNPs (rs1047840, rs1776148, rs4149867, rs4149963 and rs1776181) was associated with reducing the HCC risk (Table 11).
Table 10. Gene-environment interaction.
| Factors | β | S.E. | Wald χ2 | OR (95%CI) a | P |
|---|---|---|---|---|---|
| rs1047840 × Smoking | 0.428 | 0.093 | 21.020 | 1.534(1.278∼1.842) | 0.000 |
| rs1047840 × Alcohol Consumption | 0.336 | 0.096 | 12.225 | 1.399(1.159∼1.689) | 0.000 |
| rs1047840 × HBV Infection | 2.102 | 0.074 | 809.417 | 8.818(7.078∼9.456) | 0.000 |
| rs1047840 × HCC family history | 0.229 | 0.235 | 0.949 | 1.257(0.793∼1.293) | 0.330 |
| rs1776148 × Smoking | 0.424 | 0.093 | 20.652 | 1.527(1.272∼1.833) | 0.000 |
| rs1776148 × Alcohol Consumption | 0.318 | 0.095 | 11.066 | 1.374(1.139∼1.657) | 0.001 |
| rs1776148 × HBV Infection | 2.080 | 0.073 | 820.029 | 8.003(6.941∼9.288) | 0.000 |
| rs1776148 × HCC family history | 0.236 | 0.223 | 1.022 | 1.266(0.801∼2.001) | 0.312 |
| rs3754093 × Smoking | 0.421 | 0.103 | 16.756 | 1.524(1.245∼1.864) | 0.000 |
| rs3754093 × Alcohol Consumption | 0.367 | 0.105 | 12.101 | 1.443(1.174∼1.775) | 0.001 |
| rs3754093 × HBV Infection | 2.326 | 0.092 | 641.686 | 10.235(8.549∼12.253) | 0.000 |
| rs3754093 × HCC family history | 0.186 | 0.269 | 0.477 | 1.204(0.711∼2.041) | 0.490 |
| rs4149867 × Smoking | 0.462 | 0.124 | 13.852 | 1.587(1.244∼2.023) | 0.000 |
| rs4149867 × Alcohol Consumption | 0.385 | 0.134 | 8.240 | 1.469(1.130∼1.910) | 0.004 |
| rs4149867 × HBV Infection | 3.238 | 0.126 | 664.917 | 25.485(19.925∼32.597) | 0.000 |
| rs4149867 × HCC family history | 0.391 | 0.340 | 1.324 | 1.478(0.760∼2.876) | 0.250 |
| rs4149963 × Smoking | 0.610 | 0.149 | 16.769 | 1.840(1.374∼2.464) | 0.000 |
| rs4149963 × Alcohol Consumption | 0.454 | 0.159 | 8.276 | 1.580(1.157∼2.158) | 0.004 |
| rs4149963 × HBV Infection | 3.707 | 0.134 | 762.612 | 40.703(31.296∼52.965) | 0.000 |
| rs4149963 × HCC family history | 0.350 | 0.401 | 0.760 | 1.419(0.646∼3.113) | 0.383 |
| rs1776181 × Smoking | 0.497 | 0.110 | 20.283 | 1.644(1.324∼2.042) | 0.000 |
| rs1776181 × Alcohol Consumption | 0.347 | 0.115 | 9.139 | 1.414(1.130∼1.771) | 0.003 |
| rs1776181 × HBV Infection | 2.578 | 0.103 | 620.820 | 13.175(10.756∼16.137) | 0.000 |
| rs1776181 × HCC family history | 0.372 | 0.316 | 1.384 | 1.450(0.781∼2.695) | 0.239 |
a: Adjusted by logistic regression for age, gender, nations, smoking, alcohol consumption, HBV infection, and HCC family history.
Table 11. SNP-SNP interaction on HCC risk.
| Factors | β | S.E. | Wald χ2 | OR (95%CI) a | P |
|---|---|---|---|---|---|
| rs1047840 × rs1776148 | -0.202 | 0.113 | 3.190 | 0.817(0.654∼1.020) | 0.074 |
| rs1047840 × rs3754093 | -0.148 | 0.065 | 5.262 | 0.862(0.759∼0.979) | 0.022 |
| rs1047840 × rs4149867 | -0.055 | 0.069 | 0.627 | 0.947(0.826∼1.084) | 0.429 |
| rs1047840 × rs4149963 | -0.098 | 0.096 | 1.045 | 0.906(0.751∼1.094) | 0.307 |
| rs1047840 × rs1776181 | -0.061 | 0.063 | 0.924 | 0.941(0.831∼1.065) | 0.337 |
| rs1776148 × rs3754093 | -0.177 | 0.065 | 7.302 | 0.838(0.737∼0.953) | 0.007 |
| rs1776148 × rs4149867 | -0.052 | 0.070 | 0.547 | 0.950(0.828∼1.089) | 0.460 |
| rs1776148 × rs4149963 | -0.151 | 0.098 | 2.367 | 0.860(0.709∼1.042) | 0.124 |
| rs1776148 × rs1776181 | -0.067 | 0.063 | 1.104 | 0.937(0.827∼1.059) | 0.293 |
| rs3754093 × rs4149867 | -0.134 | 0.065 | 4.193 | 0.875(0.769∼0.994) | 0.041 |
| rs3754093 × rs4149963 | -0.171 | 0.084 | 4.153 | 0.843(0.715∼0.993) | 0.042 |
| rs3754093 × rs1776181 | -0.128 | 0.059 | 4.660 | 0.880(0.783∼0.988) | 0.031 |
| rs4149867 × rs4149963 | -0.104 | 0.098 | 1.129 | 0.901(0.744∼1.092) | 0.288 |
| rs4149867 × rs1776181 | -0.090 | 0.060 | 2.301 | 0.914(0.813∼1.027) | 0.129 |
| rs4149963 × rs1776181 | -0.066 | 0.083 | 2.628 | 0.936(0.795∼1.102) | 0.428 |
a: Adjusted by logistic regression for age, gender, nations, smoking, alcohol consumption, HBV infection, and HCC family history.
DISCUSSION
In this study, we investigated six SNPs of the hEXO1 gene and their association with hereditary susceptibility for HCC in the population of Guangxi, China. Among these six SNPs, we found rs3754093 was significantly associated with a higher susceptibility, and exhibited a protective activity in HCC patients.
hEXO1 is an endonuclease which plays a critical role in both 5′-3′ and 3′-5′ mispair-dependent excision repair to maintain the overall integrity of the MMR protein complex in human MMR system [33, 34]. Alteration of MMR gene can cause cell DNA mismatch repair by increasing cell spontaneous mutation frequency, down-regulating tumor suppressor genes and up-regulating oncogenes, eventually leading to the occurrence of tumors [35]. Therefore, hEXO1 gene is an important target gene in association with the risk of various malignancies [23, 25, 36]. Moreover, hEXO1 activates mutations of target genes through cutting DNA of telomere [43], which is a kind of special protein-DNA structure that exists in eukaryotic cell linear chromosomes [37] and plays an important role in maintaining cell proliferation ability and protecting chromosome integrity [38]. Through its role in these recombinational events, such as repairing of DNA double-strand breaks and maintaining of telomere stabilization, hEXO1 functions in carcinogenesis of various tumors [18, 26].
In consistent with its key role in carcinogenesis, hEXO1 rs3754093 showed a protective activity and decreased the risk for death in HCC patients in our study. Furthermore, function prediction showed hEXO1 rs3754093 was a transcription factor binding site (TFBS). Previous reports showed that SNPs on TFBS could alter the affinity between transcription factors and specific DNA sequences, then cause expression alterations of specific genes [39–41]. That is to say, the polymorphism of rs3754093 could alter the expression level of hEXO1 through changing its binding affinity to transcription factors. And then the expression change of hEXO1 might either alter the procedure of MMR to result in gene mutations [42], or increase the risk of cancer through telomere cutting to cause genomic instability [13]. This might be the mechanism of association between hEXO1 rs3754093 and HCC. In addition, we did not find the other 5 SNPs associated with HCC pathogenesis, indicating that the 5 SNPs didn't alter expression level of hEXO1.
From gene-environment interaction analysis, we found the six hEXO1 SNPs were associated with such environment factors, smoking, alcohol consumption and HBV infection. These environmental factors are well documented risk factors of HCC and each of them is an independent strong cause of HCC as previous studies showed. Meta-analysis of epidemiologic studies suggested tobacco use was associated with the increasing of HCC risk [43]. Koh also reported that the tobacco use increased the risk of HCC in the Singapore Chinese population [44]. Both alcohol and tobacco are synergistic risk factors for HCC [45]. In addition, the HBV and HCV infection also plays an important role in increasing the risk of HCC [46], and also is a major factor of HCC risk in Guangxi, China [47]. The chronic HBV infection resulted in the hepatic inflammation reaction, fibrosis, hepatocirrhosis and finally induced liver cancer [46]. We draw a similar conclusion on the effects of these environment factors in development of HCC with previous reports. Moreover, all the six hEXO1 SNPs in the study have interaction effects with some environment factors on HCC risk.
Previous studies have reported the SNP-SNP interaction was significantly associated with the pathological process of breast cancer [48], colon cancer [49], oesophageal squamous cell carcinoma [50] and HCC [51], respectively. We found the interaction between rs3754093 and other SNPs (rs1047840, rs1776148, rs4149867, rs4149963 and rs1776181) was significantly associated with reduced the risk of HCC. These results suggested the SNP-SNP interaction may decrease the HCC risk.
HCC is a serious cancer with a complicated pathogenesis related to lots of genes and environmental factors. However, the mechanism of HCC incidence has not been totally understood. In this study, we found the hEXO1 rs3754093 was a protective factor to reduce the risk of HCC. Moreover, the combined effects of SNP-SNP interactions may decrease the risk of HCC in Guangxi, which has a high incidence of HCC in China. As the association between rs3754093 and the risk of HCC is only an epidemiological link right now, further function study on the SNP should be conducted to validate the current finding.
MATERIALS AND METHODS
Study subjects
The cases (n=1,199) were newly diagnosed as HCC patients without chemotherapy and radiotherapy who recruited from the Affiliated Tumor Hospital of Guangxi Medical University between June 2007 and March 2014. All HCC patients were diagnosed according to the World Health Organization (WHO) Classification of HCC diagnostic criteria. The controls (n=1,196) were non-tumor patients from the First Affiliated Hospital of Guangxi Medical University during the same period. Inclusion and exclusion criteria for participants: 1. Inclusion criteria: (1) Cases: new HCC patients hospitalized in the period of study, and were pathologically confirmed. (2) Controls: patients without HCC hospitalized in the same period with the cases. 2. Exclusion criteria: (1) Cases: have been treated, HCV infected, with other tumors. (2) Controls: suffering from cancer, HCV infected. This study was respectively approved by the Institutional Review Board of the Affiliated Tumor Hospital of Guangxi Medical University and First Affiliated Hospital of Guangxi Medical University. The informed consent was obtained from each subject in this study.
Questionnaire survey and blood sample collection
Questionnaires were designed after consulting experts. Data of questionnaire survey was collected from a structured interview, and conducted by trained interviewers after a pre-investigation, which including the demographic characteristics (name, nation, gender, age), smoking alcohol consumption, hepatitis B virus (HBV) infection and HCC family history. 2 mL peripheral blood sample was collected from each study object. Genome DNA was extracted from the blood samples. All laboratory and questionnaire data were coded, entered by two investigators with a logical and consistency test, and verified using EpiData 3.1 (www.epidata.dk/download.php). Neither the laboratory nor the data entry personnel had any knowledge of the subjects’ case-control status.
DNA extraction and genotyping
The blood was collected from the all subjects using an EDTA-K2 anticoagulant blood vessel. The DNA was extracted according to the phenol-chloroform method and stored at -80°C. The genotyping was performed in an Applied Biosystems 7500 Fast Real-Time quantitative polymerase chain reaction (qPCR) system with the following TaqMan SNP Genotyping Assays kit (Applied Biosystems) according to the manufacturer's instructions. Each PCR reaction mixture (10 μL) contained DNA template (0.4 μL), ddH2O (4.35 μL), 2×TaqMan Universal PCR Master Mix (5 μL), and 20× SNP Genotyping Assay Mix (0.25 μL). The genomic DNA was amplified at 95°C for 10 min, followed by 40 cycles of 95°C for 15s and 60°C for 60s. All genotyping reagents and analytical software were purchased from Applied Biosystems. Approximately 5% of the samples were randomly repeated to validate the genotyping procedures, and the concordance rate was 100%. The results of the genotyping were analyzed with 7500 Fast System V1.4.0 SDS software.
Survival analysis
356 HCC patients were brought into a survival analysis cohort. The patients were followed up from July 2007 to May 2016. Survival time was counted the first day after surgery and end up when the patients appeared metastasis and recurrence or death. Survival time was calculated in months. Till the end of follow-up, a total of 31 patients were lost to follow-up, 325 patients with complete follow-up data, follow-up rate was 91.3%.
Statistical analysis
Data entry and consistency check were conducted on the EpiData3.1 software. The SPSS 20.0 software was used for the statistical analyses. Logistic regression models were used to estimate odds ratio (OR) and 95% confidence interval (CI). Gene-environment interactions and gene-gene interactions were performed through multiplicative model of binary logistic regression. Hardy-Weinberg Equilibrium (HWE) in the controls was tested by Haploview 4.2 software. Kaplan–Meier method, Log–rank test and COX regression were used on survival analysis.
SUPPLEMENTARY TABLES
Acknowledgments
This study was supported by the grant from the National Natural Science Foundation of China (81160359).
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1.Bosetti C, Turati F, La Vecchia C. Hepatocellular carcinoma epidemiology. Best practice & research. 2014;28:753–770. doi: 10.1016/j.bpg.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 2.Ding J, Wang H. Multiple interactive factors in hepatocarcinogenesis. Cancer letters. 2014;346:17–23. doi: 10.1016/j.canlet.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 3.Zhang CY, Huang TR, Yu JH, Zhang ZQ, Li JL, Deng W, Ye SY, Zhou DN, He ZF. Epidemiological analysis of primary liver cancer in the early 21st century in Guangxi province of China. Chinese journal of cancer. 2010;29:545–550. doi: 10.5732/cjc.009.10510. [DOI] [PubMed] [Google Scholar]
- 4.Flemming JA, Yang JD, Vittinghoff E, Kim WR, Terrault NA. Risk prediction of hepatocellular carcinoma in patients with cirrhosis: the ADRESS-HCC risk model. Cancer. 2014;120:3485–3493. doi: 10.1002/cncr.28832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jun CH, Hong HJ, Chung MW, Park SY, Cho SB, Park CH, Joo YE, Kim HS, Choi SK, Rew JS. Risk factors for hepatocellular carcinoma in patients with drug-resistant chronic hepatitis B. World journal of gastroenterology. 2013;19:6834–6841. doi: 10.3748/wjg.v19.i40.6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paranagua-Vezozzo DC, Ono SK, Alvarado-Mora MV, Farias AQ, Cunha-Silva M, Franca JI, Alves VA, Sherman M, Carrilho FJ. Epidemiology of HCC in Brazil: incidence and risk factors in a ten-year cohort. Annals of hepatology. 2014;13:386–393. [PubMed] [Google Scholar]
- 7.Toyoda H, Kumada T, Tada T, Kiriyama S, Tanikawa M, Hisanaga Y, Kanamori A, Kitabatake S, Ito T. Risk factors of hepatocellular carcinoma development in non-cirrhotic patients with sustained virologic response for chronic hepatitis C virus infection. Journal of gastroenterology and hepatology. 2015;30:1183–1189. doi: 10.1111/jgh.12915. [DOI] [PubMed] [Google Scholar]
- 8.Muro Y, Sugiura K, Mimori T, Akiyama M. DNA mismatch repair enzymes: genetic defects and autoimmunity. Clinica chimica acta; international journal of clinical chemistry. 2015;442:102–109. doi: 10.1016/j.cca.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 9.Schmutte C, Marinescu RC, Sadoff MM, Guerrette S, Overhauser J, Fishel R. Human exonuclease I interacts with the mismatch repair protein hMSH2. Cancer research. 1998;58:4537–4542. [PubMed] [Google Scholar]
- 10.Tishkoff DX, Amin NS, Viars CS, Arden KC, Kolodner RD. Identification of a human gene encoding a homologue of Saccharomyces cerevisiae EXO1, an exonuclease implicated in mismatch repair and recombination. Cancer research. 1998;58:5027–5031. [PubMed] [Google Scholar]
- 11.Wilson DM, Carney JP, 3rd, Coleman MA, Adamson AW, Christensen M, Lamerdin JE. Hex1: a new human Rad2 nuclease family member with homology to yeast exonuclease 1. Nucleic acids research. 1998;26:3762–3768. doi: 10.1093/nar/26.16.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orans J, McSweeney EA, Iyer RR, Hast MA, Hellinga HW, Modrich P, Beese LS. Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell. 2011;145:212–223. doi: 10.1016/j.cell.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jager AC, Rasmussen M, Bisgaard HC, Singh KK, Nielsen FC, Rasmussen LJ. HNPCC mutations in the human DNA mismatch repair gene hMLH1 influence assembly of hMutLalpha and hMLH1-hEXO1 complexes. Oncogene. 2001;20:3590–3595. doi: 10.1038/sj.onc.1204467. [DOI] [PubMed] [Google Scholar]
- 14.Rasmussen LJ, Rasmussen M, Lee B, Rasmussen AK, Wilson DM, Nielsen FC, 3rd, Bisgaard HC. Identification of factors interacting with hMSH2 in the fetal liver utilizing the yeast two-hybrid system. In vivo interaction through the C-terminal domains of hEXO1 and hMSH2 and comparative expression analysis. Mutation research. 2000;460:41–52. doi: 10.1016/s0921-8777(00)00012-4. [DOI] [PubMed] [Google Scholar]
- 15.Schmutte C, Sadoff MM, Shim KS, Acharya S, Fishel R. The interaction of DNA mismatch repair proteins with human exonuclease I. The Journal of biological chemistry. 2001;276:33011–33018. doi: 10.1074/jbc.M102670200. [DOI] [PubMed] [Google Scholar]
- 16.Tishkoff DX, Boerger AL, Bertrand P, Filosi N, Gaida GM, Kane MF, Kolodner RD. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tran PT, Simon JA, Liskay RM. Interactions of Exo1p with components of MutLalpha in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:9760–9765. doi: 10.1073/pnas.161175998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei K, Clark AB, Wong E, Kane MF, Mazur DJ, Parris T, Kolas NK, Russell R, Hou H, Jr., Kneitz B, Yang G, Kunkel TA, Kolodner RD, Cohen PE, Edelmann W. Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes & development. 2003;17:603–614. doi: 10.1101/gad.1060603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang JS, Yeh RF, Wiencke JK, Wiemels JL, Smirnov I, Pico AR, Tihan T, Patoka J, Miike R, Sison JD, Rice T, Wrensch MR. Pathway analysis of single-nucleotide polymorphisms potentially associated with glioblastoma multiforme susceptibility using random forests. Cancer Epidemiol Biomarkers Prev. 2008;17:1368–1373. doi: 10.1158/1055-9965.EPI-07-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen LT, Thykjaer T, Orntoft TF, Rasmussen LJ, Keller P, Spang-Thomsen M, Edmonston TB, Schmutte C, Fishel R, Petersen LN. The role of mismatch repair in small-cell lung cancer cells. Eur J Cancer. 2003;39:1456–1467. doi: 10.1016/s0959-8049(03)00306-x. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen FC, Jager AC, Lutzen A, Bundgaard JR, Rasmussen LJ. Characterization of human exonuclease 1 in complex with mismatch repair proteins, subcellular localization and association with PCNA. Oncogene. 2004;23:1457–1468. doi: 10.1038/sj.onc.1207265. [DOI] [PubMed] [Google Scholar]
- 22.Sun X, Zheng L, Shen B. Functional alterations of human exonuclease 1 mutants identified in atypical hereditary nonpolyposis colorectal cancer syndrome. Cancer research. 2002;62:6026–6030. [PubMed] [Google Scholar]
- 23.Thompson E, Meldrum CJ, Crooks R, McPhillips M, Thomas L, Spigelman AD, Scott RJ. Hereditary non-polyposis colorectal cancer and the role of hPMS2 and hEXO1 mutations. Clinical genetics. 2004;65:215–225. doi: 10.1111/j.1399-0004.2004.00214.x. [DOI] [PubMed] [Google Scholar]
- 24.Wang HC, Chiu CF, Tsai RY, Kuo YS, Chen HS, Wang RF, Tsai CW, Chang CH, Lin CC, Bau DT. Association of genetic polymorphisms of EXO1 gene with risk of breast cancer in Taiwan. Anticancer research. 2009;29:3897–3901. [PubMed] [Google Scholar]
- 25.Wu Y, Berends MJ, Post JG, Mensink RG, Verlind E, Van Der Sluis T, Kempinga C, Sijmons RH, van der Zee AG, Hollema H, Kleibeuker JH, Buys CH, Hofstra RM. Germline mutations of EXO1 gene in patients with hereditary nonpolyposis colorectal cancer (HNPCC) and atypical HNPCC forms. Gastroenterology. 2001;120:1580–1587. doi: 10.1053/gast.2001.25117. [DOI] [PubMed] [Google Scholar]
- 26.Bau DT, Wang HC, Liu CS, Chang CL, Chiang SY, Wang RF, Tsai CW, Lo YL, Hsiung CA, Lin CC, Huang CY. Single-nucleotide polymorphism of the Exo1 gene: association with gastric cancer susceptibility and interaction with smoking in Taiwan. The Chinese journal of physiology. 2009;52:411–418. doi: 10.4077/cjp.2009.amh076. [DOI] [PubMed] [Google Scholar]
- 27.Hsu NY, Wang HC, Wang CH, Chiu CF, Tseng HC, Liang SY, Tsai CW, Lin CC, Bau DT. Lung cancer susceptibility and genetic polymorphisms of Exo1 gene in Taiwan. Anticancer research. 2009;29:725–730. [PubMed] [Google Scholar]
- 28.Tsai MH, Tseng HC, Liu CS, Chang CL, Tsai CW, Tsou YA, Wang RF, Lin CC, Wang HC, Chiu CF, Bau DT. Interaction of Exo1 genotypes and smoking habit in oral cancer in Taiwan. Oral oncology. 2009;45:e90–94. doi: 10.1016/j.oraloncology.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 29.Jin G, Wang H, Hu Z, Liu H, Sun W, Ma H, Chen D, Miao R, Tian T, Jin L, Wei Q, Huang W, Lu D, Shen H. Lung cancer. Vol. 60. Amsterdam, Netherlands: 2008. Potentially functional polymorphisms of EXO1 and risk of lung cancer in a Chinese population: A case-control analysis; pp. 340–346. [DOI] [PubMed] [Google Scholar]
- 30.Luo X, Hong XS, Xiong XD, Zeng LQ, Lim CE. A single nucleotide polymorphism in EXO1 gene is associated with cervical cancer susceptibility in Chinese patients. Int J Gynecol Cancer. 2012;22:220–225. doi: 10.1097/IGC.0b013e318234fd8a. [DOI] [PubMed] [Google Scholar]
- 31.Bayram S, Akkiz H, Bekar A, Akgollu E, Yildirim S. The significance of Exonuclease 1 K589E polymorphism on hepatocellular carcinoma susceptibility in the Turkish population: a case-control study. Molecular biology reports. 2012;39:5943–5951. doi: 10.1007/s11033-011-1406-x. [DOI] [PubMed] [Google Scholar]
- 32.Kabzinski J, Mucha B, Cuchra M, Markiewicz L, Przybylowska K, Dziki A, Dziki L, Majsterek I. Efficiency of Base Excision Repair of Oxidative DNA Damage and Its Impact on the Risk of Colorectal Cancer in the Polish Population. Oxidative medicine and cellular longevity. 2016;2016:3125989. doi: 10.1155/2016/3125989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knudsen NO, Nielsen FC, Vinther L, Bertelsen R, Holten-Andersen S, Liberti SE, Hofstra R, Kooi K, Rasmussen LJ. Nuclear localization of human DNA mismatch repair protein exonuclease 1 (hEXO1) Nucleic acids research. 2007;35:2609–2619. doi: 10.1093/nar/gkl1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liberti SE, Rasmussen LJ. Is hEXO1 a cancer predisposing gene? Mol Cancer Res. 2004;2:427–432. [PubMed] [Google Scholar]
- 35.Bak ST, Sakellariou D, Pena-Diaz J. The dual nature of mismatch repair as antimutator and mutator: for better or for worse. Frontiers in genetics. 2014;5:287. doi: 10.3389/fgene.2014.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jagmohan-Changur S, Poikonen T, Vilkki S, Launonen V, Wikman F, Orntoft TF, Moller P, Vasen H, Tops C, Kolodner RD, Mecklin JP, Jarvinen H, Bevan S, et al. EXO1 variants occur commonly in normal population: evidence against a role in hereditary nonpolyposis colorectal cancer. Cancer research. 2003;63:154–158. [PubMed] [Google Scholar]
- 37.Martinez P, Blasco MA. Replicating through telomeres: a means to an end. Trends in biochemical sciences. 2015;40:504–515. doi: 10.1016/j.tibs.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 38.Konvalinova H, Dvorakova Z, Renciuk D, Bednarova K, Kejnovska I, Trantirek L, Vorlickova M, Sagi J. Diverse effects of naturally occurring base lesions on the structure and stability of the human telomere DNA quadruplex. Biochimie. 2015;118:15–25. doi: 10.1016/j.biochi.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 39.Buroker NE. Regulatory SNPs and transcriptional factor binding sites in ADRBK1, AKT3, ATF3, DIO2, TBXA2R and VEGFA. Transcription. 2015;5:e964559. doi: 10.4161/21541264.2014.964559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hesp K, Smant G, Kammenga JE. Caenorhabditis elegans DAF-16/FOXO transcription factor and its mammalian homologs associate with age-related disease. Experimental gerontology. 2015;72:1–7. doi: 10.1016/j.exger.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 41.Mundade R, Ozer HG, Wei H, Prabhu L, Lu T. Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism, epigenetic marks and beyond. Cell cycle (Georgetown, Tex. 2014;13:2847–2852. doi: 10.4161/15384101.2014.949201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nogueira GA, Lourenco GJ, Oliveira CB, Marson FA, Lopes-Aguiar L, Costa EF, Lima TR, Liutti VT, Leal F, Santos VC, Rinck-Junior JA, Lima CS. Association between genetic polymorphisms in DNA mismatch repair-related genes with risk and prognosis of head and neck squamous cell carcinoma. International journal of cancer. 2015;137:810–818. doi: 10.1002/ijc.29435. [DOI] [PubMed] [Google Scholar]
- 43.Lee YC, Cohet C, Yang YC, Stayner L, Hashibe M, Straif K. Meta-analysis of epidemiologic studies on cigarette smoking and liver cancer. International journal of epidemiology. 2009;38:1497–1511. doi: 10.1093/ije/dyp280. [DOI] [PubMed] [Google Scholar]
- 44.Koh WP, Robien K, Wang R, Govindarajan S, Yuan JM, Yu MC. Smoking as an independent risk factor for hepatocellular carcinoma: the Singapore Chinese Health Study. British journal of cancer. 2011;105:1430–1435. doi: 10.1038/bjc.2011.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marrero JA, Fontana RJ, Fu S, Conjeevaram HS, Su GL, Lok AS. Alcohol, tobacco and obesity are synergistic risk factors for hepatocellular carcinoma. Journal of hepatology. 2005;42:218–224. doi: 10.1016/j.jhep.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 46.Chuang SC, La Vecchia C, Boffetta P. Liver cancer: descriptive epidemiology and risk factors other than HBV and HCV infection. Cancer letters. 2009;286:9–14. doi: 10.1016/j.canlet.2008.10.040. [DOI] [PubMed] [Google Scholar]
- 47.Yeh FS, Yu MC, Mo CC, Luo S, Tong MJ, Henderson BE. Hepatitis B virus, aflatoxins, and hepatocellular carcinoma in southern Guangxi, China. Cancer research. 1989;49:2506–2509. [PubMed] [Google Scholar]
- 48.Sapkota Y, Mackey JR, Lai R, Franco-Villalobos C, Lupichuk S, Robson PJ, Kopciuk K, Cass CE, Yasui Y, Damaraju S. Assessing SNP-SNP interactions among DNA repair, modification and metabolism related pathway genes in breast cancer susceptibility. PloS one. 2013;8:e64896. doi: 10.1371/journal.pone.0064896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goodman JE, Mechanic LE, Luke BT, Ambs S, Chanock S, Harris CC. Exploring SNP-SNP interactions and colon cancer risk using polymorphism interaction analysis. International journal of cancer. 2006;118:1790–1797. doi: 10.1002/ijc.21523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Vogelsang M, Schafer G, Matejcic M, Parker MI. MicroRNA polymorphisms and environmental smoke exposure as risk factors for oesophageal squamous cell carcinoma. PloS one. 2013;8:e78520. doi: 10.1371/journal.pone.0078520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bei CH, Bai H, Yu HP, Yang Y, Liang QQ, Deng YY, Tan SK, Qiu XQ. Combined effects of six cytokine gene polymorphisms and SNP-SNP interactions on hepatocellular carcinoma risk in Southern Guangxi, China. Asian Pac J Cancer Prev. 2014;15:6961–6967. doi: 10.7314/apjcp.2014.15.16.6961. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


